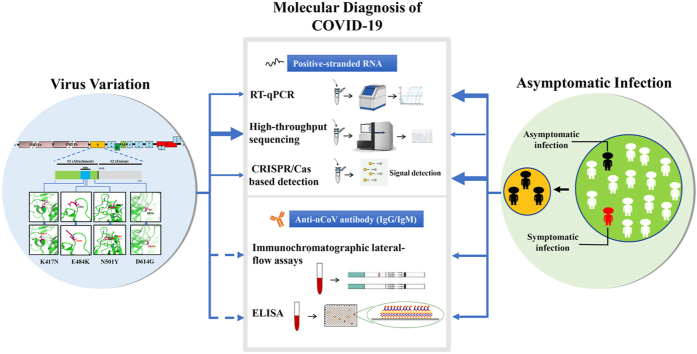
Abstract
Coronavirus disease 2019 (COVID-19) has been a pandemic for more than a year. With the expanding second wave of the pandemic in winter, the continuous evolution of SARS-CoV-2 has brought new issues, including the significance of virus mutations in infection and the detection of asymptomatic infection. In this review, we first introduced several major SARS-CoV-2 mutations since the COVID-19 outbreak and then mentioned the widely used molecular detection techniques to diagnose COVID-19, primarily focusing on their strengths and limitations. We further discussed the effects of viral genetic variation and asymptomatic infection on the molecular detection of SARS-CoV-2 infection. The review finally summarized useful insights into the molecular diagnosis of COVID-19 under the special situation being challenged by virus mutation and asymptomatic infection.
Keywords: SARS-CoV-2, Virus variation, Nucleic acid detection, Asymptomatic infection
Graphical abstract
Highlights
-
•
The widely used molecular diagnostic techniques for COVID-19 are reviewed, with a special focus on their strengths and limitations.
-
•
The genetic variation of the viral genome and silent asymptomatic infection during the global battle against the nCoV epidemic are discussed.
-
•
Some diagnosis strategies for those within the quarantine period at home or the assembly site are recommended.
1. Introduction
Novel coronavirus (nCoV, also named as SARS-CoV-2)-induced pneumonia, officially termed as the coronavirus disease 2019 (COVID-19), has become the top troubling epidemic since its outbreak last winter [1]. According to the publication of Johns Hopkins University statistics, up to February 24th, 2021, the number of COVID-19 cases had reached 112, 108, 217, and more than 2.48 million patients died of the disease worldwide. Hence, earlier and efficient diagnosis of the disease is highly crucial for controlling the pandemic. The current diagnosis of COVID-19 depends comprehensively on the epidemiological history, clinical symptoms, and crucial medical inspection, including computed tomography (CT) imaging and molecular testing such as nucleic acid detection and immunological testing on IgM/IgG [2]. Among them, molecular detection is the most powerful technology for detecting SARS-CoV-2 so far. The virus has been transmitting in humans for more than a year and huge numbers of the virus proliferations could lead to genome variation and diversity. Several SARS-CoV-2 mutations have been reported since COVID-19 outbreak [3,4]. Besides the virus mutation, another issue impeding controlling the pandemics is the number of asymptomatic cases. Both influence the various aspects of the epidemic, especially the diagnosis. Hence, the sensitive and specific detection techniques of SARS-CoV-2 are crucial for the early diagnosis of COVID-19 and play significant roles in maintaining public health. Here we review the molecular detection techniques for SARS-CoV-2 during last year, with the primary focus on their strengths, limitations, and application in uncovering viral variation and asymptomatic infection. In order to understand easily we start by introducing the molecular structure and diversity of virus genome.
2. SARS-CoV-2 variation
SARS-CoV-2 is a positive-stranded RNA virus belonging to the β coronavirus genus. Due to the highly contagious nature, the virus has attracted significant attention from researchers, and the genome information of SARS-CoV-2 was quickly reported in January 2020 [5]. However, SARS-CoV-2 has undergone many mutations throughout the pandemic, which has made the control of the epidemic more complicated. A considerable effort is currently being devoted to assessing whether these mutations affect the SARS-CoV-2 detection and transmissibility, and the effectiveness of the vaccine.
2.1. SARS-CoV-2 genome
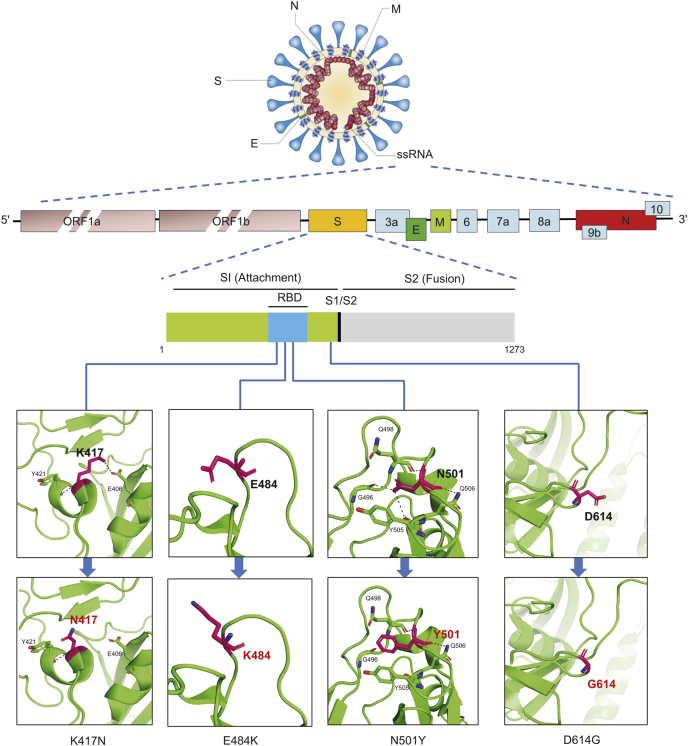
The genome size of SARS-CoV-2 is 29.99 kb, which encodes a variety of non-structural and structural proteins. ORF1a/b encodes non-structural proteins for viral RNA replication and transcription, accounting for about two-thirds of the total genome. The remaining one-third encodes the four essential proteins of coronavirus including membrane (M), nucleocapsid (N), envelope (E), and spike (S) proteins, as well as other non-structural proteins [2]. S protein directly binds to the angiotensin-converting enzyme 2 (ACE2) receptor to mediate SARS-CoV-2 into host cells [1]. In addition to ACE2 receptors, tyrosine-protein kinase receptor UFO (AXL) [6], the high-density lipoprotein (HDL) scavenger receptor B type 1 (SR-B1) [7], etc. are also identified as novel candidate receptors involved in SARS-CoV-2 entry. The S protein comprises two subunits, receptor binding subunit S1 and membrane fusion subunit S2, respectively [8]. The N protein, one of the most abundant viral proteins, combines with viral genomic RNA to form a ribonucleoprotein (RNP) complex [9]. It is involved in viral mRNA transcription, replication, cytoskeletal and immune regulation of host cells [10]. E protein relates to the virus pathogenicity and may activate the host’s inflammatory response. In some coronaviruses, the E protein deletion could reduce the virus’s toxicity [11]. The M protein of SARS-CoV-2 can inhibit IFN-β promoter activation and participate in evading host anti-viral immunity [12]. The S protein has received extreme attention among these functional proteins due to its receptor binding and membrane fusion functions, and it has also become a significant target protein for vaccine and antibody drug development.
2.2. SARS-CoV-2 molecular phylogenetics
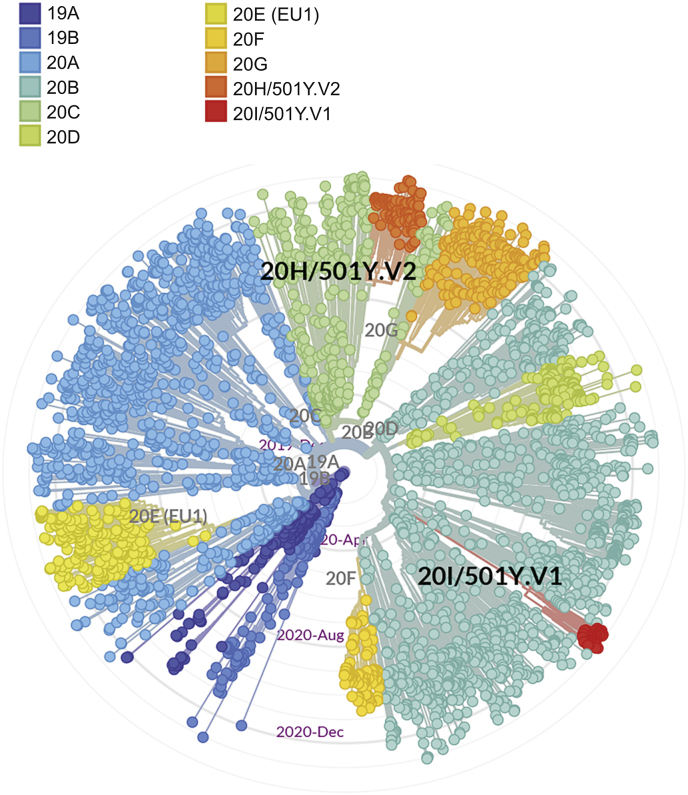
Viral mutations can occur in many different ways. Some mutations are random natural mutations, and some occur to adapt to the human immune microenvironment. The estimated mutation rate in human CoVs is medium to high compared to that of other single-stranded RNA viruses, and the average substitution rate for CoVs is ~10−4 substitutions per site per year [13]. SARS-CoV-2 belongs to nidoviruses, which can proofread genes during gene replication and recombination through an RNA polymerase enzyme to maintain high replication accuracy [14]. Although this gene proofreading function could make the mutation rate of SARS-CoV-2 lower than that of influenza A viruses, the SARS-CoV-2 genome has over 10,000 single nucleotide polymorphisms (SNP) variants [15]. These mutations may lead to changes in structure of some essential proteins and affect transmissibility, virulence and host immune evasion of the virus, further contributing to the large-scale global spread of COVID-19. As the mutations continue to appear, new subtypes of SARS-CoV-2 are increasingly observed. Up to January 2021, the Nextstrain platform had collected a total of 3,819 SARS-CoV-2 genome sequences from different countries and regions around the world. These mutant viruses can be classified into 11 lineages by cluster analysis (Fig. 1). Among these SARS-CoV-2 variants, the 20H/501Y V2 lineage recently appeared in South Africa, and 20H/501Y V1 lineage recently appeared in the United Kingdom and spread rapidly [16,17]. The common feature of these two lineages is the mutation of N501Y in the receptor-binding domain (RBD) of S protein. However, there is limited evidence on the exact relationship between the N501Y mutation and the spread of COVID-19 to date. The role of this mutation in RBD needs to be investigated further.
Fig. 1.
The phylogenetic tree of SARS-CoV-2 (global subsampling complete genome sequences).
The phylogenetic tree of complete genome sequences of 3,819 SARS-CoV-2 was obtained and analyzed with Nextstrain (https://nextstrain.org/nCoV/global?l=unrooted).
2.3. Epidemic SARS-CoV-2 variation
It has been found that the genome of the currently epidemic SARS-CoV-2 virus has undergone significant changes compared to the reference genome obtained in January 2020. The four major epidemic SARS-CoV-2 variations since COVID-19 outbreak were officially notified by the World Health Organization (WHO) on the last day of 2020 (https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en). In addition to the SARS-CoV-2 VOC 202012/01 (belongs to 20H/501Y V1 lineages) variant that caused the recent rapid deterioration of the United Kingdom pandemic, it also includes the current global major infectious virus lineage D614G variant, as well as the Cluster 5 mutation found in Denmark, and the 501Y.V2 variant found in South Africa.
A variant of SARS-CoV-2 appeared at the end of January to early February 2020, with D614G substitution in the gene encoding the S protein. Within a few months, the D614G mutation replaced the original SARS-CoV-2 strain found in China, and by June 2020, it had become the main form of the virus in the global pandemic. Experimental evidence shows that the D614G mutation of SARS-CoV-2 is associated with greater infectivity and higher infectious titers in vitro, while it increases the neutralization sensitivity of SARS-CoV-2 [4,18]. The pseudovirus with the D614G mutation is more likely neutralized by the RBD monoclonal antibody and the convalescent serum of people infected with any virus [19]. So far, there is no evidence that infection with the D614G variant of SARS-CoV-2 might cause more severe diseases [20] (Fig. 2).
Fig. 2.
Location and association of four major SARS-CoV-2 mutations notified by the WHO on December 31st, 2020.
In August and September 2020, a variant of SARS-CoV-2 was discovered in North Jutland, Denmark, which was related to the infection of farmed mink and subsequently spread to human beings. This variant is called the “Cluster 5” by the Danish authority and has a genetic sequence variation that had not been observed before. Up to January 3rd, 2021, Denmark had only found 12 cases of human infection with Cluster 5 in September 2020, and it has not spread widely. The impact of Cluster 5 on the infection rate or the scope and duration of immune protection after vaccination is still under evaluation.
SARS-CoV-2 VOC 202012/01 (Variant of Concern, the year 2020, month 12, variant 01) is a new SARS-CoV-2 variant. Davies et al. [17] uploaded a study to the MedRxiv website, describing this new variant that appeared recently in the United Kingdom. It initially appeared in Southeast England, but has begun to replace other virus lineages in this geographic area and London within a few weeks. On December 26th, 2020, SARS-CoV-2 VOC 202012/01 was identified from routine sampling and genomic testing conducted across the United Kingdom (Fig. 2). The gene sequence of SARS-CoV-2 VOC 202012/01 has 17 mutations, including 14 non-synonymous mutations and 3 deletions, of which 8 mutations locate in the S protein. At least three mutations have potential biological significance [17]. The mutation N501Y occurs at the critical contact residues between RBD and ACE2, and it may enhance the binding affinity of SARS-CoV-2 to human ACE2 [21]. The P681H mutation locates near the S protein’s furin cleavage site, which is a significant region of known infection and spread, but the specific effect of this mutation on SARS-CoV-2 is unclear [17,22]. Besides, the deletion of the two amino acids at positions 69-70 of S protein in SARS-CoV-2 VOC 202012/01 is related to the immune escape of immunocompromised patients and enhances the infectivity of the virus in vitro [23].
501Y.V2 variant of SARS-CoV-2 was named by South Africa because of the N501Y mutation. Although 501Y.V1 from the United Kingdom also has N501Y mutation, phylogenetic analysis shows that 501Y.V2 from South Africa is a different virus variant (Fig. 1). By early November 2020, the 501Y.V2 lineage had replaced the three leading South African virus lineages (B.1.1.54, B.1.1.56 and C.1) that spread during the first pandemic wave and quickly spread in the Eastern Cape, Western Cape and KwaZulu-Natal provinces of South Africa [16]. On December 22nd, 2020, Tegally et al. [16] uploaded information about the new SARS-CoV-2 lineage 501Y.V2 in MedRxiv. 501Y.V2 is characterized by 8 lineage-defining mutations in the S protein, including 3 crucial residues (K417 N, E484K, and N501Y) in the RBD that may have functional significance. E484 interacts with the K31 interaction hotspot residue of human ACE2. There is some evidence that the E484K mutation may moderately enhance binding affinity [21]. K417 of the S protein in SARS-CoV-2 is a unique residue that interacts with D30 of human ACE2 to form a salt bridge in the central contact area [24]. This area is the most significant difference in the RBD-ACE2 complex between SARS-CoV-2 and SARS-CoV, which will enhance the binding affinity of SARS-CoV-2 and human ACE2 [24,25]. Deep mutational scanning showed that K417 N mutation has little effect on human ACE2 binding affinity [21]. At present, there is no clear evidence that the new variants are associated with more severe disease or worse outcomes (Fig. 2).
Most of the mutations observed in SARS-CoV-2 have no significant effect on the virus. Only a few can change the infecting ability of the virus and affect the human immune response, resulting in different severity of the disease. According to the mutations observed in four major epidemic SARS-CoV-2 variations, significant mutations K417 N, E484K, N501Y, D614G and P681H all occur in the S protein, which changes the virulence. In the context of the continuous emergence of new mutations, it is difficult to predict whether any mutation is significant when it first appears. Hence, we should continue to monitor the genome mutation of the SARS-CoV-2 and establish an accurate nucleic acid detection method when the mutated viruses have been found as soon as possible, trace the source of the mutated viruses, and try to prevent further spread of the mutated viruses.
The SARS-CoV-2 S protein sequence was obtained on the website (https://cov3d.ibbr.umd.edu) and analyzed by PyMol software.
3. Nucleic acid detection of SARS-CoV-2
Up to February 22nd, 2021, based on the FIND database analysis of currently existing SARS-CoV-2 diagnostic kits, there are 1131 assays in the database, of which 436 are described as detection kits for SARS-CoV-2 RNA (https://www.finddx.org/covid-19/pipeline). Nucleic acid detection is the primary way to find the specific nucleic acid sequence of SARS-CoV-2 virus in patient samples. There are many detection techniques, including polymerase chain reaction (PCR), real-time quantitative polymerase chain reaction (RT-qPCR), CRISPR-based technologies, high-throughput sequencing, and hybridization technologies [26]. An analysis of 112 detection assays for SARS-CoV-2 RNA found that 90% of these detection assays use PCR or RT-PCR technology, 6% use isothermal amplification technology, 2% use CRISPR technology, and 2% use hybridization technology [26]. Rapid and accurate molecular tools detecting the SARS-CoV-2 viral nucleic acid are in great need for efficient public health responses to the viral threat and are considered as the first step to combat COVID-19.
3.1. RT-qPCR
RT-qPCR uses specific primers to amplify and identify trace amounts of viral genetic material in samples, which is considered as the gold standard for SARS-CoV-2 virus identification due to its shorter assay time, high sensitivity, and specificity. Based on the SARS-CoV-2 genome information, various RT-qPCR assays were designed to detect certain specific viral gene regions with the RNA extracted from clinical samples [27], and some of them were validated and recommended by authorities such as the Centers for Disease Control and Prevention (CDCP) in China. Its subordinate National Institute for Viral Disease Control and Prevention released its validated RT-qPCR strategy and recommended it to the entire global public on January 21st, 2020 (http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html). Within this strategy, two PCR assays (Taqman probe-based fluorescent detection) focusing on the open reading frame (ORF) 1 ab and nucleoprotein (N) gene region of the SARS-CoV-2 genome were established based on the primer sets and probes in the ORF1ab and N gene region. The detected individual was determined as “SARS-CoV-2 positive” when both PCR reactions were found positive. Numerous international teams worked at a high speed to distribute reliable RT-qPCR kits for better diagnostics with different techniques, specifications, and turnaround time all along [28].
In the daily clinical practice of SARS-CoV-2 screening in multiple countries, RT-qPCR plays a vital role as a game-changer and has been so effectively, extensively, and massively employed (by vast orders of magnitude) that even for many medical staff members it is referred to as “nucleic acid detection” itself. However, on some occasions, the RT-qPCR could be troubled with its false-negative effect. It was reported that some individuals with the signature CT change of ground-glass opacity was not initially identified as SARS-CoV-2 positive until the second repeat of RT-qPCR detection with their swab samples [29]. According to the information from 81, 554 reported confirmed SARS-CoV-2 cases of COVID-19 by March 31st, 2020 in mainland China, a single round of RT-qPCR could only help diagnose 61.8% of the total cases. The high false-negative rate of nearly 40% mainly came from the entire sampling process to the end of PCR testing within the diagnosis, but not the false-negative rate of PCR test kits themselves [30]. Hence the inpatients with high clinical suspicion of COVID-19 should participate in the second round of test, even when they get a negative result on the initial RT-qPCR test. Such protocol is presently widely applied. For example, in China, the medical workers follow such rule of 2 RT-qPCR swabs and 2 chest radiographs (CXRs) with more than 48 h apart for inpatients and 2 RT-qPCR swabs (at the beginning and the end of quarantine) for the population during the 14-day quarantine period at home or the assembly site [31].
Suitable samples for RT-qPCR could come from diverse origins, including nasopharyngeal swabs, oropharyngeal swabs, sputum, feces [30], blood, pleural fluid [32], bronchoalveolar lavage (BAL) [33], and breastmilk [34] to detect the existence of SARS-CoV-2 in patients’ and environmental samples. The nasopharyngeal and oropharyngeal swabs are chosen among various clinical samples and frequently used in the present standard protocol. The PCR kit developed by Beijing Genomics Institution could detect every positive sample when the input viral genome load is 3200 copies/mL, and the sensitivity is still as high as 97.1% if 640 copies/mL virus is loaded [30]. However, primer-probe sets with huge diversity are now employed in various RT-qPCR assays developed by different companies. Most of them could help reach the detection limit of 500 copies with high sensitivity and specificity [35]. Moreover, the airborne speech droplets have already been considered as a possible SARS-CoV-2 transmission carrier and do not disappear within 8 min from the window [36]. Meanwhile, perfect anti-viral disinfection should be emphasized at full time for safety concerns and to prevent cross-contamination of samples. Besides, it takes a couple of hours to fulfill the reaction before the results could come out.
Many scientists are working on RT-qPCR optimization, and there is some good news. For example, microneedle-based oropharyngeal swabs are introduced to improve the quality and quantity of virus collection for COVID-19 testing [37]. For high-scale screening in regions with a low prevalence, pooled tests could be applied for financial consideration [38]. Similar techniques such as reverse transcription loop-mediated isothermal amplification (RT-LAMP) have been developed to overcome certain RT-qPCR shortcomings. This assay could detect as many as 50 copies per μL in viral transport medium within 30 min [39]. In an RT-LAMP assay on surplus RNA samples isolated from 768 pharyngeal swabs after COVID-19 testing, an excellent performance with a sensitivity of 97.5% and a specificity of 99.7% was validated [40]. The reverse transcription-recombinase polymerase amplification (RT-RPA) method also exhibits its excellent potential. The inexpensive enhanced RT-RPA could detect as many as 5 copies within 45 min, does not require RNA purification, and supports the high-scale detection [41]. It was optimized to one-copy sensitivity, field-deployable, and could simultaneously detect N and S genes from SARS-CoV-2 [42]. Droplet digital PCR is also established in a small sample volume of 50 nasopharyngeal swabs, and the results showed that this direct quantitation could reach a sensitivity of 93.33% and specificity of 100% [43]. Such methods might be promising if validated with a larger number of specimens.
3.2. Sequencing
High-throughput sequencing of the genome is a powerful technique to obtain the full-length genome sequences of SARS-CoV-2 isolated from patients, which offers critical clues for potential transmission and virus evolution [1,44]. Although the equipment dependency and high-cost limit its application, its application is irreplaceable as a potent detection tool. For example, SARS-CoV-2 viral genome might stay persistently detectable in swabs from the upper respiratory tract via RT-qPCR even after several weeks since the patients were fully cured [45]. Such RT-qPCR re-positive results are confusing since it is difficult to tell whether these individuals are still actively infectious or have been cured. This confusing phenomenon could be interpreted by the nanopore sequencing result of the SARS-CoV-2 genome from individuals [35], which showed the degraded viral genome could be responsible for a relatively lower transmission risk for those re-positive outpatients [46].
3.3. CRISPR/Cas based detection
There have been several CRISPR/Cas-based methods established for SARS-CoV-2 detection. Generally speaking, such CRISPR-Cas-based strategy used Cas proteins’ nuclease activity after the gRNA was specifically bound to viral genes such as ORF1ab, and the produced fluorescent signals released from the cleaved single-stranded (ss) RNA reporter probe help reflect the existing SARS-CoV-2 [47]. Multiplex CRISPR/Cas13a-based diagnosis methods have been developed for SARS-CoV-2, and some of them could reach a nearly 100% (single-copy) sensitivity [48]. Its fulfill period of 40 min is very time-saving. No thermo-cycle helps reduce the potential cross-sample aerosol contamination within the equipment and detection facilities. In Cas13-based detection system optimized by Arizti-Sanz et al. [49], it was validated with 90% sensitivity and 100% specificity with a turnaround time of 50 min. It is more exciting that CRISPR-Cas12a assay could bring about an ultrasensitive, instrument-free, and visual detection (for few copies) within as short as 20 min [50]. For improving disease control, many point-of-care assays are optimized all along. For example, a smartphone-read saliva test combing the CRISPR-Cas12a and RT-RPA techniques has been developed for potential portable use [51].
Besides the RNA detection system, some DNA-based platform using DNA nanoswitch with very reasonable cost and detection speed has been designed in labs [52]. So far, multiple platforms and strategies with a high diversity of assay systems were developed for nucleic acid detection of SARS-CoV-2, including in a tube, a Chip, a box, a cartridge, and even on a drone.
Based on the SARS-CoV-2 genome information, some SARS-CoV-2 nucleic acid detection methods had been verified and recommended by the WHO and various countries [53]. However, the targets of SARS-CoV-2 nucleic acid detection recommended by each country could be different. The Chinese CDCP recommended ORF1ab and N gene regions as target genes for detection, while three targets in N gene were recommended as target genes by the CDC of United States and the National Institute of Health in Thailand. RNA dependent RNA polymerase (RdRP), E, and N genes were recommended as target genes in Germany, and two targets in RdRP are recommended as target genes by Institut Pasteur of France. The National Institute of Infectious Diseases in Japan recommended ORF1a, N and S genes as target genes for detection (https://www.who.int/publications/m/item/molecular-assays-to-diagnose-covid-19-summary-table-of-available-protocols). Different target genes and SARS-CoV-2 detection kits from different manufacturers can be chosen according to their own purposes, while it may cause differences in the sensitivity of SARS-CoV-2 detection.
4. Immunological testing
Immunochemical testing is a powerful technique for detecting viral infections. Although RT-qPCR is the most commonly used detection method for SARS-CoV-2 virus identification [54], it also leads to potential misdiagnosis. So other effective complementary detection methods are needed considering the false negative and false positive results of RT-qPCR.
4.1. Antigen detection of the SARS-CoV-2
For medical surveillance, the need for serologic tests on SARS-CoV-2 antigen and antibody emerged. A multiplex diagnostic pipeline named ReScan was developed to perform proteome-wide profiling of SARS-CoV-2 antigens enriched by pan-98 patients’ sera; two essential antigens, including spike and nucleocapsid proteins, have been found and are widely used in SARS-CoV-2 serologic assays [55]. Besides, some wireless electrochemical platforms were established to detect SARS-CoV-2 antigen using two-dimensional monoelemental materials (Xenes), named as SARS-CoV-2 Rapid-Plex [56]. Such electrochemical antigen detection method brings us a quick, low-cost way for SARS-CoV-2 detection.
4.2. Antibody detection of the SARS-CoV-2
Infection of SARS-CoV-2, just like many other viruses, could stimulate the human immune system for defense. Antibodies such as IgG and IgM are produced during the infection process with a well-recognized pattern and specific binding to N and S viral proteins [57], which could become the molecular basis of the present SARS-CoV-2 antibody detection. The ELISA assay of 2019-SARS-CoV-2 antibodies such as IgG and IgM was one of the earliest validated methods to explore individuals’ infection state [1]. The COVID-2019 patients displayed significantly evaluated IgG and IgM, with an outstanding cut-off value compared with that of healthy controls in ELISA assay. SARS-CoV-2-specific IgM and IgG antibodies could be detectable in serum between the first to second weeks after the onset of symptoms, and the IgG specific to SARS-CoV-2 spike protein stays detectable in serum up to 2 months after symptom onset [58]. Seropositivity for IgG and IgM was detected in 32% of patients with mild symptoms after 2 weeks of symptom onset and 3% of healthy blood donors [59]. A study on 175 once-infected outpatients and asymptomatic individuals found that their SARS-CoV-2 antibodies, including IgG, are quite long-lasting until they progressively decrease after 5 months post-infection [60]. IgM antibodies reduced sharply, while serum and saliva IgG antibodies stably remained in most COVID-19 patients for at least 3 months post symptom onset [61]. As early as in February 2020, a rapid IgM-IgG combined antibody test was developed and clinically applied for SARS-CoV-2 diagnosis. Its performance within a whole process of only 15 min ended up with a sensitivity of 88.66% and a specificity of 90.63%, validated in 397 independently-collected PCR confirmed COVID-19 patients and 128 negative patients, which is very promising [62]. In a test with 43 RT-qPCR-confirmed SARS-CoV-2 infection and 40 negative control subjects, the IgG/IgM immunochromatographic card method displayed an excellent specificity of 100% for IgG and IgM [63]. So far, there are more than 100 different serological antibody tests available for SARS-CoV-2 antibody detection currently, such as enzyme immunoassay, immunofluorescence test, dot blot/Western blotting, virus neutralization test, immunochromatographic lateral-flow assays [64], biolayer interferometry immunosorbent assay [65], and antigen microarray [66].
There is a piece of very shocking news that in some regions, such as Kenyan in Africa, the seroprevalence analysis of anti-SARS-CoV-2 IgG antibodies suggested that about 90% of the SARS-CoV-2 antibodies-positive population was not detected through the authorities’ RT-qPCR tests [67]. The serological tests on IgG and IgM were demonstrated to be potent supplementary methods for COVID-19 pandemic surveillance, considering the vast potentially infected population. However, it is not recommended to apply to the entire general population just as we did with RT-qPCR tests due to its high false-positive ratio against real positive results [68]. SARS-CoV-2 serology is very complicated; for example, the cross-reactivity with seasonal (non-severe acute respiratory syndrome) coronaviruses [69] as well as autoantibodies in autoimmune diseases [70] could contribute to the false-positive result, while immunodeficiency can lead to false-negative reaction [71].
Besides, there is still much uncertainty about the situation when the result of SARS-CoV-2 nucleic acid is negative, anti-SARS-CoV-2 IgM is negative, and anti-SARS-CoV-2 IgG is positive. He/she might be once-infected while recovering. Any false-negative for nucleic acid may leave out an infected individual experiencing an infection with few or mild symptoms. The individual could obtain the preexisting immunity (anti-SARS-CoV-2 IgG) through vaccine inoculation [72], and a few individuals might even produce preexisting neutralizing antibodies without SARS-CoV-2 pathological infection after naturally exposed to deactivated zoonotic SARS-CoV-2. Hence, it is vital to distinguish the situation of preexisting and de novo immunity before and/or after the serological antibody tests [73]. Indeed, such exceptional immunological condition of healthy individuals makes the management of pandemic surveillance more complicated than we could ever imagine. The test results of healthy and safe individuals with antibody protection could be very similar to those of most worrying asymptotic infected ones. Therefore, we suggest that the molecular diagnosis strategy should always combine the RT-qPCR based nucleic acid detection and the serological antibody tests whenever financial conditions permit.
5. Perspective
Although people from all walks of life have made huge efforts and great advances against the pandemic, COVID-19 has been becoming rampant to threaten human society. The reasons are complicated from both sides, human and virus. But two things probably contribute much more to the situation, namely, virus mutations and existence of asymptomatic cases.
5.1. Detecting SARS-CoV-2 mutations
SARS-CoV-2 variations have such a significant impact on virus transmission and bring a big challenge for virus detection. Up to now, genome sequencing is the standard method for detecting SARS-CoV-2 mutations. RT-qPCR, CRISPR/Cas, RT-LAMP, surface-enhanced Raman spectroscopy (SERS), denaturing high-performance liquid chromatography (dHPLC), and mass spectrometry (MS) can also be used to detect mutations of viruses [25,74]. However, some traditional detection methods are often complicated and time-consuming. Another significant effect of SARS-CoV-2 mutation is that it might decrease the detection sensitivity. Artesi et al. [75] reported that the failed detection of SARS-CoV-2 E protein gene in 8 patients is associated with the C-to-U transition at position 26340 of the SARS-CoV-2 genome. A study uploaded to bioRxiv in January 2021 conducted an in silico survey on SARS-CoV-2 sequence variability within the binding regions of primer/probe and performed RT-qPCR detection using synthetic RNA containing these mutations [76]. It highlights the necessity of genomic monitoring for SARS-CoV-2 and selection for RT-qPCR primers. Moreover, there is an urgent need for more efficient, easy-to-operate, and straightforward detection methods to monitor SARS-CoV-2 mutations.
As far as we are concerned, the presently reported mutations in virus variants are mainly located in the S gene region. As we mentioned before, for many of the RT-qPCR strategies, PCR primers are designed focusing on other gene regions such as ORF1ab and N gene region of the SARS-CoV-2 genome. These gene regions are considered to be free from mutations so far. Hence, RT-qPCR test is still instrumental in helping detect SARS-CoV-2, even faced with the possibility of global viral mutations. Various antibodies (IgM and IgG) specifically binding to both the spike and nucleocapsid protein are commonly used in antibody screening assays. Although there is some good news that antibody screening is not affected by so far identified SARS-CoV-2 spike mutations [77], we prefer to choose the kits with the antibody specific for the less frequently mutated nucleocaspid protein if possible. In addition, considering many of the commonly used detecting methods do not distinguish the virus variants from the wild type, it is recommended to apply additional deep sequencing on the “SARS-CoV-2 positive” individuals for any mutations.
5.2. Detecting asymptomatic infected individuals
In addition to SARS-CoV-2 variations, asymptomatic infection is another challenge for virus detection. According to the information from 81, 554 reported confirmed SARS-CoV-2 cases of COVID-19 by March 31st, 2020 in mainland China, 1.2% of cases were so-called asymptomatic infected [30]. In many other countries, the ratio is as high as 10%–30% [78]. For some of the asymptomatic infected individuals, their virus copies within the upper respiratory tract might still be accumulating below the limit of nucleic acid detection with the specific anti-SARS-CoV-2 IgG and IgM also undetectable. This is why the “false negative” results are very likely to be found in asymptomatic infected individuals. However, it was also believed that most transmission incidences could come from the pre-symptomatic stage and the asymptomatic infections. Hence, it is even more vital to determine asymptomatic infections with high specificity and sensitivity.
Hence, establishing efficient high-throughput tests for the detection of asymptomatic carriers is more urgent. Moreover, effective screening depends mostly on the testing frequency and reporting speed, and secondarily on high test sensitivity. Thus, extensive, frequent, and large-scale screening with short sample-to-answer time should be above all [79]. Accordingly, we consider that RT-qPCR with even insufficient sensitivity could still play its colossal role. A further recommendation is choosing a combination of appropriately timed and multiple rounds of RT-qPCR (e.g., once per week on Day 0, 7, and 14 within the quarantine period at home or at the assembly site) and serological antibody testing for anti-SARS-CoV-2 IgM and IgG (e.g., on Day 0 and Day 14 within the quarantine period at home or at the assembly site). In addition, for those who just passed the 14-day quarantine period at home or at the assembly site, a self-disciplined isolated lifestyle with daily health checking for any abnormal subclinical signs, mask-wearing, and minimum public social contact is beneficial to public health. Any of their voluntary tests for RT-qPCR and serological antibody testing should always be highly recommended (encouraged at moral and financial levels).
Based on the strengths mentioned above and limitations of various methods to diagnose SARS-CoV-2, a comparison chart of their turnaround time cost, dependency on sophisticated equipment, requirements for the capacity of reagent supplies, and the diagnosis value are analyzed and displayed in Table 1 [[1], [27], [28], [30], [35], [39], [40], [41], [42], [44], [46], [48], [49], [50], [51], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [68], [69], [71], [77], [80], [81]].
Table 1.
Strengths and limitations of various methods to diagnose SARS-CoV-2 (evaluated for each sample).
| Method | Turnaround time | Equipment requirement | Reagent requirement | Diagnosis value |
Refs. | ||
|---|---|---|---|---|---|---|---|
| Sensitivity/specificity | Capacity to diagnose asymptomatic infection | Potential for mutation detection | |||||
| RT-qPCR | 2 h | Medium | High | High/High | High | Medium | [27,28,30,35,80] |
| RT-LAMP, RT-RPA, etc. |
30-45 min | Low | High | High/High | High | Medium | [[39], [40], [41], [42]] |
| Sequencing | 8 h | High | Low | Low/High | Low | High | [1,44,46] |
| CRISPR based strategy | 20-50 min | Low | High | High/High | High | Medium | [[48], [49], [50], [51],81] |
| Antibody detection | Huge diversity (some <1 h) | Low | Medium | Medium/Medium (due to immunodeficiency and cross-activity) | Medium | Low | [1,[57], [58], [59], [60], [61], [62], [63], [64], [65], [66],68,69,71,77] |
6. Conclusion
Based on the recent advances, we reviewed the widely used molecular diagnostic techniques for SARS-CoV-2, with a primary focus on their strengths and limitations in discussing several concerning issues such as genetic variation of virus and asymptomatic infection during our global battle against COVID-19. Some more sensitive, efficient, easy-to-operate, and straightforward detection methods are still in urgent demand for COVID-19 due to the virus variation and asymptomatic infection.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (project No. 81970029), Fundamental Research Funds for the Central Universities of China (The Emergency Projects on COVID-19, xzy032020042), and Qinnong Bank-XJTU special project for COVID-19 (qnxjtu-12).
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Contributor Information
Liesu Meng, Email: mengliesu@xjtu.edu.cn.
Shemin Lu, Email: lushemin@xjtu.edu.cn.
References
- 1.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X.W., Geng M.M., Peng Y.Z. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheikh J.A., Singh J., Singh H. Emerging genetic diversity among clinical isolates of SARS-CoV-2: lessons for today. Infect. Genet. Evol. 2020;84:104330. doi: 10.1016/j.meegid.2020.104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korber B., Fischer W.M., Gnanakaran S. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu F., Zhao S., Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S., Qiu Z., Hou Y. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei C., Wan L., Yan Q. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metab. 2020;2:1391–1400. doi: 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]
- 8.Wrapp D., Wang N.S., Corbett K.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng Y., Du N., Lei Y.Q. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020;39 doi: 10.15252/embj.2020105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni L., Ye F., Cheng M.L. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandala V.S., McKay M.J., Shcherbakov A.A. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020;27:1202–1208. doi: 10.1038/s41594-020-00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Y., Zhuang M.W., Han L.L. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct. Target Ther. 2020;5:299. doi: 10.1038/s41392-020-00438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su S., Wong G., Shi W.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferron F., Subissi L., De Morais A.T.S. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J.H., Wang R., Wang M.L. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tegally H., Wilkinson E., Giovanetti M. Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature. 2021;592 doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 17.Davies N.G., Barnard R.C., Jarvis C.I. Estimated transmissibility and severity of novel SARS-CoV-2 variant of concern 202012/01 in England. MedRxiv. 2020 doi: 10.1101/2020.12.24.20248822. [DOI] [Google Scholar]
- 18.Plante J.A., Liu Y., Liu J.Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weissman D., Alameh M.G., de Silva T. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe. 2021;29:23–31.e4. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grubaugh N.D., Hanage W.P., Rasmussen A.L. Making sense of mutation: what D614G means for the COVID-19 pandemic remains unclear. Cell. 2020;182:794–795. doi: 10.1016/j.cell.2020.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starr T.N., Greaney A.J., Hilton S.K. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann M., Kleine-Weber H., Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemp S.A., Meng B., Ferriera I.A.T.M. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion ΔH69/V70. SSRN Electron. J. 2021 doi: 10.2139/ssrn.3780277. [DOI] [Google Scholar]
- 24.Lan J., Ge J.W., Yu J.F. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 25.Yi C.Y., Sun X.Y., Ye J. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020;17:621–630. doi: 10.1038/s41423-020-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carter L.J., Garner L.V., Smoot J.W. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corman V.M., Landt O., Kaiser M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:23–30. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheridan C. Coronavirus and the race to distribute reliable diagnostics. Nat. Biotechnol. 2020;38:382–384. doi: 10.1038/d41587-020-00002-2. [DOI] [PubMed] [Google Scholar]
- 29.Xie X.Z., Zhong Z., Zhao W. Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296:E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu F., Huang W.S. COVID-19 diagnostic process in mainland China: the math beyond pneumonia. J. Allergy Clin. Immunol. 2020;146:64–66. doi: 10.1016/j.jaci.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tavare A.N., Braddy A., Brill S. Managing high clinical suspicion COVID-19 inpatients with negative RT-PCR: a pragmatic and limited role for thoracic CT. Thorax. 2020;75:537–538. doi: 10.1136/thoraxjnl-2020-214916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mei F., Bonifazi M., Menzo S. First detection of SARS-CoV-2 by real-time reverse transcriptase-polymerase chain reaction assay in pleural fluid. Chest. 2020;158:E143–E146. doi: 10.1016/j.chest.2020.05.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harkin T.J., Rurak K.M., Martins J. Delayed diagnosis of COVID-19 in a 34-year-old man with atypical presentation. Lancet Respir. Med. 2020;8:644–646. doi: 10.1016/S2213-2600(20)30232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Groß R., Conzelmann C., Müller J.A. Detection of SARS-CoV-2 in human breastmilk. Lancet. 2020;395:1757–1758. doi: 10.1016/S0140-6736(20)31181-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogels C.B.F., Brito A.F., Wyllie A.L. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat. Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stadnytskyi V., Bax C.E., Bax A. The airborne lifetime of small speech droplets and their potential importance in SARS-CoV-2 transmission. Proc. Natl. Acad. Sci. U.S.A. 2020;117:11875–11877. doi: 10.1073/pnas.2006874117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen W., Cai B., Geng Z. Reducing false negatives in COVID-19 testing by using microneedle-based oropharyngeal swabs. Matter. 2020;3:1589–1600. doi: 10.1016/j.matt.2020.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mutesa L., Ndishimye P., Butera Y. A pooled testing strategy for identifying SARS-CoV-2 at low prevalence. Nature. 2021;589:276–280. doi: 10.1038/s41586-020-2885-5. [DOI] [PubMed] [Google Scholar]
- 39.Ganguli A., Mostafa A., Berger J. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2020;117:22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thi V.L.D., Herbst K., Boerner K. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian J., Boswell S.A., Chidley C. An enhanced isothermal amplification assay for viral detection. Nat. Commun. 2020;11:5920. doi: 10.1038/s41467-020-19258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia S.M., Chen X. Single-copy sensitive, field-deployable, and simultaneous dual-gene detection of SARS-CoV-2 RNA via modified RT-RPA. Cell Discov. 2020;6:37. doi: 10.1038/s41421-020-0175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deiana M., Mori A., Piubelli C. Assessment of the direct quantitation of SARS-CoV-2 by droplet digital PCR. Sci. Rep. 2020;10:18764. doi: 10.1038/s41598-020-75958-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bull R.A., Adikari T.N., Ferguson J.M. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat. Commun. 2020;11:6272. doi: 10.1038/s41467-020-20075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wajnberg A., Mansour M., Leven E. Humoral response and PCR positivity in patients with COVID-19 in the New York City region, USA: an observational study. Lancet Microbe. 2020:e283–e289. doi: 10.1016/S2666-5247(20)30120-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu J., Peng J.J., Xiong Q.L. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMedicine. 2020;59:102960. doi: 10.1016/j.ebiom.2020.102960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu R.S., Cui B.M., Duan X.B. Saliva: potential diagnostic value and transmission of 2019-nCoV. Int. J. Oral Sci. 2020;12:11. doi: 10.1038/s41368-020-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou T.Y., Zeng W.Q., Yang M.L. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arizti-Sanz J., Freije C.A., Stanton A.C. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020;11:5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding X., Yin K., Li Z.Y. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020;11:4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ning B., Yu T., Zhang S. A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou L.F., Chandrasekaran A.R., Punnoose J.A. Programmable low-cost DNA-based platform for viral RNA detection. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abc6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng W., Newbigging A.M., Le C. Molecular diagnosis of COVID-19: challenges and Research needs. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- 54.Wiersinga W.J., Rhodes A., Cheng A.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): A review. JAMA J. Am. Med. Assoc. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 55.Zamecnik C.R., Rajan J.V., Yamauch K.A. ReScan, a multiplex diagnostic pipeline, pans human sera for SARS-CoV-2 antigens. Cell Rep. Med. 2020;1:100123. doi: 10.1016/j.xcrm.2020.100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z., Tang Z., Farokhzad N. Sensitive, rapid, low-cost, and multiplexed COVID-19 monitoring by the wireless telemedicine platform. Matter. 2020;3:1818–1820. doi: 10.1016/j.matt.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang H.W., Li Y., Zhang H.N. SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat. Commun. 2020;11:3581. doi: 10.1038/s41467-020-17488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vabret N., Britton G.J., Gruber C. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grzelak L., Temmam S., Planchais C. A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Röltgen K., Powell A.E., Wirz O.F. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Isho B., Abe K.T., Zuo M. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Z.T., Yi Y.X., Luo X.M. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nuccetelli M., Pieri M., Grelli S. SARS-CoV-2 infection serology: a useful tool to overcome lockdown? Cell Death Dis. 2020;6:38. doi: 10.1038/s41420-020-0275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Özçürümez M.K., Ambrosch A., Frey O. SARS-CoV-2 antibody testing-questions to be asked. J. Allergy Clin. Immunol. 2020;146:35–43. doi: 10.1016/j.jaci.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dzimianski J.V., Lorig-Roach N., O’Rourke S.M. Rapid and sensitive detection of SARS-CoV-2 antibodies by biolayer interferometry. Sci. Rep. 2020;10:21738. doi: 10.1038/s41598-020-78895-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Assis R.R., Jain A., Nakajima R. Analysis of SARS-CoV-2 antibodies in COVID-19 convalescent blood using a coronavirus antigen microarray. Nat. Commun. 2021;12:6. doi: 10.1038/s41467-020-20095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uyoga S., Adetifa I.M.O., Karanja H.K. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Kenyan blood donors. Science. 2021;371:79–82. doi: 10.1126/science.abe1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peeling R.W., Wedderburn C.J., Garcia P.J. Serology testing in the COVID-19 pandemic response. Lancet Infect. Dis. 2020;20:e245–e249. doi: 10.1016/S1473-3099(20)30517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Houlihan C.F., Beale R. The complexities of SARS-CoV-2 serology. Lancet Infect. Dis. 2020;20:1350–1351. doi: 10.1016/S1473-3099(20)30699-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y.S., Sun S.H., Shen H. Cross-reaction of SARS-CoV antigen with autoantibodies in autoimmune diseases. Cell. Mol. Immunol. 2004;1:304–307. [PubMed] [Google Scholar]
- 71.Goetz L., Yang J., Greene W. A COVID-19 petient with repeatedly undetectable SARS-CoV-2 antibodies. J. Appl. Lab. Med. 2020;5:1401–1405. doi: 10.1093/jalm/jfaa137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ewer K.J., Barrett J.R., Belij-Rammerstorfer S. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat. Med. 2021;27:270–278. doi: 10.1038/s41591-020-01194-5. [DOI] [PubMed] [Google Scholar]
- 73.Ng K.W., Faulkner N., Cornish G.H. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mosko M.J., Nakorchevsky A.A., Flores E. Ultrasensitive detection of multiplexed somatic mutations using MALDI-TOF mass spectrometry. J. Mol. Diagn. 2016;18:23–31. doi: 10.1016/j.jmoldx.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 75.Artesi M., Bontems S., Göbbels P. A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01598-20. e01598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakabayashi T., Kawasaki Y., Murashima K. Evaluation of the effects of SARS-CoV-2 genetic mutations on diagnostic RT-PCR assays. bioRxiv. 2021 doi: 10.1101/2021.01.19.426622. [DOI] [Google Scholar]
- 77.Barnes C.O., West A.P., Huey-Tubman K.E. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shental N., Levy S., Wuvshet V. Efficient high-throughput SARS-CoV-2 testing to detect asymptomatic carriers. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abc5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Larremore D.B., Wilder B., Lester E. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chu D.K.W., Pan Y., Cheng S.M.S. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patchsung M., Jantarug K., Pattama A. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020;4:1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]