Abstract
Non-small cell lung cancer (NSCLC) is the number one cancer killer and its early detection can reduce mortality. Accumulating evidences suggest an etiopathogenic role of microorganisms in lung tumorigenesis. Certain bacteria are found to be associated with NSCLC. Herein we evaluated the potential use of microbiome as biomarkers for the early detection of NSCLC. We used droplet digital PCR to analyze 25 NSCLC-associated bacterial genera in 31 lung tumor and the paired noncancerous lung tissues and sputum of 17 NSCLC patients and ten cancer-free smokers. Of the bacterial genera, four had altered abundances in lung tumor tissues, while five were aberrantly abundant in sputum of NSCLC patients compared with their normal counterparts (all p < 0.05). Acidovorax and Veillonella were further developed as a panel of sputum biomarkers that could diagnose lung squamous cell carcinoma (SCC) with 80% sensitivity and 89% specificity. The use of Capnocytophaga as a sputum biomarker identified lung adenocarcinoma (AC) with 72% sensitivity and 85% specificity. The use of Acidovorax as a sputum biomarker had 63% sensitivity and 96% specificity for distinguishing between SCC and AC, the two major types of NSCLC. The sputum biomarkers were further validated for the diagnostic values in a different cohort of 69 NSCLC cases and 79 cancer-free controls. Sputum microbiome might provide noninvasive biomarkers for the early detection and classification of NSCLC.
Keywords: microbiome, bacteria, biomarkers, lung cancer, sputum
1. Introduction
Lung cancer is the leading cause of cancer-related deaths in men and women [1]. Over 85% lung cancers are non-small cell lung cancer (NSCLC), which mainly consists of squamous cell carcinoma (SCC) and adenocarcinoma (AC). The early detection of NSCLC by low-dose CT (LDCT) followed by the effective treatments can reduce mortality [1]. However, many positive LDCT scans are false alarms and result in multiple examinations and invasive biopsies that carry their own morbidity [1]. The development of noninvasive biomarkers that can accurately diagnose early stage lung cancer remains clinically imperative.
Microbiota is the group of a wide-ranging array of microorganisms, including bacteria, archaea, fungi, and viruses that inhabit various body sites [2]. The microbiome, defined as the collection of microbiota and their genes, plays an important role in health and disease [2]. Microbial agents could cause approximately 20% of the overall cancer burden [2]. For example, infection of Human Papilloma virus, Epstein–Barr virus, Helicobacter pylori (H. pylori), Escherichia coli and Fusobacterium nucleatum lead to a variety of malignancies [2]. In the respiratory tract, there are more than 500 different species of bacteria [3]. Changes of the airway microbiome are attributed to lung tumorigenesis through different mechanisms, such as damage of the local immune barrier, production of bacterial toxins that alter host genome stability, and release of cancer-promoting microbial metabolites [4]. Furthermore, intratumoral microbes may directly affect the growth and metastatic spread of tumor cells [2]. Lung cancer patients have lower microbial diversity and altered abundances of particular bacteria compared with cancer-free individuals [5]. 16S rRNA gene sequencing-based studies [6,7,8,9,10,11,12,13] have identified a set of genera with either higher or lower abundances in lung tumors vs. normal lung tissues. For example, Acidovorax was especially abundant in patients with TP53 mutation-positive lung SCC specimens [8]. The abundances of Streptococcus and Veillonella were associated with upregulation of the ERK and PI3K signaling pathways in NSCLC cells [12]. In addition, saliva of lung cancer patients possessed an elevated abundance of Capnocytophaga, Selenomonas, Veillonella, Sphingomonas, and Blastomonas [14,15]. Moreover, Granulicatella, Abiotrophia, and Streptococcus, Adiacens, Intermedius, and Mycobacterium tuberculosis were enriched in the sputum of lung cancer patients [16,17,18]. Importantly, since the microbiota of an individual is stable long-term, they might provide biomarkers for lung cancer [2,3,11,17,19,20].
Two major types of specimens, lower (e.g., bronchoalveolar lavage (BAL and lung tumor tissues)) and upper (saliva and buccal samples) respiratory tract samples, are used for the development of lung cancer biomarkers. BAL and lung tumor tissues are invasively collected via a bronchoscopy or surgery, and thus are not suitable for the development of noninvasive biomarkers. Saliva and buccal samples are obtained less invasively. However, when used for diagnosis of lung cancer, the microbiome in the specimens will be confounded by oral contamination. In contrast, sputum is readily available and can be self-collected. Furthermore, since sputum is secreted from bronchi and bronchioles of the lower respiratory tract, it is more objective and representative than saliva and buccal samples in reflecting the microbial environment of the lungs. Thus, sputum is a viable option for sampling of the lung microbiome without oral contamination. In addition, since molecular changes detected in sputum could reflect those in low respiratory tract [21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40], sputum can be substituted for the lower-airway fluids (e.g., BAL and surgical tissues), which are more invasively collected, for sensitive detection of lung cancer. Therefore, sputum has the advantages as surrogate material and overcomes the obstacles of the commonly used specimens for diagnosis of lung cancer. Taking advantage of the lung cancer-associated genera identified by previous studies [3,5,6,7,8,10,11,12,13,14,15,17,19,41,42], herein we aimed to evaluate the potential of microbiome as sputum biomarkers for lung cancer.
2. Materials and Methods
2.1. Study Population
The study protocol was approved by the Institutional Review Board of the University of Maryland Medical Center (IRB HP-00040666). From a tissue bank, we obtained 31 frozen lung tumor and the matched noncancerous lung tissues of stage I NSCLC patients who had either a lobectomy or a pneumonectomy. Tumor tissues were intraoperatively dissected from the surrounding lung parenchyma. Paired normal lung tissues were obtained from the same patients at an area distant from their tumors. Of the 31 cases, 16 cases were diagnosed with SCC and 15 were AC of the lungs. We collected sputum samples from participants between the ages of 55–79 at the point of their referral for suspected lung cancer. A total of 27 subjects including 17 lung cancer patients and ten cancer-free smokers were recruited. The 17 lung cancer patients were diagnosed with NSCLC consisting of five stage I cases, five stage II cases, and seven stage III-IV cases. The NSCLC cases consisted of ten AC and seven SCC of lungs (Table 1). The ten cancer-free patients were smokers who had either granulomatous inflammation (n = 5), nonspecific inflammatory changes (n = 3) or pulmonary infections (n = 2).
Table 1.
Cohort 1 of NSCLC patients and cancer-free smokers from whom sputum specimens were collected.
| NSCLC Cases (n = 17) | Controls (n = 10) | p-Value | |
|---|---|---|---|
| Age | 66.37 (SD 9.05) | 61.27 (SD 9.46) | 0.36 |
| Sex | 0.43 | ||
| Female | 7 | 4 | |
| Male | 10 | 6 | |
| Race | 0.39 | ||
| African Americans | 5 | 3 | |
| White American | 12 | 7 | |
| Smoking pack-years (median) | 32.17 | 28.38 | 0.26 |
| Stage | |||
| Stage I | 5 | ||
| Stage II | 5 | ||
| Stage III-VI | 7 | ||
| Histological type | |||
| Adenocarcinoma | 10 | ||
| Squamous cell carcinoma | 7 | ||
| Location of primary lung tumors | |||
| Peripheral location | 10 | ||
| Central location | 7 |
Abbreviations: NSCLC, non-small cell lung cancer; Central location: limited to the trachea, bronchi, or segmental bronchi. Peripheral location: limited more to the periphery than the subsegmental bronchi.
Sputum samples of 69 lung cancer patients and 79 cancer-free smokers were obtained from Dr. Ruth L Katz’s laboratory of The University of Texas M.D. Anderson Cancer Center. As shown in Table 2, the 69 NSCLC patients consisted of 22 stage I cases, 24 stage II cases, and 23 stage III–IV cases. Thirty-six cases were AC and 33 were SCC of the lungs. The 79 cancer-free patients who were smokers and had either granulomatous inflammation (n = 39), nonspecific inflammatory changes (n = 22) or lung infections (n = 18).
Table 2.
Cohort 2 of NSCLC patients and cancer-free smokers from whom sputum specimens were collected.
| NSCLC Cases (n = 69) | Controls (n = 79) | p-Value | |
|---|---|---|---|
| Age | 64.18 (SD 6.25) | 63.29 (SD 6.24) | 0.32 |
| Sex | 0.35 | ||
| Female | 26 | 30 | |
| Male | 43 | 49 | |
| Smoking pack-years (median) | 35.25 | 33.29 | 0.41 |
| Stage | |||
| Stage I | 22 | ||
| Stage II | 24 | ||
| Stage III–VI | 23 | ||
| Histological type | |||
| Adenocarcinoma | 36 | ||
| Squamous cell carcinoma | 33 | ||
| Location of primary lung tumors | |||
| Peripheral location | 36 | ||
| Central location | 33 |
Abbreviations: NSCLC, non-small cell lung cancer; Central location: limited to the trachea, bronchi, or segmental bronchi. Peripheral location: limited more to the periphery than the subsegmental bronchi.
2.2. Collection and Preparation of Sputum
Sputum was collected from the participants before they received any treatment as described in our previous studies [29,30,31,32,33,34,35,36,37,38,43,44,45]. To reduce the percentage of oral epithelial cells in sputum, the participants were asked to blow their nose, rinse their mouth, and swallow water to minimize contamination of squamous cells from postnasal drip and saliva. Sputum samples were then coughed into a sterile container and processed within 2 h. To further minimize oral squamous cell contamination, opaque or dense portions that looked different from saliva under the inverted microscope were selected using blunt forceps from expectorate. The samples were processed on ice in four volumes of 0.1% dithiothreitol (Sigma-Aldrich, St. Louis, Mo) followed by four volumes of phosphate-buffered saline (Sigma-Aldrich). We centrifuged the samples at 1500× g for 15 min and removed the supernatant. The remaining cell pellets were collected and stored at −80 °C until use.
2.3. Genomic DNA Isolation
We used QIAGEN-DNeasy Blood & Tissue Kit (QIAGEN, Germantown, MD, USA) to isolate DNA from the cell pellets or tissue specimens according to manufacturer’s instructions [23,46,47]. We determined the purity by taking the optical density (OD) of the sample at 280 nm for protein concentration and at 260 nm for DNA concentration. The ratio OD260 /OD280 was calculated and DNA sample within the range of 1.6–2 was considered as pure.
2.4. Detection and Quantification of Bacterial Abundances Using Droplet Digital PCR (Ddpcr)
We preformed ddPCR to detect DNA of 25 bacterial genera (Table 3) by using a QX100 Droplet Digital PCR System and 2× ddPCR Supermix (Bio-Rad, California, CA, USA) with a protocol developed in our previous studies with modification [23,30,40,46,48,49,50,51,52]. The 25 bacterial genera were suggested to be associated with lung cancer by previous studies (references in Table 3) and thus tested in this study. To design genus-specific primers of PCR test for determining their bacterial abundances, we first aligned 16S rRNA sequences for the maximum number of species for the specific genus to identify consensus regions at genus level. We then use the Primer3 primer design program to design specific primers as previous described [53,54]. Sequences of PCR primers to amplify DNA of the bacterial genera are shown in Table 3. To generate the droplets, we inserted 20 µL of PCR reaction and 70 µL of Droplet Generation oil for Probes (Bio-Rad) in an eight-well cartridge using a QX100 droplet generator (Bio-Rad). We then transferred 40 µL of the generated droplet emulsion in a 96-well PCR plate (Eppendorf, Hamburg, Germany). Amplification reaction was conducted in a T100™ thermal cycler (Bio-Rad) with the following conditions: initial denaturation at 95 °C for 5 min followed by 35 cycles of 15 s at 95.0 °C, 30 s at 55.3 °C, 5 min at 4 °C, and, finally, 5 min at 90 °C for signal stabilization. After thermal cycling, we transferred plates to a droplet reader (Bio-Rad). We used the software provided with the ddPCR system for data acquisition to calculate the concentration of target DNA in copies/µL from the fraction of positive reactions using Poisson distribution analyses.
Table 3.
Twenty-five bacterial genera tested by ddPCR and their primers.
| Name | Target Region (Accession #) | Forward (5′-3′) | Reward (5′-3′) | References |
|---|---|---|---|---|
| Acidovorax | NZ_LJGO01000014.1 | GTCATCCTCCACCAACCAATAC | GTCTATACCGGACCAACAACAA | [8] |
| Akkermansia | NZ_AP021898.1 | CAGCACGTGAAGGTGGGGAC | CCTTGCGGTTGGCTTCAGAT | [55] |
| Bacteroides | NZ_VKLY01000054.1 | GACCGCATGGTCTTGTTATT | CGTAGGAGTTTGGACCGTGT | [56] |
| Bifidobacterium | NZ_AKCA01000001.1 | CCACATGATCGCATGTGATTG | CCGAAGGCTTGCTCCCAAA | [56] |
| Bilophila | NZ_KE150238.1 | CGTGTGAATAATGCGAGGG | TCTCCGGTACTCAAGCGTG | [57] |
| Blautia | NZ_NQOF01000001.1 | GTGAAGGAAGAAGTATCTCGG | TTGGTAAGGTTCTTCGCGTT | [58] |
| Bradyrhizobium | NZ_VSSR01000023.1 | ATCGACGTGCTGCCAATAA | GCCGATAACAAGACGGAAATAAC | [13] |
| Capnocytophaga | NZ_BLBC01000010.1 | TGGWCAATGGTCGGAAGACTG | CCGCTACACTACACATTCCA | [9] |
| Curvibacter | NZ_CP022389.1 | GAGCCTTTACCTCACCAACTAC | CGTAGCGAAAGCTACGCTAATA | [59] |
| Enterococcus | NZ_CP023011.2 | GGCATATTTATCCAGCACTAG | TAGCGTACGAAAAGGCATCC | [17] |
| Escherichia | NC_000913.3 | CATGCCGCGTGTATGAAGAA | CGGGTAACGTCAATGAGCAAA | [17] |
| Faecalibacterium | NZ_CP030777.1 | GGAGGAAGAAGGTCTTCGG | AATTCCGCCTACCTCTGCACT | [60] |
| Fusobacterium | NZ_LT608325.1 | AAGCGCGTCTAGGTGGTTATGT | TGTAGTTCCGCTTACCTCTCCAG | [61] |
| Haemophilus | NZ_LS483458.1 | AGCGGCTTGTAGTTCCTCTAACA | CAACAGAGTATCCGCCAAAAGTT | [62] |
| Helicobacter | NC_017379.1 | GCGCATGTCTTCGGTTAAAAA | TTCCATAGGCTATAATGTGATCCAAA | [63] |
| Klebsiella | NZ_CP023478.1 | CGGGCGTAGCGCGTAA | GATACCCGCATTCACATTAAACAG | [64] |
| Lactobacillus | NZ_MWIK01000038.1 | CGCCACTGGTGTTCYTCCATATA | AGCAGTAGGGAATCTTCCA | [65] |
| Mycobacterium | NZ_UATA01000019.1 | CAAGCGGTGGAGCATGTG | CTAAGATGTCAAACGCTGGTAAGG | [66] |
| Neisseria | NZ_UGRT01000005.1 | CTGTTGGGCARCWTGAYTGC | GATCGGTTTTRTGAGATTGG | [7] |
| Prevotella | NZ_BAKG01000039.1 | CCTACGATGGATAGGGGTT | CACGCTACTTGGCTGGTTCAG | [5] |
| Pseudomonas | NZ_BMDE01000022.1 | CAGCCATGCCGCGTGTGTGA | GTTGGTAACGTCAAAACAGCAAGG | [67] |
| Ruminococcus | NZ_QRIH01000002.1 | GCTTAGATTCTTCGGATGAAGAGGA | AGTTTTTACCCCCGCACCA | [68] |
| Selenomonas | NZ_JH376859.1 | ACRCGTAGRCAACCTGCCG | CGATCCGAAGACCTTCTTCAC | [15] |
| Streptococcus | NZ_UYIP01000002.1 | ACGCAATCTAGCAGATGAAGCA | TCGTGCGTTTTAATTCCAGC | [7,16] |
| Veillonella | NZ_AUAN01000022.1 | CGGGTGAGTAACGCGTAATCA | CCAACTAGCTGATGGGACGC | [15] |
2.5. Statistical Analysis
We used statistical system software version 6.12 (SAS Institute, Cary, NC) and GraphPad Prism version 7 (GraphPad Software, La Jolla, CA) for data analysis. The results were graphed and plotted by GraphPad Prism version 7. Mann–Whitney U test was used to determine whether bacterial abundances were significantly different between lung cancer patients and healthy controls. Furthermore, Pearson’s correlation coefficient test was used to determine the associations of bacterial abundances with clinicopathologic and demographic characteristics of the participants. Spearman correlation test was carried out to analyze the correlation between abundances of bacterial genera. Logistic regression was used to generate prediction models. To evaluate diagnostic significance of potential biomarkers, we used receiver-operator characteristic (ROC) curve analysis and computed the area under ROC (AUC) value by numerical integration of the ROC curve.
3. Results
3.1. Bacterial Genera Displayed Different Abundances between Lung Tumor and Noncancerous Lung Tissues
We have proven that ddPCR is a direct method for absolutely and quantitatively measuring nuclear acids without requiring internal control genes and calculating standard curves, which simplifies experimentation and data comparability [23,30,40,46,48,51]. Furthermore, ddPCR had a higher sensitivity compared with conventional PCR for detection and quantification of nuclear acids [23,30,40,46,48,51]. Therefore, in this study, we used ddPCR to determine DNA abundances of 25 bacterial genera (Table 2), whose changes were suggested to be associated with lung cancer [3,5,6,7,8,10,11,12,13,14,15,17,19,41,42]. All the bacterial genera tested by ddPCR generated at least 10,000 droplets in each well of reaction and, therefore, were successfully “read” by ddPCR for the absolute quantification in the tissue specimens.
As shown in Figure 1, Acidovorax was overrepresented in SCC tissues compared with noncancerous lung tissues and AC tissues (p = 0.0051). Capnocytophaga DNA was enriched in AC tissues compared with noncancerous lung tissues and SCC tissues (p = 0.0049) (Figure 1). However, the abundances of Haemophilus and Fusobacterium were lower in AC tissues compared with noncancerous lung tissues and SCC tissues (p = 0.049 and 0.039), respectively (Figure 1).
Figure 1.
Four bacterial genera show different abundances between lung AC and SSC tissues and the matched normal lung tissues. Solid red line indicates median, while black line indicates quartiles of abundance (copies of bacterial DNA/µL) of each genera in the different types of specimens. * shows that the p-value is under 0.05 by a Mann–Whitney U test.
3.2. Bacterial Genera Displayed Different Abundances in Sputum of Lung Cancer Patients vs. Cancer-Free Smokers
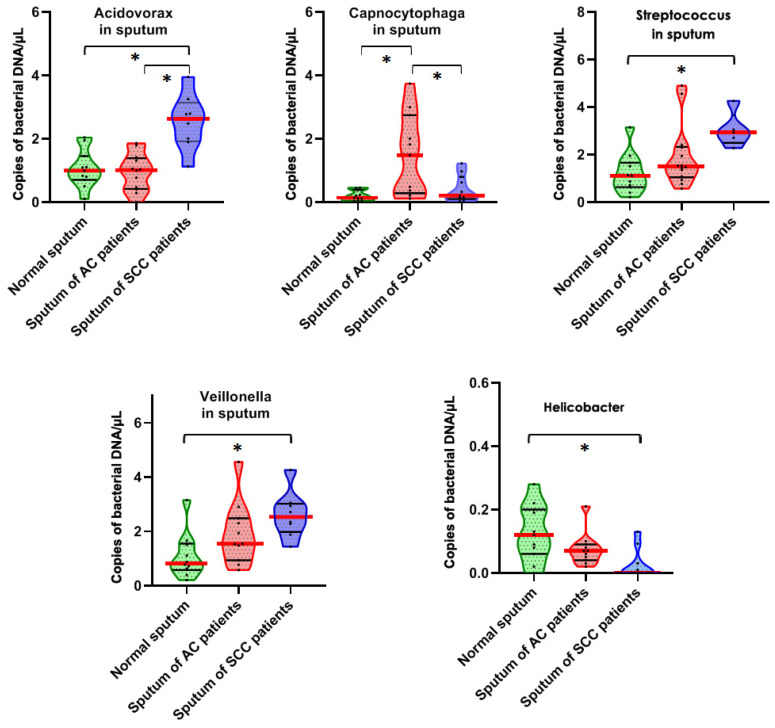
All the 25 bacteria produced more than 10,000 droplets in each reaction, and thus were also readily detected in the sputum specimens by ddPCR. Of the bacteria, Acidovorax, Streptococus, and Veillonella were overrepresented in sputum of lung SCC patients compared with lung AC patients and cancer-free smokers (pall p < 0.05) (Figure 2). The abundance of Helicobacter was underrepresented in sputum of lung SCC patients compared with lung AC patients and cancer-free smokers (p= 0.018 (Figure 2). Capnocytophaga was enriched in sputum of lung AC patients compared with lung SCC patients and cancer-free smokers (p = 0.046) (Figure 2).
Figure 2.
Five bacterial genera show different abundances in sputum of lung AS or SCC patients and cancer-free individuals. The solid red line indicates median, while the black line indicates quartiles of abundance (copies of bacterial DNA/µL) of each genera in the specimens. *, p < 0.05 determined by a Mann–Whitney U test).
Furthermore, both Acidovorax and Capnocytophaga displayed significantly different abundances in sputum of lung AC vs. SCC patients (all p < 0.05). In addition, abundances of the five sputum bacteria were not associated with the age, gender, ethnic group, tumor stage, and smoking status of the patients (all p > 0.05), except histology and location of primary lung tumors (all p > 0.05).
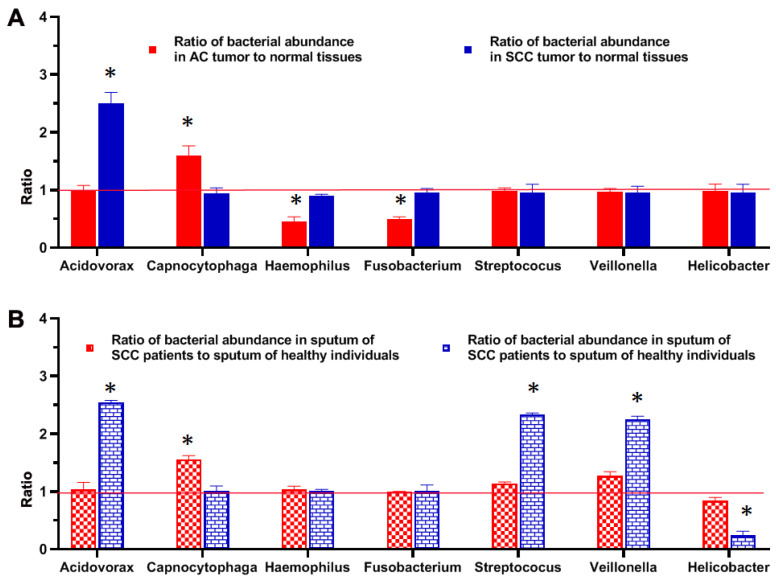
Comparison of abundances of bacteria in tumor tissues of lung cancer patients and sputum of lung cancer patients and cancer-free smokers.
The change of Acidovorax abundance had a similar trend in SCC tissues as in sputum of lung SCC patients (Figure 3) (Spearman correlation test, p = 0.023). Furthermore, Capnocytophaga had a similar trend in AC tissues as in sputum of lung AC patients. (Spearman correlation test, p = 0.017). The altered abundances of the two bacterial genera (Acidovorax and Capnocytophaga) in sputum might directly reflect those in lung tumor tissues. However, the reduced abundances of Haemophilus and Fusobacterium were only observed in lung AC tissue specimens compared with their normal counterparts (Figure 3A). The increased abundances of the Streptococcus and Veillonella were solely discovered in sputum of lung SCC patients and decreased abundances of Helicobacter were found in sputum of lung AC patients, as compared with their normal counterparts (Figure 3B).
Figure 3.
Comparison of abundances of bacteria in tumor tissues of lung cancer patients and sputum of lung cancer patients and controls. Relative abundances of bacterial genera in tumor tissues (A) and sputum of lung cancer patients (B) as compared with their normal counterparts. High abundances of bacterial genera in tumor tissue or sputum specimens are shown above ratio of 1 (red line), whereas low abundances in tumor tissue or sputum specimens are shown below ratio of 11 (red line). *, p < 0.05 determined by a Mann–Whitney U test).
3.3. Development of Sputum Bacteria Biomarkers for NSCLC
Sputum is noninvasively obtained body fluid. It contains bronchial epithelial cells from the lungs and lower respiratory tract and, thus, has the advantages as surrogate material for specifically diagnosing lung cancer. We evaluated if the five bacteria, which were readily detected in sputum and associated with lung cancer, could be used as noninvasive biomarkers for NSCLC. In the cohort 1 of sputum specimens, the five bacteria exhibited AUC values of 0.56–0.88 in distinguishing NSCLC patients from controls (Table 4).
Table 4.
The five bacterial genera display different levels in sputum samples of NSCLC patients vs. cancer-free controls of cohort 1.
| Genera |
p Value of Ac Patients vs. Controls |
p Value of Scc Patients vs. Controls |
AUC of AC Patients vs. Controls |
AUC of SCC Patients vs. Controls |
|---|---|---|---|---|
| Acidovorax | 0.7090 | 0.0015 | 0.5636 | 0.8814 |
| Capnocytophaga | 0.0455 | 0.3194 | 0.8502 | 0.6833 |
| Helicobacter | 0.0705 | 0.0175 | 0.7273 | 0.8070 |
| Streptococcus | 0.2775 | 0.0042 | 0.6753 | 0.8117 |
| Veillonella | 0.1086 | 0.0098 | 0.7062 | 0.8286 |
Abbreviations: NSCLC, non-small cell lung cancer; AC, adenocarcinoma; SCC, squamous cell carcinoma; AUC, the area under receiver-operator characteristic curve.
We used a stepwise logistic regression analysis to select the optimal panels of biomarkers. Two bacteria consisting of Acidovorax and Veillonella were selected as the best biomarkers for lung SCC. The two bacterial biomarkers used in combination produced 0.91 AUC (Figure 4A) in diagnosis of lung SCC with 80.00% sensitivity and 89.26% specificity (Table 4). The estimated correlations among levels of the two bacteria were very low (Spearman correlation test, p = 0.53), implying that the integration of the two biomarkers has complementary classification. Furthermore, the use of Capnocytophaga as a sputum biomarker could detect lung AC with 0.85 AUC (Figure 4B), 72.73% sensitivity and 85.19% specificity (Table 4).
Figure 4.
Receiver-operator characteristic (ROC) curve analysis of abundances of bacterial genera in sputum of cohort 1 comprising NSCLC patients and cancer-free controls. The area under the ROC curve (AUC) for bacterial genera conveys its accuracy for diagnosis and classification of NSCLC. (A) The combined use of Acidovorax and Veillonella produced 0.91 AUC. (B) The use of Capnocytophaga as a sputum biomarker could detect lung AC with 0.85 AUC. (C) The use of Acidovorax as a sputum biomarker had 0.85 AUC for differentiating between SCC and SC of the lungs.
In addition, the use of Acidovorax as a sputum biomarker had 0.86 AUC (Figure 4C) with 63.64% sensitivity and 96.30% specificity for distinguishing between SCC and AC of the lungs (Table 5). The bacterial biomarkers had no association with age, gender, and smoking status of the participants, and stages of lung tumors (Pearson’s correlation coefficient test, all p > 0.05), except location and histology of primary lung tumors (Supplementary Table S1).
Table 5.
The diagnostic values of the sputum bacterial biomarkers in contorts 1 and 2.
| Cohort 1 of 17 NSCLC Patients and 10 Cancer-Free Controls | Cohort 2 of 69 NSCLC Patients and 79 Cancer-Free Controls | |||
|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | |
| Combined Acidovorax and Veillonella for SCC | 80.79% | 89.08% | 75.76% | 88.61% |
| Capnocytophaga for AC | 72.70% | 85.28% | 69.44% | 84.42% |
| Acidovorax for distinguishing SCC from AC | 63.64% | 96.30% | 66.67% | 89.86% |
3.4. Validating the Bacterial Biomarkers in an Independent Set of Lung Cancer Patients and Controls
The sputum bacterial biomarkers developed from the cohort 1 were tested using the same procedures to diagnose lung cancer in cohort 2 consisting of 69 NSCLC patients and 79 controls. Consistent with findings in the cohort 1, abundances of Acidovorax, Streptococus, and Veillonella were higher in sputum of lung SCC patients compared with lung AC patients and cancer-free smokers (all p < 0.05). The abundance of Helicobacter was lower in sputum of lung SCC patients compared with lung AC patients and cancer-free smokers (p = 0.018). Capnocytophaga was overrepresented in sputum of lung AC patients compared with lung SCC patients and cancer-free smokers (p = 0.046).
Furthermore, the bacterial biomarkers displayed similar diagnostic values in the cohort 2 as did in the cohort 1 (Figure 5). Particularly, Acidovorax and Veillonella used in combination could diagnose lung SCC with 0.89 AUC, producing 75.76% sensitivity and 88.61% specificity (Table 5). In addition, sputum Capnocytophaga biomarker could detect lung AC with 0.83 AUC, yielding 69.44% sensitivity and 84.42% specificity (Table 5). Moreover, the use of Acidovorax as a sputum biomarker had 0.83 AUC with 66.67% sensitivity and 89.86% specificity for distinguishing between SCC and AC of the lungs. There was no association of sputum bacterial genera with age, gender, and smoking status of the participants, and stages of lung tumors (all p > 0.05), except location and histology of primary lung tumors (Supplementary Table S2). There was no statistical difference of sensitivity and specificity of combined use of Acidovorax and Veillonella for diagnosis of SCC and using Capnocytophaga for detection of AC (all p > 0.05) in the cohort 1 and cohort 2. There was also no statistical difference of sensitivity and specificity of using Capnocytophaga for detection of AC (all p > 0.05) in the cohort 1 and cohort 2. However, the use of sputum Acidovorax had a lower specificity in cohort 2 compared with cohort 1 for distinguishing between SCC and AC of the lungs (89% vs. 96%, p = 0.02), while maintaining a similar sensitivity (63% vs. 66%) (Table 5).
Figure 5.
ROC curve analysis of abundances of bacterial genera in sputum of cohort 2. (A) The combined use of Acidovorax and Veillonella produced 0.89 AUC. (B) The use of Capnocytophaga as a sputum biomarker could detect lung AC with 0.83 AUC. (C) The use of Acidovorax as a sputum biomarker had 0.83 AUC for differentiating between SCC and SC of the lungs.
4. Discussion
Our present study confirms that certain microbes, at genus level, are differentially abundant in lung tumor vs. normal lung tissues. Furthermore, we demonstrate the abundances of genera could be quantitatively measured in sputum by using ddPCR and the altered abundances of some sputum bacteria are associated with lung cancer. We further develop Acidovorax and Veillonella as a sputum biomarker panel for lung SCC, regardless of the stages. In addition, a single sputum bacterial biomarker, Capnocytophaga, could be used for detection of lung AC. Moreover, the use of Acidovorax as a sputum biomarker could distinguish between SCC and AC, the two major histological types of NSCLC. Therefore, the sputum microbiota might have the potential use as noninvasive biomarkers for diagnosis and classification of lung cancer at the early stage.
Previous studies have shown that diverse airway microbial profiles exist at different airway sites of lung cancer patients [42,68,69]. However, the comparison of bacterial profiles in primary tumor tissues and sputum of lung cancer patients has not been performed. Our findings in comparison of bacterial abundances in tumor tissues and sputum of lung cancer patients suggest that the altered bacterial genera could be classified into three categories. (1) Lung tumor microbes, which comprise Capnocytophaga and Haemophile. Aberrant abundances of the bacterial genera were exclusively found within lung tumors. The intratumoral microbes of lung cancer might be directly involved in the development and progressions of NSCLC, however, the imbalance in their abundance is not detectable in sputum. (2) Sputum microbes of lung cancer patients, such as Streptococcus, Veillonella, and H. pylori, whose aberrations were solely observed in sputum of NSCLC patients. Changes of these microbes in sputum might not simply mirror those in primary lung tumors. The discovery is in line with the previous observation [70]. The analysis of bladder tumors and the paired urine samples showed that aberrations of certain bacteria existed in urine rather than the tumor tissues, however, they had diagnostic significance for malignancy [70]. This category of microbiota might indirectly prompt tumor susceptibility and development via altering respiratory bacterial environment and modulating inflammation, inducing DNA damage, and producing metabolites involved in oncogenesis or tumor suppression [2]. (3) Bacterial genera whose changes in sputum were consistent with those in tumor tissues in the same direction, including Acidovorax and Capnocytophaga. The aberrant bacterial abundances in sputum could directly reflect those in primary lung tumors. We have also found that altered abundances of the bacterial genera in sputum are histologically dependent. Particularly, the abundances of sputum Acidovorax, Streptococcus, H. pylori, and Veillonella in sputum are related with lung SCC, whereas increased Capnocytophaga abundance in sputum is related to lung AC. Nevertheless, an extensive and deep investigation of the microbiota is needed to have a better understanding of the pathogenesis of NSCLC and provide new diagnostic and therapeutic targets for the disease.
Overall, the potential sputum bacterial biomarkers have a higher sensitivity for lung SCC compared with lung AC (80.00% vs. 72.73% p = 0.032). The findings are in good agreement with our previous studies [6,23,27,30,32,34,40,46,50,52]. We have shown that sputum-based molecular biomarkers have a higher sensitivity in identifying central SCC compared with peripheral AC of the lungs. The possible reason might be that sputum is secreted from large airways or main bronchi where SCC more commonly exists. Conversely, lung AC tumors often arise in peripheral lung tissue and originate from the smaller airways of the lungs. Future integration of the sputum-based assay with LDCT could overcome the weakness of the imaging analysis by improving accuracy for the early detection of lung SCC.
Among the bacterial genera analyzed, Acidovorax was found by Greathouse et al. to have an elevated abundance in lung SCC tissues with TP53 mutation [8]. Furthermore, there was a significant increase in lung tumor volume in mice inoculated with Acidovorax temperans. Acidovorax temperans could contribute to lung carcinogenesis in the presence of activated Kras and mutant p53 and, thus, act as a promoter in the development and progression of the disease [8]. Our current study supports this early report [8], and more importantly, suggests that Acidovorax might provide a sputum biomarker for lung SCC. Capnocytophaga species were proposed to be involved in lung carcinogenesis and lower respiratory tract infections [15]. Furthermore, Capnocytophaga might induce long-term immune response/infection to the organ or cancer growth environment, which favors the growth of these bacteria in the airways [68]. Our study also suggests that Capnocytophaga abundance is significantly higher in NSCLC vs. normal lung tissues. Tsay et al. found an increased abundance of Streptococcus and Veillonella in the lower respiratory tract of NSCLC patients, which was associated with upregulation of the ERK and PI3K signaling pathways [12]. It has been well accepted that H. pylori is a risk factor for gastric and several other cancers [2]. Our present study demonstrates a close association of H. pylori with lung SCC. However, rigorous investigations regarding the H. pylori-lung cancer association remain to be performed.
Smoking causes most lung cancers, but lung cancer can be found in never smokers [1]. Interestingly, the abundances of the genera tested in this present study were not associated with the smoking status of the patients. The result suggests that the microbiota aberrations might play an important role in lung tumorigenesis of nonsmokers. The observation is in line with previous studies [16,17], in which lung cancer patients who were never smokers had a long history of bacterial respiratory tract infection. Dysregulation of the genera could be involved in the development and progression of NSCLC via a specific manner that is beyond tobacco-smoking-related carcinogenesis.
The sputum biomarkers were further tested in an independent (validation) cohort of cases and controls. The diagnostic significance of the bacterial biomarkers for diagnosis of SCC and AC of the lungs was confirmed. However, although the use of sputum Acidovorax for distinguishing between SCC and AC had a similar sensitivity, its specificity was reduced in the validation cohort. Possible explanation for the difference might be that the sputum specimens of the validation cohort were collected five years ago, whereas sputum samples in cohort 1 were fresh and collected within six months. DNA quality might significantly decline with long storage duration, leading to a lower specificity of the sputum biomarker for distinguishing between SCC and AC of the lungs.
This study may suffer some limitations. (1), the sample size is small. We will prospectively validate the sputum biomarkers in a large cohort. (2), we only assessed 25 bacterial genera whose changes were previously suggested to be associated with lung cancer. Although the results show promise, the sensitivity and specificity of the biomarkers are not enough in routine laboratory settings. We will evaluate more lung tumor-bacteria to identify additional bacterial biomarkers that can be added to the current ones so that the diagnostic efficacy of the sputum tests could be improved.
5. Conclusions
We show that aberrant microbial composition, at genus level, is present in lung tumor and sputum of lung cancer patients. We have for the first time developed sputum bacterial biomarkers that could be potentially used for the early detection and classification of lung cancer, though a larger sample study is needed to validate the findings.
Acknowledgments
This work was supported in part by National Cancer Institute grant 1UH2CA251139-01, 5R21CA240556-02, and VA merit Award I01 CX000512 (F.J.). We thank Ruth L Katz, The University of Texas M.D. Anderson Cancer Center for providing sputum specimens.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4418/11/3/407/s1, Table S1. The association of abundances of bacterial genera in sputum of cohort 1 with the age, gender, ethnic group, tumor stageand location, and smoking status of the patients determined by Pearson’s correlation coefficient test. A p-value < 0.05 is statistically significant; Table S2. The association of abundances of bacterial genera in sputum of cohort 2 with the age, gender, ethnic group, tumorstageand location, and smoking status of the patients determined by Pearson’s correlation coefficient test. A p-value < 0.05 is statistically significant.
Author Contributions
Q.L. and F.J. designed research approaches, conducted the experiments, and participated in data interpretation. V.K.H., J.D., and N.W.T. consented patients and collected specimens. V.K.H. and F.J. prepared the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by National Cancer Institute grant 1UH2CA251139-01, 5R21CA240556-02, and VA merit Award I01 CX000512 (F.J.).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of the University of Maryland Baltimore.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon a reasonable request.
Conflicts of Interest
No conflicts of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.The National Lung Screening Trial Research Team Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garrett W.S. Cancer and the microbiota. Science. 2015;348:80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kovaleva O.V., Romashin D., Zborovskaya I.B., Davydov M.M., Shogenov M.S., Gratchev A. Human Lung Microbiome on the Way to Cancer. J. Immunol. Res. 2019;2019:1394191. doi: 10.1155/2019/1394191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao Q., Jiang F., Yin R., Wang J., Xia W., Dong G., Ma W., Yang Y., Xu L., Hu J. Interplay between the lung microbiome and lung cancer. Cancer Lett. 2018;415:40–48. doi: 10.1016/j.canlet.2017.11.036. [DOI] [PubMed] [Google Scholar]
- 5.Peters B.A., Hayes R.B., Goparaju C., Reid C., Pass H.I., Ahn J. The microbiome in lung cancer tissue and recurrence-free survival. Cancer Epidemiol. Biomark. Prev. 2019;28:731–740. doi: 10.1158/1055-9965.EPI-18-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu G., Gail M.H., Consonni D., Carugno M., Humphrys M., Pesatori A.C., Caporaso N.E., Goedert J.J., Ravel J., Landi M.T. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17:163. doi: 10.1186/s13059-016-1021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H.-X., Tao L.-L., Zhang J., Zhu Y.-G., Zheng Y., Liu D., Zhou M., Ke H., Shi M.-M., Qu J.-M. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int. J. Cancer. 2018;142:769–778. doi: 10.1002/ijc.31098. [DOI] [PubMed] [Google Scholar]
- 8.Greathouse K.L., White J.R., Vargas A.J., Bliskovsky V.V., Beck J.A., von Muhlinen N., Polley E.C., Bowman E.D., Khan M.A., Robles A.I., et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19:123. doi: 10.1186/s13059-018-1501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apopa P.L., Alley L., Penney R.B., Arnaoutakis K., Steliga M.A., Jeffus S., Bircan E., Gopalan B., Jin J., Patumcharoenpol P., et al. PARP1 Is Up-Regulated in Non-small Cell Lung Cancer Tissues in the Presence of the Cyanobacterial Toxin Microcystin. Front. Microbiol. 2018;9:1757. doi: 10.3389/fmicb.2018.01757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., O’Brien J.L., Ajami N.J., E Scheurer M., Amirian E.S., Armstrong G., Tsavachidis S., Thrift A.P., Jiao L., Wong M.C., et al. Lung tissue microbial profile in lung cancer is distinct from emphysema. Am. J. Cancer Res. 2018;8:1775–1787. [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S.H., Sung J.Y., Yong D., Chun J., Kim S.Y., Song J.H., Chung K.S., Kim E.Y., Jung J.Y., Kang Y.A., et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer. 2016;102:89–95. doi: 10.1016/j.lungcan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Tsay J.-C.J., Wu B.G., Badri M.H., Clemente J.C., Shen N., Meyn P., Li Y., Yie T.-A., Lhakhang T., Olsen E., et al. Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am. J. Respir. Crit. Care Med. 2018;198:1188–1198. doi: 10.1164/rccm.201710-2118OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin J., Gan Y., Liu H., Wang Z., Yuan J., Deng T., Zhou Y., Zhu Y., Zhu H., Yang S., et al. Diminishing microbiome richness and distinction in the lower respiratory tract of lung cancer pa-tients: A multiple comparative study design with independent validation. Lung Cancer. 2019;136:129–135. doi: 10.1016/j.lungcan.2019.08.022. [DOI] [PubMed] [Google Scholar]
- 14.Yang J., Mu X., Wang Y., Zhu D., Zhang J., Liang C., Chen B., Wang J., Zhao C., Zuo Z., et al. Dysbiosis of the Salivary Microbiome Is Associated With Non-smoking Female Lung Cancer and Correlated With Immunocytochemistry Markers. Front. Oncol. 2018;8:520. doi: 10.3389/fonc.2018.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan X., Yang M., Liu J., Gao R., Hu J., Li J., Zhang L., Shi Y., Guo H., Cheng J., et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am. J. Cancer Res. 2015;5:3111–3122. [PMC free article] [PubMed] [Google Scholar]
- 16.Hosgood H.D., 3rd, Sapkota A.R., Rothman N., Rohan T., Hu W., Xu J., Vermeulen R., He X., White J.B., Wu G., et al. The potential role of lung microbiota in lung cancer attributed to house-hold coal burning exposures. Environ. Mol. Mutagen. 2014;55:643–651. doi: 10.1002/em.21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cameron S.J.S., Lewis K.E., Huws S.A., Hegarty M.J., Lewis P.D., Pachebat J.A., Mur L.A.J. A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PLoS ONE. 2017;12:e0177062. doi: 10.1371/journal.pone.0177062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Druzhinin V.G., Matskova L.V., Demenkov P.S., Baranova E.D., Volobaev V.P., Minina V.I., Apalko S.V., Churina M.A., Romanyuk S.A., Shcherbak S.G., et al. Taxonomic diversity of sputum microbiome in lung cancer patients and its relationship with chromosomal aberrations in blood lymphocytes. Sci. Rep. 2020;10:9681. doi: 10.1038/s41598-020-66654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng C., Wang Z., Wang J., Ding C., Sun C., Liu P., Xu X., Liu Y., Chen B., Gu B. Characterization of the lung microbiome and exploration of potential bacterial biomarkers for lung cancer. Transl. Lung Cancer Res. 2020;9:693–704. doi: 10.21037/tlcr-19-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manzoor S.S., Doedens A., Burns M.B. The promise and challenge of cancer microbiome research. Genome Biol. 2020;21:131. doi: 10.1186/s13059-020-02037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta C., Su J., Zhan M., Stass S.A., Jiang F. Sputum long non-coding RNA biomarkers for diagnosis of lung cancer. Cancer Biomark. 2019;26:219–227. doi: 10.3233/CBM-190161. [DOI] [PubMed] [Google Scholar]
- 22.Su J., Leng Q., Lin Y., Ma J., Jiang F., Lee C.-J., Fang H., Jiang F. Integrating Circulating Immunological and Sputum Biomarkers for the Early Detection of Lung Cancer. Biomark. Cancer. 2018;10 doi: 10.1177/1179299X18759297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su Y., Fang H., Jiang F. Integrating DNA methylation and microRNA biomarkers in sputum for lung cancer detection. Clin. Epigenet. 2016;8:109. doi: 10.1186/s13148-016-0275-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su Y., Guarnera M.A., Fang H., Jiang F. Small non-coding RNA biomarkers in sputum for lung cancer diagnosis. Mol. Cancer. 2016;15:36. doi: 10.1186/s12943-016-0520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su J., Anjuman N., Guarnera M.A., Zhang H., Stass S.A., Jiang F. Analysis of Lung Flute-collected Sputum for Lung Cancer Diagnosis. Biomark. Insights. 2015;10:55–61. doi: 10.4137/BMI.S26883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su J., Liao J., Gao L., Shen J., Guarnera M.A., Zhan M., Fang H., Stass S.A., Jiang F. Analysis of small nucleolar RNAs in sputum for lung cancer diagnosis. Oncotarget. 2016;7:5131–5142. doi: 10.18632/oncotarget.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xing L., Su J., Guarnera M.A., Zhang H., Cai L., Zhou R., Stass S.A., Jiang F. Sputum microRNA biomarkers for identifying lung cancer in indeterminate solitary pul-monary nodules. Clin. Cancer Res. 2015;21:484–489. doi: 10.1158/1078-0432.CCR-14-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu L., Shen J., Mannoor K., Guarnera M., Jiang F. Identification of ENO1 as a potential sputum biomarker for early-stage lung cancer by shotgun proteomics. Clin. Lung Cancer. 2014;15:372–378.e1. doi: 10.1016/j.cllc.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen J., Liao J., Guarnera M.A., Fang H., Cai L., Stass S.A., Jiang F. Analysis of MicroRNAs in sputum to improve computed tomography for lung cancer diagnosis. J. Thorac. Oncol. 2014;9:33–40. doi: 10.1097/JTO.0000000000000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li N., Ma J., Guarnera M.A., Fang H., Cai L., Jiang F. Digital PCR quantification of miRNAs in sputum for diagnosis of lung cancer. J. Cancer Res. Clin. Oncol. 2013;140:145–150. doi: 10.1007/s00432-013-1555-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anjuman N., Li N., Guarnera M., A Stass S., Jiang F. Evaluation of lung flute in sputum samples for molecular analysis of lung cancer. Clin. Transl. Med. 2013;2:15. doi: 10.1186/2001-1326-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu L., Todd N.W., Xing L., Xie Y., Zhang H., Liu Z., Fang H., Zhang J., Katz R.L., Jiang F. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int. J. Cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang F., Todd N.W., Li R., Zhang H., Fang H., Stass S.A. A Panel of Sputum-Based Genomic Marker for Early Detection of Lung Cancer. Cancer Prev. Res. 2010;3:1571–1578. doi: 10.1158/1940-6207.CAPR-10-0128. [DOI] [PubMed] [Google Scholar]
- 34.Xing L., Todd N.W., Yu L., Fang H., Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod. Pathol. 2010;23:1157–1164. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 35.Xie Y., Todd N.W., Liu Z., Zhan M., Fang H., Peng H., Alattar M., Deepak J., Stass S.A., Jiang F. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67:170–176. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang F., Todd N.W., Qiu Q., Liu Z., Katz R.L., Stass S.A. Combined genetic analysis of sputum and computed tomography for noninvasive diagnosis of non-small-cell lung cancer. Lung Cancer. 2009;66:58–63. doi: 10.1016/j.lungcan.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu Q., Todd N.W., Li R., Peng H., Liu Z., Yfantis H.G., Katz R.L., Stass S.A., Jiang F. Magnetic enrichment of bronchial epithelial cells from sputum for lung cancer diagnosis. Cancer. 2008;114:275–283. doi: 10.1002/cncr.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li R., Todd N.W., Qiu Q., Fan T., Zhao R.Y., Rodgers W.H., Fang H.-B., Katz R.L., Stass S.A., Jiang F. Genetic Deletions in Sputum as Diagnostic Markers for Early Detection of Stage I Non–Small Cell Lung Cancer. Clin. Cancer Res. 2007;13:482–487. doi: 10.1158/1078-0432.CCR-06-1593. [DOI] [PubMed] [Google Scholar]
- 39.Liao J., Shen J., Leng Q., Qin M., Zhan M., Jiang F. MicroRNA-based biomarkers for diagnosis of non-small cell lung cancer (NSCLC) Thorac. Cancer. 2020;11:762–768. doi: 10.1111/1759-7714.13337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H., Jiang Z., Leng Q., Bai F., Wang J., Ding X., Li Y., Zhang X., Fang H., Yfantis H.G., et al. A prediction model for distinguishing lung squamous cell carcinoma from adenocarcinoma. Oncotarget. 2017;8:50704–50714. doi: 10.18632/oncotarget.17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bingula R., Filaire M., Radosevic-Robin N., Berthon J.Y., Bernalier-Donadille A., Vasson M.P., Thivat E., Kwiatkowski F., Filaire E. Characterisation of gut, lung, and upper airways microbiota in patients with non-small cell lung carcinoma: Study protocol for case-control observational trial. Medicine. 2018;97:e13676. doi: 10.1097/MD.0000000000013676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goto T. Airway Microbiota as a Modulator of Lung Cancer. Int. J. Mol. Sci. 2020;21:3044. doi: 10.3390/ijms21093044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romeo M.S., Sokolova I.A., Morrison L.E., Zeng C., Barón A.E., Hirsch F.R., Miller Y.E., Franklin W.A., Varella-Garcia M. Chromosomal abnormalities in non-small cell lung carcinomas and in bron-chial epithelia of high-risk smokers detected by multi-target interphase fluorescence in situ hybridization. J. Mol. Diagn. 2003;5:103–112. doi: 10.1016/S1525-1578(10)60459-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varella-Garcia M., Kittelson J., Schulte A.P., Vu K.O., Wolf H.J., Zeng C., Hirsch F.R., Byers T., Kennedy T., Miller Y.E., et al. Multi-target interphase fluorescence in situ hybridization assay increases sensitivity of sputum cytology as a predictor of lung cancer. Cancer Detect. Prev. 2004;28:244–251. doi: 10.1016/j.cdp.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 45.Katz R.L., Zaidi T.M., Fernandez R.L., Zhang J., He W., Acosta C., Daniely M., Madi L., A Vargas M., Dong Q., et al. Automated detection of genetic abnormalities combined with cytology in sputum is a sensitive predictor of lung cancer. Mod. Pathol. 2008;21:950–960. doi: 10.1038/modpathol.2008.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su Y., Bin Fang H., Jiang F. An epigenetic classifier for early stage lung cancer. Clin. Epigenet. 2018;10:68. doi: 10.1186/s13148-018-0502-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang F., Qiu Q., Khanna A., Todd N.W., Deepak J., Xing L., Wang H., Liu Z., Su Y., Stass S.A., et al. Aldehyde Dehydrogenase 1 Is a Tumor Stem Cell-Associated Marker in Lung Cancer. Mol. Cancer Res. 2009;7:330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma J., Li N., Guarnera M., Jiang F. Quantification of Plasma miRNAs by Digital PCR for Cancer Diagnosis. Biomark. Insights. 2013;8:127–136. doi: 10.4137/BMI.S13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma J., Mannoor K., Gao L., Tan A., Guarnera M.A., Zhan M., Shetty A., Stass S.A., Xing L., Jiang F. Characterization of microRNA transcriptome in lung cancer by next-generation deep sequencing. Mol. Oncol. 2014;8:1208–1219. doi: 10.1016/j.molonc.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin Y., Holden V., Dhilipkannah P., Deepak J., Todd N.W., Jiang F. A Non-Coding RNA Landscape of Bronchial Epitheliums of Lung Cancer Patients. Biomedicines. 2020;8:88. doi: 10.3390/biomedicines8040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Y., Leng Q., Zhan M., Jiang F. A Plasma Long Noncoding RNA Signature for Early Detection of Lung Cancer. Transl. Oncol. 2018;11:1225–1231. doi: 10.1016/j.tranon.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leng Q., Tsou J.-H., Zhan M., Jiang F. Fucosylation genes as circulating biomarkers for lung cancer. J. Cancer Res. Clin. Oncol. 2018;144:2109–2115. doi: 10.1007/s00432-018-2735-0. [DOI] [PubMed] [Google Scholar]
- 53.Sambo F., Finotello F., Lavezzo E., Baruzzo G., Masi G., Peta E., Falda M., Toppo S., Barzon L., Di Camillo B. Optimizing PCR primers targeting the bacterial 16S ribosomal RNA gene. BMC Bioinform. 2018;19:343. doi: 10.1186/s12859-018-2360-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhi X.Y., Tang S.K., Li W.J., Xu L.H., Jiang C.L. New genus-specific primers for the PCR identification of novel isolates of the genus Strep-tomonospora. FEMS Microbiol. Lett. 2006;263:48–53. doi: 10.1111/j.1574-6968.2006.00402.x. [DOI] [PubMed] [Google Scholar]
- 55.Zheng Y., Fang Z., Xue Y., Zhang J., Zhu J., Gao R., Yao S., Ye Y., Wang S., Lin C., et al. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes. 2020;11:1030–1042. doi: 10.1080/19490976.2020.1737487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katayama Y., Yamada T., Shimamoto T., Iwasaku M., Kaneko Y., Uchino J., Takayama K. The role of the gut microbiome on the efficacy of immune checkpoint inhibitors in Japanese responder patients with advanced non-small cell lung cancer. Transl. Lung Cancer Res. 2019;8:847–853. doi: 10.21037/tlcr.2019.10.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang D., Su X., Yuan M., Zhang S., He J., Deng Q., Qiu W., Dong H., Cai S. The characterization of lung microbiome in lung cancer patients with different clinicopathology. Am. J. Cancer Res. 2019;9:2047–2063. [PMC free article] [PubMed] [Google Scholar]
- 58.Bingula R., Filaire E., Molnar I., Delmas E., Berthon J.-Y., Vasson M.-P., Bernalier-Donadille A., Filaire M. Characterisation of microbiota in saliva, bronchoalveolar lavage fluid, non-malignant, peritumoural and tumour tissue in non-small cell lung cancer patients: A cross-sectional clinical trial. Respir. Res. 2020;21:129. doi: 10.1186/s12931-020-01392-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Demirci M., Tokman H., Uysal H., Demiryas S., Karakullukcu A., Saribas S., Cokugras H., Kocazeybek B. Reduced Akkermansia muciniphila and Faecalibacterium prausnitzii levels in the gut microbiota of children with allergic asthma. Allergol. Immunopathol. 2019;47:365–371. doi: 10.1016/j.aller.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 60.Abed J., Maalouf N., Parhi L., Chaushu S., Mandelboim O., Bachrach G. Tumor Targeting by Fusobacterium nucleatum: A Pilot Study and Future Perspectives. Front. Cell. Infect. Microbiol. 2017;7:295. doi: 10.3389/fcimb.2017.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wypych T.P., Wickramasinghe L.C., Marsland B.J. The influence of the microbiome on respiratory health. Nat. Immunol. 2019;20:1279–1290. doi: 10.1038/s41590-019-0451-9. [DOI] [PubMed] [Google Scholar]
- 62.Mounika P. Helicobacter pylori Infection and Risk of Lung Cancer: A Meta-Analysis. Lung Cancer Int. 2013;2013:131869. doi: 10.1155/2013/131869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ding L., Yang Z., Lu J., Ma L., Liu Y., Wu X., Yao W., Zhang X., Zhu K. Characterization of Phenotypic and Genotypic Traits of Klebsiella pneumoniae from Lung Cancer Patients with Respiratory Infection. Infect. Drug Resist. 2020;13:237–245. doi: 10.2147/IDR.S229085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin C., Lagoudas G.K., Zhao C., Bullman S., Bhutkar A., Hu B., Ameh S., Sandel D., Liang X.S., Mazzilli S., et al. Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell. 2019;176:998–1013.e16. doi: 10.1016/j.cell.2018.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamura A., Hebisawa A., Kusaka K., Hirose T., Suzuki J., Yamane A., Nagai H., Fukami T., Ohta K., Takahashi F. Relationship Between Lung Cancer and Mycobacterium Avium Complex Isolated Using Bronchoscopy. Open Respir. Med. J. 2016;10:20–28. doi: 10.2174/1874306401610010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.García-Núñez M., Millares L., Pomares X., Ferrari R., Pérez-Brocal V., Gallego M., Espasa M., Moya A., Monsó E. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J. Clin. Microbiol. 2014;52:4217–4223. doi: 10.1128/JCM.01967-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao Z., Fei K., Bai H., Wang Z., Duan J., Wang J. Metagenome association study of the gut microbiome revealed biomarkers linked to chemo-therapy outcomes in locally advanced and advanced lung cancer. Thorac. Cancer. 2021;12:66–78. doi: 10.1111/1759-7714.13711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gomes S., Cavadas B., Ferreira J.C., Marques P.I., Monteiro C., Sucena M., Sousa C., Rodrigues L.V., Teixeira G., Pinto P., et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci. Rep. 2019;9:12838. doi: 10.1038/s41598-019-49195-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu N.-N., Ma Q., Ge Y., Yi C.-X., Wei L.-Q., Tan J.-C., Chu Q., Li J.-Q., Zhang P., Wang H. Microbiome dysbiosis in lung cancer: From composition to therapy. NPJ Precis. Oncol. 2020;4:33. doi: 10.1038/s41698-020-00138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mansour B., Monyók Á., Makra N., Gajdács M., Vadnay I., Ligeti B., Juhász J., Szabó D., Ostorházi E. Bladder cancer-related microbiota: Examining differences in urine and tissue sam-ples. Sci. Rep. 2020;10:11042. doi: 10.1038/s41598-020-67443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon a reasonable request.