Graphical abstract
Keywords: SARS-CoV-2, Pseudotyped particles assay, Support vector machine (SVM), Consensus prediction, COVID-19
Abstract
In response to the pandemic caused by SARS-CoV-2, we constructed a hybrid support vector machine (SVM) classification model using a set of publicly posted SARS-CoV-2 pseudotyped particle (PP) entry assay repurposing screen data to identify novel potent compounds as a starting point for drug development to treat COVID-19 patients. Two different molecular descriptor systems, atom typing descriptors and 3D fingerprints (FPs), were employed to construct the SVM classification models. Both models achieved reasonable performance, with the area under the curve of receiver operating characteristic (AUC-ROC) of 0.84 and 0.82, respectively. The consensus prediction outperformed the two individual models with significantly improved AUC-ROC of 0.91, where the compounds with inconsistent classifications were excluded. The consensus model was then used to screen the 173,898 compounds in the NCATS annotated and diverse chemical libraries. Of the 255 compounds selected for experimental confirmation, 116 compounds exhibited inhibitory activities in the SARS-CoV-2 PP entry assay with IC50 values ranged between 0.17 µM and 62.2 µM, representing an enrichment factor of 3.2. These 116 active compounds with diverse and novel structures could potentially serve as starting points for chemistry optimization for COVID-19 drug discovery.
1. Introduction:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel coronavirus from the same family as SARS-CoV and Middle East respiratory syndrome (MERS) coronavirus.1 The disease caused by infection of SARS-CoV-2, coronavirus disease 2019 (COVID-19) remains a significant issue for global health, economics and society. It presents typical flu-like symptoms with a dry cough, sore throat, high fever, and breathing problems, which can become lethal in high-risk individuals.2 Up to January 6, 2021, COVID‐19 has caused the death of 1,876,100 individuals worldwide and infected nearly 86 million people over 222 countries, areas or territories.3 Although numerous potential therapies, including antiviral therapy, supportive intervention, immunomodulatory agents, and convalescent plasma transfusion, have been tentatively applied in clinical settings, there is still lack of a specific treatment for COVID-19.4 Currently intensive effort is ongoing worldwide to establish effective treatments for COVID-19 in addition to vaccine development.
Coronavirus entry into host cells is an important determinant of viral infectivity and pathogenesis.5, 6 It is also a major target for host immune surveillance and human intervention strategies.7, 8 One of the strategies for SARS-CoV-2 drug development is to develop a rapid and sensitive reporter assay for testing agents that block viral entry such as small molecule inhibitors and neutralizing antibodies. The SARS-CoV-2 pseudotyped particles (PP) are non-replicating, as they contain reporter RNA instead of a viral genome, and can be used in biosafety level 2 (BSL-2) facilities for high-throughput screening. There have been several reports on generating SARS-CoV and SARS-CoV-2 viral pseudotypes with glycoprotein-defective MLV, HIV, and VSV particles.9, 10, 11 More recently, Johnson et al reported on the optimized pseudotyping conditions for the SARS-CoV-2 spike glycoprotein,12 and Chen et al discussed the identification of SARS-CoV-2 entry inhibitors through drug repurposing screens of SARS-S and MERS-S PP.13
In this study, we have developed support vector machine (SVM) classification models based on a SARS-CoV-2 PP drug repurposing screen dataset to identify potent lead compounds and novel chemical matters as the starting point for drug development to treat COVID-19 patients. To evaluate the performance of SVM classification models, we conducted a parallel analysis using atom typing descriptors, 3D fingerprints, and consensus prediction based on two sets of descriptors. The consensus prediction outperformed the other two models with significantly improved area under the curve of receiver operating characteristic (AUC-ROC) of 0.91, where the compounds with inconsistent classifications were excluded. The consensus model was further used to screen the 173,898 compounds in three additional NCATS libraries, Sytravon, Genesis, and NPACT, none of which have been screened experimentally in the SARS-CoV-2 PP assay. Of the top 255 compounds cherry-picked and assayed, 116 compounds showed inhibitory activities in the PP entry assay with IC50 values ranged between 0.17 µM and 62.2 µM, representing an enrichment factor of 3.2. The 116 PP active compounds are structurally diverse. They are also structurally dissimilar to the active compounds in the training set, which offers a favorable starting point to jumpstart medicinal chemistry optimization for COVID-19 drug discovery.
2. Material and methods
2.1. Molecular descriptors
Molecular fingerprints (FPs) have been widely used in drug discovery and virtual screening in the last few decades.14 Molecular FPs are easy-to-use, and capable of handling vast number of molecules efficiently. There are many kinds of FPs including substructure- or functional group-based FPs, topological or path-based FPs, circular FPs, and pharmacophore FPs.14 Most FPs are calculated from SMILES or 2D structures, while some FPs require 3D structural information, and thus are more demanding on computational power and software resources. Atom-pair 2D FPs, computed from 2D structures, were reported to outperform 3D molecular descriptors in virtual screening, partly due to the observation that these 2D-based FPs may encode 3D features.15 However, interatomic distance through bonds in 2D space is not equivalent to the distance through 3D space, which is especially true for globular molecules. On the other hand, molecular shape based FPs,16 such as ROCS, are powerful in 3D similarity analysis, yet they are not designed for QSAR construction. In this study, we developed 3D atom-pair FPs to incorporate 3D structural information into QSAR development. 3D atom-pair FPs are to count atom pairs at increasing spatial distances in 3D structures. Atoms in a molecule are assigned into 7 categories, i.e. aromatic carbon, aliphatic carbon, positively charged atom, negatively charged atom, hydrogen bond donor, hydrogen bond acceptor, and polar atom. There are 28 different atom-pairs in total. For each atom-pair, the occurrence was computed at 13 distance ranges sampled between 1.5 Å and 7.5 Å, increasing at an interval of 0.5 Å. The count of each atom type in a molecule is appended to the FP, thus each 3D atom-pair FP is the vector of 7 + 28 * 13 = 371 numerical numbers.
Atom types are assigned according to the properties of an atom and its chemical environment. An atom type casting tree was designed to assign atom types, based on the fact of whether the atom is aromatic, whether the atom is in a ring, whether the atom is next to different functional groups, etc. This original tree, largely based on a medicinal chemist’s intuition, was subject to a recursive optimization cycles in terms of where to further split the tree, where to stop splitting, and where to combine the branches, in order to make the best prediction of logP values in the Starlist dataset containing about 11,000 structurally diverse compounds.17 The optimized tree output 218 atom types, featuring 88 different carbon types, 7 hydrogen types, 55 nitrogen types, 31 oxygen types, 8 halide types, 23 sulfur types, and 6 phosphorus types (Suppl. Table 1). Forty-six correction factors are appended to catch a number of whole molecule features, such as the molecular globularity, molecular rigidity, lipophilicity, and etc (Suppl. Table 2).17 In total, a series of 264 numerical values comprise the final set of the atom type molecular descriptors.
2.2. Datasets
The SARS-CoV-2 PP entry assay was performed in 1536-well plate format. This dataset, along with the cytotoxicity counter screen dataset, are publicly available on the NCATS OpenData Portal (https://opendata.ncats.nih.gov/covid19/).18 The primary assay screened two NCATS compound libraries, MIPE (Mechanism Interrogation Plate), containing 2,480 compounds, and NPC (the NCATS Pharmaceutical Collection) with 2,678 compounds.19 The active compounds were selected by two criteria, compounds showing activity in PP assay with curve class of −1.1, −1.2, −1.3, −2.1, −2.2, and maximum response over 60%, while showing no activity in the cytotoxicity assay. There were 415 compounds assigned as active. There were 2,527 compounds exhibiting no activity in the PP assay, which were assigned inactive. The rest compounds were inconclusive, which were not included in model construction. The hit rate was 14.1%. The compounds in the dataset were processed using Pipeline Pilot to strip salts, redundant and heavy metal containing compounds. The preprocessed data sets were randomly split into training (80%) and test (20%) sets.
2.3. SVM
As one of the few machine learning algorithms to address the generalization problem, support vector machine (SVM) is an elegant algorithm that has been successfully applied to many pattern recognition problems.20 The SVM classification algorithm υ-SVC, proposed by Schölkopf et al. and implemented by Chang and Lin in LIB-SVM, was employed in this study to construct predictive models for PP activity. The parameterization was performed on a grid-based search to minimize the mean standard error (MSE) of 5-fold cross-validation (CV) on the training data for υ, and γ, the non-linearity parameter in the kernel function of a Gaussian Radial Basis Function (RBF). Since the dataset in this study was severely imbalanced with majority of compounds being PP inactive, Jack-knife under-sampling strategy were applied to rebalance the heavily skewed training data. In Jack-knifing under-sampling, the majority class, the inactive compounds, was randomly divided into six equal-sized subgroups, and each subgroup was combined with the entire minority class to generate six downsized training sets. The averaged probabilities of the six models produced the final prediction. The AUC-ROC curve was applied to evaluate the performance of the binary classifiers.
2.4. SARS-CoV-2 PP assay
Expi293F cells with stable expression of human ACE2 (HEK293-ACE2) cells were cultured in DMEM, 10% FBS, 1x l-glutamine, 1x Pen/Strep, 1 μg/ml puromycin in a 1536-well format. Exogenous expression of ACE2 receptor is known to be necessary for SARS-CoV-2 pseudotyped virus/particle entry in HEK293 cells.11, 12 Cells were seeded at 1500 cells/well in 2 µL medium, and incubated at 37 °C, 5% CO2 overnight (~16 h). Compounds were titrated in DMSO, and 23 nL/well was dispensed via an automated pintool workstation (Wako Automation). Plates were incubated for 1 h at 37 °C, 5% CO2, and 2 µL/well of SARS-CoV-2 PP were dispensed. Plates were spinoculated by centrifugation at 1500 rpm (453 xg) for 45 min, and incubated for 48 h at 37 °C, 5% CO2. After the incubation, the supernatant was removed with gentle centrifugation using a Blue Washer (BlueCat Bio). Then, 4 µL/well of Bright-Glo (Promega) was dispensed, incubated for 5 min at room temperature, and luminescence signal was measured using a ViewLux plate reader (PerkinElmer). All data was normalized with wells containing SARS-CoV-2 PP as 100%, and bald PP as 0% entry.
2.5. ATP content cytotoxicity counter screen
HEK293-ACE2 cells were seeded at 1500 cells/well in 2 µL/well medium in 1536-well plates, and incubated at 37 °C, 5% CO2 overnight (~16 h). Compounds were titrated in DMSO, 23 nL/well was dispensed via an automated pintool workstation (Wako Automation). Plates were incubated for 1 h at 37 °C, 5% CO2, before 2 µL/well of medium was added. Plates were incubated for 48 h at 37 °C, 5% CO2. Then, 4 µL/well of ATPLite (PerkinElmer) was dispensed, incubated for 15 min at room temperature, and luminescence signal was measured using a ViewLux plate reader (PerkinElmer). Data was normalized with wells containing cells as 100%, and wells containing no cells (media only control) as 0% viability.
3. Results and discussion:
3.1. Improved performance with consensus models
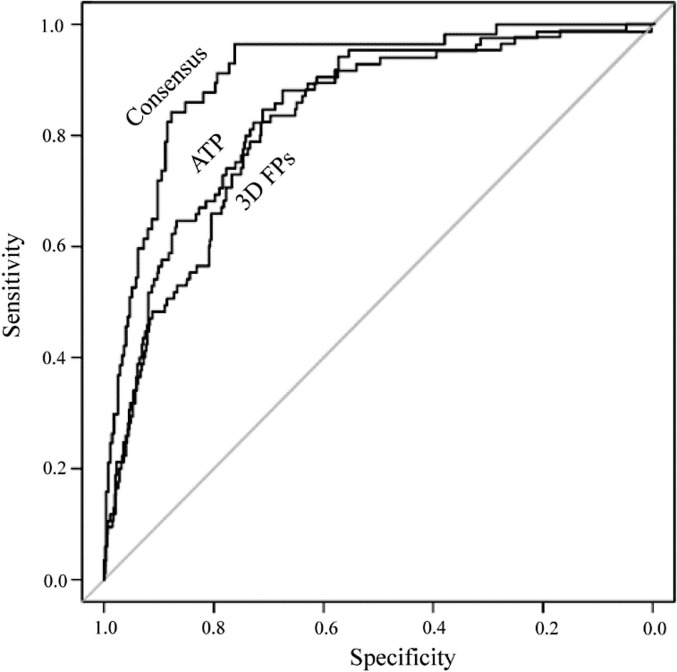
The SVC models built on the basis of atom type molecular descriptors and 3D atom-pair FPs both performed well on predictions of PP activity for the compounds in the test set, with AUC-ROC of 0.84 and 0.82, respectively. However, a performance boost was observed in the consensus model, where the AUC-ROC value was significantly improved to 0.91 (Figure 1 ).
Fig. 1.
The receiver operating characteristic (ROC) curves of the atom-type-based (ATP) model (AUC = 0.84), 3D atom-pair FP-based model (AUC = 0.82) and the consensus model (AUC = 0.91).
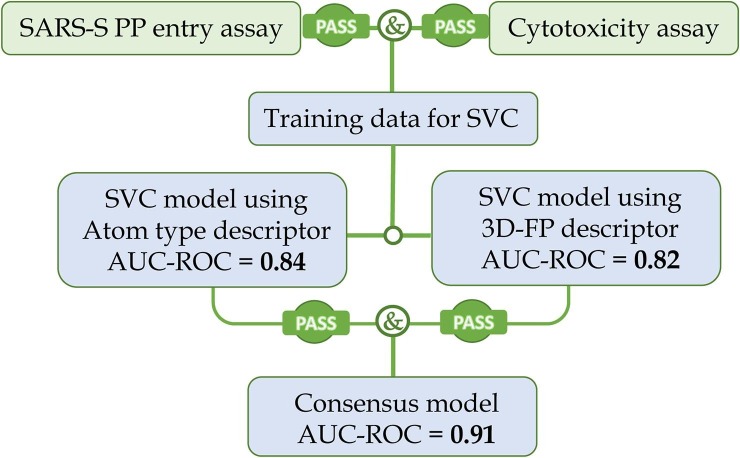
Atom-type-based FPs are rich in information of chemical features of each atom in a molecule, but poor in spatial relations among the atoms. Whereas 3D atom-pair FPs, on the contrary, are rich in interatomic spatial information, but weak in defining atomic features. Consensus models built on the two very different molecular descriptors achieved complimentary advantages by combining the strengths of both descriptor systems, resulting in a boost in predictivity. Only those VS hits that were confirmed by both SVC models were predicted as final hits in the consensus model (Figure 2 ).
Fig. 2.
The flowchart of data processing and model construction. The consensus model was achieved by excluding the disagreed hits of both SVC models.
3.2. Consensus model leading to successful VS
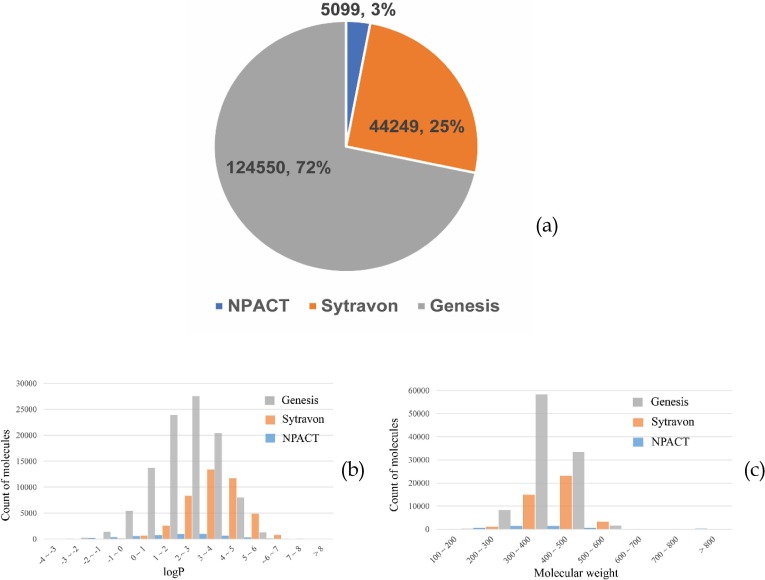
The consensus model was then applied in screening the compounds in three additional NCATS libraries, Sytravon, Genesis, and NPACT (the NCATS Pharmacologically Active Chemical Toolbox), none of which have been screened experimentally. These three compound libraries consisted of marketed drugs and molecules of pharmaceutical interest. According to the distribution of logP and molecular weight (MW) of the molecules in the three libraries, compounds in Genesis are more fragment-like, with smaller MW and higher hydrophilicity (Figure 3 ). The top 255 compounds from the consensus model were plated and assayed in SARS-CoV-2 PP entry assay and cytotoxicity assay.
Fig. 3.
Library composition and physicochemical property distribution. (a) Pie chart of library composition in the number and percentage of compounds and histogram of (b) calculated logP and (c) molecular weight for the three compound libraries, Genesis in gray, Sytravon in orange, and NPACT in blue.
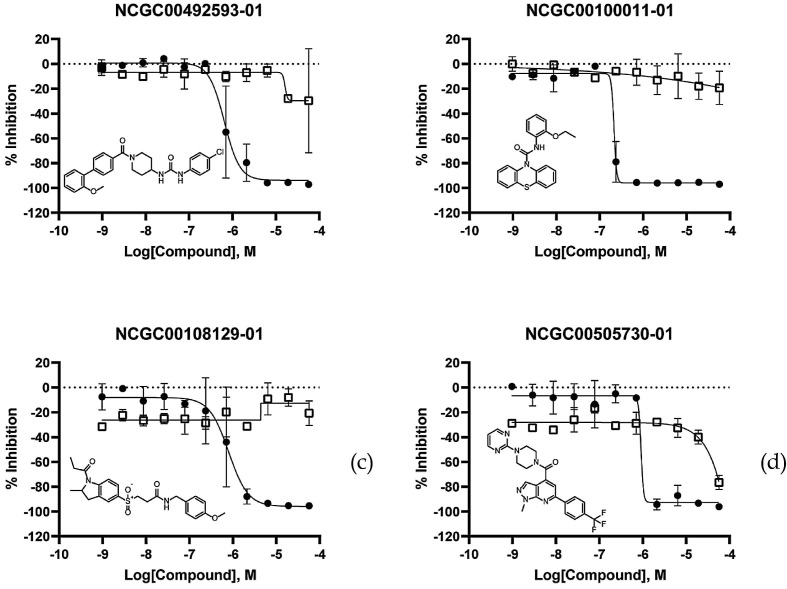
There were 116 compounds exhibiting measurable activities in the PP entry assay, representing an enrichment factor of 3.2. The IC50 values ranged between 0.17 µM and 62.2 µM. More interestingly, most of the hits of PP entry assay were either inactive or weakly active in cytotoxicity assay (Figure 4 and S1). Although cytotoxicity was not explicitly incorporated in the model construction, exclusion of the cytotoxic compounds from the hit list for training implied for the cytotoxicity requirements, and in turn reflected in the models and the observed assay results.
The 116 PP active compounds are structurally diverse, as shown in Figure 4 . They are also structurally dissimilar to the active compounds in the training set. The average Tanimoto similarities of the 46 most active compounds with their closest analogues in the training set was 0.29, as measured by FCFP4, one of the popular circular FPs.
Fig. 4.
Concentration-response curves for top ranking promising hits. Solid circles represent data from PP entry assay, blank squares represent data from cytotoxicity counter screen.
4. Conclusions
Based on the primary SARS-CoV-2 PP entry assay and cytotoxicity assay results, two SVC models were built to predict antiviral potency from chemical structures, by using two different molecular descriptor systems, atom type descriptors and 3D fingerprints (FPs). Both models achieved reasonable predictivity, with AUC-ROC of 0.84 and 0.82, respectively. Complementary advantage was revealed in the consensus model, where the compounds with inconsistent classifications were excluded. The performance boost of the consensus model, as indicated by its high AUC of 0.91, was presumably due to the diversity of the two molecular descriptor systems. The consensus model was recruited to screen the 173,898 compounds in three NCATS libraries, Sytravon, Genesis, and NPACT, which have not been screened experimentally. Of the 255 compounds plated and assayed, 116 compounds exhibited measurable activities in the PP entry assay with IC50 values ranging between 0.17 µM and 62.2 µM, representing an enrichment factor of 3.2. The 116 hit compounds are not only structurally diverse to each other, but also structurally dissimilar to the active compounds in the training set. The most potent compounds in the PP entry assay showed no/or weak cytotoxicity in the counter assay. The PP entry assay was conducted in HEK293-ACE2 cells, which is known to have a largely cathepsin protease driven, and endocytosis dependent, entry mechanism for SARS-CoV-2 entry.21 The PP entry assay is a phenotypic assay and confirmed inhibitors could act at various entry steps, including, but not limited to, cell surface receptor binding, PP endocytosis, protease cleavage, and membrane fusion. While more follow up work is needed, the novel active compounds could provide a great starting point for COVID-19 drug discovery.
Declaration of Competing Interest
The authors declared that there is no conflict of interest.
Acknowledgement
This work was supported by the Intramural Research Programs of the National Center for Advancing Translational Sciences, National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bmc.2021.116119.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lu R., Zhao X., Li J., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N., Zhou M., Dong X., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 4.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du L., Yang Y., Zhou Y., Lu L., Li F., Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin Ther Targets. 2017;21(2):131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giroglou T., Cinatl J., Jr., Rabenau H., et al. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J Virol. 2004;78(17):9007–9015. doi: 10.1128/JVI.78.17.9007-9015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millet J.K., Tang T., Nathan L., et al. Production of Pseudotyped Particles to Study Highly Pathogenic Coronaviruses in a Biosafety Level 2 Setting. J Vis Exp. 2019;145 doi: 10.3791/59010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford K.H.D., Eguia R., Dingens A.S., et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12(5) doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson M.C., Lyddon T.D., Suarez R., et al. Optimized Pseudotyping Conditions for the SARS-COV-2 Spike Glycoprotein. J Virol. 2020;94(21) doi: 10.1128/JVI.01062-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C.Z., Xu M., Pradhan M., et al. Identifying SARS-CoV-2 entry inhibitors through drug repurposing screens of SARS-S and MERS-S pseudotyped particles. bioRxiv. 2020 doi: 10.1021/acsptsci.0c00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cereto-Massague A., Ojeda M.J., Valls C., Mulero M., Garcia-Vallve S., Pujadas G. Molecular fingerprint similarity search in virtual screening. Methods. 2015;71:58–63. doi: 10.1016/j.ymeth.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Awale M., Reymond J.L. Atom pair 2D-fingerprints perceive 3D-molecular shape and pharmacophores for very fast virtual screening of ZINC and GDB-17. J Chem Inf Model. 2014;54(7):1892–1907. doi: 10.1021/ci500232g. [DOI] [PubMed] [Google Scholar]
- 16.Haigh J.A., Pickup B.T., Grant J.A., Nicholls A. Small molecule shape-fingerprints. J Chem Inf Model. 2005;45(3):673–684. doi: 10.1021/ci049651v. [DOI] [PubMed] [Google Scholar]
- 17.Sun H. Elsevier; Cambridge: 2015. A Practical Guide to Rational Drug Design. [Google Scholar]
- 18.Brimacombe, K. R.; Zhao, T.; Eastman, R. T.; Hu, X.; Wang, K.; Backus, M.; Baljinnyam, B.; Chen, C. Z.; Chen, L.; Eicher, T.; Ferrer, M.; Fu, Y.; Gorshkov, K.; Guo, H.; Hanson, Q. M.; Itkin, Z.; Kales, S. C.; Klumpp-Thomas, C.; Lee, E. M.; Michael, S.; Mierzwa, T.; Patt, A.; Pradhan, M.; Renn, A.; Shinn, P.; Shrimp, J. H.; Viraktamath, A.; Wilson, K. M.; Xu, M.; Zakharov, A. V.; Zhu, W.; Zheng, W.; Simeonov, A.; Mathe, E. A.; Lo, D. C.; Hall, M. D.; Shen, M., An OpenData portal to share COVID-19 drug repurposing data in real time. bioRxiv 2020.
- 19.Huang R., Zhu H., Shinn P., et al. The NCATS Pharmaceutical Collection: a 10-year update. Drug Discov Today. 2019;24(12):2341–2349. doi: 10.1016/j.drudis.2019.09.019. [DOI] [PubMed] [Google Scholar]
- 20.Cortes C., Vapnik V.N. Support vector networks. Machine Learning. 1995;20:273–297. [Google Scholar]
- 21.Ou X., Liu Y., Lei X., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.