Abstract
Objective
This study evaluated the differences in the appearance of COVID-19 pneumonia on chest computed tomography (CT) images of outpatient and cases that developed during hospitalisation.
Method
Chest CT images of 66 patients (median age, 76 years; range, 29–94 years) who underwent the severe acute respiratory syndrome coronavirus-2 reverse-transcription polymerase chain reaction (RT-PCR) test were included in this retrospective study. The chest CT appearance was categorised as “typical,” “indeterminate,” “atypical,” or “negative” in accordance with the recommendations of the Radiological Society of North America for COVID-19 pneumonia and compared among the following four subgroups: PCR-positive outpatient (n = 14); PCR-positive hospitalised (n = 7); PCR-negative outpatient (n = 9); and PCR-negative hospitalised (n = 36).
Findings
The frequency of “typical” findings in the PCR-positive outpatient cases (13/14, 92.9%) was significantly higher than that of those in the PCR-positive hospitalised cases (2/7, 28.6%, P = 0.022). There was no significant difference between the frequency of the “typical” appearance in PCR-positive hospitalised cases and that of those in the PCR-negative hospitalised cases (1/36, 2.8%, P = 0.192).
Conclusions
When COVID-19 patients acquire infections while hospitalised, their chest CT images are less likely to show typical findings than those of outpatient cases. Comprehensive and careful assessments of CT findings and consideration of the possibility of concomitant infections with other pathogens and clinical information, such as underlying diseases, background lung structure, and time course of the infection, are required for the management of such cases.
Keywords: COVID-19 pneumonia, SARS-CoV-2, Computed tomography, Nosocomial infection, Hospital-acquired infection, Concomitant infection
1. Introduction
Numerous healthcare institutions have been impacted by community-acquired infections and clusters of nosocomial infections as a result of the coronavirus disease (COVID-19) pandemic. During the first wave of the pandemic in Japan, 16,285 infectious cases had been confirmed by 17 May 2020, and 1570 of those cases involved nosocomial infections, which accounted for 9.6% of all cases of infection in Japan. The percentage of polymerase chain reaction (PCR)-positive healthcare workers (1330) was 8.2% [1]. Therefore, a total of 17.8% of all COVID-19 infections in Japan occurred in the healthcare sector. COVID-19 is also associated with a poorer prognosis, especially for inpatients [2].
Chest computed tomography (CT) has a high sensitivity for diagnosing COVID-19 [[3], [4], [5]]. Furthermore, it can be used as a diagnostic tool for patients presenting with acute symptoms or those in close contact with infected patients [[6], [7], [8], [9], [10]]. Typically, chest CT images of COVID-19 patients show areas of ground-glass opacity (GGO) with bilateral peripheral involvement in multiple lobes progressing to “crazy-paving” patterns and consolidation; therefore, several criteria for diagnosing COVID-19 by CT have already been proposed and have contributed to early diagnosis [11,12]. However, in hospitalised cases, CT diagnosis can often be difficult due to factors such as background lung modification due to the underlying disease or concomitant hospital-acquired pneumonia.
In this study, we evaluated the differences in the appearance of COVID-19 pneumonia on chest CT images of outpatient cases and cases that developed during hospitalisation because of other reasons.
2. Materials and methods
2.1. Patients
This multi-centric retrospective cohort study included patients who had undergone chest CT between 1 February and 15 May 2020 at two secondary care centres under the same governing body. In one of these hospitals, a cluster of nosocomial COVID-19 infections occurred in April 2020. Four-hundred and thirty-eight patients (325 outpatients, 113 hospitalised patients) who underwent severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) reverse-transcription polymerase chain reaction (RT-PCR) tests for symptoms suspicious for COVID-19 or screening because of close contact with someone with COVID-19 infection were included. Of these, 79 patients underwent chest CT. Sixty had symptoms suspicious for COVID-19 and 19 had no symptoms but underwent chest CT for screening because of close contact with PCR-positive patients. Thirteen patients with an extended interval between the PCR test and CT (more than 2 weeks) were excluded based on previous studies of the time course of CT findings of COVID-19 pneumonia [13,14]. Finally, CT images of 66 patients were included in this study. The patient selection chart is shown in Fig. 1 .
Fig. 1.
Flow diagram of the study sample.
SARS-CoV-2 RT-PCR, severe acute respiratory syndrome coronavirus-2 reverse transcription-polymerase chain reaction.
Patients were separated into PCR-positive and PCR-negative groups based on their PCR test results. For patients who underwent repeated PCR tests, those who showed positive results at least once were classified as part of the PCR-positive group. In the PCR-negative group, 29 patients had undergone PCR testing only once, 12 had undergone testing twice, and 4 had undergone testing three times. For the PCR-positive and PCR-negative groups, two subgroups, the outpatient group and the hospitalised group, were created based on the setting of the patient at the time of onset. Because the incubation period of SARS-CoV-2 is long and varied, in some cases, it is unclear whether the virus was contracted in the hospital or the community. Patients who were hospitalised at the time of onset were classified as part of the hospitalised group, and those who were in the community at the time of onset were classified as part of the outpatient group. The outpatient group also included patients associated with a cluster infection acquired on the cruise ship “Diamond Princess”. The demographic and clinical information, including medical history, symptoms, underlying diseases, and mortality, for both groups were recorded.
2.2. CT data acquisition and image assessment
All patients underwent a chest CT examination for evaluation of pulmonary lesions due to COVID-19. We used a 64-row or 32-row multidetector CT scanner (Canon Medical Systems Corporation, Otawara, Japan) and an 80-row multidetector CT scanner (GE Healthcare, Buckinghamshire, UK). The following scan parameters were used: tube potential, 120 kVp; gantry rotation time, 500 ms; and tube current, 50–370 mAs determined by auto exposure control (a predetermined level of image noise was set at a standard deviation [SD] of 8); and section thickness, 5 mm. The reconstruction algorithm of the lung window was used to reconstruct a thin layer with a thickness of 1.3 mm. The protocol is the routine institutional protocol for chest CT. It does not deviate from a previous study on the radiation dose of chest CT based on a nationwide questionnaire-based survey [15] and from a review article on chest CT parameter for COVID-19 pneumonia [16].
The CT findings were assessed by two board-certified diagnostic radiologists (Y.S. and G.S., with 6 and 11 years of experience with chest CT, respectively) using a Picture Archiving and Communication System. The lung lesion findings were classified as follows: “typical” for viral pneumonia; “indeterminate” for possible viral pneumonia; “atypical” for viral pneumonia (preferred alternate diagnosis); and “negative” for pneumonia; these classifications are in accordance with the recommendations of the Radiological Society of North America (RSNA) [11]. A third board-certified diagnostic radiologist (D.I., with 20 years of experience with chest CT) made the final decision if there were any disagreements between the two observers. All three observers were blinded to the clinical information.
Other findings, including emphysema, pulmonary fibrosis, opacities with peribronchovascular distribution, atelectasis, insufficient inspiration, and pleural effusion, were also recorded. Insufficient inspiration was defined as bilateral dorsal non-segmental GGO accompanied by volume loss.
2.3. Evaluation of concomitant infections
To compare the frequency of concomitant infections other than COVID-19 in each subgroup, the results of sputum culture tests, sputum staining for Mycobacteria, sputum PCR test for M. tuberculosis, urinary Pneumococcal antigen tests, urinary Legionella antigen tests, rapid influenza antigen tests (nasal swab), and serum β-d-Glucan levels (cut-off value, 20 pg/mL) for all patients who underwent any of these tests were retrospectively reviewed.
2.4. Statistical analysis
Weighted Cohen's kappa coefficients were calculated for CT appearance to examine the interobserver agreement for the initial assessment. A weighted Cohen's kappa coefficient of <0.4 was considered to indicate poor agreement, values ≥0.4 and < 0.6 indicated moderate agreement, values ≥0.6 and < 0.8 indicated good agreement and values ≥0.8 indicated excellent agreement. Continuous variables are presented as mean ± SD; they were compared using the Wilcoxon signed-rank test with Bonferroni multiple-testing correction. Qualitative variables are presented as percentages; they were compared using the Fisher exact test with Hochberg multiple-testing correction. P < 0.05 was considered statistically significant. R version 3.3.1 (The R Foundation for Statistical Computing, Vienna, Austria, http://www.rproject.org/) was used for all analyses.
2.5. Ethical considerations
This study received Institutional Review Board approval (approval number 20061103), and the need for written informed consent was waived because of the retrospective design of the study. We published a research plan and guaranteed an opt-out opportunity on the website of the institution. All procedures were performed in accordance with the Declaration of Helsinki.
3. Results
3.1. Demographics and clinical data
The CT images of 66 patients (age, 29–94 years; median age, 76 years; 43 males and 23 females) were included in this study. The average interval between CT and the initial PCR test was 4.6 days. Of the 66 cases, 21 cases were confirmed positive for SARS-CoV-2 by using RT-PCR and 45 cases were confirmed negative. In the PCR-positive group, 14 cases were classified as part of the outpatient subgroup and 7 cases were classified as part of the hospitalised subgroup. In the PCR-negative group, 9 cases were classified as part of the outpatient subgroup, while 36 cases were classified as part of the hospitalised subgroup. The demographic data of each group are shown in Table 1 . Ten out of 36 cases in the PCR-negative hospitalised subgroup were asymptomatic; however, in the other subgroups, all cases were symptomatic. Demographic and clinical data of each case in the PCR-positive subgroups are shown in Table 2, Table 3 .
Table 1.
Demographic and clinical data.
| PCR-positive (n = 21) |
PCR-negative (n = 45) |
|||
|---|---|---|---|---|
| Outpatient (subgroup a) | Hospitalised (subgroup b) |
Outpatient (subgroup c) | Hospitalised (subgroup d) |
|
| N | 14 | 7 | 9 | 36 |
| Agea range (mean ± S·D) |
29–92 (58.6 ± 14.5) |
50–87 (73.4 ± 12.7) |
35–88 (66.4 ± 22.1) |
44–94 (77.9 ± 11.6) |
| Sex (M:F)b | 10:4 | 2:5 | 6:3 | 24:12 |
| PCR-CT interval (days)c range (mean ± S·D) |
0–11 (3.4 ± 3.4) |
0–12 (3.0 ± 4.2) |
0–14 (2.2 ± 4.5) |
0–13 (6.1 ± 3.6) |
| Onset-CT interval (days)d range (mean ± S·D) |
1–14 (4.6 ± 4.0) |
0–12 (4.3 ± 4.5) |
1–8
|
0–15 (7.2 ± 4.6) |
| Symptoms (if any) | 14/14 | 7/7 | 9/9 | 26/36 |
| Hyperthermia | 14 | 7 | 7 | 25 |
| Cough | 5 | 1 | 6 | 1 |
| Dyspnoea | 0 | 0 | 4 | 6 |
| Diarrhoea | 3 | 0 | 0 | 0 |
| Dysosmia | 1 | 0 | 0 | 0 |
| Arthralgia | 1 | 0 | 0 | 0 |
CT, computed tomography; PCR, polymerase chain reaction; SD, standard deviation.
P = 0.182 (subgroup a vs. subgroup b), 1.000 (a vs. c), <0.001⁎ (a vs. d), 1.000 (b vs. c), 1.000 (b vs. d), 1.000 (c vs. d), Wilcoxon signed-rank test with Bonferroni correction.
P = 0.794 (subgroup a vs. subgroups b), 1.000 (a vs. c), 1.000 (a vs. d), 1.000 (b vs. c), 0.561 (b vs. d), 1.000 (c vs. d), Fisher exact test with Hochberg multiple-testing correction.
P = 1.000 (subgroup a vs. subgroup b), 0.688 (a vs. c), 0.075 (a vs. d), 1.000 (b vs. c), 0.190 (b vs. d), 0.013⁎ (c vs. d), Wilcoxon signed-rank test with Bonferroni correction.
P = 1.00 (subgroup a vs. subgroup b), 1.00 (a vs. c), 0.69 (a vs. d), 1.00 (b vs. c), 1.00 (b vs. d), 1.00 (c vs. d), Wilcoxon signed-rank test with Bonferroni correction.
Significant difference.
Table 2.
Demographics and CT findings of the PCR-positive outpatient group.
| Case | Age (years) |
Sex | PCR-CT interval (days) |
Onset-CT interval (days) |
Symptom | Comorbidity | Concomitant infection | CT findings (RSNA category) |
CT findings (additional findings |
Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 29 | F | 0 | 6 | Hyperthermia, cough | – | Typical | – | Discharged on day 25 | |
| 2 | 43 | M | 1 | 1 | Hyperthermia, diarrhoea | – | Typical | – | Discharged on day 16 | |
| 3 | 47 | M | 3 | 9 | Hyperthermia | – |
Haemophilus parainfluenzae (sputum) |
Typical | Atelectasis | Discharged on day 9 |
| 4 | 53 | M | 7 | 7 | Hyperthermia | DM | Typical | Peribronchovascular distribution | Discharged on day 27 | |
| 5 | 54 | M | 11 | 14 | Hyperthermia | DM, chronic pyodermas | Typical | Atelectasis | Discharged on day 26 | |
| 6 | 58 | M | 1 | 1 | Hyperthermia, dysosmia | Allergic rhinitis | Typical | – | Discharged on day 30 | |
| 7 | 59 | F | 1 | 1 | Hyperthermia, arthralgia | DM | Typical | – | Discharged on day 45 | |
| 8 | 59 | M | 1 | 1 | Hyperthermia, diarrhoea | DM | Typical | – | Discharged on day 20 | |
| 9 | 62 | M | 2 | 2 | Hyperthermia, diarrhoea | Gout | Typical | – | Discharged on day 32 | |
| 10 | 62 | F | 9 | 9 | Hyperthermia, cough | Hypertension | Typical | – | Discharged on day 30 | |
| 11 | 63 | F | 3 | 3 | Hyperthermia, cough | Hypertension | Typical | – | Discharged on day 16 | |
| 12 | 71 | M | 5 | 5 | Hyperthermia | Hypertension | Typical | – | Discharged on day 19 | |
| 13 | 69 | M | 3 | 3 | Hyperthermia, cough | Oesophageal cancer after surgery | Indeterminate | Peribronchovascular distribution, atelectasis | Discharged on day 24 | |
| 14 | 92 | M | 0 | 2 | Hyperthermia, cough | CKD | Typical | – | Death on day 16 |
CT, computed tomography; PCR, polymerase chain reaction; RSNA, Radiological Society of North America; DM, diabetes mellitus; CKD, chronic kidney disease.
Table 3.
Demographics and CT findings of the PCR-positive hospitalised group.
| Case | Age (years) |
Sex | Admission- onset interval (days) |
PCR-CT interval (days) |
Onset-CT interval (days) |
Symptom | Comorbidity | Concomitant infection | CT findings (RSNA category) |
CT findings (additional findings |
Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 50 | F | 44 | 0 | 0 | Hyperthermia | Thyrotoxic crisis, heart failure | Negative | – | Discharged on day 23 | |
| 16 | 65 | F | 7 | 12 | 12 | Hyperthermia | Femoral neck fracture, pH: ICH, heart failure, MRSA pneumonia | MRSA (sputum) | Indeterminate | Peribronchovascular distribution, insufficient inspiration | Tracheostomy on day 21, ongoing hospitalisation on day 50 |
| 17 | 71 | M | 35 | 1 | 4 | Hyperthermia | Ileus, after rectectomy for rectal cancer | Typical | Discharged on day 45 | ||
| 18 | 77 | F | 18 | 1 | 2 | Hyperthermia | Acute pancreatitis, cirrhosis | Indeterminate | Peribronchovascular distribution, atelectasis, pleural effusion | Death on day 19 | |
| 19 | 81 | F | 66 | 3 | 1 | Hyperthermia | ASO, DM | Normal flora (sputum) | Typical | Pulmonary fibrosis | Death on day 50 |
| 20 | 83 | M | -⁎ | 0 | -⁎ | Hyperthermia | Thoracic empyema | MRCoNS (sputum) | Indeterminate | Atelectasis, pleural effusion | Discharged on day 33 |
| 21 | 87 | F | 8 | 4 | 7 | Hyperthermia, cough | Rectal cancer (before surgery), ischaemic heart disease, BA, DM |
Serratia marcescens (sputum) |
Indeterminate | Peribronchovascular distribution, atelectasis, insufficient inspiration, pleural effusion | Discharged on day 40 |
CT, computed tomography; PCR, polymerase chain reaction; RSNA, Radiological Society of North America; ICH, intracranial haemorrhage; MRSA, methicillin-resistant Staphylococcus aureus; ASO, arteriosclerosis obliterans; BA, bronchial asthma; DM, diabetes mellitus; MRCoNS, methicillin-Resistant Coagulase-Negative Staphylococci.
In case 20, because the hyperthermia caused by a pustule had continued from the time of admission, the hyperthermia caused by COVID-19 may have been masked, thus making it difficult to determine the date of onset.
3.2. Chest CT images
Interobserver agreement for the assessment of CT appearance was good (weighted Cohen's kappa, 0.788). Chest CT findings, according to the RSNA category, are shown in Table 4 . During the subgroup analysis of PCR-positive cases, the frequency of “typical” findings in outpatient cases (92.9%) was significantly higher than that of those in hospitalised cases (28.6%, P = 0.022). Four PCR-positive hospitalised cases showed an “indeterminate” appearance. There was no significant difference between the frequency of the “typical” appearances found for PCR-positive and PCR-negative hospitalised groups (P = 0.192).
Table 4.
Chest CT findings (RSNA category)
| PCR-positive (n = 21) |
PCR-negative (n = 45) |
|||
|---|---|---|---|---|
| Outpatient | Hospitalised | Outpatient | Hospitalised | |
| N (%) | 14 (100) | 7 (100) | 9 (100) | 36 (100) |
| CT findings (RSNA category [11]) |
||||
| Typical * | 13 (93) | 2 (29) | 0 (0) | 1 (3) |
| Indeterminate | 1 (7) | 4 (57) | 2 (22) | 4 (11) |
| Atypical | 0 (0) | 0 (0) | 3 (33) | 23 (64) |
| Negative | 0 (0) | 1 (14) | 4 (44) | 8 (22) |
CT, computed tomography; PCR, polymerase chain reaction; RSNA, Radiological Society of North America.
*P values of the Fisher exact test with Hochberg multiple-testing correction among subgroups are as follows:
PCR-positive/outpatient vs. PCR-positive/hospitalised 0.022 ⁎
PCR-positive/outpatient vs. PCR-negative/outpatient < 0.001⁎
PCR-positive/outpatient vs. PCR-negative/hospitalised < 0.001⁎
PCR-positive/hospitalised vs. PCR-negative/outpatient 0.350
PCR-positive/hospitalised vs. PCR-negative/hospitalised 0.192
PCR-negative/outpatient vs. PCR-negative/hospitalised 1.000
⁎ Significant difference.
The frequency of additional CT findings was lower in PCR-positive outpatient cases than in PCR-positive hospitalised cases, although there was no significant difference between these subgroups (Table 5 ). Chest CT images of cases 16, 18, and 21 (PCR-positive hospitalised) showed peribronchovascular distribution which is considered not typical in COVID-19 [12]. In case 16, on day 12 from the onset, CT already showed acute respiratory distress syndrome (ARDS)-like extensive opacities, and it was difficult to determine whether these were due to COVID-19 or other disorders, including concomitant infection. In this case, as mentioned above, methicillin-resistant Staphylococcus aureus (MRSA) was detected from sputum collected 6 days prior to chest CT, and the patient was suspected of having concomitant pneumonia caused by MRSA.
Table 5.
Chest CT findings (additional findings).
| PCR-positive (n = 21) |
PCR-negative (n = 45) |
|||
|---|---|---|---|---|
| Outpatient | Hospitalised | Outpatient | Hospitalised | |
| N (%) | 14 (100) | 7 (100) | 9 (100) | 36 (100) |
| CT findings (additional findings) |
||||
| Emphysema | 0 (0) | 0 (0) | 1 (11) | 15 (42) |
| Pulmonary fibrosis | 0 (0) | 1 (14) | 0 (0) | 10 (28) |
| Peribronchovascular distribution | 2 (14) | 3 (43) | 4 (44) | 17 (47) |
| Atelectasis | 3 (21) | 3 (43) | 1 (11) | 17 (47) |
| Insufficient inspiration | 0 (0) | 2 (29) | 2 (22) | 20 (56) |
| Pleural effusion | 0 (0) | 3 (43) | 3 (33) | 22 (61) |
| Additional findings * (if any) |
4 (29) | 5 (71) | 6 (67) | 34 (94) |
CT, computed tomography; PCR, polymerase chain reaction.
⁎Significant differences in the frequency of additional findings of the following combination of subgroups (Fisher exact test with Hochberg multiple-testing correction):
PCR-positive/outpatient vs PCR-negative/hospitalised P < 0.001.
No significant differences between other combinations of subgroups.
In case 15 (hospitalised PCR-positive), chest CT showed negative findings for pneumonia. No CT was performed during the course of treatment in this case.
Fig. 2, Fig. 3 show examples of chest CT images of PCR-positive outpatient cases, and Fig. 4, Fig. 5 show the corresponding images of PCR-positive hospitalised cases.
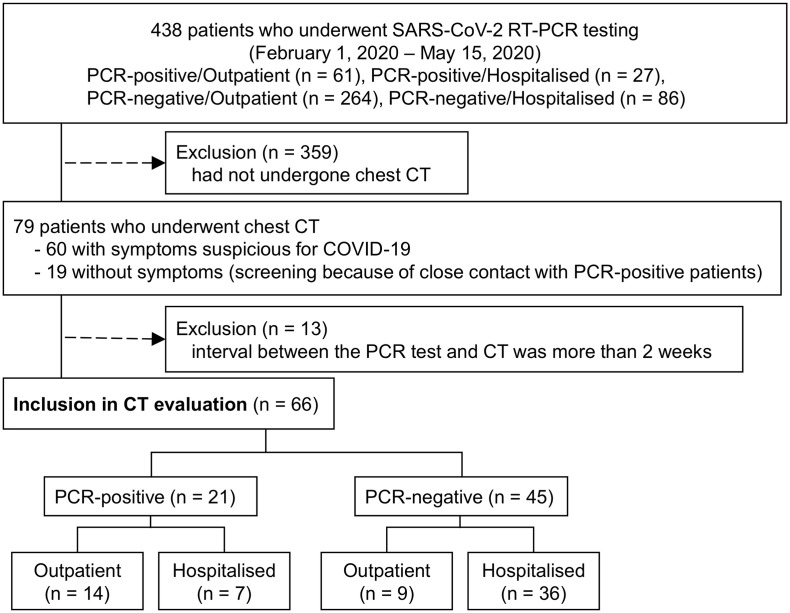
Fig. 2.
Chest CT images of a 43-year-old male patient (case 2, COVID-19 PCR-positive, outpatient, 1 day after onset). CT shows peripheral ground-glass opacities in the lower lobes of both lungs (RSNA category: typical appearance).
PCR, polymerase chain reaction; RSNA, Radiological Society of North America.
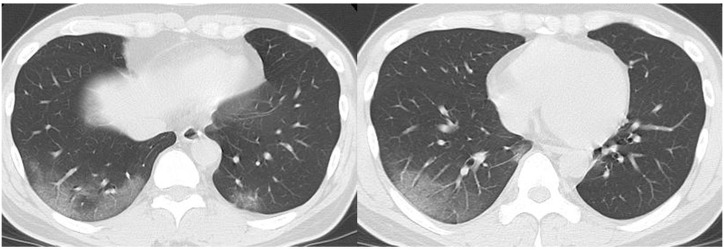
Fig. 3.
Chest CT images of a 62-year-old male patient (case 9, COVID-19 PCR-positive, outpatient, 2 days after onset). CT shows multi-focal peripheral dominant ground-glass opacities (some are round-shaped) in both lungs (RSNA category: typical appearance).
PCR, polymerase chain reaction; RSNA, Radiological Society of North America.
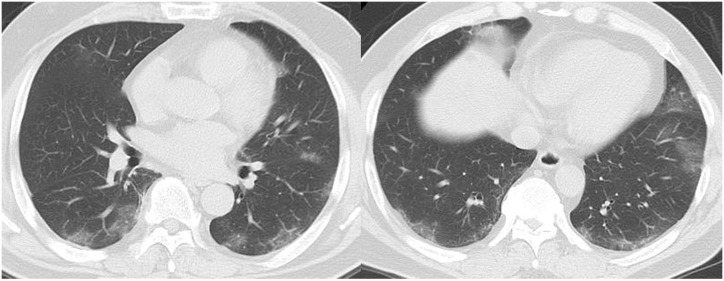
Fig. 4.
Chest CT images of a 65-year-old female patient (case 16, COVID-19 PCR-positive, hospitalised, 12 days after onset) SARS-CoV-2 that developed during hospitalisation for a femoral neck fracture with comorbidity of intracranial haemorrhage (ICH) and heart failure as comorbidities. CT shows extensive consolidation and ground-glass opacities along with the bronchovascular bundles in both lungs, which is difficult to distinguish from acute respiratory distress syndrome (ARDS) (RSNA category: indeterminate appearance). In this case, methicillin-resistant Staphylococcus aureus (MRSA) was detected in the sputum collected 6 days before chest CT. Concomitant pneumonia caused by MRSA was suspected.
PCR, polymerase chain reaction; RSNA, Radiological Society of North America.
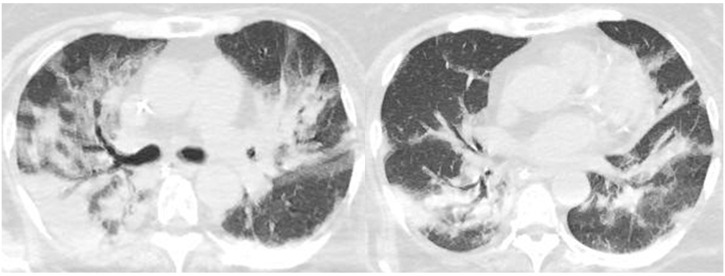
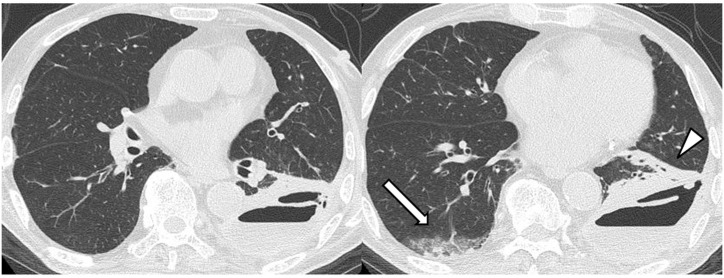
Fig. 5.
Chest CT images of an 83-year-old male patient (case 20, COVID-19 PCR-positive, hospitalised) with SARS-CoV-2 that developed during hospitalisation for thoracic empyema. CT shows non-segmental sub-pleural ground-glass opacity with consolidation (white arrow), which is compatible with COVID-19 pneumonia in the right lower lobe. Left thoracic empyema accompanied by atelectasis in the left lower lobe is evident (white arrowhead). Note that the left lower lobe lacks findings compatible with COVID-19 pneumonia (RSNA category: indeterminate appearance).
PCR, polymerase chain reaction; RSNA, Radiological Society of North America.
3.3. Concomitant infections
Results of sputum culture tests, sputum staining for Mycobacteria, sputum PCR tests for M. tuberculosis, urinary Pneumococcal antigen tests, urinary Legionella antigen tests, rapid influenza antigen tests (nasal swab), and serum β-d-Glucan levels (cut-off value, 20 pg/mL) were available for 31, 6, 6, 9, 7, 13, and 16 patients, respectively. No patient had positive results for the sputum PCR tests for M. tuberculosis, urinary Pneumococcal antigen tests, urinary Legionella antigen tests, and rapid influenza antigen tests (nasal swab). In the PCR-positive hospitalised subgroup, sputum culture tests of four patients revealed three positive results for causative pathogens of pneumonia (methicillin-resistant coagulase-negative Staphylococci [MRCoNS] for 1 patient, MRSA for 1 patient, and Serratia marcescens for 1 patient). Chest CT images of these three patients showed an indeterminate appearance. In the PCR-positive outpatient subgroup, sputum culture testing results of a patient revealed Haemophilus parainfluenzae. Chest CT images of this patient showed a typical appearance. Results of concomitant infections in each subgroup are summarised in Table 6 .
Table 6.
Concomitant infections of each subgroups.
| PCR-positive (n = 21) |
PCR-negative (n = 45) |
|||
|---|---|---|---|---|
| Outpatient | Hospitalised | Outpatient | Hospitalised | |
| N (%) | 14 | 7 | 9 | 36 |
| Sputum culture test | 1/1a | 3/4b | 4/5c | 18/21d |
| Sputum staining for Mycobacteria | NA | NA | 0/1 | 0/5 |
| Sputum PCR test for M. tuberculosis | NA | 0/1 | 0/1 | 0/4 |
| Urinary Pneumococcal antigen test | 0/2 | 0/1 | 0/4 | 0/2 |
| Urinary Legionella antigen test |
NA | 0/1 | 0/4 | 0/2 |
| Rapid influenza antigen test (nasal swab) | NA | NA | 0/6 | 0/7 |
| Serum β-d-Glucane | NA | 0/3 | 1/2 | 1/11 |
The numerator represents the number of patients with positive results for each test and the denominator represents the number of patients tested (numerator/denominator). PCR, polymerase chain reaction; NA, no data applicable.
Sputum culture testing of a patient in the PCR (+)/outpatient subgroup yielded positive results for Haemophilus parainfluenzae.
Sputum culture testing of 4 patients in the PCR (+)/hospitalised subgroup yielded 3 positive results for causative pathogens of pneumonia (methicillin-resistant coagulase-negative Staphylococci [MRCoNS] in 1 patient, methicillin-resistant Staphylococcus aureus [MRSA] in 1 patient, and Serratia marcescens in 1 patient).
Sputum culture testing of 5 patients in the PCR(−)/outpatient subgroup yielded four positive results for causative pathogens of pneumonia (methicillin-susceptible Staphylococcus aureus [MSSA] in 1 patient, Haemophilus parainfluenzae in 3 patients, and Citrobacter koseri in 1 patient).
Sputum culture testing of 21 patients in the PCR-negative/hospitalised subgroup yielded 18 positive results for causative pathogens of pneumonia (Pseudomonas aeruginosa in 5 patients, Haemophilus parainfluenzae in 3 patients, methicillin-susceptible Staphylococcus aureus [MSSA] in 3 patients, Klebsiella pneumoniae in 2 patients, Streptococcus agalactiae in 1 patient, methicillin-resistant Staphylococcus aureus [MRSA] in 1 patient, Aeromonas caviae in 1 patient, Burkholderia cepacia in 1 patient, and yeast-like fungi in 1 patient).
Cut-off value for the serum β-d-Glucan test was set at 20 pg/mL.
4. Discussion
During nosocomial outbreaks of COVID-19, chest CT scans of suspected patients at an appropriate timing complement other investigations such as PCR and antibody tests. Since PCR tests can yield false-negative findings [[17], [18], [19], [20]], some previous studies have reported that CT is more sensitive than PCR [3,4,21,22]. However, the present study revealed that the chest CT images of COVID-19 patients differs between hospitalised and outpatient cases. PCR-positive hospitalised cases were significantly less likely to present with “typical” findings compared to those of PCR-negative outpatient cases. In addition, there was no significant difference between the frequency of “typical” findings in the PCR-positive and PCR-negative hospitalised cases. Therefore, when hospitalised patients exhibit symptoms of infection, it is difficult to distinguish COVID-19 pneumonia from other disorders. Several factors can explain this discrepancy. One of the greatest contributors to the discrepancies in the CT image appearance of COVID-19 patients in the hospitalised group and those in the outpatient group is the underlying risk of concomitant infections due to host factors (such as immunological deterioration to underlying diseases) and environmental factors (hospital-acquired infections). In particular, geriatric patients may experience concomitant aspiration pneumonia because of their low performance status and respiratory function disabilities [23,24]. In our cohort, sputum culture tests of four PCR-positive hospitalised patients revealed three positive results for causative pathogens of pneumonia (MRCoNS, MRSA, and Serratia marcescens). Chest CT images of these three patients showed an indeterminate appearance; furthermore, the images of two of these patients showed peribronchovascular distribution, which is considered not typical in COVID-19 [12]. The chest CT findings of COVID-19 in these patients might have been masked by concomitant bacterial infections. According to a meta-analysis of studies of chest CT findings in patients with COVID-19, the pooled prevalence of normal chest CT imaging findings was 10.6% (95% confidence interval [CI], 7.6%–13.7%) [25]; however, chest CT images of patients with concomitant infections reflect only bacterial pneumonia without findings of COVID-19 pneumonia. Therefore, chest CT images with indeterminate or atypical findings cannot rule out COVID-19, especially when there is a risk of concomitant infections, and PCR or other tests confirming COVID-19 should be performed when COVID-19 is clinically suspected. Clinicians should also rule out the possibility of concomitant infections, especially in the hospital setting.
Another reason for the discrepancies in the CT appearance of COVID-19 pneumonia in hospitalised and outpatient cases could be the differences in the background lung structure. In the present study, all PCR-positive outpatient cases with typical appearance showed no other additional CT findings. Case 17 (PCR-positive hospitalised) that showed had no other additional findings due to underlying pulmonary diseases also showed a typical appearance for COVID-19 pneumonia. In contrast, in the hospitalised group, most cases had underlying diseases that affect lung structure, including chronic heart failure, bronchial asthma, or thoracic empyema as comorbidities [26]. For example, in case 20, CT showed sub-pleural GGO with consolidation compatible with COVID-19 pneumonia in the right lower lobe, while the left lung showed atelectasis due to thoracic empyema without any findings compatible with COVID-19 pneumonia. This case suggests that the typical pulmonary lesion due to COVID-19 pneumonia is unlikely to occur in a lobe in which the existing structure has been destroyed. Differences in local inspiratory volume or blood flow may also result in local differences in CT findings of the same individual. Studies on the pathological mechanism underlying viral pneumonia have revealed that it first affects the terminal bronchioles and their surrounding pulmonary parenchyma and then develops into the infiltration of pulmonary lobules [27,28]. More investigations on the radiological-pathological correlation for COVID-19 pneumonia are necessary. Clinically, if available, CT images scanned before the onset of infection should always be referred to, and any new findings compatible with COVID-19 pneumonia should be carefully considered.
Insufficient inspiration due to low performance status and prolonged bed rest, especially in geriatric patients, often causes GGOs on the dorsal side of the lung. This makes the diagnosis of COVID-19 pneumonia more difficult. In our cohort, two patients in the PCR-positive hospitalised group (28.6%) and 20 patients in the PCR-negative hospitalised group (55.6%) showed dorsal GGOs attributable to insufficient inspiration. In such patients, chest CT images may yield false-positive or false-negative results when screening of COVID-19 pneumonia, which shows dorsal dominant non-segmental GGOs on CT.
Another factor influencing the discrepancies in the CT appearance of COVID-19 pneumonia between hospitalised cases and outpatient cases is the interval between onset and CT scans. When an outbreak of nosocomial infection is discovered in a facility, symptomatic patients are more likely to undergo CT scans earlier than in cases of community-acquired infection. In some cases, CT scans may have been performed before the typical chest CT findings of COVID-19 pneumonia become apparent. Case 15 in our cohort involved a patient who underwent CT on the day of onset and showed negative findings; however, the PCR test already showed positive findings on the same day, which may correspond to the aforementioned situation. According to previous studies on the time course of chest CT changes of COVID-19 pneumonia, four stages of evolution on chest CT scans were identified from symptom onset: early stage, 0–4 days; progressive stage, 5–8 days; peak stage, 9–13 days; and absorption stage, ≥14 days [13]. In case 15 of our cohort, because no CT scans were performed during the course of treatment, it is unclear whether the imaging findings of pneumonia occurred subsequently. However, in case 16, on day 12 after onset, CT already showed acute respiratory distress syndrome (ARDS)-like extensive opacities, which were difficult to attribute to COVID-19 or other disorders including concomitant infection. The appearance of different imaging manifestations at different stages may be associated with the pathological mechanism of viral pneumonia, which is initially prone to affect the terminal bronchioles and their surrounding pulmonary parenchyma, cause infiltration of pulmonary lobules and result in diffuse alveolar damage [27,28]. More investigations are necessary to reveal the appropriate timing of CT scans to screen and diagnose COVID-19 pneumonia in a hospital environment.
This study had several limitations. First, the sample size of the PCR-positive group was small. Although 66 cases were analysed, the majority were COVID-19-negative and the numbers were too small for statistical analyses. Further investigations of larger numbers of cases are necessary to confirm the findings of the present study. Second, CT scans were not performed for all cases with proven or suspected COVID-19 pneumonia. During the first wave of the pandemic, because of insufficient knowledge of and experience with infection precautions during CT scanning, we were not able to perform CT for all cases. This might have caused a selection bias due to differences in the indications for CT scans among cases. Third, although sputum culture tests and other tests should be performed for all patients to investigate the effects of concomitant infections on the chest CT image appearance, they were not performed for all patients. Additionally, in the PCR-negative group, 29 out of 45 patients had undergone PCR testing only once. Because of the relatively low sensitivity of the PCR test, there is a possibility of including pseudo-negative cases into PCR-negative groups, especially for those who underwent PCR testing only once.
5. Conclusions
Because of various factors, chest CT is less likely to show typical findings in hospitalised patients with COVID-19 than in outpatient cases, and it is difficult to distinguish COVID-19 pneumonia from other disorders. During nosocomial outbreaks of COVID-19, patients with suspicious symptoms should be screened by CT at an appropriate time, and a comprehensive and careful assessment of the CT appearance that considers the possibility of concomitant infections of other pathogens and clinical information, such as underlying disease, background lung structure, and the time course of infection, is required.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
None.
Acknowledgments
The authors wish to acknowledge the contribution of Dr. Katsunori Oikado, Diagnostic Imaging Center, Cancer Institute Hospital of JFCR, Tokyo, Japan for his advice on the chest CT findings of COVID-19 pneumonia and Dr. Yui Hanabusa, Nerima Hikarigaoka Hospital, Tokyo, Japan for her support in patient data collection.
References
- 1.Nikkei Healthcare . Nikkei Shimbun; 2020. Large clusters in healthcare institutions (article in Japanese) [Google Scholar]
- 2.Moliere S., Veillon F. COVID-19 in post-operative patients: imaging findings. Surg Infect (Larchmt) 2020 doi: 10.1089/sur.2020.169. [DOI] [PubMed] [Google Scholar]
- 3.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020:200432. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbasi-Oshaghi E., Mirzaei F., Farahani F., Khodadadi I., Tayebinia H. Diagnosis and treatment of coronavirus disease 2019 (COVID-19): laboratory, PCR, and chest CT imaging findings. Int J Surg. 2020 doi: 10.1016/j.ijsu.2020.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Tawfiq J.A., Memish Z.A. Diagnosis of SARS-CoV-2 infection based on CT scan vs. RT-PCR: reflecting on experience from MERS-CoV. J Hosp Infect. 2020;105:154–155. doi: 10.1016/j.jhin.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inui S., Fujikawa A., Jitsu M., Kunishima N., Watanabe S., Suzuki Y., et al. Chest CT findings in cases from the cruise ship “diamond princess” with coronavirus disease 2019 (COVID-19) Radiol Cardiothorac Imag. 2020;2 doi: 10.1148/ryct.2020200110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Y., Guo D., Li C., Chen T., Li R. High-resolution CT features of the COVID-19 infection in Nanchong City: initial and follow-up changes among different clinical types. Radiol Infect Dis (Beijing, China) 2020 doi: 10.1016/j.jrid.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai H.X., Hsieh B., Xiong Z., Halsey K., Choi J.W., Tran T.M.L., et al. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020:200823. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpson S., Kay F.U., Abbara S., Bhalla S., Chung J.H., Chung M., et al. Radiological Society of North America expert consensus statement on reporting chest CT findings related to COVID-19. Endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA. Radiol Cardiothorac Imag. 2020;2 doi: 10.1148/ryct.2020200152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19) imaging reporting and data system (COVID-RADS) and common lexicon: a proposal based on the imaging data of 37 studies. Eur Radiol. 2020:1–13. doi: 10.1007/s00330-020-06863-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan F., Ye T., Sun P., Gui S., Liang B., Li L., et al. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 2020:200370. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N., et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020 doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsunaga Y., Kawaguchi A., Kobayashi K., Kobayashi M., Asada Y., Minami K., et al. Effective radiation doses of CT examinations in Japan: a nationwide questionnaire-based study. Br J Radiol. 2016;89 doi: 10.1259/bjr.20150671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azadbakht J., Khoramian D., Lajevardi Z.S., Elikaii F., Aflatoonian A.H., Farhood B., et al. A review on chest CT scanning parameters implemented in COVID-19 patients: bringing low-dose CT protocols into play. Egypt J Radiol Nucl Med. 2021;52:1–10. doi: 10.1186/s43055-020-00400-1. [DOI] [Google Scholar]
- 17.Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020 doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao A.T., Tong Y.X., Zhang S. False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J Med Virol. 2020 doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li D., Wang D., Dong J., Wang N., Huang H., Xu H., et al. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based ct diagnosis and insights from two cases. Korean J Radiol. 2020;21:505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skriver E.B., Olsen T.S. Transient disappearance of cerebral infarcts on CT scan, the so-called fogging effect. Neuroradiology. 1981;22:61–65. doi: 10.1007/BF00344775. [DOI] [PubMed] [Google Scholar]
- 21.Long C., Xu H., Shen Q., Zhang X., Fan B., Wang C., et al. Diagnosis of the coronavirus disease (COVID-19): rRT-PCR or CT? Eur J Radiol. 2020;126 doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng H., Liu Y., Lv M., Zhong J. A case report of COVID-19 with false negative RT-PCR test: necessity of chest CT. Jpn J Radiol. 2020;38:409–410. doi: 10.1007/s11604-020-00967-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandell L.A., Niederman M.S. Aspiration pneumonia. N Engl J Med. 2019;380:651–663. doi: 10.1056/NEJMra1714562. [DOI] [PubMed] [Google Scholar]
- 24.Bowerman T.J., Zhang J., Waite L.M. Antibacterial treatment of aspiration pneumonia in older people: a systematic review. Clin Interv Aging. 2018;13:2201–2213. doi: 10.2147/CIA.S183344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams H.J.A., Kwee T.C., Yakar D., Hope M.D., Kwee R.M. Chest CT imaging signature of coronavirus disease 2019 infection: in pursuit of the scientific evidence. Chest. 2020;158:1885–1895. doi: 10.1016/j.chest.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salehi S., Abedi A., Radmard A.R., Sorouri M., Gholamrezanezhad A. Chest computed tomography manifestation of coronavirus disease 2019 (COVID-19) in patients with cardiothoracic conditions. J Thorac Imaging. 2020:90–96. doi: 10.1097/rti.0000000000000531. Publish Ah. [DOI] [PubMed] [Google Scholar]
- 27.Dai W.C., Zhang H.W., Yu J., Xu H.J., Chen H., Luo S.P., et al. CT imaging and differential diagnosis of COVID-19. Can Assoc Radiol J. 2020;71:195–200. doi: 10.1177/0846537120913033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bassetti M., Vena A., Giacobbe D.R. The novel Chinese coronavirus (2019-nCoV) infections: challenges for fighting the storm. Eur J Clin Invest. 2020;50 doi: 10.1111/eci.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]