Abstract
Background
The coronavirus disease 2019 (COVID-19) pandemic is a major public health concern. Accurate and rapid diagnosis of COVID-19 is critical for disease control. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) is a nucleic acid amplification assay similar to reverse transcription-polymerase chain reaction (RT-PCR), the former being a simple, low cost, and rapid method.
Objectives
This study aimed to compare the RT-LAMP assay with RT-PCR using the Loopamp™ SARS-CoV-2 Detection Kit.
Study design
One hundred and fifty-one nasopharyngeal swab and 88 sputum samples obtained from individuals with suspected or confirmed COVID-19 were examined.
Results
RT-LAMP had high specificity (98.5 % (95 % CI: 96.9–100 %)), sensitivity (87.0 % (95 % CI: 82.8–91.3 %)), positive predictive value (97.9 % (95 % CI: 96.1–99.7 %)), negative predictive value (90.2 % (95 % CI: 86.4–94.0 %)), and concordance rate (93.3 % (95 % CI: 90.1–96.5 %)). Nasopharyngeal and sputum samples positive in RT-LAMP contained as few as 10.2 and 23.4 copies per 10 μL, respectively. RT-LAMP showed similar performance to RT-PCR for samples with cycle threshold value below 36.
Conclusions
These results indicate that RT-LAMP is a highly reliable and at least equivalent to RT-PCR in utility, and potentially applicable in settings that are more diverse as a point-of-care tool.
Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; RT-LAMP, reverse transcription loop-mediated isothermal amplification; RT-PCR, reverse transcription-polymerase chain reaction; NIID, the National Institute of Infectious Diseases
Keywords: SARS-CoV-2, COVID-19, Polymerase chain reaction, RT-LAMP, Sputum, Diagnosis
1. Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was first reported in Wuhan, Hubei Province, China, and rapidly spread worldwide [1,2]. Approximately 2 months after the first report, the World Health Organization declared a COVID-19 pandemic on March 11, 2020. By November 3, 2020, nearly 50 million confirmed cases of COVID-19, including more than one million deaths, had been reported in 219 countries [3]. A serious consequence of the pandemic is that the medical-care system has become overwhelmed. Therefore, the accurate and rapid diagnosis of COVID-19, as well as the development of effective vaccines and treatments, is critical to control this disease and its spread within populations.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis of upper and lower respiratory swab samples is the current standard for the diagnosis of COVID-19 [2,4]. However, it relies on sophisticated equipment, specific skills, and requires 1.5–2 h to complete the reaction [5,6]. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) does not require expensive equipment such as thermal cyclers, is easy to perform, and only requires 1 h to obtain results [[7], [8], [9]]. Hence, RT-LAMP is regarded as a point-of-care testing tool, with a reported detection limit of 0.4–500 viral copies/10 μL, which varies with different enzymes and master mixes [10].
In this study, we evaluated the clinical performance of RT-LAMP for detecting SARS-CoV-2, including specificity, sensitivity, and the minimum amount of RNA detected, comparing and contrasting it with that of RT-PCR using the same viral RNA specimens.
2. Materials and methods
2.1. Study approval
The study protocol was approved by the institutional review board of Osaka Habikino Medical Center on April 30, 2020 (Approval No. 1017). Written informed consent was obtained from all enrolled patients.
2.2. Patient characteristics, specimen collection, and RNA extraction
We enrolled patients with suspected or confirmed COVID-19, who had been admitted to the Osaka Habikino Medical Center between April 1 and July 31, 2020. Nasopharyngeal swab samples were collected in 1 mL of sterile saline, with sputum samples collected by the patients themselves. All samples were stored at −30 °C until analyzed. RNA was extracted from the specimens using the QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany) in accordance with the manufacturer’s instructions. The extracted RNA was aliquoted and stored at −80 °C until it was used for amplification by RT-LAMP and RT-PCR. A flowchart of the study protocol is provided in Fig. 1 .
Fig. 1.
Study protocol. We enrolled patients with suspected or confirmed COVID-19; all provided written informed consent. Nasopharyngeal swab samples were collected, viral RNA was extracted, and divided into two aliquots for amplification by RT-LAMP and RT-PCR.
2.3. RT-LAMP assay
Assays were performed using a Loopamp™ SARS-CoV-2 Detection Kit (Eiken Chemical, Tokyo, Japan), targeting genes encoding the nucleocapsid (N) and RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2, in accordance with the manufacturer’s instructions. The reaction mixture consisted of RNA sample (10 μL) and the provided master mix including a set of primers (15 μL). The mixture was incubated at 62.5 °C for 35 min, with turbidity measured every 6 s using a real-time Loopamp EXIA turbidity meter (Eiken Chemical). Assays were scored as positive when the differential value approached 0.05, at which point the threshold time (Tt) was recorded. In addition, LAMP positive reactions can also be detected visually, through fluorescence, using calcein staining; therefore, we reassessed RT-LAMP reactions using this visual endpoint detection [9].
2.4. RT-PCR
RT-PCR was performed using a protocol devised by the National Institute of Infectious Diseases (NIID), which is a national recommendation for SARS-CoV-2 detection in Japan [11]. In this assay, the QuantiTect® Probe RT-PCR kit (QIAGEN, Hilden, Germany) and LightCycler® 480 (Roche, Penzberg, Germany) were used with the following primers and probe targeting the SARS-CoV-2 N gene: 500 nM of forward primer, 5′-AAA TTT TGG GGA CCA GGA AC-3′; 700 nM of reverse primer, 5′-TGG CAG CTG TGT AGG TCA AC-3′; and 200 nM of the probe, 5′-FAM- ATG TCG CGC ATT GGC ATG GA -BHQ-3′ [12]. The extracted RNA (5 μL) was added, and the thermal cycling conditions were 50 °C for 30 min; initial denaturation at 95 °C for 15 min; 45 cycles of 95 °C for 15 s and 60 °C for 60 s. Artificial RNA for the sequence of SARS-CoV-2 N gene described in a protocol devised by the NIID was used as the positive control for the RT-PCR [11]. The concentration of RNA was calculated based on the OD260 values, and 10-fold serial dilutions from 5 × 103 to 5 × 10° copies per 5 μL were prepared. The standard curve was included in every RT-PCR run.
Test outcomes were evaluated based on the cycle threshold (Ct) value. Viral loads were determined using the Ct value and a calibration curve.
3. Results
3.1. Positivity rates
In total, 151 nasopharyngeal samples and 88 sputum samples were assayed. Using the conventional RT-PCR assay, 79 of 151 (52.3 %) nasopharyngeal and 29 of 88 (33.0 %) sputum samples were positive. Using the RT-LAMP assay, 71 of 151 (47.0 %) nasopharyngeal and 25 of 88 (28.4 %) sputum samples were positive (Table 1 ). We obtained the same RT-LAMP results using both turbidity and visual fluorescence detection.
Table 1.
Correlation between RT-PCR and RT-LAMP assays.
|
RT-LAMP |
|||||
|---|---|---|---|---|---|
| + | – | Total | |||
| Nasopharyngeal swab and sputum samples | RT-PCR | + | 94 | 14 | 108 |
| – | 2 | 129 | 131 | ||
| total | 96 | 143 | 239 | ||
| Nasopharyngeal swab samples | RT-PCR | + | 70 | 9 | 79 |
| – | 1 | 71 | 72 | ||
| total | 71 | 80 | 151 | ||
| Sputum samples | RT-PCR | + | 24 | 5 | 29 |
| – | 1 | 58 | 59 | ||
| total | 25 | 63 | 88 | ||
The specificities for nasopharyngeal samples, sputum samples, and total samples between the RT-PCR and RT-LAMP assays were 98.6 (71/72, 95 % CI: 97.1–100 %), 98.3 (58/59, 95 % CI: 95.6–100 %), and 98.5 % (129/131, 95 % CI: 96.9–100 %), respectively. The sensitivities for nasopharyngeal samples, sputum samples, and total samples between the RT-PCR and RT-LAMP assays were 88.6 (70/79, 95 % CI: 83.5–93.7 %), 82.8 (24/29, 95 % CI: 74.9–90.7 %), and 87.0 % (94/108, 95 % CI: 82.8–91.3 %), respectively. The positive predictive values for nasopharyngeal samples, sputum samples, and total samples between the RT-PCR and RT-LAMP assays were 96.0 (24/25, 95 % CI: 91.9–100 %), 98.6 (70/71, 95 % CI: 96.7–100 %), and 97.9 % (94/96, 95 % CI: 96.1–99.7 %), respectively. The negative predictive values for nasopharyngeal samples, sputum samples, and total samples between the RT-PCR and RT-LAMP assays were 92.1 (58/63, 95 % CI: 86.4–97.7 %), 88.8 (71/80, 95 % CI: 83.7–93.8 %), and 90.2 % (129/143, 95 % CI: 86.4–94.0 %), respectively. The concordance rates for nasopharyngeal samples, sputum samples, and total samples between RT-PCR and RT-LAMP assays were 93.4 (141/151, 95 % CI: 89.4–97.3 %), 93.2 (82/88, 95 % CI: 87.9–98.4 %), and 93.3 % (223/239, 95 % CI: 90.1–96.5 %), respectively (Table 1).
3.2. RT-LAMP Tt, RT-PCR Ct, and viral load
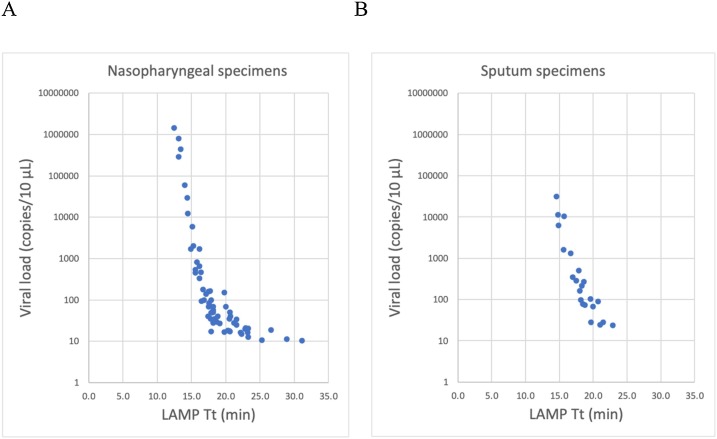
Nasopharyngeal and sputum samples positive in RT-LAMP contained as few as 10.2 and 23.4 copies/10 μL, respectively (Fig. 2 ). Nine nasopharyngeal samples and five sputum specimens were positive in RT-PCR, but negative in RT-LAMP (Table 1, Table 2 ). The 14 samples that were negative in RT-LAMP exhibited high Ct values (≥ 35.7) in RT-PCR, with relatively low viral RNA copy numbers: 18.9 (1 sputum specimen), 12.6 (1 nasopharyngeal specimen), or < 10 copies/10 μL (4 sputum and 8 nasopharyngeal specimens; Table 2).
Fig. 2.
Association between threshold time (Tt) of RT-LAMP and viral loads for (A) nasopharyngeal swabs and (B) sputum specimens.
Table 2.
Nasopharyngeal and sputum specimens assayed as positive in RT-PCR and negative in RT-LAMP and vice versa.
| RT-PCR |
RT-LAMP | ||||
|---|---|---|---|---|---|
| Sample source | Sample number | Result | Ct value | Viral load (copies/10 μL) | Result (Tt value)a |
| Nasopharyngeal | 1 | + | 36.0 | 12.6 | – |
| 2 | + | 36.7 | <10 | – | |
| 3 | + | 36.7 | <10 | – | |
| 4 | + | 37.0 | <10 | – | |
| 5 | + | 37.0 | <10 | – | |
| 6 | + | 37.1 | <10 | – | |
| 7 | + | 37.2 | <10 | – | |
| 8 | + | 37.6 | <10 | – | |
| 9 | + | 38.3 | <10 | – | |
| Sputum | 1 | + | 35.7 | 18.9 | – |
| 2 | + | 36.8 | <10 | – | |
| 3 | + | 37.1 | <10 | – | |
| 4 | + | 37.2 | <10 | – | |
| 5 | + | 37.6 | <10 | – | |
| Nasopharyngeal | 1 | – | NA | NA | + (22.2) |
| Sputum | 1 | – | NA | NA | + (24.9) |
Tt, threshold time; NA, not applicable.
When RT-LAMP and RT-PCR positive outcomes were stratified based on the viral load, 47 of 47, 37 of 39, and 10 of 22 samples with >60, 10–60, and <10 RNA copies/10 μL, respectively, were positive in the RT-LAMP and RT-PCR assays.
4. Discussion
Because of its transmissibility and rapid spread, COVID-19 is a major public health concern. Although PCR is considered the gold standard for the diagnosis of COVID-19, early and accurate means of diagnosis, such as RT-LAMP, are urgently required. Nasopharyngeal samples analyzed by RT-LAMP have a documented concordance rate of 76.2–97.4 %, a sensitivity of 80–100 %, and a specificity of 72.7–100 %, compared to analysis by RT-PCR [[13], [14], [15]].
In this study, we evaluated the difference between RT-PCR and RT-LAMP for the diagnosis of COVID-19. The key differences in this study, compared to the previously published studies, were the sample size (239 in this study vs 21, 56, and 76 in the previous studies), and the evaluation of the sputum samples in addition to the nasopharyngeal swab samples.
The concordance rate for all samples in our study between RT-PCR and RT-LAMP was 93.3 % (223/239, 95 % CI: 90.1–96.5 %). RT-LAMP displayed a high degree of specificity, sensitivity, positive predictive value, and negative predictive value for SARS-CoV-2 detection.
When samples were classified based on viral load, the divergence of outcomes between RT-PCR and RT-LAMP was associated with viral load. These results indicate that RT-LAMP is a highly suitable tool for detection of SARS-CoV-2 in patients harboring >60 copies/10 μL, but the sensitivity of the method is insufficient for patients harboring <10 copies/10 μL. The minimum amount of RNA detected by RT-LAMP found herein was similar to that reported previously [10]. RT-LAMP may be suitable for patients with relatively high viral load, in terms of detecting the virus, while RT-PCR would be a better test for detection of SARS-CoV-2, especially in asymptomatic patients, who may have low viral loads.
Interestingly, one sample each from the nasopharyngeal and sputum samples tested negative for SARS-CoV-2 in RT-PCR, but positive in RT-LAMP (Table 2). Any association with Tt values is unclear. This discrepancy may depend on the characteristics of the RT-LAMP method, which uses a set of four types of primers based on six distinct regions of the target gene, as opposed to the two primers used in RT-PCR [7].
The primary limitation of this study is the small number of samples analyzed. In addition, samples were obtained from patients hospitalized with symptoms. Hence, further studies with larger sample sizes are required. In conclusion, this study supports the applicability of the RT-LAMP assay for nasopharyngeal swabs and sputum samples for the detection of SARS-CoV-2. RT-LAMP is a simple, low-cost, rapid, and highly reliable method, equivalent to RT-PCR, especially for patients with relatively high viral loads. It is also adaptable to more diverse diagnostic settings, similar to the ones during the current pandemic.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
The authors thank the patients, their families, and all members of the COVID-19 care team at Osaka Habikino Medical Center.
References
- 1.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2.Sharfstein J.M., Becker S.J., Mello M.M. Diagnostic testing for the novel coronavirus. JAMA. 2020;323:1437–1438. doi: 10.1001/jama.2020.3864. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization . 2020. WHO Coronavirus Disease (COVID-19) Dashboard.https://covid19.who.int/ [Google Scholar]
- 4.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu R., Wu X., Wan Z., Li Y., Zuo L., Qin J., Zhang C. Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virol. Sin. 2020;35:344–347. doi: 10.1007/s12250-020-00218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang M., Pan W., Arasthfer A., Fang W., Ling L., Fang H., Daneshnia F., Yu J., Liao W., Pei H., Li X., Lass-Flörl C. Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. Front. Cell. Infect. Microbiol. 2020;10:331. doi: 10.3389/fcimb.2020.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehme C.C., Nabeta P., Henostroza G., Raqib R., Rahim Z., Gerhardt M., Sanga E., Hoelscher M., Notomi T., Hase T., Perkins M.D. Operational feasibility of using loop-mediated isothermal amplification for diagnosis of pulmonary tuberculosis in microscopy centers of developing countries. J. Clin. Microbiol. 2007;45:1936–1940. doi: 10.1128/JCM.02352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori Y., Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson D., Lei Y. Mini review: recent progress in RT-LAMP enabled COVID-19 detection. Sens. Actuators Rep. 2020;2 doi: 10.1016/j.snr.2020.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.2019. National Institute of Infectious Diseases, Tokyo. Manual for the Detection of Pathogen 2019-nCoV ver.2.6.https://www.niid.go.jp/niid/images/epi/corona/2019-nCoVmanual20200217-en.pdf (Accessed 3 November 2020) [Google Scholar]
- 12.Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y., Kageyama T., Matsuyama S., Takeda M. Development of genetic diagnostic methods for detection for novel coronavirus 2019(nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020;73:304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 13.Kitagawa Y., Orihara Y., Kawamura R., Imai K., Sakai J., Tarumoto N., Matsuoka M., Takeuchi S., Maesaki S., Maeda T. Evaluation of rapid diagnosis of novel coronavirus disease (COVID-19) using loop-mediated isothermal amplification. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R., Wu X., Wan Z., Li Y., Jin X., Zhang C. A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int. J. Mol. Sci. 2020;21:2826. doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Österdahl M.F., Lee K.A., Lochlainn M.N., Wilson S., Douthwaite S., Horsfall R., Sheedy A., Goldenberg S.D., Stanley C.J., Spector T.D., Steves C.J. Detecting SARS-CoV-2 at point of care: preliminary data comparing loop-mediated isothermal amplification (LAMP) to polymerase chain reaction (PCR) BMC Infect. Dis. 2020;2:783. doi: 10.1186/s12879-020-05484-8. [DOI] [PMC free article] [PubMed] [Google Scholar]