Abstract
Pseudomonas aeruginosa is one of the most commonly isolated bacteria from clinical specimens, with an increasing isolation frequency in nosocomial outbreaks. The hypothesis tested was whether carbapenem-resistant P. aeruginosa strains display an altered carriage of the virulence factor genes, depending on the type of carbapenem resistance. The aim of the study was to investigate, by PCR, the frequency of 10 chosen virulence factors genes (phzM, phzS, exoT, exoY, exoU, toxA, exoS, algD, pilA and pilB) and the genotype distribution in 107 non-duplicated carbapenem-resistant P. aeruginosa isolates. P. aeruginosa genes involved in phenazine dyes and exoenzyme T synthesis were noted with the highest frequency (100%). Fimbriae-encoding genes were detected with the lowest incidence: 15.9% and 4.7% for pilin A and B, respectively. The differences observed between the exoS gene prevalence amongst the carbapenemase-positive and the carbapenemase-negative strains and the pilA gene prevalence amongst the strains of different origins were statistically significant. Virulence genes’ prevalence and the genotype distribution vary amongst P. aeruginosa strains resistant to carbapenems, especially in terms of their carbapenemase synthesis ability and the strain origin.
Keywords: biofilm, exotoxins, Pseudomonas aeruginosa biofilm, Pseudomonas aeruginosa virulence, toxins, virulence, virulence genes
1. Introduction
Pseudomonas aeruginosa is a ubiquitous Gram-negative non-fermenting rod, a typical bacterial opportunistic pathogen often found in a hospital environment. P. aeruginosa strains are responsible for a wide range of human infections, occurring mostly amongst hospitalized patients. Thus, P. aeruginosa rods are one of the most often isolated bacteria from clinical specimens. Their increasing resistance to carbapenems and their role in nosocomial outbreaks of different localizations are underlined by their involvement in severe respiratory and urinary tract infections, as well as skin and soft tissue infections or bacteremia [1].
P. aeruginosa rods are intrinsically resistant to numerous antimicrobials and also easily acquire antibiotic resistance mechanisms to the majority of antibiotic groups, including carbapenems. These beta-lactam representatives (e.g., imipenem and meropenem) are often the last-resort antibiotics. The enzymatic mechanism of carbapenem resistance usually results from metallo-β-lactamase (MBL) synthesis. The most prevalent of these carbapenemases among P. aeruginosa strains worldwide are Verona-Integron-Metallo-beta-lactamases (VIM) and imipenemases (IMP-type enzymes), encoded by blaVIM and blaIMP genes, respectively. P. aeruginosa resistance to carbapenems may also result from a loss of OprD protein porins responsible for carbapenem transport into bacterial cells, overexpression of efflux pumps that actively pump out antimicrobials outside the cell or some other carbapenem-hydrolyzing enzymes.
It has been previously found, both within our unit as well as other units worldwide, that the incidences of carbapenem-resistant P. aeruginosa (CRPA) strain isolation, and their contribution to nosocomial infections, have increased with time [2]. Additionally, it has been shown that CRPA strains may clonally spread worldwide with some dominant sequence types [3].
The pathogenicity of P. aeruginosa is based on the synthesis of several cell- and biofilm-associated compounds (e.g., flagella, pili, alginate capsule and lipopolysaccharide). P. aeruginosa is also capable of secreting proteins that may play a potential role in its pathogenicity, typically extracellular virulence factors, mostly toxins (e.g., toxin A, exotoxins T, U and Y and exoenzyme S) and enzymes (e.g., elastases and phospholipases) [4]. Exotoxin A, encoded by the toxA gene, is responsible for the cytotoxic capacity of P. aeruginosa. It is a protein involved in pro-inflammatory cytokine synthesis stimulation. The algD gene encodes GDP-mannose 6-dehydrogenase, an enzyme involved in the alginate biosynthesis pathway. This compound is one of the key components of P. aeruginosa’s biofilm. The pilA and pilB genes encode bacterial pilins—the structural proteins synthesized by P. aeruginosa which are strongly involved in bacterial adhesion, as well as biofilm formation. The phzM and phzS genes encode phenazine-1-carboxylate, an enzyme involved in pyocyanin biosynthesis. The role of pyocyanin is to increase the level of reactive oxygen species (ROS) within a eukaryotic cell. An increased concentration of ROS within neutrophils could result in apoptosis of human cells. The exoS, exoT, exoU and exoY genes in P. aeruginosa encode components of the type III secretion system (TTSS). They are believed to be some of the most important virulence determinants of P. aeruginosa.
The above-mentioned virulence factors usually cooperate during P. aeruginosa colonization of the host cell and biofilm formation, allowing for further infection and additionally increasing pathogenicity. Most of the virulence factors are chromosomally encoded and their gene sequence has been determined in the P. aeruginosa PAO1 strain genome [5]. However, P. aeruginosa genome elasticity has been previously confirmed by Ramsay et al. [6].
Although the synthesis of the mentioned virulence factors is controlled by regulatory systems (e.g., bacterial quorum sensing signaling) [4,7], the presence of virulence genes is a key feature of the strains’ pathogenicity. Additionally, it has been shown that the co-existence of antimicrobial resistance and virulence determinants in P. aeruginosa isolates is becoming an alarming threat. This underlines the importance of the mentioned issue and demands continuous monitoring of multidrug-resistant (MDR) pathogens in order to determine therapeutic options for the successful treatment of such infections [8].
The aim of this study was to determine the genetic characteristics of 107 non-repeatable CRPA strains by assessing the distribution of two genes encoding MBL (blaVIM and blaIMP) and 10 genes of the virulence factors or the enzymes involved in their biosynthesis (exoS, toxA, algD, pilA, pilB, exoT, exoY, exoU, phzM and phzS). The mentioned genes were chosen based on previous studies. An additional purpose of this study was to check whether the virulence potential of CRPA strains differs depending on the type of mechanism of carbapenem resistance.
2. Results
2.1. The MBL Genes
The prevalence of carbapenem resistance mechanism genes amongst the examined strains was as follows: the blaVIM gene was detected in 32 (29.9%) of the examined strains while none of the examined strains were positive for the blaIMP gene.
2.2. The Virulence Factor Genes
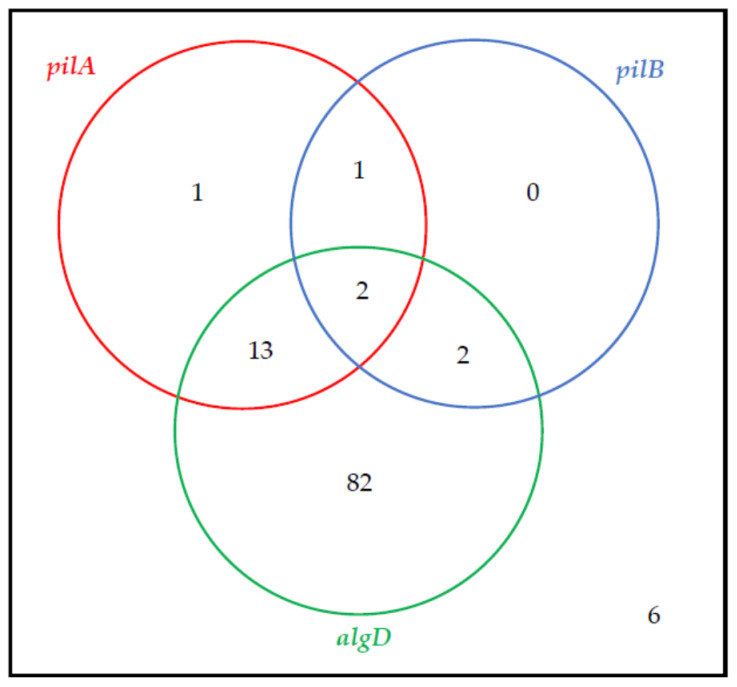
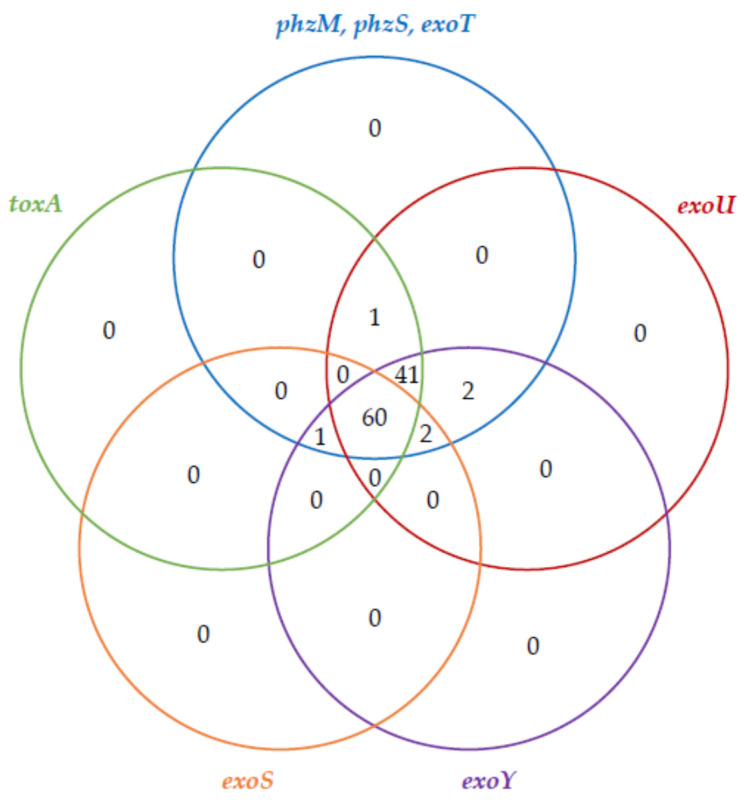
Assessment of the virulence background revealed a wide gene distribution variety. The prevalence of virulence genes amongst the examined strains was as follows: the exoT, phzM and phzS genes were noted in all of the tested strains, whereas the pilA and pilB genes were observed with the lowest frequency—amongst 15.9% and 4.7% of the examined isolates, respectively. The occurrence of the remaining virulence genes was as follows: 99.1% for exoY and exoU; 96.3%—toxA; 92.5%—algD; and 58.9% for exoS. Venn diagrams representing the co-existence of the biofilm-associated genes and toxin genes amongst the examined P. aeruginosa strains are shown in Figure 1 and Figure 2, respectively.
Figure 1.
The co-existence of biofilm synthesis-associated genes (Venn diagram) amongst the tested strains (n = 107).
Figure 2.
The co-existence of toxin synthesis-associated genes (Venn diagram) amongst the examined P. aeruginosa strains (n = 107).
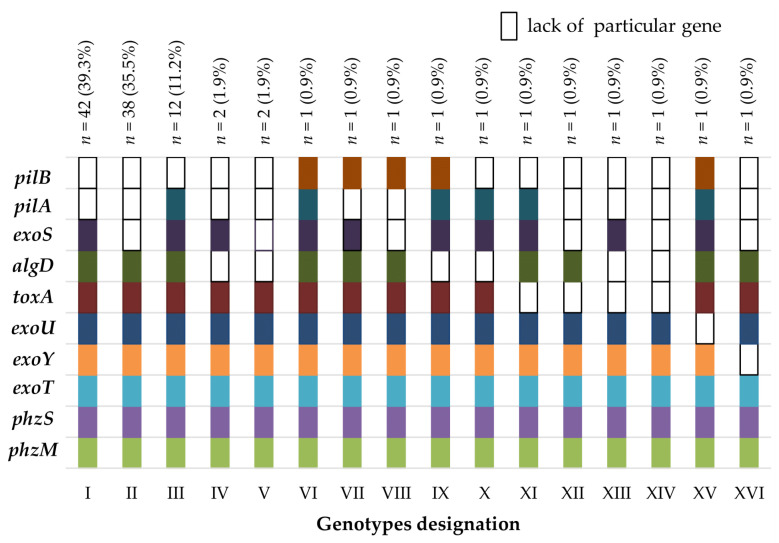
Sixteen observed genotypes (named I–XVI) and their prevalence with the distribution amongst the examined strains are shown in Figure 3. The most prevalent genotype, including all the genes detected (except for the pilA and pilB genes), was observed among 42 (39.3%) of the isolates. Eleven (10.3%) strains presented distinctive genotypes (numbered VI–XVI). The distribution and co-existence of the genotypes noted amongst P. aeruginosa strains derived from surgery, intensive care units (ICU) and rehabilitation clinics are presented in Table S1 and Figure S1. The corresponding genotypes’ distributions and co-existence with respect to the specimens that the strains were isolated from are presented in Table S2 and Figure S2. Of note, genotype II was the most dominant amongst the strains derived from the ICU patients, while for the remaining wards and the whole group of strains included in the study, the most prevalent was genotype I. A statistically significant difference (χ2 test) was additionally observed between the exoS gene frequency when compared to the rest of the tested genes prevalence (p < 0.001).
Figure 3.
Genotypes’ distribution detected amongst the examined P. aeruginosa strains (n = 107).
The differences between the virulence factor genes’ frequency noted between carbapenemase-positive and carbapenemase-negative CRPA strains are indicated in Table 1. The greatest differences in the positive results of these genes in terms of the carbapenemase synthesis ability were observed for the exoS gene (77.0% vs 18.2%—statistically significant difference, p < 0.001) and the pilA gene (20.3% vs 6.1%).
Table 1.
The prevalence of the virulence factor genes with respect to the presence of carbapenem resistance mechanisms (n = 107); (*)—a statistically significant difference (χ2 test) noted between the exoS gene frequency in the two strain subgroups is indicated (p < 0.001).
| Subgroup | Gene | phzM | phzS | exoT | exoY | exoU | toxA | algD | exoS * | pilA | pilB |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbapenemase (−) | n = 74 | 74 | 74 | 74 | 73 | 73 | 72 | 69 | 57 | 15 | 4 |
| % | 100.0 | 100.0 | 100.0 | 98.6 | 98.6 | 97.3 | 93.2 | 77.0 | 20.3 | 5.4 | |
| Carbapenemase (+) | n = 33 | 33 | 33 | 33 | 33 | 33 | 31 | 30 | 6 | 2 | 1 |
| % | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 93.9 | 90.9 | 18.2 | 6.1 | 3.0 |
A statistical analysis also revealed a positive correlation for genes’ coexistence, noted for the following pairs of genes: toxA/algD (0.34471) and exoS/pilA (0.340511).
Additionally, the pilA gene was noted more frequently among the strains derived from gastrointestinal tract infections when compared to the strains isolated from respiratory (0.0250), bloodstream (0.0229) or skin and soft tissue infections (0.0394), and when compared to the pilB gene frequency among the strains isolated from the gastrointestinal tract (0.0389).
Moreover, the pilA gene was noted less frequently when compared to the exoS gene among the strains derived from urinary (0.0094), respiratory tract (<0.0000), bloodstream (0.0184) or skin and soft tissue infections (0.0352). As well as this, the pilB gene was noted statistically less frequently than the exoS gene, regardless of the clinical specimen type that the strains were isolated from.
Of note, in the genotypes distribution analysis, with respect to the strains’ origin, the only statistically significant relation was observed for the strains with genotype III that were noted more often from the gastrointestinal tract when compared to those of respiratory tract origin.
3. Discussion
In recent years, an increased number of carbapenem-resistant strains among P. aeruginosa isolates has been observed. Infections due to CRPA are currently one of the most dangerous healthcare-associated infections, since carbapenems are often the last-choice drugs in the treatment of infections with this etiology [9]. In the available literature, numerous researchers have described the genetic features of P. aeruginosa with respect to different variables (e.g., strain origin, susceptibility to antimicrobials, clinical specimen and patients’ hospitalization length). However, little information on the virulence gene prevalence and genotype distribution amongst the CRPA strains can be found in the relevant literature [10,11].
In the available literature, the only significant relationship between virulence factors and beta-lactams resistance was observed for the strains with oxacillinases (OXA)- and IMP-like beta-lactamase genes [12]. The association between the CRPA phenotype and virulence genes carriage is still unclear and needs further studies.
Relatively much is known about the association between P. aeruginosa strains type III secretion, their antibiotic resistance and the clinical outcome of the patients infected by them [13]. However, to our knowledge, the only correlation between antibiotic resistance and a specific virulence genotype, in a group of CRPA strains, was observed between the exoU gene and the mechanism of resistance to fluoroquinolones [14].
In the present study, the prevalence of the two genes encoding MBL (blaVIM and blaIMP) and 10 genes of the virulence factors or enzymes involved in their biosynthesis (exoS, toxA, algD, pilA, pilB, exoT, exoY, exoU, phzM and phzS) was evaluated. Interestingly, there was only one strain among the examined isolates group that carried all of the genes for the studied virulence factors. Of note, the mentioned strain was negative for both of the investigated carbapenemase genes. Further studies are needed to decipher the hyper-virulent genetic potential of this particular isolate and its pathogenic properties.
In our study, 32 (29.9%) of the isolates were blaVIM-positive, while none of the strains were blaIMP-positive. This finding is quite different from another study carried out in Poland among pediatric patient-derived strains, which showed that only 10.9% (6/55) of P. aeruginosa isolates were blaVIM-positive and 1.8% (1/55) blaIMP-positive [15]. Our result is also different from the observation noted by Sharifi et al. [16], in which the predominant MBLs (84.5%) were IMP-like enzymes. In the cited study, VIM-like carbapenemases were observed amongst 4.8% of the investigated strains only. In turn, Pobiega et al. obtained similar results to ours for CRPA strains isolated from patients with urinary tract infections (UTIs) in southern Poland [17]. Interestingly, in different countries, P. aeruginosa carrying the blaIMP gene is also more prevalent than that with blaVIM [18,19,20]. Since the pathogenic strains are derived mainly from hospitalized patients, these results could confirm an endemic intra-hospital spread of this species.
The resistance to carbapenems among P. aeruginosa may also be conditioned by other mechanisms (e.g., loss of OprD protein, overexpression of efflux pumps and other carbapenem-hydrolyzing enzymes) [9]. Therefore, the mechanism of the antibiotic resistance type, resulting from blaVIM gene presence, was found only amongst 29.9% of the strains. The genetic background (e.g., lack of OprD porins or efflux pumps) of the carbapenem resistance among the remaining strains needs further investigation.
In this study, the frequency of the chosen 10 virulence factor genes in CRPA was examined and the strains selected for this study show a relatively high percentage of virulence factor genes. One of the most common P. aeruginosa virulence factor genes detected in our study, amongst 103 (96.3%) analyzed strains, was the toxA gene. To our knowledge, the only research on toxA gene carriage among CRPA was conducted by Goncalves et al. [11]. The mentioned authors obtained similar results—in their study, 87.5% of the CRPA strains carried the toxA gene. Other researchers who analyzed P. aeruginosa isolates derived from respiratory tract infections, but not CRPA exclusively, also showed a high percentage of the toxA gene (80.3%) [21]. Our results confirm the observation revealed in previous studies: the toxA gene is more common among MDR strains and, thus, may lead, additionally, to their increased virulence [22]. Noteworthily, a positive correlation was observed for the toxA and algD genes’ existence, suggesting that there might be a relationship between the carriage of toxins and biofilm genes in P. aeruginosa, but this observation requires further studies.
The majority of pathogenic P. aeruginosa are capable of biofilm formation. As a result, they tend to be more difficult to remove from the hospital environment, as well as from medical and diagnostic equipment. This virulence determinant is common in P. aeruginosa strains [8]. To our knowledge, the only phenotypic study on biofilm-involved virulence factors of CRPA was conducted by El-Mahdy and El-Kannishy [23]. The mentioned property was found amongst 77.5% of the examined strains. In our study on the algD gene’s presence, 99 (92.5%) CRPA strains were positive. These results are consistent with the results of the research carried out by Ellappan et al. [8], showing that 92.9% of CRPA strains carry the algD gene. The percentage of the algD gene reached 87.5% in a group of P. aeruginosa isolates (not CRPA exclusively) examined by Kamali et al. [24], with 88.1% and 84.6% for biofilm producer and non-biofilm strains, respectively.
The second determinants of bacterial biofilm formation are pilins. However, relatively low percentages of the pilA (15.9%) and the pilB (4.7%) genes were observed among the studied P. aeruginosa strains. These results are comparable with the second research study from Poland among P. aeruginosa strains derived from patients with a UTI (not CRPA exclusively). In their study, 19.2% of the isolates carried the pilA gene, while none of the strains were positive for the pilB gene [17]. Another study showed that the percentage of the pilB gene reached 8.3% in a group of MDR P. aeruginosa isolates, examined by Sharifi et al. [16]. Only three of the strains studied in the present work carried both of the mentioned genes. These results indicate that for the evaluated strains group, the potential for alginate synthesis, observed at least on a molecular level, might be one of the major determinants involved in biofilm formation. Interestingly, a positive correlation was observed for the pilA and exoS genes’ co-existence.
pilA gene carriage was noted most frequently among the strains derived from the gastrointestinal tract when compared to strains of other origin. In our opinion, this finding is very interesting, suggesting that this particular gene’s presence is crucial for human gastrointestinal tract colonization or infection. Thus, there may be a potential relationship between the selected virulence genes’ carriage and P. aeruginosa’s ability to form biofilm or colonize a particular place, but this finding needs further studies.
All of the investigated CRPA isolates were positive for the phzM and phzS genes (n = 107). This may suggest that pyocyanin is one of the most prevalent virulence determinants of CRPA. Noteworthily, Fuse et al. [25] showed that in a group of MDR P. aeruginosa strains, the synthesis of pyocyanin decreases. Since our study results show that all of the CRPA strains carry the phzM and phzS genes, it is possible that either the decreased production of pyocyanin is associated with resistance to other groups of antibiotics or that its synthesis is more likely controlled at the expression level, not carriage itself [25]. It has also been previously shown that there is a significant difference in the phzS gene’s presence between environmental and clinical P. aeruginosa strains [26]. Noteworthily, in the relevant literature, the first mentioned group is considered as more sensitive to antimicrobials, while the second one—more resistant or even multidrug-resistant [26].
In our study, a relatively high prevalence of the TTSS genes (exoT—100%; exoU and exoY—99.1%; exoS—58.9%) was noted. The data from a recent meta-analysis on P. aeruginosa isolates with toxin potency indicated relatively high prevalence of exotoxins in P. aeruginosa clinical isolates. In turn, the ability to synthesize toxins is of fundamental importance from a clinical aspect. It was found that the exoT gene had the highest prevalence rate among P. aeruginosa exotoxins (0.83 (CI95%: 0.64–0.96)), reaching up to 100% amongst CRPA isolates exclusively [27]. It is consistent with the results of the present study.
The results of the exoS gene presence in our study are also concordant with the observations made by Khodayary et al. [28] and relatively close to those of Sharifi et al. [16], in which the percentage of the exoS gene in a group of the examined MDR P. aeruginosa isolates (but not for CRPA exclusively) reached 59.0% and 44.0%, respectively. The statistically significant difference observed in the exoS gene incidence in terms of carbapenemase synthesis, as the only TTSS gene, is an interesting and unclear finding that also needs further research.
Khodayary’s study also showed a high frequency of TTSS genes among MDR P. aeruginosa strains—all of the strains possessed the exoT and exoY genes, while the exoU gene was noted among 41.0% of the isolates [28]. As previously indicated, the occurrence of a particular TTSS gene is associated with increased resistance to antibiotics [28], at least amongst the strains derived from burn patients. It was even proposed that the mentioned association of MDR and the presence of specific virulence genes might be a predictive marker for a worse clinical condition of the infected patients [28].
Our study results also showed that among blaVIM-positive isolates, 81.8% (n = 27) presented the exoU+/exoS− genotype. It has also been shown that the exoU+/exoS− genotype predominates in a high-risk blaVIM-positive ST235 clone [29], which is the most disseminated clone worldwide [30]. Further studies would be necessary to confirm the mentioned strains’ subgroup classification to the most prevalent sequence type.
The results of the present study show, in turn, that among the blaVIM-negative isolates (n = 74), 57 carried the exoS gene. Only one strain carrying the exoU−/exoS+ genotype was negative for the blaVIM gene.
In the relevant literature, the association between antimicrobial resistance (or MDR phenotype) and genes carriage was investigated the most in terms of the exoU gene. As it has been previously indicated, the occurrence of this particular gene is associated with increased resistance to some antibiotics [28,31], and the exoU gene is more often found amongst MDR P. aeruginosa strains [32]. On the contrary, the exoU gene’s presence reached only 26.0% in a group of MDR P. aeruginosa isolates examined by Sharifi et al. [16]. These discrepancies may also result from the different definitions of multi-drug resistance applied by different researchers.
To summarize, higher antimicrobial resistance is generally observed in P. aeruginosa isolates possessing the exoU gene. The researchers propose that understanding the prevalence of the exoU-positive and the mRNA overexpression of resistance genes may help to select empirical therapy for the treatment of nosocomial infections caused by P. aeruginosa [31]. Noteworthily, 99.1% of the carbapenem-resistant isolates in this study also carried the exoU gene, and this finding is concordant with the results of previous research. In the meta-analysis on P. aeruginosa toxin genes, the lowest prevalence for the exoU gene reached 0.32 (CI95%: 0.24–0.41) [27].
In summary, a great genetic diversity is observed amongst CRPA strains. This variability is based on the existence of a particular plasmid or chromosomal genes set. The cause of this diversity might be the genetic material organization or “management”—the property of attaining and conserving the genes crucial for survival in the hospital environment (e.g., genes of carbapenem-resistance) and losing the currently less important virulence factor genes (e.g., pilin-encoded genes). However, the mentioned properties can also be achieved through the particular expression of relevant genes. As it has been previously shown, massive changes in the transcriptome of the strains are observed [33], including metabolic changes and the alteration of virulence—for example, during bloodstream infections [34]. These results suggest that, as part of its survival strategy and in response to changes in the patients’ blood, P. aeruginosa enhances the expression of certain virulence genes and reduces the expression of others [33,34].
Taken together, this study is believed to be one of the first and largest evaluations of virulence factor genes’ carriage in a group of CRPA strains exclusively. It reports on the diverse virulence potential of P. aeruginosa strains, at least on a genetic level, also with respect to carbapenemases synthesis ability. Further research is required to understand the mechanism of P. aeruginosa virulence genes’ carriage and the involvement of other contributing factors to this phenomenon, e.g., the mentioned genes’ expression evaluation, as previously shown [35].
The results of the simultaneous evaluation of exotoxins and antimicrobial resistance can be used to develop treatment policies against P. aeruginosa infections in hospitals and amongst hospitalized patients [27]. In effect, it would provide the necessary tools for the rapid diagnosis of hyper-virulent strains, as well as allowing us to monitor their spread. It could, finally, help to establish better strategies for the treatment [36,37] or control of infections caused by P. aeruginosa. For example, searching for the quorum sensing inactivation compounds [38,39,40] and selected virulence gene expression (down)regulation [40,41,42] may be achieved either by using natural agents [39,40,43] or attempting to increase the strains’ sensitivity to existing antimicrobials [44].
4. Materials and Methods
4.1. Origin of Strains and Selection Criteria
In total, 107 clinical isolates of imipenem- and meropenem-resistant P. aeruginosa strains were included in the study. The strains were initially identified based on typical growth on a selective medium (Cetrimide agar, bioMérieux, France) and biochemical reaction results. The final identification was made using a MALDI-TOF MS device (Bruker, Mannheim, Germany). All the strains were also initially screened for the presence of MBLs, OXA-like and Klebsiella pneumoniae carbapenemases (KPC) enzymes (data not shown). All of them were isolated in the Clinical Microbiology Laboratory of Dr Antoni Jurasz University Hospital No. 1 in Bydgoszcz, Poland. Most of the strains were derived from patients in the Anesthesiology and Intensive Care Unit (45–42.1%), different surgery clinics (26–24.3%) as well as the Rehabilitation Ward (16–15.0%). Each strain was isolated from a different patient. All the strains were also initially tested genotypically using the polymerase chain reaction—random amplified polymorphic DNA (PCR-RAPD) technique to exclude repeated isolates [45]. According to Mahenthiralingam et al.’s methodology, PCR-RAPD primers 208 and 272 were applied to confirm the absence of duplicate strains in the further study and analysis. The strains with an identical PCR-RAPD genotypic pattern were excluded from the further study (data not shown). The strains included in the study were isolated mostly from respiratory tract- (31, 29.0%), urinary tract- (25, 23.4%), wounds- (21, 19.6%), bloodstream- (12, 11.2%) and gastrointestinal tract-derived (10, 9.3%) specimens.
4.2. The Susceptibility to Carbapenems and Carbapenemase Activity Evaluation
During the antimicrobial susceptibility testing step, the following were used: the diffusion method on Mueller–Hinton agar (Becton Dickinson, Germany), imipenem (10 µg) and meropenem (10 µg) discs (Becton Dickinson, Germany) as well as Etest® strips (bioMérieux, France). The results of the antimicrobial susceptibility tests were interpreted according to the European Committee on Antimicrobial Susceptibility Testing recommendations (EUCAST, Breakpoint tables for bacteria, Clinical breakpoints—bacteria v 9.0). P. aeruginosa obtained from the American Type Culture Collection (ATCC 27853) as well as Escherichia coli ATCC 25922 strains served as a susceptibility testing quality control.
All of the examined strains were also checked phenotypically for MBL synthesis, using the method described by Lee et al. [46] and by Yong et al. [47]. Among the examined strains, only the blaVIM gene-positive isolates were positive for the detection of an enzymatic resistance mechanism to carbapenems, regardless of the methodology applied (data not shown). For all of the strains with positive carbapenemases tests, VIM-like enzymes were detected. Of note, this particular type of enzymatic resistance to carbapenems is the only mechanism recognized among the P. aeruginosa collection from our department. They were subsequently called carbapenemase-positive, while the rest were noted as carbapenemase-negative.
4.3. Bacterial DNA Isolation
A Genomic Mini kit (A&A Biotechnology, Gdynia, Poland) was applied for the DNA isolation, according to the manufacturer’s protocol. The DNA samples were stored at 4 °C before their further use for the purpose of PCR.
4.4. MBL Genes Detection
PCR was performed to detect the genes for the most common carbapenemases in Poland (VIM- and IMP-like enzymes) in the duplex version, as previously described [48]. The PCR reaction products were separated by electrophoresis on 1% agarose gel in a 1 × Tris-Boric Acid-EDTA (TBE) running buffer at 9 V/cm for one hour in a MINI SUBTM DNA CELL (BioRad, Feldkirchen, Germany) device. Their pictures were recorded and documented in Gel Doc 2000 system using the Quantity One (BioRad, Feldkirchen, Germany) program. P. aeruginosa strains carrying the blaIMP or blaVIM genes served as a positive control, while P. aeruginosa ATCC 27853 as a negative control, of the PCR (Figure S3).
4.5. Virulence Factor Genes Detection
The prevalence of 10 virulence factor genes was determined by PCR, in a separate reaction for each gene. The genes were amplified with primers selected on the basis of the published PAO1 strain genome sequence and the P. aeruginosa ATCC 27853 isolate. The amplification procedure was carried out as previously described [49,50]. The primers’ sequences and the PCR annealing temperatures for each gene amplification are presented in Table 2. The DNA isolated from the P. aeruginosa PAO1 strain (kindly provided by the National Medicines Institute in Warsaw, Poland) and the P. aeruginosa ATCC 27853 isolate served as a PCR positive control. In the amplification procedure, a thermal cycler GeneAmp® PCR System 2700 (Applied Biosystems, Foster City, CA, USA) was applied.The presence of amplicons for the particular genes was evaluated visually with the application of gel electrophoresis, based on the product size and control strain DNA amplification (Figures S4–11). The duplicates of PCR were performed for the selected strain–gene pairs to confirm the repeatability of the results, giving the consent results in each case (data not shown).
Table 2.
The PCR primers’ specification and the PCR parameters applied in the present study.
| Gene Detected | PCR Primer Name |
Manufacturer | Primer Sequence 5′→3′ | Tm (°C) |
Annealing Temperature (°C) | Product Size (bp) |
|---|---|---|---|---|---|---|
| phzS | phzS F | Sigma | TCGCCATGACCGATACGCTC | 55.9 | 63 | 1752 |
| phzS R | ACAACCTGAGCCAGCCTTCC | 55.9 | ||||
| exoT | exoT F | AATCGCCGTCCAACTGCATGCG | 58.6 | 64 | 152 | |
| exoT R | TGTTCGCCGAGGTACTGCTC | 55.9 | ||||
| exoY | exoY F | CGGATTCTATGGCAGGGAGG | 55.9 | 64 | 289 | |
| exoY R | GCCCTTGATGCACTCGACCA | 55.9 | ||||
| exoU | exoU F | CCGTTGTGGTGCCGTTGAAG | 55.9 | 64 | 134 | |
| exoU R | CCAGATGTTCACCGACTCGC | 55.9 | ||||
| toxA | toxA F | Integrated DNA Technologies | GGTAACCAGCTCAGCCACAT | 57.4 | 52 | 352 |
| toxA R | TGATGTCCAGGTCATGCTTC | 54.8 | ||||
| algD | algD F | ATGCGAATCAGCATCTTTGGT | 55.2 | 50 | 1310 | |
| algD R | CTACCAGCAGATGCCCTCGGC | 62.5 | ||||
| exoS | exoS F | CTTGAAGGGACTCGACAAGG | 55.2 | 53 | 504 | |
| exoS R | TTCAGGTCCGCGTAGTGAAT | 56.2 | ||||
| phzM | phzM F | Genomed | ATGGAGAGCGGGATCGACAG | 55.9 | 54 | 875 |
| phzM R | ATGCGGGTTTCCATCGGCAG | 55.9 | ||||
| pilA | pilA F | ACAGCATCCAACTGAGCG | 50.3 | 59 | 1675 | |
| pilA R | TTGACTTCCTCCAGGCTG | 50.3 | ||||
| pilB | pilB F | TCGAACTGATGATCGTGG | 48.0 | 56 | 408 | |
| pilB R | CTTTCGGAGTGAACATCG | 48.0 |
4.6. Statistical Methods
Statistical analysis was performed in the StatSoft Inc. (2017) STATISTICA 13.1 (data analysis software system) program using the standard chi-square test (χ2) with α ≤ 0.05 to determine the significance of the differences between the investigated genes’ prevalence amongst the examined strains and the genes’ distribution in the two distinct examined strain subgroups (carbapenemase-positive and carbapenemase-negative). The chi-square test (with Yates’ correction for small groups tested) was applied to investigate the differences in the distribution of the virulence genes and genotypes with respect to the origin of the strains. The Spearman’s rank-order correlation test was applied to investigate the association of the particular genes’ co-existence (α ≤ 0.05).
5. Conclusions
High genetic diversity was noted amongst the CRPA strains—all or most of the strains possess genes involved in phenazine synthesis and the genes for the exoenzymes released in the TTSS. Genes encoding P. aeruginosa pilins were found at the lowest frequency, suggesting that, at least for this particular strain group, alginate synthesis is a more likely factor of biofilm-associated virulence. The prevalence of the exoS gene varied the most with respect to the ability of carbapenemase synthesis. It is likely that the decreased virulence of CRPA strains, at least in some cases, may result from a particular gene loss, especially for carbapenemase producers.
Acknowledgments
This research was financially supported by Nicolaus Copernicus University funds for the basic research activity of the Microbiology Department Ludwik Rydygier Collegium Medicum in Bydgoszcz, Poland (PDB WF 839).
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/10/3/241/s1, Figure S1: Coexistence of particular genotypes (Venn diagram) noted amongst P. aeruginosa strains derived from surgery, ICU and rehabilitation clinics; Figure S2: Co-existence of particular genotypes (Venn diagram) noted amongst P. aeruginosa strains derived from clinical specimens collected for the diagnostics of respiratory tract infection (RTI), urinary tract infection (UTI), gastrointestinal tract infections (GTI), bloodstream infection (BSI) and skin and soft tissue infection (SSTI); Figure S3: Picture of an electrophoretic gel showing the amplicons of duplex PCR for MBL genes; Figure S4: Picture of an electrophoretic gel showing the amplicons of PCR for the algD gene; Figure S5: Picture of an electrophoretic gel showing the amplicons of PCR for the exoS gene; Figure S6: Picture negative of an electrophoretic gel showing the amplicons of PCR for the exoT, exoU and exoY genes; Figure S7: Picture of an electrophoretic gel showing the amplicons of PCR for the phzM gene; Figure S8: Picture of an electrophoretic gel showing the amplicons of PCR for the phzS gene; Figure S9: Picture of an electrophoretic gel showing the amplicons of PCR for the pilA gene; Figure S10: Picture of an electrophoretic gel showing the amplicons of PCR for the pilB gene; Figure S11: Picture of an electrophoretic gel showing the amplicons of PCR for the toxA gene. Table S1: Distribution of the genotypes noted amongst P. aeruginosa strains derived from surgery, ICU and rehabilitation clinics; Table S2: Distribution of the genotypes noted amongst P. aeruginosa strains derived from clinical specimen collected for the diagnostics of respiratory tract infection (RTI), urinary tract infection (UTI), gastrointestinal tract infection (GTI), bloodstream infection (BSI) and skin and soft tissue infections (SSTI).
Author Contributions
Conceptualization, T.B.; Data curation, T.B., D.D., M.R.; Formal analysis, T.B., D.D., M.R., J.K.-P.; Funding acquisition, T.B.; Investigation, T.B., D.D.; Methodology, T.B., D.D., M.R., J.K.-P.; Project administration, T.B.; Resources, T.B., D.D.; Software, T.B., J.K.-P.; Supervision, E.G.-K.; Validation, T.B., D.D., M.R., J.K.-P.; Writing–original draft, T.B., D.D.; Writing–review & editing, T.B., D.D., M.R., J.K.-P., E.G.-K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kucisec-Tepes N. Pseudomonas aeruginosa--a significant hospital pathogen and resistance to carbapenem. Acta Med. Croat. Cas. Hravatske Akad. Med. Znan. 2004;58:313–321. [PubMed] [Google Scholar]
- 2.Bogiel T., Mikucka A., Skalski T., Gospodarek E. Occurrence and susceptibility to antibiotics of carbapenem-resistant Pseudomonas aeruginosa strains between 1998 and 2009. Med. Dosw. Mikrobiol. 2010;62:221–229. [PubMed] [Google Scholar]
- 3.Papagiannitsis C.C., Medvecky M., Chudejova K., Skalova A., Rotova V., Spanelova P., Jakubu V., Zemlickova H., Hrabak J. Czech Participants of the European Antimicrobial Resistance Surveillance Network Molecular Characterization of Carbapenemase-Producing Pseudomonas Aeruginosa of Czech Origin and Evidence for Clonal Spread of Extensively Resistant Sequence Type 357 Expressing IMP-7 Metallo-β-Lactamase. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.01811-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Wrafy F., Brzozowska E., Górska S., Gamian A. Pathogenic Factors of Pseudomonas Aeruginosa—The Role of Biofilm in Pathogenicity and as a Target for Phage Therapy. Postepy Hig. Med. Dosw. 2017;71:78–91. doi: 10.5604/01.3001.0010.3792. [DOI] [PubMed] [Google Scholar]
- 5.Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., et al. Complete Genome Sequence of Pseudomonas Aeruginosa PAO1, an Opportunistic Pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 6.Ramsay K.A., Wardell S.J.T., Patrick W.M., Brockway B., Reid D.W., Winstanley C., Bell S.C., Lamont I.L. Genomic and Phenotypic Comparison of Environmental and Patient-Derived Isolates of Pseudomonas Aeruginosa Suggest That Antimicrobial Resistance Is Rare within the Environment. J. Med. Microbiol. 2019;68:1591–1595. doi: 10.1099/jmm.0.001085. [DOI] [PubMed] [Google Scholar]
- 7.Van Delden C., Iglewski B.H. Cell-to-Cell Signaling and Pseudomonas Aeruginosa Infections. Emerg. Infect. Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellappan K., Belgode Narasimha H., Kumar S. Coexistence of Multidrug Resistance Mechanisms and Virulence Genes in Carbapenem-Resistant Pseudomonas Aeruginosa Strains from a Tertiary Care Hospital in South India. J. Glob. Antimicrob. Resist. 2018;12:37–43. doi: 10.1016/j.jgar.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 9.Morita Y., Tomida J., Kawamura Y. Responses of Pseudomonas Aeruginosa to Antimicrobials. Front. Microbiol. 2014;4:422. doi: 10.3389/fmicb.2013.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pobiega M., Maciąg J., Chmielarczyk A., Romaniszyn D., Pomorska-Wesolowska M., Ziolkowski G., Heczko P.B., Bulanda M., Wojkowska-Mach J. Molecular Characterization of Carbapenem-Resistant Pseudomonas Aeruginosa Strains Isolated from Patients with Urinary Tract Infections in Southern Poland. Diagn. Microbiol. Infect. Dis. 2015;83:295–297. doi: 10.1016/j.diagmicrobio.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 11.Rossi Gonçalves I., Dantas R.C.C., Ferreira M.L., Batistão D.W.d.F., Gontijo-Filho P.P., Ribas R.M. Carbapenem-Resistant Pseudomonas Aeruginosa: Association with Virulence Genes and Biofilm Formation. Braz. J. Microbiol. 2017;48:211–217. doi: 10.1016/j.bjm.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dehbashi S., Tahmasebi H., Arabestani M.R. Association between Beta-Lactam Antibiotic Resistance and Virulence Factors in AmpC Producing Clinical Strains of P. Aeruginosa. Osong Public Health Res. Perspect. 2018;9:325–333. doi: 10.24171/j.phrp.2018.9.6.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawa T., Shimizu M., Moriyama K., Wiener-Kronish J.P. Association between Pseudomonas Aeruginosa Type III Secretion, Antibiotic Resistance, and Clinical Outcome: A Review. Crit. Care. 2014;18:668. doi: 10.1186/s13054-014-0668-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho H.H., Kwon K.C., Kim S., Koo S.H. Correlation between Virulence Genotype and Fluoroquinolone Resistance in Carbapenem-Resistant Pseudomonas Aeruginosa. Ann. Lab. Med. 2014;34:286–292. doi: 10.3343/alm.2014.34.4.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wołkowicz T., Patzer J.A., Kamińska W., Gierczyński R., Dzierżanowska D. Distribution of Carbapenem Resistance Mechanisms in Pseudomonas Aeruginosa Isolates among Hospitalised Children in Poland: Characterisation of Two Novel Insertion Sequences Disrupting the OprD Gene. J. Glob. Antimicrob. Resist. 2016;7:119–125. doi: 10.1016/j.jgar.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Sharifi H., Pouladfar G., Shakibaie M.R., Pourabbas B., Mardaneh J., Mansouri S. Prevalence of β-Lactamase Genes, Class 1 Integrons, Major Virulence Factors and Clonal Relationships of Multidrug-Resistant Pseudomonas Aeruginosa Isolated from Hospitalized Patients in Southeast of Iran. Iran. J. Basic Med. Sci. 2019;22:806–812. doi: 10.22038/ijbms.2019.35063.8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pobiega M., Maciag J., Pomorska-Wesolowska M., Chmielarczyk A., Romaniszyn D., Ziolkowski G., Heczko P.B., Wojkowska-Mach J., Bulanda M. Urinary Tract Infections Caused by Pseudomonas Aeruginosa among Children in Southern Poland: Virulence Factors and Antibiotic Resistance. J. Pediatr. Urol. 2016;12:36.e1–36.e6. doi: 10.1016/j.jpurol.2015.05.034. [DOI] [PubMed] [Google Scholar]
- 18.Rostami S., Farajzadeh Sheikh A., Shoja S., Farahani A., Tabatabaiefar M.A., Jolodar A., Sheikhi R. Investigating of Four Main Carbapenem-Resistance Mechanisms in High-Level Carbapenem Resistant Pseudomonas Aeruginosa Isolated from Burn Patients. J. Chin. Med. Assoc. 2018;81:127–132. doi: 10.1016/j.jcma.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Al-Charrakh A.H., Al-Awadi S.J., Mohammed A.S. Detection of Metallo-β-Lactamase Producing Pseudomonas Aeruginosa Isolated from Public and Private Hospitals in Baghdad, Iraq. Acta Med. Iran. 2016;54:107–113. [PubMed] [Google Scholar]
- 20.Fan X., Wu Y., Xiao M., Xu Z.-P., Kudinha T., Bazaj A., Kong F., Xu Y.-C. Diverse Genetic Background of Multidrug-Resistant Pseudomonas Aeruginosa from Mainland China, and Emergence of an Extensively Drug-Resistant ST292 Clone in Kunming. Sci. Rep. 2016;6:26522. doi: 10.1038/srep26522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al Dawodeyah H.Y., Obeidat N., Abu-Qatouseh L.F., Shehabi A.A. Antimicrobial Resistance and Putative Virulence Genes of Pseudomonas Aeruginosa Isolates from Patients with Respiratory Tract Infection. Germs. 2018;8:31–40. doi: 10.18683/germs.2018.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amirmozafari N., Fallah Mehrabadi J., Habibi A. Association of the Exotoxin A and Exoenzyme S with Antimicrobial Resistance in Pseudomonas Aeruginosa Strains. Arch. Iran. Med. 2016;19:353–358. [PubMed] [Google Scholar]
- 23.El-Mahdy R., El-Kannishy G. Virulence Factors of Carbapenem-Resistant Pseudomonas Aeruginosa in Hospital-Acquired Infections in Mansoura, Egypt. Infect. Drug Resist. 2019;12:3455–3461. doi: 10.2147/IDR.S222329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamali E., Jamali A., Ardebili A., Ezadi F., Mohebbi A. Evaluation of Antimicrobial Resistance, Biofilm Forming Potential, and the Presence of Biofilm-Related Genes among Clinical Isolates of Pseudomonas Aeruginosa. BMC Res. Notes. 2020;13:27. doi: 10.1186/s13104-020-4890-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuse K., Fujimura S., Kikuchi T., Gomi K., Iida Y., Nukiwa T., Watanabe A. Reduction of Virulence Factor Pyocyanin Production in Multidrug-Resistant Pseudomonas Aeruginosa. J. Infect. Chemother. 2013;19:82–88. doi: 10.1007/s10156-012-0457-9. [DOI] [PubMed] [Google Scholar]
- 26.Liew S.M., Rajasekaram G., Puthucheary S.A., Chua K.H. Antimicrobial Susceptibility and Virulence Genes of Clinical and Environmental Isolates of Pseudomonas Aeruginosa. PeerJ. 2019;7:e6217. doi: 10.7717/peerj.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Javanmardi F., Emami A., Pirbonyeh N., Keshavarzi A., Rajaee M. A Systematic Review and Meta-Analysis on Exo-Toxins Prevalence in Hospital Acquired Pseudomonas Aeruginosa Isolates. Infect. Genet. Evol. 2019;75:104037. doi: 10.1016/j.meegid.2019.104037. [DOI] [PubMed] [Google Scholar]
- 28.Khodayary R., Nikokar I., Mobayen M.R., Afrasiabi F., Araghian A., Elmi A., Moradzadeh M. High Incidence of Type III Secretion System Associated Virulence Factors (Exoenzymes) in Pseudomonas Aeruginosa Isolated from Iranian Burn Patients. BMC Res. Notes. 2019;12:28. doi: 10.1186/s13104-019-4071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellés A., Bueno J., Rojo-Bezares B., Torres C., Javier Castillo F., Sáenz Y., Seral C. Characterisation of VIM-2-Producing Pseudomonas Aeruginosa Isolates from Lower Tract Respiratory Infections in a Spanish Hospital. Eur. J. Clin. Microbiol. Infect. Dis. 2018;37:1847–1856. doi: 10.1007/s10096-018-3318-3. [DOI] [PubMed] [Google Scholar]
- 30.Oliver A., Mulet X., López-Causapé C., Juan C. The Increasing Threat of Pseudomonas Aeruginosa High-Risk Clones. Drug Resist. Updat. 2015;21–22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Takata I., Yamagishi Y., Mikamo H. Association of the ExoU Genotype with a Multidrug Non-Susceptible Phenotype and MRNA Expressions of Resistance Genes in Pseudomonas Aeruginosa. J. Infect. Chemother. 2018;24:45–52. doi: 10.1016/j.jiac.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 32.Rodulfo H., Arcia A., Hernández A., Michelli E., Martinez D.D.V., Guzman M., Sharma A., Donato M.D. Virulence Factors and Integrons Are Associated with MDR and XDR Phenotypes in Nosocomial Strains of Pseudomonas Aeruginosa in a Venezuelan University Hospital. Rev. Inst. Med. Trop Sao Paulo. 2019;61:e20. doi: 10.1590/s1678-9946201961020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruczek C., Kottapalli K.R., Dissanaike S., Dzvova N., Griswold J.A., Colmer-Hamood J.A., Hamood A.N. Major Transcriptome Changes Accompany the Growth of Pseudomonas Aeruginosa in Blood from Patients with Severe Thermal Injuries. PLoS ONE. 2016;11:e0149229. doi: 10.1371/journal.pone.0149229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elmassry M.M., Mudaliar N.S., Kottapalli K.R., Dissanaike S., Griswold J.A., San Francisco M.J., Colmer-Hamood J.A., Hamood A.N. Pseudomonas Aeruginosa Alters Its Transcriptome Related to Carbon Metabolism and Virulence as a Possible Survival Strategy in Blood from Trauma Patients. mSystems. 2019;4 doi: 10.1128/mSystems.00312-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golpayegani A., Nodehi R.N., Rezaei F., Alimohammadi M., Douraghi M. Real-Time Polymerase Chain Reaction Assays for Rapid Detection and Virulence Evaluation of the Environmental Pseudomonas Aeruginosa Isolates. Mol. Biol. Rep. 2019;46:4049–4061. doi: 10.1007/s11033-019-04855-y. [DOI] [PubMed] [Google Scholar]
- 36.Pang Z., Raudonis R., Glick B.R., Lin T.-J., Cheng Z. Antibiotic Resistance in Pseudomonas Aeruginosa: Mechanisms and Alternative Therapeutic Strategies. Biotechnol. Adv. 2019;37:177–192. doi: 10.1016/j.biotechadv.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 37.Rezzoagli C., Archetti M., Mignot I., Baumgartner M., Kümmerli R. Combining Antibiotics with Antivirulence Compounds Can Have Synergistic Effects and Reverse Selection for Antibiotic Resistance in Pseudomonas Aeruginosa. PLoS Biol. 2020;18:e3000805. doi: 10.1371/journal.pbio.3000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soto-Aceves M.P., Cocotl-Yañez M., Merino E., Castillo-Juárez I., Cortés-López H., González-Pedrajo B., Díaz-Guerrero M., Servín-González L., Soberón-Chávez G. Inactivation of the Quorum-Sensing Transcriptional Regulators LasR or RhlR Does Not Suppress the Expression of Virulence Factors and the Virulence of Pseudomonas Aeruginosa PAO1. Microbiology. 2019;165:425–432. doi: 10.1099/mic.0.000778. [DOI] [PubMed] [Google Scholar]
- 39.Hnamte S., Parasuraman P., Ranganathan S., Ampasala D.R., Reddy D., Kumavath R.N., Suchiang K., Mohanty S.K., Busi S. Mosloflavone Attenuates the Quorum Sensing Controlled Virulence Phenotypes and Biofilm Formation in Pseudomonas Aeruginosa PAO1: In Vitro, in Vivo and in Silico Approach. Microb. Pathog. 2019;131:128–134. doi: 10.1016/j.micpath.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Ahmed S.A.K.S., Rudden M., Smyth T.J., Dooley J.S.G., Marchant R., Banat I.M. Natural Quorum Sensing Inhibitors Effectively Downregulate Gene Expression of Pseudomonas Aeruginosa Virulence Factors. Appl. Microbiol. Biotechnol. 2019;103:3521–3535. doi: 10.1007/s00253-019-09618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong W., Dong M., Yan R., Liang Q., Zhang H., Luo W., Zhang Y., Liang H., Duan K. A Unique ATPase, ArtR (PA4595), Represses the Type III Secretion System in Pseudomonas Aeruginosa. Front. Microbiol. 2019;10:560. doi: 10.3389/fmicb.2019.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casciaro B., Lin Q., Afonin S., Loffredo M.R., de Turris V., Middel V., Ulrich A.S., Di Y.P., Mangoni M.L. Inhibition of Pseudomonas Aeruginosa Biofilm Formation and Expression of Virulence Genes by Selective Epimerization in the Peptide Esculentin-1a(1-21)NH2. FEBS J. 2019;286:3874–3891. doi: 10.1111/febs.14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Islamieh D.I., Afshar D., Esmaeili D. Effect of Satureja Khuzistanica Essential Oil (SKEO) Extract on Expression of LasA and LasB Genes in Pseudomonas Aeruginosa. Iran. J. Microbiol. 2019;11:55–59. doi: 10.18502/ijm.v11i1.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lloyd M.G., Vossler J.L., Nomura C.T., Moffat J.F. Blocking RpoN Reduces Virulence of Pseudomonas Aeruginosa Isolated from Cystic Fibrosis Patients and Increases Antibiotic Sensitivity in a Laboratory Strain. Sci. Rep. 2019;9:6677. doi: 10.1038/s41598-019-43060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahenthiralingam E., Campbell M.E., Foster J., Lam J.S., Speert D.P. Random Amplified Polymorphic DNA Typing of Pseudomonas Aeruginosa Isolates Recovered from Patients with Cystic Fibrosis. J. Clin. Microbiol. 1996;34:1129–1135. doi: 10.1128/JCM.34.5.1129-1135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee K., Lim Y.S., Yong D., Yum J.H., Chong Y. Evaluation of the Hodge Test and the Imipenem-EDTA Double-Disk Synergy Test for Differentiating Metallo-β-Lactamase-Producing Isolates of Pseudomonas Spp. and Acinetobacter Spp. J. Clin. Microbiol. 2003;41:4623–4629. doi: 10.1128/JCM.41.10.4623-4629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yong D., Lee K., Yum J.H., Shin H.B., Rossolini G.M., Chong Y. Imipenem-EDTA Disk Method for Differentiation of Metallo-β-Lactamase-Producing Clinical Isolates of Pseudomonas Spp. and Acinetobacter Spp. J. Clin. Microbiol. 2002;40:3798–3801. doi: 10.1128/JCM.40.10.3798-3801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pitout J.D.D., Gregson D.B., Poirel L., McClure J.-A., Le P., Church D.L. Detection of Pseudomonas Aeruginosa Producing Metallo-Beta-Lactamases in a Large Centralized Laboratory. J. Clin. Microbiol. 2005;43:3129–3135. doi: 10.1128/JCM.43.7.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lanotte P., Watt S., Mereghetti L., Dartiguelongue N., Rastegar-Lari A., Goudeau A., Quentin R. Genetic Features of Pseudomonas Aeruginosa Isolates from Cystic Fibrosis Patients Compared with Those of Isolates from Other Origins. J. Med. Microbiol. 2004;53:73–81. doi: 10.1099/jmm.0.05324-0. [DOI] [PubMed] [Google Scholar]
- 50.Finnan S., Morrissey J.P., O’Gara F., Boyd E.F. Genome Diversity of Pseudomonas Aeruginosa Isolates from Cystic Fibrosis Patients and the Hospital Environment. J. Clin. Microbiol. 2004;42:5783–5792. doi: 10.1128/JCM.42.12.5783-5792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.