Abstract
Background
In Marseille, France, following a first severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outbreak in March–May 2020, a second epidemic phase occurred from June, involving 10 new variants. The Marseille-4 variant caused an epidemic that started in August and is still ongoing.
Methods
The 1038 SARS-CoV-2 whole genome sequences obtained in our laboratory by next-generation sequencing with Illumina technology were analysed using Nextclade and nextstrain/ncov pipelines and IQ-TREE. A Marseille-4-specific qPCR assay was implemented. Demographic and clinical features were compared between patients with the Marseille-4 variant and those with earlier strains.
Results
Marseille-4 harbours 13 hallmark mutations. One leads to an S477N substitution in the receptor binding domain of the spike protein targeted by current vaccines. Using a specific qPCR, it was observed that Marseille-4 caused 12–100% of SARS-CoV-2 infections in Marseille from September 2020, being involved in 2106 diagnoses. This variant was more frequently associated with hypoxemia than were clade 20A strains before May 2020. It caused a re-infection in 11 patients diagnosed with different SARS-CoV-2 strains before June 2020, suggesting either short-term protective immunity or a lack of cross-immunity.
Conclusions
Marseille-4 should be considered as a major SARS-CoV-2 variant. Its sudden appearance points towards an animal reservoir, possibly mink. The protective role of past exposure and current vaccines against this variant should be evaluated.
Keywords: SARS-CoV-2, COVID-19, Variant, Marseille-4, Mutations, Spike, Molecular epidemiology
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemic that started in Wuhan, China in December 2019 has spread rapidly around the world (https://coronavirus.jhu.edu/map.html). At the Méditerranée Infection Institute (IHU) in Marseille, routine diagnosis of SARS-CoV-2 by PCR was set up in January 2020 (Lagier et al., 2020, Colson et al., 2020a). The first SARS-CoV-2-infected patient was diagnosed at the IHU on February 27, 2020 (Colson et al., 2020c) (https://www.mediterranee-infection.com/covid-19/). Since then, more than 450 000 SARS-CoV-2 PCR tests have been performed at IHU, 2000 virus isolates have been obtained by cell culture, whole genome sequencing has been performed on 2000 isolates, and care has been given to 14 000 SARS-CoV-2-positive patients.
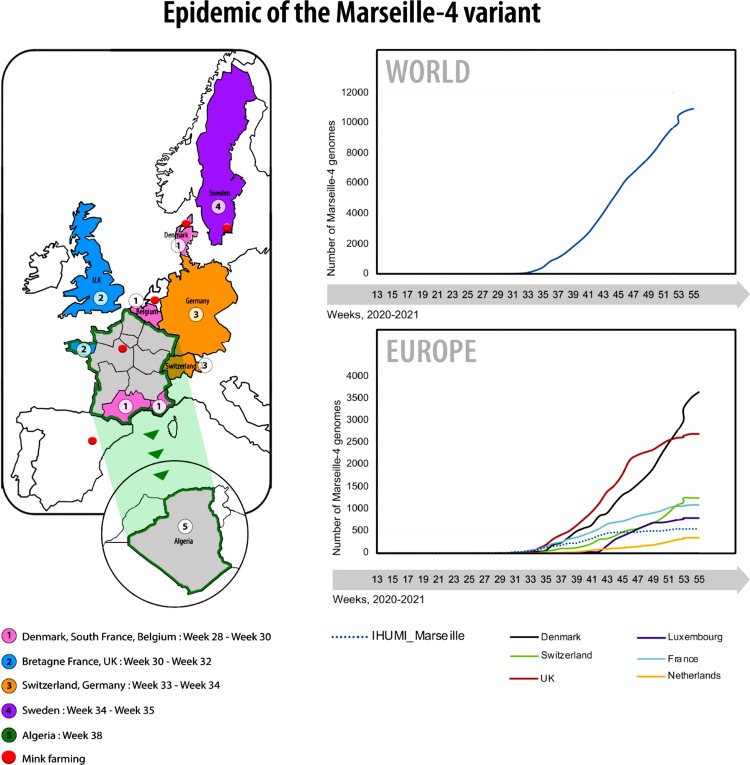
In Europe, SARS-CoV-2 circulation has so far been characterized by two major episodes. The first one, referred to herein as phase 1, started in February and almost ended in May (Colson et al., 2020d; Colson et al., 2021). However, a second phase (phase 2) suddenly occurred at the end of June, exhibiting an atypical epidemic curve, which led us to suspect that the two episodes were caused by distinct viral variants. Hence, whole genome sequencing of SARS-CoV-2 strains was performed over time to characterize their genetic diversity. This enabled us to identify 10 distinct genomic patterns that successively or concomitantly spread in the Marseille area (Colson et al., 2021, Fournier et al., 2021). Of these, two variants were identified at high frequency in the population of individuals diagnosed at the IHU. The Marseille-1 variant caused mild infections in younger patients and predominated from the end of June to the end of July 2020 (Colson et al., 2021). Evidence was accumulated indicating that this variant originated in Africa and was brought to Marseille by ferry boat travellers and sailors from North Africa. In France, it did not spread outside Marseille and it vanished rapidly. On July 29, 2020, a new variant was identified and named Marseille-4 (Figure 1, Figure 2 ). The aim of this study was to examine the virological, clinical, and epidemiological characteristics of this variant.
Figure 1.
Schematic diagram of the evolution of the SARS-CoV-2 Marseille-4 variant in Europe.
Figure 2.
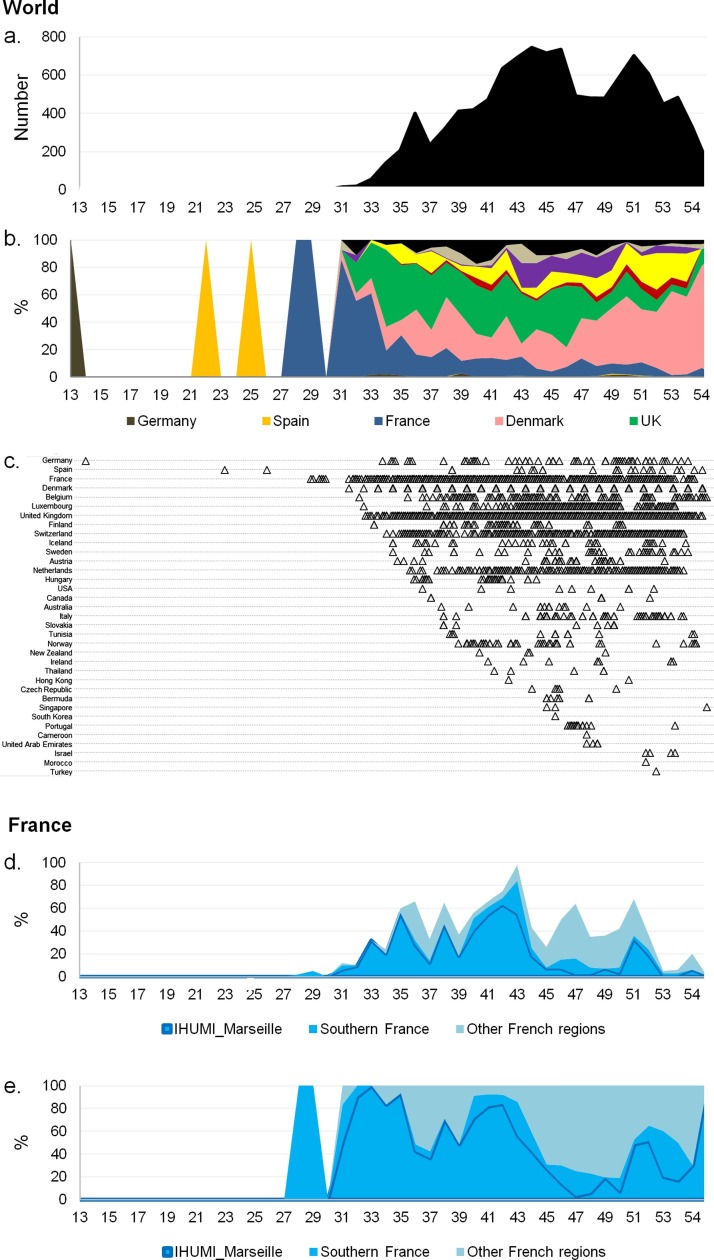
Evolution of the Marseille-4 variant over time. (a) Weekly number of genomes of the Marseille-4 variant worldwide. (b) Weekly frequency normalized to 100% of the countries where genomes of the Marseille-4 variant were obtained. (c) Time distribution of the daily number of genomes of the Marseille-4 variant per country. (d) Weekly number of genomes of the Marseille-4 variant in French regions. (e) Weekly frequency normalized to 100% of the French regions where genomes of the Marseille-4 variant were obtained.
Materials and methods
Genome sequencing
Viral genomes were obtained from nasopharyngeal swab fluid using next-generation sequencing (NGS) and the Illumina Nextera XT paired-end strategy on a MiSeq instrument (Illumina Inc., San Diego, CA, USA), as described previously (Colson et al., 2021). Genome consensus sequences were assembled by mapping on the SARS-CoV-2 genome of GenBank accession number NC_045512.2(Wuhan-Hu-1 isolate) using CLC Genomics workbench v.7, with thresholds of 80% for nucleotide sequence coverage and 90% for nucleotide similarity. SARS-CoV-2 sequences obtained in the study institute have been submitted to the GISAID database (https://www.gisaid.org).
Genome analysis
The 1038 SARS-CoV-2 whole genome sequences obtained in our laboratory were analysed using the Nextclade tool (https://clades.nextstrain.org/) (Hadfield et al., 2018) and an in-house script written in Python. Viral clades were defined on the basis of at least five available genomes sharing the same pattern of mutations. Phylogenetic trees were reconstructed using the nextstrain/ncov tool (https://github.com/nextstrain/ncov) and visualized with Auspice software (https://docs.nextstrain.org/projects/auspice/en/stable/). In addition, the SARS-CoV-2 genomes obtained in our laboratory were integrated into another phylogenetic analysis together with sequences from the GISAID database (https://www.gisaid.org) that were recovered from humans and mink. All of these genomes were aligned using MAFFT v.7 (Katoh and Standley, 2013). Then, phylogeny reconstruction was performed using IQ-TREE software with the GTR Model and 1000 ultrafast bootstrap repetitions (http://www.iqtree.org) (Minh et al., 2020), and the tree was visualized with iTOL (Interactive Tree Of Life) software (https://itol.embl.de/) (Letunic and Bork, 2016).
PCR detection of the SARS-CoV-2 Marseille-4 variant
A qPCR system was designed that targets the nsp4 gene at nucleotide positions 9460–9543 in reference to genome GenBank accession number NC_045512.2(Wuhan-Hu-1 isolate). The primers and probe are described in Supplementary material Table S1. This qPCR was run on an LC480 thermocycler (Roche Diagnostics, Mannheim, Germany). The reaction mixture contained 5 μl of 4X TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific, Grand Island, NY, USA), 0.5 μl of forward primer (10 pmol/μl), 0.5 μl of reverse primer (10 pmol/μl), 0.4 μl of probe (10 pmol/μl), and 8.6 μl of water, and it was completed with 5 μl of extracted viral RNA. PCR conditions were as follows: a reverse transcription step for 10 min at 50 °C, then 20 s at 95 °C followed by 40 cycles comprising a denaturation step at 95 °C for 15 s and a hybridization and elongation step at 60 °C for 60 s.
Comparisons of epidemiological and clinical features of patients diagnosed during phases 1 and 2
The demographic and clinical features of patients infected with the Marseille-4 variant were compared to those of patients infected with clade 20A strains during phase 1, between March and May 2020. Statistical analyses were conducted using R version 4.0.2. (R Core Team, R Foundation for Statistical Computing, Vienna, Austria, 2020; https://www.Rproject.org/).
Results
Identification and circulation of the Marseille-4 variant
The highly transmissible SARS-CoV-2 Marseille-4 variant identified in Marseille at the end of July 2020 rapidly became predominant, reaching 100% of identified viral strains in the geographical area on November 2, 2020. Using genome sequences available through the GISAID database (https://www.gisaid.org/), the outbreaks of this variant were traced back in different countries. The first case of infection with the Marseille-4 variant, named 20A.EU2 in the Nextstrain classification (https://clades.nextstrain.org/) (Hodcroft et al., 2020), was detected in a German patient on March 24, 2020. Then, two cases were detected on a Balearic island, Spain, on May 29 and June 18, 2020. Additional cases were detected in Southwestern France from July 9, then in Denmark, and from August 1 in other European countries and other regions of France (Figure 1, Figure 2; Supplementary material Figure S1). The Marseille-4 variant was detected from September in North America (Canada, then USA), Australia, and New Zealand, from October in Asia (Thailand, Hong Kong, Singapore, and South Korea) and Africa (Tunisia and Morocco), and from December in Israel. In Marseille, 269 Marseille-4 complete genomes were sequenced from infected patients, and a Marseille-4-specific qPCR (Supplementary material Table S1) was designed that enabled rapid identification of an additional 1579 cases. Overall, this variant caused 2106 cases and accounted for about two-thirds of all SARS-CoV-2 viruses tested from September 2020 to January 2021 at IHU.
Genomic features
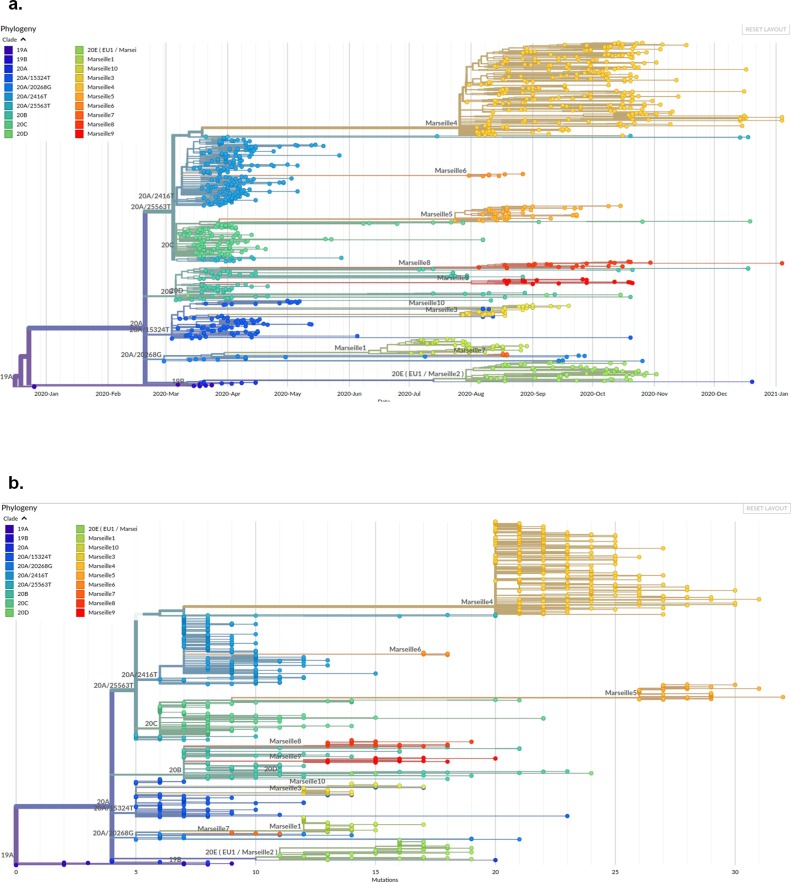
The Marseille-4 variant evolved from clade 20A strains (Figure 3 ) and is characterized by a combination of 20 mutations compared to the Wuhan-Hu-1 strain. Among these mutations, 13 are hallmarks of this variant (C4543T, G5629T, G9526T, C11497T, G13993T, G15766T, A16889G, G17019T, G22992A, C25710T, T26876C, G28975C, and G29399A) (Supplementary material Figure S2). The Marseille-4 variant was provisionally subdivided into 11 subgroups (Marseille-4-A1 to Marseille-4-J), with a genetic drift ranging from 21 to 24 mutations compared to the Wuhan-Hu-1 strain (Table 1 ). Strikingly, comparative genomics showed that the set of 13 hallmark mutations appeared altogether. They are losses of a G in seven cases and of a C in three cases, and are scattered along the viral genome. Seven (46%) are non-synonymous mutations, including two located in the RNA-dependent RNA polymerase (RdRP) (nsp12; A176S and V767L), two in the NTPase/helicase (nsp13; K1141R and E1184D), two in the nucleocapsid (N; M234I and A376T), and one in the spike glycoprotein (S; S477N). Fifteen additional mutations were observed in ≥5 viral genomes obtained in the study institute (C222U, C503U, G2600U, A2647G, C8937U, G18105U, C23191U, G25534U, U26442C, G26720U, G27877U, C27942U, G28086U, G29701A, and G29511U). Overall, 283 nucleotide positions were found to be mutated in ≥1 Marseille-4 genomes, mostly in the nsp3 and S genes. They were most frequently C > U (36%), G > U (25%), U > C (8%), G > A (6%), and A > G (5%) mutations, and U>− deletions (6%). Phylogenetically, the Marseille-4 variant was found to fall within a group of viruses from Europe only (Supplementary material Figure S3).
Figure 3.
Genome sequence-based phylogenetic trees showing the evolution of SARS-CoV-2 Marseille-4 variant strains. (a) Time-scale phylogenetic tree. (b) Phylogenetic tree based on mutational events.
Full-length genome sequences obtained in this study were compared to those available in the GISAID database (https://www.gisaid.org/). Phylogenetic trees were reconstructed and visualized using the Nextstrain pipeline (https://github.com/nextstrain/ncov/) (Hadfield et al., 2018).
Table 1.
Nucleotide mutations and amino acid substitutions in the genomes of SARS-CoV-2 Marseille-4 variant.
 |
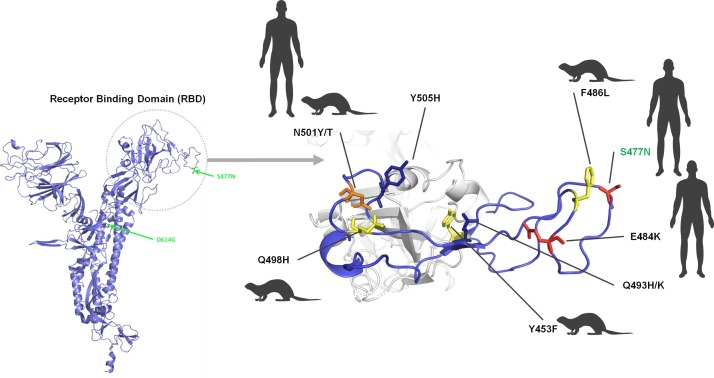
The Marseille-4 variant harbours the S477N substitution within the receptor binding domain (RBD) of the spike glycoprotein. This RBD attaches the virion to the cell membrane by binding to the viral receptor ACE2, and mediates viral entry (Lan et al., 2020). The spike is a major target of neutralizing antibodies (Barnes et al., 2020) and the current vaccines (Dai and Gao, 2020) (Figure 4 ). The S477N substitution has been reported to be associated with broad resistance to monoclonal neutralizing antibodies (Liu et al., 2020). These data could explain the lack of resistance to infection by this Marseille-4 variant among people previously infected with different strains that circulated earlier, during the first phase of the 2020 pandemic. This substitution lies between substitutions observed in viruses infecting humans and others seen in viruses infecting mink (Figure 4) (Garry, 2021). It adds to the D614G substitution that was reported to increase the stability of spike trimers and to confer greater affinity for ACE2 (Korber et al., 2020). It is worth noting that the first genome available in the GISAID database (EPI_ISL_7079562020-03-24), originating from Germany on March 24, 2020, does not harbour this S477N substitution, which may explain why it did not apparently spread further. Other critical mutations may be substitution Q57H in ORF3a, a viroporin that forms ion channels and was reported as required for viral replication, virulence, and release, and is also predicted to be a pro-apoptotic protein (Bianchi et al., 2021, Law et al., 2005), and substitutions A176S in the RdRP and K1141R and E1184D in the NTPase/helicase.
Figure 4.
Three-dimensional structure of the spike protein showing the amino acid substitutions in the receptor-binding motif of the Marseille-4 variant and of other variants detected in humans and/or mink.
The structure was predicted using the Phyre2 web portal (http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index) (Kelley et al., 2015) and visualized using the Pymol tool v.1.8 (https://pymol.org/2/) (Janson and Paiardini, 2020). Amino acids where a substitution was observed in humans are shown in red, those where a substitution was observed in mink are shown in yellow, and those where a substitution was observed in humans and mink are shown in orange.
In search of the origin of the Marseille-4 variant
The origin of the Marseille-4 variant is currently unknown. It emerged abruptly with its block of specific mutations, with no known intermediate form, at a time when the SARS-CoV-2 epidemic had almost ended in France and Europe (Figure 1, Figure 2, Figure 3). This apparently discontinuous evolution of SARS-CoV-2 genomes is abnormal, particularly if we consider that after its first detection this variant showed a subsequent mutation rate similar to that of other lineages (e.g., mutation in the RdRP did not alter the polymerase fidelity). Although the existence of a missing intermediate that has not so far been sequenced from coronavirus disease 2019 (COVID-19) patients cannot be excluded, this could also suggest that there is an overlooked reservoir in which the virus was submitted to a selection pressure that favoured a particular increase in mutation accumulation.
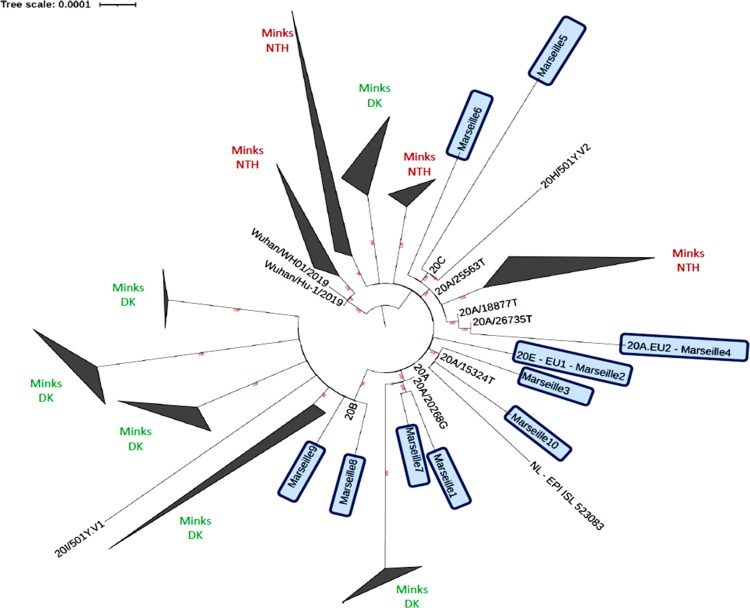
Interestingly, among the 10 516 sequences from the Marseille-4 variant in the GISAID database (on January 24, 2021), the 272 genomes from our laboratory had close relatives with those originating from Northern Europe, mostly Denmark (n = 3366), the UK (n = 2652), and Switzerland (n = 1147) (Supplementary material Figure S1). A phylogenetic tree was constructed that included genomes from mink and human SARS-CoV-2 strains. Mink strains were divided into four and six main groups for the samples from the Netherlands and Denmark, respectively (Figure 5 ). A common phylogenetic node between mink strains, the Marseille-4, Marseille-5, Marseille-6 variants, and the 20 H/501Y.V2 variant was observed. This node pointed to the common mutation Q57H in ORF3a described above.
Figure 5.
Phylogenetic tree based on SARS-CoV-2 full-length genomes.
A total of 744 genomes of SARS-CoV-2 were integrated in a phylogenetic analysis. All genomes were aligned using MAFFT version 7 (Katoh and Standley, 2013). The phylogenetic tree was reconstructed using IQ-TREE with the GTR model, with 1000 ultrafast bootstrap repetitions (Minh et al., 2020), and was visualized with iTOL (Interactive Tree Of Life; https://itol.embl.de/) (Letunic and Bork, 2016). DK, Denmark; NTH, The Netherlands.
The rapid emergence of the Marseille-4 variant during the summer of 2020, after the end of the first epidemic phase, may point towards an animal reservoir. Mink farms were identified as reservoirs and sources of SARS-CoV-2 mutants in the Netherlands in April (Oude Munnink et al., 2021) and in Denmark in June 2020 (Hammer et al., 2021). In France, one of the four mink farms was infected and animals were culled. SARS-CoV-2 is an epizootic agent that caused an outbreak in humans before being transferred to mink in which it spread rapidly through densely caged animals and subsequently became a source for human infection. To date, more than 800 human infections from mink have been reported (Oude Munnink et al., 2021). One hypothesis could be that a human SARS-CoV-2 from infected caregivers infected mink, then the frequency of viral mutations changed in the mink due to a different host selection pressure, and this mink-adapted virus (with multiple mutations) became a new viral source to infect humans.
The genome obtained from a German patient sampled on March 24, 2020 (EPI_ISL_7079562020-03-24) is atypical as it is devoid of the S477N substitution, one of the Marseille-4 hallmark mutations, but harbours more mutations (n = 31) than the other Marseille-4 strains, including in the Nsp2, Nsp3, S, and N proteins, and in ORF1b, particularly the Nsp14 exonuclease, which has proofreading activity (Shannon et al., 2020). The evolutionary relationships of this genome with other Marseille-4 genomes warrants further investigation with the availability of other genomes obtained from samples collected during the same period.
Clinical findings: the Marseille-4 variant may escape immunity conferred by a first SARS-CoV-2 infection
Compared to the clade 20A strains that predominated during phase 1 between March and May 2020, the Marseille-4 variant was associated with a lower frequency of cough, rhinitis, and olfactory and gustatory disorders (Table 2 ). By contrast, hypoxemia was more frequent in patients infected with the Marseille-4 variant. It has been reported that differences observed in COVID-19 severity may in part be associated with the dysfunction of cellular immune responses to SARS-CoV-2 and/or a weakness of the neutralizing humoral response (Moderbacher et al., 2020).
Table 2.
Demographics, outcomes, and clinical symptoms in patients infected with different SARS-CoV-2 variants.
| Demographics and outcomes (N = 759) | 20A (n = 339) |
Marseille-4 (n = 420) |
P-valuea | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Sex | 0.059 | ||||
| Female | 188 | 55.5 | 204 | 48.6 | |
| Male | 151 | 44.5 | 216 | 51.4 | |
| Age (years), mean ± SD | 50.2 ± 22.3 | 48.9 ± 23.1 | 0.41 | ||
| Hospitalization | 53 | 15.6 | 68 | 16.2 | 0.835 |
| Transfer to intensive care unit | 5 | 1.5 | 10 | 2.4 | 0.44 |
| Death | 10 | 2.9 | 16 | 3.8 | 0.52 |
| Symptoms (N = 444) | 20A (n = 254) |
Marseille-4 (n = 190) |
P-valuea | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Cough | 123 | 48.4 | 73 | 38.4 | 0.036* |
| Rhinitis | 106 | 41.7 | 37 | 19.5 | <0.0001* |
| Anosmia | 76 | 29.9 | 35 | 18.5 | 0.006* |
| Ageusia | 71 | 27.9 | 34 | 18.0 | 0.015* |
| Dyspnoea | 72 | 28.3 | 42 | 22.1 | 0.136 |
| SpO2 <96% | 37 | 14.6 | 42 | 22.1 | 0.04* |
SD, standard deviation; SpO2, oxygen saturation.
Chi-square test or Fisher’s exact test for qualitative variables; Student t-test for quantitative variables.
Statistically significant.
We diagnosed two successive cases of COVID-19, separated by more than 4 months, in 11 patients. The first infection was diagnosed before June 2020 when Marseille-4 was not circulating in Marseille (Colson et al., 2020b, Brouqui et al., 2021), and genomic or qPCR (one and 10 patients, respectively) confirmation that the second episode was caused by the Marseille-4 variant was obtained. This suggests either short protective immunity (only a few weeks or months), as observed previously with seasonal coronaviruses (Edridge et al., 2020), or a lack of cross-immunity between different SARS-CoV-2 variants, allowing Marseille-4 to evade immune protection elicited by another earlier variant. This may be related to the S477N mutation, which could change the affinity of RBD for ACE2 and decrease the sensitivity of the variant virus to anti-RBD-specific neutralizing antibodies (Andreano et al., 2020).
Discussion
The recent evolution of the SARS-CoV-2 epidemics reflects the generation of new variants in different ecosystems that have spread with globalization and have replaced the original variants arising from Wuhan. Some can be associated with different clinical features, as in the case of the Marseille-4 variant. The ecosystems allowing this selection may consist of human groups isolated for a while, or animal reservoirs such as mink in large farms. Large concentrations of farmed mink have been infected by human SARS-CoV-2 (Oude Munnink et al., 2021). Under these conditions, sub-speciation may occur (Darwin, 1859). The re-connection of isolated ecosystems (either countries and/or farmed animals) where different variants have developed, has generated new outbreaks in countries that were exposed to incoming populations such as travellers.
Several reasons led us to believe that mink were the source of the Marseille-4 variant. First, this variant carries a new set of several mutations that seems to have appeared suddenly based on the analysis of all the genomes available worldwide, and not gradually. This suggests that this brutal genome evolution was overlooked. Secondly, there was no SARS-CoV-2 epidemic in France at the time of the emergence of this variant, except in a region near the city of Laval (Mayenne, Western France) located between the most dense area for wild mink (Brittany) and a mink farm (Eure-et-Loire) where 30% of mink were proved to be SARS-CoV-2-positive by qPCR and 97% had antibodies against the virus. As a consequence, the entire mink population of the farm was slaughtered (https://www.plateforme-esa.fr/article/covid-19-et-animaux-mise-a-jour-au-05-01-2021) (Fenollar et al., 2021). Progressively, this SARS-CoV-2 epidemic spread in France during the summer, and we observed the first cases of Marseille-4 infections in Marseille when French tourists arrived in our region. For unknown reasons, the sequence of the virus obtained from the farmed mink infected in mid-November is not yet available.
In conclusion, overall we believe that the segregation of viral strains in isolated geographical areas and in animal reservoirs may contribute to explain the differences observed among the epidemic curves around the world. This would help to understand the mechanism of the second episode of SARS-CoV-2 circulation that developed in Marseille, initially caused by an African variant that disappeared (Colson et al., 2021), and then by emerging new variants linked to different areas of Europe, including those hosting huge mink farms. Finally, the role of the treatments of COVID-19 with remdesivir or hyperimmune plasma (Choi et al., 2020, Kemp et al., 2020) in generating and selecting variants should be considered as they may also have contributed to the new outbreaks observed in the most developed countries.
Since the final acceptance of this article, the sequence of the SARS-CoV-2 genome obtained from a farm mink sampled the 15th of November, 2020 in Eure-et-Loire was eventually released the 29th of March, 2021 (EPI_ISL_1392906). As we suspected and stated in the present article, this genome is strictly identical to the genome of a Marseille-4 variant confirming our hypothesis of a common source of this variant between French minks and humans.
Funding
This work was supported by the French Government under the “Investments for the Future” program managed by the National Agency for Research (ANR), Méditerranée-Infection 10-IAHU-03, and was also supported by Region Provence-Alpes-Côte d’Azur and European funding FEDER PRIMMI (Fonds Européen de Developpement Regional-Plateformes de Recherche et d’Innovation Mutualisées Mediterranée Infection), FEDER PA 0000320 PRIMMI.
Ethical approval
The study was approved by the Ethics Committee of the Méditerranée Infection Institute (Reference No. 2020-016-3).
Availability of data and materials
Data underlying the study are available from the GISAID database (https://www.gisaid.org/) or from the corresponding author upon request.
Conflict of interest
The authors have no conflicts of interest to declare. Funding sources had no role in the design and conduct of the study, in the collection, management, analysis, and interpretation of the data, or in the preparation, review, and approval of the manuscript.
Author contributions
Conceived and designed the experiments: DR, PEF, PC and PG. Contributed materials/analysis tools: PEF, PC, AL, CD, PG, MB, JD, LB, LP, JCL and FF. Analysed the data: PEF, PC, AL, PG, JD, JCL, FF and DR. Wrote the paper: PEF, PC, CD, PG and DR. All authors approved the final version of the manuscript.
Acknowledgements
We are grateful to Olivia Ardizzoni, Vincent Bossi, Madeleine Carrera, Vera Esteves-Vieira, Laurence Thomas, Priscilla Jardot, and Raphael Tola for their technical help, and to Audrey Giraud-Gatineau and Léa Luciani for their help with the data analysis. The manuscript text has been edited by a native English speaker.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2021.03.068.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Andreano E., Piccini G., Licastro D., Johnson N.V., Paciello I., Monego S.D. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M., Borsetti A., Ciccozzi M., Pascarella S. SARS-Cov-2 ORF3a: mutability and function. Int J Biol Macromol. 2021;170:820–826. doi: 10.1016/j.ijbiomac.2020.12.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouqui P., Colson P., Melenotte C., Houhamdi L., Bedotto M., Devaux C. COVID-19 re-infection. Eur J Clin Invest. 2021;(March) doi: 10.1111/eci.13537. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X. Persistence and evolution of SARS-CoV-2 in an immunocompromised Host. N Engl J Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Esteves-Vieira V., Giraud-Gatineau A., Zandotti C., Filosa V., Chaudet H. Temporal and age distributions of SARS-CoV-2 and other coronaviruses, southeastern France. Int J Infect Dis. 2020;101:121–125. doi: 10.1016/j.ijid.2020.09.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Finaud M., Levy N., Lagier J.C., Raoult D. Evidence of SARS-CoV-2 re-infection with a different genotype. J Infect. 2020;(November) doi: 10.1016/j.jinf.2020.11.011. S0163-4453(20)30706-4. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Lagier J.C., Baudoin J.P., Bou K.J., La Scola B., Raoult D. Ultrarapid diagnosis, microscope imaging, genome sequencing, and culture isolation of SARS-CoV-2. Eur J Clin Microbiol Infect Dis. 2020;39:1601–1603. doi: 10.1007/s10096-020-03869-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Levasseur A., Delerce J., Chaudet H., Bossi V., Ben Khedher M. IHU Pre-prints; 2020. Dramatic increase in the SARS-CoV-2 mutation rate and low mortality rate during the second epidemic in summer in Marseille. [DOI] [Google Scholar]
- Colson P., Levasseur A., Gautret P., Fenollar F., Hoang V.T., Delerce J. Introduction into the Marseille geographical area of a mild SARS-CoV-2 variant originating from sub-Saharan Africa. Travel Med Infect Dis. 2021;40 doi: 10.1016/j.tmaid.2021.101980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2020:1–10. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. John Murray; London: 1859. On the origin of species. [Google Scholar]
- Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K. Seasonal coronavirus protective immunity is short-lasting. Nat Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Fenollar F., Mediannikov O.Y., Maurin M., Devaux C.A., Colson P., Levasseur A. Mink, SARS-CoV-2, and the human-animal interface. Front Microbiol. 2021;(April) doi: 10.3389/fmicb.2021.663815. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier P.E., Colson P., Levasseur A., Gautret P., Bedotto M., Filosa V. IHU Pre-prints; 2021. Genome sequence analysis enabled deciphering the atypical evolution of COVID-19 epidemics in Marseille, France. [DOI] [Google Scholar]
- Garry R.F. Virological org; 2021. Mutations arising in SARS-CoV-2 spike on sustained human-to-human transmission and human-to-animal passage.https://virological.org/t/mutations-arising-in-sars-cov-2-spike-on-sustained-human-to-human-transmission-and-human-to-animal-passage/578 [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer A.S., Quaade M.L., Rasmussen T.B., Fonager J., Rasmussen M., Mundbjerg K. SARS-CoV-2 transmission between Mink (Neovison vison) and humans, Denmark. Emerg Infect Dis. 2021;27:547–551. doi: 10.3201/eid2702.203794. Epub 2020 November 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft E.B., Zuber M., Nadeau S., Comas I., Gonzalez Candelas F., SeqCOVID-SPAIN Consortium Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. medRxiv. 2020 doi: 10.1101/2020.10.25.20219063. [DOI] [Google Scholar]
- Janson G., Paiardini A. PyMod 3: a complete suite for structural bioinformatics in PyMOL. Bioinformatics. 2020;(October):btaa849. doi: 10.1093/bioinformatics/btaa849. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp S.A., Collier D.A., Datir R., Ferreira I., Gayed S., Jahun A. Neutralising antibodies in Spike mediated SARS-CoV-2 adaptation. medRxiv. 2020 doi: 10.1101/2020.12.05.20241927. [DOI] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier J.C., Million M., Gautret P., Colson P., Cortaredona S., Giraud-Gatineau A. Outcomes of 3,737 COVID-19 patients treated with hydroxychloroquine/azithromycin and other regimens in Marseille, France: a retrospective analysis. Travel Med Infect Dis. 2020;36 doi: 10.1016/j.tmaid.2020.101791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Law P.T.W., Wong C.H., Au T.C.C., Chuck C.P., Kong S.K., Chan P.K.S. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J Gen Virol. 2005;86:1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv. 2020 doi: 10.1101/2020.11.06.372037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von H.A. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moderbacher C.R., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A., Le N.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.C. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 exonuclease active-sites. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104793.:104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data underlying the study are available from the GISAID database (https://www.gisaid.org/) or from the corresponding author upon request.