ABSTRACT
Dengue virus, the etiological agent of dengue fever (DF) occurs in four genetically distinct serotypes (DENV1-4), being transmitted by female Aedes mosquitoes. DF incidence is increasing in Brazil, following vector dispersal, proliferation and DENV serotypes introduction, co-circulation and substitution. Medium- and small-sized cities in Sao Paulo State, such as Marilia (Midwest region), have been affected by huge epidemics. To understand the evolution of DENV epidemics in medium-sized cities, in this study a historical data on DENV incidence (2000-2015) in Marilia, was evaluated. Previous studies disclosed regional and specific DF outcomes associated with 2007 outbreak in that city, when co-circulating DENV1 and DENV3 presented different hematological profiles. In this study, characteristics of 2007 DENV epidemics were compared to the epidemiological, hematological and demographic outlines of the major outbreak of DENV1 in Marilia in 2015. DENV1 genetic diversity was assessed through capsid and pre-membrane junction encoding gene (CprM) sequencing. The results revealed circulation of DENV1 serotype from 2007 to 2015, with epidemics occurring every three-years until 2013 and then, increasing yearly. There were significant differences in hematological profiles of DENV1 patients between 2015 and 2007. CprM showed DENV1 genetic variability in 2015, contrasting with the unique sequence pattern in 2007. These results reinforce the regional and temporal characteristics of DENV epidemics that need local public health research to improve care for people and to limit the spread of new serotypes/genotypes to uninfected areas.
Keywords: Dengue fever, Dengue serotype 1, Epidemiology, Hematological profile, Serotyping
INTRODUCTION
Four dengue virus (DENV; Flavivirus: Flaviviridae) serotypes are transmitted to humans by female Aedes aegypti and A. albopictus mosquitoes and cause dengue fever (DF)1. Clinical forms of DF range from asymptomatic cases or nonspecific symptoms, like other feverish arboviruses, to severe forms such as Hemorrhagic Dengue Fever (DHF) and Dengue Shock Syndrome (DSS)2.
According to the World Health Organization (WHO), dengue virus has worldwide distribution and is endemic in over 100 tropical and subtropical nations3. Brazil is one of the countries with the largest number of DF cases and an annual incidence rate higher than 100/100,000 inhabitants4. Since 2010, the four dengue serotypes circulate in all Brazilian regions, being DENV1 the most prevalent over the last five years5. In 2015, Brazil faced the largest dengue epidemics of the last decade, with a total of 1,700,324 reported cases, but only 0.4% were confirmed by laboratory tests6. Located in the Southeastern region, Sao Paulo State accounted for 72% of such cases. In that year, the number of notified DF cases in the city of Marilia, located in the Midwest region of Sao Paulo State, reached 5,000/100,000 inhabitants, being the largest municipal epidemics in the country. Thus, epidemiological investigation of DENV epidemics in Marilia could help to understand transmission dynamics of DF and related diseases, which in turn can be used as a model for other regions with similar characteristics.
Previous studies on DENV epidemiology in Marilia revealed the maintenance of hematological patterns of DF patients associated with specific serotypes7,8. These characteristics indicated a regional specificity and, in a country as large as Brazil, investigation of the initial burden of a DENV epidemic could help in DF control by local health institutions during the transmission/propagation period. The history of DENV incidence and identification of circulating serotypes in Marilia can also contribute to predict epidemic outcomes. According to publicly available data, DENV epidemics in Marilia are characterized by periods of higher and lower intensity, presenting the largest epidemics in the years 2007, 2010, 2014 and 20159. However, information about serotypes circulation and the number of laboratory-checked cases are only available in local municipal sources, being restricted to public access.
The Public Health Institution of Sao Paulo recommends the laboratory confirmation of DF by viral isolation techniques, serological methods, molecular biology and immunohistochemistry. However, when the incidence of DF exceeds 150/100,000 inhabitants, the guideline is to suspend laboratory confirmation and use the clinical-epidemiological diagnosis10. This strategy is important in view of technical and economic constraints to perform specific and expensive tests during large epidemics. However, it prevents the detection of regional particularities: the frequency of cases with nonspecific symptoms and even asymptomatic infections, the wide variation in clinical presentation of the disease and the number of circulating serotypes. There are also factors that may obscure the occurrence of other seasonal infectious diseases, in addition to underestimating the real number of cases during an epidemic11.
In 2015, several Brazilian States, including Sao Paulo, reported the occurrence of other arboviruses as Chikungunya virus and Zika virus6. The transmission of such viruses occurs by the same DF vector, thus spreading in regions that are also affected by DENV12. Moreover, these diseases present clinical similarities of early symptoms, such as the presence of fever, joint pain and skin rash, making differential diagnosis troublesome12. Thus, characterization of hematological profiles associated with specific arboviruses could help to identify different etiological agents co-circulating with DENV.
The four DENV serotypes are antigenically distinct but they embrace several genotypes and lineages, with differences in virulence and severity of infections13-15. Infections provide life-long protective immunity for the serotype of the given agent and cross-protection immunity against other serotypes for a short period of time, probably due to an improved antibody-dependent enhancement process16. Different genomic regions are used to identify genetic variants of DENV17-20 and their role in determining the molecular diversity of DENV, associated with the disease outcome in human populations. In addition, such protocols are employed to detect dispersal and virulence patterns related to the outbreaks, besides the geographic origin of a given lineage. The DENV capsid and pre-membrane junction encoding gene (CprM) is used to characterize the genetic variation of the virus in endemic areas of Brazil21.
Considering the importance to investigate regional, epidemiological characteristics of DF and to understand the DENV transmission dynamics, in this study, the historical incidence and circulating serotypes in Marilia from 2000 to 2015, are described. Hematological and demographic profiles of DENV1 patients in 2015 and both DENV1 and DENV3 patients in 2007 outbreaks were compared. DENV1 genetic diversity was assessed in both groups by the CprM encoding gene. The results obtained can be used by local health institutions to improve the care of people and limit the spread of DENV new serotypes/genotypes to uninfected areas.
MATERIAL AND METHODS
Dengue historical incidence, patients and laboratory data
Data on the historical incidence of laboratory-confirmed DENV cases in Marilia from 2000 to 2015 were obtained from epidemiological surveillance notification systems: SINAN (Sistema de Informacao de Agravos de Notificacao, from 2000 to 2006); SINANNET (2007-2013), and SINAN online (2014-2015). Percentages of DENV-positive patients were calculated using confirmed cases among those submitted to dengue serological test to detect IgM. Information on DENV circulating serotypes in each year was available for a few samples, and serotyping was carried out by using RT-PCR and virus isolation.
Human samples used in this study corresponded to discarded peripheral blood samples, collected between January and February 2015 for monitoring hematological characteristics of patients with suspected DENV infection. These subjects were attended at the Hemocenter of Marilia Medical School during the 2015 DF epidemics, and they were diagnosed only by clinical-epidemiological criteria. Demographic data (gender and age) and hematological parameters at the first consultation (leukocyte and platelet counts, and percentage of atypical lymphocytes) were obtained for each patient included in the study. Data from patients of the 2007 outbreak were described elsewhere7.
RNA extraction and serotyping
RNA extraction from serum samples was performed using the Mini-Kit RNA Purelink (ThermoFisher Scientific®, Waltham, MA, USA) according to manufacturer’s instructions. Then, the reverse transcription (RT) was performed followed by the polymerase chain reaction (PCR). RT was performed using the reverse transcriptase enzyme M-MLV (ThermoFisher Scientific®, Waltham, MA, USA) in addition to primer D2 (100 μM). PCR master mix containedTaq DNA polimerase (ThermoFisher Scientific®, Waltham, MA, USA) and primers D1/D2 for the amplification of a 511bp fragment from the CprM gene. Next, PCR products were submitted to a multiplex nested PCR with previously described specific primers for DENV1, DENV2, DENV3 and DENV4 (Table 1)22.
Table 1. Oligonucleotide primers used in RT-PCR and Nested PCR to amplify dengue viruses.
| Primers | Sequence (5’-3’) | Molecular weight | Serotype |
|---|---|---|---|
| D1 | TCAATATGCTGAAACGCGCGAGAAACC' | 511 bp | All |
| D2 | TTGCACCAACAGTCAATGTCTTCAGGTTC | 511 bp | All |
| TS1 | CGTCTCAGTGATCCGGGGG | 482 bp | DENV1 |
| TS2 | CGCCACAAGGGCCATGAACAG' | 219 bp | DENV2 |
| TS3 | TAACATCATCATGAGACAGAGC' | 290 bp | DENV3 |
| Den4 | TGTTGTCTTAAACAAGAGAGG' | 394 bp | DENV4 |
| Bp = base pair. | |||
Sequencing of the CprM amplicon, alignment and genetic divergence analysis
Positive amplicons were sequenced by the Sanger method using the Big Dye Terminator® version 3.1 (ABI, Foster City, CA, USA) and primer D1 and D2, according to the manufacturer’s specifications (forward and reverse strands) The specificity of the sequences was assessed by the Basic Local Alignment Sequence Tool (BLAST) software available on the GeneBank website23. Only good quality chromatographic sequences were used for the genetic divergence analysis through the alignment of nucleotides and in silico translated amino acid sequences by the ClustalX software from the Molecular Evolutionary Genetics Analysis (MEGA) software (version 4.0)24.DENV1 nucleotide and amino acid-translated sequences from 2015 were compared to DENV1 sequences from Sao Jose do Rio Preto obtained in 2012, 2013 and 2014; Ribeirao Preto obtained in 2018; Jundiai obtained in 2013; and from Marilia during the 2007 epidemic7 which were used as external groups in the Neighbour-joining method analysis.
Demographic and hematological analyses
Based on molecular typing, DENV1-positive subjects were assembled according to the year of sampling (2007 or 2015; using the same protocol) in Marilia, Southeastern Brazil. Also based on molecular typing, DENV3-positive subjects of the year 2007 were also included in the following comparison. These three groups were firstly compared with respect to gender (the categorical variable, SEX; males coded as 0, females as 1, age (AGE, in years), leukocyte and platelet counts (LEU and PLA, both in Log10 cells/mL), and percentage of atypical lymphocytes (LYM). Another binary variable, LYM2, reflects the fraction of patients with atypical lymphocytes in each group. Subjects considered as univariate outliers for PLA (|z| > 1.96) were previously removed. The X2 test was used to assess a “SEX: Group” contingency table, as well as a “LYM2: Group” contingency table. ANOVA was used for comparisons between groups regarding all the remaining variables. Generalized linear models using the identity function were used to assess PLA as response variables for the explaining factors: Group, SEX, AGE, and the other two hematological variables: LEU e LYM (LYM2 not included). In turn, LEU and LYM were also used as response variables including the remaining hematological variables in the block of explaining factors (LYM2 also discarded) for each additional model. Other models included the interaction among Group and SEX and Group and SEX on the response variables PLA, LEU and LYM. Critical P-values were considered after Bonferroni correction based on the number of similar tests. Analysis were performed with the help of the R Program.
Ethical approval
Human samples evaluation protocols were approved by the Ethics Committee on Human Research of the Marilia School of Medicine (Protocol Nº 069/03) and analysis of Marilia municipal databank information protocol was also approved by the same Ethics Committee under the protocol Nº 45126715.9.0000.5413.
RESULTS
Dengue historical incidence and serotypes circulation
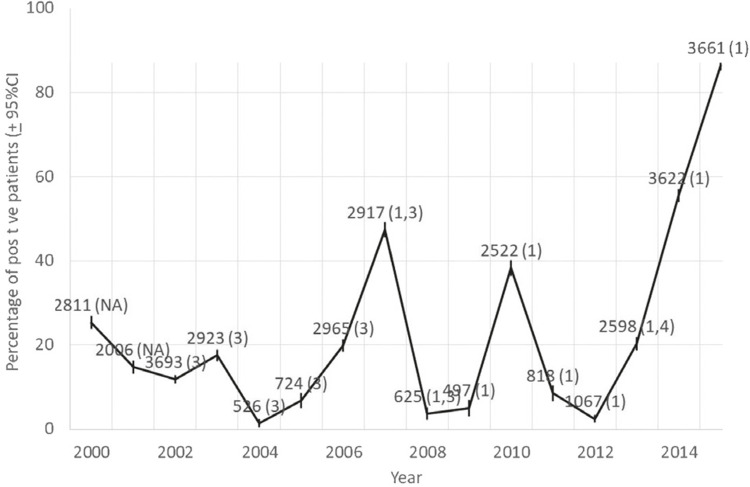
The historical incidence of serological IgM-confirmed dengue cases in Marilia is shown in Figure 1. The circulation of DENV1 serotype was detected from 2007 to 2015, with epidemics occurring every three-years until 2013 and then, increasing yearly.
Figure 1. Dengue yearly incidence based on IgM-confirmed cases in Marilia, Sao Paulo State, Brazil, from 2000 to 2015. Integer numbers are the number of patients tested, followed by the serotypes found (in parenthesis; NA = not available). Confidence intervals were based on the normal approximation to the binomial distribution.
Molecular serotype diagnosis based on the CprM encoding gene
Among 150 patients submitted to molecular diagnosis by RT-PCR22, 65 (43.3%) were positive for DENV1 serotype and the amplicons were submitted to Sanger sequencing, confirming the serotype (data not shown).
Demographic and hematological profiles in 2007 and 2015
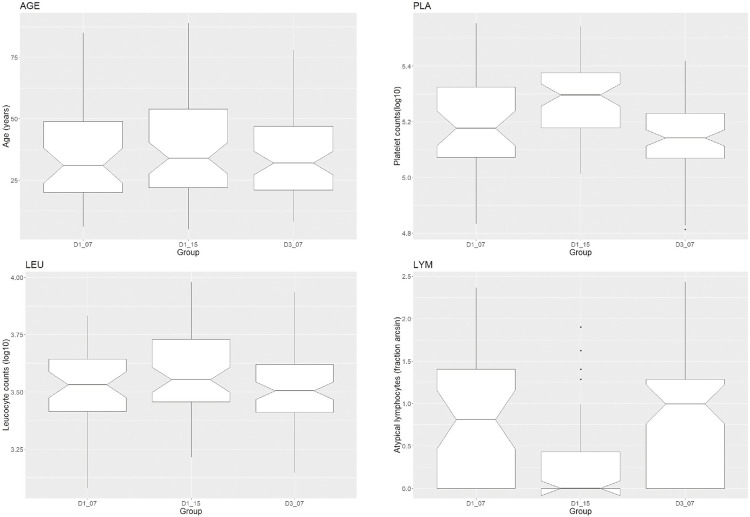
There were 41 DENV1- and 76 DENV3-positive patients in 2007, besides 62 DENV1-positive patients in 2015, after removing eight outliers based on PLA distribution. There was no difference between groups regarding sex, age or leukocyte counts (Table 2). Groups significantly differed in platelet counts (Table 2 and Figure 2). DENV1 patients in the year-group 2015 have the highest scores, presenting 40,000 platelets more than DENV1 patients in 2007. Groups significantly differed also with respect to atypical lymphocytes (Table 2 and Figure 2), both, in the (squared arc sin) fraction of such cells by patient counts (LYM) and in the fraction of patients who presented these abnormal cells (LYM2).
Table 2. Demographic and hematological status of DENV-positive subjects based on molecular typing, detected in Marilia during 2007 and 2015. Groups are labeled by combining the serotype (DENV1 or DENV3) and year. They were compared by gender (SEX; males coded as 0, females as 1), age (AGE, in years), platelet and leukocyte counts (PLA and LEU, both in Log10 cells/mL), and atypical lymphocytes (LYM = squared arc sin; LYM2 = frequency of patients presenting abnormal cells). X2 test was used for SEX: Group and LYM2: Group contingency tables and ANOVA for the remaining variables. P-values < 0.05 are indicated by asterisks. Considering the number of comparisons for the Bonferroni correction, the critical P = 0.05/6 = 0.0083.
| Variables | D1_07 (n = 41) | D1_15 (n = 62) | D3_07 (n = 76) | Test | P | |||
|---|---|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | Mean | SE | |||
| SEX | 0.439 | 0.078 | 0.565 | 0.063 | 0.539 | 0.057 | 1.67 | 0.4344 |
| AGE | 32.732 | 0.645 | 38.694 | 0.591 | 36.145 | 0.497 | 1.16 | 0.3160 |
| PLA | 5.180 | 0.065 | 5.285 | 0.045 | 5.139 | 0.041 | 19.07 | *0.0001 |
| LEU | 3.527 | 0.064 | 3.591 | 0.056 | 3.520 | 0.049 | 2.81 | 0.0631 |
| LYM | 0.760 | 0.137 | 0.287 | 0.093 | 0.860 | 0.091 | 14.94 | *0.0001 |
| LYM2 | 0.561 | 0.065 | 0.258 | 0.042 | 0.724 | 0.045 | 29.96 | *0.0001 |
Figure 2. Demographic and hematological status of DENV-positive subjects based on molecular typing and detected in Marilia during 2007 and 2015. Groups are labeled by combining the serotype (DENV1 or DENV3) and year. They were compared by age (AGE, in years), platelet and leukocyte counts (PLA and LEU, both in Log10 cells/mL), and frequency of atypical lymphocytes (LYM = squared arc sin). ANOVA pointed significant differences among groups with respect to PLA and LYM (Table 2). Post-hoc tests indicated that the 2015 group accounts for the contrasts in these two variables.
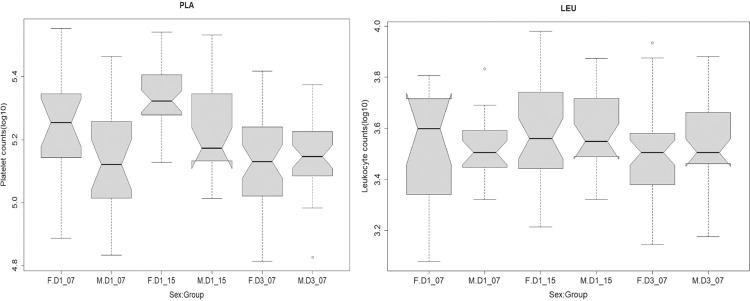
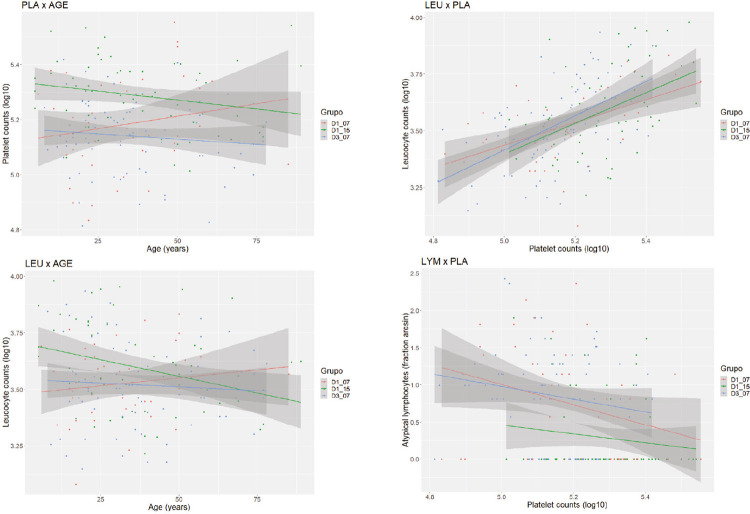
The generalized linear models using, each in its own time, platelet counts, leukocyte counts and the fraction of atypical lymphocytes as response variables and the remaining variables as additional explaining factors indicated that, in 2015, the group of DENV1 patients had an increased platelet count when compared to the reference level of DENV1 patients in 2007 (Table 3). DENV3 patients did not differ from the reference level, and this has also been indicated by post-hoc ANOVA tests (Table 2, Figure 2) among groups; females showed an increased contribution in platelet counts (Table 3, Figure 3); platelet counts were positively and strongly related to leukocyte counts (Table 3, Figure 4); females showed a slight decreased in leukocyte counts (Table 3). However, this finding did not persist after the application of the Bonferroni correction for the number of similar tests; leukocyte counts were negatively and strongly related to the fraction of atypical lymphocytes (Table 3, Figure 4); and the group of DENV1 patients, in 2015, had a decreasing effect on the fraction of atypical lymphocytes when compared to the reference level of DENV1 patients in 2007. DENV3 patients did not differ from the reference level (Table 3, Figure 4).
Table 3. Generalized linear models using the identity function to detect the response of individual hematological variables to other additional demographic and hematological factors in DENV-positive patients detected by molecular typing in Marilia during 2007 and 2015. Groups are labeled by combining the serotype (DENV1 or DENV3) and year. D1_07 was used as the reference group. Platelet and leukocyte counts (PLA and LEU, both in Log10 cells/mL), and frequency of atypical lymphocytes (LYM = squared arc sin) alternated as response variables. Gender (SEX; males coded as 0, females as 1) and age (AGE, in years) were the demographic explaining factors. B is the regression coefficient; SE is its standard error. P < 0.05 indicate significant t-tests. Bonferroni correction for the number of tests comparisons indicates the critical P-value = 0.05/3 = 0.0167.
| Response | Factor | B | SE | t | P |
|---|---|---|---|---|---|
| PLA | Group D1_15 | 0.075 | 0.025 | 2.96 | *0.0035 |
| Group D3_07 | -0.044 | 0.023 | -1.87 | 0.0626 | |
| SEX | 0.060 | 0.018 | 3.27 | *0.0013 | |
| AGE | -0.001 | 0.001 | -0.50 | 0.6199 | |
| LEU | 0.373 | 0.053 | 6.98 | *0.0001 | |
| LYM | -0.001 | 0.015 | -0.07 | 0.9461 | |
| LEU | Group D1_15 | -0.027 | 0.032 | -0.84 | 0.4022 |
| Group D3_07 | 0.034 | 0.030 | 1.15 | 0.2508 | |
| SEX | -0.049 | 0.023 | -2.11 | *0.0364 | |
| AGE | -0.001 | 0.001 | -1.40 | 0.1637 | |
| PLA | 0.592 | 0.085 | 6.98 | *0.0001 | |
| LYM | -0.085 | 0.018 | -4.63 | *0.0001 | |
| LYM | Group D1_15 | -0.360 | 0.124 | -2.90 | *0.0042 |
| Group D3_07 | 0.110 | 0.116 | 0.95 | 0.3449 | |
| SEX | -0.144 | 0.092 | -1.57 | 0.1181 | |
| AGE | -0.001 | 0.002 | -0.59 | 0.5581 | |
| PLA | -0.025 | 0.375 | -0.07 | 0.9461 | |
| LEU | -1.301 | 0.281 | -4.63 | *0.0001 |
Figure 3. Sex: Group interaction acting on hematological variables in DENV-positive patients detected by molecular typing in Marilia during 2007 and 2015. Groups are labeled by combining the serotype (DENV1 or DENV3) and year. Platelet and leukocyte counts (PLA and LEU, both in Log10 cells/mL) and frequency of atypical lymphocytes (LYM = squared arc sin) alternated as response variables to gender (SEX; males coded as M, females as F) in each group. See Table 3 for results of generalized linear models.
Figure 4. Age effects (left side) and relationship among hematological variables (right side) in DENV-positive patients detected by molecular typing in Marilia during 2007 and 2015. Groups are labeled by combining the serotype (DENV1 or DENV3) and year. D1_07 was used as the reference group. Platelet and leukocyte counts (PLA and LEU, both in Log10 cells/mL), and frequency of atypical lymphocytes (LYM = squared arc sin) alternated as response variables. See Tables 4 and 5 for results of generalized linear models.
Despite the significantly increased platelet and leukocyte counts in DENV1 patients in 2015, when compared to DENV1 patients in 2007, the increment of age reduced both scores in the first group (Table 4; Figure 4, left side). However, this finding did not persist after the Bonferroni correction for the number of similar tests. DENV1 patients, in 2015, had a significant decrease in the fraction of atypical lymphocytes, when compared to DENV1 patients in 2007, without any significant effect of age. There was no significant interaction of the group effect on the relationship among the three hematological variables (Table 5; Figure 4, right side).
Table 4. Generalized linear models using the identity function to detect age effects on hematological variables in DENV-positive patients detected by molecular typing in Marilia during 2007 and 2015. Groups are labeled by combining the serotype (DENV1 or DENV3) and year. Platelet and leukocyte counts (PLA and LEU, both in Log10 cells/mL) and frequency of atypical lymphocytes (LYM = squared arc sin) alternated as response variables to age (AGE; in years) in each group. Groups are labeled by combining the serotype (DENV1 or DENV3) and year. D1_07 was used as the reference group. B is the regression coefficient; SE is its standard error. P < 0.05 indicates significant t-tests. Bonferroni correction for the number of tests comparisons indicates the critical P-value = 0.05/3 = 0.0167.
| Response | Factor: Interaction | B | SE | t | P |
|---|---|---|---|---|---|
| PLA | AGE | 0.002 | 0.001 | 1.40 | 0.1637 |
| Group D1_15 | 0.215 | 0.060 | 3.59 | *0.0004 | |
| Group D3_07 | 0.046 | 0.059 | 0.77 | 0.4413 | |
| AGE: Group D1_15 | -0.003 | 0.002 | -2.03 | *0.0436 | |
| AGE: Group D3_07 | -0.003 | 0.002 | -1.66 | 0.0996 | |
| LEU | AGE | 0.001 | 0.002 | 0.83 | 0.4055 |
| Group D1_15 | 0.224 | 0.078 | 2.87 | *0.0047 | |
| Group D3_07 | 0.063 | 0.077 | 0.82 | 0.4113 | |
| AGE: Group D1_15 | -0.004 | 0.002 | -2.17 | *0.0311 | |
| AGE: Group D3_07 | -0.002 | 0.002 | -1.02 | 0.3085 | |
| LYM | AGE | -0.004 | 0.006 | -0.76 | 0.4502 |
| Group D1_15 | -0.794 | 0.273 | -2.91 | *0.0041 | |
| Group D3_07 | 0.069 | 0.268 | 0.26 | 0.7980 | |
| AGE: Group D1_15 | 0.009 | 0.007 | 1.29 | 0.1989 | |
| AGE: Group D3_07 | 0.001 | 0.007 | 0.18 | 0.8556 |
Table 5. Generalized linear models using the identity function to detect relationships among hematological variables of DENV-positive patients detected by molecular typing in Marilia during 2007 and 2015. Groups are labeled by combining the serotype (DENV1 or DENV3) and year. Platelet and leukocyte counts (PLA and LEU, both in Log10 cells/mL) and frequency of atypical lymphocytes (LYM = squared arc sin) alternated as responses to the remaining variables in each group. Groups are labeled by combining the serotype (DENV1 or DENV3) and year. D1_07 was used as the reference group. B is the regression coefficient; SE is its standard error. P < 0.05 indicates significant t-tests. Bonferroni correction for the number of tests comparisons indicates the critical P-value = 0.05/3 = 0.0167.
| Response | Factor: Interaction | B | SE | t | P |
|---|---|---|---|---|---|
| LEU | PLA | 0.503 | 0.147 | 3.41 | *0.0008 |
| Group D1_15 | -0.917 | 1.148 | -0.80 | 0.4255 | |
| Group D3_07 | -1.290 | 1.057 | -1.22 | 0.2242 | |
| PLA: Group D1_15 | 0.176 | 0.219 | 0.80 | 0.4239 | |
| PLA: Group D3_07 | 0.254 | 0.205 | 1.24 | 0.2171 | |
| LYM | PLA | -1.359 | 0.570 | -2.38 | *0.0182 |
| Group D1_15 | -4.371 | 4.443 | -0.98 | 0.3266 | |
| Group D3_07 | -2.533 | 4.092 | -0.62 | 0.5367 | |
| PLA: Group D1_15 | 0.765 | 0.848 | 0.90 | 0.3685 | |
| PLA: Group D3_07 | 0.502 | 0.793 | 0.63 | 0.5278 | |
| LYM | LEU | -1.827 | 0.563 | -3.24 | *0.0014 |
| Group D1_15 | -3.271 | 2.435 | -1.34 | 0.1809 | |
| Group D3_07 | -1.805 | 2.374 | -0.76 | 0.4481 | |
| LEU: Group D1_15 | 0.812 | 0.686 | 1.18 | 0.2379 | |
| LEU: Group D3_07 | 0.538 | 0.673 | 0.80 | 0.4251 |
Comparison of CprM genetic diversity between DENV1 circulating in 2007 and 2015 DF epidemics
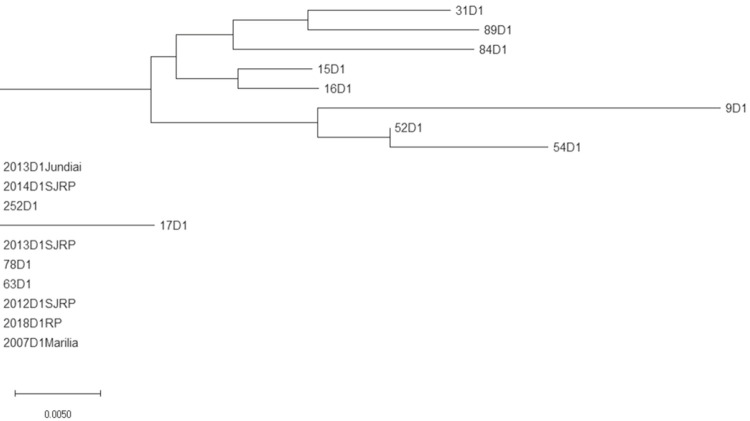
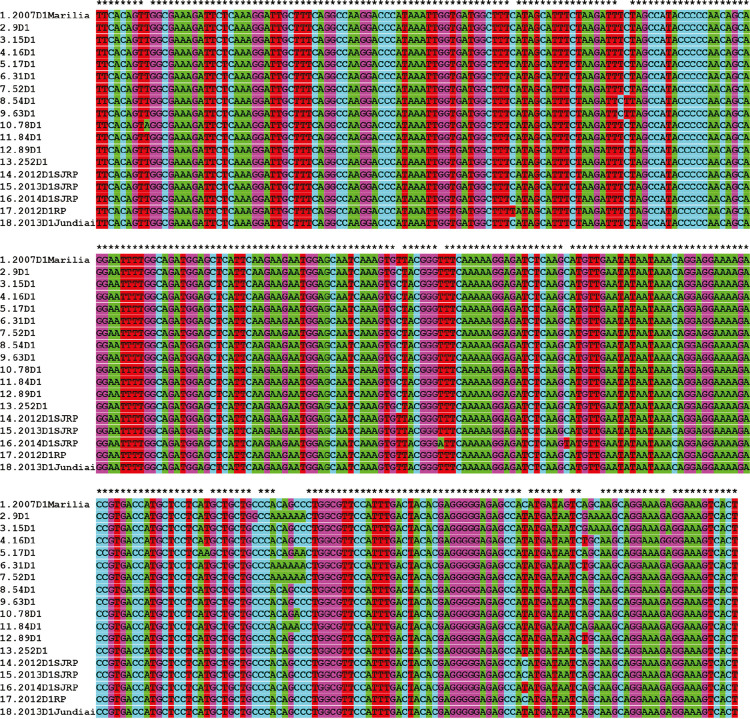
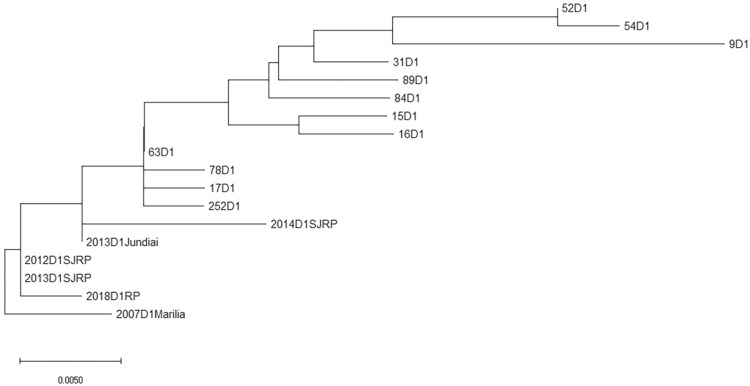
From 65 DENV1 amplicon sequences obtained from patients attended at the Hemocenter from Marilia Medical School in 2015, 12 sequences presented with excellent quality and were used in the genetic diversity evaluation. The corresponding CprM DENV1 unique sequence from 2007 was used as the external group. The results revealed a genetic variability of DENV1 in 2015 (Figure 3), concentrated in the 3’ end portion of the CprM DENV1 fragment (Supplementary Figures 1 and 2).
The genetic variability and divergence of DENV1 CprM evolution in 2015 compared to 2007 was confirmed by the translation of amino acids sequences (Figures 5 and 6).
Figure 5. DENV1 CprM amino acid-translated sequences obtained from 12 Marilia patients in 2015 plus the unique correspondent sequence described in 2007, and from DENV1 retrieved from GenBank of patients of Sao Jose do Rio Preto (SJRP), Ribeirao Preto (RP) and Jundiai in the year 2012, 2013, 2014 and 2018.
Figure 6. Marilia DENV 1 amino acid phylogenetic analysis by Neighbour Joining. The unique sequence obtained in 2007 was used as an outgroup for the 12 remaining, obtained in 2015. CprM amino acid sequences from DENV1 obtained from patients of Sao Jose do Rio Preto (SJRP), Ribeirao Preto (RP) and Jundiai in the year 2012, 2013, 2014 and 2018 were included in the analysis.
DISCUSSION
DENV reemergence in Brazil followed the reintroduction of Aedes aegypti in the 1980’s. Nowadays, all Brazilian States are endemic and the four DENV serotypes circulated differentially in space and time25. As a large country, Brazil has regional peculiarities such as environmental characteristics, population genetic background, social and economic conditions that can contribute distinctly to DENV and its associated diseases evolution. Monitoring and notification26-28 of DENV cases in Brazil are performed in three governmental levels. The Federal and State-owned public databases harbor data generated by the Brazilian State Central Reference Laboratories (LACENs) through serological diagnostic methods and municipal random sampling serotyping. Marilia is a medium sized city of Sao Paulo State (220,000 inhabitants) affected by major dengue epidemics. According to health institutional local rules, laboratory diagnosis is required to confirm the disease (serology and/or viral isolation, exceptionally, by PCR and/or immunohistochemistry) until the incidence reaches 150/100,000 inhabitants. After this mark, a clinical-epidemiological criterion is used. Cases with symptoms compatible with DF are notified to the State-owned public health database, without a laboratory-checked diagnostic. Only cases of DHF, SCD and dengue with complications are laboratory analyzed. At the municipal level, the public health institution tests a high number of patients through DENV IgM detection-based method and the results are registered in the local database, our source for the historical DF incidence and DENV serotypes circulation in Marilia from 2000 to 2015 (Figure 1). The observed pattern of DENV circulation in Marilia city agrees with that expected for the disease in endemic American regions, with epidemics occurring every three to five years, at least until 20131.
DENV serotype entry and substitution are usually associated with increased DF incidence and greater severity of the disease29. According to LACENs, DENV3 predominated in several Brazilian States between 2002 and 2006 and from 2007 to 2009, DENV2 replaced DENV3 as the predominant serotype, which in turn was replaced by DENV1 in 200930. The historical investigation of DENV cases in Marilia revealed a regional specificity. Between 2013 and 2015, DENV incidence increased annually without a main serotype changing. The conspicuous DENV epidemics showed no correlation with serotype introduction and DENV1 entered the city in 2007. DENV1 substituted DENV3 serotype after co-circulation in 2007-2008 and DENV4 after co-circulation in 2013. Thus, DENV1 serotype circulated for at least nine years and was identified in the largest DF epidemics of the historical series (2007, 2010, 2014 and the top-most relevant epidemics in the year 2015, with 3,162 confirmed cases).
There is not a biorepository of biological samples available for retrospective studies on DENV serotyping. Thus, the relationship between DENV1 intra-serotype genetic variation and the DF increase in Marilia was not investigated. However, the partial sequence of the CprM encoding gene presented high genetic variability (at least eleven different variants) in the 2015 circulating DENV1, when compared to the only available DENV1 homologous sequence obtained in 20077 (Supplementary Figures 1 and 2). These results indicate a high evolution rate of DENV1 in Marilia, which corroborate the regional specificity of DF, suggesting that the local health institution epidemic management obtained an efficient control.
The number of suspected and confirmed DENV cases in the historical series of Marilia varies among years (Figure 1). Unconfirmed cases may result from diseases with the same symptoms of DF, in the period of other arboviruses, including Chikungunya and Zika fever, as well as leptospirosis31,32, which could be tested in a differential diagnosis. In addition, identification of demographic and hematological profiles associated with DENV infection risk (such as platelet and leukocyte counts) in the initial epidemic burden could assist in the diagnosis when the clinical-epidemiological criteria is in use, while laboratory tests recommended by State-owned public health institutions are underway8.
During DF epidemics, patients diagnosed by clinical-epidemiological criteria are submitted to hematological exams for platelet count and plasma concentration monitoring in order to detect DHF signals. In 2015, the Hemocenter of Marilia Medical School concentrated part of the Marilia DF cases which were used in the DENV molecular diagnosis and typing, in accordance to the local Human Experimental Ethical Committee approved protocol. From all samples tested for DENV through RT-PCR, 65 were positive and belonged to serotype 1. Among these, 62 patients had complete data on age, gender, platelet and leucocyte counts, and percentage of atypical lymphocytes and were included in the comparative analysis with DENV1-positive patients investigated in 20077.
In the 2007 cohort, there was a positive correlation between atypical lymphocytes, that can be associated with the viremia titer33, and decrease in platelets, while in 2015, increase in atypical lymphocytes was correlated to the decrease in leukocytes (Figure 4, Table 5). These results suggest a change in the DENV1 physio-pathological pattern.
Symptomatic acute dengue infections are associated with the decrease in leukocyte and platelet counts, which are also conditions to the evolution of DF to severe cases34. In 2015 and 2007, both parameters of DENV1-infected patients showed a direct correlation in the bivariate analysis (Figure 4). Age and gender did not correlate to platelet and leukocyte counts in 2007, while in 2015, decreasing in both counts were marginally associated with the increasing of age. These results revealed that, in Marilia, older people can suffer slightly more severe disease outcomes, a different picture from that detected in other Brazilian regions35.
In 2015, despite the circulation of only one serotype, there was a major dengue epidemic in the city of Marilia. Sequencing of the CprM encoding gene from DENV1 in 2015 in positive samples showed a high genetic variability (Supplementary Figures 1 and 2). Besides, the divergence investigation of the CprM encoding gene in 2015 circulating DENV1 revealed a genetic variation, contrasting with the unique DENV1 CprM DENV1 sequence in 2007, as well as resulting in amino acid substitutions in the translated sequences (Figures 5 and 6). This can partially be explained by the high genetic variability presented by RNA genome viruses, which may be associated with the low fidelity of the RNA-dependent RNA polymerase enzyme. It allows the incorporation of mutations into the RNA strand that is being synthesized, together with the absence of a repair mechanism, resulting in the generation of many viral variants36. Thus, even if a serotype has been found in previous regional epidemics, the genetic variation within the serotype itself can affect the previously affected population, increasing the incidence rates of the disease37. The CprM has an important role in the immune response of infected individuals38. Research conducted with human monoclonal antibodies generated after primary or secondary natural DENV infections showed a cross-reactivity with epitopes of different antigens, including the prM protein, which could contribute to increased risks of clinical evolution of DF to DHF39.
The epidemiological study of dengue outbreaks and epidemics is important for understanding the association of genetic characteristics of circulating serotypes and regional aspects, including age, gender and history of the disease in the affected population. It is expected that knowledge of regional features of circulating DENV genetic variants associated with demographic and the hematological profiles of the infected population can contribute to improve the management of DF in Marilia and can also be used as a model for other cities.
CONCLUSION
The historical evaluation of serotypes circulation and the incidence of DENV in Marilia from 2000 to 2015 revealed epidemics occurring in three-years intervals until 2013, and annually afterwards. DENV1 serotype circulated from 2007 to 2015, after co-circulation with DENV3 in 2007-2008 and with DENV4 in 2013. There were significant differences in hematological profiles between 2007 and 2015 DENV1 patients and even between patients with different serotypes circulating in 2007. In 2015, platelet and lymphocyte counts marginally decreased with age, indicating a slightly severe disease in older people. The percentage of atypical lymphocytes, used as a marker for the viral titer, increased with the decreasing in platelets, in 2007, and lymphocytes, in 2015, showing temporal differences in DENV1 physio-pathologic mechanisms. The CprM encoding gene from 2015 DENV1 showed a genetic variability contrasting with the unique sequence obtained in 2007. These results reinforce the regional and temporal characteristics of DENV epidemics, which needs a local public health investigation to improve people’s medical care and to limit DENV new serotypes/genotypes spreading to uninfected areas.
ACKNOWLEDGMENTS
We would like to thank all patients and the Faculdade de Medicina de Marília (FAMEMA) for gently contributing with biological samples, laboratory and demographic data. This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (scholarship to Luana Prado Rolim de Oliveira, fellow Nº 2019/11384-0); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), finance code 001; Universidade Federal do ABC (UFABC). Conselho Nacional de Desenvolvimento Científico e Tecnológico/CAPES and Ministry of Health of Brazil (CNPq/MCTIC/FNDCT grant Nº 440812/2016-0).
SUPPLEMENTARY MATERIAL.
Supplementary Figure 1. DENV1 CprM nucleotide sequences obtained from 12 Marilia patients in 2015 plus the unique correspondent sequence described in 2007; and from DENV1 retrieved from GenBank of patients of Sao Jose do Rio Preto (SJRP), Ribeirao Preto (RP) and Jundiai in the year 2012, 2013, 2014 and 2018.
Supplementary Figure 2. Marilia DENV 1 nucleotide phylogenetic analysis by Neighbour Joining. The unique sequence obtained in 2007 was used as an outgroup for the 12 remaining, obtained in 2015. CprM nucleotide sequences from DENV1 obtained from patients of Sao Jose do Rio Preto (SJRP), Ribeirao Preto (RP) and Jundiai in the year 2012, 2013, 2014 and 2018 were included in the analysis.
REFERENCES
- 1.Salles TS, Encarnação Sá-Guimarães T, Alvarenga ES, Guimarães-Ribeiro V, Meneses MD, Castro-Salles PF, et al. History, epidemiology and diagnostics of dengue in the American and Brazilian contexts: a review. 264Parasit Vectors. 2018;11 doi: 10.1186/s13071-018-2830-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin CY, Huang CH, Chen YH. Classification of dengue: the clinical use of World Health Organization 2009 guideline. J Formos Med Assoc. 2013;112:61–63. doi: 10.1016/j.jfma.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization Dengue control: epidemiology. [cited 2021 Jan 15]. https://www.who.int/denguecontrol/epidemiology/en/
- 4.San Martin JL, Brathwaite O, Zambrano B, Solórzano JO, Bouckenooghe A, Dayan GH, et al. The epidemiology of dengue in the Americas over the last three decades: a worrisome reality. Am J Trop Med Hyg. 2010;82:128–135. doi: 10.4269/ajtmh.2010.09-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruycker-Nogueira F, Souza TM, Chouin-Carneiro T, Costa Faria NR, Santos JB, Torres MC, et al. DENV-1 Genotype V in Brazil: spatiotemporal dispersion pattern reveals continuous co-circulation of distinct lineages until 2016. 17160Sci Rep. 2018;8 doi: 10.1038/s41598-018-35622-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brasil. Ministério da Saúde. Secretaria de Vigilância em Saúde Monitoramento dos casos de dengue, febre de chikungunya e febre pelo vírus Zika até a Semana Epidemiológica 48, 2015. Bol Epidemiol. 2015;46:1–9. https://antigo.saude.gov.br/images/pdf/2016/janeiro/07/2015-svs-be-pncd-se48.pdf [Google Scholar]
- 7.Carmo AM, Suzuki RB, Cabral AD, Costa RT, Massari GP, Riquena MM, et al. Co-circulating serotypes in a dengue fever outbreak: differential hematological profiles and phylogenetic relationships among viruses. J Clin Virol. 2017;90:7–13. doi: 10.1016/j.jcv.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Santos Carmo AM, Suzuki RB, Riquena MM, Eterovic A, Sperança MA. Maintenance of demographic and hematological profiles in a long-lasting dengue fever outbreak: implications for management. 84Infect Dis Poverty. 2016;5 doi: 10.1186/s40249-016-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Araujo RV, Albertini MR, Costa-da-Silva AL, Suesdek L, Franceschi NC, Bastos NM, et al. São Paulo urban heat islands have a higher incidence of dengue than other urban areas. Braz J Infect Dis. 2015;19:146–155. doi: 10.1016/j.bjid.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.São Paulo. Secretaria da Saúde . Grupo Técnico Assessor em Arboviroses no Estado de São Paulo. Plano de contingência estadual contra arboviroses urbanas. São Paulo: Secretaria da Saúde; 2018. [cited 2021 Jan 15]. http://docs.bvsalud.org/biblioref/2020/08/1116798/arboviroses18_plano_contingencia_out18.pdf [Google Scholar]
- 11.Passos SR, Bedoya SJ, Hökerberg YH, Maia SC, Georg I, Nogueira RM, et al. Clinical and laboratory signs as dengue markers during an outbreak in Rio de Janeiro. Infection. 2008;36:570–574. doi: 10.1007/s15010-008-7334-6. [DOI] [PubMed] [Google Scholar]
- 12.Estofolete CF, Terzian AC, Colombo TE, Freitas Guimarães G, Ferraz HC, Junior, Silva RA, et al. Co-infection between Zika and different dengue serotypes during DENV outbreak in Brazil. J Infect Public Health. 2019;12:178–181. doi: 10.1016/j.jiph.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Vicente CR, Herbinger KH, Froschl G, Malta Romano C, Souza Areias Cabidelle A, Cerutti C., Junior Serotype influences on dengue severity: a cross-sectional study on 485 confirmed dengue cases in Vitoria, Brazil. 320BMC Infect Dis. 2016;16 doi: 10.1186/s12879-016-1668-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vicente CR, Lauar JC, Santos BS, Cobe VM, Cerutti C., Junior Factors related to severe dengue during an epidemic in Vitória, State of Espírito Santo, Brazil, 2011. Rev Soc Bras Med Trop. 2013;46:629–632. doi: 10.1590/0037-8682-1579-2013. [DOI] [PubMed] [Google Scholar]
- 15.Guilarde AO, Turchi MD, Siqueira JB, Junior, Feres VC, Rocha B, Levi JE, et al. Dengue and dengue hemorrhagic fever among adults: clinical outcomes related to viremia, serotypes, and antibody response. J Infect Dis. 2008;197:817–824. doi: 10.1086/528805. [DOI] [PubMed] [Google Scholar]
- 16.Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res. 2003;60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- 17.Gallichotte EN, Baric TJ, Nivarthi U, Delacruz MJ, Graham R, Widman DG, et al. Genetic variation between dengue virus type 4 strains impacts human antibody binding and neutralization. Cell Rep. 2018;25:1214–1224. doi: 10.1016/j.celrep.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fontaine A, Lequime S, Moltini-Conclois I, Jiolle D, Leparc-Goffart I, Reiner RC, et al. Epidemiological significance of dengue virus genetic variation in mosquito infection dynamics. PLoS Pathog. 2018;14:e1007187. doi: 10.1371/journal.ppat.1007187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castro MG, Nogueira FB, Nogueira RM, Lourenço-de-Oliveira R, Santos FB. Genetic variation in the 3’ untranslated region of dengue virus serotype 3 strains isolated from mosquitoes and humans in Brazil. Virol J. 2013;10:3. doi: 10.1186/1743-422X-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore PR, van den Hurk AF, Mackenzie JS, Pyke AT. Dengue viruses in Papua New Guinea: evidence of endemicity and phylogenetic variation, including the evolution of new genetic lineages. Emerg Microbes Infect. 2017;6:e114. doi: 10.1038/emi.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bona AC, Twerdochlib AL, Navarro-Silva MA. Genetic diversity of dengue virus serotypes 1 and 2 in the State of Paraná, Brazil, based on a fragment of the capsid/premembrane junction region. Rev Soc Bras Med Trop. 2012;45:297–300. doi: 10.1590/s0037-86822012000300003. [DOI] [PubMed] [Google Scholar]
- 22.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowd SE, Zaragoza J, Rodriguez JR, Oliver MJ, Payton PR. Windows.NET Network Distributed Basic Local Alignment Search Toolkit (W.ND-BLAST) 93BMC Bioinformatics. 2005;6 doi: 10.1186/1471-2105-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 25.Tauil PL. Urbanização e ecologia do dengue. Cad Saude Publica. 2001;17(Suppl):99–102. [PubMed] [Google Scholar]
- 26.Coffey LL, Mertens E, Brehin AC, Fernandez-Garcia MD, Amara A, Despres P, et al. Human genetic determinants of dengue virus susceptibility. Microbes Infect. 2009;11:143–156. doi: 10.1016/j.micinf.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Sessions OM, Wilm A, Kamaraj US, Choy MM, Chow A, Chong Y, et al. Analysis of dengue virus genetic diversity during human and mosquito infection reveals genetic constraints. PLoS Negl Trop Dis. 2015;(9):e0004044. doi: 10.1371/journal.pntd.0004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanley KA, Guerbois M, Kautz TF, Brown M, Whitehead SS, Weaver SC, et al. Infection dynamics of sylvatic dengue virus in a natural primate host, the African Green Monkey. Am J Trop Med Hyg. 2014;91:672–676. doi: 10.4269/ajtmh.13-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nunes PC, Sampaio SA, Costa NR, Mendonça MC, Lima MR, Araujo SE, et al. Dengue severity associated with age and a new lineage of dengue virus-type 2 during an outbreak in Rio de Janeiro, Brazil. J Med Virol. 2016;88:1130–1136. doi: 10.1002/jmv.24464. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Barraquer I, Cordeiro MT, Braga C, Souza WV, Marques ET, Cummings DA. From re-emergence to hyperendemicity: the natural history of the dengue epidemic in Brazil. PLoS Negl Trop Dis. 2011;5:e935. doi: 10.1371/journal.pntd.0000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Figueiredo LT. Saint Louis encephalitis virus and other arboviruses in the differential diagnosis for dengue. Rev Soc Bras Med Trop. 2014;47:541–542. doi: 10.1590/0037-8682-0197-2014. [DOI] [PubMed] [Google Scholar]
- 32.Chaurasia R, Thresiamma KC, Eapen CK, Zachariah BJ, Paul R, Sritharan M. Pathogen-specific leptospiral proteins in urine of patients with febrile illness aids in differential diagnosis of leptospirosis from dengue. Eur J Clin Microbiol Infect Dis. 2018;37:423–433. doi: 10.1007/s10096-018-3187-9. [DOI] [PubMed] [Google Scholar]
- 33.Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- 34.Nascimento EJ, Hottz ED, Garcia-Bates TM, Bozza F, Marques ET, Barratt-Boyes SM. Emerging concepts in dengue pathogenesis: interplay between plasmablasts, platelets, and complement in triggering vasculopathy. Crit Rev Immunol. 2014;34:227–240. doi: 10.1615/critrevimmunol.2014010212. [DOI] [PubMed] [Google Scholar]
- 35.Cavalcanti LP, Vilar D, Souza-Santos R, Teixeira MG. Change in age pattern of persons with dengue, northeastern Brazil. Emerg Infect Dis. 2011;17:132–134. doi: 10.3201/eid1701.100321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clyde K, Kyle JL, Harris E. Recent advances in deciphering viral and host determinants of dengue virus replication and pathogenesis. J Virol. 2006;80:11418–11431. doi: 10.1128/JVI.01257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carneiro AR, Cruz AC, Vallinoto M, Melo DV, Ramos RT, Medeiros DB, et al. Molecular characterisation of dengue virus type 1 reveals lineage replacement during circulation in Brazilian territory. Mem Inst Oswaldo Cruz. 2012;107:805–812. doi: 10.1590/s0074-02762012000600016. [DOI] [PubMed] [Google Scholar]
- 38.Wirawan M, Fibriansah G, Marzinek JK, Lim XX, Ng TS, Sim AY, et al. Mechanism of enhanced immature dengue virus attachment to endosomal membrane induced by prM antibody. Structure. 2019;27:253–258. doi: 10.1016/j.str.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith SA, Nivarthi UK, Alwis R, Kose N, Sapparapu G, Bombardi R, et al. Dengue virus prM-specific human monoclonal antibodies with virus replication-enhancing properties recognize a single immunodominant antigenic site. J Virol. 2015;90:780–789. doi: 10.1128/JVI.01805-15. [DOI] [PMC free article] [PubMed] [Google Scholar]