Accumulation of trimethylamine N-oxide improves both growth and survival of deep-sea bacteria under high hydrostatic pressure.
Abstract
High hydrostatic pressure (HHP) is a characteristic environmental factor of the deep ocean. However, it remains unclear how piezotolerant bacteria adapt to HHP. Here, we identify a two-step metabolic pathway to cope with HHP stress in a piezotolerant bacterium. Myroides profundi D25T, obtained from a deep-sea sediment, can take up trimethylamine (TMA) through a previously unidentified TMA transporter, TmaT, and oxidize intracellular TMA into trimethylamine N-oxide (TMAO) by a TMA monooxygenase, MpTmm. The produced TMAO is accumulated in the cell, functioning as a piezolyte, improving both growth and survival at HHP. The function of the TmaT-MpTmm pathway was further confirmed by introducing it into Escherichia coli and Bacillus subtilis. Encoded TmaT-like and MpTmm-like sequences extensively exist in marine metagenomes, and other marine Bacteroidetes bacteria containing genes encoding TmaT-like and MpTmm-like proteins also have improved HHP tolerance in the presence of TMA, implying the universality of this HHP tolerance strategy in marine Bacteroidetes.
INTRODUCTION
The deep ocean (with water depth more than 1000 m) biosphere represents one of the largest ecosystems on Earth and breeds a variety of life forms. It is estimated that more than 1029 microbial cells exist there (1). In addition, a lot of microorganisms passively migrate from surface seawater to deep seawater every day with marine snow, whale falls, and ocean currents (2, 3). The migration of these microorganisms is accompanied by increasing pressure and decreasing temperature. High hydrostatic pressure (HHP) has a large inhibitory effect on many physiological activities of microbial cells (4), such as membrane fluidity, RNA synthesis, motility, nutrient uptake, cell division, protein synthesis, and replication (5, 6). While many of the sinking microbes are eliminated because of their failure to adapt to the deep-sea environment, no doubt some microbes can survive in deep sea and even grow and reproduce at HHP environment. On the basis of their hydrostatic pressure tolerance ability, bacteria are divided into piezosensitive bacteria, piezotolerant bacteria, piezophiles, and hyperpiezophiles (7). Among them, some piezophiles have been well studied, which are mainly distributed in α-proteobacteria, δ-proteobacteria, and γ-proteobacteria (7, 8). In piezophiles of the family Vibrionaceae, the ToxR/ToxS two-component system is a pressure sensor, which controls the expression of the ompH/ompL genes and many others (4, 9). However, for widely distributed piezotolerant bacteria, it remains unclear whether their adaption to HHP has driven the evolution of specific gene sets to cope with HHP stress.
Trimethylamine (TMA) and trimethylamine N-oxide (TMAO) are nitrogen-containing organic compounds widely dispersed in the ocean. Two functions of TMAO in marine microbes have been described. First, TMAO is used as a nitrogen source by the SAR11 clade and marine Roseobacter clade of α-proteobacteria (10). Second, TMAO is used as an electron acceptor by some γ-proteobacteria under anaerobic conditions (11), such as Shewanella (12–14) and Vibrio (11, 15). In addition, the functions of TMAO in marine animals are also studied. One well-known function is related to HHP tolerance of animals in deep sea. Deep-sea animals can enrich TMAO in their cells, which is used to stabilize intracellular proteins and cell structure for their survival under HHP (16). In teleost fish, muscle TMAO contents range from less than 50 mmol/kg in shallow species to more than 260 mmol/kg in a deep-sea species from 4850-m depth (17). It is believed that in the deep ocean with a water depth of more than 5 km, fish cannot survive without TMAO (18). However, it is yet unknown whether deep-sea bacteria use TMAO to cope with HHP stress.
Myroides profundi D25T (hereafter referred to as strain D25) is a member of the phylum Bacteroidetes, which was isolated from a deep-sea sediment (1245-m water depth) in the southern Okinawa Trough (19). Comparative genomic and transcriptomic analyses have revealed the strategy adopted by this species during its evolution from land to sea (20). In the current study, we found that strain D25 is a piezotolerant bacterium, which uses TMAO as a piezolyte to cope with HHP stress. Strain D25 takes up TMA through a TMA transporter, TmaT, then oxidizes TMA into TMAO using a TMA monooxygenase (MpTmm). The resulting accumulation of intracellular TMAO enables growth at high pressure. The function of the TmaT-MpTmm pathway was also confirmed by introducing it in Escherichia coli and Bacillus subtilis. Further analysis showed that the presence of TMA also improved the growth of other bacteria in the phylum Bacteroidetes containing genes encoding the TmaT and MpTmm homologs under HHP, suggesting that this may be a common strategy adopted by deep-sea Bacteroidetes. This study reveals a new function of TMA/TMAO in marine bacteria and provides a direct link between a unique metabolic pathway and HHP adaptation in deep-sea bacteria.
RESULTS
Accumulation of intracellular TMAO facilitates the survival of strain D25 under HHP
Quaternary amines TMA and TMAO are widespread in the ocean, which can be taken up and metabolized as nutrients by marine bacteria of the Roseobacter clade, e.g., Ruegeria pomeroyi (21). A gene (MPR_3295, Mptmm) in the genome of strain D25 was predicted to encode a putative TMA monooxygenase (Tmm). Tmm has been reported to convert TMA to TMAO in cosmopolitan marine bacteria such as members of the Roseobacter clade (e.g., R. pomeroyi) and the SAR11 clade (Pelagibacter ubique), which is essential for the utilization of TMA as a nutrient source in bacteria (21). To test whether it can use TMA or TMAO, strain D25 was cultured with TMA or TMAO as the sole nitrogen source, and R. pomeroyi DSS-3 (hereafter referred to as strain DSS-3) was used as a positive control. Unlike strain DSS-3 that could grow with TMA or TMAO as nitrogen source, strain D25 could not grow with either TMA or TMAO as nitrogen source (Fig. 1A). However, when strain D25 was cultured in a 2216E medium containing TMA, it was found that TMA was consumed and TMAO was accumulated intracellularly (Fig. 1B), which suggests that strain D25 can transport TMA into the cell and metabolize TMA into TMAO in the cell but cannot further metabolize TMAO as nutrient. In addition, we searched the genome of strain D25 for genes encoding key enzymes involved in the downstream catabolism of TMAO, i.e., TMAO demethylase (tdm) and dimethylamine monooxygenase (dmmABC); however, none of such genes were found. This supports our observation that strain D25 cannot metabolize TMAO (Fig. 1B).
Fig. 1. Effect of methylamine on the growth and survival of strain D25 under HHP.
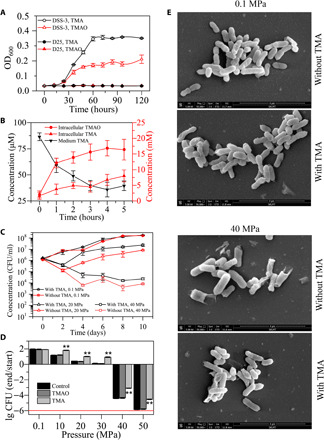
(A) Growth curves of strains D25 and DSS-3 cultured with TMA or TMAO as the sole nitrogen source. (B) Changes of the concentrations of medium TMA and intracellular TMA and TMAO of strain D25 cultured in 2216E medium with an initial TMA concentration of 85 μM. (C) Growth curves of strain D25 at 4°C at 0.1, 20, and 40 MPa with or without 10 μM TMA. (D) Effect of 10 μM TMAO or TMA on the growth and survival of strain D25 at different pressures. The cell number at the start of the incubation (start) was ca. 1 × 106 CFU/ml. The cultures were incubated at 4°C for 10 days, and then CFU was counted and compared with that of the starting culture (end/start). The red line refers to a complete death of cells in the culture. Control: Strain D25 cultured without TMAO or TMA. (E) Morphological observation of strain D25 cultivated at different pressures with or without 10 μM TMA. The error bars represent SDs from triplicate experiments. The asterisks show statistical difference from control (**P < 0.01, two-tailed t test). ETD, Everhart-Thornley detector; WD, working distance. SKLMT, State Key Laboratory of Microbial Technology.
Because TMAO is a well-known protein stabilizer that can counteract the effects of HHP in marine animals (18), we postulated that strain D25 may accumulate TMAO in the cell to cope with HHP stress. To test this postulation, we investigated the growth of strain D25 with or without TMA in the medium at the atmospheric pressure, 20 and 40 MPa. The result showed that the addition of TMA in the culture medium improved the growth of strain D25 at 20 MPa and the survival of strain D25 at 40 MPa, and 10 μM TMA showed the best effect, but such an effect was not observed at the atmospheric pressure (Fig. 1C and fig. S1A). We then investigated the growth and survival of strain D25 under different HHP conditions and found that the presence of 10 μM TMA significantly improved the growth of strain D25 under 10 to 30 MPa and increased the survival of strain D25 even under 50 MPa (Fig. 1D). In contrast, neither TMAO nor any other quaternary amines added in the medium could improve the growth of strain D25 at HHP condition (Fig. 1D and fig. S1B). In these experiments, after incubation of strain D25 under different pressures, the remained dissolved oxygen concentration in the medium was all more than 4.1 mg/liter, indicating that dissolved oxygen was sufficient to support the growth of strain D25 during cultivation and that HHP was the main factor to impair the cell growth. In addition, it is noteworthy that the presence of TMA also helped maintain the cell integrity and prevent cells from collapsing at 40 MPa (Fig. 1E). Together, these results suggest that strain D25 can take up TMA from the environment and accumulate TMAO in the cell to cope with HHP stress.
Identification and characterization of the TMA monooxygenase responsible for oxidizing TMA into TMAO in strain D25
Because strain D25 can convert TMA into TMAO in its cell, we speculate that the putative Tmm encoded by the gene Mptmm (MPR_3295) may catalyze this reaction in strain D25. This putative Tmm has 55% sequence identity and 98% sequence coverage with the Tmm of strain DSS-3. To analyze the function of gene Mptmm, we overexpressed this gene in E. coli BL21 (fig. S2A), and the recombinant protein was active to oxidize TMA into TMAO at both 25° and 4°C (Fig. 2A), indicating that gene Mptmm encodes a Tmm, named MpTmm. Moreover, the presence of TMA in the culture medium significantly induced the transcription of gene Mptmm (Fig. 2B), suggesting that gene Mptmm is functional in converting TMA to TMAO in strain D25. HHP also induced the higher expression level of gene Mptmm in strain D25 (Fig. 2C), suggesting that gene Mptmm may be involved in coping with HHP stress in strain D25.
Fig. 2. Characterization of MpTmm.
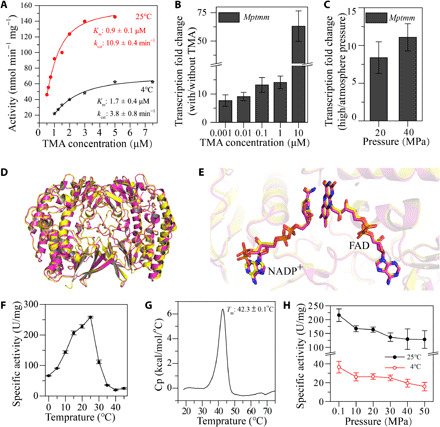
(A) Nonlinear fit curves for the oxidation of TMA by recombinant MpTmm at 4° and 25°C. (B) Effect of different concentrations of TMA on the expression of gene Mptmm in strain D25. Strain D25 was cultured in 2216E medium with or without TMA for 1 hour. (C) Effect of hydrostatic pressure on the expression of gene Mptmm in strain D25. Strain D25 was cultured in 2216E medium at atmosphere pressure, 20 or 40 MPa for 1 hour. (D) Comparison of the structures of MpTmm and RnTmm. MpTmm is colored in yellow, and RnTmm in purple. (E) Comparison of the locations of NADP+ (left) and FAD (right) in the crystal structures of MpTmm and RnTmm. The FAD and NADP+ molecules are shown in sticks colored in yellow for MpTmm and in purple for RnTmm. (F) Effect of temperature on the activity of MpTmm. (G) Measurement of the Tm value of MpTmm by DSC. (H) Effect of pressure on the activity of MpTmm. The enzymatic activities at different pressures were measured at 25° and 4°C, respectively. The error bars represent SDs from triplicate experiments.
We further characterized the MpTmm of strain D25 using the recombinant protein. The crystal structure of MpTmm was solved to 1.69 Å (table S1). The overall structure of MpTmm is similar to that of RnTmm (22), an MpTmm homolog from Roseovarius nubinhibens ISM (hereafter referred to as strain ISM), with a root mean square deviation between these two structures of 0.8 Å (Fig. 2D). The cofactors flavin adenine dinucleotide (FAD) and nicotinamide adenine dinucleotide phosphate (NADP+) in these two structures are also located in the similar positions (Fig. 2E). In addition, MpTmm has 52.8% sequence identity and 98% sequence coverage with RnTmm. These data further indicate that MpTmm is a Tmm. Strain ISM is a marine Roseobacter clade strain isolated from the surface water of the Caribbean Sea (23). Compared with RnTmm (22), MpTmm displays typical characteristics of a cold-adapted enzyme. It has a low optimum temperature of 25°C, retains 20 to 40% of the maximum activity at 0° to 5°C (Fig. 2F), and has a low Tm (denaturing temperature) of 42° ± 0.05°C (Fig. 2G). In addition, even at 50 MPa pressure, MpTmm still retained 59% (25°C) and 43% (4°C) of its activity at atmosphere pressure (Fig. 2H). Together, these results indicate that MpTmm is a cold-adapted and HHP tolerant enzyme, which can be functional in the in situ deep-sea environment with low temperature and high pressure. Thus, it can be predicted that strain D25 is capable of converting TMA into TMAO via MpTmm and accumulating TMAO in its cell in the in situ deep-sea environment.
Identification and characterization of the TMA-specific transporter in strain D25
The result in Fig. 1B showed that TMA in the 2216E medium of strain D25 was consumed during cultivation, indicating that strain D25 can transport TMA into its cell. We tried to find the gene encoding the TMA transporter by searching the genome of strain D25, but failed. Then, we sequenced the transcriptomes of strain D25 cultured under different pressures to uncover the potential TMA transporter and to investigate the expression and regulation of the genes involved in TMA metabolism. When the pressure increased, there are 533 and 430 genes significantly up- and down-regulated, respectively, and the transcription level of the gene Mptmm was up-regulated 22.2-fold, consistent with the result in Fig. 2C. We then screened the transporter genes whose transcription levels were up-regulated at HHP condition (38 genes were found), and a gene (MPR_0426, tmaT) encoding a predicted membrane protein belonging to the betaine, carnitine, and choline transporter (BCCT) family was found to be up-regulated 7.9-fold. The quantitative real-time polymerase chain reaction (PCR) result also confirmed that HHP induced higher expression level of this gene (Fig. 3A), suggesting that gene tmaT may be involved in coping with HHP stress in strain D25.
Fig. 3. Characterization of TmaT.
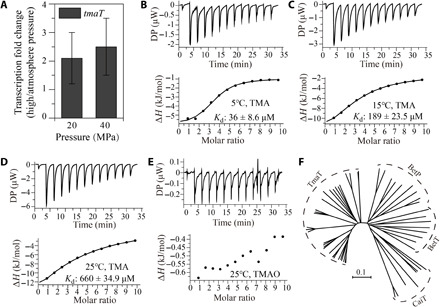
(A) Effect of hydrostatic pressure on the expression of gene tmaT in strain D25 detected by quantitative real-time PCR. Strain D25 was cultured in 2216E medium at atmosphere pressure, 20 or 40 MPa for 1 hour. The error bars represent SDs from triplicate experiments. (B to E) ITC curves for titrations of TMA or TMAO into TmaT. ITC traces (top) and integrated binding isotherms (bottom) are shown. The titration substrates and temperatures are shown in the pictures. DP, differential power. (F) Molecular phylogenetic analysis of TmaT and its homologs with other BCCT transporters. Sequences are from the IMG/JGI database. BetP, BetT, and CaiT are BCCT transporters for glycine betaine, choline, and carnitine, respectively.
Transporters of the BCCT family are responsible for the uptake of several quaternary amines and other organic osmolytes in bacteria, including glycine betaine, choline, carnitine, and dimethylsulfoniopropionate (DMSP) (24). To ascertain whether gene tmaT encodes a TMA transporter, we overexpressed this gene in E. coli BL21 (fig. S2B) and tested the substrate binding specificity of the recombinant protein using isothermal titration calorimetry (ITC) assays. The result showed that the recombinant protein had a high binding affinity to TMA (Fig. 3, B to D) and did not bind TMAO (Fig. 3E), glycine betaine, carnitine, choline, or DMSP (fig. S3), which indicates that gene tmaT encodes a TMA-specific transporter, which is named TmaT. Because no TMA transporter has ever been reported, TmaT represents a new member of the BCCT family, which is different in substrate specificity from the other BCCT family proteins including transporters BetP, BetT, and CaiT for glycine betaine, choline, and carnitine, respectively. This is further supported by phylogenetic analysis, which showed that TmaT and its homologs are clustered in a separate branch from the other BCCT family sequences (Fig. 3F). In addition, we found that the dissociation constant (Kd) of TmaT for TMA binding at 5°C was lower than that at 15° or 25°C (Fig. 3, B to D), suggesting that TmaT can function well at deep-sea in situ temperature. Together, these results suggest that TmaT is a TMA-specific transporter, which is likely responsible for TMA uptake from the environment into strain D25 cells.
Analysis of the in vivo function of TmaT and MpTmm in recombinant E. coli and B. subtilis
During our study, we made numerous attempts to disrupt the genes tmaT and Mptmm in strain D25. Unfortunately, all our attempts failed. Alternatively, we tested the function of TmaT and MpTmm in vivo in recombinant E. coli (strain DH5α) in which TmaT and MpTmm were constitutively expressed. We investigated whether the introduction of both genes tmaT and Mptmm can improve the survival and growth of E. coli under HHP. As shown in fig. S4A, heterologous expression of TmaT conferred upon E. coli the ability to transport exterior TMA into its cells, indicating that TmaT is a functional TMA transporter. Coexpression of both genes tmaT and Mptmm in E. coli resulted in the accumulation of TMAO intracellularly in E. coli when TMA was present in the medium (fig. S4B), indicating that TmaT is responsible for TMA uptake and that MpTmm is responsible for intracellular TMA oxidation to produce TMAO. At the atmospheric pressure, coexpression of both genes did not affect the growth of E. coli regardless of whether or not TMA was present (Fig. 4A and fig. S4C). However, coexpression of both genes improved the growth of E. coli at HHP conditions (20 and 40 MPa) when TMA was supplied, while expression of TmaT or MpTmm alone with addition of TMA, or coexpression of both genes without TMA in the medium, did not improve the growth or survival of E. coli at HHP (Fig. 4A and fig. S4C). This indicates that the TMA metabolism involved in TmaT and MpTmm in E. coli can improve its HHP tolerance.
Fig. 4. The in vivo function of TmaT and MpTmm in recombinant E. coli DH5α, E. coli mutant ΔtorCAD, and B. subtilis 168.
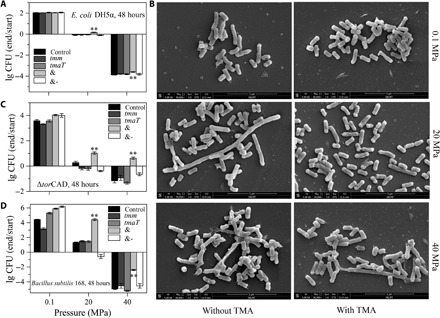
(A, C, and D) Effect of expression of TmaT and MpTmm on the growth and survival of E. coli DH5α (A), ΔtorCAD (C), and B. subtilis 168 (D) cultured at different pressures for 48 hours. Control, strain containing the empty plasmid cultured with TMA; tmm, strain expressing MpTmm cultured with TMA; tmaT, strain expressing TmaT cultured with TMA; &, strain coexpressing MpTmm and TmaT cultured with TMA; and &-, strain coexpressing MpTmm and TmaT cultured without TMA. The cell number at the start of the incubation (start) was ca. 1 × 105 CFU/ml. The strains were cultivated at 25°C for 48 hours with or without 20 μM TMA, and then CFU were counted and compared with those of the starting culture (end/start). The asterisks show statistical difference from control (**P < 0.01, two-tailed t test). The error bars represent SDs from triplicate experiments. (B) Morphological observation of cells of ΔtorCAD coexpressing TmaT and MpTmm cultured at 25°C for 48 hours under different pressures with or without 20 μM TMA. ETD, Everhart-Thornley detector; WD, working distance. SKLMT, State Key Laboratory of Microbial Technology.
Because E. coli contains a TMAO reductase gene in the torCAD cluster (including the genes encoding a pentaheme c-type cytochrome, TMAO reductase, and molecular chaperone) that may further metabolize intracellular TMAO to eliminate the effect of TMAO reductase on the pressure tolerance of the recombinant E. coli strain, we knocked out the torCAD cluster from E. coli and constructed the mutant strain ΔtorCAD. The growth of ΔtorCAD containing tmaT and Mptmm at HHP conditions (20 and 40 MPa) was much better than that of the wild-type E. coli containing tmaT and Mptmm when TMA was present in the medium (Fig. 4C and fig. S4D). This further indicates that accumulation of intracellular TMAO is beneficial for E. coli to cope with HHP stress.
It has been reported that HHP can impair cell division of E. coli, leading to elongated filaments (25, 26). We then investigated the cell morphological changes of ΔtorCAD containing genes tmaT and Mptmm under HHP conditions. When TMA was absent, significantly longer filaments were observed in the cells of ΔtorCAD under 20 MPa, and some cells began to collapse under 40 MPa (Fig. 4B). When TMA was present in the medium, coexpression of TmaT and MpTmm reduced the impact of HHP on the cell morphology and cell division of ΔtorCAD, especially under 20 MPa (Fig. 4B and fig. S4E).
To further confirm that TmaT and MpTmm can improve the HHP tolerant ability of bacteria, the tmaT and Mptmm genes were introduced into B. subtilis 168. As anticipated, the introduction of both genes significantly improved the growth of this strain at HHP conditions (20 and 40 MPa) when TMA was supplied (Fig. 4D and fig. S4F), indicating the improvement of its HHP tolerance.
Overall, these data corroborate the in vivo function of the TMA metabolism pathway mediated by TmaT and MpTmm in bacteria, that is, accumulating TMAO to cope with HHP stress. Thus, TMAO, the product of this pathway, is a piezolyte for bacteria.
Universality of genes tmaT and Mptmm in marine bacteria
TMA in marine sediments can reach up to 50 μM (27), which is used by sedimentary bacteria. On the basis of the above results, it can be concluded that the deep-sea sedimentary strain D25 can use the environmental TMA to cope with HHP stress via a TMA metabolism pathway mediated by TmaT and MpTmm. Strain D25 transports environmental TMA into its cell through the TMA transporter TmaT, and the TMA monooxygenase MpTmm oxidizes the intracellular TMA into TMAO, which is accumulated in the cell without further metabolism and used as a piezolyte by strain D25 to cope with HHP stress (Fig. 5A). Thus, the TmaT-MpTmm system provides a simple and economic strategy for strain D25 to cope with HHP stress.
Fig. 5. The HHP tolerance strategy of strain D25 and its universality in Bacteroidetes.
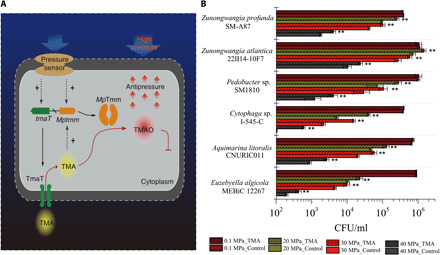
(A) The proposed model for HHP tolerance of strain D25 via the TMA metabolism pathway mediated by TmaT and MpTmm. Under HHP stress in deep sea, strain D25 expresses genes tmaT and Mptmm and produces the TMA transporter TmaT and the TMA monooxygenase MpTmm. Strain D25 takes up environmental TMA into its cell by TmaT and oxidizes the intracellular TMA into TMAO by MpTmm. TMAO is accumulated in the cell without further metabolism and is used as a piezolyte by strain D25 to cope with HHP stress. (B) Effect of the presence of TMA in medium on the growth of other Bacteroidetes strains at different pressures. Strains were cultured at 15°C for 10 days in the medium containing 3% artificial sea salt, 0.05% peptone, 0.01% yeast powder, and 20 μM TMA. The error bars represent SDs from triplicate experiments. Strains E. algicola MEBiC 12267, A. litoralis CNURIC011, and Cytophaga sp. I-545-C were all isolated from surface seawater in our lab. The asterisks show statistical difference from control (**P < 0.01, two-tailed t test). The error bars represent SDs from triplicate experiments.
To better understand the ecological significance of the TmaT-MpTmm system in marine bacteria, genes encoding TmaT-like and MpTmm-like proteins were searched in the Integrated Microbial Genome Database of the Joint Genome Institute (the IMG/JGI database) (28, 29). TmaT-like and MpTmm-like proteins are found in the marine metagenome dataset (table S2) from many sites across a range of sampling depths, indicating that the two genes are widespread. When we searched the IMG/JGI database with gene tmaT as the query, all the tmaT homologs (>40% identity) are from Bacteroidetes. Therefore, it is most likely that the TmaT-MpTmm system exists only in Bacteroidetes. We then investigated the distribution of genes encoding TmaT-like and MpTmm-like proteins in the genomes of Bacteroidetes from marine environments, including shallow sea (seawater or sediments from <1000-m depth or depth unreported) and deep sea (seawater or sediments from >1000-m water depth). In the IMG/JGI database, 337 genomes of Bacteroidetes were found from shallow sea, and only 7 from deep sea. Among them, 44 contain both tmaT and Mptmm homologs, including 41 from shallow sea and 3 from deep sea (table S3). These strains are distributed in four classes of Bacteroidetes (fig. S5), and there does not appear to be a distinct evolutionary lineage within the phylum Bacteroidetes that has this TMA metabolism pathway. The three deep-sea strains are Zunongwangia profunda SM-A87 from 1234-m water depth (30), Pedobacter sp. SM1810 from 3268-m water depth (31), and Zunongwangia atlantica 22II14-10F7 from 2927-m water depth (table S3) (32). We investigated the effect of TMA on the growth and survival of these strains under 20, 30, and 40 MPa. Consistent with the observation on strain D25, the presence of TMA in the medium significantly improved the growth and survival of all these strains under high pressure (Fig. 5B). Thus, accumulation of TMAO in the cell by assimilating environmental TMA to cope with HHP stress may be a common strategy adopted by deep-sea Bacteroidetes bacteria containing the TmaT-MpTmm system.
In addition, because some Bacteroidetes strains from shallow sea also contain both tmaT and Mptmm homologs (table S3), we tested the effect of TMA on the growth and survival of three Bacteroidetes strains that originated from surface seawater under 20, 30, and 40 MPa, and the result indicated that exterior TMA can improve the survival and growth of all these strains under HHP (Fig. 5B). This suggests that Bacteroidetes strains containing the TmaT-MpTmm system are likely able to cope with HHP stress when they migrate to deep sea, provided that TMA is present. However, it cannot be excluded that there are other functions of this TMA metabolism pathway in bacteria, especially non–deep-sea bacteria, which needs further study.
DISCUSSION
TMA and TMAO are ubiquitous in the oceans, and their concentrations range from nanomolar in surface seawater to micromolar in deep-sea sediments (27), and the content of TMAO in the muscle of deep-sea teleost fish can reach as high as 260 mmol/kg (17). TMA/TMAO metabolism in heterotrophic marine bacteria has been extensively studied over the last decade. On the basis of previous results and the results in this study, it can be concluded that heterotrophic marine bacteria use TMA/TMAO via three pathways (Fig. 6): (i) Many marine α-proteobacteria including members of the cosmopolitan Roseobacter and SAR11 contain genes tmm, tdm, and tmoX (table S4) and metabolize TMA/TMAO as a carbon, nitrogen, or energy source (10, 27, 33); (ii) some γ-proteobacteria, such as Shewanella and Vibrio, contain gene torA (table S4) and use TMAO as a terminal electron acceptor in anaerobic conditions (13, 15, 34); (iii) and the results in this study reveal that the deep-sea Bacteroidetes strain D25 has a new TMA metabolism pathway mediated by genes tmaT and Mptmm, which takes up environmental TMA and converts it to TMAO to counteract HHP stress (Figs. 5A and 6). This novel HHP adaptation strategy appears to be nonspecific in strain D25 because heterologous expression of the TmaT-MpTmm system in E. coli and B. subtilis also leads to their HHP tolerance, and other marine Bacteroidetes bacteria containing genes encoding TmaT-like and MpTmm-like also have improved HHP tolerance in the presence of TMA. Thus, this study uncovers a novel physiological function of TMA/TMAO in marine bacteria and a simple pathway to cope with HHP stress in deep-sea piezotolerant Bacteroidetes bacteria. Bacteroidetes bacteria are widespread in the ocean and considered to play an important role, especially in the degradation of high–molecular weight organic matters in marine ecosystem. This study offers a better understanding of how the generalist deep-sea bacterial lineage, Bacteroidetes bacteria, survives in deep-sea environments.
Fig. 6. Three TMA/TMAO metabolism pathways in marine bacteria.
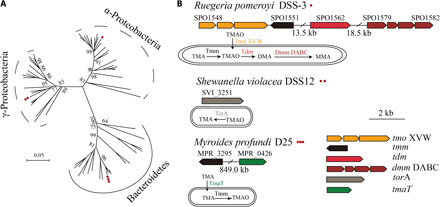
(A) A neighbor-joining phylogenetic tree showing the bacterial strains, which are capable of metabolizing TMA/TMAO. The tree was constructed on the basis of the 16S ribosomal RNA gene sequences of the strains. The red dot(s) indicates the phylogenetic position of the representative stains illustrated in (B). The detailed information of each strain is listed in table S4. (B) Schemes of the TMA/TMAO metabolism pathways and related genomic regions in R. pomeroyi DSS-3 (α-proteobacteria), Shewanella violacea DSS12 (γ-proteobacteria), and M. profundi D25 (Bacteroidetes). MpTmm, TMA monooxygenase gene; tdm, TMAO demethylase gene; tmaT, TMA transporter gene; dmm DABC, dimethylamine monooxygenase gene; tmo XVW, adenosine 5′-triphosphate–dependent TMAO transporter gene; and torA, TMAO reductase gene.
METHODS
Bacterial strains
Marine bacterial strains M. profundi D25, R. pomeroyi DSS-3, E. coli DH5α, E. coli BL21, B. subtilis 168, Z. profundi SM-A87, Pedobacter sp. SM1810, Euzebyella algicola MEBiC 12267, Aquimarina litoralis CNURIC011, and Cytophaga sp. I-545-C were previously maintained in our lab. Z. atlantica 22II14-10F7 was obtained from the Marine Culture Collection of China. The gene knockout mutant of E. coli DH5α, ΔtorCAD, was constructed as described previously (35). Marine strains D25, DSS-3, SM-A87, SM1810, 22II14-10F7, MEBiC 12267, CNURIC011, and I-545-C were routinely cultivated in the 2216E medium.
The effect of exterior TMA on the growth of strain D25 at HHP
A defined medium containing 3% (m/v) artificial sea salt, vitamin complex, 1 mM TMA or TMAO, 10 mM glucose, 0.2 mM Na3PO4, 10 mM Hepes, and 5 μM FeCl3 (36) was used to test whether TMA or TMAO can be used as a nitrogen source by strains D25 and DSS-3. Automatic growth curve analyzer (Bioscreen C MBR, Finland) was used to record the growth of both strains cultured at 25°C.
To test whether TMA can facilitate the growth of strain D25 at HHP, the medium containing 3% (m/v) artificial sea salt, 0.05% (m/v) peptone, 0.01% (m/v) yeast powder, and TMA (0 to 1 mM) was used. After 1% (v/v) cell suspension of strain, D25 was inoculated into the medium, 4 ml of culture was filled into a 5-ml sterile syringe with a polyethylene pipe cap, which was then put into the high-pressure vessel, and the hydrostatic pressure was increased to 10, 20, 30, 40, or 50 MPa using a microbial high-pressure culture device (Feiyu Science and Technology Exploitation Co. Ltd., Nantong, China). After incubation at 4°C for 10 days, the syringes were taken out of the high-pressure culture device, and the bacterial cells in each syringe were fully resuspended in the medium by vortexing and shaking. The culture was then diluted using 2216E medium to an appropriate concentration, and 100 μl of the diluted culture was plated onto a 2216E agar plate for counting colony-forming units (CFU).
Scanning electron microscopy imaging
The bacterial cells cultivated at different pressures were collected by centrifugation at 7000g for 3 min. After being fixed with 2.5% (v/v) glutaraldehyde solution, the bacterial cells were washed with ethanol at concentrations of 30, 50, 70, 80, and 90%, respectively, and eventually resuspended in pure ethanol, which were then dried out in Automated Critical Point Dryer (Leica EM CPD300, Germany) with the critical-point drying method and coated with gold using a sputter coater (Cressington 108, England). The surface morphological structures of the bacterial cells were captured in an environmental mode on a scanning electron microscope (FEI Quanta 250 FEG, American) under 5-kV voltage. The length of the bacterial cells was calculated using ImageJ, and Student’s t test was used to determine the difference in bacterial cell length between different cultivation conditions.
Quantification of TMA and TMAO
Strain D25 was cultivated at 25°C to an OD600 (optical density at 600 nm) of ~1.0 in the medium containing 3% (m/v) artificial sea salt, 0.05% (m/v) peptone, and 0.01% (m/v) yeast powder. Then, a final concentration of 85 μM TMA was added into the medium, which was further cultured. Every 1 hour, 1000 ml of culture was centrifuged at 7000g for 3 min, and the supernatant was used for measuring the concentrations of extracellular TMA and TMAO (37). The cell pellet was resuspended in 100 ml of deionized water. After 0.4 M trichloroacetic acid (final concentration) was added, the cells were lysed by sonication. Cell debris was then removed via centrifugation (17,000g, 10 min), and the resultant supernatant was used for measuring the concentrations of intracellular TMA and TMAO. The TMA and TMAO concentrations of all samples were measured on a cation exchange ion chromatograph (ICS-1100, Dionex, USA) with nonsuppressed conductivity detection (38). The optimized eluent solution contained 4 mM nitric acid and 3% (v/v) acetonitrile solution. The separation was carried out on a Metrosep C 4-250/4.0 separation column (Metrohm, Switzerland) under isocratic condition at a flow rate of 0.9 ml/min and a column temperature of 30°C. To determine the intracellular concentrations of TMA/TMAO, the average volume of a cell was first calculated by measuring the average length and diameter of the bacterial cells based on scanning electron microscope observation, assuming a cylinder shape of each bacterial cell. The total volume of the cells in a sample was calculated on the basis of the cell number.
Transcriptome analysis and quantitative real-time PCR
Strain D25 was cultivated in the 2216E medium to an OD600 of 0.6 at atmospheric pressure. The culture was then divided into two aliquots and incubated at 30 MPa and atmospheric pressure, respectively, for 1 hour. The cells were collected by centrifugation, and the total RNA was extracted using an RNeasy Protect Bacteria Mini Kit (QIAGEN, Germany). The transcriptome sequencing was performed by Majorbio Co. Ltd. (Shanghai, China), and the reads were mapped to the genome of strain D25. Gene expression was normalized using the RPKM (reads per kilobases per million mapped reads) method, and the RPKM value of each gene was used to compare levels of expression under different conditions. A gene was considered to be differentially regulated when it showed a >4-fold change in expression and displayed a false discovery rate–adjusted P value of <0.01.
For quantitative real-time PCR, DNA-free RNA was reverse transcribed into cDNA by using TransScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech, China). The cDNA was used for quantitative PCR analysis using the LightCycler 1.5 system (Roche, Switzerland), and SYBR Green fluorescence (Takara, Japan) was used for detection. The relative expression of the target gene was normalized to the reference gene of recA that was not regulated by HHP.
Analysis of the in vivo function of TmaT and MpTmm
To analyze the in vivo function of TmaT and MpTmm, recombinant E. coli DH5α mutant strain ΔtorCAD and strain B. subtilis 168 that can constitutively express genes tmaT and Mptmm were constructed. The genes Mptmm and tmaT were amplified from the genome DNA of strain D25. BBa-J23111 (http://parts.igem.org), a constitutive expression promotor, was introduced in the front primer. The gene Mptmm or tmaT was recombined with the plasmid pET-22b, and the recombinant plasmid was transformed into ΔtorCAD and B. subtilis 168 by electroporation to construct the MpTmm expression strain and the TmaT expression strain. To construct an MpTmm-TmaT coexpression strain, the gene Mptmm was recombined into the TmaT expression plasmid, which was then transformed into ΔtorCAD and B. subtilis 168.
To test the in vivo function of TmaT, the ΔtorCAD strain containing a TmaT expression plasmid and the ΔtorCAD strain harboring an empty plasmid were, respectively, cultivated in the medium containing 1% (m/v) NaCl, 0.5% (m/v) peptone, and 0.1% (m/v) yeast powder to an OD600 of ~1.0. The cells were collected by centrifugation and resuspended in the medium containing 1% (m/v) NaCl, 0.05% (m/v) peptone, 0.01% (m/v) yeast powder, and 30 μM TMA, which were then incubated at 25°C, and TMA concentration in the medium was measured every 30 min.
To test the in vivo function of the TmaT-MpTmm metabolism pathway, the ΔtorCAD strain containing a TmaT-MpTmm coexpression plasmid was cultured in 1 liter of medium containing 1% (m/v) NaCl, 0.5% (m/v) peptone, and 0.1% (m/v) yeast powder to an OD600 of ~1.0. Then, a final concentration of 85 μM TMA was added into the medium, which was further cultured. Concentrations of medium TMA and intracellular TMA and TMAO of ΔtorCAD were measured every 1 hour as described above.
To test whether TMA can improve the HHP tolerance of strains ΔtorCAD and B. subtilis 168 expressing MpTmm and/or TmaT, the medium containing 1% (m/v) NaCl, 0.05% (m/v) peptone, and 0.01% (m/v) yeast powder and 20 μM TMA was used. After incubation under atmosphere pressure, 20 or 40 MPa at 25°C for 40 hours, the CFUs of ΔtorCAD were counted, and the cell morphology was observed using a scanning electron microscope as described above.
Expression and purification of MpTmm and TmaT of strain D25
The full-length genes Mptmm and tmaT were amplified from the genome DNA of strain D25 and were cloned into the pET-22b vector with a C-terminal His tag, respectively, which were then transformed into E. coli BL21. The recombinant E. coli strains were cultured at 37°C in Luria-Bertani medium to an OD600 of 0.8 to 1.0 and then incubated at 20°C for 16 hours with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) as an inducer. The recombinant MpTmm protein was purified using the method described by Li et al. (39). The recombinant TmaT protein was purified by a membrane protein purification method (40).
Crystallization, data collection, structure determination, and refinement
The purified recombinant MpTmm (10 mg/ml) was mixed with reduced form of NADP (NADPH) (5 mg/ml) and was crystallized at 18°C using the hanging drops vapor diffusion method. The crystals were obtained in a buffer containing 0.1 M magnesium acetate, 0.1 M MES (pH 6.5), and 10% (w/v) polyethylene glycol 10,000. X-ray diffraction data were collected on BL17U1 beamline at the Shanghai Synchrotron Radiation Facility using detector ADSC Quantum 315r (41). The initial diffraction datasets were processed by HKL2000.
The crystals of MpTmm belong to the P212121 space group. The crystal structure of MpTmm was determined by molecular replacement using the CCP4 Program Phaser (42) with the crystal structure of RnTmm (Protein Data Bank code: 5IPY) as the search model. The refinement of the structure was performed using Coot (43) and PHENIX (44). All the structure figures were processed using the program PyMOL (www.pymol.org/).
ITC measurements
ITC measurements to test the substrate binding specificity of TmaT were performed at 5°, 15°, and 25°C using the MicroCal iTC200 system (GE Healthcare, USA). The sample cell was loaded with 250 μl of protein sample (~10 μM), and the reference cell was filled with distilled water. The syringe contained 40 μl of tested compound (100 μM). The proteins and tested compounds were dissolved in the same buffer containing 10 mM tris-HCl (pH 7.0) and 100 mM NaCl. Titrations were carried out by adding 0.4 μl of tested compound for the first injection and 3 μl for the following 12 injections, with stirring at 800 revolutions per minute.
Enzyme assay and characterization
The enzymatic activity of MpTmm toward TMA was measured by following the decrease in absorbance at 340 nm of substrate-dependent oxidation of NADPH as described previously (45). To determine the optimal temperature for MpTmm activity, 300 μl of buffer containing 10 mM tris-HCl (pH 8.0), 100 mM NaCl, 0.25 mM NADPH, and 1 mM TMA was preincubated at different temperatures (0° to 45°C) for 30 min. After incubation, 1 μM MpTmm was added into the buffer, and the mixture was further incubated at different temperatures for 3 min before detection by Multiskan Spectrum (SpectraMax 384 plus, Molecular Devices, America). To determine the optimal pressure for MpTmm activity, 1 ml of the reaction mixture containing 10 mM tris-HCl (pH 8.0), 100 mM NaCl, 0.25 mM NADPH, 1 mM TMA, and 1 nM MpTmm was incubated at different pressures (0.1 to 50 MPa) for 3 hours before detection by Multiskan Spectrum. The control group had the same reaction system, except that MpTmm was not added. One enzyme activity unit was defined as the amount of enzyme required to consume 1 nM NADPH per minute for TMA oxidation. To determine the Km of MpTmm, substrate (TMA, dimethylamine, dimethyl sulfide, or monomethylamine) of different concentrations from 500 to 10 μM was used. The Km values of MpTmm were determined by nonlinear analysis based on the initial rates, and the measurements were performed at the optimal temperature. All enzyme assays were carried out in triplicate.
Differential scanning calorimetry (DSC) measurements were carried out using a MicroCal VP-DSC (GE Healthcare) to measure the Tm value of MpTmm. The sample cell was loaded with 250 μl of MpTmm (~15 μM), and the reference cell contained distilled water. The temperature was raised from 25° to 90°C at a constant heating rate of 1°C/min.
Phylogenetic analyses of TmaT
Using the TmaT of strain D25 as the query, a BLASTP (E value 1 × 10−50) search was performed against all genomes in the IMG/JGI database (https://img.jgi.doe.gov/cgi-bin/m/main.cgi) (28, 29), and TmaT homologs were retained. Sequences retrieved from the IMG/JGI dataset were aligned with other characterized BCCT transporters and visualized using Molecular Evolutionary Genetics Analysis version 7.0 (MEGA7) (46).
Distribution of TmaT-like and MpTmm-like proteins in marine metagenomes and marine Bacteroidetes genomes
Metagenomes from different water depths were chosen from the IMG/JGI database. Using MpTmm and TmaT of strain D25 as the query, BLASTP analysis was performed using a stringency of 30% identity and a cutoff value of 10−50. The number of retrieved sequences for each protein was normalized by dividing the retrieved number by genome size (Gb). The metagenomes used in this study are listed in table S2.
Genomes of Bacteroidetes isolated from deep-sea water and marine sediments in the IMG/JGI database were screened for TmaT-like and MpTmm-like sequences using TmaT and MpTmm of strain D25 as the query, respectively (28, 29). Marine Bacteroidetes genomes containing both TmaT-like and MpTmm-like sequences are listed in table S3.
Analysis of enzyme genes involved in the metabolism of methylated amines in marine microbial genomes
All available defined marine bacterial genomes in the IMG/JGI database were screened for enzymes catalyzing degradation of methylated amines using BLASTP analysis with MpTmm and TmaT from strain D25, Tdm (Spo1562), and TmoX (Spo1548) from R. pomeroyi DSS-3, and TorA (swp_5031) from Shewanella piezotolerans WP3 as query sequences using a stringent cutoff value of 10−50. Marine bacterial genomes containing genes encoding these proteins are listed in table S4. A phylogenetic tree was constructed by a maximum likelihood approach with 500 bootstrap replicates and using a maximum parsimony tree derived from neighbor joining as the initial tree.
Acknowledgments
We thank the staff from BL18U1 and BL19U1 beamlines of the National Facility for Protein Sciences Shanghai (NFPS) and Shanghai Synchrotron Radiation Facility for assistance during data collection. We also thank C. Sun, Z. Li, S. Wang, H. Yu, X. Zhao, and Y. Guo from State Key laboratory of Microbial Technology of Shandong University for help and guidance in ion chromatography, ITC, DSC, and SEM. Funding: This work was supported by the National Natural Science Foundation of China (31870101, U1706207, 31630012, and 91851205), the National Key R&D Program of China (2018YFC1406700), the Program of Shandong for Taishan Scholars (tspd20181203), and the Young Scholars Program of Shandong University (2016WLJH36). Author contributions: Q.-L.Q. and Z.-B.W. performed the majority of the experiments and data interpretation. Z.-B.W. and X.-J.W. purified and crystallized the enzyme, and C.-Y.L. solved the structure. H.-N.S. performed the SEM observation. X.-L.C., J.M., P.-Y.L., and X.-Y.Z. helped in the experiments. I.L. and W.Z. performed the bioinformational analyses. Z.-B.W., J.F., Y.C., and Y.-Z.Z. wrote the manuscript. M.W., X.-H.Z., X.-Y.Z., and G.-P.Y. revised the manuscript. Y.-Z.Z. designed and supervised the study. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. All the RNA sequencing read data have been deposited in NCBI’s Sequence Read Archive (SRA) under project accession number PRJNA540220. The crystal structure of MpTmm has been deposited in PDB with the code number of 6KBW.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/13/eabf9941/DC1
REFERENCES AND NOTES
- 1.Kallmeyer J., Pockalny R., Adhikari R. R., Smith D. C., D’Hondt S., Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 109, 16213–16216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lundsten L., Schlining K. L., Frasier K., Johnson S. B., Kuhnz L. A., Harvey J. B. J., Clague G., Vrijenhoek R. C., Time-series analysis of six whale-fall communities in Monterey Canyon, California, USA. Deep Sea Res. Part I Oceanogr. Res. Papers 57, 1573–1584 (2010). [Google Scholar]
- 3.Newell C. R., Pilskaln C. H., Robinson S. M., MacDonald B. A., The contribution of marine snow to the particle food supply of the benthic suspension feeder, Mytilus edulis. J. Exp. Mar. Biol. Ecol. 321, 109–124 (2005). [Google Scholar]
- 4.Oger P. M., Jebbar M., The many ways of coping with pressure. Res. Microbiol. 161, 799–809 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Jebbar M., Franzetti B., Girard E., Oger P., Microbial diversity and adaptation to high hydrostatic pressure in deep-sea hydrothermal vents prokaryotes. Extremophiles 19, 721–740 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y., Li X., Bartlett D. H., Xiao X., Current developments in marine microbiology: High-pressure biotechnology and the genetic engineering of piezophiles. Curr. Opin. Biotechnol. 33, 157–164 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Fang J., Zhang L., Bazylinski D. A., Deep-sea piezosphere and piezophiles: Geomicrobiology and biogeochemistry. Trends Microbiol. 18, 413–422 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Lauro F. M., Bartlett D. H., Prokaryotic lifestyles in deep sea habitats. Extremophiles 12, 15–25 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Welch T. J., Bartlett D. H., Isolation and characterization of the structural gene for OmpL, a pressure-regulated porin-like protein from the deep-sea bacterium Photobacterium species strain SS9. J. Bacteriol. 178, 5027–5031 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lidbury I., Mausz M. A., Scanlan D. J., Chen Y., Identification of dimethylamine monooxygenase in marine bacteria reveals a metabolic bottleneck in the methylated amine degradation pathway. ISME J. 11, 1592–1601 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Proctor L. M., Gunsalus R. P., Anaerobic respiratory growth of Vibrio harveyi, Vibrio fischeri and Photobacterium leiognathi with trimethylamine N-oxide, nitrate and fumarate: Ecological implications. Environ. Microbiol. 2, 399–406 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Dos Santos J. P., Iobbi-Nivol C., Couillault C., Giordano G., Méjean V., Molecular analysis of the trimethylamine N-oxide (TMAO) reductase respiratory system from a Shewanella species. J. Mol. Biol. 284, 421–433 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Lemaire O. N., Honoré F. A., Jourlin-Castelli C., Méjean V., Fons M., Iobbi-Nivol C., Efficient respiration on TMAO requires TorD and TorE auxiliary proteins in Shewanella oneidensis. Res. Microbiol. 167, 630–637 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Tranier S., Mortier-Barrière I., Ilbert M., Birck C., Iobbi-Nivol C., Méjean V., Samama J.-P., Characterization and multiple molecular forms of TorD from Shewanella massilia, the putative chaperone of the molybdoenzyme TorA. Protein Sci. 11, 2148–2157 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin Q.-J., Zhang W.-J., Qi X.-Q., Zhang S.-D., Jiang T., Li X.-G., Chen Y., Santini C.-L., Zhou H., Chou I.-M., Wu L.-F., High hydrostatic pressure inducible trimethylamine N-oxide reductase improves the pressure tolerance of piezosensitive bacteria Vibrio fluvialis. Front. Microbiol. 8, 2646 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yancey P. H., Blake W. R., Conley J., Unusual organic osmolytes in deep-sea animals: Adaptations to hydrostatic pressure and other perturbants. Comp. Biochem. Physiol. A 133, 667–676 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Samerotte A. L., Drazen J. C., Brand G. L., Seibel B. A., Yancey P. H., Correlation of trimethylamine oxide and habitat depth within and among species of teleost fish: An analysis of causation. Physiol. Biochem. Zool. 80, 197–208 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Yancey P. H., Gerringer M. E., Drazen J. C., Rowden A. A., Jamieson A., Marine fish may be biochemically constrained from inhabiting the deepest ocean depths. Proc. Natl. Acad. Sci. U.S.A. 111, 4461–4465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X.-Y., Zhang Y.-J., Chen X.-L., Qin Q.-L., Zhao D.-L., Li T.-G., Dang H.-Y., Zhang Y.-Z., Myroides profundi sp. nov., isolated from deep-sea sediment of the southern Okinawa Trough. FEMS Microbiol. Lett. 287, 108–112 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y.-Z., Li Y., Xie B.-B., Chen X.-L., Yao Q.-Q., Zhang X.-Y., Kempher M. L., Zhou J., Oren A., Qin Q.-L., Nascent genomic evolution and allopatric speciation of Myroides profundi D25 in its transition from land to ocean. MBio 7, e01946-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y., Patel N. A., Crombie A., Scrivens J. H., Murrell J. C., Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 108, 17791–17796 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C.-Y., Chen X.-L., Zhang D., Wang P., Sheng Q., Peng M., Xie B.-B., Qin Q.-L., Li P.-Y., Zhang X.-Y., Su H.-N., Song X.-Y., Shi M., Zhou B.-C., Xun L.-Y., Chen Y., Zhang Y.-Z., Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide. Mol. Microbiol. 103, 992–1003 (2017). [DOI] [PubMed] [Google Scholar]
- 23.González J. M., Covert J. S., Whitman W. B., Henriksen J. R., Mayer F., Scharf B., Schmitt R., Buchan A., Fuhrman J. A., Kiene R. P., Moran M. A., Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., dimethylsulfoniopropionate-demethylating bacteria from marine environments. Int. J. Syst. Evol. Microbiol. 53, 1261–1269 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Ziegler C., Bremer E., Krämer R., The BCCT family of carriers: From physiology to crystal structure. Mol. Microbiol. 78, 13–34 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Welch T. J., Farewell A., Neidhardt F. C., Bartlett D. H., Stress response of Escherichia coli to elevated hydrostatic pressure. J. Bacteriol. 175, 7170–7177 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Black S. L., Dawson A., Ward F. B., Allen R. J., Genes required for growth at high hydrostatic pressure in Escherichia coli K-12 identified by genome-wide screening. PLOS ONE 8, e73995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J., Mausz M. A., Chen Y., Giovannoni S. J., Microbial trimethylamine metabolism in marine environments. Environ. Microbiol. 21, 513–520 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Chen I.-M. A., Chu K., Palaniappan K., Pillay M., Ratner A., Huang J., Huntemann M., Varghese N., White J. R., Seshadri R., Smirnova T., Kirton E., Jungbluth S. P., Woyke T., Eloe-Fadrosh E. A., Ivanova N. N., Kyrpides N. C., IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Katta H. Y., Mojica A., Chen I.-M. A., Kyrpides N. C., Reddy T., Genomes OnLine database (GOLD) v.7: Updates and new features. Nucleic Acids Res. 47, D649–D659 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin Q.-L., Zhang X.-Y., Wang X.-M., Liu G.-M., Chen X.-L., Xie B.-B., Dang H.-Y., Zhou B.-C., Yu J., Zhang Y.-Z., The complete genome of Zunongwangia profunda SM-A87 reveals its adaptation to the deep-sea environment and ecological role in sedimentary organic nitrogen degradation. BMC Genomics 11, 247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He X.-y., Li N., Chen X.-l., Zhang Y.-z., Zhang X.-y., Song X.-y., Pedobacter indicus sp. nov., isolated from deep-sea sediment. Antonie Van Leeuwenhoek 113, 357–364 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Shao R., Lai Q., Liu X., Sun F., Du Y., Li G., Shao Z., Zunongwangia atlantica sp. nov., isolated from deep-sea water. Int. J. Syst. Evol. Microbiol. 64, 16–20 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Lidbury I., Murrell J. C., Chen Y., Trimethylamine N-oxide metabolism by abundant marine heterotrophic bacteria. Proc. Natl. Acad. Sci. U.S.A. 111, 2710–2715 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee K.-M., Park Y., Bari W., Yoon M. Y., Go J., Kim S. C., Lee H.-I., Yoon S. S., Activation of cholera toxin production by anaerobic respiration of trimethylamine N-oxide in Vibrio cholerae. J. Biol. Chem. 287, 39742–39752 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Z.-C., Zhao D.-L., Ran L.-Y., Mi Z.-H., Wu Z.-Y., Pang X., Zhang X.-Y., Su H.-N., Shi M., Song X.-Y., Xie B.-B., Qin Q.-L., Zhou B.-C., Chen X.-L., Zhang Y.-Z., Development of a genetic system for the deep-sea psychrophilic bacterium Pseudoalteromonas sp. SM9913. Microb. Cell Fact. 13, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y., Comparative genomics of methylated amine utilization by marine Roseobacter clade bacteria and development of functional gene markers (tmm, gmaS). Environ. Microbiol. 14, 2308–2322 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Wekell J. C., Barnett H., New method for analysis of trimethylamine oxide using ferrous sulfate and EDTA. J. Food Sci. 56, 132–135 (1991). [Google Scholar]
- 38.Erupe M. E., Liberman-Martin A., Silva P. J., Malloy Q. G. J., Yonis N., Cocker D. R. III, Purvis-Roberts K. L., Determination of methylamines and trimethylamine-N-oxide in particulate matter by non-suppressed ion chromatography. J. Chromatogr. A 1217, 2070–2073 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Li C.-Y., Chen X.-L., Qin Q.-L., Wang P., Zhang W.-X., Xie B.-B., Su H.-N., Zhang X.-Y., Zhou B.-C., Zhang Y.-Z., Structural insights into the multispecific recognition of dipeptides of deep-sea gram-negative bacterium Pseudoalteromonas sp. strain SM9913. J. Bacteriol. 197, 1125–1134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu F., Li S., Jiang Y., Jiang J., Fan H., Lu G., Deng D., Dang S., Zhang X., Wang J., Yan N., Structure and mechanism of the uracil transporter UraA. Nature 472, 243–246 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Wang Q.-S., Zhang K.-H., Cui Y., Wang Z.-J., Pan Q.-Y., Liu K., Sun B., Zhou H., Li M.-J., Xu Q., Xu C.-Y., Yu F., He J.-H., Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl. Sci. Tech. 29, 68 (2018). [Google Scholar]
- 42.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol.Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Struct. Biol. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alfieri A., Malito E., Orru R., Fraaije M. W., Mattevi A., Revealing the moonlighting role of NADP in the structure of a flavin-containing monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 105, 6572–6577 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/13/eabf9941/DC1


