Abstract
Background
Cancer patients are considered highly vulnerable to the COVID-19 pandemic. However, delaying cancer-specific therapies could have a deleterious effect on survival. The potential suppressive effects of chemotherapies or cancer-related microenvironment raised the question on how cancer patients’ immune system responds to SARS-CoV-2 virus.
Methods
We have started a prospective monocentric trial entitled COV-CREM (NCT04365322) in April 2020. The primary objective of the trial was to assess specific immune response's intensity and diversity to SARS-CoV-2 in infected patients.
Results
In this study, we showed that cancer patients (28 solid tumours, 11 haematological malignancies) exposed to SARS-CoV-2 produced a high rate of specific antibodies, as observed in patients without a cancer history (n = 29). However, our results highlight a lack in the generation of T-cell responses against CoV–N, M and S proteins from the SARS-CoV-2 virus, suggesting that cancer patients failed to mount a protective T-cell immunity. Nevertheless, SARS-CoV-2 infection did not impair established immune memory since specific responses against common viruses were not hampered in cancer patients.
Conclusion
Given the severity and the unknown evolution of the ongoing COVID-19 pandemic, it is of fundamental importance to integrate cancer patients in vaccination programs.
Keywords: SARS-CoV-2 infection, Cancer patients, Immune T cell response, SARS-CoV-2 antibodies
1. Introduction
Today's fight against the SARS-CoV-2 virus generates a scientific emulation that brings together researchers from all horizons. Older age and medical comorbidities are the main risk factors for severe outcomes and higher mortality from COVID-19 infection [1]. This is why cancer patients are considered highly vulnerable [2,3]. Comorbidities and active cancer are the major factors for high risk COVID-19-related severe events [[4], [5], [6]]. In a SARS-CoV context, T-cells are critical in virus clearance by eliminating virus-infected cells both in mice models and human [[7], [8], [9]]. SARS-CoV viruses are composed of structural proteins namely N (nucleocapsid), M (membrane), E (envelope) and S (spike). Recently, Grifoni et al. reported the presence of SARS-CoV-2 specific T-cells, using peptides derived from these structural proteins, in convalescent patients who presented a mild illness form [9]. The supposed vulnerability of cancer patients in the COVID-19 pandemic raised the questions of how the immune system of these patients responds against the SARS-CoV-2. However, delaying treatment is not recommended during the COVID-19 pandemic. Cancer patients must receive antitumour treatment under a vigorous SARS-CoV-2 screening. Growing evidence sustains the rationale for the development of a vaccine against SARS-CoV-2 [7,8,10]. Thus, it is critical to understand how the immune system of the supposedly vulnerable cancer patients would respond against the SARS-CoV-2 to anticipate the impact of a possible vaccine on these patients.
2. Materials and methods
2.1. Patients
We have started a prospective monocentric trial entitled COV-CREM (NCT04365322) in April 2020. All patients were enrolled after the signature of informed consent in accordance with the French regulation. Three cohorts of patients were enrolled: i) patients with mild illness COVID-19 infection without cancer, whose clinical parameters have been described elsewhere [11], ii) patients with solid tumours and iii) patients with haematological malignancies. The analysed population was diagnosed with COVID-19 between March 10 and May 20. Patients with cancers were included if they presented a COVID-19 infection diagnosed based on RT-PCR performed on a nasopharyngeal swab or positive serology. The primary trial objective was to assess specific immune response's intensity and diversity to SARS-CoV-2 in infected patients. In this study, we presented the results of 28 patients with solid tumours and 11 patients with haematological malignancies. Results are comparatively analysed against immune analyses performed in healthy donor samples and patients treated for mild illness COVID-19 infection without a cancer history [11]. A detailed description of the methods can be found in the supplementary methods.
2.2. Immune responses analysis
Peripheral blood mononuclear cells (PBMCs) from patients and healthy donors were isolated by density centrifugation on Ficoll gradient (Eurobio). We assessed spontaneous T-cell responses against SARS-CoV-2 by using IFNγ ELISpot assay. Humoral responses were assessed by using ELISA assay to detect IgG antibodies to SARS-CoV-2. A detailed description of the laboratory analysis methods can be found in the supplementary methods.
2.3. Statistical analysis
Statistical analyses were performed using GraphPad Prism 6 software (San Diego, CA). The level of significance was set at p < 0.05 for all tests (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 and ∗∗∗∗p ≤ 0.0001). Variables were expressed as a median and interquartile range (IQR) and tested with the Mann–Whitney U test. Variables were expressed as frequencies and tested with Fisher's exact test.
3. Results
Cancer patients’ characteristics are reported in Table 1 and Table 2 . All enrolled patients presented a mild illness form of COVID-19. The viral load detected in nasal swabs was available for 16 patients. For the majority of them (88%), the viral load was high (mean 19.5 ct (15.29–36.76)). We notice that four patients were hospitalised for a preventive approach. The median age was 69. When diagnosed with COVID-19, 82% had a fever. At the time of infection, 61.5% of patients had a metastatic or advanced stage of cancer, of which 79.2% had ongoing anticancer therapies (excluding hormonotherapy) and thirteen (68.4%) displayed lymphopenia (lymphocyte count under 1G/L).
Table 1.
Cancer patient's characteristics.
| With T cell responses |
Without T cell response |
|||
|---|---|---|---|---|
| Solid tumours (n = 14) | Haematological malignancies (n = 4) | Solid tumours (n = 14) | Haematological malignancies (n = 7) | |
| Patient characteristics n (%) | ||||
| Age - median (year) | 70 | 73 | 69 | 59 |
| [Range] | [51–75] | [54–80] | [48–84] | [23–73] |
| <65 | 4 (28.6) | 1 (25.0) | 6 (42.9) | 4 (57.1) |
| 65–74 | 8 (57.1) | 2 (50.0) | 5 (35.7) | 3 (42.9) |
| >75 | 2 (14.3) | 1 (25.0) | 3 (21.4) | 0 (0) |
| Male | 7 (50.0) | 4 (100.0) | 5 (35.7) | 4 (57.1) |
| History of cancer n (%) | ||||
| Metastatic or late stage | 9 (64.3) | 1 (25.0) | 12 (85.7) | 2 (28.6) |
| Active anticancer treatment during 4 weeks before COVID infection | 10 (71%) | 3 (75.0) | 12 (85.7) | 1 (50) |
| Chemotherapy | 6 (42.9) | 0 (0.0) | 6 (42.9) | 0 (0.0) |
| Chemotherapy + immunotherapy | 0 (0.0) | 1 (25.0) | 1 (7.1) | 0 (0.0) |
| Immunotherapy | 2 (14.3) | 1 (25.0) | 1 (7.1) | 0 (0.0) |
| Targeted therapy | 1 (7.1) | 1 (25.0) | 4 (28.6) | 1 (50) |
| Hormonotherapy | 1 (7.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| COVID-19 n (%) | ||||
| Symptomatic | 12 (85.7%) | 4 (100.0) | 14 (100.0) | 7 (100.0) |
| Diagnostic of COVID-19 | ||||
| PCR | 8 (57.1) | 4 (100.0) | 11 (78.6) | 7 (100.0) |
| PCR+ >1 month | 0 (0.0) | 0 (0.0) | 3 (21.4) | 1 (14.3) |
| Serology | 6 (42.9) | 0 (0.0) | 3 (21.4) | 0 (0.0) |
| Time between COVID-19 infection and blood sample - median [range] in days | 70.5 [48–109] | 76.5 [49–92] | 62.5 [41–85] | 81.0 [48–119] |
| Immunological data | ||||
| PNN median (mm3) [Range] | 2 205 [1370–5250] | 2 935 [1410–5980] | 3 075 [902–9000] | 5 550 [900–35 000] |
| Missing | 4 | 0 | 0 | 1 |
| LDH median (UI/I) [Range] | 417 [207–464] | 199 [177–235] | 239 [61–341] | 444 [206–750] |
| Missing | 8 | 1 | 3 | 3 |
| Lymphopenia at the moment of COVID-19 infection (G/L) n (%) | 3 (42.8) | 2 (50.0) | 9 (64.3) | 2 (28.6) |
| Median [range] | 1 340 [600––2060] | 1 505 [690–2320] | 840 [300–1920] | 1 300 [580–1 900] |
| Missing | 7 | 0 | 0 | 1 |
| CD3 (%) [range] | 54.1 [26.4–89.4] | 65 [14.7–94.5] | 61.1 [31.7–87.5] | 54.4 [5.2–91.1] |
| CD4 (%) [range] | 51.3 [16.2–91.5] | 31.1 [4.3–51.4] | 52.2 [20.8–83.7] | 46.7 [8.3–66.2] |
| CD8 (%) [range] | 38.5 [7.1–69] | 58.9 [39.4–92.3] | 35.9 [13.8–63.3] | 48.4 [24.7–90.3] |
| CD4/CD8 (%) [range] | 2.4 [0.3–12.8] | 0.6 [0.04–1.3] | 2.0 [0.4–5.8] | 1.3 [0.09–2.6] |
| CD19 (%) [range] | 23.9 [1.5–58.3] | 27.3 [1–87.8] | 27.6 [2.9–75.1] | 18.7 [0.1–59.1] |
| Missing | 0 | 0 | 2 | 2 |
Table 2.
Clinical characteristics of individual cancer patients.
|
Solid tumours | |||||||||||||||
|
Patient number |
Sex |
Age |
PS |
Diabetes |
BMI |
HTA |
Other comorbidities |
Concurrent ongoing therapies |
Cancer localisation |
Metastatic disease Y/N |
Active anticancer treatment during 4 weeks before COVID infection |
Time between COVID-19 infection and blood sample in days |
COVID-19 serology |
At least one specific T-cell response (S, M or N) |
CEF specific T-cell responses |
| 03–01 | M | 72 | 1 | NO | 27.7 | YES | NO | NO | Hepatocarcinoma | YES | Sorafenib | 64,0 | POS | NO | YES |
| 03–02 | F | 53 | 0 | NO | 26.5 | NO | NO | NO | Breast | YES | Trastuzumab Pertuzumab Hormonotherapy | 72,0 | POS | YES | YES |
| 03–03 | M | 55 | 3 | NO | 23.8 | NO | Arrhythmia | Amlodipine Cordarone Perindopril |
Pulmonary | NO | Durvalumab | 85,0 | POS | YES | YES |
| 03–05 | F | 75 | 1 | NO | 26.5 | YES | NO | NO | Breast | YES | Hormonotherapy | 70,0 | POS | YES | YES |
| 03–06 | M | 62 | 3 | NO | 24.3 | YES | Arrhythmia | Sotalol Apixaban |
HNC | YES | NO | 54,0 | NEG | NO | YES |
| 03–07 | M | 69 | 2 | NO | 19.8 | YES | NO | NO | HNC | YES | Methotrexate | 48,0 | POS | YES | YES |
| 03–10 | F | 69 | 0 | NO | 24.8 | NO | Dyslipidemia | Pravastatine | Colorectal | YES | FOLFOX | 63,0 | POS | YES | YES |
| 03–14 | F | 75 | 1 | NO | 21.5 | NO | NO | NO | Breast | YES | Capecitabine Lapatinib | 85,0 | POS | NO | YES |
| 03–15 | M | 73 | 1 | NO | 18 | NO | NO | NO | Pulmonary | YES | Cisplatin Pemetrexed Prembrolizumab | 58,0 | POS | NO | NO |
| 03–16 | F | 48 | 1 | NO | 24.4 | NO | NO | NO | Breast | NO | Trastuzumab | 61,0 | POS | NO | NO |
| 03–17 | F | 74 | 0 | NO | 18.6 | NO | NO | NO | Glioblastoma | YES | NO | 59,0 | POS | NO | YES |
| 03–18 | F | 62 | 0 | NO | 22.3 | NO | NO | NO | Breast | YES | Tucatinib Capecitabine Trastuzumab | 85,0 | POS | NO | YES |
| 03–19 | M | 67 | 1 | NO | 20.5 | YES | NO | NO | Stomach | YES | FOLFIRI | 67,0 | POS | NO | YES |
| 03–20 | F | 64 | 1 | NO | 21 | NO | NO | NO | Ovarian | YES | Liposomal doxorubicin | 60,0 | POS | NO | NO |
| 03–21 | F | 57 | 1 | NO | 40 | YES | NO | NO | Ovarian | YES | Carboplatin Paclitaxel | 83,0 | POS | YES | YES |
| 03–22 | F | 71 | 0 | NO | 22 | NO | NO | NO | Breast | NO | NO | 91,0 | POS | YES | YES |
| 03–23 | M | 72 | 2 | NO | 21 | NO | NO | NO | Colorectal | YES | FOLFOX Bevacizumab | 71,0 | POS | YES | YES |
| 03–24 | M | 70 | 0 | NO | 21 | NO | NO | NO | Vesical | NO | NO | 81,0 | POS | YES | YES |
| 03–25 | M | 74 | 1 | NO | 19.8 | NO | NO | NO | HNC | NO | DCF | 69,0 | POS | YES | NO |
| 03–26 | M | 84 | 1 | NO | 22.6 | NO | NO | NO | Renal | YES | Nivolumab | 65,0 | POS | NO | NO |
| 03–27 | F | 80 | 0 | YES | 26 | NO | NO | NO | Hepatocarcinoma | YES | Sorafenib | 56,0 | POS | NO | YES |
| 03–28 | F | 62 | 0 | NO | 19.3 | NO | NO | NO | Breast | NO | Docetaxel | 75,0 | POS | NO | NO |
| 03–29 | F | 70 | 0 | NO | 31 | YES | NO | NO | Breast | YES | Palbociclib Hormonotherapy | 85,0 | POS | NO | NO |
| 03–31 | M | 75 | 2 | NO | 20 | NO | Ischemia Cardiopathy - Chronic Renal Failure |
Sotalol Perindopril Metformin Lercanidipine | Pulmonary | YES | NO | 70,0 | NEG | YES | YES |
| 03–34 | F | 64 | 1 | NO | 25 | NO | NO | NO | Renal | YES | Nivolumab | 101,0 | NEG | YES | YES |
| 03–40 | M | 74 | 1 | YES | 24.7 | YES | NO | NO | Prostate | YES | Cabazitaxel | 89 | POS | YES | NO |
| 03–42 | F | 51 | 2 | NO | 20 | YES | NO | NO | Sarcoma | NO | NO | 109 | POS | YES | YES |
| 03–44 |
F |
58 |
1 |
NO |
19 |
YES |
NO |
Hydrochlorothiazide |
Pancreas |
YES |
Gemcitabine |
41 |
POS |
NO |
YES |
|
Haematological malignancies | |||||||||||||||
|
Patient number |
Sex |
Age |
PS |
Diabetes |
BMI |
HTA |
Other comorbidities |
Concurrent ongoing therapies |
Cancer localisation |
Disease stage |
Active anticancer treatment during 4 weeks before COVID infection |
Time between COVID-19 infection and blood sample in months |
COVID-19 serology |
At least one T cell response (S, M or N) |
CEF specific T-cell responses |
| 03–12 | M | 73 | 1 | NO | 1.72 | NO | NO | NO | DLBCL | Early | R–CHOP | 49 | NEG | YES | YES |
| 03–13 | M | 23 | 1 | NO | 2.5 | NO | NO | Valaciclovir Tacrolimus Prednisone |
ALL | Early | NO (allograft in October 2019) | 48 | NEG | NO | YES |
| 03–32 | M | 80 | 1 | NO | 2 | YES | NO | Sotalol | CLL | Early | R-Venetoclax | 80 | POS | YES | NO |
| 03–35 | F | 48 | 2 | YES | 1.4 | NO | Pulmonary transplantation | Tacrolimus | AML | Early | NO (Allograft in 2006) | 48 | NEG | NO | NO |
| 03–36 | M | 54 | 1 | NO | 2 | NO | NO | Tacrolimus | AML | Early | NO (Allograft in august 2019) | 54 | POS | NO | NO |
| 03–37 | M | 54 | 2 | NO | 1.9 | NO | NO | Prednisone Polyvalent Intravenous Immunoglobulin |
AML | Early | NO (Allograft in 2007) | 54 | POS | YES | YES |
| 03–38 | M | 72 | 1 | NO | 1.8 | YES | Hypothyroidism | Levothyroxine | Multiple myeloma | Late | Isatuximab Pomalidomide Dexamethasone |
72 | POS | YES | YES |
| 03–39 | F | 73 | 2 | NO | 1.7 | NO | Stroke | Valaciclovir | AML | Late | Venetoclax | 73 | POS | NO | YES |
| 03–41 | M | 67 | 1 | NO | 2.1 | NO | Arrhythmia | Prednisone | Myelodysplastic syndrome | Late | NO | 67 | NEG | NO | YES |
| 03–43 | F | 52 | 1 | NO | 2.2 | NO | Hypothyroidism | Levothyroxine | CML | Early | Nilotinib | 52 | POS | NO | NO |
| 03–46 | M | 66 | 1 | NO | 2.1 | NO | NO | Valaciclovir Irbesartan | Bone marrow failure | Early | Cyclosporine | 66 | NEG | NO | YES |
Abbreviation: ALL, Acute Lymphocytic Leukaemia; AML, Acute Myelogenous Leukaemia; CLL, Chronic Lymphocytic Leukaemia; CML, Chronic Myelogenous Leukaemia; DCF, Docetaxel Cisplatin Fluorouracil; DLBC, Diffuse Large B Cell Lymphoma; F, Female; HNC, Head and Neck Cancer; M, Male; PS, Performance Status.
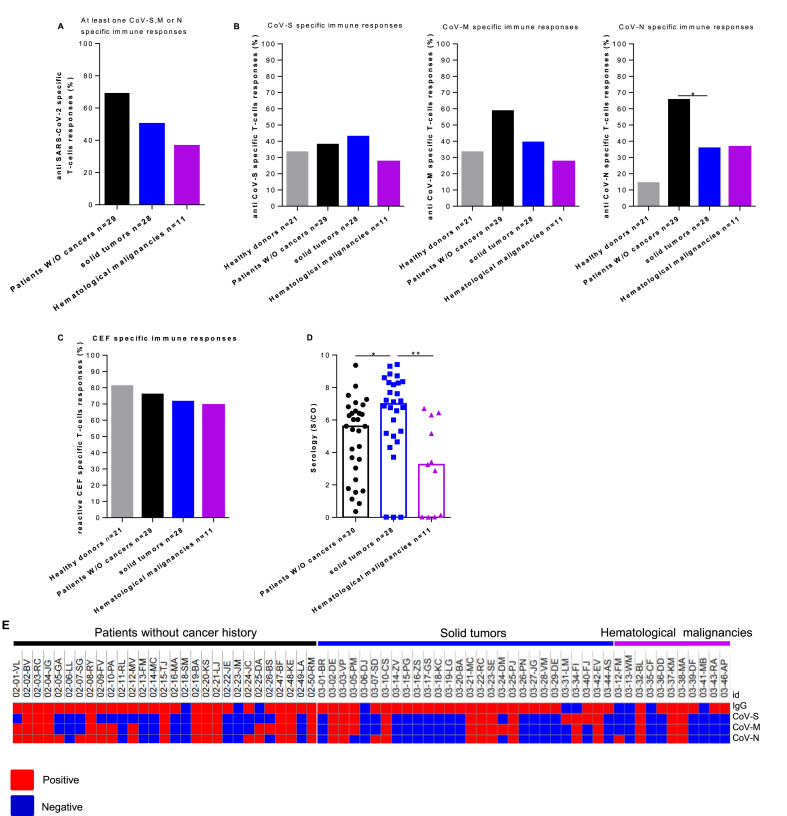
Among patients with cancers, 14 out of 28 patients with solid tumours (50.0%) and 4 out of 11 patients with haematological malignancies (36.4%) exhibited T-cell responses against at least one of the SARS-CoV-2 proteins (CoV–S, CoV-M or CoV–N) (Fig. 1 A). In line with a potential cross-reactivity with previous coronavirus exposition, recognition of CoV–S, M and N proteins by T-cells was observed in respectively 33.3%, 33.3% and 14.3% of the healthy donors tested (Fig. 1B). Furthermore, SARS-CoV-2 elicited specific immune responses against CoV-M and CoV–N in 58.6 and 65.5% of the patients without a cancer history (p ≤ 0.05 and p ≤ 0.01 respectively) (Fig. 1B, middle and right panel). The T-cell immune responses’ intensity against SARS-CoV-2 proteins was investigated. No statistically significant differences were observed between the responses of patients without a cancer history and both cohorts of cancer patients. This provides evidence that immune responses during SARS-CoV-2 infection do not sustain antigen memory T-cells detectable in the PBMCs. There were no significant frequency differences in CoV–S T-cell responses between patients with solid tumour (42.8%) or haematological malignancies (27.3%), patients without a cancer history (37.9%) or healthy donors (33.3%) (Fig. 1B, left panel). Specific CoV-M and N T-cell response frequencies tend to be lower in patients with haematological malignancies compared to patients without a cancer history (27.3% versus 58.6% (p = 0.15) and 36.4% versus 65.5% (p = 0.15) for SARS-CoV-M and N respectively) (Fig. 1B middle and right panel). Specific CoV–N T-cell response frequencies were statistically lower in patients with solid tumours compared to patients without a cancer history (35.7% versus 58.6%, p = 0.0348). The one against CoV-M tends to be lower (39.3% versus 58.6%, p = 0.12) (Fig. 1B). Interestingly, two cancer patients without peripheral SARS-CoV-2 specific T-cells had prolonged virus RNA detection after symptoms resolution, while no prolonged RNA production was reported in patients with specific adaptive immunity. The SARS-CoV-2 specific T-cells absence was not correlated to clinical determinants. Similarly, lymphopenia and lymphocyte subset distribution did not account for the lack of SARS-CoV-2-specific immunity (Table 1).
Fig. 1.
SARS-CoV-2 specific T-cell responses were increased in COVID-19 patients without a cancer history compared to COVID-19 patients with solid or haematological malignancies. PBMCs from 21 healthy donors and COVID-19 patients without a cancer history (n = 29), solid tumour (n = 28) or haematological malignancies were analysed for SARS-CoV-2 and antiviral-specific T-cell responses by IFNγ ELISpot assay. A. Frequency of patients with specific responses to at least one SARS-CoV-2 protein. B. Frequencies (%) of positive SARS-CoV-2 specific immune responses in healthy donors and COVID-19 patients (Mann–Whitney test, ∗p > 0.05). C. Frequency (%) of positive antiviral memory CD8 T-cell responses in healthy donors and COVID-19 patients. D. The median serology index (S/CO) in COVID-19 patients without a cancer history, solid tumour or haematological malignancies (Mann–Whitney test, ∗p > 0.05, ∗∗p > 0.01). E. Heatmap showing the positivity or the negativity of the serology index, T-cell immune responses to SARS-CoV-S, M and N proteins for each patient included in the study (online Morpheus software). Healthy donors' population is represented by light gray, COVID-19 patients without a cancer history by black, COVID-19 patients with solid tumour by blue and haematological malignancies by violet. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The other immunological issue for cancer patients is the potential risk that SARS-CoV-2 virus-mediated infection dampens pre-existing memory T-cell repertoire. To investigate this issue, we concomitantly measured the reactivity against common viruses using IFNγ ELISpot assay. Median intensities and frequencies were similar across all patient groups (Fig. 1C). This demonstrated that the recent history of COVID-19 did not hamper viral memory T-cell pool in patients with or without cancers. It suggests that the lack of SARS-CoV-2 specific T-cells is related to a weak immune adaptive response initiation rather than a global T-cell dysfunction.
We then analysed the SARS-CoV-2 humoral responses. We compared the serology index between our patients’ cohorts (Fig. 1D). The median serology index in patients without a cancer history was 5.6 S/CO [IQR: 2.9–6.6] and it was statistically lower compared to that of patients with solid tumours 6.9 S/CO [IQR: 5.0–8.3] (p = 0.016). In contrast, patients with haematological malignancies had a median serology index equal to 3.25 S/CO [IQR 0.1–6.3] that was statistically lower when compared to both patients with solid tumours and without a cancer history (p = 0.0017 and p = 0.052). A heatmap was generated, using the online Morpheus software (https://software.broadinstitute.org/morpheus/), to correlate all immunological parameters investigated (Fig. 1E). These results unravel that SARS-CoV-2 infection is associated with a lack of adaptive T-cell immunity in cancer patients. In addition, a discrepancy was unravelled, in the context of cancer history, between SARS-CoV-2 seroconversion and the presence of memory T-cells.
4. Discussion
To date, few studies have analysed specific SARS-CoV-2 cellular and humoral responses in cancer patients. This study, based on patients with solid tumours treated mostly at a metastatic stage, showed a high rate of SARS-CoV-2 antibody production. The specific T-cell response frequency in cancer patients was similar to the one in healthy donors and significantly lower than the frequency in COVID-19 patients without a cancer history underlying the weakness of SARS-CoV-2 immunogenicity in cancer patients. The presence of SARS-CoV-2 specific T-cells in healthy donors might be explained by the sequence homology between structural proteins from various coronaviruses suggesting the existence of cross-reactive memory T-cells [12,13]. Indeed, high degrees of similarities between coronaviruses and SARS-CoV-2 concerning S, M and N structural proteins have been recently described [9,14].
This study is intended to be descriptive but not comparative. The small cohort of cancer patients studied is heterogeneous in terms of cancer type and anticancer treatment at the time of COVID-19 infection, which make it difficult to statistically explain confounding factors of non-response. However, both cohorts of patients were comparable in terms of COVID-19 severity and included patients with mild disease. The T-cell responses against common viruses (Cytomegalo-, Epstein–Barr and Flu-virus) were used as a control of memory T-cell responses. The level of T-cell responses against CEF was similar between the two cohorts excluding a possible abnormality of the peripheral memory T-cells compartment. Nevertheless, the effects of the previous line of anticancer treatment and lymphopenia at the time of the infection remain to be investigated in a larger cohort of patients.
The impairment of adaptive immunity due to immune-suppressing therapies might alter cancer patients’ outcome following COVID-19. Patients with haematological malignancies have usually long-lasting immunodeficiency because of the malignancy itself but also because of anticancer treatments such as rituximab or haematopoietic stem-cell transplantations [15]. The first report on COVID-19 in chronic lymphocytic leukaemia (CLL) patients receiving venetoclax showed a substantial adaptive immune deficiency that might lead to an impairment of T-cell responses and more COVID-19 severity in infected CLL patients [16]. The cases of severe COVID-19 have also been reported in patients treated with anti-CD20 targeted therapy supporting the impact of B-cell depletion thereby the incapacity to produce neutralising antibodies on the COVID-19 course [17,18]. Nevertheless, in a recent multicentric study including 177 patients with lymphoma, active treatment and the number of prior lines did not show significant differences in mortality due to the infection [19]. Furthermore, in a study including 423 cases of patients with solid tumours, the active anticancer treatment did not show a significant association with the COVID-19 severity [20]. The potential effects of immunosuppressive treatments on the severity of COVID-19 disease and outcome need future investigations in a larger cohort of patients.
The induction of anti-SARS-CoV-2 protective immunity by vaccine in cancer patients is an important medical need. However, the heterogeneity of SARS-CoV-2 specific-T-cell responses among cancer patients emphasises the interest to investigate anti-SARS-CoV-2 immune responses following vaccination in this population.
To summarise, this study raises three major concerns to be considered for further research programs: i) adaptive T-cell immunity targeting SARS-CoV-2 is weak in cancer patients compared to common viruses; ii) specific immunoglobulin monitoring is not sufficient to characterise anti-SARS-CoV-2 immunity and assays to monitor specific T-cell responses should be developed, and iii) SARS-CoV-2 infection does not alter the common virus's memory T-cell responses level in cancer patients. Even if those first data need to be confirmed in a larger cohort, the present results support the promotion of specific SARS-CoV-2 vaccination programs for cancer patients given the ongoing COVID-19 pandemic severity.
Authors’ contribution
L. Mansi, L. Spehner and M. Kroemer had full access to all data in the study and take responsibility for the integrity of the data and accuracy of data analysis. Study concept and design: M. Kroemer, K. Bouiller, L. Mansi, L. Spehner, E. Daguindeau, C. Borg and O. Adotévi. Acquisition, analysis or interpretation of data: all authors. Drafting of the manuscript: L. Mansi, M. Kroemer, L. Spehner and C. Borg. Statistical analysis: L. Mansi and L. Spehner. Study supervision: L. Mansi, C. Borg, O. Adotévi and M. Kroemer. We are grateful to the Department of Oncological Clinical Research (A. Martin, J. Meier, A. Sauget and D. Berthod) and the nurses team (D. Bourdenet, M. Pagnot and A. Terzibachian).
Fundings
No funding directly supported this work.
Conflict of interest statement
The authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejca.2021.03.033.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L., Zhu F., Xie L., Wang C., Wang J., Chen R., et al. Clinical characteristics of COVID-19-infected cancer patients: a retrospective case study in three hospitals within Wuhan, China. Ann Oncol. 2020 doi: 10.1016/j.annonc.2020.03.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang W., Guan W., Chen R., Wang W., Li J., Xu K., et al. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21:335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai M., Liu D., Liu M., Zhou F., Li G., Chen Z., et al. Patients with cancer appear more vulnerable to SARS-COV-2: a multi-center study during the COVID-19 outbreak. Canc Discov. 2020 doi: 10.1158/2159-8290.CD-20-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee L.Y.W., Cazier J.B., Starkey T., Turnbull C.D., Kerr R., Middleton G. COVID-19 mortality in patients with cancer on chemotherapy or other anticancer treatments: a prospective cohort study. Lancet. 2020;395:1919–1926. doi: 10.1016/S0140-6736(20)31173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuderer N.M., Choueiri T.K., Shah D.P., Shyr Y., Rubinstein S.M., Rivera D.R., et al. Clinical impact of COVID-19 on patients with cancer (CCC19): a cohort study. Lancet. 2020;395:1907–1918. doi: 10.1016/S0140-6736(20)31187-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vardhana S.A., Wolchok J.D. The many faces of the anti-COVID immune response. J Exp Med. 2020;217 doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., et al. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12 doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroemer M., Spehner L., Vettoretti L., Bouard A., Eberst G., Pili Floury S., et al. COVID-19 patients display distinct SARS-CoV-2 specific T-cell responses according to disease severity. J Infect. 2020 doi: 10.1016/j.jinf.2020.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., et al. Immunology of COVID-19: current state of the science. Immunity. 2020 doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Passamonti F., Cattaneo C., Arcaini L., Bruna R., Cavo M., Merli F., et al. Clinical characteristics and risk factors associated with COVID-19 severity in patients with haematological malignancies in Italy: a retrospective, multicentre, cohort study. Lancet Haematol. 2020;7:e737–e745. doi: 10.1016/S2352-3026(20)30251-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fürstenau M., Langerbeins P., De Silva N., Fink A.M., Robrecht S., von Tresckow J., et al. COVID-19 among fit patients with CLL treated with venetoclax-based combinations. Leukemia. 2020;34:2225–2229. doi: 10.1038/s41375-020-0941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tepasse P., Hafezi W., Lutz M., Kühn J., Wilms C., Wiewrodt R., et al. Persisting SARS-CoV-2 viremia after rituximab therapy: two cases with fatal outcome and a review of literature. Br J Haematol. 2020 doi: 10.1111/bjh.16896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hueso T., Pouderoux C., Péré H., Beaumont A.-L., Raillon L.-A., Ader F., et al. Convalescent plasma therapy for B-cell–depleted patients with protracted COVID-19. Blood. 2020;136:2290–2295. doi: 10.1182/blood.2020008423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Regalado-Artamendi I., Jiménez-Ubieto A., Hernández-Rivas J.Á., Navarro B., Núñez L., Alaez C., et al. Risk factors and mortality of COVID-19 in patients with lymphoma: a multicenter study. Hemasphere. 2021;5 doi: 10.1097/HS9.0000000000000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robilotti E.V., Babady N.E., Mead P.A., Rolling T., Perez-Johnston R., Bernardes M., et al. Determinants of severity in cancer patients with COVID-19 illness. Nat Med. 2020;26:1218–1223. doi: 10.1038/s41591-020-0979-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.