Several studies attempting to dissect the molecular and cellular aspects of the SARS-CoV-2 virus pathophysiology have investigated the host immune responses and the roles of complement in viral sensing and the inflammation dynamics during infection (Yuki et al., 2020; Kaneko et al., 2020; Wilk, 2020; Yu et al., 2020; Java et al., 2020). Aspects of the pandemic COVID-19 pathology resemble, from moderate to severe cases, clinical features traditionally associated with complementopathies (Marcos-Jiménez et al., 2021; Joseph et al., 2020; Baines and Brodsky, 2017). Some of these are associated with disease phenotypes characterised by the deregulated activation of the Alternative pathway resulting in complement C3 consumption and deprivation of the downstream activation processes (Fig. 1 ) (Łukawska et al., 2018; Sim and Tsiftsoglou, 2004; Roversi et al., 2011). Therefore, there is a rationale for targeting complement in COVID-19 (Polycarpou et al., 2020).
Fig. 1.
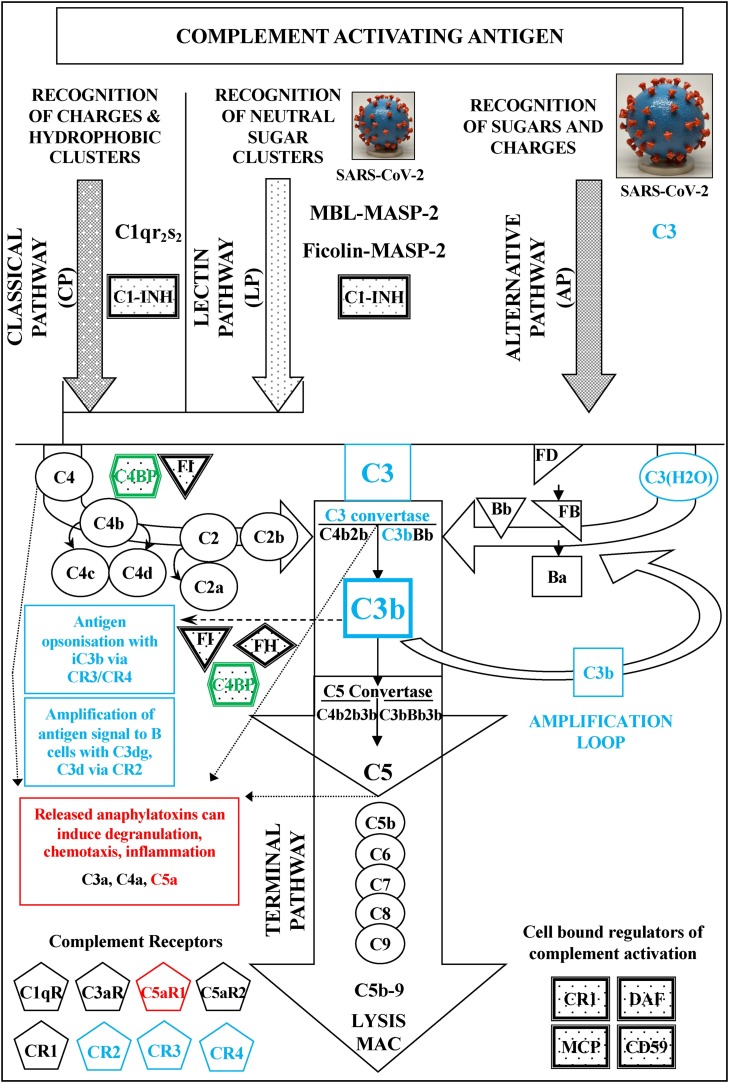
A schematic representation of the human complement system. The three activation pathways of the system (Classical, Lectin and Alternative) are represented together with the Terminal pathway that eventually leads to the formation of the Membrane Attack Complex (MAC) after complement activation. Regulatory inhibitory proteins are shown in shaded boxes; complement receptors in pentagons. One positive regulator, properdin, which stabilizes C3bBb is not shown. Missense polymorphic variants of the genes encoding for C3  , C4BPα
, C4BPα  and C5aR1
and C5aR1  have been recently associated with a susceptibility to SARS-CoV-2 virus infection and establishment. The highlighted complement components, as well as their coloured associated products and processes, are important for the homeostasis of complement and its interactions with cellular immune surveillance and defence. Recent studies have suggested that the SARS-CoV-2 virus is exploiting the Alternative pathway to gradually drain the host of complement and disrupt associated cellular immune responses (Yu et al., 2020; Kulkarni and Atkinson, 2020). C3
have been recently associated with a susceptibility to SARS-CoV-2 virus infection and establishment. The highlighted complement components, as well as their coloured associated products and processes, are important for the homeostasis of complement and its interactions with cellular immune surveillance and defence. Recent studies have suggested that the SARS-CoV-2 virus is exploiting the Alternative pathway to gradually drain the host of complement and disrupt associated cellular immune responses (Yu et al., 2020; Kulkarni and Atkinson, 2020). C3 and C4BPαfx2polymorphic variants: During SARS-CoV-2 virus infection specific point mutations in the C3 and C4BP proteins can hypothetically, independently or synergistically, lead to the strengthening of the relevant dynamics of the AP amplification loop facilitating a faster consumption of C3 and inefficient opsonisation. Turnover of C3 can gradually cripple the activation of the downstream terminal complement components. This can eventually lead to reduction of C5a anaphylatoxin release and limited or no formation of the terminal MAC attack complex. In terms of the point mutations discussed in the text, faster consumption of C3 can occur either through the weakening of the inactivation of the viral surface deposited C3/C3b by complement factor I in the presence of a cofactor such as FH or C4BP, or through the strengthening of the interaction of C3b with the activated FB (Bb) in the C3bBb C3 convertase of the Alternative pathway. The weakening of the C3/C3b inactivation leads not only to poor opsonisation and enhanced masking of the viral particles from phagocytic cells expressing the CR3 and/or CR4 receptors, but also from any cross-reactive B cells expressing the CR2 receptor. C5aR1
and C4BPαfx2polymorphic variants: During SARS-CoV-2 virus infection specific point mutations in the C3 and C4BP proteins can hypothetically, independently or synergistically, lead to the strengthening of the relevant dynamics of the AP amplification loop facilitating a faster consumption of C3 and inefficient opsonisation. Turnover of C3 can gradually cripple the activation of the downstream terminal complement components. This can eventually lead to reduction of C5a anaphylatoxin release and limited or no formation of the terminal MAC attack complex. In terms of the point mutations discussed in the text, faster consumption of C3 can occur either through the weakening of the inactivation of the viral surface deposited C3/C3b by complement factor I in the presence of a cofactor such as FH or C4BP, or through the strengthening of the interaction of C3b with the activated FB (Bb) in the C3bBb C3 convertase of the Alternative pathway. The weakening of the C3/C3b inactivation leads not only to poor opsonisation and enhanced masking of the viral particles from phagocytic cells expressing the CR3 and/or CR4 receptors, but also from any cross-reactive B cells expressing the CR2 receptor. C5aR1 polymorphic variants: During the early stages of the SARS-CoV-2 virus infection and prior to any exploitation of the Alternative pathway amplification loop, any downstream assembly of the C5 convertase can support the release of C5a which is regarded as the most potent complement anaphylatoxin. Its immune pleiotropic potency however can be hampered by any mutations in the C5aR1 that weaken the interaction of the ligand with the receptor. In terms of the point mutations discussed in the text, the lack of efficient C5a-C5aR1 interaction can actually weaken critical immune defence responses at sites of infections that include chemoattraction of specialized immune cell types and subsets of granulocytes and neutrophils. Fig. 1 was adapted from (Sim and Tsiftsoglou, 2004) with minor modifications. The SARS-CoV-2 3D print picture is from the NIH 3D Print Exchange: https://flic.kr/p/2ixx3G3.
polymorphic variants: During the early stages of the SARS-CoV-2 virus infection and prior to any exploitation of the Alternative pathway amplification loop, any downstream assembly of the C5 convertase can support the release of C5a which is regarded as the most potent complement anaphylatoxin. Its immune pleiotropic potency however can be hampered by any mutations in the C5aR1 that weaken the interaction of the ligand with the receptor. In terms of the point mutations discussed in the text, the lack of efficient C5a-C5aR1 interaction can actually weaken critical immune defence responses at sites of infections that include chemoattraction of specialized immune cell types and subsets of granulocytes and neutrophils. Fig. 1 was adapted from (Sim and Tsiftsoglou, 2004) with minor modifications. The SARS-CoV-2 3D print picture is from the NIH 3D Print Exchange: https://flic.kr/p/2ixx3G3.
Targeting complement activation in COVID-19 has been recently discussed more extensively, as a couple of studies have showed that the SARS-CoV-2 surface proteins N (Wilk, 2020) and S (Yu et al., 2020) can activate the Lectin and Alternative pathways, respectively (Fig. 1). The activation of the Alternative pathway by the spike glycoprotein S has been of particular interest as it promotes entry of the virus into cells and is a major antigenic target for B cell responses (Walls et al., 2020). Furthermore, the potential exploitation of the activation of the Alternative pathway via the amplification loop by SARS-CoV-2 is of even greater particular interest, because it can explain to a considerable extent the gradual diminishment of complement responses during the systemic establishment of the virus (Marcos-Jiménez et al., 2021; Kulkarni and Atkinson, 2020).
A very recently published study aiming to associate genetic variation at the chromosome multigene cluster 3p21.31 and the ABO blood group with complement activation and COVID-19 severity, identified a variant (rs11385942) that predisposes to severe COVID-19 and is associated with enhanced complement activation, as manifested by circulating C5a and sC5-C9 and C5a only in the non-O blood group (Valenti et al., 2021). Although, the particular variant is not directly mapped in any complement gene coding loci, it may be linked with enhanced complement activation indirectly at subsequent stages of the disease during systemic inflammation and tissue damage and/or organ failure (Bianco et al., 2021).
The potential exploitation of the activation of the Alternative pathway via the by SARS-CoV-2 is an attractive working hypothesis that prompted me to explore further published advanced complement genotyping datasets from hospitalised COVID-19 patients in order to identify clinical genetic evidence supporting this notion (Ramlall et al., 2020). In the Supplementary Table S2 of their study, Ramlall et al. had reported among all, 23 study-wide significant SNPs that lie within 60Kb of 12 complement genes in at least one of the reported analyses examined (Ramlall et al., 2020). My online database search using the NCBI dbSNP database (Sherry et al., 2001) (https://www.ncbi.nlm.nih.gov/snp/) revealed that the majority of those SNPs, 19 (∼83 %), are genomically located in introns or in upstream or downstream untranslated regions that may contain elements or harbour sites important for epigenetic regulation. The remaining 4 SNPs (∼17 %), are located within exons of the 3 complement genes C3, C4BPA and C5AR1 and encode for missense amino acid substitutions in the expressed proteins (Table 1 ). Given the nature of the protein-protein interactions among complement components and the influences that polymorphisms may contribute to the relevant protein complex structures, I investigated further in the NCBI ClinVar (Landrum et al., 2018) (https://www.ncbi.nlm.nih.gov/clinvar/) and PubMed (https://pubmed.ncbi.nlm.nih.gov/) databases for published evidence reporting altered properties of the polymorphic variants corresponding to the missense mutations of interest (Roversi et al., 2011). In ClinVar I found some relevant clinical evidence only for the R102G and the P314L amino acid substitutions of C3, but none for the others (Table 1).
Table 1.
Four SNPs in three human complement genes that encode for missense polymorphic variants associated with SARS-CoV-2 susceptibility.
| Gene symbol, ID1 | rsID2 | Position3 | dbSNP4 | ClinVar5 | UniProt Entry6 |
|---|---|---|---|---|---|
| C3, 718 | rs2230199 | chr19:6718376 | R102G, R102S | R102G | P01024, CO3_HUMAN |
| rs1047286 | chr19:6713251 | P314L, P314R | P314L | ||
| C4BPA, 722 | rs45574833 | chr1:207126725 | R240H, R240P | N/A | P04003, C4BPA_HUMAN |
| C5AR1, 728 | rs4467185 | chr19:47319781 | D2H, D2N, D2Y | N/A | P21730, C5AR1_HUMAN |
NCBI Gene database.
Reference SNP cluster ID.
Anchor position in the human genome primary assembly GRCh38.p12.
Encoded amino acid substitutions reported in the NCBI dbSNP database.
Amino acid substitutions of clinical significance reported in the NCBI ClinVar database.
UniProt Entry for each complement component.
In terms of protein structure, the R80 amino acid residue is close to the N-terminal end of the C3 beta chain and located in the MG1 domain near the TED domain of the C3 protein (Janssen et al., 2006). The three positively charged amino acids R72, R80 and K82 of the MG1 domain, along with the four negatively charged amino acids D1007, E1008, E1010, E1013 of the TED domain, form the internal thioester bond that is critical for the reactivity of C3 with nucleophilic groups (Law and Dodds, 1997). The rs2230199 introduced R102G (R80G) polymorphism was first reported in 1967 and has been described in the literature to distinguish the C3S (Slow, R80) and the C3F (Fast, G80) variants (Wieme and Demeulenaere, 1967; Alper and Propp, 1968; Botto et al., 1990). The C3F variant, containing neutral glycine instead of the positively charged arginine has potentially reduced properties with regards to the stability of its thioester and its capacity to react with potential complement activator surfaces. It also exhibits a weaker ability to bind to complement factor H (FH) which is the major circulating cofactor for the complement factor I (FI) mediated downregulation of the C3bBb AP convertase (Sim and Tsiftsoglou, 2004; Janssen et al., 2006; Torreira et al., 2009; Roversi et al., 2011). The common C3S/F comprises approximately 98 % of all C3 phenotypes (Łukawska et al., 2018). The frequency of the F allele varies among different populations, and it is the highest in Caucasian (18 %) and the lowest in Asian (1%) (Bazyar et al., 2012). Carriers of the C3F allele have been reported to be at higher risk for developing age-related macular degeneration (AMD) (Yates et al., 2007) and various types of glomerulonephritis namely membranoproliferative glomerulonephritis type II (MPGN II) that is presently known as dense deposit disease (DDD) (Abrera-Abeleda et al., 2011), IgA nephropathy (IgAN) (Rambausek et al., 1987) and systemic vasculitis (SV) (Persson et al., 2013).
The second C3 point mutation corresponding to rs1047286 in the exon 9 of the beta chain, the P314L (P292L) has also been historically characterised as HAV4−1+/- (Table 1) (Koch and Behrendt, 1986; Botto et al., 1990). The P292 (HAV 4−1-) residue is located in the MG3 domain of C3 that contains the major binding site for the activated complement factor B (Bb). Published structural studies have showed that the polymorphic variant HAV4−1+ (L292) exhibits stronger ability to bind to Bb (Torreira et al., 2009; Forneris et al., 2010). This practically suggests that the HAV4-1+ (L292) gain-of-function variant may form a hyperactive C3bBb AP convertase that may be also more resilient to inactivation by complement factor I (FI) in the presence of a cofactor such as complement factor H (FH) or C4BP (Blom et al., 2003). It has been estimated that 98 % of C3S allele carriers is HAV4−1- and only 2% is HAV4−1+; in contrast, 90 % carriers of the C3F allele is HAV4−1+ and 10 % is HAV4−1- (Botto et al., 1990). For both C3 SNPs discussed above, the hypothetical impaired downregulation of any formed C3bBb AP convertase, can practically result in lower generation of iC3b, C3dg and C3d which are major opsonins of the complement system (Fig. 1) (Carroll, 2004). Impaired opsonisation during infection can contribute to poor immune synergies of complement with the various professional immune surveillance cell populations that include phagocytes, neutrophils and B cells (Carroll, 2004). Thus, impaired downregulation of any C3bBb AP convertase formed during the Alternative pathway of complement activation, can enable the virus to exploit the alternative activation pathway amplification loop introducing C3 consumption (Fig. 1). Based on the findings above, in theory, individuals carrying combinations of C3 alleles harbouring the C3F and/or the HAV4−1+ polymorphisms must be considerably more susceptible to SARS-CoV-2 infection based on the structural interactions of C3 with various antigen surfaces and the Bb, FH, FI and C4BP complement components.
The R240 (R192) residue that corresponds to the rs45574833 SNP is located in the 4th N-terminal CCP (Sushi) domain of the C4BP alpha chain (C4BPα) (Table 1). C4BP can act as a cofactor for the inactivation of C3b (Blom et al., 2003) and C4b (Blom et al., 1999) by complement FI (Sim and Tsiftsoglou, 2004). Refined mutagenesis studies in C4BPα had mapped the C3b binding sites to CCP1−4 (Blom et al., 2003) and the C4b binding sites to CCP1−3 (Blom et al., 1999). Given the ionic nature of these interactions and the position of R240, the R240H or R240P substitution may reduce the downregulation of the C3bBb AP convertase by FI in the presence of C4BP, especially in cases where the interaction of C3 with FH is weakened as discussed earlier for the R80G polymorphism. The inactivation of C4b by FI should not be directly affected (Blom et al., 1999). Overall, in theory the allelic co-existence of the missense mutations encoding for the C3 R80, P292 and C4BPα R192 polymorphisms, can create synergistic thresholds of enhanced susceptibility in the deregulation of the activation Alternative pathway by SARS-CoV-2.
Last, but not least is the rs4467185 SNP that encodes for a missense mutation at the extracellular 2nd N-terminal end residue of the C5aR1 receptor. C5a is regarded as the most potent complement anaphylatoxin with a wide effect of pleiotropic effects (Java et al., 2020; Carroll, 2004). Mutagenesis studies had showed that the N-terminus accounts for at least 45 % of the total energy required for the binding of C5a. When the region 2–30 containing aspartic acids was deleted, the C5a binding avidity diminished by 45,000-fold (DeMartino et al., 1994). The candidate D2H, D2N, D2Y encoded missense point mutations at position 2 can thus theoretically reduce the binding dynamics of C5a to its receptor, thus possibly affecting neutrophil recruitment and/or other complement interactions with specialized immune cells during the inflammation of infection (Woodruff and Shukla, 2020; Carvelli et al., 2020).
Overall, I consider these findings important, because at the protein level they may provide a hypothetical, but rational, frame proposing the exploitation of the activation of the Alternative pathway by SARS-CoV-2. In the context of the COVID-19 molecular pathology, this can gradually lead to complement deregulation, poor opsonisation, weakening of the immune clearance mechanisms and sustained inflammation resulting in various host tissue damage responses in severe cases of infection.
Author contributions statement
SAT conceived the study, searched the relevant databases and literature and wrote the manuscript.
Declaration of Competing Interest
None.
Footnotes
I wish to dedicate this paper to the memory of my Graduate school supervisor in Oxford University Dr. Robert B. Sim, a pioneer in the field of Complement Biochemistry, who sadly passed away during the reviewing of this manuscript. I feel grateful for all his interest, care and inspirational mentorship. I am also grateful for all the decades-long outstanding contributions from basic research scientists and physicians that helped enrich our modern understanding of humoral immunity.
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.molimm.2021.03.021.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Abrera-Abeleda M.A., Nishimura C., Frees K., Jones M., Maga T., Katz L.M., Zhang Y., Smith R.J.H. Allelic variants of complement genes associated with dense deposit disease. J. Am. Soc. Nephrol. 2011;22:1551–1559. doi: 10.1681/ASN.2010080795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper C.A., Propp R.P. Genetic polymorphism of the third component of human complement (C’3) J. Clin. Invest. 1968;47:2181–2191. doi: 10.1172/JCI105904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines A.C., Brodsky R.A. 2017. Complementopathies. Blood Rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazyar N., Azarpira N., Khatami S.R., Galehdari H. The investigation of allele and genotype frequencies of human C3 (rs2230199) in south Iranian population. Mol. Biol. Rep. 2012;39:8919–8924. doi: 10.1007/s11033-012-1759-9. [DOI] [PubMed] [Google Scholar]
- Bianco C., Baselli G., Malvestiti F., Santoro L., Pelusi S., Manunta M., Grasselli G., Bandera A., Scudeller L., Prati D., Valenti L. Genetic insight into COVID-19-related liver injury. Liver Int. 2021 doi: 10.1111/liv.14708. [DOI] [PubMed] [Google Scholar]
- Blom A.M., Webb J., Villoutreix B.O., Dahlbäck B. A cluster of positively charged amino acids in the C4BP α-chain is crucial for C4b binding and factor I cofactor function. J. Biol. Chem. 1999;274:19237–19245. doi: 10.1074/jbc.274.27.19237. [DOI] [PubMed] [Google Scholar]
- Blom A.M., Kask L., Dahlbäck B. CCP1-4 of the C4b-binding protein α-chain are required for factor I mediated cleavage of complement factor C3b. Mol. Immunol. 2003;39:547–556. doi: 10.1016/S0161-5890(02)00213-4. [DOI] [PubMed] [Google Scholar]
- Botto M., Fong K.Y., So A.K., Koch C., Walport M.J. Molecular basis of polymorphisms of human complement component C3. J. Exp. Med. 1990;172:1011–1017. doi: 10.1084/jem.172.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M.C. The complement system in regulation of adaptive immunity. Nat. Immunol. 2004 doi: 10.1038/ni1113. [DOI] [PubMed] [Google Scholar]
- Carvelli J., Demaria O., Vély F., Batista L., Chouaki Benmansour N., et al. Association of COVID-19 inflammation with activation of the C5a–C5aR1 axis. Nature. 2020;588:146–150. doi: 10.1038/s41586-020-2600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMartino J.A., Van Riper G., Siciliano S.J., Molineaux C.J., Konteatis Z.D., Rosen H., Springer M.S. The amino terminus of the human C5a receptor is required for high affinity C5a binding and for receptor activation by C5a but not C5a analogs. J. Biol. Chem. 1994;269:14446–14450. doi: 10.1016/s0021-9258(17)36643-7. [DOI] [PubMed] [Google Scholar]
- Forneris F., Ricklin D., Wu J., Tzekou A., Wallace R.S., Lambris J.D., Gros P. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science (80-.) 2010;330:1816–1820. doi: 10.1126/science.1195821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen B.J.C., Christodoulidou A., McCarthy A., Lambris J.D., Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- Java A., Apicelli A.J., Kathryn Liszewski M., Coler-Reilly A., Atkinson J.P., Kim A.H.J., Kulkarni H.S. The complement system in COVID-19: Friend and foe? JCI Insight. 2020 doi: 10.1172/jci.insight.140711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph A., Zafrani L., Mabrouki A., Azoulay E., Darmon M. Acute kidney injury in patients with SARS-CoV-2 infection. Ann. Intensive Care. 2020 doi: 10.1186/s13613-020-00734-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N., Kuo H.H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., et al. Loss of Bcl-6-Expressing t follicular helper cells and germinal centers in COVID-19. Cell. 2020 doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C., Behrendt N. A novel polymorphism of human complement component C3 detected by means of a monoclonal antibody. Immunogenetics. 1986;23:322–325. doi: 10.1007/BF00398796. [DOI] [PubMed] [Google Scholar]
- Kulkarni H.S., Atkinson J.P. Targeting complement activation in COVID-19. Blood. 2020 doi: 10.1182/BLOOD.2020008925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., Karapetyan K., Katz K., Liu C., Maddipatla Z., Malheiro A., McDaniel K., Ovetsky M., Riley G., Zhou G., Holmes J.B., Kattman B.L., Maglott D.R. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law S.K.A., Dodds A.W. The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 1997 doi: 10.1002/pro.5560060201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łukawska E., Polcyn-Adamczak M., Niemir Z.I. The role of the alternative pathway of complement activation in glomerular diseases. Clin. Exp. Med. 2018 doi: 10.1007/s10238-018-0491-8. [DOI] [PubMed] [Google Scholar]
- Marcos-Jiménez A., Sánchez-Alonso S., Alcaraz-Serna A., Esparcia L., López-Sanz C., Sampedro-Núñez M., Mateu-Albero T., Sánchez-Cerrillo I., Martínez-Fleta P., Gabrie L., del Campo Guerola L., Rodríguez-Frade J.M., Casasnovas J.M., Reyburn H.T., Valés-Gómez M., López-Trascasa M., Martín-Gayo E., Calzada M.J., Castañeda S., de la Fuente H., González-Álvaro I., Sánchez-Madrid F., Muñoz-Calleja C., Alfranca A. Deregulated cellular circuits driving immunoglobulins and complement consumption associate with the severity of COVID-19 patients. Eur. J. Immunol. 2021 doi: 10.1002/eji.202048858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson U., Gullstrand B., Pettersson Å., Sturfelt G., Truedsson L., Segelmark M. A candidate gene approach to ANCA-associated vasculitis reveals links to the C3 and CTLA-4 genes but not to the IL1-Ra and Fcγ-RIIa genes. Kidney Blood Press. Res. 2013;37:641–648. doi: 10.1159/000355744. [DOI] [PubMed] [Google Scholar]
- Polycarpou A., Howard M., Farrar C.A., Greenlaw R., Fanelli G., Wallis R., Klavinskis L.S., Sacks S. Rationale for targeting complement in COVID‐19. EMBO Mol. Med. 2020 doi: 10.15252/emmm.202012642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambausek M., Van den Wall Bake A.W.L., Schumacher-Ach R., Spitzenberg R., Rother U., Van Es L.A., Ritz E. Genetic polymorphism of C3 and Bf in IgA nephropathy. Nephrol. Dial. Transplant. 1987;2:208–211. doi: 10.1093/oxfordjournals.ndt.a091543. [DOI] [PubMed] [Google Scholar]
- Ramlall V., Thangaraj P.M., Meydan C., Foox J., Butler D., Kim J., May B., De Freitas J.K., Glicksberg B.S., Mason C.E., Tatonetti N.P., Shapira S.D. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat. Med. 2020;26:1609–1615. doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roversi P., Johnson S., Caesar J.J.E., McLean F., Leath K.J., Tsiftsoglou S.A., Morgan B.P., Harris C.L., Sim R.B., Lea S.M. Structural basis for complement factor I control and its disease-associated sequence polymorphisms. Proc. Natl. Acad. Sci. U. S. A. 2011 doi: 10.1073/pnas.1102167108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. DbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim R.B., Tsiftsoglou S.A. Proteases of the complement system. Biochem. Soc. Trans. 2004 doi: 10.1042/BST0320021. [DOI] [PubMed] [Google Scholar]
- Torreira E., Tortajada A., Montes T., De Córdoba S.R., Llorca O. 3D structure of the C3bB complex provides insights into the activation and regulation of the complement alternative pathway convertase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:882–887. doi: 10.1073/pnas.0810860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti L., Griffini S., Lamorte G., Grovetti E., Uceda Renteria S.C., Malvestiti F., Scudeller L., Bandera A., Peyvandi F., Prati D., Meroni P., Cugno M. Chromosome 3 cluster rs11385942 variant links complement activation with severe COVID-19. J. Autoimmun. 2021;117 doi: 10.1016/j.jaut.2021.102595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and Antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieme R.J., Demeulenaere L. Genetically determined electrophoretic variant of the human complement component C′3. Nature. 1967;214:1042–1043. doi: 10.1038/2141042a0. [DOI] [PubMed] [Google Scholar]
- Wilk C.M. Coronaviruses hijack the complement system. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff T.M., Shukla A.K. The complement C5a-C5aR1 GPCR Axis in COVID-19 therapeutics. Trends Immunol. 2020 doi: 10.1016/j.it.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J.R.W., Sepp T., Matharu B.K., Khan J.C., Thurlby D.A., Shahid H., Clayton D.G., Hayward C., Morgan J., Wright A.F., Armbrecht A.M., Dhillon B., Deary I.J., Redmond E., Bird A.C., Moore A.T. Complement C3 variant and the risk of age-related macular degeneration. N. Engl. J. Med. 2007;357:553–561. doi: 10.1056/nejmoa072618. [DOI] [PubMed] [Google Scholar]
- Yu J., Yuan X., Chen H., Chaturvedi S., Braunstein E.M., Brodsky R.A. Direct activation of the alternative complement pathway by SARS-CoV-2 spike proteins is blocked by factor D Inhibition. Blood. 2020 doi: 10.1182/BLOOD.2020008248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020 doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.