Abstract
Several existing drugs are currently being tested worldwide to treat COVID-19 patients. Recent data indicate that SARS-CoV-2 is rapidly evolving into more transmissible variants. It is therefore highly possible that SARS-CoV-2 can accumulate adaptive mutations modulating drug susceptibility and hampering viral antigenicity. Thus, it is vital to predict potential non-synonymous mutation sites and predict the evolution of protein structural modifications leading to drug tolerance. As two FDA-approved anti-hepatitis C virus (HCV) drugs, boceprevir, and telaprevir, have been shown to effectively inhibit SARS-CoV-2 by targeting the main protease (Mpro), here we used a high-throughput interface-based protein design strategy to identify mutational hotspots and potential signatures of adaptation in these drug binding sites of Mpro. Several mutants exhibited reduced binding affinity to these drugs, out of which hotspot residues having a strong tendency to undergo positive selection were identified. The data further indicated that these anti-HCV drugs have larger footprints in the mutational landscape of Mpro and hence encompass the highest potential for positive selection and adaptation. These findings are crucial in understanding the potential structural modifications in the drug binding sites of Mpro and thus its signatures of adaptation. Furthermore, the data could provide systemic strategies for robust antiviral design and discovery against COVID-19 in the future.
Keywords: Drug tolerance, Fitness, Main protease, Protein design, Adaptable mutations, SARS-CoV-2, Signatures of adaptation, Drug resistance
1. Introduction
The global death toll from COVID-19 has crossed two million. Vaccines are being developed with record-breaking speed; however, the production and widespread distribution of COVID-9 vaccines will likely take 12–18 months, whereas de novo development of novel antivirals usually requires 10–17 years. Thus, the repurposing of known drugs could substantially accelerate the deployment of new therapies to manage COVID-19. The use of new therapeutic alternatives for existing approved drugs has allowed a faster and efficient response to tackle COVID-19 [1]. As of now, more than one dozen existing drugs have already been clinically tested for the treatment of COVID-19.
SARS-CoV-2 encodes a chymotrypsin-like cysteine protease, also called the main protease (Mpro) or 3CLpro. Mpro catalyzes the proteolytic cleavage of the viral polyproteins into nonstructural (nsp4-nsp16) proteins required for viral packaging, maturation, and replication. Therefore, the inhibition of Mpro prevents virus maturation and replication, and is thus central for virus survival. Mpro is a homodimeric protein with each subunit (306 residues) consisting of three domains (I–III). The active site is located within domains I and II that fold into a six-stranded β-barrel. Domain III is involved in the dimerization of Mpro and forms a cluster of five antiparallel α-helices. Domain II and domain III are connected by a flexible loop. The active site contains a Cys-His catalytic dyad (H41 and C145) and several binding pockets (denoted as P1, P1′, P2, P3, and P4) [2,3]. Among the key drugs currently used as Mpro inhibitors are the FDA-approved anti-hepatitis C virus (HCV) drugs, boceprevir and telaprevir [[4], [5], [6], [7], [8]]. Recent studies have shown that boceprevir and telaprevir effectively inhibit SARS-CoV-2 by targeting Mpro [5,6,9,10]. Several complex structures of Mpro with boceprevir and telaprevir are now available [6,8,11]. Boceprevir and telaprevir are β-ketoamide inhibitors that bind to the highly flexible active site of Mpro and change the geometry and conformation of the active site cavity.
With the progress of the pandemic, the SARS-CoV-2 is also continually mutating. SARS-CoV-2 has spread farther and faster than any other virus in the last century. This may also be attributed to the natural selection that has optimized the mutation rate of RNA viruses. In such scenarios, the virus may undergo positive selection and adapt the pressure of antivirals used as the first line of defense [12]. Recently, we identified key residues in RNA dependent RNA polymerase (RdRp) that could contribute to remdesivir and molnupiravir resistance in SARS-CoV-2 [13]. In the current study, we used multiparametric, high-throughput computational analyses to identify potential signatures of drug susceptibility in the boceprevir and telaprevir binding sites of SARS-CoV-2 Mpro. Prior knowledge of the mutation-prone hotspot residues will be of great importance to predict the potential structural modifications in the Mpro as well as the viral adaptation and fitness under drug pressure. Furthermore, the data could guide scientists to better modify the antivirals and develop newer effective therapeutics.
2. Materials and methods
2.1. Identification of interacting residues between Mpro and drugs boceprevir and telaprevir
The X-ray crystal structures of SARS-CoV-2 Mpro bound with boceprevir and telaprevir with PDB ID 7BRP and 7C7P, respectively, were used to obtain the interactions between Mpro and the respective drugs. The binding pocket residues from each complex structure were identified and subjected to further analysis. As a control, the Mpro structure in an apo form (PDB ID: 6M03) was used. The complex structures were prepared using the Molecular Operating Environment (MOE, version 2021.03).
2.2. High-throughput residue scan design methodology
To identify potential single point mutations of Mpro that exhibit the highest potential for mutation, positive selection, and adaptability against these drugs, MOE’s high-throughput resistance scan methodology was employed (MOE version 2021.03) [14]. The mutations were limited to single nucleotide polymorphisms (SNPs) of the wild-type sequence to emulate the mutations that are more likely to occur naturally during protein evolution. The prepared complex structures from the above step were used as input, and 19 Mpro residues that interacted with boceprevir and 20 Mpro residues that interacted with telaprevir were designed with naturally sampled SNPs. During each independent design experiment, the catalytic dyad residues H41 and C145 were not considered in the design, as they carry out the acylation-deacylation reaction and cleavage of the substrates between Q at the P1 position and G/A/S at the P1′ position [15,16]. The rotamer explorer option of the ensemble protein design protocol was utilized due to the flexibility of the designed site. The RMSD limit was set to 0.5 Å, and the “energy window,” “conformation limit,” and “fix residues farther than” were set to default values at 10 kcal/mol, 25 K, and 4.5 Å, respectively. A total of 302 designs from the two independent runs were generated, and various parameters such as affinity and stability between the designed Mpro-boceprevir and Mpro-telaprevir complexes were analyzed. The relative binding affinity of the mutation to the wild-type protein (dAffinity) was computed, where dAffinity is equal to the Boltzmann average of the relative affinities of the ensemble. This methodology indirectly served as a computational fitness test model to sample only those mutations that are not deleterious to the virus and are more likely to evolve naturally.
2.3. Validation of the design methodology
We validated the design methodology using two computational control experiments. First, the accuracy and predictive ability of our interface design protocol were validated on a well-known remdesivir-resistant V557L mutant of nsp12 of SARS-CoV [17]. The MOE resistance scan methodology was used, where V557 was designed with naturally sampled SNP residues (V557 [A, D, E, G, I, L, M, F]), and then the affinities between remdesivir and designed nsp12 were computed. Second, the adaptive mutations computed from our design calculations were compared with the known sequences of SARS-CoV-2 deposited in the GISAID and CoV-GLUE databases [18,19]. The frequency of the mutations obtained from these databases were compared with the boceprevir and telaprevir bound Mpro designs to demonstrate the accuracy of our design methodology.
2.4. Calculation of intermolecular interactions between Mpro-boceprevir and Mpro-telaprevir designs
The intermolecular interactions between top-ranked affinity-attenuating and affinity-enhancing Mpro-boceprevir and Mpro-telaprevir designs were obtained using the Arpeggio web-server [20]. The total number of interactions represented the sum of the number of polar contacts, hydrogen bonds, proximal contacts, van der Waals interactions, hydrophobic contacts, aromatic contacts, and carbonyl interactions. The interaction diagrams between Mpro-boceprevir and Mpro-telaprevir were created using MOE (version 2021.03).
2.5. Analyses of the normal mode and other energetics of the SARS-CoV-2 Mpro
The normal modes, eigenvalues of the 50 lowest-frequency non-trivial modes, average deformation energies, normalized fluctuations for the modes, the correlation matrix of Mpro, and squared atomic displacements were calculated using the WEBnm@ server [21,22]. Profile alignment- and covariance similarity-based comparative analyses were carried out using Comparative NMA utility.
3. Results
3.1. Analysis of interacting residues between Mpro-boceprevir, Mpro-telaprevir, and their selection for design experiments
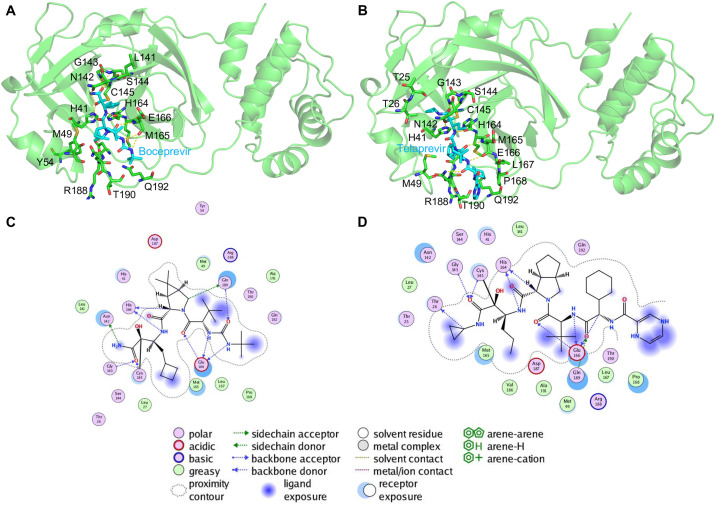
The interacting residues between Mpro-boceprevir and Mpro-telaprevir were identified from their respective crystal structures (Fig. 1 A and B). A total of 39 residues interacting with the two drugs (19 residues of Mpro-boceprevir and 20 residues of Mpro-telaprevir) were identified and subjected to designing. Most of the residues were located within 4 Å distance of the bound drugs. In the boceprevir bound pocket of Mpro, certain critical residues, such as N142, G143, C145, H164, E166, and Q189, formed hydrogen bonds with boceprevir (Fig. 1C). However, in the Mpro-telaprevir complex, an additional hydrogen bond interaction between Thr26 and telaprevir was present (Fig. 1D).
Fig. 1.
Structure of the SARS-CoV-2 Mpro-boceprevir and Mpro-telaprevir bound complexes. The crystal structures of SARS-CoV-2 Mpro in complex with (A) boceprevir and (B) telaprevir are shown. The Mpro is shown in a green cartoon, and the residues within 4 Å of boceprevir (cyan stick) and telaprevir (cyan stick) are shown as stick models and labeled. Interacting hydrogen bonds are shown as yellow dashes. In (C) and (D), various types of intermolecular interactions between boceprevir and telaprevir with Mpro are shown and specified in the legend, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Hotspot residues, adaptive mutations, and associated features from ligand-based interface design
We designed a total of 39 shortlisted residues of Mpro using resistance mutation scan methodology, where each residue was mutated with only an SNP of the wild-type sequence. Therefore, the mutations were limited to SNPs to mimic the variations that were more likely to occur naturally during protein evolution. A list of SNPs sampled and designed for 39 drug-interacting residues of Mpro is presented in Table 1 . A total of 302 single point mutants were generated and the affinities between the designed proteins with respective drugs were computed. For clarity and ease of analysis, the relative binding affinity of the mutation to the wild-type protein (dAffinity) was computed, where a more positive value signified that the mutation had a lower affinity with the bound drugs, thus indicating the mutant could become easily tolerated to the ligand.
Table 1.
Mpro-boceprevir and Mpro-telaprevir interacting residues that are designed with corresponding SNPs of the wild-type sequence to emulate the mutations that are more likely to happen naturally over the evolution of the protein. Mutations reported in GISAID hCoV-19 with number of sequences from the pandemic are presented in brackets.
| S. No | Mpro-boceprevir interacting and designed wild-type residuesa | Sampled SNPs in designs | Mutations reported in GISAID hCoV-19 with number of sequences from the pandemic | S. No | Mpro-telaprevir interacting and designed wild-type residuesa | Sampled SNPs in designs | Mutations reported in GISAID hCoV-19 with number of sequences from the pandemic |
|---|---|---|---|---|---|---|---|
| 1 | T26 | ARNIKMPS | T26I (7), T26A (4) | 1 | T25 | ARNIKMPS | T25I (11), T25A (4) |
| 2 | L27 | RQHIMFPSWV | L27F (4), L27H (1) | 2 | T26 | ARNIKMPS | T26I (7), T26A (4) |
| 3 | M49 | RILKTV | M49I (68), M49T (12), M49V (4), M49R (1) | 3 | L27 | RQHIMFPSWV | L27F (4), L27H (1) |
| 4 | Y54 | NDCHFS | Y54F (1), Y54 N (1) | 4 | M49 | RILKTV | M49I (68), M49T (12), M49V (4), M49R (1) |
| 5 | L141 | RQHIMFPSWV | L141I (3), L141F (2), L141P (1) | 5 | L141 | RQHIMFPSWV | L141I (3), L141F (2), L141P (1) |
| 6 | N142 | DHIKSTY | N142S (18), N142I (8), N142L (5), N142D (4), N142K (1) | 6 | N142 | DHIKSTY | N142S (18), N142I (8), N142L (5), N142D (4), N142K (1) |
| 7 | G143 | ARDCESWV | G143S (11), G143V (5), G143C (4), G143D (1) | 7 | G143 | ARDCESWV | G143S (11), G143V (5), G143C (4), G143D (1) |
| 8 | S144 | ACLFPTWY | S144L (5), S144E (3), S144K (1), S144P (1) | 8 | S144 | ACLFPTWY | S144L (5), S144E (3), S144K (1), S144P (1) |
| 9 | H164 | RNDQLPY | H164 N (3), H164Y (2), H164P (1) | 9 | H164 | RNDQLPY | H164 N (3), H164Y (2), H164P (1) |
| 10 | M165 | RILKTV | M165I (10), M165K (4) | 10 | M165 | RILKTV | M165I (10), M165K (4) |
| 11 | E166 | ADQGKV | E166D (3), E166K (3), E166G (1) | 11 | E166 | ADQGKV | E166D (3), E166K (3), E166G (1) |
| 12 | K167 | RNQEIMT | L167S (2), L167F (1), L167I (1) | 12 | K167 | RNQEIMT | L167S (2), L167F (1), L167I (1) |
| 13 | P168 | ARQHLST | P168S (49), P168L (3), P168R (1) | 13 | P168 | ARQHLST | P168S (49), P168L (3), P168R (1) |
| 14 | D187 | ANEGHYV | D187Y (6), D187G (2), D187T (1) | 14 | V186 | ADEGILMF | V186F (55), V186I (5), V186A (3), V186L (2), V186D (1) |
| 15 | R188 | NCQGHILKMPSTW | R188K (22), R188S (15), R188 M (7), R188A (1), R188G (1) |
15 | D187 | ANEGHYV | D187Y (6), D187G (2), D187T (1) |
| 16 | Q189 | REHLKP | Q189K (43) | 16 | R188 | NCQGHILKMPSTW | R188K (22), R188S (15), R188 M (7), R188A (1), R188G (1) |
| 17 | T190 | ARNIKMPS | T190I (134), T190A (2), T190S (1) | 17 | Q189 | REHLKP | Q189K (43) |
| 18 | A191 | DEGPSTV | A191V (1240), A191T (9), A191S (7), A191E (1) | 18 | T190 | ARNIKMPS | T190I (134), T190A (2), T190S (1) |
| 19 | Q192 | REHLKP | Q192L (1) | 19 | A191 | DEGPSTV | A191V (1240), A191T (9), A191S (7), A191E (1) |
| 20 | Q192 | REHLKP | Q192L (1) |
The catalytic dyad residues H41 and C145 of Mpro were not considered in the design, as they carry out the acylation-deacylation reaction and cleavage of the substrates.
3.2.1. Design of boceprevir binding pocket in Mpro
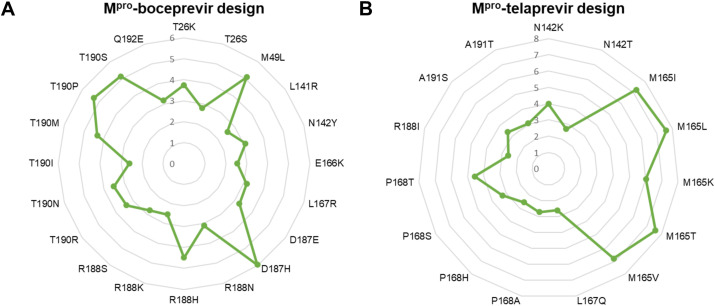
First, designs of 19 boceprevir binding pocket in Mpro comprising 146 designs revealed that the dAffinity varied from −7.97 to 5.96 kcal/mol (Supplementary Figure 1A). Among the 146 designs, 60 Mpro designs retained positive dAffinity values, suggesting they could compromise their affinity toward boceprevir (Supplementary Figure 2A). The dAffinity values of each of the 19 boceprevir-interacting residues with their corresponding single point resistant mutations were computed and shown in Supplementary Figure 1A. Further, a stringent dAffinity cut-off showed that 20 mutants exhibited dAffinities higher than 2.5 kcal/mol (Fig. 2 A). These mutants exhibited the highest dAffinity values, suggesting their significance in developing tolerance against boceprevir. Interestingly, these 20 mutants originated predominantly from ten Mpro residues, namely T26, M49, L141, N142, E166, L167, D187, R188, T190, and Q192 (Fig. 2A). Moreover, certain residues such as M49, D187, R188, T190, and their sampled mutations were more susceptible to positive selection and developing adaptation during evolution, as revealed from their affinity profiles (Fig. 2A and Supplementary Figure 2A).
Fig. 2.
Relative binding affinities of the most plausible positive selection mutant designs from the Mpro-boceprevir and Mpro-telaprevir complexes. Radar plots showing the computed relative binding affinities (dAffinities) for the most plausible positive selection (A) Mpro-boceprevir and (B) Mpro-telaprevir binding pocket residues and their single point mutant designs, respectively. The radar plots for interacting residues and their corresponding mutated designs are labeled, and dAffinity values are highlighted.
3.2.2. Design of telaprevir binding pocket in Mpro
Second, designs of 20 telaprevir binding pocket in Mpro comprising 156 designs revealed that the dAffinity varied from −9.27 to 7.60 kcal/mol (Supplementary Figure 1B). Among the designs, 80 Mpro designs retained positive dAffinity values, suggesting they could compromise their affinity toward telaprevir (Supplementary Figure 2B). The dAffinity values of each of the 20 telaprevir-interacting residues with their corresponding single point resistant mutations were computed and are shown in Supplementary Figure 1B. Further, a stringent dAffinity cut-off showed that 15 mutants exhibited dAffinities higher than 2.5 kcal/mol (Fig. 2B). These mutants exhibited the highest dAffinity values, suggesting their significance in developing tolerance against telaprevir. Interestingly, these 15 mutants originated from mutations predominantly in six Mpro residues, namely N142, M165, L167, P168, R188, and A191 (Fig. 2B and Supplementary Figure 2B). A comparison between Mpro-boceprevir and Mpro-telaprevir designs and their mutational landscapes revealed that certain conserved residues, including N142, L167, and R188, are commonly occurring and are more prone to mutations and positive selection, as revealed from their affinity profiles (Fig. 2A and B).
Using this high-throughput targeted approach, we performed a computational fitness test to sample only those mutations that are not lethal to the virus and more likely to evolve naturally over time to preserve the structural and functional integrity of Mpro. This sequence-specific conservation and diversity of the crucial drug-bound Mpro designs suggested that these hotspot residues will have the highest tendency to undergo selective mutations and adaptation in the future to render tolerance and make these key drugs ineffective, facilitating the spread and survival of SARS-CoV-2 or a newly evolving-related virus. Interestingly, several mutations are already reported in the GISAID and CoV-GLUE databases at the Mpro active sites, drug binding sites, and other functionally important regions. This suggests that SARS-CoV-2 is already actively mutating (>1000 unique mutations at the Mpro), however, it is probably maintaining its viral fitness and catalytic functions by acquiring compensatory mutations.
3.3. Validation of the interface design methodology
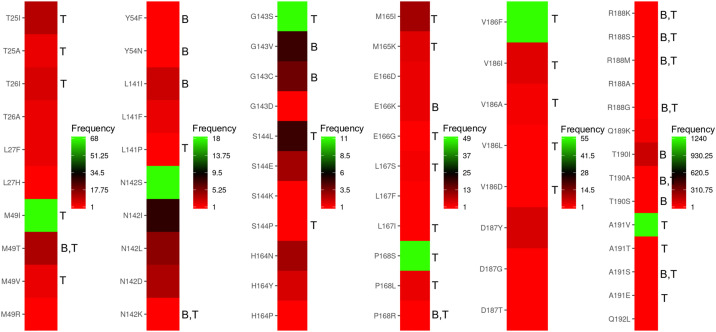
First, to validate the predictive ability and accuracy of our design methodology, a known remdesivir-resistant V557L mutant of SARS-CoV was evaluated. The HCV has been shown to develop resistance against boceprevir. However, because of the unavailability of their crystal structures, we validated our findings with the SARS-CoV V557L mutant, as V557 is conserved in the nsp12 of SARS-CoV-2. To examine whether the design methodology could rank L557 among the low-affinity designs, we scanned residues in the 557th position with (A, D, E, G, I, L, M, F) residues and found that V557L was ranked as a low-affinity mutant in the design computations (Supplementary Figure 3). Second, mutations at the boceprevir and telaprevir binding sites of Mpro were retrieved from GISAID and CoV-GLUE databases, and their frequency of occurrence was obtained. Mutations reported in GISAID hCoV-19 with their corresponding number of sequences from the pandemic are presented in Table 1. It was found that out of 64 mutations, 42 mutations were predicted as tolerant and positively selected based on our design calculations, thus achieving ∼65% correlation and match with the sequencing data (Fig. 3 ). Interestingly, several high-frequency mutations such as M49I, G143S, P168S, V186F, and A191V were already found to be tolerant against telaprevir in our design calculations (Fig. 3). However, it is important to note here that the frequencies of the mutations reported in GISAID and CoV-GLUE databases are not directly correlated with the computed dAffinities. As the pandemic progresses, drug pressure mounts and more sequencing data is available, it is possible that the other sampled mutations from our designs may appear and correlate with the sequencing data. Overall, these control experiments validated our design methodology in scoring and rank-ordering the affinity-attenuating designs, leading to the emergence of susceptible mutations for possible drug tolerance in SARS-CoV-2.
Fig. 3.
Heatmaps showing the mutations and their frequencies at the boceprevir and telaprevir binding sites of Mproretrieved from GISAID and CoV-GLUE databases. Mutations at the boceprevir and telaprevir binding site of SARS-CoV-2 Mpro obtained from GISAID and CoV-GLUE databases are shown, where the frequencies of the mutations among the COVID-19 infected cases ranged from lower to higher numbers are denoted from red to green colors respectively. The computed mutants that developed tolerance and adaptation towards boceprevir and telaprevir are denoted as ’B’ and ‘T’ respectively adjacent to the mutants. It was found that out of 64 mutations, 42 mutations were predicted as tolerant and positively selected from our design calculations, thus achieving ∼65% correlation and match with the sequencing data. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Intermolecular interactions between Mpro-boceprevir and Mpro-telaprevir in affinity-attenuating designs
We visualized several interactions and computed the intermolecular interactions between Mpro-boceprevir and Mpro-telaprevir, which exhibited reduced binding in the affinity-attenuating designs. In the Mpro-boceprevir design, although the top-scored affinity-enhancing design formed 376 interactions, the affinity-attenuating design formed only 318 interactions (Supplementary Table 1). Similarly, in the Mpro-telaprevir design, while the affinity-enhancing design formed 449 interactions, the affinity-attenuating design formed only 416 interactions (Supplementary Table 1). In both these cases, the van der Waals, proximal, and hydrophobic interactions played a major role in governing the reduced affinity in the affinity-attenuating designs between Mpro and the drugs. While in the Mpro-boceprevir, loss of arene-H interaction with His41 was found to play a major role in reduced affinity in the affinity-attenuating design, in the Mpro-telaprevir design, the absence of hydrogen bond with His164 appeared to play a significant role in reduced affinity in the affinity-attenuating design (Supplementary Figure 4).
3.5. Analysis of the local flexibility of Mpro using normal mode
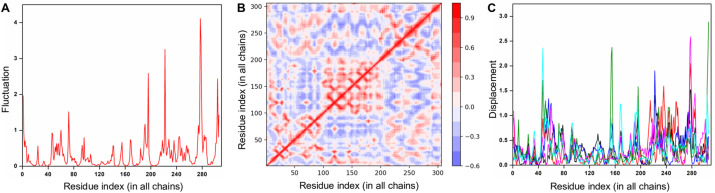
To understand the structural dynamics of SARS-CoV-2 Mpro contributed by its regions/sites, a normal mode analysis was performed. First, the eigenvalues of the 50 lowest-frequency non-trivial modes were determined and the normalized fluctuations for the modes with respect to the residues were computed (Fig. 4 A). Despite Mpro exhibiting interdomain motions, residues 1 to 200 (comprising the drug-binding pockets) were found to be largely stable (Fig. 4A). Second, the residue correlation matrix depicting the correlated movement of C⍺ atoms in Mpro emphasized that residues 100 to 200 experienced a highly correlated coupled motion (Fig. 4B). Here, both axes denote the C⍺ atoms of Mpro, with each cell in the plot showing the coupling of two residues ranging from -1 (anti-correlated, blue) to 0 (uncorrelated) to 1 (correlated, red), thereby representing correlated motions. Finally, the atomic displacement results were computed, which referred to the normalized square of displacement of each C⍺ atom (for modes 1 to 6) so that the sum of all residues was 100 (Fig. 4C). As the highest values corresponded to the highly displaced regions, clusters of peaks on the plots identified significantly displaced regions, confirming that residues 60 to 140 and 160 to 240 were less displaced and underwent local flexibility in Mpro (Fig. 4C).
Fig. 4.
Normal mode and energetic analysis of SARS-CoV-2 Mpro. (A) The normalized fluctuations for the lowest frequency non-trivial modes with respect to Mpro residues are shown. (B) The residue correlation matrix highlighting the correlated movement of the C⍺ atom in Mpro is shown, where each cell in the plot shows the coupling of two residues ranging from -1 (anti-correlated, blue) to 0 (uncorrelated) to 1 (correlated, red), thereby representing correlated motions. (C) The atomic displacement, i.e., the square of displacement of each C⍺ atom for modes 1 to 6 of Mpro are shown in black, red, blue, magenta, green and cyan colors respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
COVID-19 has triggered the biggest scientific adventure in the history of modern science. With remarkable dedication, scientists have produced a string of lifesaving breakthroughs at an unprecedented pace. However, the job is not finished yet. With the origin and spread of new SARS-CoV-2 variants, a dangerous new phase of the COVID-19 pandemic is unfolding. The emergence of SARS-CoV-2 strains in the UK, Ireland, South Africa, Brazil, Hong Kong, France, and several other countries suggests a higher risk of morbidity and mortality. For successful management of COVID-19, strengthening the existing control measures and the use of repurposed drugs accompanying the vaccines are essential. Curtailing infections will sharply reduce the chances for the virus to mutate further. However, continuous and long-term usage of these drugs may also lead to viral adaptation and the development of more infective and pathogenic variants [12,13]. Further, in the long term, certain mutations could impend the efficacy of vaccines. Perhaps the only positive part is that most of the mutations currently reported are synonymous mutations and rare non-synonymous mutations. However, the non-synonymous mutations can be positively selected and will increase in frequency if they confer an intrinsic fitness advantage concerning viral replication, transmission, or immune escape [23,24]. The SARS- and MERS-CoVs have evolved in humans for several years, and they have ample opportunity to explore the sequence space in the future. Additionally, since SARS-CoV-2 entered the human population recently, it may still be adapting to its human host. Recent evidence has demonstrated a parallel, convergent pattern of SARS-CoV-2 antigenic evolution that has induced resistance against neutralizing antibodies [[25], [26], [27], [28]].
The development of drug tolerance in SARS-CoV-2 viruses is expected. As the use of antiviral drugs continues, reports of drug tolerance and adaptable viruses are sure to emerge. Typically, such strains spread rapidly in the community and are a matter of great concern and importance to public health. Besides, given that several repurposed drugs are being used for the treatment of COVID-19, sequential development of multiple resistant strains is also possible. Reports of boceprevir and telaprevir resistant viruses are sporadic, with yet to be confirmed circulation of boceprevir and telaprevir tolerance SARS-CoV-2 strains within communities or worldwide. However, with the threat of new SARS-CoV-2 mutations now clearly identified, we have to be equipped to respond and to anticipate the challenges that may develop in the future. Our present data is significant with information on the potential structural and residue-specific sites in the Mpro that are susceptible to mutation under drug pressure and lead to viral adaptation with fitness advantages.
Nonetheless, we acknowledge certain limitations of our study. The computationally predicted positively selected and plausible tolerance mutations of Mpro against boceprevir and telaprevir need to be validated using functional assays. A focused biochemical experiment could be used to quantify the gain/loss and strength of interactions between Mpro designs and key drugs to obtain a comprehensive picture of the emergence of adaptable and positively selected residues. However, we performed appropriate control experiments to ensure that our design methodology correctly predicts the affinity-attenuating designs representing the tolerance mutations and positively selected residues of Mpro. Our high-throughput design methodology was used to ensure that the most plausible single point mutations representing the tolerance mutations and positively adapted Mpro residues are identified and predicted quantitatively. Finally, lessons learned from HIV-1 protease demonstrated that mutants distal from the active site could also cause drug resistance. Accordingly, such an event may also occur in the case of SARS-CoV-2, and hence a systematic design methodology should be developed to address this scenario.
Author Contributions
AKP carried out all the design experiments, data generation, and analysis. AKP and TT conceived the study, participated in its design and coordination, and drafted the manuscript. Both authors read and approved the final manuscript.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
The authors are grateful to Dr. Kam Y.J. Zhang (Laboratory for Structural Bioinformatics, RIKEN, Yokohama) for his continuous support and valuable suggestions for improving the manuscript. The authors acknowledge RIKEN ACCC for the Hokusai supercomputing resources.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2021.03.118.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Mishra S.K., Tripathi T. One year update on the COVID-19 pandemic: where are we now? Acta Trop. 2021;214:105778. doi: 10.1016/j.actatropica.2020.105778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L., Lin D., Sun X., Curth U., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Z., Du X., Xu Y., Deng Y., et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 4.Kumar N., Sharma S., Kumar R., Tripathi B.N., et al. Host-directed antiviral therapy. Clin. Microbiol. Rev. 2020;33 doi: 10.1128/cmr.00168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oerlemans R., Ruiz-Moreno A.J., Cong Y., Dinesh Kumar N., et al. Repurposing the HCV NS3–4A protease drug boceprevir as COVID-19 therapeutics. RSC Med. Chem. 2021 doi: 10.1039/D0MD00367K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu L., Ye F., Feng Y., Yu F., et al. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker J.D., Uhrich R.L., Kraemer G.C., Love J.E., et al. A drug repurposing screen identifies hepatitis C antivirals as inhibitors of the SARS-CoV2 main protease. PloS One. 2021;16 doi: 10.1371/journal.pone.0245962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kneller D.W., Galanie S., Phillips G., O’Neill H.M., et al. Malleability of the SARS-CoV-2 3CL Mpro active-site cavity facilitates binding of clinical antivirals. Structure. 2020;28:1313–1320. doi: 10.1016/j.str.2020.10.007. e1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma C., Sacco M.D., Hurst B., Townsend J.A., et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y., Ma C., Szeto T., Hurst B., et al. Boceprevir, calpain inhibitors II and XII, and GC-376 have broad-spectrum antiviral activity against coronaviruses. ACS Infect. Dis. 2021 doi: 10.1021/acsinfecdis.0c00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Günther S., Reinke P.Y.A., Fernández-García Y., Lieske J., et al. bioRxiv; 2020. Inhibition of SARS-CoV-2 Main Protease by Allosteric Drug-Binding. 2020.2011.2012.378422. [DOI] [Google Scholar]
- 12.Salpini R., Alkhatib M., Costa G., Piermatteo L., et al. Key genetic elements, single and in clusters, underlying geographically dependent SARS-CoV-2 genetic adaptation and their impact on binding affinity for drugs and immune control. J. Antimicrob. Chemother. 2021;76:396–412. doi: 10.1093/jac/dkaa444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Padhi A.K., Shukla R., Saudagar P., Tripathi T. High-throughput rational design of the remdesivir binding site in the RdRp of SARS-CoV-2: implications for potential resistance. iScience. 2021;24:101992. doi: 10.1016/j.isci.2020.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vilar S., Cozza G., Moro S. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008;8:1555–1572. doi: 10.2174/156802608786786624. [DOI] [PubMed] [Google Scholar]
- 15.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramos-Guzmán C.A., Ruiz-Pernía J.J., Tuñón I. Unraveling the SARS-CoV-2 main protease mechanism using multiscale methods. ACS Catal. 2020;10:12544–12554. doi: 10.1021/acscatal.0c03420. [DOI] [PubMed] [Google Scholar]
- 17.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. eabb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to msc3 health. Glob Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singer J., Gifford R., Cotten M., Robertson D. Preprints; 2020. CoV-GLUE: A Web Application for Tracking SARS-CoV-2 Genomic Variation. [DOI] [Google Scholar]
- 20.Jubb H.C., Higueruelo A.P., Ochoa-Montaño B., Pitt W.R., et al. Arpeggio: a web server for calculating and visualising interatomic interactions in protein structures. J. Mol. Biol. 2017;429:365–371. doi: 10.1016/j.jmb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollup S.M., Salensminde G., Reuter N. WEBnm@: a web application for normal mode analyses of proteins. BMC Bioinf. 2005;6:52. doi: 10.1186/1471-2105-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiwari S.P., Fuglebakk E., Hollup S.M., Skjærven L., et al. WEBnm@ v2.0: web server and services for comparing protein flexibility. BMC Bioinf. 2014;15:427. doi: 10.1186/s12859-014-0427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Callaway E. Making sense of coronavirus mutations. Nature. 2020;585:174–177. doi: 10.1038/d41586-020-02544-6. [DOI] [PubMed] [Google Scholar]
- 24.Padhi A.K., Tripathi T. Can SARS-CoV-2 accumulate mutations in the S-protein to increase pathogenicity? ACS Pharmacol. Transl. Sci. 2020;3:1023–1026. doi: 10.1021/acsptsci.0c00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weisblum Y., Schmidt F., Zhang F., DaSilva J., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q., Wu J., Nie J., Zhang L., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. e1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57. doi: 10.1016/j.chom.2020.11.007. e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.