Abstract
This work is aimed to elucidate the prevalence and characteristics of antimicrobial resistance, virulence, and molecular typing in Klebsiella pneumoniae from clinical companion animals in Beijing, China. In total, 105 K. pneumoniae (2.0%) isolates were recovered from 5359 samples (dogs, n = 3356; cats, n = 2003). All tested isolates exhibited high resistance to amoxicillin-clavulanate (74.3%). Moreover, resistance rates in dog isolates (2.1%) were significantly higher than in cat isolates (0.9%); however, the rate of multidrug-resistance (MDR) was 57.1% and the MDR prevalence in cats was significantly higher than dogs. Whole-genome sequencing demonstrated plasmids IncX4 and IncFIA (HI1)/FII(K) carried mcr-1 (n = 1) and mcr-8 (n = 1), but blaOXA-181 (n = 1) and blaNDM-5 (n = 4) were harbored in IncX3-type plasmids, and the above genes were in different isolates. The most prevalent sequence types (STs) in companion animals were ST1 (n = 9) and ST37 (n = 9). Compared to National Center for Biotechnology Information (NCBI) data on human K. pneumoniae, resistance genes blaCTX-M and blaTEM were more prevalent in human isolates; however, aac(6′)-Ib-cr and oqxAB showed a higher prevalence in companion animals. Hypermucoviscosity was reported in 9 (8.6%) isolates, whereas 64 isolates (61.0%) were hypervirulent K. pneumoniae (hvKP) via the Galleria mellonella. These findings validate the high risk of K. pneumonia and necessitate its relevant control in pet clinics.
Keywords: Klebsiella pneumonia, companion animals, carbapenem-resistant, hypervirulent, multidrug-resistant
1. Introduction
Klebsiella pneumoniae is an opportunistic pathogen that colonizes the skin, upper respiratory tract, and digestive tract of healthy asymptomatic subjects [1]. Also, it is a primary cause of diseases in neonates, elderly and immunocompromised humans and animals, including pneumonia, wound infections, urinary tract infections (UTIs), sepsis and meningitis [2]. K. pneumoniae is inherently resistant to penicillin; members of this population, in most cases, acquire resistance to multiple antibiotics. Thus, it is associated with resistance to important antimicrobial agents, thereby a significant threat to public health [3].
In recent years, K. pneumoniae has rapidly become a multidrug-resistant (MDR) pathogen. It develops resistance to third-generation cephalosporins, fluoroquinolones, and aminoglycosides. Of concern, K. pneumoniae has increasingly become resistant to carbapenems by acquiring carbapenemases [4]. The World Health Organization recognizes extended-spectrum β-lactam (ESBL)-producing and carbapenem-resistant K. pneumoniae (CRKP) as a serious public health threat [5]. Colistin and tigecycline were considered the “last resort” in managing CRKP-related critical carcinogens. However, with the increase in CRKP prevalence, various carbapenem antibiotics have proven ineffective [6]. Companion animals had been reported to harbor blaNDM, which could quickly contaminate wild birds through dogs, flies, and livestock as hosts; this phenomenon is a human health threat [7]. The first mobile colistin resistance gene, mcr-1, was found in Enterobacteriaceae (mainly Escherichia coli and Klebsiella pneumoniae) [8], other variants of mcr were also discovered, and have spread widely in different hosts and regions [9,10].
Based on previous reports, hypervirulent K. pneumoniae (hvKP) can cause acquired liver abscess with severe organ failure. Notably, 22.8% of K. pneumoniae clinical isolates were identified as hvKP in China [11]. Various hypervirulence-associated factors are critical in hvKP isolates, including capsular serotypes (K1 and K2), sequence types (ST23), a virulence plasmid, a pathogenicity island, and several virulence factors [12]. The hypermucoviscosity K. pneumoniae (hmKP) is attributed to the elevated production of capsular polysaccharides encoded by specific virulence genes, rmpA and rmpA2 [13]. Otherwise, a genetic and phenotypic convergent clone, CR-hvKP, simultaneously exhibits carbapenem resistance and hypervirulence has emerged in recent years [14].
K. pneumoniae isolates had been shown to elevate the risk of antibiotic treatment failure both in humans and companion animals [2]. Similarly, if the isolates are transmitted to humans through pets, the antimicrobial bacteria present in companion animals may significantly impact human public health. The K. pneumoniae isolate that infects companion animals potentially belongs to high-risk clonal lineages found in humans [15]. As a result, there is an urgent need to understand the molecular and genetic characteristics of K. pneumoniae isolates from a veterinary medicine and public health perspective. However, the current prevalence and characteristics of K. pneumoniae in clinical companion animals in China is unknown. In this study, we clarified the prevalence and characteristics of antimicrobial resistance, virulence, and molecular typing in K. pneumoniae from clinical companion animals in Beijing, China.
2. Results
2.1. Pets Samples and K. pneumoniae Isolates
We collected 5359 samples from 3356 dogs and 2003 cats, and isolated 105 K. pneumoniae isolates (2.0%, 95% confidence interval (CI): 1.6–2.4, n = 105/5359); 85 from dogs (2.5%, 95% CI: 2.0–3.1, n = 85/3356) and 20 from cats (1.0%, 95% CI: 0.6–1.5, n = 20/2003). A significant difference was noted in the separation rate between dogs and cats (p < 0.05). Throat swabs (6.5%, 95% CI: 3.5–10.9, n = 13/200), nasal swabs (4.6%, 95% CI: 2.1–8.6, n = 9/194), and tracheal lavage (6.3%, 95% CI: 2.8–12.1, n = 8/126) exhibited higher rates of bacterial detection than the total rates (p < 0.05) (Table 1).
Table 1.
The distribution of K. pneumoniae samples and isolates.
| Parameters | Category | No. of Samples (%) | Kp Isolates (%, 95% CI) |
|---|---|---|---|
| Origin | Dog | 3356 (62.6) | 85 (2.5, 2.0–3.1) |
| Cat | 2003 (37.4) | 20 (1.0, 0.6–1.5) | |
| Gender | Male | 3352 (62.5) | 58 (1.7, 1.3–2.2) |
| Female | 2007 (37.5) | 47 (2.3, 1.7–3.1) | |
| Source | Urine | 2879 (53.7) | 37 (1.3, 0.9–1.8) |
| Throat swabs | 200 (3.7) | 13 (6.5, 3.5–10.9) | |
| Nasal swabs | 194 (3.6) | 9 (4.6, 2.1–8.6) | |
| Abscess | 391 (7.3) | 10 (2.6, 1.2–4.7) | |
| Tracheal lavage | 126 (2.4) | 8 (6.3, 2.8–12.1) | |
| Ear swabs | 267 (5.0) | 8 (3.0, 1.3–5.8) | |
| Pyometra | 158 (2.9) | 7 (4.4, 1.8–8.9) | |
| Coelomic fluid | 367 (6.8) | 4 (1.1, 0.3–2.8) | |
| Skin | 452 (8.4) | 3 (0.7, 0.1–1.9) | |
| Oral swabs | 21 (0.4) | 2 (9.5, 1.2–30.4) | |
| Prostatic fluid | 22 (0.4) | 1 (4.5, 0.1–22.8) | |
| Surgical infection | 70 (1.3) | 1 (1.4, 0–7.7) | |
| Anal swabs | 32 (0.6) | 1 (3.1, 0.1–16.2) | |
| Eye secretion | 41 (0.8) | 1 (2.4, 0.1–12.9) | |
| Others | 139 (2.6) | 0 (0, 0–2.6) | |
| Total | —— | 5359 | 105 (2.0, 1.6–2.4) |
2.2. Antimicrobial Resistance
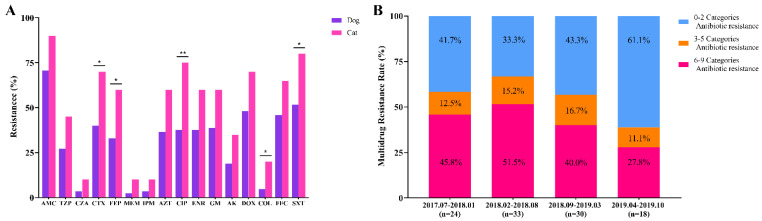
K. pneumoniae isolates exhibited the highest (74.3%, n = 78/105) resistance to amoxicillin-clavulanate, high resistance to doxycycline (52.4%, n = 55/105) and trimethoprim-sulfamethoxazole (57.1%, n = 60/105). However, K. pneumoniae isolates demonstrated relatively low resistance to colistin (7.6%, n = 8/105), ceftazidime-avibactam (4.8%, n = 5/105), imipenem (4.8%, n = 5/105), and meropenem (3.8%, n = 4/105) (Table S1). Of these isolates, resistance of isolates in dogs (2.1%, 95% CI: 1.6–2.6, n = 70/3356) was significantly higher than that of cat isolates (0.9%, 95% CI: 0.6–1.5, n = 19/2003) (p < 0.05). Notably, the resistance of cat isolates to five antimicrobial agents tested was significantly higher than that of dogs (p < 0.05) (Figure 1A, Table S1). Minimum inhibitory concentrations MIC50 and MIC90 of cats were generally above or equal to that of dogs. Overall, 60 isolates showed MDR (57.1%, n = 60/105). The MDR prevalence rate of cats was significantly higher than dogs (p < 0.05). Compared to the MDR rate between February and August 2018, that between April to October 2019 was significantly lower (p < 0.05) (Figure 1B).
Figure 1.
Antimicrobial resistance of K. pneumoniae isolates. (A) The resistance of isolates to different antibiotics from dogs and cats, respectively. AMC, Amoxicillin-clavulanate; TZP, Piperacillin-tazobactam; CZA, Ceftazidime-avibactam; CTX, Cefotaxime; FEP, Cefepime; MEM, Meropenem; IPM, Imipenem; AZT, Aztreonam; CIP, Ciprofloxacin; ENR, Enrofloxacin; GM, Gentamicin; AK, Amikacin; DOX, Doxycycline; COL, Colistin; FFC, Florfenicol; SXT, Trimethoprim-sulfamethoxazole. p-values as determined by Chi-square (χ2) and Fisher’s exact test in SPSS Statistics. * p < 0.05, ** p < 0.01. (B) Multi-drug-resistant rates of 105 isolates based on the antimicrobial categories from July 2017 to October 2019.
2.3. Antibiotic Resistance Genes and Virulence Genes
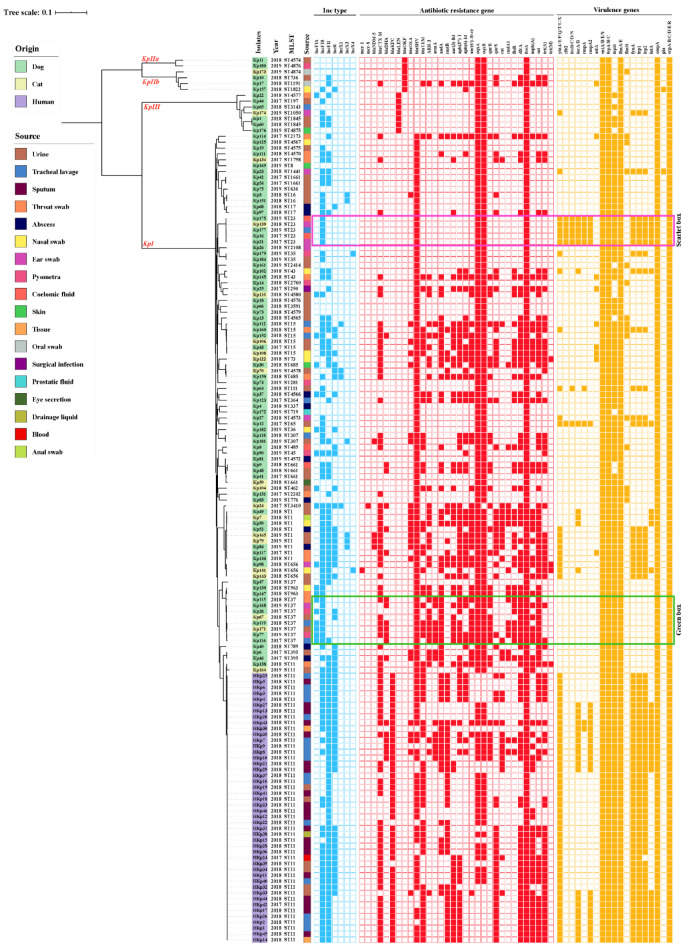
The mobile colistin resistance gene mcr-1 (Kp141) and mcr-8 (Kp24) were detected in K. pneumoniae from tracheal lavage and urine from different cats. Carbapenem resistance genes blaOXA-181 and blaNDM-5 were harbored in one (Kp3), and four (Kp79, Kp84, Kp165, Kp181) isolates, respectively. Additionally, we analyzed the sequence read archive (SRA) sequences (n = 46) of human K. pneumoniae (HKp) from National Center for Biotechnology Information (NCBI) to demonstrate the similarities, differences, and relevance of the molecular characteristics of K. pneumoniae from humans and companion animals. Resistance genes, mcr and blaNDM, were not present in the genomes of K. pneumoniae isolated from humans; other variants were also absent. blaKPC (95.7%, n = 44/46) was harbored in human isolates, whereas none was found in companion animals. Resistance genes blaCTX-M and blaTEM were more prevalent among K. pneumoniae isolates from humans, compared to companion animals (p < 0.01). Meanwhile, isolates from companion animals showed higher rates of aac(6′)Ib-cr and oqxAB than those from humans (p < 0.01). Moreover, the prevalence of blaCTX-M and rmtB from cats was higher than in dogs (p < 0.05); notably, blaSHV (87.6%, n = 92/105) was the most prevalent β-lactamase resistance gene in companion animals (Figure 2). Besides, we found eight CTX-M-genotypes (-3, -14, -15, -27, -55, -64, -65, -104) in companion animals, dominated by blaCTX-M-55 (14.3%, n = 15/105). blaCTX-M-65 (60.9%, n = 28/46) was the largest proportion of four CTX-M-genotypes (-14, -15, -65, -147) in human isolates. Among them, companion animals harbored blaCTX-M-55 (p < 0.05) and blaCTX-M-65 (p < 0.01), which were significantly more and less prevalent than humans, respectively. The fosfomycin resistance gene fosA showed full coverage in companion animals and human isolates (Figure 2).
Figure 2.
Distribution of K. pneumoniae phylogroups, year, multilocus sequence typing (MLST), Inc-type plasmid, antibiotic-resistance genes, and virulence-associated genes among isolates from companion animals and humans across the phylogenetic tree. Scarlet box: ST23-hvKP with the most virulence genes harboring the least resistance genes; green box: ST37-hvKP carrying only the common virulence factors while taking along relatively more resistance genes.
Similarly, virulence-associated genes enterotoxins (entA/B/E/S), ferrienterochelin receptor (fepA/B/C), fimbriae (fimA/E), outer membrane protein (ompA), and common pili (ecpA/B/C/D/E/R) were harbored by all isolates from humans and companion animals, as revealed using abricate (Figure 2). The ybtA/E/P/Q/T/U/X, iucA/B, rmpA2, fepD, fyuA, irp1/2, and iutA from human isolates were significantly more prevalent (p < 0.05) than those from companion animals; however, virulence genes of dogs and cats exhibited no difference. Interestingly, the ST23-hvKP (n = 5) isolates harbored ybt, clb, iro, iuc, ent, rmpA, and rmpA2 virulence genes, with the least resistance genes. Moreover, ST37-hvKP isolates harbored common virulence factors ent, fep, fim, ompA, and ecp but simultaneously took along more resistance genes (Figure 2).
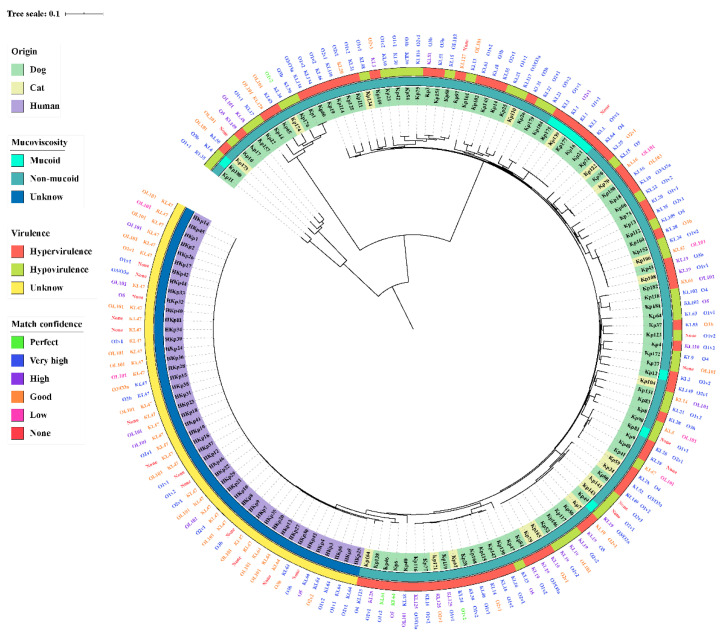
2.4. Hypervirulent, Hypermucoviscosity, Capsule Serotype, and O-Antigen of K. Pneumonia
Of the 105 isolates, 64 (61.0%, n = 64/105) demonstrated higher or equal pathogenicity to the positive control ATCC43816 after injection for 72 h in G. mellonella, demonstrating that these isolates were hypervirulent (Figure S1). ST37 (12.5%, n = 8/64), ST1 (7.8%, n = 5/64) and ST23 (7.8%, n = 5/64) were predominately among hvKP. There were 50 (58.8%, n = 50/85) and 14 (70%, n = 14/20) hvKP isolates from dogs and cats, respectively, but showed no significant difference. Mucoid was observed in nine (8.6%, n = 9/105) isolates, evaluated by mucoviscosity assay (Figure 3); however, three of them were not among hvKP. There was no association between hvKP and hmKP (p > 0.05). The resistance and virulence of mucoid and non-mucoid isolates had no significant difference in this study (p > 0.05). In addition, there was no significant difference in antibiotic resistance between hypervirulence and hypovirulence isolates from companion animals (p > 0.05). MDR-hvKP accounted for 39.0% (n = 41/105) of total K. pneumonia isolates, harboring four CR-hvKP. Additionally, capsule serotype and O-antigen type were obtained from Kleborate. Among the six hv-KP harboring ybt, clb, iro, iuc and rmpA, sequence types and capsular serotypes of Kp16, Kp21, Kp139, Kp175, and Kp177 were ST23 and KL1, but Kp12 was ST65 and KL2, respectively (Figure 3). The KL1 and KL2 isolates were hmKP, and the O-antigen type of 77.8% (n = 7/9) hmKP was O1v2. In companion animals, we obtained 47 capsule serotypes and 10 O-antigen types; the most prevalent serotypes were KL19 (10.5%, n = 11/105) and O1v2 (20.0%, n = 21/105). In human isolates, the most abundant serotypes were KL47 (65.2%, n = 30/46) and O2v1 (43.5%, n = 20/46), respectively (Figure 3). O loci named OL101 onwards were defined based on gene content and are not yet associated with a specific serologically defined O type.
Figure 3.
Distribution of K. pneumoniae mucoviscosity, virulence evaluation by Galleria mellonella, capsular serotype, and O-antigen type among isolates from companion animals and humans across the phylogenetic tree. The match confidences of serotypes were perfect, very high, high, good, low, and none.
2.5. Diversity Genotypes of the K. pneumoniae Isolates
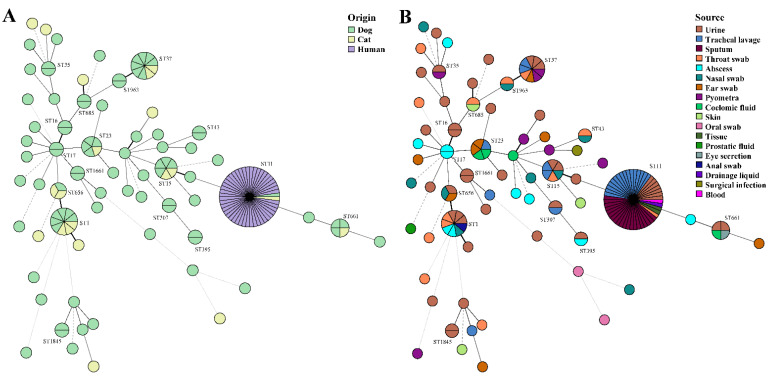
Most of the 105 isolates belonged to the KpI phylogroup (87.6%, n = 92/105), with only a few isolates belonging to KpIIa (1.9%, n = 2/105), KpIII (6.7%, n = 7/105) and KpIIb (3.8%, n = 4/105) in companion animals. MLST presented a diverse distribution, 89 out of 105 K. pneumoniae isolates were assigned to 48 known STs, whereas 16 isolates represented 16 (25.0%, n = 16/64) novel STs (ST4565-ST4567, ST4570, ST4572-ST4580, ST4874-ST4876) (Figure 2 and Figure 4). However, STs of all human isolates were classified as ST11, indicating that ST11 is of great concern and widely reported in humans. We also reported two ST11 animal isolates shared by the dog (Kp138) and cat (Kp164), they were not in the same evolutionary branch as human isolates in the core SNP-based phylogenetic tree (Figure 2). In companion animals, the most prevalent STs were ST1 (8.6%, n = 9/105) and ST37 (8.6%, n = 9/105), followed by ST15 (n = 6) and ST23 (n = 5). ST1 and ST37 were prevalent in cats (n = 3) and dogs (n = 7), respectively. The minimum spanning trees of MLST further validated the commonness of K. pneumoniae isolates from humans and companion animals with the same STs (Figure 4). In this study, the STs of four blaNDM-5-positive K. pneumoniae were ST1 (Kp79, Kp84, Kp165) and ST307 (Kp181), but the isolates harboring mcr-1 (Kp141), mcr-8 (Kp24) and blaOXA181 (Kp3) were ST656, ST3410, and ST16.
Figure 4.
Minimum spanning trees of MLST typing. (A) STs colored based on different origins of dog, cat, and human. (B) STs colored based on different sources, such as urine, tracheal lavage, sputum, throat swab, abscess, nasal swab, ear swab, pyometra, coelomic fluid, skin, oral swab, tissue, prostatic fluid, eye secretion, anal swab, drainage liquid, surgical infection, and blood.
2.6. Characterization of Plasmids
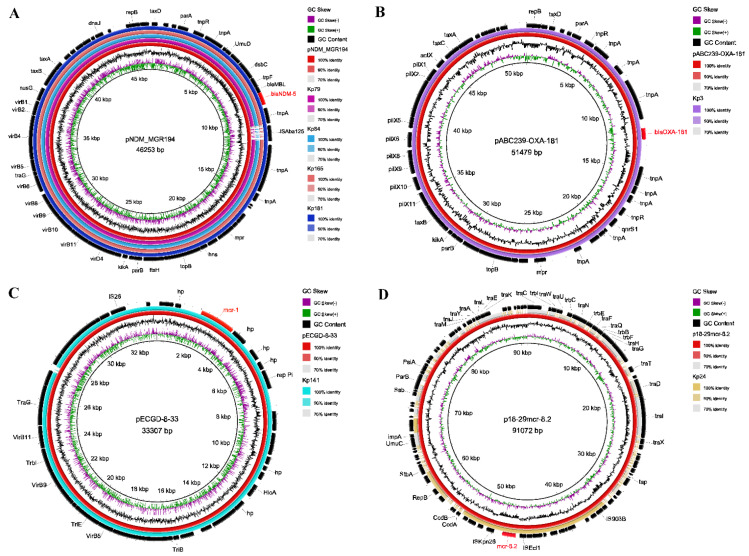
After assembling backbone sequences, all contigs and gaps were identified by whole-genome analysis. Among the Inc-type plasmids, IncFIB (67.6%, n = 71/105) was prevalent in K. pneumoniae isolates of companion animals, whereas human isolates were covered by IncFII (100%, n = 46/46). Of these, IncFIA was significantly more prevalent in companion animals than that of humans (p < 0.01), but the prevalence of IncFII and IncR were significantly higher in human isolates (p < 0.01). All Inc-type plasmids exhibited no difference between dogs and cats (Figure 2). Plasmids IncX3, IncX4, and IncFIA (HI1)/FII(K) harbored blaOXA-181 (Kp3), mcr-1 (Kp141) and mcr-8 (Kp24) respectively. Moreover, BLASTn results demonstrated that blaNDM-5 was harbored in four isolates of companion animals (Figure 2 and Figure 5). The blaNDM-5 gene was distributed among dogs (n = 2) and cats (n = 2), source from urine (n = 3) and abscess (n = 1). The complete genome sequences of four blaNDM-5-positive isolates contained regions showing > 99% nucleotide sequence homology to the reference plasmid pNDM_MGR194 (46253bp, accession No. KF220657), suggesting that blaNDM-5 were likely located on IncX3-type plasmids (Figure 5A). Also, the blaNDM-5 gene was included in an insertion sequence (IS) cassette (∆ISAba125-IS5-blaNDM-ble-trpF-dsbC-IS26) compared using ISfinder. Similarly, the plasmids of IncX3 and IncX4 which harbored blaOXA-181 and mcr-1 were consistent with reference plasmids pABC239-OXA-181 (51479bp, accession No. MK412916) and pECGD-8-33 (33307bp, accession No. KX254343), respectively (Figure 5B,C). Interestingly, the mcr-8 gene was harbored by IncFIA (HI1)/FII(K), similar to plasmid p18-29mcr-8.2 (91072bp, accession No. MK262711) within the structure of ISEcl1-mcr-8.2-orf-ISKpn26 (Figure 5D).
Figure 5.
BLAST ring comparison of plasmids in K. pneumoniae isolates, each ring represents a separate isolate. The comparison of plasmids carrying (A) blaNDM-5 (IncX3), (B) blaOXA-181 (IncX3), (C) mcr-1 (IncX4), and (D) mcr-8 (IncFIA). The internal ring is the reference sequence of pNDM_MGR194 (46253bp, accession No. KF220657), pABC239-OXA-181 (51479bp), pECGD-8-33 (33307bp), and p18-29mcr-8.2 (91072bp) respectively, and the outside rings are isolates from this study, which are similar to the reference plasmid.
3. Discussion
K. pneumoniae is an important host and transmission carrier of clinically important antimicrobial resistance genes in humans and animals [16]. Typically, humans have close contact with pets; therefore, there is a close association between the health of both. However, there is currently a dearth of clinical research on K. pneumoniae from companion animals in China. Herein, we report the prevalence of antibiotic resistance, virulence, molecular typing, and phylogroups in K. pneumoniae from companion animals in Beijing, China. Previous findings demonstrated that the overall prevalence of recovered (2.0%) K. pneumoniae was slightly lower than 3.53% in Italy [17]. K. pneumoniae is the primary pathogen of UTIs, and is usually associated with resistance to the most significant antibiotics [15]. Urine (53.7%) accounted for the largest proportion (1.3%) of samples, from which the largest number of K. pneumoniae isolates (n = 37) was isolated in this study, this was similar with a previous study in Japan [18].
Among the K. pneumoniae isolates from companion animals, the resistance rate in dogs was significantly higher than in cat isolates, but showed no difference in South Korea [19]. This discordance in results could be explained by different types and quantities of tested drugs. An overall MDR rate of 57.1% was slightly higher than that previously reported in Singapore (50.0%) [20]. Otherwise, the MDR prevalence in cats was significantly higher than in dogs in this study (p < 0.05). As a result, this aggravates the threat to human health, as owners are more likely to exhibit intimate behavior with companion animals, increasing the probability of mutual transmission. Colistin had been used as an animal feed contributing to the prevalence of mcr in China; similarly, studies have reported wide prevalence in Vietnam and other South Asian countries. However, the prevalence of mcr-1 in food animals in Europe and America is low [21], which may be attributed to the fact that countries in these regions have not approved colistin use as an antibacterial growth-promoting agent. Elsewhere, a previous study demonstrated low prevalence (<1%) of mcr-1 of K pneumoniae from humans [22]. Moreover, mcr-1 in K. pneumoniae from companion animals; thus, the pet food industry was speculated to be a source of mcr-1 [23]. Since the first description of mcr-8, with IncFII-type plasmid as the carrier, it has been widely disseminated among K. pneumoniae isolates of livestock origin [6]. In the current study, mcr-1 (n = 1) and mcr-8 (n = 1) were carried by plasmids IncX4 and IncFIA (HI1)/FII(K) of K. pneumoniae. To the best of our knowledge, we present the first report on the isolation of the two isolates from tracheal lavage and urine-derived from different cats. Of concern, having found five CRKP, which also appeared in other countries was considered a severe situation [19,20]. In this study, carbapenem resistance genes, blaOXA-181 and blaNDM-5, were harbored in IncX3-type plasmids of different CRKP but no blaOXA-48 and blaKPC was found in companion animals, while blaOXA-48 was prevalent in other countries [24]. Resistance genes, blaCTX-M and blaTEM, of ESBL, showed a lower prevalence among companion animals, but the prevalence was higher than Japan (34.8%) [18], Italy (21.4%) [17], and other European countries (11.2%) [25]. In addition, CTX-M-genotypes were diverse in different species and countries. Herein, blaCTX-M-65 and blaCTX-M-55 predominated in the isolates of humans and companion animals, a phenomenon similar to findings from a previous study of pets in South Korea [19] and China [26]. Moreover, K. pneumoniae of type CTX-15 was prevalent in companion animals in Japan [18]. These findings demonstrate that the mutations of K. pneumoniae resistance genes are not limited to specific hosts or regions, and highlights the necessity of coordinated control in One Health. Furthermore, the genes aac(6′)Ib-cr and oqxAB of quinolone from companion animals showed higher resistance rates than those from human isolates; the rates were similar to those reported previously [18,27]. Otherwise, the most prevalent STs were ST1 and ST37, which concur with a previous report in China [26], but contrasts from reports in companion animals from Portugal [15] and Japan [18].
Based on the current understanding, virulence factors are encoded by several paragenes and can further increase the severity and/or pathogenicity of K. pneumoniae infection. Researches proved that any three of four siderophore systems (ent, ybt, iuc, and iro) could enhance virulence in murine models [3]. The ybt and iuc of human isolates were significantly more prevalent than that of companion animals. However, virulence factor-encoding genes of dogs and cats exhibited no difference in the current study. Previous reports had indicated that hvKP is mainly associated with ST23 and CRKP primarily belonging to ST11, which are considered the two major clinically important pathogens in China [28]. In this study, hvKP isolates (61.0%) from companion animals were less than the previously reported rates (76.4%) in humans [12]. Unfortunately, five ST23 isolates harbored all siderophore systems, identified as hvKP by G. mellonella model, but contained the least resistance genes. In companion animals, the four CR-hvKP were ST1 (n = 3) and ST16 (n = 1), whereas the CR-hvKP isolates were common genetic types such as ST11 and ST23 [14]. Hypermucoviscosity may be the most dominant virulence factor of K. pneumoniae, but its genetic basis and pathogenic factors are not direct. Due to the presence of one or two of the para-regulatory genes rmpA or rmpA2, this phenotype is usually associated with the overproduction of capsules [29]. Meanwhile, six of nine hypermucoviscous isolates were hvKP, but only one hypermucoviscous isolate in companion animals showed the MDR phenotype. Additionally, MDR-hvKP accounted for 68.3% of MDR isolates, 64.1% of hvKP, and 39.0% of total K. pneumonia isolates. The above findings demonstrate the high-risk of K. pneumonia, creating a huge treatment limitation in pet clinics.
In conclusion, the present study did a large-scale investigation of antimicrobial resistance and molecular genetic analysis in K. pneumoniae from clinical companion animals in Beijing, China. A high prevalence of MDR and hypervirulent K. pneumoniae isolates were found from dogs and cats. The wide distribution of amoxicillin-clavulanic and third-generation cephalosporins in veterinary hospitals may contribute to the ESBL resistance in these isolates. The presence of mcr, blaOXA181 and blaNDM in K. pneumoniae demonstrates that the pathogen is a potential reservoir of colistin and carbapenem resistance genes in pet clinics. Meanwhile, the emergence of MDR-hvKP and epidemic clones elevates the risks of veterinarians; however, the predominant clones of CRKP are scarce in human-related ST clones. These findings emphasize the importance of managing K. pneumonia comorbidities and scientifically conducting antimicrobial susceptibility tests for more accurate treatments. This would reduce the spread of such high-risk clonal lineages to ensure the safety of companion animal practitioners and public health.
4. Materials and Methods
4.1. Samples Collection and Bacterial Characterization
All samples of companion animals were collected aseptically from the Veterinary Teaching Hospital of China Agricultural University (VTH-CAU), Lpet Veterinary Diagnostic Center (LVDC-Beijing), and North China (Tianjin) Testing Center, between July 2017 and October 2019. Sampling was conducted following the principles of the Beijing Municipality Review of Welfare and Ethics of Laboratory Animals and approved by the China Agricultural University Animal Ethics Committee document (No. AW01017102-2). K. pneumoniae isolates were isolated using 5% sheep blood agar and MacConkey Inositol Adonitol Agar medium (HopeBio, Qingdao, China) containing 100 mg/L carbenicillin following aerobic incubation at 37 °C overnight. The DNA of individual clones with the red centre was extracted by TIANamp Bacteria DNA Kit (Tiangen, Beijing, China) with the protocol as stipulated by the manufacturer. Subsequently, the DNA was used as templates for polymerase chain reaction (PCR) amplification of 16S rDNA gene sequencing (16SrRNA-F: AGAGTTTGATCCTGGCTCAG, 16SrRNA-R: ACGGCTACCTTGTTACGACTT) and conditions consisted 95 °C (10 min), 30 cycles of [95 °C (30 s), 55 °C (30 s), 72 °C (90 s)], and 72 °C (10 min) as previously described [30]. Amplicons were sequenced to reveal bacterial genus using the BLAST algorithm.
4.2. Antimicrobial Susceptibility Testing
For this experiment, we adopted agar/broth microdilution method with two-fold dilutions for the commonly used antimicrobial agents in human and/or companion animal clinics, including 16 antimicrobials belonging to 10 different categories, such as β-lactams combination agents (amoxicillin-clavulanate, piperacillin-tazobactam, and ceftazidime-avibactam), cephalosporins (cefotaxime and cefepime), carbapenems (meropenem and imipenem), monobactams (aztreonam), fluoroquinolones (ciprofloxacin and enrofloxacin), aminoglycosides (gentamicin and amikacin), tetracyclines (doxycycline), lipopeptides (colistin), phenicols (florfenicol), folate pathway antagonists (trimethoprim-sulfamethoxazole). All antibiotics were purchased from China Institute of Veterinary Drug Control and Solarbio (Beijing, China). Results of minimum inhibitory concentrations (MICs) were expressed according to breakpoint tables of Clinical and Laboratory Standards Institute (CLSI) documents VET08-ED4:2018/M100-ED30:2020 and European Commission on Antimicrobial Susceptibility Testing (EUCAST) documents (version 9.0, 2019). E. coli ATCC 25922 served as a quality control organism. Based on Standardized International Terminology; the drug resistance was classified as MDR for three or more categories of antimicrobial agents.
4.3. Mucoviscosity Assay
Because mucoviscous cells remain in suspension, whilst non-mucoid cells could form pellets after centrifugation, measurement of the turbidity after low-speed centrifugation can serve as an indicator of hypermucoviscosity. In short, bacterial isolates were grown in Lysogeny broth (LB) broth at 37 °C after 6 h incubation with shaking and then centrifuged at 1000× g for 5 min. The optical density at 600 nm (OD600) values of the supernatant were determined and measured. Mucoviscosity isolates were more difficult to pellet, so the supernatants have higher absorbance readings.
4.4. Galleria Mellonella Virulence Assay
Here, we tested the in vivo virulence using the G. mellonella infection model to indicate hypervirulence, as previously described [31]. In total, 10 randomly selected G. mellonella larvae approximately 250 to 350 mg for each isolate were purchased from Huiyude Biotech Company, Tianjin, China. Bacteria cells were cultured in a mid-log-phase and pelleted via centrifugation at 3500 rpm, washed twice, and resuspended in 0.01 M phosphate-buffered saline (PBS, pH6.5). Larvae were injected with 10 μL bacterial suspension (with 106 colony-forming units, CFU) via the rear left proleg using a micro-sample syringe. PBS and ATCC43816 were used as negative and positive control groups, respectively. After injection, larvae were placed in 90 mm Petri-dishes and kept at 37 °C in the dark. Insects were considered dead when they did not respond to physical stimuli. We monitored death at 6 h intervals during 72 h. Experiments were performed in triplicate.
4.5. Whole-Genome Sequencing and Molecular Analysis
To extract the genomic DNA of the isolates, the TIANamp Bacteria DNA Kit was used following the manufacturer’s manuals. Indexed DNA libraries were constructed using the KAPA Hyper Prep Kit Illumina platforms (Roche, Basel, Switzerland) following the instruction, then sequenced on the Illumina Hiseq X Ten platform via the 150-bp paired-end strategy (Annoroad, Beijing, China). The draft genomes were assembled using SPAdes (version 3.9.0) [32]. All whole-genome sequencing data for this work are deposited in the GenBank and under BioProject accession no. PRJNA684769. Plasmid types, antibiotic resistance genes, and virulence genes were identified using abricate (https://github.com/tseemann/abricate, accessed on 13 August 2020), whereas the multilocus sequence typing (MLST) was obtained using the SRST2 toolkit (version 0.2.0) [33]. Capsule serotype (KL) and O-antigen (O) were analyzed using Kleborate (https://github.com/katholt/Kleborate, accessed on 16 September 2020). All draft genomes were used for core-genome alignments, after which we constructed a phylogenetic tree using parsnp in the Harvest package (version 1.1.2) [34]. The tree was visualized using the online tool Interactive Tree of Life (iTOL, http://itol.embl.de/, accessed on 19 September 2020) with the corresponding features of each isolate. The minimum spanning tree for all STs was generated by BioNumerics version 7.6 (Applied Maths, Sint-Martens-Latem, Belgium) using the BURST algorithm between different backgrounds. To compare the genetic context in the different plasmids, BLAST Ring Image Generator (BRIG) was applied [35]. Additionally, we analyzed the SRA of K. pneumoniae derived from humans in the NCBI database, which was collected in Beijing between July 2017 and October 2019.
4.6. Statistical Analysis
Statistical significance was determined using Chi-square (χ2) and Fisher’s exact test in SPSS Statistics (version 22, IBM Corporation, Armonk, NY, USA). The level of significance was set at p < 0.05.
Acknowledgments
Thanks to Weiwei Zhang and Yue Wang for their help in collecting samples. We thank the team of curators of the Institut Pasteur MLST system (Paris, France) for importing novel alleles, profiles, and isolates at http://bigsdb.web.pasteur.fr (accessed on 14 November 2019).
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/10/3/271/s1, Figure S1. K. pneumoniae infection of Galleria mellonella lethality. (A-M) Larvae injected with phosphate-buffered saline (PBS) or 106 colony-forming units (CFU) of 105 K. pneumoniae isolates from companion animals, and survival was monitored over 72 h after infection. (N) Mortality of larvae infected with PBS or ATCC43816 (104, 105, 106, 107 CFU) was dose-dependent, and survival was monitored over 72 h after infection. PBS and ATCC43816 are the negative and positive control groups, respectively. Table S1. Minimum inhibitory concentration (MIC, μg/mL) of antimicrobial agents for clinical K. pneumoniae isolates from companion animals.
Author Contributions
Conceptualization, Y.W. and Z.X.; Data curation, Z.Z. and L.L. (Lei Lei); Formal analysis, Z.Z., H.Z., H.D. and Y.S.; Funding acquisition, Z.X.; Investigation, Z.Z., H.D., Y.S. and L.L. (Lei Li); Methodology, Z.Z., H.D., Y.S. and L.L. (Lei Li); Resources, H.Z. and Z.X.; Software, Z.Z. and L.L. (Lei Lei); Supervision, Y.W.; Writing—original draft, Z.Z.; Writing—review and editing, L.L. (Lei Lei), Y.W. and Z.X. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Beijing Science and Technology Planning Project (Z171100001517008).
Institutional Review Board Statement
Sampling was conducted following the principles of the Beijing Municipality Review of Welfare and Ethics of Laboratory Animals and approved by the China Agricultural University Animal Ethics Committee document (No. AW01017102-2).
Informed Consent Statement
Not applicable.
Data Availability Statement
All whole-genome sequencing data for this work are deposited in the GenBank and under BioProject accession no. PRJNA684769.
Conflicts of Interest
All authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Holt K.E., Wertheim H., Zadoks R.N., Baker S., Whitehouse C.A., Dance D., Jenney A., Connor T.R., Hsu L.Y., Severin J., et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA. 2015;112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyres K.L., Holt K.E. Klebsiella pneumoniae Population Genomics and Antimicrobial-Resistant Clones. Trends Microbiol. 2016;24:944–956. doi: 10.1016/j.tim.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Wyres K.L., Lam M.M.C., Holt K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020;18:344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 4.Parisi S.G., Bartolini A., Santacatterina E., Castellani E., Ghirardo R., Berto A., Franchin E., Menegotto N., De Canale E., Tommasini T., et al. Prevalence of Klebsiella pneumoniae strains producing carbapenemases and increase of resistance to colistin in an Italian teaching hospital from January 2012 To December 2014. BMC Infect. Dis. 2015;15:244. doi: 10.1186/s12879-015-0996-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 6.Wang X., Wang Y., Zhou Y., Li J., Yin W., Wang S., Zhang S., Shen J., Shen Z., Wang Y. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 2018;7:122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Zhang R., Li J., Wu Z., Yin W., Schwarz S., Tyrrell J.M., Zheng Y., Wang S., Shen Z., et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2017;2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y.-Y., Wang Y., Walsh T.R., Yi L.-X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 9.Liu L., Feng Y., Zhang X., McNally A., Zong Z. New Variant of mcr-3 in an Extensively Drug-Resistant Escherichia coli Clinical Isolate Carrying mcr-1 and blaNDM-5. Antimicrob. Agents Chemother. 2017;61:e01757-17. doi: 10.1128/AAC.01757-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H., Yang L., Liu Z., Yin W., Liu D., Shen Y., Walsh T., Shao B., Wang Y. Molecular Insights into Functional Differences between mcr-3- and mcr-1-Mediated Colistin Resistance. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.00366-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Y., Wang S., Zhan L., Jin Y., Duan J., Hao Z., Lv J., Qi X., Chen L., Kreiswirth B.N., et al. Microbiological and Clinical Characteristics of Hypermucoviscous Klebsiella pneumoniae Isolates Associated with Invasive Infections in China. Front. Cell. Infect. Microbiol. 2017;7:24. doi: 10.3389/fcimb.2017.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Jin L., Ouyang P., Wang Q., Wang R., Wang J., Gao H., Wang X., Wang H., Kang H., et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: A multicentre, molecular epidemiological analysis. J. Antimicrob. Chemother. 2020;75:327–336. doi: 10.1093/jac/dkz446. [DOI] [PubMed] [Google Scholar]
- 13.Lee C.-R., Lee J.H., Park K.S., Jeon J.H., Kim Y.B., Cha C.-J., Jeong B.C., Lee S.H. Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front. Cell. Infect. Microbiol. 2017;7:483. doi: 10.3389/fcimb.2017.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong N., Yang X., Zhang R., Chan E.W., Chen S. Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerg. Microbes Infect. 2018;7:146. doi: 10.1038/s41426-018-0146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marques C., Menezes J., Belas A., Aboim C., Cavaco-Silva P., Trigueiro G., Gama L.T., Pomba C. Klebsiella pneumoniae causing urinary tract infections in companion animals and humans: Population structure, antimicrobial resistance and virulence genes. J. Antimicrob. Chemother. 2019;74:594–602. doi: 10.1093/jac/dky499. [DOI] [PubMed] [Google Scholar]
- 16.Hu Y., Liu C., Shen Z., Zhou H., Cao J., Chen S., Lv H., Zhou M., Wang Q., Sun L., et al. Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg. Microbes Infect. 2020;9:1771–1779. doi: 10.1080/22221751.2020.1799721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donati V., Feltrin F., Hendriksen R.S., Svendsen C.A., Cordaro G., García-Fernández A., Lorenzetti S., Lorenzetti R., Battisti A., Franco A. Extended-Spectrum-Beta-Lactamases, AmpC Beta-Lactamases and Plasmid Mediated Quinolone Resistance in Klebsiella spp. from Companion Animals in Italy. PLoS ONE. 2014;9:e90564. doi: 10.1371/journal.pone.0090564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harada K., Shimizu T., Mukai Y., Kuwajima K., Sato T., Usui M., Tamura Y., Kimura Y., Miyamoto T., Tsuyuki Y., et al. Phenotypic and Molecular Characterization of Antimicrobial Resistance in Klebsiella spp. Isolates from Companion Animals in Japan: Clonal Dissemination of Multidrug-Resistant Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae. Front. Microbiol. 2016;7:1021. doi: 10.3389/fmicb.2016.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong J.S., Song W., Park H.-M., Oh J.-Y., Chae J.-C., Shin S., Jeong S.H. Clonal Spread of Extended-Spectrum Cephalosporin-Resistant Enterobacteriaceae Between Companion Animals and Humans in South Korea. Front. Microbiol. 2019;10:1371. doi: 10.3389/fmicb.2019.01371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartantyo S.H.P., Chau M.L., Fillon L., Ariff A., Kang J.S.L., Aung K.T., Gutiérrez R.A. Sick pets as potential reservoirs of antibiotic-resistant bacteria in Singapore. Antimicrob. Resist. Infect. Control. 2018;7:106. doi: 10.1186/s13756-018-0399-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun J., Zhang H., Liu Y.-H., Feng Y. Towards Understanding MCR-like Colistin Resistance. Trends Microbiol. 2018;26:794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Tian G.-B., Zhang R., Shen Y., Tyrrell J.M., Huang X., Zhou H., Lei L., Li H.-Y., Doi Y., et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1 -positive Enterobacteriaceae in patients and healthy adults from China: An epidemiological and clinical study. Lancet Infect. Dis. 2017;17:390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 23.Lei L., Wang Y., Schwarz S., Walsh T.R., Ou Y., Wu Y., Li M., Shen Z. mcr-1 in Enterobacteriaceae from Companion Animals, Beijing, China, 2012–2016. Emerg. Infect. Dis. 2017;23:710–711. doi: 10.3201/eid2304.161732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pulss S., Stolle I., Stamm I., Leidner U., Heydel C., Semmler T., Prenger-Berninghoff E., Ewers C. Multispecies and Clonal Dissemination of OXA-48 Carbapenemase in Enterobacteriaceae From Companion Animals in Germany, 2009–2016. Front. Microbiol. 2018;9:1265. doi: 10.3389/fmicb.2018.01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ewers C., Stamm I., Pfeifer Y., Wieler L.H., Kopp P.A., Schønning K., Prenger-Berninghoff E., Scheufen S., Stolle I., Günther S., et al. Clonal spread of highly successful ST15-CTX-M-15 Klebsiella pneumoniae in companion animals and horses. J. Antimicrob. Chemother. 2014;69:2676–2680. doi: 10.1093/jac/dku217. [DOI] [PubMed] [Google Scholar]
- 26.Xia J., Fang L.-X., Cheng K., Xu G.-H., Wang X.-R., Liao X.-P., Liu Y.-H., Sun J. Clonal Spread of 16S rRNA Methyltransferase-Producing Klebsiella pneumoniae ST37 with High Prevalence of ESBLs from Companion Animals in China. Front. Microbiol. 2017;8:529. doi: 10.3389/fmicb.2017.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Jong A., Muggeo A., El Garch F., Moyaert H., De Champs C., Guillard T. Characterization of quinolone resistance mechanisms in Enterobacteriaceae isolated from companion animals in Europe (ComPath II study) Vet. Microbiol. 2018;216:159–167. doi: 10.1016/j.vetmic.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Struve C., Roe C.C., Stegger M., Stahlhut S.G., Hansen D.S., Engelthaler D.M., Andersen P.S., Driebe E.M., Keim P., Krogfelt K.A. Mapping the Evolution of Hypervirulent Klebsiella pneumoniae. mBio. 2015;6:e00630-15. doi: 10.1128/mBio.00630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wacharotayankun R., Arakawa Y., Ohta M., Tanaka K., Akashi T., Mori M., Kato N. Enhancement of extracapsular polysaccharide synthesis in Klebsiella pneumoniae by RmpA2, which shows homology to NtrC and FixJ. Infect. Immun. 1993;61:3164–3174. doi: 10.1128/IAI.61.8.3164-3174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou Z.-Y., Lei L., Chen Q.-Y., Wang Y.-Q., Cai C., Li W.-Q., Zhang Z., Shao B., Wang Y. Prevalence and dissemination risk of antimicrobial-resistant Enterobacteriaceae from shared bikes in Beijing, China. Environ. Int. 2019;132:105119. doi: 10.1016/j.envint.2019.105119. [DOI] [PubMed] [Google Scholar]
- 31.Insua J.L., Llobet E., Moranta D., Pérez-Gutiérrez C., Tomás A., Garmendia J., Bengoechea J.A. Modeling Klebsiella pneumoniae Pathogenesis by Infection of the Wax Moth Galleria mellonella. Infect. Immun. 2013;81:3552–3565. doi: 10.1128/IAI.00391-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J., Bi Z., Hulth A., Wang Y., Shen Z., Wang S., Wu C., Nilsson L.E., Walsh T.R., Börjesson S., et al. Inter-host Transmission of Carbapenemase-Producing Escherichia coli among Humans and Backyard Animals. Environ. Health Perspect. 2019;127:107009. doi: 10.1289/EHP5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treangen T.J., Ondov B.D., Koren S., Phillippy A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:1–15. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alikhan N.-F., Petty N.K., Ben Zakour N.L., Beatson S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All whole-genome sequencing data for this work are deposited in the GenBank and under BioProject accession no. PRJNA684769.