SUMMARY
The intestinal epithelium senses nutritional and microbial stimuli using epithelial sensory enteroendocrine cells (EECs). EECs communicate nutritional information to the nervous system, but whether they also relay signals from intestinal microbes remains unknown. Using in vivo real-time measurements of EEC and nervous system activity in zebrafish, we discovered that the bacteria Edwardsiella tarda activate EECs through the receptor transient receptor potential ankyrin A1 (Trpa1) and increase intestinal motility. Microbial, pharmacological, or optogenetic activation of Trpa1+EECs directly stimulates vagal sensory ganglia and activates cholinergic enteric neurons by secreting the neurotransmitter 5-hydroxytryptamine (5-HT). A subset of indole derivatives of tryptophan catabolism produced by E. tarda and other gut microbes activates zebrafish EEC Trpa1 signaling. These catabolites also directly stimulate human and mouse Trpa1 and intestinal 5-HT secretion. These results establish a molecular pathway by which EECs regulate enteric and vagal neuronal pathways in response to microbial signals.
Graphical Abstract
In Brief
The gut communicates nutritional information to the nervous system through epithelial sensory enteroendocrine cells (EECs). Ye et al. reveal that EECs also relay signals from gut microbes. They find that gut bacteria produce tryptophan catabolites that activate Trpa1 channels on EECs causing rapid activation of enteric and vagal neurons.
INTRODUCTION
The intestine harbors complex microbial communities that shape intestinal physiology, modulate systemic metabolism, and regulate brain function. These effects on host biology are often evoked by distinct microbial stimuli including microbe-associated molecular patterns (MAMPs) and microbial metabolites derived from digested carbohydrates, proteins, lipids, and bile acid (Brown and Hazen, 2015, Liu et al., 2020, Coleman and Haller, 2017). The intestinal epithelium is the primary interface that mediates this host-microbe communication (Kaiko and Stappenbeck, 2014). The mechanisms by which the intestinal epithelium senses distinct microbial stimuli and transmits that information to the rest of the body remains incompletely understood.
The intestinal epithelium has evolved specialized enteroendocrine cells (EECs) that exhibit conserved sensory functions in insects, fishes, and mammals (Guo et al., 2019, Ye et al., 2019, Furness et al., 2013). Distributed along the entire digestive tract, EECs are activated by diverse luminal stimuli to secrete hormones or neuronal transmitters in a calcium dependent manner (Furness et al., 2013). Recent studies have revealed that EECs form synaptic connections with sensory neurons (Kaelberer et al., 2018, Bellono et al., 2017, Bohorquez et al., 2015). The connection between EECs and neurons forms a direct route for the intestinal epithelium to transmit nutrient sensory information to the brain (Kaelberer et al., 2018). EECs are classically known for their ability to sense nutrients (Symonds et al., 2015) but whether they can be directly stimulated by microbes or microbially derived products is less clear. Limited examples include the observation that short chain fatty acids and branched chain fatty acids from microbial carbohydrate and amino acid catabolism activate EECs via G-protein coupled receptors (Bellono et al., 2017, Lu et al., 2018). Indole, a microbial catabolite of the amino acid tryptophan, has also been reported to activate EECs, but the EEC receptor that mediates the effect remains unidentified (Chimerel et al., 2014). With the growing understanding of gut microbiota and their metabolites, identifying the EEC receptors that recognize distinct microbial stimuli as well as the downstream pathways by which EECs transmit microbial stimuli to regulate local and systemic host physiology, has emerged as an important goal.
The vertebrate intestine is innervated by the intrinsic enteric nervous system (ENS) and extrinsic neurons from autonomic nerves, including sensory nerve fibers from the nodose vagal ganglia and dorsal root ganglia in the spinal cord (Furness et al., 1999). Both vagal and spinal sensory nerve fibers transmit visceral stimuli to the central nervous system and modulate a broad spectrum of brain functions (Brookes et al., 2013). A previous study demonstrated that stimulating EECs with the microbial metabolite isovalerate activates spinal sensory nerves through 5-hydroxytryptamine (5-HT) secretion (Bellono et al., 2017). Whether and how gut microbial stimuli modulate ENS or vagal sensory activity through EECs is still unknown.
RESULTS
Edwardsiella tarda activates EECs through Trpa1
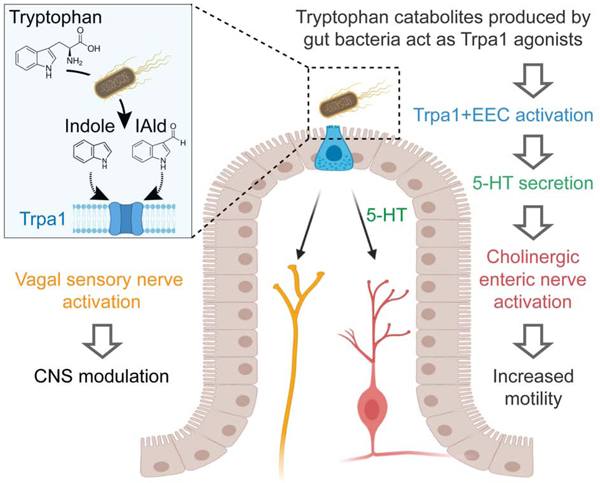
To identify stimuli that activate EECs in live animals, we developed a transgenic zebrafish line that permits recording of EEC activity by expressing the calcium modulated photoactivatable ratiometric integrator (CaMPARI) protein in EECs under control of the neurod1 promoter (Fig. 1A, Fig. S1A–F; see Methods for details). When exposed to 405nm light, CaMPARI protein irreversibly photoconverts from a configuration that emits green light to one that emits red in a manner positively correlated with intracellular calcium levels [Ca2+]i. A high red:green CaMPARI ratio thus reports high intracellular calcium (Fosque et al., 2015). This EEC-CaMPARI system therefore enables imaging of the calcium activity history of intestinal EECs in the intact physiologic context of live free-swimming animals (Fig. 1A–B, Fig. S1G–J). To test the validity of this EEC-CaMPARI system, we first stimulated larvae with different nutrients known to activate zebrafish EECs (Ye et al., 2019). Exposure to only water as a vehicle control revealed an expected low basal red:green CaMPARI ratio (Fig. 1C, E–F). Following long-chain fatty acid stimulation with linoleate, a subpopulation of EECs displayed high red:green CaMPARI ratio (Fig. 1D, E–F). EECs with a high red:green CaMPARI ratio were classified as “activated EECs”. The percentage of activated EECs significantly increased in response to chemical stimuli known to activate EECs, including linoleate, oleate, laurate, and glucose (Fig. 1F), but not in response to the short chain fatty acid butyrate, consistent with our previous finding (Fig. 1F) (Ye et al., 2019).
Figure 1. E. tarda activates zebrafish EECs in vivo.
(A) Experimental approach for measuring EEC activity in free-swimming zebrafish. (B) Method for recording EEC responses to chemical and microbial stimulants in the EEC-CaMPARI model. (C-D) Confocal projection of mid-intestinal EECs upon water (C, negative control) or linoleate (D) stimulation in Tg(neurod1:CaMPARI) following UV-photoconversion. (E) Frequency distribution of EECs’ red:green CaMPARI fluorescence intensity ratio in water or linoleate-stimulated zebrafish. n=177 for water group and n=213 for linoleate group. (F) Percent EEC response in Tg(neurod1:CaMPARI) zebrafish. (G-H) Confocal projection of mid-intestinal EECs upon Aeromonas sp. (G) or E. tarda (H) stimulation in Tg(neurod1:CaMPARI) following UV-photoconversion. (I) Frequency distribution of EECs’ red:green CaMPARI fluorescence intensity ratio in zebrafish treated with water or E. tarda. n=117 for water group and n=156 for E. tarda group. (J) Representative heatmap image showing Aeromonas sp., B. subtilis and E. tarda stimulated EEC red:green CaMPARI fluorescence ratio. (K) EEC activation in Tg(neurod1:CaMPARI) zebrafish stimulated with different bacterial strains. (L) Representative Tg(neurod1:Gcamp6f) zebrafish intestine stimulated with E. tarda. One-way ANOVA with Tukey’s post-test was used in F and K. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
We next applied the EEC-CaMPARI system to ask whether EECs acutely respond to live bacterial stimulation in vivo. We exposed Tg(neurod1:CaMPARI) zebrafish to individual bacterial strains for 20 mins in zebrafish housing water (GZM), followed by photoconversion and imaging of CaMPARI fluorescence. For these experiments, we selected a panel of 11 bacterial strains including 3 model species (P. aeruginosa, E. coli, B. subtilis), 7 commensal strains isolated from the zebrafish intestine (Rawls et al., 2006, Stephens et al., 2016), and the pathogen E. tarda FL6–60 (also called E. piscicida (Abayneh et al., 2013, Bujan et al., 2018); Fig. 1K and Key Resources Table). Within this panel, the only strain that induced a high red:green EEC-CaMPARI signal was E. tarda (Fig. 1G–K). We further confirmed that E. tarda directly activates EECs using an alternative reporter of EEC activity based on the [Ca2+]i-sensitive fluorescent protein Gcamp6f (neurod1:Gcamp6f) (Fig. 1L, Fig. S1K–P) (Ye et al., 2019). Although E. tarda has been reported to infect zebrafish (Abayneh et al., 2013, Flores et al., 2020), we observed no overt pathogenesis in these acute exposure experiments.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti-GFP Polyclonal antibody | Aves Lab | Cat# GFP-1010, RRID:AB_2307313 |
| Living Colors® anti DsRed Polyclonal Antibody | TAKARA | Cat# 632496, RRID:AB_10013483 |
| Mouse anti-zebrafish gut secretory cell epitopes Monoclonal antibody [FIS 2F11/2] | Abcam | ab71286 |
| Mouse anti-zebrafish Zn-12 Monoclonal antibody | ZIRC | AB_10013761 |
| Rabbit anti-serotonin whole Polyclonal antibody | Sigma | Cat# 5545 |
| Mouse anti-p44/42 MAPK (Erk1/2) (L34F12) Monoclonal antibody | Cell Signaling | Cat# 4696 |
| Rabbit anti-Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) Monoclonal antibody | Cell Signaling | Cat# 4370T |
| Rabbit anti-chicken desmin Polyclonal Antibody | Sigma | Cat# D8281 |
| Rabbit anti-mouse PYY antibody | PMID: 28614796 | Custom antibody generated in Liddle Laboratory, aa4–21 (mouse) |
| Bacterial and Virus Strains | ||
| Edwardsiella tarda FL6–60 | PMID: 22003892 | N/A |
| Edwardsiella tarda LSE40; pmkb:mCherry | Gift by Mark Cronan | N/A |
| Edwardsiella tarda 23685 | ATCC | ATCC23685 |
| Edwardsiella tarda 15974 | ATCC | ATCC15974 |
| Acinetobacter sp. ZOR0008 | PMID: 26339860 | N/A |
| Aeromonas veronii ZOR0002 | PMID: 26339861 | N/A |
| Shewanella sp. ZOR0012 | PMID: 26339862 | N/A |
| Enterobacter sp. ZOR0014 | PMID: 26339863 | N/A |
| Vibrio sp. ZWU0020 | PMID: 26339864 | N/A |
| Chryseobacterium sp. ZOR0023 | PMID: 26339865 | N/A |
| Exiguobacterium acetylicum ZWU0009 | PMID: 26339866 | N/A |
| Bacillus subtilis 168 | PMID: 18723616 | N/A |
| Pseudomonas aeruginosa PAK | PMID: 30524971 | N/A |
| Plesiomonas sp. ZOR0011 | PMID: 26339861 | N/A |
| Escherichia coli MG1655 | PMID: 26339862 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Optovin | Tocris Biotech | 4901 |
| HC030031 | Sigma | 377430–5g |
| Allyl isothiocyanate | Sigma | H4415–10MG |
| Atropine | Sigma | A0132 |
| 4-DAMP | Sigma | SML0255 |
| Clozapine | Sigma | C6305 |
| α-Bungarotoxin | Sigma | 203980 |
| Indole-3-carboxaldehyde | Sigma | 129445 |
| Indole-acetaldehyde | Ambeed | A626636 |
| Indole | Sigma | I3408–25G |
| L-Tryptophan | Sigma | T8941–10mg |
| Indole-3-acetic acid sodium salt | Sigma | I5148–2G |
| Indole-3-pyruvic acid | Sigma | I7017 |
| Indole-3-acetamide | Sigma | 286281–1G |
| Tryptophol | Sigma | T90301–5G |
| CH030031 | Sigma | C8124–5M |
| Folic acid | Sigma | F7876–1G |
| Deposited Data | ||
| Raw and analyzed data | This paper | GSE151711 |
| Experimental Models: Cell Lines | ||
| HEK-293T cells | ATCC | ATCC-CRL-1573 |
| Experimental Models: Organisms/Strains | ||
| Zebrafish: Tg(neurod1:CaMPARI)rdu78 | This paper | N/A |
| Zebrafish: Tg(−5kbneurod1:Gcamp6f)icm05 | PMID: 27231612 | N/A |
| Zebrafish: TgBAC(cldn15la:EGFP)pd1034 | PMID: 24504339 | N/A |
| Zebrafish: Tg(−5kbneurod1:TagRFP)w69 | PMID: 22738203 | N/A |
| Zebrafish: TgBAC(trpa1b:EGFP)a129 | PMID: 22190641 | N/A |
| Zebrafish: Tg(neurod1:cre; cmlc2:EGFP)rdu79 | This paper | N/A |
| Zebrafish: TgBAC(gata5:loxp-mcherry-stop-loxp-DTA)pd315 | This paper | N/A |
| Zebrafish: trpa1b mutantvu197 | PMID: 18829968 | N/A |
| Zebrafish: trpa1a mutanthu2163 | PMID: 18829968 | N/A |
| Zebrafish: TgBAC(neurod1:EGFP)nl1 | PMID: 19424431 | N/A |
| Zebrafish: Tg(−5kbneurod1:Gal4; cmlc2:EGFP)rdu71 | PMID: 31793875 | N/A |
| Zebrafish: Tg(−5kbneurod1:Lifeact-EGFP)rdu70 | PMID: 31793875 | N/A |
| Zebrafish: Tg(UAS:ChR2(H134R)-mCherry)s1985 | PMID: 26752076 | N/A |
| Zebrafish: Tg(NBT:DsRed)zf148 | PMID: 29628374 | N/A |
| Zebrafish: ret mutanthu2846 | PMID: 21490065 | N/A |
| Zebrafish: sox10 mutantt3 | PMID: 28207737 | N/A |
| Zebrafish: TgBAC(chata:Gal4)mpn202 | PMID: 28701772 | N/A |
| Zebrafish: Tg(UAS:Gcamp6s)mpn101 | PMID: 28701772 | N/A |
| Zebrafish: Tg(UAS:NTR-mCherry)c264 | PMID: 24496182 | N/A |
| Zebrafish: Tg(isl1:EGFP)rw0 | PMID: 23850871 | N/A |
| Zebrafish: tph1b mutant pd249 | PMID: 28811310 | N/A |
| Zebrafish: Tg(tph1b:mCherry-NTR)pd275 | This paper | N/A |
| Zebrafish: Tg(β-act2:Brainbow1.0L)pd50 | PMID: 22538609 | N/A |
| Zebrafish: Tg(neurod1:CaMPARI)rdu78 | This paper | N/A |
| Zebrafish: Tg(−5kbneurod1:Gcamp6f)icm05 | PMID: 27231612 | N/A |
| Zebrafish: TgBAC(cldn15la:EGFP)pd1034 | PMID: 24504339 | N/A |
| Zebrafish: Tg(−5kbneurod1:TagRFP)w69 | PMID: 22738203 | N/A |
| Zebrafish: Tg(−5kbneurod1:mitoEos)Y586 | PMID: 30410881 | N/A |
| Mouse: C57BL/6J | Jackson Laboratory | 000664 |
| Oligonucleotides | ||
| RT-qPCR for zebrafish gene: pyyb NM_001327895 |
Eurofins Genomics | AGCGTATCCACCCAAACCTG |
| GCCGGATGTCCTGTTCATCA | ||
| RT-qPCR for zebrafish gene: ccka XM_001346104 |
Eurofins Genomics | AACCAAAGGCTCATACCGCA |
| TCATATTCCTCGGCGCTTCG | ||
| RT-qPCR for zebrafish gene: adcyap1a NM_152885 |
Eurofins Genomics | GGGGTTTTCACGGACAGCTA |
| TGTGTCACAAAGCCGGGAAT | ||
| RT-qPCR for zebrafish gene: insl5a NM_001037669 |
Eurofins Genomics | TGCTGTAAGCAGACGAGACC |
| AGCAGAGGAACGTCAGGTCA | ||
| RT-qPCR for zebrafish gene: fabp2 NM_131431 |
Eurofins Genomics | TGGAAAGTCGACCGCAATGA |
| TGAACTTGTCTCCGGTCTGC | ||
| RT-qPCR for zebrafish gene: muc5.3 XM_021477626 |
Eurofins Genomics | ATGCGAACCATGGGGCTTTA |
| TTGTTCGCGTTCCCGTCATA | ||
| RT-qPCR for zebrafish gene: sypa NM_001143977 |
Eurofins Genomics | GATCGTGGCACCGTTTATGC |
| ATTGTAGCCTTGCTGGCTGT | ||
| RT-qPCR for zebrafish gene: sypb NM_001030242 |
Eurofins Genomics | ATCCTATGGGGAGGCAACCT |
| ACCTTCCTGTCCATAGCCCT | ||
| RT-qPCR for zebrafish gene: agr2 NM_001012481 |
Eurofins Genomics | AGTGCTCTTGGTCATGGTGG |
| AGGGGCTTGTTCTTGGATCG | ||
| RT-PCR for zebrafish gene: trpa1a NM_001007065 |
Eurofins Genomics | TACCAACATGTCGTGTTTTCAGTG |
| GATTGCACACAACCGGTTTACA | ||
| RT-PCR for zebrafish gene: trpa1b NM_001007066 |
Eurofins Genomics | CTCATTTGTCTTGGAAAGGGAGC |
| GGAGGAAGTTGCGACCTGTT | ||
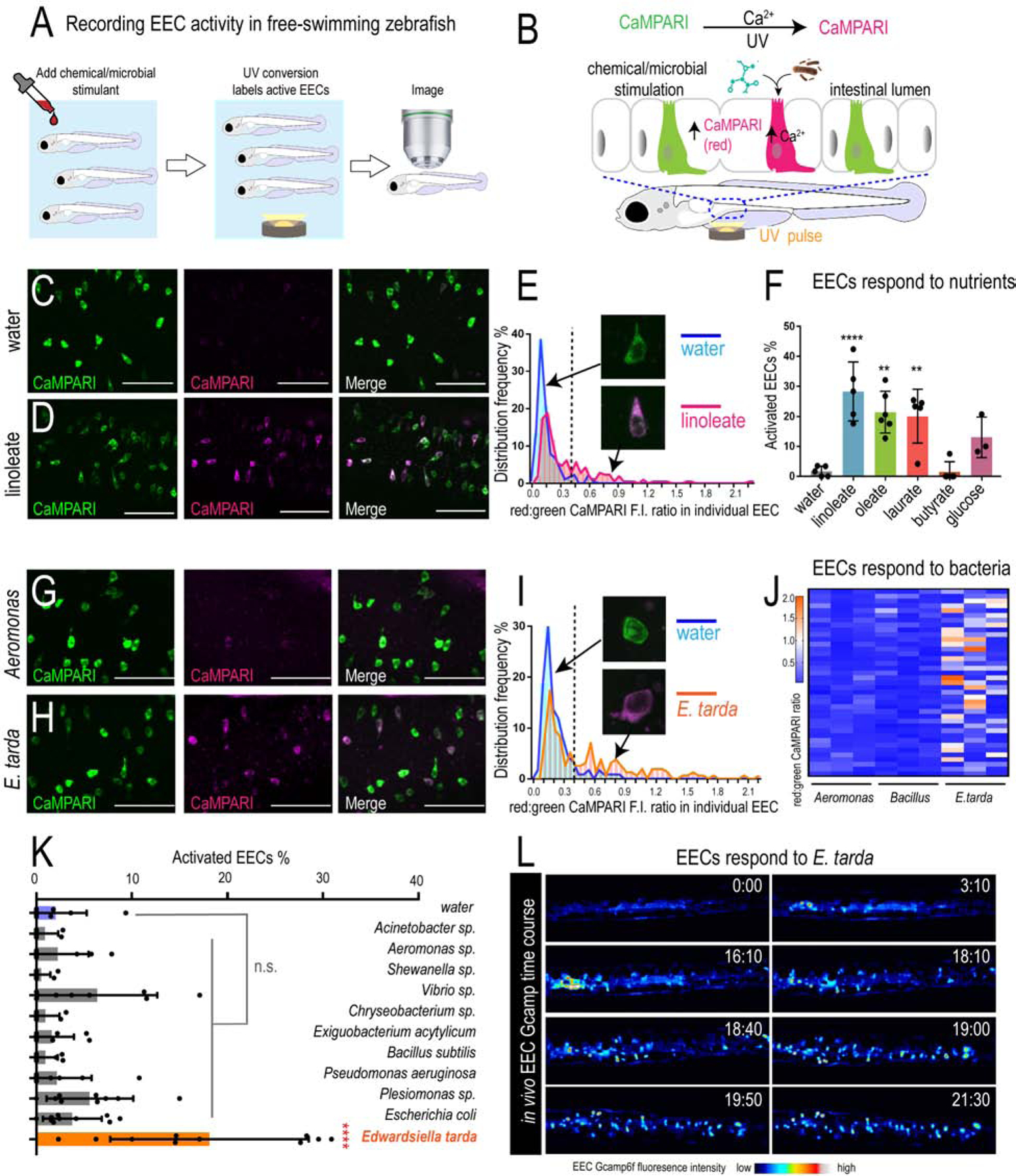
EECs express a variety of sensory receptors that can be activated by different environmental stimuli. To investigate the mechanisms by which EECs perceive E. tarda stimulation, we isolated EECs from zebrafish larvae and performed RNA-seq analysis. Transcript levels in FACS-sorted EECs (cldn15la:EGFP+; neurod1:TagRFP+) were compared to all other intestinal epithelial cells (IECs) (cldn15la:EGFP+; neurod1:TagRFP−) (Fig. 2A). We identified 192 zebrafish transcripts that were significantly enriched in EECs by DESeq2 using PFDR<0.05 (Fig. 2B and Table S1A–D). Gene Ontology analysis revealed that those zebrafish genes are enriched for processes like hormone secretion, chemical synaptic transmission and neuropeptide signaling (Table S1A–D). To identify gene homologs enriched in EECs in both zebrafish and mammals, we compared these 192 genes to published RNA-seq data from Neurod1+ EECs from mouse duodenum and CHGA+ EECs from human jejunum (Roberts et al., 2019). Despite the evolutionary distance and differences in tissue origin, we found that 24% of zebrafish EEC-enriched gene homologs (46 out of 192) were shared among zebrafish, human, and mouse, and that 40% of zebrafish EEC-enriched genes (78 out of 192) were shared between zebrafish EECs and human jejunal EECs (Table S2E–I). The genes with conserved EEC expression include those encoding hormones, transcription factors, G-protein coupled receptors, and ion channels that regulate membrane potential (Fig. 2C and Table S3J–N). Using published data from mouse intestinal epithelial single-cell RNA-seq data that revealed different EEC subtypes (Haber et al., 2017), we found that many of the signature genes in mouse enterochromaffin cells (EC), which are identified by their 5-HT synthesis, are enriched in zebrafish EECs (Table S3J–N). Among these conserved EEC-enriched genes, one of the genes with the highest expression in zebrafish EECs is transient receptor potential ankyrin 1 (Trpa1) (Fig. 2C and Table S1A–I).
Figure 2. E. tarda activates EECs through Trpa1.
(A) Schematic diagram of zebrafish EEC RNA-seq. (B) Clustering of genes that are significantly enriched in zebrafish EECs and other IECs (Padj<0.05). (C) Comparison of zebrafish and mouse EEC enriched genes. Mouse EEC RNA-seq data was obtained from GSE114913 (Roberts et al., 2019). (D) Fluorescence image of TgBAC(trpa1b:EGFP)(Pan et al., 2012). Zoom-in view shows the expression of trpa1b+ cells in intestine. (E) Confocal projection of a TgBAC(trpa1b:EGFP);Tg(neurod1:TagRFP) zebrafish intestine. Yellow arrows indicate zebrafish EECs that are trpa1b:EGFP+. (F) Quantification of EEC Gcamp responses to Trpa1 agonist AITC stimulation in trpa1b+/+, trpa1b+/− and trpa1b−/− zebrafish. (G) Experimental design. (H-I) Confocal projection of Tg(neurod1:CaMPARI) zebrafish intestine stimulated with E. tarda with or without the Trpa1 antagonist HC030031. (J) Quantification of activated EECs in control and HC030031 treated zebrafish treated with water or E. tarda. (K) Experimental approach. (L-M) Confocal projection of trpa1b+/+ or trpa1b−/− Tg(neurod1:CaMPARI) intestine after stimulation with water or E. tarda. (N) Quantification of activated EEC percentage in WT and trpa1b−/− zebrafish treated with water or E. tarda. (O) Experimental design. (P-Q) Timed images of trpa1b+/+ or trpa1b−/− Tg(neurod1:Gcamp6f) zebrafish stimulated with E. tarda. (R) Quantification of relative EEC Gcamp6f fluorescence intensity in WT or trpa1b−/− zebrafish treated with E. tarda. One-way ANOVA with Tukey’s posttest was used in F, J, N. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
Trpa1 is an excitatory calcium-permeable non-selective cation channel that has important roles in pain sensation and neurologic inflammation (Bautista et al., 2013, Lapointe and Altier, 2011). A broad spectrum of chemical irritants, including AITC and other compounds derived from food spices, activate Trpa1 (Nilius et al., 2011). In addition to chemical irritants, certain bacterial products, including lipopolysaccharide (LPS) and hydrogen sulfide (H2S), stimulated nociceptive neurons in a Trpa1-dependent manner (Meseguer et al., 2014). The zebrafish genome encodes two trpa1 paralogs, trpa1a and trpa1b (Prober et al., 2008). Our data establish that trpa1b, but not trpa1a, is expressed by a subset of zebrafish EECs and is required for EEC activation by Trpa1 agonist AITC (Fig. 2D–F, Fig. S2B–G). Since the expression of classic microbial pattern recognition receptors is very low in zebrafish EECs (Table S1J–N), we tested if Trpa1 mediated E. tarda-induced EEC activation. We first observed that treatment of wild-type (WT) Tg(neurod1:CaMPARI) fish with the Trpa1 antagonist HC030031 significantly inhibited E. tarda’s ability to induce EEC activation (Fig. 2G–J). The ability of E. tarda to induce EEC activity in the EEC-CaMPARI model was similarly blocked in trpa1b mutant zebrafish (Fig. 2K–N). In accord, experiments in Tg(neurod1:Gcamp6f) zebrafish confirmed that Gcamp6f fluorescence increased in EECs in response to E. tarda stimulation in WT, but not trpa1b mutant zebrafish (Fig. 2O–R). Therefore live E. tarda bacteria stimulate EECs in a Trpa1-dependent manner, suggesting that EEC Trpa1 signaling may play an important role in mediating microbe-host interactions.
EEC Trpa1 signaling is important to maintain microbial homeostasis by regulating intestinal motility
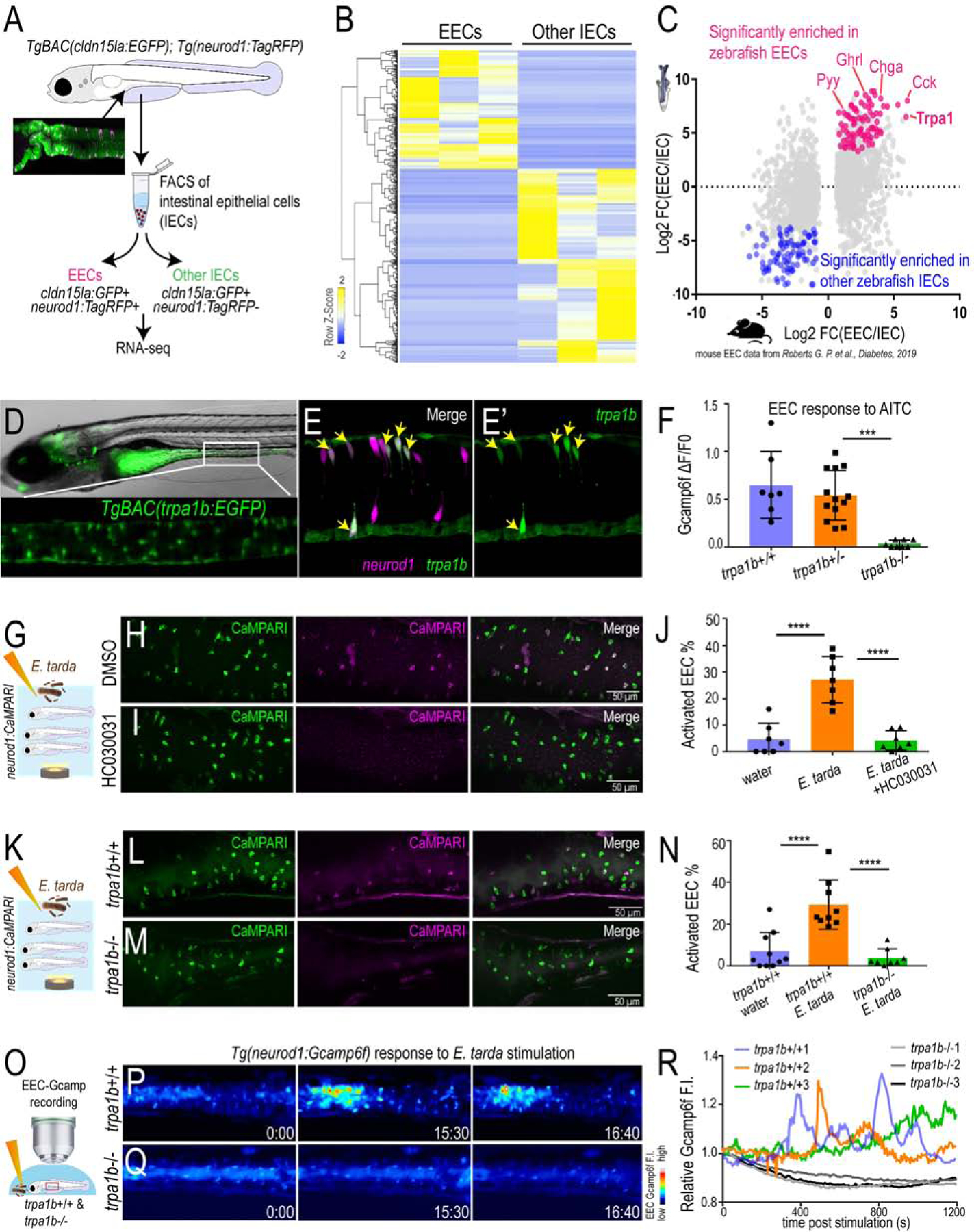
To determine how E. tarda-induced Trpa1 signaling in EECs affects the host, we exposed trpa1b+/+ and trpa1b−/− zebrafish larvae to an E. tarda strain expressing mCherry fluorescent protein. High-dose (107 CFU/mL) E. tarda exposure for 3 days decreased survival rate and caused gross pathology (Fig. S2M–N), consistent with its reported activity as a zebrafish pathogen (Abayneh et al., 2013, Flores et al., 2020). To investigate the interaction between E. tarda and Trpa1+EECs under relatively normal physiological conditions, we exposed zebrafish with a low E. tarda dose (106 CFU/mL) that did not significantly affect survival rate or cause gross pathology (Fig. S2M–N and Fig. 3A). When reared under conventional conditions in the absence of E. tarda, we observed no significant difference in the abundance of culturable gut microbes between trpa1b+/+ and trpa1b−/− zebrafish (Fig. S2H–I). However, upon 3-day treatment with E. tarda, there was significant accumulation of E. tarda mCherry+ bacteria in the intestinal lumen in trpa1b−/− but not trpa1b+/+ larvae (Fig. 3A–C). This accumulation could be observed by either quantifying intestinal E. tarda mCherry fluorescence (Fig. 3D) or counting E. tarda colony forming units (CFU) from digestive tracts dissected from E. tarda treated trpa1b+/+ and trpa1b−/− zebrafish (Fig. 3E). This suggests that Trpa1 signaling may act as a host defense mechanism to facilitate clearance of specific types of bacteria such as E. tarda.
Figure 3. Activation of EEC Trpa1 signaling facilitates enteric E. tarda clearance.
(A) Schematic of zebrafish E. tarda treatment. (B-C) Representative image of trpa1b+/+ or trpa1b−/− zebrafish treated with E. tarda expressing mCherry (E. tarda mCherry). (D) Quantification of E. tarda mCherry fluorescence in trpa1b+/+ or trpa1b−/− zebrafish intestine. (E) Quantification of intestinal E. tarda CFU in trpa1b+/+ or trpa1b−/− zebrafish. (F) Schematic of genetic model in which EECs are ablated via Cre-induced Diptheria Toxin (DTA) expression. (G) Representative image of Tg(neurod1:cre; cmlc2:EGFP) and Tg(neurod1:cre; cmlc2:EGFP); TgBAC(gata5:RSD) with EECs that are labelled by Tg(neurod1:EGFP). (H) Quantification of intestinal E. tarda CFU in WT or EEC ablated zebrafish. Student’s t-test was used in D, E, H. *p<0.05; ****p<0.0001.
In addition to EECs, Trpa1 is also expressed in mesenchymal cells within the intestine (Fig.2D–E and Fig. S4O) and nociceptive sensory neurons (Yang et al., 2019, Holzer, 2011). To test if the phenotype we observed above is specifically mediated by EECs, we generated a Cre-loxP transgenic system that permits specific ablation of EECs without affecting other IECs or other neurod1 expressing cells like CNS or pancreatic islets (Fig. 3F, Fig. S2J). Quantitative RT-PCR and immunofluorescence confirmed a reduction of EEC hormones but not non-EEC marker genes (Fig. S2K–L). Establishing this EEC ablation system allowed us to define the specific role of EECs in mediating E. tarda-host interaction. As with trpa1b−/− zebrafish, we did not detect significant differences in gut microbial abundance between unexposed WT and EEC-ablated zebrafish (Fig. S2O). However, in response to E. tarda exposure, a significantly higher amount of E. tarda mCherry accumulated in EEC-ablated zebrafish compared to WT sibling controls (Fig. 3H and Fig. S2P–Q). Together, these data establish that EEC Trpa1 signaling maintains gut microbial homeostasis by facilitating host clearance of specific types of bacteria like E. tarda. To understand the mechanisms by which EEC Trpa1 regulates gut microbial homeostasis, we used an opto-pharmacological approach that permits temporal control of EEC Trpa1 activation through UV light exposure (Fig. 4A). We pretreated zebrafish with Optovin, a chemical that specifically activates Trpa1 only in the presence of UV light (Kokel et al., 2013) (Fig. S3A). To specifically activate Trpa1 in EECs, we mounted zebrafish larvae pretreated with Optovin and restricted UV light exposure specifically to the intestinal epithelium using a confocal laser (Fig. S3A). UV light activation significantly increased [Ca2+]i in a subpopulation of EECs in WT larvae, as measured by Gcamp6f fluorescence (Fig. S3B–C). The same UV light exposure in trpa1b−/− larvae pretreated with Optovin did not increase EEC [Ca2+]i (Fig. 4B–C), indicating that EEC activation induced by Optovin-UV was dependent on Trpa1. Next, we used this approach to examine the effect of EEC Trpa1 activation on intestinal motility. Trpa1 activation in EECs within the middle intestine via UV light application in WT larvae produced a propulsive movement of the intestine from anterior to posterior, and the velocity of intestinal motility increased accordingly (Fig. 4D–F, Fig. S3D–E and Video 1). In contrast, Optovin treatment and UV activation failed to induce intestinal motility in EEC-ablated zebrafish (Fig. 4D–F and Video 1). These results indicate that intestinal motility triggered by Trpa1 activation is dependent on EECs. To further test if signaling from Trpa1+EECs is sufficient to activate intestinal motility, we developed an optogenetic system in which a mCherry tagged Channelrhodopsin (ChR2-mCherry) is expressed in EECs from the neurod1 promoter (Fig. 4G–H). Blue light activation of ChR2 causes cation influx and plasma membrane depolarization, and [Ca2+]i then increases through the activation of voltage-dependent calcium channels (Nagel et al., 2003) which are abundantly expressed in zebrafish EECs (Fig. 4I–J, Table S3J–N). This tool permits selective activation of the ChR2-mCherry+ EECs using a confocal laser, without affecting the activity of nearby EECs (see Methods and Fig. S3F). We therefore used Tg(neurod1:Gal4); Tg(UAS:ChR2-mCherry); TgBAC(trpa1b:EGFP) larvae to selectively activate ChR2-mCherry expressing EECs that are either trpa1b+ or trpa1b−. We found that activation of trpa1b+ EECs but not trpa1b− EECs consistently increased intestinal velocity magnitude (Fig. 4K–L, Fig. S3F–H and Video 2), again indicating a unique role for Trpa1+EECs in regulating intestinal motility. Consistent with the Optovin-UV result, stimulating Trpa1+ChR2+ EECs in the middle intestine resulted in anterograde intestinal movement (Video 8). Interestingly, stimulating Trpa1+ChR2+ EECs in the proximal intestine initiated a retrograde intestinal movement (Video 2). This is consistent with previous findings that the zebrafish proximal intestine typically exhibits a retrograde motility pattern whereas the middle and distal intestine display antegrade motility (Fig. S3D) (Roach et al., 2013). Finally, we tested whether microbial activation of Trpa1 signaling in EECs also increased intestinal motility. Using microgavage (Cocchiaro and Rawls, 2013), we found that delivery of live E. tarda into the intestinal lumen significantly promoted intestinal peristalsis and motility compared to PBS-gavaged controls (Fig. 4M–O and Video 3). E. tarda induced intestinal motility was significantly reduced in trpa1b−/− zebrafish (Fig. S3I). When we gavaged zebrafish with Aeromonas or Bacillus, two of the tested bacterial strains that do not activate EECs (Fig. 1), no significant change of intestinal motility was observed (Fig. 4M–O and Video 3). These experiments together establish that activation of Trpa1 in EECs directly stimulates intestinal motility, and provide a potential physiologic mechanism underlying Trpa1-dependent clearance of E. tarda from the intestinal lumen.
Figure 4. Activation of EEC Trpa1 signaling promotes intestinal motility.
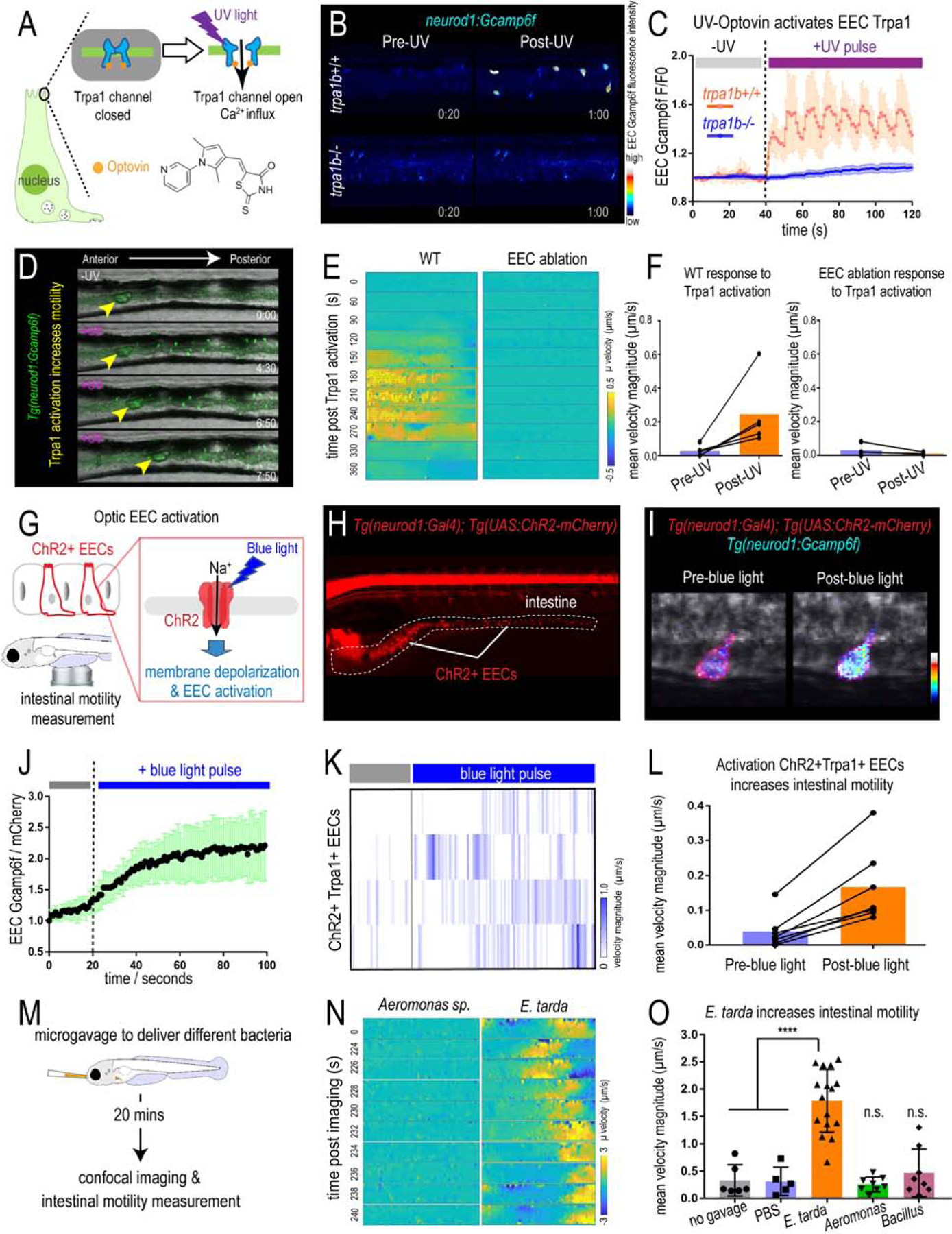
(A) Illustration of EEC Trpa1 activation using an Optovin-UV platform. (B) Confocal image of trpa1b+/+ and trpa1b−/− Tg(neurod1:Gcamp6f) zebrafish EECs before and after UV activation. (C) Quantification of EEC Gcamp6f fluorescence changes in trpa1b+/+ and trpa1b−/− zebrafish before and after UV induction. (D) Representative images of Tg(neurod1:Gcamp6f) zebrafish intestine before and after UV-induced Trpa1 activation. Yellow arrowheads indicate the movement of intestinal luminal contents from anterior to posterior following EEC activation. (E) PIV-Lab velocity analysis to quantify intestinal motility in WT and EEC ablated zebrafish. Spatiotemporal heatmap series representing the µ velocity of the imaged intestinal segment at the indicated timepoint post Trpa1 activation. (F) Quantification of the mean intestinal velocity magnitude before and after UV activation in WT and EEC ablated zebrafish. (G) Model of light activation of ChR2 in EECs. (H) Fluorescence image of Tg(neurod1:Gal4); Tg(UAS:ChR2-mCherry) zebrafish that express ChR2 in EECs. (I) Confocal image of ChR2 expressing EECs in Tg(neurod1:Gcamp6f) intestine before and after blue light-induced ChR2 activation. (J) Quantification of EEC Gcamp fluorescence intensity before and after blue light-induced ChR2 activation. (K) Intestinal velocity magnitude before and after blue-light induced activation in ChR2+Trpa1+ EECs. (L) Mean velocity magnitude before and after blue light-induced activation in ChR2+Trpa1+ EECs. (M) Experimental design schematic for panels N and O. (N) Heatmap representing the µ velocity of the imaged intestinal segment at indicated timepoints following Aeromonas sp. or E. tarda gavage. (O) Mean intestinal velocity magnitude in zebrafish without gavage or gavaged with PBS or different bacterial strains. Student’s t-test was used in O. ****p<0.0001.
EEC Trpa1 signaling promotes intestinal motility by activating cholinergic enteric neurons
To test the role of the ENS in Trpa1-activated intestinal motility, we used zebrafish that lack an ENS due to mutation of the receptor tyrosine kinase gene ret (Taraviras et al., 1999). Immunofluorescence demonstrated that ret−/− zebrafish lack all identifiable enteric nerves (marked by NBT transgenes, Fig. 5B and Fig. S4A–B), whereas EECs remain intact (marked by neurod1 transgenes, Fig. 5B) and responsive to Trpa1 agonist (Fig. S4C–F). Using the Optovin-UV system (Fig. 5A), we observed that EEC Trpa1 activation increased intestinal motility in control (ret+/+ or ret+/−) but not ret−/− zebrafish (Fig. 5C–D and Fig. S4G–H). These results were confirmed using a second zebrafish mutant that lacks an ENS due to mutation of the transcription factor gene sox10 (Bondurand and Sham, 2013) (Fig. S4I–N). These data suggest that Trpa1+ EECs do not signal directly to enteric smooth muscle to promote intestinal motility, but instead signal to the ENS.
Figure 5. Activation of EEC Trpa1 signaling activates enteric cholinergic neurons and promotes intestinal motility through 5-HT.
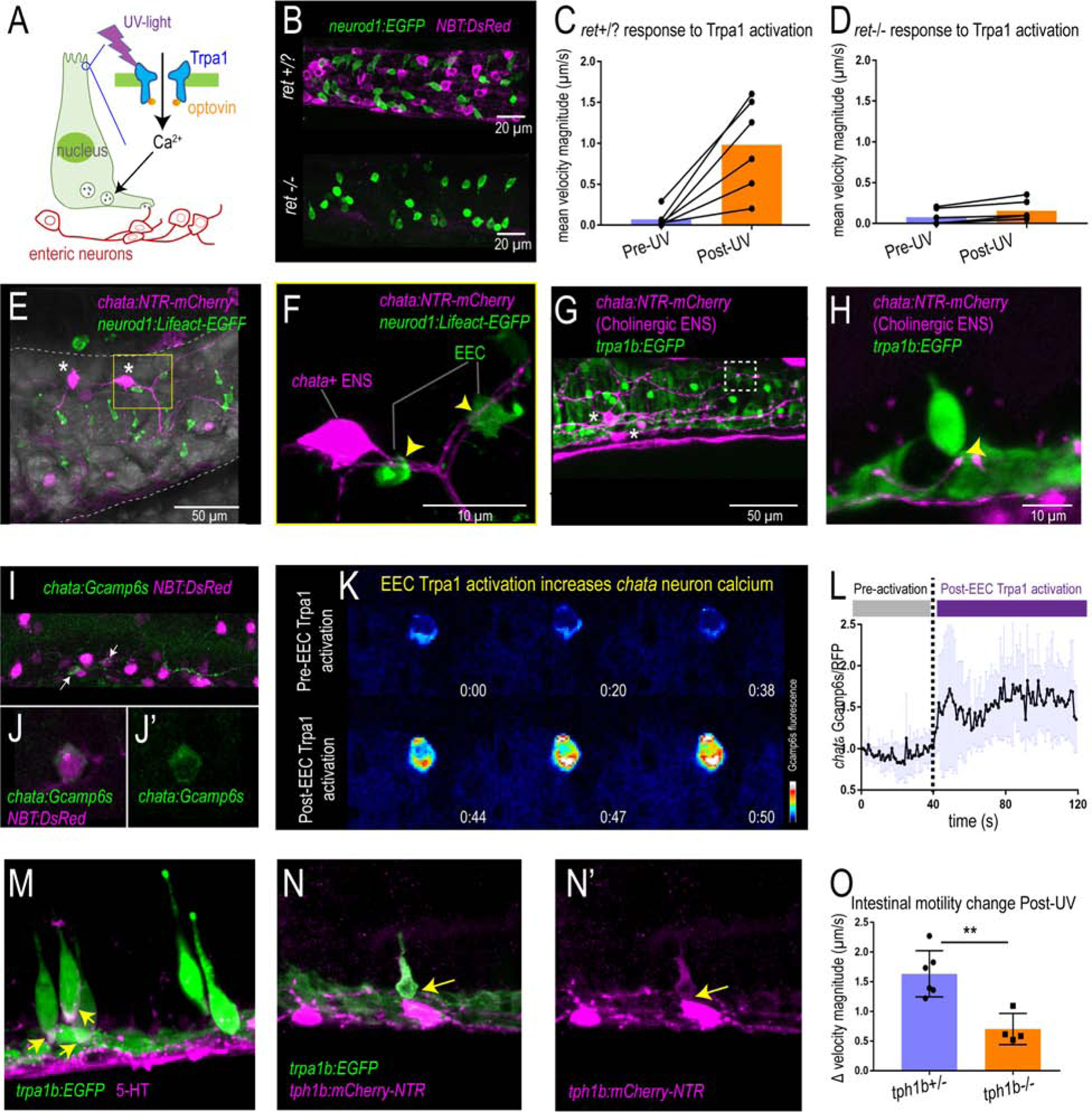
(A) Working model showing Trpa1 stimulation in EECs activates enteric neurons. (B) Confocal image of ret+/? (ret+/+ or ret+/−) and ret−/− zebrafish intestine. neurod1 labelled EECs shown in green and NBT labelled ENS shown in magenta. (C) Quantification of mean intestinal velocity magnitude before and after EEC Trpa1 activation in ret+/? zebrafish. (D) Quantification of mean intestinal velocity magnitude before and after UV activation in ret−/− zebrafish. (E) Confocal image showing EECs (neurod1+; green) and cholinergic enteric neurons (chata+; magenta) in the zebrafish intestine. Asterisks indicate Cholinergic enteric neuron cell bodies which reside on the intestinal wall. (F) Higher magnification view indicates the EECs (green) directly contact nerve fibers that are extended from the chata+ enteric neuron cell body (magenta) as indicated by yellow arrows. (G-H) Confocal image showing Trpa1+EECs (green) form direct contact with chata+ enteric neurons (magenta). (I-J) In vivo calcium imaging of cholinergic enteric neurons. All the enteric neurons are labelled as magenta by NBT:DsRed. Yellow arrow indicates a chata+ enteric neuron that express Gcamp6s. (K) In vivo calcium imaging of chata+ enteric neuron before and after EEC Trpa1 activation. (L) Quantification of chata+ enteric neuron Gcamp6s fluorescence intensity before and after EEC Trpa1 activation. (M) Confocal image of TgBAC(trpa1b:EGFP) zebrafish intestine stained for 5-HT. Yellow arrows indicate the presence of 5-HT in the basal area of trpa1b+ EECs. (N) Confocal image showing zebrafish Trpa1b+ EECs (green) express Tph1b (magenta). (O) Quantification of intestinal motility changes in response to EEC Trpa1 activation in tph1b+/− and tph1b−/− zebrafish. Student’s t test was used in O. **p<0.01
The ENS is a complex network composed of many different neuronal subtypes. Among these subtypes, cholinergic neurons secrete the excitatory neurotransmitter acetylcholine to stimulate other enteric neurons or smooth muscle (Pan and Gershon, 2000, Qu et al., 2008) and are essential for normal intestinal motility (Johnson et al., 2018). One of the key enzymes for the synthesis of acetylcholine in the ENS is choline acetyltransferase (Chat) (Furness et al., 2014). Using TgBAC(chata:Gal4); Tg(UAS:NTR-mCherry) transgenic zebrafish, we visualized the cholinergic enteric neurons in the zebrafish intestine (Fig. 5E and Fig. S5E–J). We found that chata+ neurons have smooth cell bodies which are located within the intestinal wall, many of which display multiple axons (Fig. 5E and Fig. S5E–F). Such multipolar neurons have also been classified as Dogiel type II neurons (Cornelissen et al., 2000). These Dogiel type II neurons are likely to be the intestinal intrinsic primary afferent neurons (IPANs) (Bornstein, 2006). Our results indicate that some EECs including Trpa1+EECs form direct contacts with nerve fibers extending from chata+ enteric neurons (Fig. 5F–H and Fig. S5G–J). In addition, we found that zebrafish EECs are enriched for transcripts encoding presynaptic vesicle proteins (Fig. S5P and Table S1A–D) and form neuropod structures that connect with neurons (Fig. S5A–D) similar to mouse EECs (Bohorquez et al., 2015, Bellono et al., 2017, Kaelberer et al., 2018). To test if activation of Trpa1+EECs stimulates chata+ enteric neurons, we employed TgBAC(chata:Gal4); Tg(UAS:Gcamp6s) zebrafish, which permit recording of in vivo calcium activity in chata+ neurons (Fig. 5I–J). Upon Trpa1+EEC activation, Gcamp6s fluorescence increased in chata+ enteric neurons (Fig. 5K, L). We found that Trpa1 is not expressed in chata+ enteric neurons or in any other ENS cells (Fig. S6O–R), indicating that chata+ enteric neurons cannot be directly activated by optic Trpa1 stimulation but are instead activated via stimulation by Trpa1+ EECs. In addition to the ENS, efferent vagal nerves also play an important role in modulating intestinal motility (Travagli and Anselmi, 2016). However, we observed similar Trpa1+EEC induced intestinal motility change and chata+ enteric neuron activation in zebrafish whose intestine is anatomically disconnected from the CNS (Fig. S5K–O), suggesting that vagal efferent nerves are not required for Trpa1+EEC induced intestinal motility, and that Trpa1+EEC induced intestinal motility is mediated by intrinsic enteric circuitry which likely involves chata+ enteric neurons.
Previous mouse studies demonstrated that Trpa1 mRNA is highly enriched in 5-HT-secreting EC cells in the small intestine (Nozawa et al., 2009). Immunofluorescence staining indicated that, similar to mammals, 5-HT expression in the zebrafish intestinal epithelium is also highly enriched in Trpa1+EECs (Fig. 5M). 5-HT in EECs is synthesized from tryptophan via tryptophan hydroxylase 1 (Tph1) (Li et al., 2011). Zebrafish possess two Tph1 paralogs, tph1a and tph1b (Ulhaq and Kishida, 2018), but only tph1b is expressed in zebrafish EECs (Fig. S5S). The expression of tph1b in Trpa1+EECs was also confirmed by crossing a Tg(tph1b:mCherry-NTR) transgenic line to TgBAC(trpa1b:EGFP) zebrafish (Fig. 5N and Fig. S5Q–R, T–U). To investigate whether 5-HT mediates EEC Trpa1-induced intestinal motility, we tested whether a similar response was present in tph1b+/+ and tph1b−/− zebrafish larvae (Tornini et al., 2017) using the Optovin-UV platform. Under baseline conditions, we did not observe a significant difference in intestinal motility between tph1b+/+ and tph1b−/− zebrafish (Fig. S5V). However, in response to UV stimulated EEC Trpa1 activation, intestinal motility was significantly reduced in tph1b−/− compared to tph1b+/+ zebrafish (Fig. 5O). These findings suggest a working model in which Trpa1+EECs signal to cholinergic enteric neurons through 5-HT, which in turn stimulates intestinal motor activity and promotes intestinal motility.
Chemical and microbial stimulation of EEC Trpa1 signaling activate vagal sensory ganglia
The intestine is innervated by both intrinsic ENS and extrinsic sensory nerves from the brain and spinal cord (Brookes et al., 2013). In mammals, afferent neuronal cell bodies of the vagus nerve reside in the nodose ganglia and travel from the intestine to the brainstem to convey visceral information to the CNS. However, in zebrafish, it is unknown if the vagal sensory system innervates the intestine. The zebrafish vagal sensory ganglia can be labelled using TgBAC(neurod1:EGFP) or immunofluorescence staining of the neuronal marker acetylated α Tubulin (Ac-αTub) (Fig. 6B). Using lightsheet confocal imaging, we demonstrated that not only do the vagal ganglion in zebrafish extend projections to the intestine (Fig. 6B–C and Fig. S6A–B) but vagal sensory nerve fibers directly contact a subpopulation of EECs (Fig. 6D). Using Tg(neurod1:cre); Tg(β-act2:Brainbow) transgenic zebrafish (Gupta and Poss, 2012) (Vagal-Brainbow) in which individual vagal ganglion cells are labeled with different fluorescent colors through Cre recombination (Foglia et al., 2016) (Fig. S6C), we revealed that zebrafish vagal sensory ganglia cells also directly project to the vagal sensory region in the hindbrain (Fig. 6E–F). Using this Vagal-Brainbow system, we found vagal sensory nerves that are labelled by Cre recombination in both the proximal and distal intestine (Fig. S6D–G). To further visualize the vagal sensory network in zebrafish, we used Tg(isl1:EGFP) zebrafish in which EGFP is expressed in vagal sensory ganglia and overlaps with neurod1 (Fig. 6G and Fig. S6H–J). Our data revealed that after leaving the vagal sensory ganglia, the vagus nerve travels along the esophagus and enters the intestine in the region between the pancreas and the liver (Fig. 6G and Fig. S6I–J). Direct contact of EECs and the vagus nerve could also be observed in Tg(isl1:EGFP); Tg(neurod1:TagRFP) zebrafish (Fig. 6H). These data demonstrate the existence of a vagal network in the zebrafish intestine.
Figure 6. EEC Trpa1 signaling activates vagal sensory ganglia.
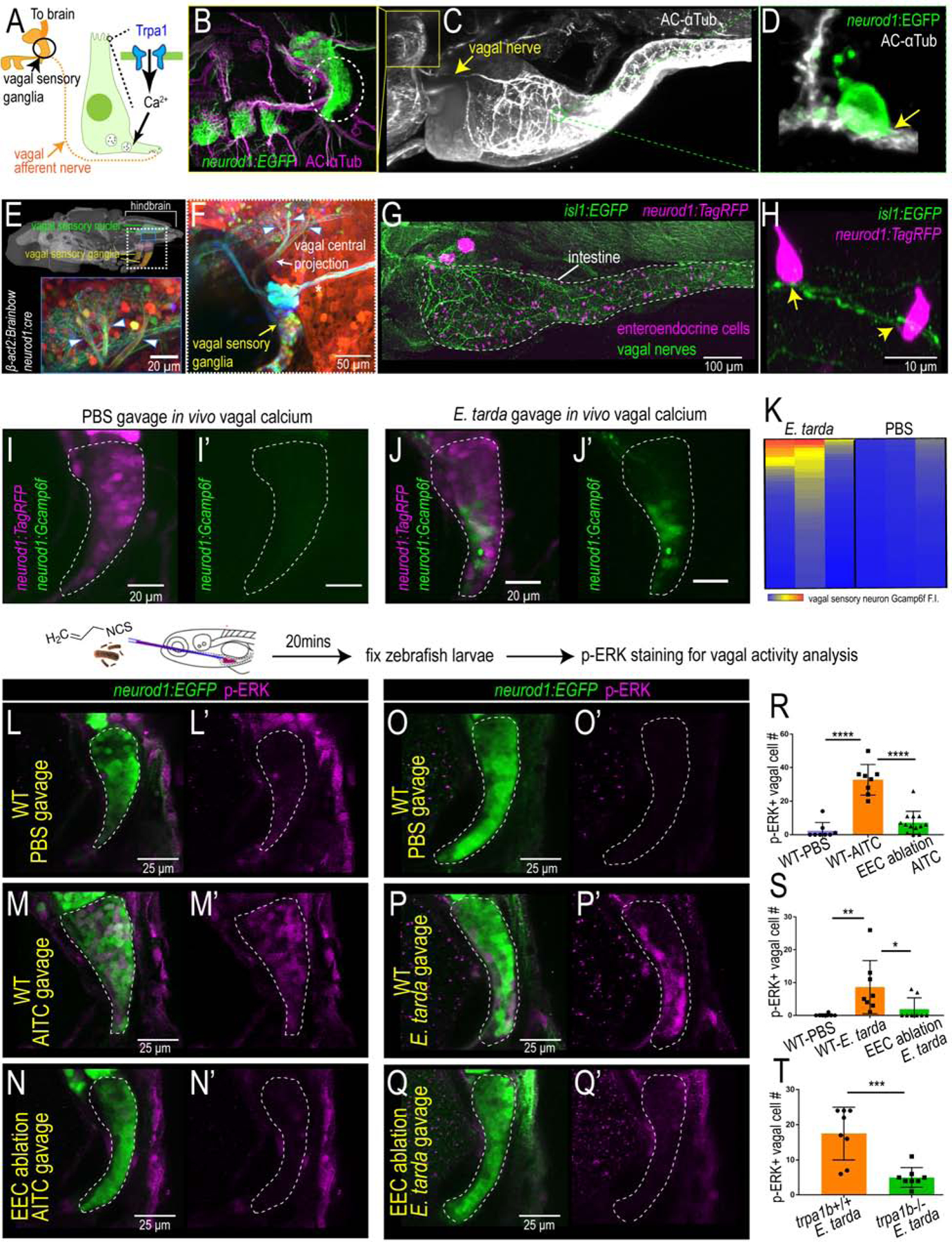
(A) Working model. (B) Confocal image of zebrafish vagal sensory ganglia labelled with Tg(neurod1:EGFP) (green) and acetylated αTubulin antibody staining (AC-αTub, magenta). (C) Lightsheet projection of zebrafish stained for AC-αTub. Yellow arrow indicates vagal nerve innervation to the intestine. (D) neurod1:EGFP+ EECs (green) directly contact vagal sensory nerve fibers labelled with αTubulin (white). (E) Confocal image of the vagal sensory nucleus in zebrafish larvae hindbrain where vagal sensory neurons project. Vagal sensory nerve fibers are labeled with different fluorophores through Cre-brainbow recombination in Tg(neurod1:cre); Tg(βact2:Brainbow) zebrafish. The 3D zebrafish brain image was generated using mapzebrain (Kunst et al., 2019). (F) Confocal image of vagal sensory ganglia in Tg(neurod1:cre); Tg(βact2:Brainbow) zebrafish. Asterisk indicates posterior lateral line afferent nerve fibers. Blue arrowheads indicate three branches from vagal sensory ganglia that project to the hindbrain. (G) Confocal image demonstrates the EEC-vagal network in zebrafish intestine. EECs are labeled as magenta by neurod1:TagRFP and the vagal nerve is labeled green by isl1:EGFP. (H) EECs (neurod1+; magenta) directly contact vagal nerve fibers (isl1+; green) as indicated by yellow arrows. (I-J) In vivo calcium imaging of vagal sensory ganglia in zebrafish gavaged with PBS (I) or E. tarda (J). (K) Quantification of individual vagal sensory neuron Gcamp6f fluorescence intensity in E. tarda or PBS gavaged zebrafish. (L-N) Confocal image of vagal ganglia (neurod1+; green) stained with p-ERK antibody (activated vagal sensory neurons; magenta) in WT or EEC ablated zebrafish gavaged with PBS or Trpa1 agonist AITC. (O-Q) Confocal projection of vagal ganglia stained with p-ERK antibody in WT or EEC ablated zebrafish gavaged with PBS or E. tarda. (R) Quantification of p-ERK+ vagal sensory neurons in WT or EEC ablated zebrafish following PBS or AITC gavage. (S) Quantification of p-ERK+ vagal sensory neurons in WT or EEC ablated zebrafish following PBS or E. tarda gavage. (T) Quantification of p-ERK+ vagal sensory neurons in WT or trpa1b−/− zebrafish following E. tarda gavage. One-way ANOVA with Tukey’s post test was used in R and S and Student’s t-test was used in T. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
We next investigated whether this vagal network is activated in response to enteric microbial stimulation with E. tarda. We gavaged Tg(neurod1:Gcamp6f); Tg(neurod1:TagRFP) zebrafish larvae with either PBS or live E. tarda bacteria. We found that 30 min after enteric stimulation with Trpa1 agonist AITC or E. tarda, but not after PBS vehicle stimulation, Gcamp6f fluorescence intensity significantly increased in a subset of vagal sensory neurons (Fig. 6I–K, Fig. S6K–L). This result indicated that acute enteric chemical or microbial stimulation directly activated vagal sensory neurons. To further investigate whether the vagal activation induced by enteric E. tarda was mediated by Trpa1+EECs, we used a published method that labels active zebrafish neurons through pERK immunofluorescence (Randlett et al., 2015) to measure vagal activity. Delivering AITC into the zebrafish intestine by microgavage (Cocchiaro and Rawls, 2013) increased the number of pERK+ vagal cells compared to PBS treatment (Fig. 6L–N, R). AITC-induced vagal activation was abrogated in the absence of EECs (Fig. 6N, R), indicating that Trpa1 signaling in the intestine increases vagal sensory activity in an EEC-dependent manner. Next, we gavaged live E. tarda into both WT and EEC-ablated zebrafish. Similar to Trpa1 chemical agonist stimulation, E. tarda gavage increased the number of activated pERK+ vagal sensory neurons in WT zebrafish (Fig. 6O–Q, S) but not in EEC ablated zebrafish (Fig. 6Q, S). Furthermore, the vagal activation induced by enteric E. tarda was dependent on Trpa,1 as pERK+ vagal cell number was significantly reduced in E. tarda treated trpa1b−/− zebrafish (Fig. 6T). Together, these results reveal that chemical or microbial stimuli in the intestine can stimulate Trpa1+ EECs, which then signal to the vagal sensory ganglia.
Tryptophan catabolites secreted from E. tarda activate the EEC Trpa1 gut-brain pathway
To identify the molecular mechanism by which E. tarda activates Trpa1 in EECs, we examined the effects of live and killed E. tarda cells and cell-free supernatant (CFS) on EEC calcium activity (Fig. 7A). Formalin-killed or heat-killed E. tarda cells failed to stimulate EECs, however, CFS, at levels comparable to live E. tarda cells, stimulated EECs (Fig. 7A–B). The ability of E. tarda CFS to activate EECs was diminished in trpa1b mutant zebrafish (Fig. 7C), suggesting that a factor secreted from E. tarda has the ability to activate Trpa1 in EECs. HPLC-MS analysis revealed that E. tarda CFS is enriched for several indole ring-containing tryptophan catabolites (Fig. 7D and Fig. S7A–C), three of the most abundant being indole, tryptophol (IEt), and indole-3-carboxyaldhyde (IAld) (Fig. 7D and Fig. S7A–C). To test if other bacteria secrete tryptophan catabolites like E. tarda, we performed similar HPLC-MS analysis of CFS from bacteria we previously tested for EEC activation (Fig.1K). Although several tested bacterial strains produced indole or IAld when cultured in nutrient-rich medium (Fig. S7D), E. tarda was the only bacteria that uniquely retained a high level of indole and IAld production when cultured in zebrafish GZM housing water (Fig. 7E), consistent with our finding that E. tarda uniquely activates zebrafish EECs when added into GZM water (Fig. 1K). To investigate whether these tryptophan metabolites were directly linked with E. tarda pathogenesis, we tested two other E. tarda strains (E. tarda 23685 and E. tarda 15974) which were isolated from human gut microbiota and do not cause fish pathogenesis (Yang et al., 2012, Srinivasa Rao et al., 2003, Nakamura et al., 2013). Both E. tarda strains activated EECs and exhibited similar indole and IAld secretion capacity as pathogenic E. tarda FL6–60 and E. tarda LSE40 (Fig. 7E and Fig. S7G–H). This result suggested that tryptophan catabolites production, EEC Trpa1 activation and its downstream consequences may be distinct from E. tarda pathogenesis in fish (Edwardsiellosis) (Park et al., 2012).
Figure 7. E. tarda derived Tryptophan catabolites activate Trpa1 and the EEC-vagal pathway.
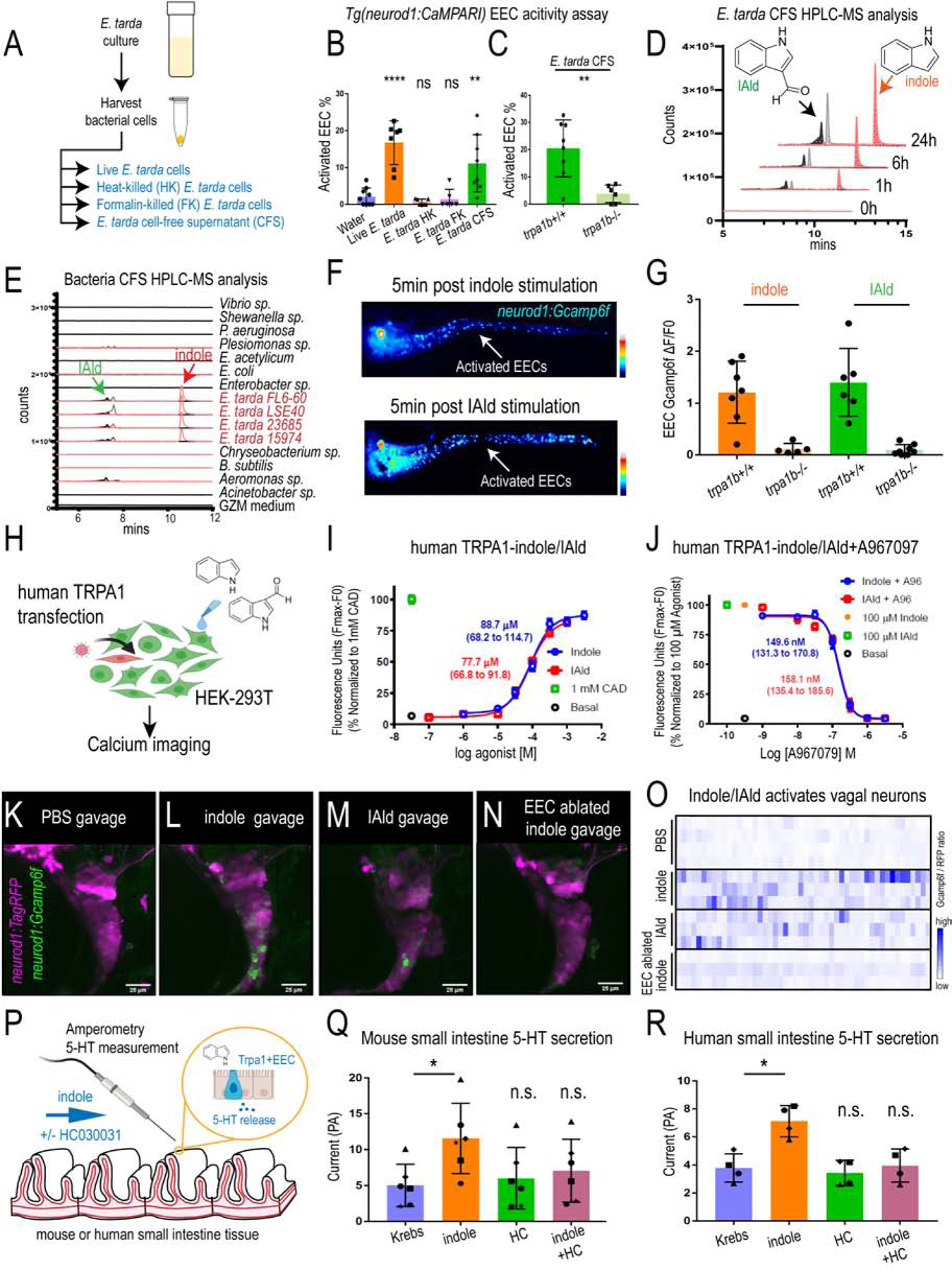
(A) Method for preparing different fractions from E. tarda GZM (zebrafish water) culture. (B) Activated EECs in Tg(neurod1:CaMPARI) zebrafish stimulated by different E. tarda fractions. (C) Activated EECs in trpa1b+/+ and trpa1b−/− Tg(neurod1:CaMPARI) zebrafish stimulated with E. tarda CFS. (D) Screening of supernatants of E. tarda in GZM culture medium by HPLC-MS. Samples were collected at 0, 1, 6, 24 h. Abbreviations are as follows: IAld, indole-3-carboxaldehyde; and IEt, tryptophol. Extracted ions were selected for IAld (m/z 145), IEt, (m/z 161), and Indole (m/z 117). (E) Chemical profiles of Trp-Indole derivatives from supernatants of various commensal bacteria in GZM medium for 1 day of cultivation. Y-axis values represent production of Trp-Indole derivatives normalized to CFU, with each strain beginning at zero. (F) Tg(neurod1:Gcamp6f) zebrafish stimulated by Indole or IAld. Activated EECs in the intestine are labelled with white arrows. (G) Quantification of EEC Gcamp activity in trpa1b+/+ and trpa1b−/− zebrafish stimulated with Indole or IAld. (H) Schematic of experimental design to test effects of indole and IAld on human or mouse Trpa1. (I) Dose-response analysis of the integrated Calcium 6 fluorescence response above baseline (Fmax-F0; maximal change in Ca2+ influx) as a function of indole and IAld concentration in human TRPA1 expressing HEK-293T cells. (EC50 = 88.7 µM, 68.2–114.7 µM 95% CI for indole; and, EC50 = 77.7 µM, 66.8–91.8 µM 95% CI for IAld). Concentration-response data were normalized to 1 mM cinnamaldehyde (CAD), a known TRPA1 agonist. Data represent the mean of 3–4 experiments, each performed with 3–4 replicates. (J) Dose-response analysis of A967079 inhibition of Indole and IAld induced Ca2+ influx. (IC50 = 149.6 nM, 131.3–170.8 nM 95% CI for Indole; and, IC50 = 158.1 nM, 135.4 – 185.6 µM 95% CI for IAld). Concentration-response data of A967079 inhibition was normalized to response elicited by 100 µM agonist (Indole or IAld). (K-N) In vivo calcium imaging of vagal sensory ganglia in WT or EEC ablated Tg(neurod1:Gcamp6f); Tg(neurod1:TagRFP) zebrafish gavaged with PBS, Indole or IAld. (O) Quantification of individual vagal sensory ganglia cell Gcamp6f fluorescence intensities in WT or EEC ablated zebrafish gavaged with PBS or 1mM Indole. (P) Schematic of amperometric measurements to examine the effects of indole on 5-HT secretion in mouse and human small intestinal tissue. (Q) Indole caused a significant increase in 5-HT secretion in mouse duodenum; however, no such effects were observed in the presence of Trpa1 antagonist HC030031. (R) Indole caused a significant increase in 5-HT secretion in human ileum; however, no such effects were observed in the presence of Trpa1 antagonist HC030031. Data in B, C, G, Q, R are presented as mean +/− SD. One-way ANOVA with Tukey’s post test was used in B and Q, Student’s t-test was used in C, H and paired one-way ANOVA with Tukey’s post test was used in P-R. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
We next tested if E. tarda tryptophan catabolites are sufficient to activate EECs. Indole and IAld, but not other tested tryptophan catabolites, strongly activated zebrafish EECs in a trpa1b-dependent manner (Fig. 7F–G, Fig. S7F and Video 4). Indole and IAld also activated the human TRPA1 receptor transfected into HEK cells (Fig. 7H–J and Fig. S7I–J). Both indole and IAld exhibited full TRPA1 agonist activity with an efficiency comparable to cinnamaldehyde (CAD), a well characterized TRPA1 activator (Fig. 7I–J and Fig. S7I–J) (Macpherson et al., 2007). Both indole and IAld also activated mouse Trpa1, but in a less potent manner (Fig. S7M). Both indole- and IAld-induced human and mouse Trpa1 activation were blocked by the TRPA1 inhibitor A967079 (Fig. 7J and Fig. S7K–L, N). These results identify the microbial tryptophan catabolites indole and IAld as evolutionarily-conserved agonists of vertebrate TRPA1.
Next, we investigated whether indole and IAld can mimic live E. tarda bacterial stimulation and activate a similar gut-brain pathway through EEC Trpa1 signaling. Our results indicate that enteric delivery of indole or IAld by microgavage increased Gcamp6f fluorescence in a subset of vagal sensory neurons (Fig. 7K–O, Fig. S8E–F) and stimulating zebrafish larvae with indole increases intestinal motility (Fig. S8A–D). This vagal sensory neuron activation induced by enteric indole was abrogated in zebrafish larvae lacking EECs (Fig. 7K–O, Fig. S8E–F). To test if the ability of microbial tryptophan metabolites to induce Trpa1+EEC signaling is conserved in mammals, we used amperometry on fresh tissue sections from human and mouse small intestine to measure the impact of acute indole exposure on 5-HT secretion. Indeed, indole was able to significantly induce 5-HT secretion from both human and mouse small intestine, and this effect was blocked by the Trpa1 inhibitor HC030031 (Fig. 7P–R). These data support our model and further suggest that these microbial tryptophan catabolites may modulate intestinal motility and gut-brain communication in humans.
DISCUSSION
Microbially derived tryptophan catabolites interact with the host through Trpa1
Trpa1 is a primary nociceptor involved in pain sensation and neuroinflammation. Trpa1 can be activated by several environmental chemical irritants and inflammatory mediators (Bautista et al., 2006), however, it was not known if and how Trpa1 might be activated by microbes. Tryptophan is an essential amino acid that is released in the intestinal lumen by dietary protein digestion or microbial synthesis. It is well known that gut microbes can catabolize tryptophan to produce a variety of metabolites, among which indole was the first discovered and often the most abundant (Smith, 1897). These tryptophan-derived metabolites secreted by gut bacteria can act as interspecies and interkingdom signaling molecules. Some microbially-derived tryptophan catabolites including indole and IAld may regulate host immune homeostasis and intestinal barrier function through ligand binding to the transcription factors, Ahr and Pxr (Venkatesh et al., 2014, Zelante et al., 2013). Another microbial tryptophan catabolite, tryptamine, activates epithelial 5-HT4R and increases anion-dependent fluid secretion in the proximal mouse colon (Bhattarai et al., 2018). Though several tryptophan metabolites including IAld can act as Ahr agonists (Zelante et al., 2013), conflicting effects of indole on AhR activation have been reported (Heath-Pagliuso et al., 1998, Hubbard et al., 2015, Jin et al., 2014). Our results indicate that E. tarda or Trpa1+EEC-induced intestinal motility is not mediated via AhR (Fig. S8G–L). Whether other host receptors can recognize microbially derived tryptophan catabolites was previously unknown. Here, we present evidence that bacteria-derived tryptophan catabolites activate Trpa1 in zebrafish, human, and mouse. A previous study suggested that indole also activates the yeast TRP channel homolog TRPY1 (John Haynes et al., 2008). This together with our findings point to an ancient role for TRP channels in microbial metabolite sensing. Our results indicate that intestinal colonization by bacteria that produce high levels of tryptophan catabolites (e.g., E. tarda) leads to detection of those catabolites by Trpa1+EECs leading to purging of those bacteria by increased intestinal motility. These discoveries were made possible because E. tarda, but none of the other tested zebrafish commensals, exhibited high capacity to produce and secrete tryptophan catabolites in zebrafish water conditions. Since we did not detect overt pathogenesis in E. tarda-treated zebrafish under those experimental conditions, and since many of the E. tarda induced responses were recapitulated by indole or IAld alone, we speculate that EEC Trpa1 activation and its downstream consequences reported here are separable from E. tarda induced pathogenesis and have broader relevance for host-microbial relationships in the gut.
Trpa1+EECs in humans and rodents are abundant in the small intestine but not in the colon (Yang et al., 2019, Nozawa et al., 2009). Our data demonstrated that microbially derived tryptophan metabolites are restricted to the colon and largely absent in small intestine under normal physiological conditions (Fig. S8G–H), suggesting that Trpa1+EECs may not play a major role to regulate intestinal motility under normal physiological conditions. Instead, Trpa1+EECs may act as a host protective mechanism that detects tryptophan catabolites accumulating in the small intestine due to aberrant microbiota overgrowth or invasion of specific microbes like E. tarda that precociously produce those catabolites, and in response increase intestinal motility to purge that particular community. On one hand, loss or impairment of that protective mechanism may result in overgrowth or dysbiosis of small intestinal microbial communities or an increased risk of enteric infection. On the other hand, excessive or chronic activation of those Trpa1+EECs may result in pathophysiological changes. One such scenario may be small intestinal bacteria overgrowth (SIBO), which is prevalent in patients suffering from diarrhea-dominant irritable bowel syndrome (IBS) (Ghoshal et al., 2017). IBS is a complicated disease that display comorbidities of both impairment of GI motility and CNS symptoms. The cause of SIBO in IBS is incompletely understood although several studies demonstrated that some of the indole producing bacteria like Escherichia coli exhibit high abundance in the small intestine of SIBO associated IBS patients (Ghoshal et al., 2014, Leite et al., 2020, Avelar Rodriguez et al., 2019). Our findings raise the possibility that SIBO leads to an increase of microbial tryptophan metabolite production in the small intestine, which then activates Trpa1+EECs to increases intestinal motility and modulate CNS activity through the vagal nerve, resulting in the complex comorbidities of intestinal and psychiatric disorders in IBS.
Gut microbiota-EEC-ENS communication
Nerve fibers do not penetrate the gut epithelium therefore, sensation is believed to be a transepithelial phenomenon as the host senses gut contents through the relay of information from EECs to the ENS (Gershon, 2004). Using an in vitro preparation of mucosa–submucosa, mechanical or electrical stimulation of mucosa was shown to activate submucosal neuronal ganglia, an effect blocked by a 5-HT1R antagonist (Pan and Gershon, 2000). Consistent with these previous findings, our zebrafish data suggest a model that 5-HT released from Trpa1+EECs stimulates intrinsic primary afferent neurons (IPANs) which then activate secondary neurons to promote intestinal motility through the local enteric EEC-ENS circuitry.
90% of 5-HT in the intestine is produced by EC cells, and therefore, EC cell 5-HT secretion was thought to be important in regulating intestinal motility (Gershon, 2013). This hypothesis, however, was challenged by recent findings that depletion of EC 5-HT production in Tph1−/− mice had only minor effects on gastric emptying, intestinal transit, and colonic motility (Li et al., 2011). Therefore, the physiological role of EC 5-HT production and secretion remains unclear. Our data suggest that EEC 5-HT production may be necessary for intestinal motility changes in response to environmental chemical or microbial stimuli, but not for intestinal motility under normal physiological conditions. Mice raised germ-free displayed lower 5-HT content in the colon, however no significant difference of 5-HT production was observed in the small intestine compared to colonized mice (Yano et al., 2015). Whether gut microbiota regulate small intestinal 5-HT secretion and signaling remains unknown. Our data suggest a model in which specific microbial communities or constituent species stimulate 5-HT secretion from Trpa1+EECs to modulate small intestinal motility by producing tryptophan catabolites. This may provide a mechanism by which gut microbiota can regulate 5-HT signaling in the small intestine. Indole, IAld and other tryptophan catabolites are produced by a wide range of gut bacteria, so we expect our results to be applicable to commensal and pathogenic bacteria and their host interactions.
Gut microbiota-EEC-CNS communication
The vagus nerve is the primary sensory pathway by which visceral information is transmitted to the CNS. Recent evidence suggests that the vagus nerve may play a role in communicating gut microbial information to the brain (Fulling et al., 2019, Breit et al., 2018, Bonaz et al., 2018). For example, the beneficial effects of Bifidobacterium longum and Lactobacillus rhamnosus in neurogenesis and behavior were abolished following vagotomy (Bercik et al., 2011, Bravo et al., 2011). However, direct evidence for whether and how vagal sensory neurons perceive and respond to gut bacteria has been lacking. Our results demonstrate that bacterial tryptophan catabolites activate vagal sensory ganglia through EEC Trpa1 signaling. Previous findings have shown that EC cells transmit microbial metabolite and chemical irritant stimuli to pelvic fibers from the spinal cord dorsal root ganglion (Bellono et al., 2017). Our findings here demonstrate that, in addition to spinal sensory nerves, EEC-vagal signaling is an important pathway for transmitting specific gut microbial signals to the CNS. The vagal ganglia project directly onto the hindbrain, and that vagal-hindbrain pathway has key roles in appetite and metabolic regulation (Grill and Hayes, 2009, Han et al., 2018, Travagli et al., 2006, Berthoud et al., 2006). Our findings raise the possibility that certain tryptophan catabolites, including indole, may directly impact these processes as well as emotional behavior and cognitive function (Jaglin et al., 2018). If so, this pathway could be manipulated to treat gut microbiota-associated neurological disorders.
STAR METHODS
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, John F. Rawls (john.rawls@duke.edu).
Materials Availability
Zebrafish strains and plasmids and bacterial strains generated and used in this study are available upon request from the Lead Contact.
Data and Code Availability
Sequencing reads generated as part of this study are available at Gene Expression Omnibus accession GSE151711.
Experimental Model and Subject Details
Zebrafish strains and husbandry
All zebrafish experiments conformed to the US Public Health Service Policy on Humane Care and Use of Laboratory Animals, using protocol numbers A115-16-05 and A096-19-04 approved by the Institutional Animal Care and Use Committee of Duke University. For experiments involving conventionally raised zebrafish larvae, adults were bred naturally in system water and fertilized eggs were transferred to 100mm petri dishes containing ~25 mL of egg water at approximately 6 hours post-fertilization. The resulting larvae were raised under a 14 h light/10 h dark cycle in an air incubator at 28°C at a density of 2 larvae/mL water. All the experiments performed in this study ended at 6 dpf unless specifically indicated. The strains used in this study are listed in Key Resources Table. All lines were maintained on a mixed Ekkwill (EKW) background.
Bacterial strains and growing conditions
All bacterial strains in this study were cultured at 30°C in Trypticase soy broth (TSB) or Gnotobiotic zebrafish medium (GZM) (Pham et al., 2008). Tryptic Soy Agar (TSA) plate was used for streaking bacterial from glycerol stock or performing colony forming unit (CFU) experiments. The antibiotic carbenicillin was used to select E. tarda LSE that express mCherry at the working concentration of 100 µg/mL.
Method Details
Generating transgenic zebrafish
The Gateway Tol2 cloning approach was used to generate the neurod1:CaMPARI and neurod1:cre plasmids (Kawakami, 2007, Kwan et al., 2007). The 5kb pDONR-neurod1 P5E promoter was previously reported (McGraw et al., 2012) and generously provided by Dr. Hillary McGraw. The pME-cre plasmid as reported previously (Cronan et al., 2016) was generously donated by Dr. Mark Cronan. The pcDNA3-CaMPARI plasmid was reported previously (Fosque et al., 2015) and obtained from Addgene. The CaMPARI gene was cloned into pDONR-221 plasmid using BP clonase (Invitrogen, 11789–020) to generate PME-CaMPARI. pDONR-neurod1 P5E and PME-CaMPARI were cloned into pDestTol2pA2 using LR Clonase (ThermoFisher,11791). Similarly, pDONR-neurod1 P5E and pME-cre were cloned into pDestTol2CG2 containing a cmlc2:EGFP marker. The final plasmid was sequenced and injected into the wild-type EKW zebrafish strain and the F2 generation of alleles Tg(neurod1:CaMPARI)rdu78 and Tg(neurod1:cre; cmlcl2:EGFP)rdu79 were used for this study.
To make transgenic lines, that permit specific EEC ablation, we used a Tg(neurod1:cre) and TgBAC(gata5:loxp-mCherry-stop-loxp-DTA) transgenic system. This system consists of two transgene alleles that we generated - one expressing Cre recombinase from the neurod1 promoter (in EECs, CNS, and islets) and a second expressing the diphtheria toxin (DTA) in gata5+ cells (in EECs, other IECs, heart, and perhaps other cell types) only in the presence of Cre (Fig. 3F). As the only cells known to co-express neurod1 and gata5 in the zebrafish larvae, EECs are ablated whereas non-EEC cell populations, including islets and the CNS, remain unaffected (Fig. 3G). A small percentage of EECs remained in the distal intestine presumably due to the low level of gata5 expression in that region (Fig. S4C). The method for generating Tg(neurod1:cre) was described above. To generate the TgBAC(gata5:loxp-mCherry-stop-loxp-DTA) transgenic line, the translational start codon of gata5 in the BAC clone DKEYP-73A2 was replaced with the loxP-mCherry-STOP-loxP-DTA (RSD) cassette by Red/ET recombineering technology (GeneBridges). For recombination with arms flanking the RSD cassette, the 5’ homologous arm used was a 716 bp fragment upstream of the start codon and the 3’ homologous arm was a 517 bp downstream fragment. The vector-derived loxP site was replaced with an I-SceI site using the same technology. The final BAC was purified using the Qiagen Midipre kit, and coinjected with I-SceI into one-cell stage zebrafish embryos. The full name of this transgenic line is Tg(gata5:loxP-mCherry-STOP-loxP-DTA)pd315.
Tg(tph1b:mCherry-NTR)pd275 zebrafish were generated using I-SceI transgenesis in an Ekkwill (EK) background. Golden Gate Cloning with BsaI-HF restriction enzyme (NEB) and T4 DNA ligase (NEB) was used to generate the tph1b:mCherry-NTR plasmid by cloning the 5kb tph1b promoter sequence (tph1bP GG F: GGTCTCGATCGGtctaaggtgaatctgtcacattc; tph1bP GG R: GGTCTCGGCTACggatggatgctcttgttttatag), mCherry (mC GG F: GGTCTCGTAGCC gccgccaccatggtgag; mC GG2 R: GGTCTCGGTACCcttgtacagctcgtccatgccgcc), a P2A polycistronic sequence and triple mutant variant nitroreductase (Mathias et al., 2014) (mutNTR GG F: GGTCTCGGTACCtacttgtacaagggaagcggagc; mutNTR GG2 R: GGTCTCCCATGC caggatcggtcgtgctcga), into a pENT7 vector backbone with a poly-A tail and I-SceI sites (pENT7 mCN GG F: GGTCTCGCATGGacacctccccctgaacctg; pENT7 mCN GG R: GGTCTCCCGATC gtcaaaggtttggggtccgc). 500 pL of 25 ng/μL plasmid, 333 U/mL I-SceI (NEB), 1x I-SceI buffer, 0.05% Phenol Red (Sigma-Aldrich) solution was injected into EK 1-cell zebrafish embryos. F0 founders were discovered by screening for fluorescence in outcrossed F1 embryos.
RNA sequencing and bioinformatic analysis
To isolate zebrafish EECs and other IECs, we crossed two transgenic zebrafish lines, one that specifically expresses enhanced green fluorescent protein (EGFP) in all intestinal epithelial cells (TgBAC(cldn15la:EGFP)) (Alvers et al., 2014) and a second that expresses red fluorescent protein (RFP) in EECs, pancreatic islets, and the central nervous system (CNS) (Tg(neurod1:TagRFP)) (McGraw et al., 2012). The FACS-isolated EECs were identified by cldn15la:EGFP+; neurod1: TagRFP+; and the other IECs were identified by cldn15la:EGFP+; neurod1:TagRFP-. Conventionalized (CV) and germ-free (GF) TgBAC(cldn15la:EGFP); Tg(neurod1:TagRFP) ZM000 fed zebrafish larvae were derived and reared using our published protocol (Pham et al., 2008) for Flow Activated Cell Sorting (FACS) to isolate zebrafish EECs and other IECs. The protocol for FACS was adopted from a previous publication (Espenschied et al., 2019). Replicate pools of 50–100 double transgenic TgBAC(cldn15la:EGFP); Tg(neurod1:TagRFP) zebrafish larvae were euthanized with Tricaine and washed with deyolking buffer (55 mM NaCl, 1.8 mM KCl and 1.25 mM NaHCO3) before they were transferred to dissociation buffer [HBSS supplemented with 5% heat-inactivated fetal bovine serum (HI-FBS, Sigma, F2442) and 10 mM HEPES (Gibco, 15630–080)]. Larvae were dissociated using a combination of enzymatic disruption using Liberase (Roche, 05 401 119 001, 5 μg/mL final), DNaseI (Sigma, D4513, 2 μg/mL final), Hyaluronidase (Sigma, H3506, 6 U/mL final) and Collagenase XI (Sigma, C7657, 12.5 U/mL final) and mechanical disruption using a gentleMACS dissociator (Miltenyi Biotec, 130-093-235). 400 μL of ice-cold 120 mM EDTA (in 1x PBS) was added to each sample at the end of the dissociation process to stop the enzymatic digestion. Following addition of 10 mL Buffer 2 [HBSS supplemented with 5% HI-FBS, 10 mM HEPES and 2 mM EDTA], samples were filtered through 30 μm cell strainers (Miltenyi Biotec, 130-098-458). Samples were then centrifuged at 1800 rcf for 15 minutes at room temperature. The supernatant was decanted, and cell pellets were resuspended in 500 μL Buffer 2. FACS was performed with a MoFlo XDP cell sorter (Beckman Coulter) at the Duke Cancer Institute Flow Cytometry Shared Resource. Single-color control samples were used for compensation and gating. Viable EECs or IECs were identified as 7-AAD negative.
Samples from three independent experimental replicates were performed. 250–580 EECs (n=3 for each CV and GF group) and 100 IECs (n=3 for each CV and GF group) from each experiment were used for library generation and RNA sequencing. Total RNA was extracted from cell pellets using the Argencourt RNAdvance Cell V2 kit (Beckman) following the manufacturer’s instructions. RNA amplification prior to library preparation had to be performed. The Clontech SMART-Seq v4 Ultra Low Input RNA Kit (Takara) was used to generate full-length cDNA. mRNA transcripts were converted into cDNA through Clontech’s oligo(dT)-priming method. Full length cDNA was then converted into an Illumina sequencing library using the Kapa Hyper Prep kit (Roche). In brief, cDNA was sheared using a Covaris instrument to produce fragments of about 300 bp in length. Illumina sequencing adapters were then ligated to both ends of the 300bp fragments prior to final library amplification. Each library was uniquely indexed allowing for multiple samples to be pooled and sequenced on two lanes of an Illumina HiSeq 4000 flow cell. Each HiSeq 4000 lane could generate >330M 50bp single end reads per lane. This pooling strategy generated enough sequencing depth (~55M reads per sample) for estimating differential expression. Sample preparation and sequencing was performed at the GCB Sequencing and Genomic Technologies Shared Resource.
Zebrafish RNA-seq reads were mapped to the danRer10 genome using HISAT2(Galaxy Version 2.0.5.1) using default settings. Normalized counts and pairwise differentiation analysis were carried out via DESeq2 (Love et al., 2014) with the web based-galaxy platform: https://usegalaxy.org/. For the purpose of this study, we only displayed the CV EEC (n=3) and CV IEC (n=3) comparison and analysis in the Results section. The default significance threshold of FDR < 5% was used for comparison. Hierarchical clustering of replicates and a gene expression heat map of RNA-seq data were generated using the online expression heatmap tool: http://heatmapper.ca/expression/. The human and mouse RNA-seq raw counts data were obtained from the NCBI GEO repository: human, GSE114853; mouse, GSE114913 (Roberts et al., 2019). Pairwise differentiation analysis of human jejunum CHGA+ (n=11) and CHGA− (n=11) and mouse duodenum Neurod1+ (n=3) and Neurod1− (n=3) was performed using DESeq2. The mouse and zebrafish ortholog Gene ID conversion was downloaded from Ensemble. The genes that were significantly enriched (PFDR<0.05) in the human and mouse EEC data sets were used to query the zebrafish EEC RNA seq dataset and data were plotted using Graphpad Prism7. RNA-seq data generated in this study can be accessed under Gene Expression Omnibus accession GSE151711.
Recording in vivo EEC activity
CaMPARI undergoes permanent green-to-red photoconversion (PC) under 405 nm light when calcium is present. This permanent conversion records the calcium activity for all areas illuminated by PC-light. Red fluorescence intensity correlates with calcium activity during photoconversion (Fosque et al., 2015). In the Tg(neurod1:CaMPARI) zebrafish line, the CaMPARI (calcium-modulated photoactivatable ratiometric integrator) transgene is expressed under control of the −5kb promoter cloned from the zebrafish neurod1 locus. CaMPARI mRNA is transcribed and the CaMPARI protein is expressed in cells that are able to activate the neurod1 promoter. There are multiple cell types in the zebrafish body that are sufficient to activate the neurod1 promoter, including all EECs in the intestine (Ye et al., 2019). CaMPARI protein is a calcium indicator protein that binds calcium and converts from green fluorescence to red fluorescence in the presence of UV light. This protein is engineered and described in detail in a previous publication (Fosque et al., 2015). We use this transgenic model to measure the level of intracellular calcium in EECs. Similar to neurons, it is well known that when extracellular stimulants act on various receptors on EECs, this leads to an increase of intracellular calcium either due to calcium influx through calcium channels in the plasma membrane or release of calcium stored in the ER. Through either of these pathways, increased intracellular calcium then directly triggers EECs to release hormone/neurotransmitter vesicles. To record in vivo EEC activity using the CaMPARI platform, conventionally raised Tg(neurod1:CaMPARI) zebrafish larvae were sorted at 3 dpf and maintained in Gnotobiotic Zebrafish Media (GZM) (Pham et al., 2008) with 1 larvae/mL density. At 6 dpf, for each experimental group, ~20 larvae were transferred into 50mL conical tubes in 2 mL GZM medium. The larvae were adjusted to the s environment for 30 mins before stimuli were added to each conical tube. For nutrient stimulation, since linoleate, oleate and laurate are not soluble in water, a bovine serum albumin (BSA) conjugated fatty acid solution was generated as described previously (Ye et al., 2019). 2 mL linoleate, oleate, laurate, butyrate or glucose was added to the testing tube containing ~20 zebrafish larvae in 3 mL GZM. The final stimulant concentrations were: linoleate (1.66 mM), oleate oleate (1.66 mM), laurate (1.66 mM), butyrate (2 mM) and glucose (500 mM). Zebrafish larvae were stimulated for 2 mins (fatty acids) or 5 mins (glucose) before the UV pulse. For bacterial stimulation, single colonies of the different bacterial strains were cultured aerobically in tryptic soy broth (TSB) at 30°C overnight (rotating 50–60 rpm, Thermo Fisher Tissue Culture Rotator CEL-GRO #1640Q)(see strains listed in Key Resources Table). O/N TSB cultured bacteria were harvested, washed with GZM and resuspended in 2 mL GZM. 2 mL bacteria were then added to a test tube containing ~20 zebrafish larvae in 3 mL GZM. The final concentration of the bacterial is ~ 108 CFU/ml. Zebrafish were then stimulated for 20mins before treated with a UV pulse. A customized LED light source (400 nm–405 nm, Hongke Lighting CO. LTD) was used to deliver a UV light pulse (100 W power, DC32–34 V and 3500 mA) for 30 seconds. Following the UV pulse, zebrafish larvae were transferred to 6-well plates. To block spontaneous intestinal motility and facilitate in vivo imaging, zebrafish larvae were incubated in 20 µM 4-DAMP (mAChR blocker), 10 µM atropine (mAChR blocker) and 20 µM clozapine (5-HTR blocker) for 30 mins. Zebrafish larvae were then anesthetized with Tricaine (1.64 mg/ml) and mounted in 1% low melting agarose and imaged using a 780 Zeiss upright confocal microscope in the Duke Light Microscope Core Facility. Z-stack confocal images were taken of the mid-intestinal region in individual zebrafish. The laser intensity and gain were set to be consistent across different experimental groups. The resulting images were then processed and analyzed using FIJI software (Schindelin et al., 2012). To quantify the number of activated EECs, the color threshold was set for the CaMPARI red channel. EECs surpassing the color threshold were counted as activated EECs. The CaMPARI green channel was used to quantify the total number of EECs in each sample. The ratio of activated EECs to the total EEC number was calculated as the percentage of activated EECs. As reported in Fig. S1A–F, in Tg(neurod1:CaMPARI) zebrafish model, in addition to EECs, CaMPARI is also expressed in other neurod1+ cells including CNS and pancreatic islet. Therefore, the Tg(neurod1:CaMPARI) model can also be used to measure the activity of the CNS and pancreatic islet. However, the method we described above permit us to specifically analyze EEC signal through restricting our image inquiry in the middle intestine, a region in which only EECs express CaMPARI.
To record in vivo EEC activity using the Tg(neurod1:Gcamp6f) system, we used our published protocol with slight modification (Ye et al., 2019). In brief, unanesthetized zebrafish larvae were gently mounted in 3% methylcellulose. Excess water was removed and zebrafish larvae were gently positioned with right side up. Zebrafish were then moved onto an upright Leica M205 FA fluorescence stereomicroscope equipped with a Leica DFC 365FX camera. The zebrafish larvae were allowed to recover for 2mins before 100 µL of test agent was pipetted directly in front of the mouth region. Images were then recorded every 10 seconds. The stimulants used in this study are listed in Supplemental Table 1. The data shown in Fig. 2O–R, depicting the EEC responses to E. tarda stimulation, were obtained by mounting unanesthetized zebrafish larvae in 1% low melting agarose. A window (5 × 5 mm) was cut to expose the head region of the zebrafish. 10 µL of E. tarda culture [~109 Colony Forming Unit (CFU)] were delivered at the zebrafish mouth area. Images were recorded every 10 secs for 20 mins. Image processing and analysis were performed using FIJI software. Time-lapse fluorescence images were first aligned to correct for experimental drift using the plugin “align slices in stack.” Normalized correlation coefficient matching and bilinear interpolation methods for subpixel translation were used for aligning slices (Tseng et al., 2012). The plugin “rolling ball background subtraction” with the rolling ball radius=10 pixels was used to remove the large spatial variation of background intensities. The Gcamp6f fluorescence intensity in the intestinal region was then calculated for each time point. The ratio of maximum fluorescence (Fmax) and the initial fluorescence (F0) was used to measure EEC calcium responses.
Immunofluorescence staining and imaging
Whole mount immunofluorescence staining was performed as previously described (Ye et al., 2019). In brief, ice cold 2.5% formalin was used to fix zebrafish larvae overnight at 4°C. The samples were then washed with PT solution (PBS+0.75%Triton-100). The skin and remaining yolk were then removed using forceps under a dissecting microscope. The deyolked samples were then permeabilized with methanol for more than 2 hrs at −20°C. Samples were then blocked with 4% BSA at room temperature for more than 1 hr. The primary antibody was diluted in PT solution and incubated at 4°C for more than 24 hrs. Following primary antibody incubation, the samples were washed with PT solution and incubated overnight with secondary antibody with Hoechst 33342 for DNA staining. Imaging was performed with Zeiss 780 inverted confocal and Zeiss 710 inverted confocal microscopes with 40× oil lens. The primary antibodies were listed in Supplemental Table 1. The secondary antibodies in this study were from Alexa Fluor Invitrogen were used at a dilution of 1:250.
To quantify vagal activity by pERK staining, we used a published protocol with slight modification (Randlett et al., 2015). Zebrafish larvae were quickly collected by funneling through a 0.75 mm cell strainer and dropped into a 5mL petri dish containing ice cold fix buffer (2.5% formalin+ 0.25% Triton 100). Larvae were fixed overnight at 4°C, then washed 3 times in PT (PBS+ 0.3% Triton 100), treated with 150 mM Tris-HCl (PH=9) for 15 mins at 70°C, washed with PT and digested with 0.05% trypsin-EDTA on ice for 45 mins. Following digestion, samples were then washed with PT and transferred into block solution [PT + 1% bovine serum albumin (BSA, Fisher) + 2% normal goat serum (NGS, Sigma) + 1% dimethyl sulfoxide (DMSO)]. The primary antibodies [pERK (Cell signaling); tERK (Cell signaling); GFP (Aves Lab)] were diluted in block solution (1:150 for pERK; 1:150 for tERK and 1:500 for GFP) and samples were incubated in 100 µl of primary antibody overnight at 4°C. Following primary antibody incubation, samples were then washed with PT and incubated with secondary antibody overnight at 4°C. Samples were then washed with PBS, mounted in 1% LMA and imaged using a Zeiss 780 upright confocal microscope.
Zebrafish E. tarda colonization
For E. tarda colonization experiments, fertilized zebrafish eggs were collected, sorted and transferred into a cell culture flask containing 80 mL GZM at 0 dpf. At 3 dpf, dead embryos and 60 mL GZM were removed and replaced with 50 mL fresh GZM in each flask. To facilitate consistent commensal gut bacterial colonization, an additional 10 mL of filtered system water (5 μm filter, SLSV025LS, Millipore) were added to each flask. Overnight E. tarda mCherry (Ampr, see details in Supplemental Table 1) culture was harvested, washed three times with GZM. 150 µL of GZM-washed E. tarda mCherry culture were inoculated into each flask. The E. tarda concentration is ~106 CFU/ml. Daily water changes (60 ml) was performed and 200 µL autoclaved solution of ZM000 food (ZM Ltd.) was added from 3 dpf to 6 dpf as previously described (Pham et al., 2008). At 6 dpf, zebrafish larvae were subjected to fluorescence imaging analysis or CFU quantification. For fluorescence imaging analysis, zebrafish larvae were anesthetized with Tricaine (1.64 mg/ml), mounted in 3% methylcellulose and imaged with a Leica M205 FA upright fluorescence stereomicroscope equipped with a Leica DFC 365FX camera. For CFU quantification, digestive tracts were dissected and transferred into 1 mL sterile PBS which was then mechanically disassociated using a Tissue-Tearor (BioSpec Products, 985370). 100 µL of serially diluted solution was then spread on a Tryptic soy agar (TSA) plate with Carbenicillin (100 µg/ml) and cultured overnight at 30°C under aerobic conditions. The mCherry+ colonies were quantified from each plate and E. tarda colony forming units (CFUs) per fish were calculated.
Zebrafish microgavage and chemical treatment
For delivering bacterial or chemicals specifically to the intestine, we adopted our established microgavage technique (Cocchiaro and Rawls, 2013). Zebrafish were anesthetized with 1 mg/mL α-Bungarotoxin (α-BTX) and the gavage procedure was performed as previously described using microinjection station (Cocchiaro and Rawls, 2013). For bacteria gavage experiments, 1ml of overnight bacterial culture was harvested, pelleted, washed with PBS and resuspended in 100ul PBS. ~ 8nl was then delivered into the zebrafish intestine using microgavage. The gavaged zebrafish was then transferred into egg water and mounted in 1% LMA for imaging. For chemical gavage experiments, ~ 8nl of AITC (100mM), indole (1mM) and IAld (1mM) was gavaged into the intestine.
For Trpa1 inhibition, Trpa1 antagonist HC030031 (280µM) was treated 2 hours before and during the 30 mins of E. tarda stimulation. For AhR inhibition, two AhR inhibitors, CH030031 and folic acid, were selected based on previous publications (Puyskens et al., 2020, Kim et al., 2020). CH030031 is a well-established specific AhR inhibitor (Choi et al., 2012). Whereas folic acid is shown to act as a competitive AhR antagonist at the concentration as low as 10ng/ml (Kim et al., 2020). For the E. tarda treatment experiment, DMSO, CH030031 (1µM) or folic acid (10µM) was added into zebrafish water at 3 dpf zebrafish at the same time as E. tarda administration. The AhR inhibitors were replenished during daily water changes, and zebrafish were analyzed at 6 dpf. For the Optovin-UV experiment, overnight Optovin treated zebrafish were treated for 2 hours with DMSO, CH030031 (10µM) or folic acid (10µM). As demonstrated by previous study, 2-hour 10µM and 3-day 0.5µM CH030031 treatment is sufficient to inhibit larve zebrafish AhR signaling (Puyskens et al., 2020, Sun et al., 2019b, Yue et al., 2017). The concentration of FA was chosen based on zebrafish tolerance and a previous study shown the treatment of early zebrafish embryos with 0.05µM FA inhibits AhR signaling (Yue et al., 2017).
Optic EEC activation
For EEC Trpa1 activation using the Optovin platform, zebrafish larvae were treated with 10 µM Optovin overnight. Following Optovin treatment, unanesthetized zebrafish were mounted in 1% LMA and imaged under a 780 upright Zeiss confocal microscope using 20× water objective lenses. For all the experiments, the mid-intestine region was imaged (Fig. S5D). The intestinal epithelium was selected as the region of interest (ROI) (Fig. S5A). Serial images were obtained at 1 s/frame. A 405 nm pulse of light was applied to the ROI at 1 pulse/10s. For some experiments (Fig. 4D–F, Fig. S5B–G), the images were obtained at 10s/frame. When measuring Optovin effects on intestinal motility in ret−/−, sox10−/− or tph1b−/− zebrafish larvae, embryos were collected from heterozygous zebrafish. ret−/− zebrafish were identified by lack of ENS and deflated swim bladder (Knight et al., 2011), sox10−/− zebrafish were identified by lack of pigment (Rolig et al., 2017), and tph1b−/− zebrafish were identified by PCR-based genotyping (Tornini et al., 2017).
Photoactivation of channelrhodopsin (ChR2) in EECs was performed in Tg(neurod1:Gal4, cmlc2;EGFP); Tg(UAS:ChR2-mCherry) transgenic zebrafish. In this model, ChR2 expression in EECs is mosaic. At 6 dpf, unanesthetized zebrafish larvae were mounted in 1% LMA. Photoactivation and imaging were performed with a Zeiss 780 upright confocal using 20× water objective lenses. Individual ChR2+ EECs were selected as ROI (Fig. S5H, I). Serial images were obtained at 1 s/frame. The 488 nm and 458 nm pulses were applied to the selected ROI at 1 pulse/s. For selectively activating trpa1b+ or trpa1b− ChR2 expressing EECs, Tg(neurod1:Gal4, cmlc2;EGFP); Tg(UAS:ChR2-mCherry) was crossed with TgBAC(trpa1b:EGFP). In each zebrafish, either Trpa1+ChR2+ EEC or Trpa1-ChR+ EEC was selected to activate and examine the motility pre and post activation. Each dot in updated Fig. 4L and Fig. S3H represent data from individual zebrafish. A snapshot of the intestinal area was obtained to determine the trpa1b+ChR2+ and trpa1b-ChR2+ EECs (Fig. S5H, I) and light pulses were applied to the selected EECs as indicated above. Due to the mosaic expression of ChR2 in the EECs in the Gal4-UAS transgenic system, the ChR2+EECs in both proximal and middle intestinal regions are selected.
To determine whether Optovin-UV or ChR2 was sufficient to activate EECs, Tg(neurod1:Gcamp6f) zebrafish were used. To facilitate EEC calcium imaging under the confocal microscope, zebrafish larvae were incubated in 20 µM 4-DAMP, 10 µM atropine and 20 µM clozapine for 30 mins before mounting in 1% LMA to reduce spontaneous motility. The Gcamp6f signal was recorded with 488nm laser intensity less than 0.5.
The zebrafish intestinal motility is quantified through recorded image series of zebrafish intestine using the method similar as previously described (Ganz et al., 2018). Intestinal µ velocity and ν velocity were used to estimate intestinal motility in zebrafish as previously described using the PIV-Lab MATLAB app (Ganz et al., 2018). A positive value of the µ velocity indicates an anterograde intestinal movement and a negative value of the µ velocity indicates a retrograde intestinal movement. The time-course µ velocity number is plotted as heatmaps. When calculating the mean velocity, only the mean velocity magnitude was calculated, which therefore doesn’t account for the movement direction. The MTrackJ FIJI plugin was used to quantify the mean velocity magnitude (Meijering et al., 2012).
To assess whether Trpa1+EEC activation induced intestinal motility change is due to the indirect communication through vagal afferent and efferent system, we anatomically disconnected zebrafish CNS with the intestine by decapitating. Optovin-treated unanesthetized zebrafish were mounted and placed on the 780 Zeiss upright confocal station as described above. The zebrafish head was then removed with a razor blade. The same imaging and 405nm activation of the mid-intestinal region was performed as described above.
Enteric cholinergic neuron and vagal ganglion calcium imaging
TgBAC(chata:Gal4); Tg(UAS:Gcamp6s); Tg(NBT:DsRed) or TgBAC(chata:Gal4); Tg(UAS:Gcamp6s); Tg(UAS:NTR-mCherry) zebrafish were used to record in vivo calcium activity in enteric cholinergic neurons. The NBT promoter labels all ENS neurons while the Chata promoter labels only cholinergic enteric neurons. DsRed or mCherry fluorescence was used as reference for cholinergic neuron Gcamp quantification. Zebrafish larvae were incubated in 20 µM 4-DAMP for 30 mins before mounting in 1% LMA to reduce spontaneous motility and facilitate in vivo imaging using a Zeiss 780 upright confocal microscope with 20× water lenses. Serial images were taken at 5 s/frame. To record cholinergic neuron calcium activation, zebrafish was pretreated with Optovin and 40 nm light was applied at the frequency of 1 pulse/5s to the intestinal epithelium ROI. The Gcamp6s to DsRed fluorescence in cholinergic neurons was calculated for recorded.
Tg(neurod1:Gcamp6f); Tg(neurod1:TagRFP) zebrafish were used to record vagal sensory ganglia calcium activity in vivo. Zebrafish were anesthetized with 1 mg/mL α-Bungarotoxin (α-BTX) and gavaged with chemical compounds or bacteria as described (Naumann et al., 2016). Zebrafish larvae were mounted in 1% LMA and imaged under a Zeiss 780 upright confocal microscope. Z-stack images of the entire vagal ganglia were collected as serial images at 10 mins/frame and processed in FIJI. Individual vagal sensory neurons were identified and the Gcamp6f to TagRFP fluorescence ratios of individual vagal sensory neurons were calculated.
Quantitative real-time PCR
Quantitative real-time PCR was performed as described previously (Murdoch et al., 2019). In brief, 20 zebrafish larvae digestive tracts were dissected and pooled into 1 mL TRIzol (ThermoFisher, 15596026). mRNA was then isolated with isopropanol precipitation and washed with 70% ethanol. 500ng mRNA was used for cDNA synthesis using the iScript kit (Bio-Rad, 1708891). Quantitative PCR was performed in triplicate 25 μL reactions using 2X SYBR Green SuperMix (PerfeCTa, Hi Rox, Quanta Biosciences, 95055) run on an ABI Step One Plus qPCR instrument using gene specific primers (Supplementary file 1). Data were analyzed with the ΔΔCt method. 18S was used as a housekeeping gene to normalize gene expression.
Mammalian TRPA1 activity analysis
HEK-293T cells were cultured in DMEM (Thermofisher Scientific, Waltham, MA) and supplemented with 10% fetal bovine serum (FBS) (Thermofisher Scientific), penicillin (100 units/mL) and streptomycin (0.1 mg/mL). Cells were plated on 100 mm tissue culture plates coated with poly-D-lysine (Sigma Aldrich, Saint Louis, MO) and grown to ~60% confluence. The cells were transiently transfected for 16–24 hours with either human or mouse orthologs of TRPA1 using Fugene 6 transfection reagents and Opti-MEM (Thermofisher Scientific) according to the manufacturer’s protocol. Subsequently, cells were trypsinized, re-suspended and replated onto poly-D-lysine coated 96-well plates (Krystal black walled plates, Genesee Scientific) at 5×105 cells/mL (100 µL/well) and allowed to grow for another 16–20 hrs prior to the experiments. Cells were maintained as monolayers in a 5% CO2 incubator at 37°C.
Measurements of changes in intracellular Ca2+ concentrations ([Ca2+]i) were performed as described previously (Caceres et al., 2017). In brief, cells in 96-well plates were loaded with Calcium 6, a no-wash fluorescent indicator, for 1.5 hrs (Molecular Devices, San Jose, CA) and then transferred to a FlexStation III benchtop scanning fluorometer chamber (Molecular Devices). Fluorescence measurements in the FlexStation were performed at 37°C (Ex:485 nm, Em: 525 nm at every 1.8 s). After recording baseline fluorescence, agonists (indole, IAld, cinnamaldehyde) were added and fluorescence was monitored for a total of 60 s. To determine the effects of TRPA1 inhibition on agonist response, TRPA1 transfected HEK-293 cells were pretreated with various concentrations of A967079 (Medchem101, Plymouth Meeting, PA), a specific antagonist of TRPA1, and then exposed to either 100 µM indole or IAld. The change in fluorescence was measured as Fmax-F0, where Fmax is the maximum fluorescence and F0 is the baseline fluorescence measured in each well. The EC50 and IC50 values and associated 95% confidence intervals for agonist (Indole and IAld) stimulation of Ca2+ influx and A967079 inhibition of agonist-induced Ca2+ influx, respectively, were determined by non-linear regression analysis with a 4-parameter logistic equation (Graphpad Prism, San Diego, CA). Indole and IAld concentration-response data was normalized to 1 mM cinnamaldehyde for EC50’s calculations and A967079 concentration-response data was normalized to 100 µM indole or IAld for IC50’s calculations.
HPLC-MS analysis of Trp-Indole derivatives
The chemical profiling of Trp-Indole derivatives was performed using 1 L culture of E. tarda. The strain was inoculated in 3 mL of TSB medium and cultivated for 1 day on a rotary shaker at 180 rpm at 30°C under aerobic conditions. After 1 day, 1 mL of E. tarda liquid culture was inoculated in 1 L of TSB medium in a 4-L Pyrex flask. The E. tarda culture was incubated at 30°C for 24 hr under aerobic conditions. For time-course screening, 10 mL from the E. tarda TSB culture was collected at 0, 6, 18, and 24 hours. Each 10 mL sample of E. tarda culture was extracted with 15 mL of ethyl acetate (EtOAc). The EtOAc layer was separated from the aqueous layer and residual water was removed by addition of anhydrous sodium sulfate. Each EtOAc fraction was dried under reduced pressure, then resuspended in 500 μL of 50% MeOH/50% H2O and 50 μL of each sample were analyzed using an Agilent Technologies 6130 quadrupole mass spectrometer coupled with an Agilent Technologies 1200–series HPLC (Agilent Technologies, Waldbron, Germany). The chemical screening was performed with a KinetexⓇ EVO C18 column (100 × 4.6 mm, 5 µm) using the gradient solvent system (10 % ACN/90 % H2O to 100 % ACN over 20 min at a flow rate of 0.7 mL/min).
For HPLC-MS analysis of E. tarda in GZM medium, the remaining 1 L culture of E. tarda in TSB culture was centrifuged at 7,000 rpm for 30 min. Pellets were transferred to 1 L of GZM medium in a 4-L Pyrex flask and cultivated on a rotary shaker at 30°C for 24 hr. For time-course screening, 10 mL from the E. tarda GZM culture was collected at 0, 1, 6, and 24 hours. Sample preparation and HPLC-MS analysis of E. tarda culture GZM medium were performed using same procedures as described above for TSB. Trp-Indole derivatives of E. tarda culture broths were identified by comparing the retention time and extracted ion chromatogram with authentic standards. Extracted ions were selected for Indole (m/z 117, Sigma-Aldrich), IAld (m/z 145, Sigma-Aldrich), IAAld (m/z 159, Ambeed), IEt (m/z 161, Sigma-Aldrich), IAM (m/z 174, Sigma-Aldrich), IAA (m/z 175, Sigma-Aldrich), and IpyA (m/z 203, Sigma-Aldrich).
For HPLC-MS analysis of Trp-indole derivatives from 15 different bacterial strains in TSB medium, each of the strains (Acinetobacter sp. ZOR0008, Aeromonas veronii ZOR0002, Bacillus subtilis 168, Chryseobacterium sp. ZOR0023, Edwardsiella tarda 15974, Edwardsiella tarda 23685, Edwardsiella tarda LSE40, Edwardsiella tarda FL6–60, Enterobacter sp. ZOR0014, Escherichia coli MG1655, Exiguobacterium acetylicum sp. ZWU0009, Plesiomonas sp. ZOR0011, Pseudomonas aeruginosa PAK, Shewanella sp. ZOR0012, and Vibrio sp. ZWU0020) were inoculated in 3 mL of TSB medium and cultivated for 1 day on a rotary shaker at 180 rpm at 30°C under aerobic conditions. After 1 day, 1 mL of each liquid culture was inoculated in 100 mL of TSB medium in 500 mL Pyrex flasks and cultivated on a rotary shaker at 30°C overnight. A 10 mL sample was taken from each culture and extracted and analyzed via HPLC-MS as explained above. CFU was calculated for each bacterial liquid culture and the HPLC-MS data was normalized to the CFU.
For HPLC-MS analysis of Trp-indole derivatives from 15 different bacterial strains in GZM medium, the remaining 100 mL culture of each strain was centrifuged at 4500 rpm for 20 min. Pellets were transferred to 100 mL of GZM medium in 500 mL Pyrex flasks and cultivated on a rotary shaker at 30°C overnight. Sample preparation and HPLC-MS analysis of each GZM culture were performed using the same procedure as described above.
For HPLC-MS analysis of Trp-indole derivatives from murine small intestine and large intestine, three 10-week old female and three 10-week old male conventionally-reared specific pathogen-free C57BL/6J mice were ordered from Jackson Lab. The mice were not fast in advance and euthanized with 5% isoflurane. The 2/5–4/5 portion of the small intestinal region and the colon caudal to cecum was collected from each mouse and transferred to a 50 mL conical tube that was placed on dry-ice. 80% methanol was then added according to the tissue weight (50 µL/mg tissue). The intestine was then homogenized with a Tissue-Tearor (BioSpec Products, 985370). Following homogenizing, the tryptophan metabolites were extracted and analyzed with HPLC-MS as explained above. The relative metabolite abundance was normalized to tissue weight. These mouse experiments conformed to the US Public Health Service Policy on Humane Care and Use of Laboratory Animals, using protocol number A170–17-07 approved by the Institutional Animal Care and Use Committee of Duke University.
Measurement of serotonin release from mouse and human small intestine
These experiments using C57BL/6J mice were approved by the Flinders University Animal Welfare Committee (number 965–19) and human ileum tissue was collected from resected small and large intestine from patients that gave written informed consent under the approval of the Southern Adelaide Clinical Human Research Ethics Committee (number 50.07) as previous (Sun et al., 2019a). Mice were euthanized at 8 to 12 weeks by isoflurane overdose followed by cervical dislocation. The duodenum was removed and placed in Krebs solution oxygenated with 95 % O2, 5 % CO2. A midline incision was made along the duodenum to create a flat sheet, the section was pinned with the mucosal side up in an organ bath lined with Sylgard and containing oxygenated Krebs solution. Serotonin release was measured using amperometry. A carbon-fibre electrode (5-μm diameter, ProCFE; Dagan Corporation, Minneapolis, MN), was lowered above the mucosa and 400 mV potential was applied to the electrode causing oxidation of serotonin (Zelkas et al., 2015). 10mM Indole and/or 50µM HC030031 were applied to tissue by constantly perfusing the bath. The change in amplitude due to serotonin oxidation was recorded using an EPC-10 amplifier and Pulse software (HEKA Electronic, Lambrecht/Pfalz, Germany), and samples at 10 kHz and low-pass filtered at 1 kHz. Data was assessed as peak current during each treatment. Data was analyzed comparing all groups using one-way ANOVA with Tukey’s post-hoc test. For the mouse experiments, 6 independent experiments were performed in 6 mouse duodenal samples. For the human experiments, 4 independent experiments were performed in 3 human samples.
Statistical analysis
The appropriate sample size for each experiment was suggested by preliminary experiments evaluating variance and effects. Using significance level of 0.05 and power of 90%, a biological replicate sample number 8 was suggested for EEC CaMPARI analysis. For each experiment, wildtype or indicated transgenic zebrafish embryos were randomly allocated to test groups prior to treatment. Individual data points, mean and standard deviation are plotted in each figure. The raw data points in each figure are represented as solid dots. Data were analyzed using GraphPad Prism 7 software. For experiments comparing just two differentially treated populations, a Student’s t-test with equal variance assumptions was used. For experiments measuring a single variable with multiple treatment groups, a single factor ANOVA with post hoc means testing (Tukey) was utilized. Statistical evaluation for each figure was marked * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001 or ns (no significant difference, P>0.05).
Supplementary Material
Table S1. Zebrafish EEC RNA-seq data analyzed and compared with mouse and human EEC data, related to Figure 2.
Video 1. Activation of EEC Trpa1 in WT zebrafish, but not EEC ablated zebrafish, increases intestinal motility, related to Figure 4. Time-course video of Tg(neurod1:Gcamp6f) WT and EEC ablated zebrafish before and post Optovin-UV induced EEC Trpa1 activation. WT zebrafish are shown first followed by EEC ablated zebrafish. Anterior is to the left, and dorsal is to the top.
Video 2. Activation of Trpa1+ChR2+EECs, but not Trpa1-ChR2+EECs, increases intestinal motility, related to Figure 4. Time-course videos of TgBAC(trpa1b:EGFP); Tg(neurod1:Gal4); Tg(UAS:ChR2-mCherry) before and post optic activation of Trpa1- EECs and Trpa1+ EECs. Anterior is to the left, and dorsal is to the bottom. Yellow light was delivered specifically to the selected EECs to activate the ChR2 channel. Note that the first frame in each video shows the EGFP channel to identify EECs that do or do not express trpa1b. Also note that the ChR2 EECs are in the intestinal bulb and an retrograde intestinal movement was observed upon Trpa1+ChR2+EEC activation. The third part of the video shows that activation of Trpa1+ChR2+EECs in the mid-intestine induces an anterograde intestinal movement.
Video 3. E. tarda increases intestinal motility, related to Figure 4. Time-course videos of WT zebrafish 30 mins post Aeromonas veronii ZOR0002 or E. tarda FL6–60 gavage. Anterior is to the left, and dorsal is to the top.
Video 4. Indole activates EECs in vivo, related to Figure 7. Time-course videos of trpa1b+/+ (WT) and trpa1b−/− Tg(neurod1:Gcamp6f) zebrafish that are stimulated with indole. Note that there is a basal amount of intestinal motility associated with this methylcellulose preparation that is retained in vehicle-only negative controls (not shown) and in trpa1b mutants. Anterior is to the right, and dorsal is to the top.
Highlights:
Intestinal bacteria activate enteroendocrine cells (EECs) through Trpa1.
Bacterial tryptophan metabolites activate zebrafish, mouse and human Trpa1.
Activated EECs transmit bacterial signals to enteric and vagal nerves in zebrafish.
Bacterial tryptophan metabolites induce mouse and human small intestinal 5-HT secretion.
ACKNOWLEDGEMENTS
This work was supported by NIH R01-DK093399, R01-DK109368, VA-BX002230, and a Pew Scholars Innovation Award from the Pew Charitable Trusts. S.V.J. and S.-E.J. were supported by cooperative agreement U01ES030672 of the NIH CounterACT Program. K.D.P. was supported by R01 GM074057 and R35 HL150713. L.Y. was supported by NIH T32-DK007568. C. D.C. was supported by NIH F32AT010415. D.J.K. was supported by the Australian Research Council, DP190103525. We are grateful to Cecelia Kelly for assistance in mouse experiments, and other members of the Rawls and Liddle labs for feedback and support. The content is solely the responsibility of the authors and does not necessarily represent the views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES:
- ABAYNEH T, COLQUHOUN DJ & SORUM H 2013. Edwardsiella piscicida sp. nov., a novel species pathogenic to fish. J Appl Microbiol, 114, 644–54. [DOI] [PubMed] [Google Scholar]
- ALVERS AL, RYAN S, SCHERZ PJ, HUISKEN J & BAGNAT M 2014. Single continuous lumen formation in the zebrafish gut is mediated by smoothened-dependent tissue remodeling. Development, 141, 1110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AVELAR RODRIGUEZ D, RYAN PM, TORO MONJARAZ EM, RAMIREZ MAYANS JA & QUIGLEY EM 2019. Small Intestinal Bacterial Overgrowth in Children: A State-Of-The-Art Review. Front Pediatr, 7, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAUTISTA DM, JORDT SE, NIKAI T, TSURUDA PR, READ AJ, POBLETE J, YAMOAH EN, BASBAUM AI & JULIUS D 2006. TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell, 124, 1269–82. [DOI] [PubMed] [Google Scholar]
- BAUTISTA DM, PELLEGRINO M & TSUNOZAKI M 2013. TRPA1: A gatekeeper for inflammation. Annu Rev Physiol, 75, 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELLONO NW, BAYRER JR, LEITCH DB, CASTRO J, ZHANG C, O’DONNELL TA, BRIERLEY SM, INGRAHAM HA & JULIUS D 2017. Enterochromaffin Cells Are Gut Chemosensors that Couple to Sensory Neural Pathways. Cell, 170, 185–198 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERCIK P, PARK AJ, SINCLAIR D, KHOSHDEL A, LU J, HUANG X, DENG Y, BLENNERHASSETT PA, FAHNESTOCK M, MOINE D, BERGER B, HUIZINGA JD, KUNZE W, MCLEAN PG, BERGONZELLI GE, COLLINS SM & VERDU EF 2011. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol Motil, 23, 1132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERTHOUD HR, SUTTON GM, TOWNSEND RL, PATTERSON LM & ZHENG H 2006. Brainstem mechanisms integrating gut-derived satiety signals and descending forebrain information in the control of meal size. Physiol Behav, 89, 517–24. [DOI] [PubMed] [Google Scholar]
- BHATTARAI Y, WILLIAMS BB, BATTAGLIOLI EJ, WHITAKER WR, TILL L, GROVER M, LINDEN DR, AKIBA Y, KANDIMALLA KK, ZACHOS NC, KAUNITZ JD, SONNENBURG JL, FISCHBACH MA, FARRUGIA G & KASHYAP PC 2018. Gut Microbiota-Produced Tryptamine Activates an Epithelial G-Protein-Coupled Receptor to Increase Colonic Secretion. Cell Host Microbe, 23, 775–785 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOHORQUEZ DV, SHAHID RA, ERDMANN A, KREGER AM, WANG Y, CALAKOS N, WANG F & LIDDLE RA 2015. Neuroepithelial circuit formed by innervation of sensory enteroendocrine cells. J Clin Invest, 125, 782–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BONAZ B, BAZIN T & PELLISSIER S 2018. The Vagus Nerve at the Interface of the Microbiota-Gut-Brain Axis. Front Neurosci, 12, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BONDURAND N & SHAM MH 2013. The role of SOX10 during enteric nervous system development. Dev Biol, 382, 330–43. [DOI] [PubMed] [Google Scholar]
- BORNSTEIN JC 2006. Intrinsic sensory neurons of mouse gut--toward a detailed knowledge of enteric neural circuitry across species. Focus on “characterization of myenteric sensory neurons in the mouse small intestine”. J Neurophysiol, 96, 973–4. [DOI] [PubMed] [Google Scholar]
- BRAVO JA, FORSYTHE P, CHEW MV, ESCARAVAGE E, SAVIGNAC HM, DINAN TG, BIENENSTOCK J & CRYAN JF 2011. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A, 108, 16050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BREIT S, KUPFERBERG A, ROGLER G & HASLER G 2018. Vagus Nerve as Modulator of the Brain-Gut Axis in Psychiatric and Inflammatory Disorders. Front Psychiatry, 9, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROOKES SJ, SPENCER NJ, COSTA M & ZAGORODNYUK VP 2013. Extrinsic primary afferent signalling in the gut. Nat Rev Gastroenterol Hepatol, 10, 286–96. [DOI] [PubMed] [Google Scholar]
- BROWN JM & HAZEN SL 2015. The gut microbial endocrine organ: bacterially derived signals driving cardiometabolic diseases. Annu Rev Med, 66, 343–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUJAN N, MOHAMMED H, BALBOA S, ROMALDE JL, TORANZO AE, ARIAS CR & MAGARINOS B 2018. Genetic studies to re-affiliate Edwardsiella tarda fish isolates to Edwardsiella piscicida and Edwardsiella anguillarum species. Syst Appl Microbiol, 41, 30–37. [DOI] [PubMed] [Google Scholar]
- CACERES AI, LIU B, JABBA SV, ACHANTA S, MORRIS JB & JORDT SE 2017. Transient Receptor Potential Cation Channel Subfamily M Member 8 channels mediate the anti-inflammatory effects of eucalyptol. Br J Pharmacol, 174, 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHIMEREL C, EMERY E, SUMMERS DK, KEYSER U, GRIBBLE FM & REIMANN F 2014. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep, 9, 1202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHOI EY, LEE H, DINGLE RW, KIM KB & SWANSON HI 2012. Development of novel CH223191-based antagonists of the aryl hydrocarbon receptor. Mol Pharmacol, 81, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COCCHIARO JL & RAWLS JF 2013. Microgavage of zebrafish larvae. J Vis Exp, e4434. [DOI] [PMC free article] [PubMed]
- COLEMAN OI & HALLER D 2017. Bacterial Signaling at the Intestinal Epithelial Interface in Inflammation and Cancer. Front Immunol, 8, 1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CORNELISSEN W, DE LAET A, KROESE AB, VAN BOGAERT PP, SCHEUERMANN DW & TIMMERMANS JP 2000. Electrophysiological features of morphological Dogiel type II neurons in the myenteric plexus of pig small intestine. J Neurophysiol, 84, 102–11. [DOI] [PubMed] [Google Scholar]
- CRONAN MR, BEERMAN RW, ROSENBERG AF, SAELENS JW, JOHNSON MG, OEHLERS SH, SISK DM, JURCIC SMITH KL, MEDVITZ NA, MILLER SE, TRINH LA, FRASER SE, MADDEN JF, TURNER J, STOUT JE, LEE S & TOBIN DM 2016. Macrophage Epithelial Reprogramming Underlies Mycobacterial Granuloma Formation and Promotes Infection. Immunity, 45, 861–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESPENSCHIED ST, CRONAN MR, MATTY MA, MUELLER O, REDINBO MR, TOBIN DM & RAWLS JF 2019. Epithelial delamination is protective during pharmaceutical-induced enteropathy. Proc Natl Acad Sci U S A, 116, 16961–16970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FLORES EM, NGUYEN AT, ODEM MA, EISENHOFFER GT & KRACHLER AM 2020. The zebrafish as a model for gastrointestinal tract-microbe interactions. Cell Microbiol, 22, e13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOGLIA MJ, CAO J, TORNINI VA & POSS KD 2016. Multicolor mapping of the cardiomyocyte proliferation dynamics that construct the atrium. Development, 143, 1688–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOSQUE BF, SUN Y, DANA H, YANG CT, OHYAMA T, TADROSS MR, PATEL R, ZLATIC M, KIM DS, AHRENS MB, JAYARAMAN V, LOOGER LL & SCHREITER ER 2015. Neural circuits. Labeling of active neural circuits in vivo with designed calcium integrators. Science, 347, 755–60. [DOI] [PubMed] [Google Scholar]
- FULLING C, DINAN TG & CRYAN JF 2019. Gut Microbe to Brain Signaling: What Happens in Vagus. Neuron, 101, 998–1002. [DOI] [PubMed] [Google Scholar]
- FURNESS JB, CALLAGHAN BP, RIVERA LR & CHO HJ 2014. The enteric nervous system and gastrointestinal innervation: integrated local and central control. Adv Exp Med Biol, 817, 39–71. [DOI] [PubMed] [Google Scholar]
- FURNESS JB, KUNZE WA & CLERC N 1999. Nutrient tasting and signaling mechanisms in the gut. II. The intestine as a sensory organ: neural, endocrine, and immune responses. Am J Physiol, 277, G922–8. [DOI] [PubMed] [Google Scholar]
- FURNESS JB, RIVERA LR, CHO HJ, BRAVO DM & CALLAGHAN B 2013. The gut as a sensory organ. Nat Rev Gastroenterol Hepatol, 10, 729–40. [DOI] [PubMed] [Google Scholar]
- GANZ J, BAKER RP, HAMILTON MK, MELANCON E, DIBA P, EISEN JS & PARTHASARATHY R 2018. Image velocimetry and spectral analysis enable quantitative characterization of larval zebrafish gut motility. Neurogastroenterol Motil, 30, e13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GERSHON MD 2004. Review article: serotonin receptors and transporters -- roles in normal and abnormal gastrointestinal motility. Aliment Pharmacol Ther, 20 Suppl 7, 3–14. [DOI] [PubMed] [Google Scholar]
- GERSHON MD 2013. 5-Hydroxytryptamine (serotonin) in the gastrointestinal tract. Curr Opin Endocrinol Diabetes Obes, 20, 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GHOSHAL UC, SHUKLA R & GHOSHAL U 2017. Small Intestinal Bacterial Overgrowth and Irritable Bowel Syndrome: A Bridge between Functional Organic Dichotomy. Gut Liver, 11, 196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GHOSHAL UC, SRIVASTAVA D, GHOSHAL U & MISRA A 2014. Breath tests in the diagnosis of small intestinal bacterial overgrowth in patients with irritable bowel syndrome in comparison with quantitative upper gut aspirate culture. Eur J Gastroenterol Hepatol, 26, 753–60. [DOI] [PubMed] [Google Scholar]
- GRILL HJ & HAYES MR 2009. The nucleus tractus solitarius: a portal for visceral afferent signal processing, energy status assessment and integration of their combined effects on food intake. Int J Obes (Lond), 33 Suppl 1, S11–5. [DOI] [PubMed] [Google Scholar]
- GUO X, YIN C, YANG F, ZHANG Y, HUANG H, WANG J, DENG B, CAI T, RAO Y & XI R 2019. The Cellular Diversity and Transcription Factor Code of Drosophila Enteroendocrine Cells. Cell Rep, 29, 4172–4185 e5. [DOI] [PubMed] [Google Scholar]
- GUPTA V & POSS KD 2012. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature, 484, 479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HABER AL, BITON M, ROGEL N, HERBST RH, SHEKHAR K, SMILLIE C, BURGIN G, DELOREY TM, HOWITT MR, KATZ Y, TIROSH I, BEYAZ S, DIONNE D, ZHANG M, RAYCHOWDHURY R, GARRETT WS, ROZENBLATT-ROSEN O, SHI HN, YILMAZ O, XAVIER RJ & REGEV A 2017. A single-cell survey of the small intestinal epithelium. Nature, 551, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAN W, TELLEZ LA, PERKINS MH, PEREZ IO, QU T, FERREIRA J, FERREIRA TL, QUINN D, LIU ZW, GAO XB, KAELBERER MM, BOHORQUEZ DV, SHAMMAH-LAGNADO SJ, DE LARTIGUE G & DE ARAUJO IE 2018. A Neural Circuit for Gut-Induced Reward. Cell, 175, 665–678 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEATH-PAGLIUSO S, ROGERS WJ, TULLIS K, SEIDEL SD, CENIJN PH, BROUWER A & DENISON MS 1998. Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry, 37, 11508–15. [DOI] [PubMed] [Google Scholar]
- HOLZER P 2011. TRP channels in the digestive system. Curr Pharm Biotechnol, 12, 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUBBARD TD, MURRAY IA, BISSON WH, LAHOTI TS, GOWDA K, AMIN SG, PATTERSON AD & PERDEW GH 2015. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep, 5, 12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAGLIN M, RHIMI M, PHILIPPE C, PONS N, BRUNEAU A, GOUSTARD B, DAUGE V, MAGUIN E, NAUDON L & RABOT S 2018. Indole, a Signaling Molecule Produced by the Gut Microbiota, Negatively Impacts Emotional Behaviors in Rats. Front Neurosci, 12, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIN UH, LEE SO, SRIDHARAN G, LEE K, DAVIDSON LA, JAYARAMAN A, CHAPKIN RS, ALANIZ R & SAFE S 2014. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol Pharmacol, 85, 777–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHN HAYNES W, ZHOU XL, SU ZW, LOUKIN SH, SAIMI Y & KUNG C 2008. Indole and other aromatic compounds activate the yeast TRPY1 channel. FEBS Lett, 582, 1514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSON CD, BARLOW-ANACKER AJ, PIERRE JF, TOUW K, ERICKSON CS, FURNESS JB, EPSTEIN ML & GOSAIN A 2018. Deletion of choline acetyltransferase in enteric neurons results in postnatal intestinal dysmotility and dysbiosis. FASEB J, 32, 4744–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAELBERER MM, BUCHANAN KL, KLEIN ME, BARTH BB, MONTOYA MM, SHEN X & BOHORQUEZ DV 2018. A gut-brain neural circuit for nutrient sensory transduction. Science, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAIKO GE & STAPPENBECK TS 2014. Host-microbe interactions shaping the gastrointestinal environment. Trends Immunol, 35, 538–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAWAKAMI K 2007. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol, 8 Suppl 1, S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM DJ, VENKATARAMAN A, JAIN PC, WIESLER EP, DEBLASIO M, KLEIN J, TU SS, LEE S, MEDZHITOV R & IWASAKI A 2020. Vitamin B12 and folic acid alleviate symptoms of nutritional deficiency by antagonizing aryl hydrocarbon receptor. Proc Natl Acad Sci U S A, 117, 15837–15845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KNIGHT RD, MEBUS K, D’ANGELO A, YOKOYA K, HEANUE T, TUBINGEN SCREEN C & ROEHL H 2011. Ret signalling integrates a craniofacial muscle module during development. Development, 138, 2015–24. [DOI] [PubMed] [Google Scholar]
- KOKEL D, CHEUNG CY, MILLS R, COUTINHO-BUDD J, HUANG L, SETOLA V, SPRAGUE J, JIN S, JIN YN, HUANG XP, BRUNI G, WOOLF CJ, ROTH BL, HAMBLIN MR, ZYLKA MJ, MILAN DJ & PETERSON RT 2013. Photochemical activation of TRPA1 channels in neurons and animals. Nat Chem Biol, 9, 257–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUNST M, LAURELL E, MOKAYES N, KRAMER A, KUBO F, FERNANDES AM, FORSTER D, DAL MASCHIO M & BAIER H 2019. A Cellular-Resolution Atlas of the Larval Zebrafish Brain. Neuron, 103, 21–38 e5. [DOI] [PubMed] [Google Scholar]
- KWAN KM, FUJIMOTO E, GRABHER C, MANGUM BD, HARDY ME, CAMPBELL DS, PARANT JM, YOST HJ, KANKI JP & CHIEN CB 2007. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn, 236, 3088–99. [DOI] [PubMed] [Google Scholar]
- LAPOINTE TK & ALTIER C 2011. The role of TRPA1 in visceral inflammation and pain. Channels (Austin), 5, 525–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEITE G, MORALES W, WEITSMAN S, CELLY S, PARODI G, MATHUR R, BARLOW GM, SEDIGHI R, MILLAN MJV, REZAIE A & PIMENTEL M 2020. The duodenal microbiome is altered in small intestinal bacterial overgrowth. PLoS One, 15, e0234906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI Z, CHALAZONITIS A, HUANG YY, MANN JJ, MARGOLIS KG, YANG QM, KIM DO, COTE F, MALLET J & GERSHON MD 2011. Essential roles of enteric neuronal serotonin in gastrointestinal motility and the development/survival of enteric dopaminergic neurons. J Neurosci, 31, 8998–9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU Y, HOU Y, WANG G, ZHENG X & HAO H 2020. Gut Microbial Metabolites of Aromatic Amino Acids as Signals in Host-Microbe Interplay. Trends Endocrinol Metab, 31, 818–834. [DOI] [PubMed] [Google Scholar]
- LOVE MI, HUBER W & ANDERS S 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU VB, GRIBBLE FM & REIMANN F 2018. Free Fatty Acid Receptors in Enteroendocrine Cells. Endocrinology, 159, 2826–2835. [DOI] [PubMed] [Google Scholar]
- MACPHERSON LJ, DUBIN AE, EVANS MJ, MARR F, SCHULTZ PG, CRAVATT BF & PATAPOUTIAN A 2007. Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines. Nature, 445, 541–5. [DOI] [PubMed] [Google Scholar]
- MATHIAS JR, ZHANG Z, SAXENA MT & MUMM JS 2014. Enhanced cell-specific ablation in zebrafish using a triple mutant of Escherichia coli nitroreductase. Zebrafish, 11, 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCGRAW HF, SNELSON CD, PRENDERGAST A, SULI A & RAIBLE DW 2012. Postembryonic neuronal addition in zebrafish dorsal root ganglia is regulated by Notch signaling. Neural Dev, 7, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEIJERING E, DZYUBACHYK O & SMAL I 2012. Methods for cell and particle tracking. Methods Enzymol, 504, 183–200. [DOI] [PubMed] [Google Scholar]
- MESEGUER V, ALPIZAR YA, LUIS E, TAJADA S, DENLINGER B, FAJARDO O, MANENSCHIJN JA, FERNANDEZ-PENA C, TALAVERA A, KICHKO T, NAVIA B, SANCHEZ A, SENARIS R, REEH P, PEREZ-GARCIA MT, LOPEZ-LOPEZ JR, VOETS T, BELMONTE C, TALAVERA K & VIANA F 2014. TRPA1 channels mediate acute neurogenic inflammation and pain produced by bacterial endotoxins. Nat Commun, 5, 3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURDOCH CC, ESPENSCHIED ST, MATTY MA, MUELLER O, TOBIN DM & RAWLS JF 2019. Intestinal Serum amyloid A suppresses systemic neutrophil activation and bactericidal activity in response to microbiota colonization. PLoS Pathog, 15, e1007381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAGEL G, SZELLAS T, HUHN W, KATERIYA S, ADEISHVILI N, BERTHOLD P, OLLIG D, HEGEMANN P & BAMBERG E 2003. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A, 100, 13940–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAKAMURA Y, TAKANO T, YASUIKE M, SAKAI T, MATSUYAMA T & SANO M 2013. Comparative genomics reveals that a fish pathogenic bacterium Edwardsiella tarda has acquired the locus of enterocyte effacement (LEE) through horizontal gene transfer. BMC Genomics, 14, 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAUMANN EA, FITZGERALD JE, DUNN TW, RIHEL J, SOMPOLINSKY H & ENGERT F 2016. From Whole-Brain Data to Functional Circuit Models: The Zebrafish Optomotor Response. Cell, 167, 947–960 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NILIUS B, PRENEN J & OWSIANIK G 2011. Irritating channels: the case of TRPA1. J Physiol, 589, 1543–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOZAWA K, KAWABATA-SHODA E, DOIHARA H, KOJIMA R, OKADA H, MOCHIZUKI S, SANO Y, INAMURA K, MATSUSHIME H, KOIZUMI T, YOKOYAMA T & ITO H 2009. TRPA1 regulates gastrointestinal motility through serotonin release from enterochromaffin cells. Proc Natl Acad Sci U S A, 106, 3408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAN H & GERSHON MD 2000. Activation of intrinsic afferent pathways in submucosal ganglia of the guinea pig small intestine. J Neurosci, 20, 3295–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAN YA, CHOY M, PROBER DA & SCHIER AF 2012. Robo2 determines subtype-specific axonal projections of trigeminal sensory neurons. Development, 139, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK SB, AOKI T & JUNG TS 2012. Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet Res, 43, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHAM LN, KANTHER M, SEMOVA I & RAWLS JF 2008. Methods for generating and colonizing gnotobiotic zebrafish. Nat Protoc, 3, 1862–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PROBER DA, ZIMMERMAN S, MYERS BR, MCDERMOTT BM JR., KIM SH, CARON S, RIHEL J, SOLNICA-KREZEL L, JULIUS D, HUDSPETH AJ & SCHIER AF 2008. Zebrafish TRPA1 channels are required for chemosensation but not for thermosensation or mechanosensory hair cell function. J Neurosci, 28, 10102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PUYSKENS A, STINN A, VAN DER VAART M, KREUCHWIG A, PROTZE J, PEI G, KLEMM M, GUHLICH-BORNHOF U, HURWITZ R, KRISHNAMOORTHY G, SCHAAF M, KRAUSE G, MEIJER AH, KAUFMANN SHE & MOURA-ALVES P 2020. Aryl Hydrocarbon Receptor Modulation by Tuberculosis Drugs Impairs Host Defense and Treatment Outcomes. Cell Host Microbe, 27, 238–248 e7. [DOI] [PubMed] [Google Scholar]
- QU ZD, THACKER M, CASTELUCCI P, BAGYANSZKI M, EPSTEIN ML & FURNESS JB 2008. Immunohistochemical analysis of neuron types in the mouse small intestine. Cell Tissue Res, 334, 147–61. [DOI] [PubMed] [Google Scholar]
- RANDLETT O, WEE CL, NAUMANN EA, NNAEMEKA O, SCHOPPIK D, FITZGERALD JE, PORTUGUES R, LACOSTE AM, RIEGLER C, ENGERT F & SCHIER AF 2015. Whole-brain activity mapping onto a zebrafish brain atlas. Nat Methods, 12, 1039–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAWLS JF, MAHOWALD MA, LEY RE & GORDON JI 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell, 127, 423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROACH G, HEATH WALLACE R, CAMERON A, EMRAH OZEL R, HONGAY CF, BARAL R, ANDREESCU S & WALLACE KN 2013. Loss of ascl1a prevents secretory cell differentiation within the zebrafish intestinal epithelium resulting in a loss of distal intestinal motility. Dev Biol, 376, 171–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBERTS GP, LARRAUFIE P, RICHARDS P, KAY RG, GALVIN SG, MIEDZYBRODZKA EL, LEITER A, LI HJ, GLASS LL, MA MKL, LAM B, YEO GSH, SCHARFMANN R, CHIARUGI D, HARDWICK RH, REIMANN F & GRIBBLE FM 2019. Comparison of Human and Murine Enteroendocrine Cells by Transcriptomic and Peptidomic Profiling. Diabetes, 68, 1062–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROLIG AS, MITTGE EK, GANZ J, TROLL JV, MELANCON E, WILES TJ, ALLIGOOD K, STEPHENS WZ, EISEN JS & GUILLEMIN K 2017. The enteric nervous system promotes intestinal health by constraining microbiota composition. PLoS Biol, 15, e2000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHINDELIN J, ARGANDA-CARRERAS I, FRISE E, KAYNIG V, LONGAIR M, PIETZSCH T, PREIBISCH S, RUEDEN C, SAALFELD S, SCHMID B, TINEVEZ JY, WHITE DJ, HARTENSTEIN V, ELICEIRI K, TOMANCAK P & CARDONA A 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods, 9, 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMITH T 1897. A Modification of the Method for Determining the Production of Indol by Bacteria. J Exp Med, 2, 543–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SRINIVASA RAO PS, LIM TM & LEUNG KY 2003. Functional genomics approach to the identification of virulence genes involved in Edwardsiella tarda pathogenesis. Infect Immun, 71, 1343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEPHENS WZ, BURNS AR, STAGAMAN K, WONG S, RAWLS JF, GUILLEMIN K & BOHANNAN BJ 2016. The composition of the zebrafish intestinal microbial community varies across development. ISME J, 10, 644–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUN EW, MARTIN AM, WATTCHOW DA, DE FONTGALLAND D, RABBITT P, HOLLINGTON P, YOUNG RL & KEATING DJ 2019a. Metformin Triggers PYY Secretion in Human Gut Mucosa. J Clin Endocrinol Metab, 104, 2668–2674. [DOI] [PubMed] [Google Scholar]
- SUN Y, TANG L, LIU Y, HU C, ZHOU B, LAM PKS, LAM JCW & CHEN L 2019b. Activation of aryl hydrocarbon receptor by dioxin directly shifts gut microbiota in zebrafish. Environ Pollut, 255, 113357. [DOI] [PubMed] [Google Scholar]
- SYMONDS EL, PEIRIS M, PAGE AJ, CHIA B, DOGRA H, MASDING A, GALANAKIS V, ATIBA M, BULMER D, YOUNG RL & BLACKSHAW LA 2015. Mechanisms of activation of mouse and human enteroendocrine cells by nutrients. Gut, 64, 618–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TARAVIRAS S, MARCOS-GUTIERREZ CV, DURBEC P, JANI H, GRIGORIOU M, SUKUMARAN M, WANG LC, HYNES M, RAISMAN G & PACHNIS V 1999. Signalling by the RET receptor tyrosine kinase and its role in the development of the mammalian enteric nervous system. Development, 126, 2785–97. [DOI] [PubMed] [Google Scholar]
- TORNINI VA, THOMPSON JD, ALLEN RL & POSS KD 2017. Live fate-mapping of joint-associated fibroblasts visualizes expansion of cell contributions during zebrafish fin regeneration. Development, 144, 2889–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRAVAGLI RA & ANSELMI L 2016. Vagal neurocircuitry and its influence on gastric motility. Nat Rev Gastroenterol Hepatol, 13, 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRAVAGLI RA, HERMANN GE, BROWNING KN & ROGERS RC 2006. Brainstem circuits regulating gastric function. Annu Rev Physiol, 68, 279–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSENG Q, DUCHEMIN-PELLETIER E, DESHIERE A, BALLAND M, GUILLOU H, FILHOL O & THERY M 2012. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc Natl Acad Sci U S A, 109, 1506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ULHAQ ZS & KISHIDA M 2018. Brain Aromatase Modulates Serotonergic Neuron by Regulating Serotonin Levels in Zebrafish Embryos and Larvae. Front Endocrinol (Lausanne), 9, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VENKATESH M, MUKHERJEE S, WANG H, LI H, SUN K, BENECHET AP, QIU Z, MAHER L, REDINBO MR, PHILLIPS RS, FLEET JC, KORTAGERE S, MUKHERJEE P, FASANO A, LE VEN J, NICHOLSON JK, DUMAS ME, KHANNA KM & MANI S 2014. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity, 41, 296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG M, LV Y, XIAO J, WU H, ZHENG H, LIU Q, ZHANG Y & WANG Q 2012. Edwardsiella comparative phylogenomics reveal the new intra/inter-species taxonomic relationships, virulence evolution and niche adaptation mechanisms. PLoS One, 7, e36987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG Y, WANG S, KOBAYASHI K, HAO Y, KANDA H, KONDO T, KOGURE Y, YAMANAKA H, YAMAMOTO S, LI J, MIWA H, NOGUCHI K & DAI Y 2019. TRPA1-expressing lamina propria mesenchymal cells regulate colonic motility. JCI Insight, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANO JM, YU K, DONALDSON GP, SHASTRI GG, ANN P, MA L, NAGLER CR, ISMAGILOV RF, MAZMANIAN SK & HSIAO EY 2015. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell, 161, 264–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YE L, MUELLER O, BAGWELL J, BAGNAT M, LIDDLE RA & RAWLS JF 2019. High fat diet induces microbiota-dependent silencing of enteroendocrine cells. Elife, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YUE C, JI C, ZHANG H, ZHANG LW, TONG J, JIANG Y & CHEN T 2017. Protective effects of folic acid on PM2.5-induced cardiac developmental toxicity in zebrafish embryos by targeting AhR and Wnt/beta-catenin signal pathways. Environ Toxicol, 32, 2316–2322. [DOI] [PubMed] [Google Scholar]
- ZELANTE T, IANNITTI RG, CUNHA C, DE LUCA A, GIOVANNINI G, PIERACCINI G, ZECCHI R, D’ANGELO C, MASSI-BENEDETTI C, FALLARINO F, CARVALHO A, PUCCETTI P & ROMANI L 2013. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity, 39, 372–85. [DOI] [PubMed] [Google Scholar]
- ZELKAS L, RAGHUPATHI R, LUMSDEN AL, MARTIN AM, SUN E, SPENCER NJ, YOUNG RL & KEATING DJ 2015. Serotonin-secreting enteroendocrine cells respond via diverse mechanisms to acute and chronic changes in glucose availability. Nutr Metab (Lond), 12, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Zebrafish EEC RNA-seq data analyzed and compared with mouse and human EEC data, related to Figure 2.
Video 1. Activation of EEC Trpa1 in WT zebrafish, but not EEC ablated zebrafish, increases intestinal motility, related to Figure 4. Time-course video of Tg(neurod1:Gcamp6f) WT and EEC ablated zebrafish before and post Optovin-UV induced EEC Trpa1 activation. WT zebrafish are shown first followed by EEC ablated zebrafish. Anterior is to the left, and dorsal is to the top.
Video 2. Activation of Trpa1+ChR2+EECs, but not Trpa1-ChR2+EECs, increases intestinal motility, related to Figure 4. Time-course videos of TgBAC(trpa1b:EGFP); Tg(neurod1:Gal4); Tg(UAS:ChR2-mCherry) before and post optic activation of Trpa1- EECs and Trpa1+ EECs. Anterior is to the left, and dorsal is to the bottom. Yellow light was delivered specifically to the selected EECs to activate the ChR2 channel. Note that the first frame in each video shows the EGFP channel to identify EECs that do or do not express trpa1b. Also note that the ChR2 EECs are in the intestinal bulb and an retrograde intestinal movement was observed upon Trpa1+ChR2+EEC activation. The third part of the video shows that activation of Trpa1+ChR2+EECs in the mid-intestine induces an anterograde intestinal movement.
Video 3. E. tarda increases intestinal motility, related to Figure 4. Time-course videos of WT zebrafish 30 mins post Aeromonas veronii ZOR0002 or E. tarda FL6–60 gavage. Anterior is to the left, and dorsal is to the top.
Video 4. Indole activates EECs in vivo, related to Figure 7. Time-course videos of trpa1b+/+ (WT) and trpa1b−/− Tg(neurod1:Gcamp6f) zebrafish that are stimulated with indole. Note that there is a basal amount of intestinal motility associated with this methylcellulose preparation that is retained in vehicle-only negative controls (not shown) and in trpa1b mutants. Anterior is to the right, and dorsal is to the top.
Data Availability Statement
Sequencing reads generated as part of this study are available at Gene Expression Omnibus accession GSE151711.


