Abstract
The new coronavirus disease known as COVID-19 is currently a pandemic that is spread out the whole world. Several methods have been presented to detect COVID-19 disease. Computer vision methods have been widely utilized to detect COVID-19 by using chest X-ray and computed tomography (CT) images. This work introduces a model for the automatic detection of COVID-19 using CT images. A novel handcrafted feature generation technique and a hybrid feature selector are used together to achieve better performance. The primary goal of the proposed framework is to achieve a higher classification accuracy than convolutional neural networks (CNN) using handcrafted features of the CT images. In the proposed framework, there are four fundamental phases, which are preprocessing, fused dynamic sized exemplars based pyramid feature generation, ReliefF, and iterative neighborhood component analysis based feature selection and deep neural network classifier. In the preprocessing phase, CT images are converted into 2D matrices and resized to 256 × 256 sized images. The proposed feature generation network uses dynamic-sized exemplars and pyramid structures together. Two basic feature generation functions are used to extract statistical and textural features. The selected most informative features are forwarded to artificial neural networks (ANN) and deep neural network (DNN) for classification. ANN and DNN models achieved 94.10% and 95.84% classification accuracies respectively. The proposed fused feature generator and iterative hybrid feature selector achieved the best success rate, according to the results obtained by using CT images.
Keywords: COVID-19, Fused residual dynamic exemplar pyramid model, RFINCA feature Selector, Deep neural network
1. Introduction
1.1. Background
Coronaviruses are ribonucleic acid (RNA) viruses that can infect many species of animals and people. Until severe acute respiratory failure syndrome (SARS) appeared, it was known as the cause of 15–30% of the common cold [1]. The common cold is usually associated with mild and self-limiting infections. New types of coronavirus began to appear in 2002. The disease manifested itself in the form of a respiratory infection, which is more severe than normal cold symptoms. The first of these is severe acute respiratory failure syndrome (SARS), which is thought to be transmitted from bats to musk cats and then from cats to people in the Guangdong region of China in February 2003. The infection has affected 8000 people in the world and approximately 800 deaths have been reported [2]. A new case report for SARSCoV was not produced after 2004. The second new coronavirus infection was the Middle East respiratory syndrome (MERS) seen in Saudi Arabia in 2012 and thought to be transmitted from camel to human. The MERS-CoV epidemic that continued until 2018 was detected in 2229 cases and 791 of the cases resulted in death [3].
In 2019, new coronavirus disease (COVID-19) appeared as an infectious disease caused by severe acute respiratory syndrome coronavirus (SARS-CoV-2) and was first reported in the Wuhan province of China [4]. The virus has spread out worldwide and has been declared a pandemic by the World Health Organization (WHO) [5]. Although the most important clinical symptoms are fever and cough, symptoms such as fatigue, headache and shortness of breath can also be seen. However, diagnostic tests are needed because all these symptoms are not specific to the disease and the disease can progress rapidly to severe pneumonia [6]. Although the real-time reverse transcription-polymerase chain reaction (RT-PCR) test for viral nucleic acids in the diagnosis of COVID-19 is the gold standard, computed tomography (CT) has become more and more important in the diagnosis [7].
1.2. Motivation
Early detection and treatment of COVID-19 is crucial. Compared to RT-PCR tests, thoracic (lung area) CT imaging is known to be a more reliable, prompt and practical method to diagnose and evaluate COVID-19, especially in the outbreak area [8]. Studies have demonstrated that the sensitivity of CT images achieves higher accuracy [[9], [10], [11], [12], [13], [14]]. According to recent research, sensitivity of the PCR (capacity to detect patients) remains only 71% for COVID-19 infection [15]. However, CT images can be used as a problem-solving approach in patients who are negative for RT-PCR but not diagnosed precisely.
Numerous studies presented in the literature on COVID-19 disease detection. Ozturk et al. [9] proposed DarkCovidNet model for classifying COVID-19, healthy and pneumonia disease. They used X-ray images and achieved 87.02% accuracy rate. Hemdan et al. [10] proposed a deep learning model, which is called COVIDX-Net. They used 25 COVID-19, and 25 healthy images. They obtained 90% success rate. Wang et al. [11] collected 1119 CT images for COVID-19 diagnosis. They proposed a new architecture called M-inception by modifying the classical inception network. In the proposed M-inception method, the features were reduced before the classification layer. In the experiments, 89.5% accuracy with 88% specificity and 87% sensitivity was achieved. Zhao and colleagues [12] proposed a method using transfer learning and data augmentation together with deep learning. In their study, they achieved an accuracy of 84.7% by using 275 CT images in total. Al-Karawi et al. [13] improved a Fast Fourier Transform-Gabor scheme, which predicted in almost real-time, the state of the patient with an average accuracy of 95.37%. They selected randomly 150 images, which were consisting of 75 COVID-19 and 75 non-COVID-19 images. 60% of the total images were used for training, the remaining 40% of the images were used for testing. In the classification step, SVM classifier was used. Loey and colleagues [14] presented a deep transfer learning model with classical data augmentation and conditional generative adversarial network (CGAN). VGGNet16, VGGNet19, AlexNet, GoogleNet, and ResNet50 were used for detecting the COVID-19 infected patient. ResNet50 was found as the most appropriate classifier for detecting the COVID-19 from the chest CT images. They obtained 82.91% testing accuracy by using the classical data augmentation and CGAN.
1.3. Contributions
The novelty of this study is to develop a new hybrid and simple handcrafted feature generation model and to achieve high performance by using fewer features. Hence, the contributions of the proposed approach are as follows:
-
-
A novel effective feature generation model is developed. This model uses dynamic-sized exemplars to generate more comprehensive features with the pyramid model. The main aim of the presented feature generator is to extract local and global features in detail.
-
-
The main problem of the most feature selectors like ReliefF and NCA is to select optimal features automatically. The RFINCA feature selector addresses this issue and uses the efficiency of both feature selectors.
-
-
A robust COVID-19 detection framework was developed, and results revealed that the proposed handcrafted features based automated COVID-19 classification method attained higher accuracies than deep learning models (See Table 5).
Table 5.
Comparison of the proposed approach with the previous studies.
| Study | Dataset reference | Proposed methods | Accuracy (%) |
|---|---|---|---|
| [25] | 777 COVID-19 708 Healthy (Private Dataset) |
DRE-Net | 86 |
| [26] | 313 COVID-19 229 Healthy (Private Dataset) |
UNetþ3D Deep Network |
90.8 |
| [27] | 219 COVID-19 175 Healthy 224 Pneumonia (Private Dataset) |
ResNet þ Location Attention |
86.7 |
| [11] | 325 COVID-19 740 Healty (Private Dataset) |
M-Inception | 89.5 |
| [12] | 349 COVID-19, 397 Healty CT images [28] | DenseNet | 84.7 |
| [13] | 275 COVID-19 195 Healty (Private Dataset) |
FFT-Gabor | 95.37 |
| [14] | 349 COVID-19, 397 Healty CT images [28] | CGAN | 82.91 |
| [29] | 496 COVID-19 1385 others (Private Dataset + [44,45]) |
CNN | 94.98 |
| [30] | 100 normal 98 lung cancer 397 other (Private Dataset + [46,47]) |
DL multitask | 93 |
| [31] | 564 COVID-19 660 non-COVID-19 [51] |
VGG16 based lesion-attention DNN | 88.6 |
| [32] | 313 COVID-19 229 without COVID-19 (Private) |
UNet | 90.1 |
| [33] | 413 COVID-19 439 non-COVID-19 [49,50] |
ResNet-50 + 2D CNN | 93.02 |
| [34] | 460 COVID-19 397 non-COVID-19 [51,52] |
SqueezeNet | 83 |
| [35] | 1029 COVID-19 1695 non-COVID-19 (Private) |
AH-Net + DenseNet121 | 90.8 |
| [36] | 53 COVID-19 97 other (Private) |
SVM | 98.71 |
| [37] | 51 COVID-19 55 control(Private) |
UNet++ | 98.85 |
| [38] | 230 COVID-19 130 normal (Private) |
AD3D-MIL | 97.9 |
| [39] | 521 COVID-19 397 normal 76 bac. pneu. (Private) |
DL ShuffleNet V2 |
85.40 |
| [40] | COVID-19/other pneu./healthy. (Private) | 3D UNet-based network | 94 |
| [41] | 284 COVID-19, 281 CAP, 293 SPT, 306 HC |
CCSHNet | 97.04 |
| [42] | 219 COVID-19, 1345 pneumonia and 1341 normal images [50] | mAlexNet + BiLSTM | 98.70 |
| [43] | 361 COVID-19, 1341 Normal and 1345 Pneumonia |
InstaCovNet-19 | 99.08 |
| Proposed method | 349 COVID-19, 397 Healty CT images [28] | FDEPFGN and RFINCA | 95.84 |
1.4. Organization
Information about the used CT image corpus is provided in section 2. The proposed image classification framework is explained in Section 3. Section 4 presents a complete experimental study of the proposed CT based COVID-19 detection framework. The overall discussion is given in section 5. Lastly, the conclusion part is given in section 6.
2. Material
Chest CT scan is an effective method for the diagnosis of COVID-19. The radiological knowledge of physicians does not always provide an exact diagnosis. At the same time, it takes a longer time to get results with the nose, throat swabs, and blood tests. Therefore, chest CT images have an important role in the management and follow-up of patients who are hospitalized. The dataset [12] used in this study is publicly available. This data set was confirmed by senior radiologists from Tongji Hospital in Wuhan, China, who diagnosed and treated a large number of COVID-19 patients during the emergence of this disease between January and April. The dataset contains 349 COVID-19, 397 healthy CT images. Fig. 1 shows the COVID-19 and healthy chest CT images.
Fig. 1.
Pictorial demonstration of the CT image dataset.
3. The proposed image classification method
The main goal of this work is to present a novel handcrafted features based CT image classification approach for COVID-19 detection. To achieve this goal, this work presents a novel residual dynamic-sized exemplars based pyramid model. The used feature generation functions are local binary pattern (LBP) and statistical features [[16], [17], [18]]. By using these functions and dynamically sized exemplars-based pyramid model together, a novel feature extraction network is developed. The proposed method is called fused dynamic sized exemplars based pyramid feature generation network (FDEPFGN). After the feature generation process, the generated features are utilized as an input of the ReliefF that selects positive weighted features and eliminates negative weighted features. In RFINCA [19] feature selector phase, the selected positive weighted features by ReliefF are forwarded to iterative NCA to select the optimum number of best features automatically. The selected features are then used as an input of the classifier. The graphical representation of this framework is shown in Fig. 2 .
Fig. 2.
Graphical representation of the proposed FRDEPFGN and RFINCA based CT image classification framework.
Fig. 2 denotes the graphical explanation of the developed CT image classification framework. This model is an iterative model, where firstly, the used input (CT) RGB image is transformed into a gray-level image. Three types of the exemplars are used for feature generation and the sizes of these exemplars are 16 × 16, 32 × 32, and 64 × 64. This model generates features using three levels. For the level creation, bilinear interpolation is used. At each level, bilinear interpolation halves the size of the image. The extracted features in each level from the exemplars are concatenated and the informative ones are selected by RFINCA. These informative features are classified by using DNN and ANN classifiers.
Although, Fig. 2 explains the proposed FRDEPFGN and RFINCA based CT image classification method graphically, in order to explain the proposed approach as a whole in a clear way, the pseudo-code of the proposed FRDEPFGN and RFINCA based CT image classification approach is given in Fig. 3 . Lines 01–19 express preprocessing, feature generation, and feature concatenation steps of the proposed approach. Actually, these lines (Lines 01–19) define the FRDEPFGN network. Line 20 shows the RFINCA feature selector and Line 21 illustrates classification phases. LBP generates 59 features and the statistical feature generation function (SF) extracts 19 features. Therefore, LBP and SF generate 78 features together as can be seen in Line 05 and Line 12. The details of the proposed framework are explained in the subsections.
Fig. 3.
Pseudo code of the proposed RDEPFGN and RFINCA based CT image classification method.
3.1. Preprocessing
The first phase of the proposed FRDEPFGN and RFINCA based CT image classification framework is preprocessing. In this phase, RGB to gray conversion is carried out to obtain a 2D image. Then, the obtained CT image is resized as 256 × 256 dimensions. The first and second steps of our method are given below.
Step 1
Transform CT image to 2D image.
Step 2
Resize the 2D CT image to 256 × 256 sized image.
3.2. Feature generation
Feature generation is one of the crucial phases of pattern recognition. A novel model, which is called as FRDEPFGN, is proposed to generate features. The proposed FRDEPFGN is inspired from deep networks. Especially, inception network uses dynamic sized convolution operator. Therefore, FRDEPFGN uses dynamic sized exemplars. FRDEPFGN uses two feature generation functions, which are explained in the subsections.
3.2.1. Statistical feature generation
Statistical feature generation has been widely used in the literature and it is one of the first known handcrafted feature generation methods. Numerous statistical moments can be used in the statistical feature generation, such as maximum, minimum, average, energy, skewness, kurtosis, variance, and standard deviation. These methods work effectively and the computational complexity is low. In this work, 19 statistical functions are used for statistical feature generation. The mathematical definitions of the used statistical function are given below.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
| (13) |
| (14) |
| (15) |
| (16) |
| (17) |
| (18) |
| (19) |
where is CT image. By using the defined 19 statistics above, the used SF is created. Therefore, SF extracts 19 features from an image or an exemplar.
3.2.2. Textural feature generation
LBP, which is one of the widely preferred feature generators, is used as a second feature generation function. There are many LBP like feature extractor in the literature. Since the time complexity of the LBP is , it extracts distinctive features and has a simple structure. Therefore, it can be programmed easily and has been used in many study areas [20]. LBP is applied to generate textural features of the CT images and the steps are as follows:
-
1
Divide image into overlapping windows with size of 3 × 3.
| (20) |
-
2
Generate bits using signum, center pixel and neighborhood pixels together.
| (21) |
| (22) |
-
3
Calculate decimal map value by using bits extracted.
| (23) |
-
4
Generate histogram and obtain feature vector.
These steps are defined for LBP feature generation function. A numerical example of LBP is shown in Fig. 4 graphically.
Fig. 4.
Graphical representation of LBP feature generation.
LBP divides the image into 3 × 3 sized overlapping blocks to generate features. In Fig. 4, the center pixel value is 52. This pixel is compared to its neighborhood pixels to generate binary features using the signum function. The generated bits are then converted into a decimal value.
3.2.3. Description of the proposed fused residual dynamic exemplar pyramid feature generation network
Steps of the proposed FRDEPFGN method are given in this section.
Step 3
Extract textural and statistical features from image by using LBP and SF function.
| (24) |
Step 4
Divide CT image into exemplars () with size of 16 × 16, 32 × 32 and 64 × 64.
| (25) |
| (26) |
| (27) |
where expresses th exemplar of the th type. Three types of exemplar are used in this work and they are categorized according to size.
Step 5
Generate features using three types of exemplars.
| (28) |
| (29) |
| (30) |
Step 6
Concatenate generated features.
Step 7
Decompose CT image by using bilinear interpolation. In this step, W x H sized images are resized to W/2 x H/2 sized images. By using this step, a multileveled feature generation method is obtained.
Step 8
Step 9
Concatenate all extracted features.
3.3. RFINCA feature selection
RFINCA aims to use the effectiveness of the ReliefF and NCA selectors together and selects the optimum number of features automatically. Because ReliefF and NCA cannot select the optimum number of features automatically alone. In the feature extraction phase, the proposed FRDEPFGN generates 34,632 features in total, since the used feature extraction functions are run 444 times in the proposed approach. In order to select the most informative features among these 34632 features, RFINCA, which is the combination of ReliefF and NCA, is used as a feature selector. ReliefF and NCA are distance-based feature selectors where ReliefF can generate negative weights, on the other hand, NCA generates non-negative weights. Generally, negative weighted features can be assigned as redundant features. Therefore, in the first step, ReliefF is applied to all features in order to calculate negative weighted features and eliminate them. In the second step, iterative NCA is applied to the selected positive weighted features [21].
To select an optimal number of features, the error calculation of the k-nearest neighbor (k-NN) classifier is used. Feature range from 20 to 1000 was also used to decrease the complexity of the RFINCA. To better explain the proposed RFINCA, the flow diagram is shown in Fig. 5 [19].
Fig. 5.

Flow diagram of the RFINCA.
According to this diagram (see Fig. 5), ReliefF selects positively weighted features from the extracted features. This step is called redundant feature elimination. The selected positive weighted features are utilized as an input of the NCA, then NCA weights are calculated. By using these weights, an iterative feature selection is realized. The error of the selected feature vector is calculated by using k-NN (or any other classifier can be used). The minimum error valued feature vector is selected as an optimal feature vector.
In order to clearly explain the steps of the RFINCA, the pseudo code of the procedure is also given in Fig. 6 .
Fig. 6.
Graphical representation of the RFINCA procedure.
Lines 01–03 define min-max normalization. Since both NCA and ReliefF are distance-based feature selectors, normalization is applied to obtain effective results. Lines 04–11 express the ReliefF phase and Lines 12–18 shows iterative NCA. The optimal number of features () selection is denoted in Lines 20–22. The 10th step of this work is given as follows:
Step 10
Select the optimal/informative features by using RFINCA.
| (31) |
RFINCA selects 989 features from the extracted 34,632 features.
3.4. Classification
In the last step, deep neural network (DNN) [[22], [23], [24]] is used with 10-fold cross-validation. DNN is the enhanced type of the artificial neural network (ANN), and has 2 or more hidden layers. The employed DNN is backward network and utilizes scaled conjugate gradient (SCG) for learning, since it needs gradient computing of functions. SCG algorithm utilizes steepest descent direction. During the implementation of the DNN approach, first weights are randomly allocated and (input of hidden layers) are computed using Eq. (32).
| (32) |
where is allocated weights, expresses inputs and is activation function. Then, the weights are recalculated by employing the backpropagation technique. In this step SCG that is a steepest optimization method is utilized and it employs orthogonal vectors to minimize the error. The mathematical notation of SCG is given in Eqs. (33), (34), (35)).
| (33) |
| (34) |
| (35) |
Herein, is multiplier, orthogonal vector and is input. By employing this optimization approach, weights are recalculated. The selected 989 features are employed as an input to the SCG based 3 hidden layered DNN to assess the performance of the proposed feature extraction and selection framework. There is no regular approach presently for constructing an optimal neural network with an appropriate number of layers and number of neurons in each layer. For this reason, we empirically construct the DNN by carrying out different experiments. We have manually adjusted a DNN in every experiment by tuning the number of hidden layers, the number of nodes creating the layer for each hidden layer, the number of learning steps, learning rate, momentum, and the activation function. We employed an SCG optimization technique for the backpropagation algorithm and set the learning rate to 0.7, momentum to 0.3, and batch size to 100. We have calculated the classification accuracy, using 10-fold cross-validation for every manual formation to look for the remaining DNN hyperparameters. This search procedure is repeated for different sizes of the hidden layer representations. After this exhausting manual process, the best performance in the classification is achieved with a DNN consisting of 3 hidden layers with 270, 150, and 50 nodes respectively. In the ANN implementation, a similar process is carried out, and the best performance is achieved with a hidden layer of 250 neurons. The scaled conjugate gradient is selected as an optimizer in this work. The tangent sigmoid is selected as an activation function. Moreover, batch normalization is employed in the model.
Step 11
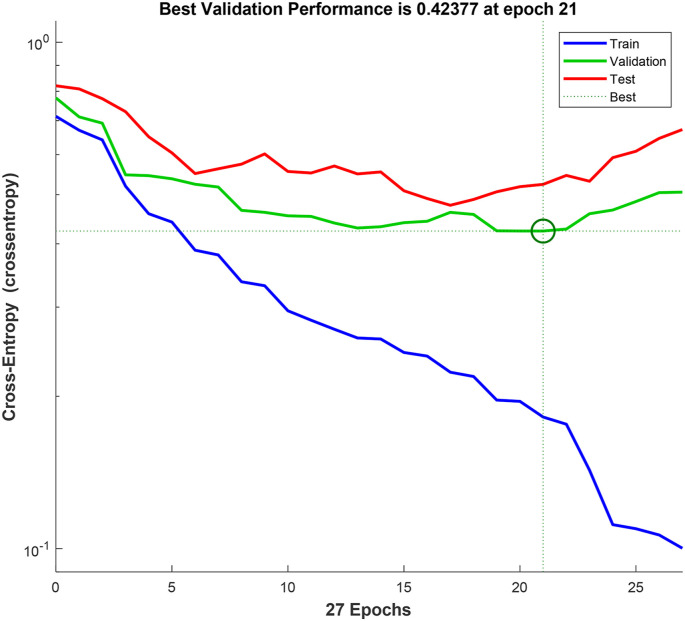
Classify the selected features by using ANN or DNN classifier with 10-fold cross-validation. Training/validation curve for ANN and DNN are shown in Fig. 7, Fig. 8 respectively.
Fig. 7.
Training/validation curve for ANN.
Fig. 8.
Training/validation curve for DNN.
4. Experimental results
The CT images are utilized as an input of the proposed framework. The details of the used dataset are provided in Section 2. These images include two categories, which are COVID-19 and healthy CT images. Ten-fold cross-validation is selected as validation and testing methodology. This method was carried out on a personal computer (PC) using MATLAB (2018a) programming environment. The used PC has simple configurations (i5 1.8 GHz CPU 8 GB RAM). This method does not require the use of a graphical processing unit or a parallel computing structure. In the classification phase, ANN and DNN classifiers are used. The obtained results are listed in Table 1 according to the folds.
Table 1.
The calculated accuracy rates (%) of the ANN and DNN according to the folds.
| Fold | ANN | DNN |
|---|---|---|
| 1 | 82.43 | 83.78 |
| 2 | 88.0 | 90.67 |
| 3 | 92.0 | 92.0 |
| 4 | 96.0 | 100.0 |
| 5 | 96.0 | 98.67 |
| 6 | 97.33 | 98.67 |
| 7 | 94.67 | 97.33 |
| 8 | 97.30 | 98.65 |
| 9 | 98.65 | 98.65 |
| 10 | 98.65 | 100.0 |
| Average | 94.10 | 95.84 |
Per Table 1, ANN and DNN achieve 94.10% and 95.84% classification accuracies respectively. To give details of these results, confusion matrices of the used classifiers are shown in Table 2 and Table 3 . Recall, precision, and accuracy parameters can be extracted from these matrices. By using recall and precision, F1-score can be calculated. Since F1-score is the harmonic mean of the precision and recall and mathematical expression of it is given in Eq. (32). Geometric mean and balanced accuracy can also be calculated using Eqs. (33), (34)).
| (36) |
| (37) |
| (38) |
Table 2.
Confusion matrix of the ANN classifier.
| Actual classes | Predicted classes |
Recall | |
|---|---|---|---|
| COVID-19 | Healthy | ||
| COVID-19 | 326 | 23 | 0.9341 |
| Healthy | 21 | 376 | 0.9471 |
| Precision | 0.9395 | 0.9423 | 0.9410 |
Table 3.
Confusion matrix of the DNN classifier.
| Actual classes | Predicted classes |
Recall | |
|---|---|---|---|
| COVID-19 | Healthy | ||
| COVID-19 | 330 | 19 | 0.9456 |
| Healthy | 12 | 385 | 0.9698 |
| Precision | 0.9649 | 0.9530 | 0.9584 |
The calculated F1-score and geometric mean and accuracy are also listed in Table 4 .
Table 4.
The calculated F1-score and geometric mean and accuracy of the ANN and DNN classifiers.
| Classifier | Class | F1-score | Geometric mean | Accuracy |
|---|---|---|---|---|
| ANN | COVID-19 | 93.68 | – | – |
| Healthy | 94.47 | – | – | |
| Average | 94.08 | 94.06 | 94.06 | |
| DNN | COVID-19 | 95.52 | – | – |
| Healthy | 96.13 | – | – | |
| Average | 95.83 | 95.76 | 95.77 |
5. Discussions
This work presents a novel CT based COVID-19 detection approach by using a new handcrafted feature generation network and a new hybrid and iterative feature selector. The proposed feature generator is inspired by the inception network and residual network. Therefore, a novel residual dynamic exemplar pyramid model is used to generate the features comprehensively. 16 × 16, 32 × 32, and 64 × 64 sized exemplars are used and the pyramid model is constructed by using bilinear interpolation. RFINCA selects the most informative or discriminative features and automatically determines the optimum features. Feature generation and feature selection are two fundamental problems in pattern recognition and classification. These problems need to be resolved to obtain high classification accuracy. This study resolved these issues using the proposed FRDEPFGN and RFINCA approaches.
The proposed framework achieves high classification accuracies (See Table 1). To indicate the success of the proposed framework, comparisons with the state-of-the-art methods are listed in Table 5.
X-ray images are generally preferred for COVID-19 diagnosis studies. But, in this study, high performance (95.84%) is obtained using chest CT images. As it can be seen from Table 5, one of the highest performance is the proposed approach with an accuracy of 95.84%. Although the obtained results are satisfactory, we believe that higher accuracy can be achieved by employing more CT images.
Moreover, the developed model uses handcrafted features, and in the classification phase, DNN classifier is used. The primary aim of this model is to reach high accuracies using handcrafted features. Since the number of samples is less, the transfer learning based models must be utilized in this case. But the accuracy of the transfer learning based models [14,33] are lower than the proposed approach. Since, the proposed model utilizes the handcrafted and simple feature generation methods, the computational cost of the proposed model is less than the transfer learning based models. Furthermore, if we compare the proposed model performance with the state-of-the-art for COVID-19 detection using CT images, most of the state-of-the-art models have achieved lower accuracy than the proposed approach (See Table 5).
The advantages of the developed FDEPFGN and RFINCA based chest CT image classification approach are given below.
-
•
A novel dynamic exemplar pyramid feature extraction method is developed.
-
•
A novel iterative feature selection method is proposed.
-
•
ANN and DNN classification results show that the developed framework successfully extracts and selects features.
-
•
The proposed method is a cognitive method, since there is no need to set millions of parameters as deep learning networks.
6. Conclusions
This paper presents a hybrid FDEPFGN and RFINCA framework for COVID-19 detection from a limited number of chest CT images. Our model is based on generating features with FDEPFGN and selecting informative features with RFINCA. DNN and ANN are employed to test the model. Evaluation of the proposed method is measured with different performance metrics such as precision, geometric mean, F1 score, and accuracy. Overall, our experiments show that the developed FRDEPFGN and RFINCA based CT image classification framework can achieve a better performance than 16 state-of-the-art approaches (See Table 5). The future work may contain the following aspects: (i) Expand the size of the dataset and test our proposed model on a larger dataset. (ii) Try to use some pretrained deep learning architectures. (iii) Some advanced or hybrid fusion rules will be tested by using pretrained deep learning architectures. (iv) Our proposed method can be embedded into other automated healthcare systems.
Funding
There is no funding source for this article.
CRediT authorship contribution statement
Fatih Ozyurt: Conceptualization, Data curation, Formal analysis, Visualization, Writing – original draft. Turker Tuncer: Methodology, Resources, Validation, Visualization, Writing – original draft. Abdulhamit Subasi: Conceptualization, Methodology, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Howley P.M., Knipe D.M. Lippincott Williams & Wilkins; 2020. Fields Virology: Emerging Viruses. [Google Scholar]
- 2.Sampathkumar P., Temesgen Z., Smith T.F., Thompson R.L. SARS: epidemiology, clinical presentation, management, and infection control measures. Mayo Clin. Proc. 2003;vol. 78(7):882–890. doi: 10.4065/78.7.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choudhry H., et al. Middle East respiratory syndrome: pathogenesis and therapeutic developments. Future Virol. 2019;vol. 14(4):237–246. doi: 10.2217/fvl-2018-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., et al. China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. In this issue, Epub 2020 Jan 24. PMID: 31978945; PMCID: PMC7092803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.C. D. C. C.-19 R. Team ‘Severe outcomes among patients with coronavirus disease 2019 (COVID-19)—United States, February 12--March 16, 2020’. MMWR Morb. Mortal. Wkly. Rep. 2020;vol. 69(12):343–346. doi: 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.cSULE Akçay M., Özlü T., Yilmaz A. Radiological approaches to COVID-19 pneumonia. Turk. J. Med. Sci. 2020;vol. 50(SI-1):604–610. doi: 10.3906/sag-2004-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. 2020. Chest CT for Typical 2019-nCoV Pneumonia: Relationship to Negative RT-PCR Testing. Radiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CT outperforms lab diagnosis for coronavirus infection. https://healthcare-in-europe.com/en/news/ct-outperforms-lab-diagnosis-for-coronavirus-infection.html
- 9.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemdan E.E.-D., Shouman M.A., Karar M.E. arXiv Prepr. arXiv; 2020. Covidx-net: A Framework of Deep Learning Classifiers to Diagnose Covid-19 in X-Ray Images. [Google Scholar]
- 11.Wang S., et al. Vol. 2.14. medRxiv; 2020. A Deep Learning Algorithm Using CT Images to Screen for Corona Virus Disease (COVID-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J., Zhang Y., He X., Xie P. arXiv Prepr. arXiv; 2020. COVID-CT-Dataset: a CT Scan Dataset about COVID-19; p. 13865. [Google Scholar]
- 13.Al-Karawi D., Al-Zaidi S., Polus N., Jassim S. medRxiv; 2020. Machine Learning Analysis of Chest CT Scan Images as a Complementary Digital Test of Coronavirus (COVID-19) Patients. [Google Scholar]
- 14.Loey M., Smarandache F., Eldeen N., Khalifa M. A deep transfer learning model with classical data augmentation and CGAN to detect COVID-19 from chest CT radiography digital images. Neural Comput. Appl. 2020:1–13. doi: 10.1007/s00521-020-05437-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang Y., et al. 2020. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuncer T., Aydemir E., Dogan S. Automated ambient recognition method based on dynamic center mirror local binary pattern: DCMLBP. Appl. Acoust. 2020;vol. 161 [Google Scholar]
- 17.Yaman O., Ertam F., Tuncer T. ‘Automated Parkinson's disease recognition based on statistical pooling method using acoustic features'. Med. Hypotheses. 2020;vol. 135 doi: 10.1016/j.mehy.2019.109483. [DOI] [PubMed] [Google Scholar]
- 18.Tuncer T., Dogan S., Ozyurt F. An automated Residual Exemplar Local Binary Pattern and iterative ReliefF based corona detection method using lung X-ray image. Chemometr. Intell. Lab. Syst. 2020 doi: 10.1016/j.chemolab.2020.104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuncer T., Akbal E., Dogan S. An automated snoring sound classification method based on local dual octal pattern and iterative hybrid feature selector. Biomed. Signal Process Contr. 2021;63 doi: 10.1016/j.bspc.2020.102173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ojala T., Pietikainen M., Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002;24(7):971–987. [Google Scholar]
- 21.Tuncer T., Dogan S., Özyurt F., Belhaouari S.B., Bensmail H. Novel multi center and threshold ternary pattern based method for disease detection method using voice. IEEE Access. 2020;8:84532–84540. [Google Scholar]
- 22.Yosinski J., Clune J., Bengio Y., Lipson H. Advances in Neural Information Processing Systems. 2014. How transferable are features in deep neural networks? pp. 3320–3328. [Google Scholar]
- 23.Özyurt F. A fused CNN model for WBC detection with MRMR feature selection and extreme learning machine. Soft Comput. 2019:1–10. [Google Scholar]
- 24.Özyurt F. Efficient deep feature selection for remote sensing image recognition with fused deep learning architectures. J. Supercomput. 2019:1–19. [Google Scholar]
- 25.Song Y., et al. medRxiv; 2020. Deep Learning Enables Accurate Diagnosis of Novel Coronavirus (COVID-19) with CT Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng C., et al. medRxiv; 2020. Deep Learning-Based Detection for COVID-19 from Chest CT Using Weak Label. [Google Scholar]
- 27.Butt C., Gill J., Chun D., Babu B.A. Deep learning system to screen coronavirus disease 2019 pneumonia. Appl. Intell. 2020:1. doi: 10.1007/s10489-020-01714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.https://github.com/UCSD-AI4H/COVID-CT
- 29.Jin C., Chen W., Cao Y., et al. MedRxiv; 2020. Development and Evaluation of an AI System for COVID-19 Diagnosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amyar A., Modzelewski R., Ruan S. medRxiv; 2020. Multi-task Deep Learning Based Ct Imaging Analysis for Covid-19: Classification and Segmentation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu B., Gao X., He M., Liu L., Yin G. Proceedings of KDD 2020, New York, NY, USA. 2020. A fast online COVID-19 diagnostic system with chest CT scans. [Google Scholar]
- 32.Wang X., Deng X., Fu Q., et al. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imag. 2020;vol. 39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 33.Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S., Shukla P.K. IRBM; 2020. Deep Transfer Learning Based Classification Model for COVID-19 Disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polsinelli M., Cinque L., Placidi G. rXiv Prepr. arXiv; 2020. A Light CNN for Detecting COVID-19 from CT Scans of the Chest; p. 12837. a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harmon S.A., Sanford T.H., Xu S., et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat. Commun. 2020;vol. 11(1):4080. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barstugan M., Ozkaya U., Ozturk S. ArXiv; 2020. Coronavirus (COVID-19) Classification Using CT Images by Machine Learning Methods.https://arXiv:2003.09424 [Google Scholar]
- 37.Chen J., Wu L., Zhang J., et al. medRxiv; 2020. Deep Learning-Based Model Fordetecting 2019 Novel Coronavirus Pneumonia on Highresolution Computed Tomography: a Prospective Study.https://medRxiv.org/abs/2020.02.25.20021568 [Google Scholar]
- 38.Han Z., Wei B., Hong Y., et al. Accurate screening of COVID-19 using attention based deep 3D multiple instance learning. IEEE Trans. Med. Imag. 2020;vol. 39(8):2584–2594. doi: 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- 39.Hu R., Ruan G., Xiang S., Huang M., Liang Q., Li J. medRxiv; 2020. Automated Diagnosis of COVID-19 Using Deep Learning and Data Augmentation on Chest CT.https://medRxiv.org/abs/2020.04.24.20078998 [Google Scholar]
- 40.Ni Q., Sun Z., Qi L., et al. A deep learning approach to characterize 2019 coronavirus disease (COVID-19) pneumonia in chest CT images. Eur. Radiol. 2020:1–11. doi: 10.1007/s00330-020-07044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S.H., Nayak D.R., Guttery D.S., Zhang X., Zhang Y.D. COVID-19 classification by CCSHNet with deep fusion using transfer learning and discriminant correlation analysis. Inf. Fusion. 2020;68:131–148. doi: 10.1016/j.inffus.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aslan M.F., Unlersen M.F., Sabanci K., Durdu A. CNN-based transfer learning–BiLSTM network: a novel approach for COVID-19 infection detection. Appl. Soft Comput. 2020;98 doi: 10.1016/j.asoc.2020.106912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta A., Gupta S., Katarya R. InstaCovNet-19: a deep learning classification model for the detection of COVID-19 patients using Chest X-ray. Appl. Soft Comput. 2020 doi: 10.1016/j.asoc.2020.106859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armato S.G., McLennan G., Bidaut L., et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med. Phys. 2011;vol. 38(2):915–931. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Depeursinge A., Vargas A., Platon A., Geissbuhler A., Poletti P.A., Müller H. Building a reference multimedia database for interstitial lung diseases. Comput. Med. Imag. Graph. 2012;vol. 36(3):227–238. doi: 10.1016/j.compmedimag.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Zhao J., Zhang Y., He X., Xie P. vol. 13865. arXiv preprint arXiv; 2003. Covid-ct-dataset: a Ct Scandataset about Covid-19. [Google Scholar]
- 47.COVID-19 CT segmentation dataset. http://medicalsegmentation.com/covid19/
- 49.Singh D., Kumar V., Vaishali M.K., Kaur M. “Classification of COVID-19 patients from chest CT images using multiobjective differential evolution–based convolutional neural networks. Eur. J. Clin. Microbiol. Infect. Dis. 2020;vol. 39(7):1379–1389. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chowdhury M.E.H., Rahman T., Khandakar A., et al. vol. 13145. arXiv preprint arXiv; 2003. (Can AI Help in Screening Viral and Covid-19 Pneumonia?). [Google Scholar]
- 51.COVID-19 Database Italian society of medical and interventional radiology (SIRM) https://www.sirm.org/en/category/articles/covid-19-database/
- 52.ChainZ http://www.ChainZ.cn