Abstract
Angiopoietin-like 3 (ANGPTL3) plays an important role in lipid metabolism in humans. Loss-of-function variants in ANGPTL3 cause a monogenic disease named familial combined hypolipidemia. However, the potential contribution of ANGPTL3 gene in subjects with familial combined hyperlipidemia (FCHL) has not been studied. For that reason, the aim of this work was to investigate the potential contribution of ANGPTL3 in the aetiology of FCHL by identifying gain-of-function (GOF) genetic variants in the ANGPTL3 gene in FCHL subjects. ANGPTL3 gene was sequenced in 162 unrelated subjects with severe FCHL and 165 normolipemic controls. Pathogenicity of genetic variants was predicted with PredictSNP2 and FruitFly. Frequency of identified variants in FCHL was compared with that of normolipemic controls and that described in the 1000 Genomes Project. No GOF mutations in ANGPTL3 were present in subjects with FCHL. Four variants were identified in FCHL subjects, showing a different frequency from that observed in normolipemic controls: c.607-109T>C, c.607-47_607-46delGT, c.835+41C>A and c.*52_*60del. This last variant, c.*52_*60del, is a microRNA associated sequence in the 3′UTR of ANGPTL3, and it was present 2.7 times more frequently in normolipemic controls than in FCHL subjects. Our research shows that no GOF mutations in ANGPTL3 were found in a large group of unrelated subjects with FCHL.
Subject terms: Disease genetics, Genetic predisposition to disease
Introduction
Angiopoietin-like 3 (ANGPTL3) is a 70 kDa-secreted (54 kDa before glycosylation) protein, mainly expressed in the liver, discovered by Conklin et al. in 19991. ANGPTL3 is an endogenous inhibitor of lipoprotein lipase (LPL) and endothelial lipase (EL)2,3. Different studies in families with hypolipemia and in general population have reported that loss-of-function (LOF) variants in ANGPTL3 gene are associated with decreased plasma levels of triglycerides (TG), low-density lipoprotein cholesterol (LDLc) and high-density lipoprotein cholesterol (HDLc)4. The N-terminal domain of ANGPTL3 containing residues from 17 to 207 is responsible for the increased plasma TG levels in mice. Loss of this region prevents the inhibition of LPL5 and EL3 by ANGPTL3. Recently, the inhibition of ANGPTL3 with a human monoclonal antibody against ANGPTL3 (evinacumab) in dyslipidemic mice and in healthy volunteers caused a dose-dependent placebo-adjusted reduction in fasting TG levels of up to 76% and LDLc levels of up to 23%4. Therefore, ANGPTL3 has been considered a potent modulator of TG2 and supports an important role of ANGPTL3 in lipid metabolism in humans.
In addition, new evidence sustains a possible role of ANGPTL3 in the progression of atherosclerosis through a lipid-independent mechanism6. Carriers of LOF mutations in ANGPTL3 associated a 34% decrease in cardiovascular events7 and ANGPTL3 plasma concentration was associated with arterial wall thickness in humans8. Moreover, a decreased expression of ANGPTL3 in apolipoprotein E null (apoE-/-) mice was protective in the development of atherosclerosis9.
Familial combined hyperlipidemia (FCHL) is a common and complex inherited disorder of lipid metabolism with important environmental influences10. FCHL is characterized by elevated very low-density lipoprotein (VLDL) and/or LDL concentrations, low HDLc levels11, and frequently, reduced LPL activity12. The FCHL genetic background is mostly polygenic and associated with the variation in at least 35 different genes, including genes related to metabolic disorders such as obesity, peripheral insulin resistance, type 2 diabetes, hypertension and metabolic syndrome13,14. However, FCHL is a genetically heterogeneous syndrome and monogenic and oligogenic cases have been also described15–17. Subjects with FCHL have high predisposition to develop premature cardiovascular disease (CVD). Actually, FCHL is the most common genetic lipid abnormality found in subjects with premature coronary heart disease18. The FCHL phenotype is quite similar to that observed after ANGPTL3 administration in mice. However, the potential involvement of the ANGPTL3 gene in FCHL has not been previously analysed in contrast with the major role of a loss-of-function mutation in ANGPTL3 in the opposite situation, familial combined hypolipidemia19,20. Therefore, the aim of this study was to identify gain-of-function (GOF) genetic variants in ANGPTL3 gene in FCHL subjects and to establish the potential contribution of ANGPTL3 in the aetiology of FCHL.
Material and methods
Subjects
Cases
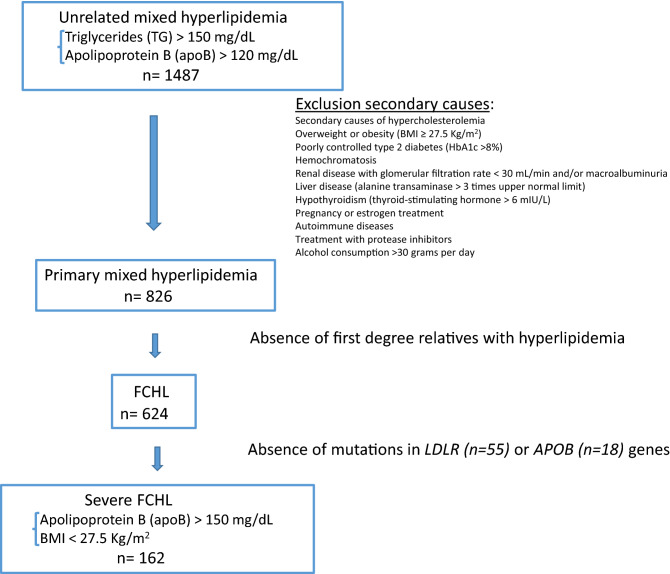
A total of 162 unrelated subjects, aged 23 to 82, with the clinical diagnosis of severe FCHL from Lipid Unit at Hospital Universitario Miguel Servet, Zaragoza, Spain, were selected for this study. Severe FCHL included: LDLc and TG > 90th percentile adjusted for age and sex, apolipoprotein B (apoB) > 150 mg/dL, body mass index (BMI) < 27.5 kg/m2 and at least one first-degree family member with mixed hyperlipidemia. Clinical exclusion criteria were: secondary causes of hypercholesterolemia including significant overweight or obesity (BMI ≥ 27.5 kg/m2), poorly controlled type 2 diabetes (HbA1c > 8%), hemochromatosis, renal disease with glomerular filtration rate < 30 mL/min and/or macroalbuminuria, liver disease (alanine transaminase > 3 times upper normal limit), hypothyroidism (thyroid-stimulating hormone > 6 mIU/L), pregnancy or estrogen treatment, autoimmune diseases, treatment with protease inhibitors and alcohol consumption > 30 g per day (Fig. 1).
Figure 1.
Flow chart of subject selection process.
Most of the subjects included in this work had been studied previously to discard severe genetic defects in the genes regulating the LPL pathway21. Subjects with LDLR, APOB or PCSK9 functional mutations causing familial hypercholesterolemia (FH) and subjects with dysbetalipoproteinemia and the APOE2/2 genotype were excluded from the study. The lipid phenotype of FH and dysbetalipoproteinemia may overlap with FCHL and with this approach both genetic hyperlipidemias were ruled out to avoid confusion with FCHL.
Controls
We selected 165 consecutive normolipemic, unrelated subjects, aged 20–79, who underwent a medical visit at our hospital as control group. Exclusion criteria for control subjects were personal or parental history of premature cardiovascular disease (before 55 years in men and 65 years in women) or personal or parental dyslipidaemia, current acute illness, or use of drugs that might influence glucose or lipid metabolism.
In all subjects, clinical and analytical variables were registered, including personal and familial risk factors, history of cardiovascular disease and intake of drugs affecting intestinal or lipid metabolism.
All experimental protocols were approved by our local ethical committee (Comité Ético de Investigación Clínica de Aragón, CEICA, Zaragoza, Spain). Informed consent was obtained from all subjects before participating in the protocol. Samples from patients included in this study were provided by the Biobank of the Aragon Health System (PT17/0015/0039), integrated in the Spanish National Biobanks Network, and they were processed following standard operating procedures with the appropriate approval of the Ethics and Scientific Committees.
Biochemical analysis
Ethylenediaminetetraacetic acid (EDTA) plasma and serum samples were collected from all participants after at least 10 h fasting, without lipid-lowering drugs for > 5 weeks, to obtain baseline biochemical characteristics. Total cholesterol (TC) and TG measurements were performed with commercially available diagnostic kits (Boehringer Mannheim, Germany), in a laboratory participating in a lipid standardisation programme. HDLc was measured directly by an enzymatic reaction using cholesterol oxidase (UniCel DxC 800; Beckman Coulter Inc., Brea, California, USA). ApoA1, apoB and lipoprotein(a)22 were determined by IMMAGE kinetic immunonephelometry (Beckman Coulter Inc., Brea, California, USA). LDLc was calculated using the Friedewald’s formula23. All methods were carried out in accordance with guidelines and regulations of Spanish Society of Clinical Biochemistry.
Genetic analysis
DNA was isolated from EDTA blood samples using the KingFisher Duo Prime System (Thermo Fisher Scientific). A previously described protocol for sequencing the exon 4 of APOE gene24 was used for disclosing APOE2/2 genotype or functional mutations in exon 4 of the APOE gene in order to rule out carrier subjects. Moreover, LDLR, APOB and PCSK9 genes were analysed for functional mutations with Lipochip platform (Progenika Grifols, Spain)25 in order to rule out subjects with any pathogenic mutation in these genes.
ANGPTL3 gene (NM_014495.4) was amplified in 7 fragments by polymerase chain reaction with primers showed in Table 1. Each amplified fragment comprised the corresponding exon and its 5′ and 3′ flanking sequences, including intron–exon boundaries. After purification with ExoSap-IT (USB), amplified fragments were sequenced by the Sanger method26 using the BigDye 3.1 sequencing kit (Applied Biosystems) in an automated ABI 3500xL sequencer (Applied Biosystems). DNA sequences were analysed using Variant Reporter software (Applied Biosystems).
Table 1.
Primers and conditions used for ANGPTL3 amplification and sequencing.
| ANGPTL3 | Primer sequence 5′ → 3′ | Annealing temperature (°C) | Product size (bp) |
|---|---|---|---|
| Fragment 1 | F: CCTTACCTTTTCTGGGCAA | 51.5 | 821 |
| R: AAATGCAAATTTTCAGTGTTTTCA | |||
| Fragment 2 | F: GCTGGGCTTTTTCTTTTAATTG | 51 | 496 |
| R: CTTCAGAGCCTGCAATTTT | |||
| Fragment 3 | F: CCGACCAATGTCTGCTTTTT | 51 | 555 |
| R: TCAAGTCCATATTTGTATTTCTCTG | |||
| Fragment 4 | F: TCCAGACTGGTGATAGAACAAG | 53.5 | 597 |
| R: GGCAATTAATGAATTTTGGCATAGT | |||
| Fragment 5 | F: TCTCCTTTTCCTCTAAAATAATCTGAA | 52.5 | 596 |
| R: TGATCATTGTAAGCCGTGG | |||
| Fragment 6 | F: ATGCATTATAGAAAGGATAATCAGACT | 52.5 | 700 |
| R: GAGGAAGATTAGAGGTAAAATACCTG | |||
| Fragment 7 | F: ACCTCTAATCTTCCTCAGATTTTC | 51 | 599 |
| R: TTTTGATTGAGAAATGTAAACGGTA |
Each amplified fragment comprises the corresponding exon and its 5′ and 3′ flanking sequences, including intron–exon boundaries.
F forward, R reverse.
To evaluate the pathogenicity of new identified genetic variants, we used PredictSNP227. The effect of variants in potential splicing sites was predicted with FruitFly28. To compare the frequency of identified variants with that of the general population, we compiled the allele frequencies of identified variants from the 1000 Genomes Project29 and genome aggregation data base (gnomAD)30. ClinVar database was used for additional information about genomic variation and its relationship to human health31. Finally, information about microRNAs was obtained from PolymiRTS Database 3.032. All methods were carried out in accordance with guidelines and regulations of Spanish Society of Human Genetics.
Statistical analysis
Analyses were performed using statistical computing software R version 3.5.033. The level of significance was set at P < 0.05. The distribution of the variables was analysed by the Shapiro test. Quantitative variables with a normal distribution were expressed as mean ± standard deviation and were analysed by the Student t test. Variables with a skewed distribution were expressed as medians and interquartile ranges and were analysed with the Mann–Whitney U test. Qualitative variables were expressed as percentages and were analysed by the Chi squared test.
Results
Study subjects
The main clinical and biochemical characteristics of both studied groups (162 FCHL subjects and 165 normolipemic controls) are presented in Table 2. FCHL subjects showed higher predominance of males (60.5%) and were significantly older than normolipemic subjects (P = 0.022 and P < 0.001, respectively). Compared with normolipemic controls, FCHL subjects had significantly higher values of BMI, TC, TG, LDLc, apoB and lipoprotein(a) (P < 0.001, P < 0.001, P < 0.001, P < 0.001, P < 0.001 and P = 0.003, respectively). FCHL subjects presented higher prevalence of hypertension, type 2 diabetes and CVD than normolipemic subjects (P = 0.009, P = 0.001 and P = 0.016, respectively). The APOE genotype distribution was homogenous between both cohorts, being E3/3 genotype the most frequent in both groups, although E3/2 genotype had a lower frequency in FCHL subjects (5.56%) in contrast to normolipemic subjects (15.2%).
Table 2.
Clinical and biochemical characteristics in FCHL subjects and normolipemic controls.
| FCHL subjects n = 162 | Normolipemic controls n = 165 | p | |
|---|---|---|---|
| Men, n (%) | 98 (60.5) | 78 (47.3) | 0.022 |
| Age (years) | 50.4 ± 11.4 | 38.5 ± 14.7 | < 0.001 |
| Body Mass Index (kg/m2) | 25.6 (24.2–26.5) | 23.6 (21.4–26.6) | < 0.001 |
| Total Cholesterol (mg/dL) | 312 ± 36.1 | 170 ± 21.0 | < 0.001 |
| Triglycerides (mg/dL) | 277 (232–373) | 64.0 (49.0–93.0) | < 0.001 |
| LDL cholesterol (mg/dL) | 204 (183–230) | 108 (91.8–117) | < 0.001 |
| HDL cholesterol (mg/dL) | 48.5 ± 12.0 | 55.7 ± 11.4 | 0.015 |
| Apolipoprotein A1 (mg/dL) | 147 ± 25.0 | 147 ± 27.4 | 0.930 |
| Apolipoprotein B (mg/dL) | 167 (165–190) | 83.0 (72.0–91.0 | < 0.001 |
| Lipoprotein(a), (mg/dL) | 39.1 (10.3–80.8) | 16.2 (7.79–44.5) | 0.003 |
| Glucose (mg/dL) | 93.0 (86.0–103) | 85.0 (80.0–92.0) | < 0.001 |
| HbA1c (%) | 5.50 (5.30–5.80) | 5.20 (5.00–5.40) | < 0.001 |
| Type 2 diabetes, n (%) | 13 (8.02) | 2 (1.21) | 0.009 |
| Hypertension, n (%) | 30 (18.5) | 10 (6.06) | 0.001 |
| Cardiovascular disease, n (%) | 7 (4.32) | 0 | 0.016 |
| Tobacco, n (%) | |||
| Non smoker | 51 (31.5) | 96 (58.1) | < 0.001 |
| Smoker | 70 (43.2) | 31 (18.8) | |
| Former smoker | 40 (24.7) | 28 (16.7) | |
| Apolipoprotein E genotype, n (%) | |||
| E3/3 | 113 (69.8) | 109 (66.1) | 0.035 |
| E3/2 | 9 (5.56) | 25 (15.2) | |
| E2/2 | 0 | 0 | |
| E3/4 | 31 (19.1) | 25 (15.2) | |
| E4/4 | 6 (3.70) | 2 (1.21) | |
| E2/4 | 3 | 4 | |
Quantitative continuous variables are expressed as mean ± standard deviation or median [percentile 25–75]. Student’s t or Mann–Whitney tests were used to assess differences between two groups. Quantitative categorical variables are expressed as n (%) and statistical differences were assessed by Chi-squared.
ANGPTL3 genetic variants
Table 3 shows all variants in the ANGPTL3 gene identified in both groups. A total of 16 genetic variants, four of them not previously described, were identified by sequencing analysis. Only four of them (c.607-109T>C, c.607-47_607-46delGT, c.835+41C>A and c.*52_*60del) presented significantly different allele frequency in normolipemic group than in FCHL subjects (P = 0.020, P = 0.031, P = 0.043 and P < 0.001, respectively). Out of the 16 variants, seven variants were located in the coding region (c.379C>T, c.565T>C, c.961T>A, c.1003T>C, c.1028A>G, c.1089T>G and c.1122G>A), and three of them were missense variants: p. (Leu127Phe), p.(Tyr321Asn) and p.(His343Arg), but only p.(Leu127Phe) was described as deleterious by bioinformatics analysis. The other four variants located in the coding region, p.(Leu189Leu), p.(Leu335Leu), p.(Val363Val) and p.(Pro374Pro) were synonymous variants. Seven variants were located in the intronic region, c.496-88T>G, c.607-120A>G, c.607-109T>C, c.607-47_607-46delGT, c.835+41C>A, c.1198+111G>A and c.1198+140T>C. All of them were described as benign or not splicing change affected by the bioinformatics analysis. Nevertheless, three of them, c.607-109T>C, c.607-47_607-46delGT and c.835+41C>A, presented significantly higher allele frequency in FCHL subjects than in the normolipemic group. Finally, two variants were located in the 3′UTR, c.*52_*60del and c.*76T>G. One of them, c.*52_*60del, showed significantly higher allele frequency in the normolipemic group than in FCHL subjects.
Table 3.
Frequency and bioinformatics analysis of identified variants in ANGPTL3 in FCHL cases and controls.
| Variant | Location | Nucleotide change | Protein change | Bioinformatics analysis | Allele frequency in the general population | Allele frequency in our study | ACMG classificatione | MicroRNAsf | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PredictSNP2a (probability) | FruitFlyb | GnomADc | 1000 Genomes Projectd | Normolipemic subjects | FCHL subjects | p | ||||||
| rs72649573 | Exon 1 | c.379C>T | p.(Leu127Phe) | Deleterious (82%) | NA | 0.00711 | 0.0020 | 0.000 | 0.003 | 0.313 | Benigng | NR |
| – | Intron 1 | c.496-88T>G | NA | Neutral (88%) | Not splicing change | – | – | 0.000 | 0.003 | 0.313 | – | – |
| rs111414963 | Exon 2 | c.565T>C | p.(Leu189Leu) | Neutral (88%) | Not splicing change | 0.00025 | 0.0008 | 0.003 | 0.000 | 0.313 | Likely benign | NR |
| rs531071581 | Intron 2 | c.607-120A>G | NA | Neutral (88%) | Not splicing change | 0.00013 | 0.0006 | 0.000 | 0.003 | 0.313 | – | NR |
| rs72649576 | Intron 2 | c.607-109T>C | NA | Neutral (88%) | Not splicing change | 0.01079 | 0.0042 | 0.024 | 0.003 | 0.020 | – | NR |
| rs72649577 | Intron 2 | c.607-47_607-46delGT | NA | Neutral (88%) | NA | 0.02222 | 0.0136 | 0.022 | 0.003 | 0.031 | – | NR |
| rs185472483 | Intron 3 | c.835+41C>A | NA | – | Not splicing change | 0.00032 | 0.0006 | 0.000 | 0.012 | 0.043 | – | NR |
| rs747725081 | Exon 6 | c.961T>A | p.(Tyr321Asn) | Neutral (88%) | NA | NR | NR | 0.000 | 0.003 | 0.313 | – | NR |
| rs12563308 | Exon 6 | c.1003T>C | p.(Leu335Leu) | Neutral (88%) | NA | 0.03550 | 0.0559 | 0.003 | 0.003 | 0.989 | VUSg | NR |
| rs199555921 | Exon 6 | c.1028A>G | p.(His343Arg) | Neutral (89%) | NA | 0.00016 | NR | 0.003 | 0.000 | 0.321 | – | NR |
| rs763259225 | Exon 6 | c.1089T>G | p.(Val363Val) | Neutral (96%) | NA | NR | NR | 0.003 | 0.000 | 0.321 | – | NR |
| rs145086916 | Exon 6 | c.1122G>A | p.(Pro374Pro) | Neutral (96%) | NA | 0.00077 | 0.0006 | 0.003 | 0.000 | 0.321 | – | NR |
| rs72651034 | Intron 6 | c.1198+111G>A | NA | – | Not splicing change | NR | NR | 0.003 | 0.003 | 0.989 | – | NR |
| rs908541128 | Intron 6 | c.1198+140T>C | NA | – | Not splicing change | 0.000 | 0.000 | 0.003 | 0.000 | 0.321 | – | NR |
| rs34483103 | 3′UTR | c.*52_*60del | NA | – | NA | 0.33531 | 0.3484 | 0.276 | 0.102 | < 0.001 | – |
hsa-miR-151a-3p hsa-miR-7702 |
| – | 3′UTR | c.*76T>G | NA | – | NA | – | – | 0.000 | 0.003 | 0.313 | – | – |
NR not reported, NA not applicable, VUS variant of uncertain significance.
aPredictSNP2 uses CADD, DANN, FATHMM and Funseq2 as predictors.
bFruitFly. New prediction score 0.87 (wild type score 0.89).
cGnomAD. https://gnomad.broadinstitute.org/
d1000 Genomes Project Consortium, Abecasis GR, Auton A, Books LD et al. An integrated map of genetic variation from 1092 human genomes. Nature 2012;491:56–65.
eRichards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 May;17(5):405–24. https://doi.org/10.1038/gim.2015.30. Epub 2015 Mar 5. PMID: 25741868; PMCID: PMC4544753.
fPolymiRTS Database 3.0: http://compbio.uthsc.edu/miRSNP/
gTikka A, Metso J, Jauhiainen M. ANGPTL3 serum concentration and rare genetic variants in Finnish population. Scand J Clin Lab Invest. 2017;77:601–609.
Discussion
We have studied the possible contribution of the gene encoding ANGPTL3 in the aetiology of FCHL. Our hypothesis was that some rare gain-of-function variants could have a major effect on the disease or, on the contrary, that common variants with minor effect on ANGPTL3 function could be in different frequency with respect to the general population. The results of our study do not support the first possibility, since the identified variants are not predictive of relevant functional changes in the protein. There are no previous ANGPTL3 sequencing studies looking for GOF mutations in subjects with FCHL. At least 5 different loci have been associated with rare cases of monogenic FCHL: LDLR16,17, LPL15, APOE34, PCSK935 and APOA536,37, but ANGPTL3 does not appear to be associated with this form of FCHL nor familial hypercholesterolemia (FH). Although FH and FCHL are different phenotypes, there is some degree of overlap between the two entities since they share many clinical aspects. Studies in subjects with genetic hypercholesterolemia of unknown origin suggestive of FH have also failed to detect causal mutations in ANGPTL3. We have not found any severe mutation neither in cases nor in controls in the total of 654 alleles investigated. This leads us to think about how well preserved is this gene probably related to the importance of this gene in human metabolism.
These results contrast with the role of ANGPTL3 in the lipid phenotype called familial combined hypolipidemia (FHBL2, OMIM #605019)20, in which LOF mutations in ANGPTL3 are responsible of reduced plasma levels of TC, TG, VLDL cholesterol, LDLc, apoB, and free fatty acids, just the opposite lipid profile found in FCHL. Furthermore, FCHL and familial combined hypolipidemia share abnormal hepatic VLDL secretion rates as the main mechanism of the lipid abnormalities, being increased in FCHL38,39 and decreased in familial combined hypolipidemia40.
Most cases of FCHL are considered as a complex disease with interaction of polygenes or multiple allele relationships with effect on TC, TG and environmental factors, mainly obesity and diets rich in saturated fat. ANGPTL3 genetic variation has not been associated with FCHL or mixed hyperlipidemia in genome-wide association studies (GWAS)13,41. Similar conclusions can be drawn from large-scale deep-coverage whole-genome sequencing42. Our study cannot rule out that the genetic variation in ANGPTL3 participates in the final phenotype of polygenic forms of FCHL. We found four variants with different allele frequency in FCHL subjects and in normolipemic controls: c.607-109T>C, c.607-47_607-46delGT, c.835+41C>A and c.*52_*60del. The first three are located in intron regions and the in silico analysis does not predict any splicing change with clinical significance, so their contribution to FCHL seems unlikely. The variant c.*52_*60del, located in 3′UTR, presented statistically significant differences in allelic frequencies between FCHL subjects and normolipemic controls: 0.276 and 0.102, respectively (P < 0.001). This variant has been previously associated with two microRNAs, hsa-miR-151a-3p and hsa-miR-7702, modulators of gene expression32. However, this is a very frequent genetic variant in the general population and this variation has not been previously associated with cholesterol and triglyceride concentrations43,44, so its implication in the FCHL pathogenesis is unlikely, although it should be confirmed in future studies.
In summary, no GOF mutations in ANGPTL3 were present in a large group of unrelated subjects with FCHL. Our results do not support a substantial role of ANGPTL3 in FCHL.
Acknowledgements
This work was supported by Grants from Gobierno de Aragon, B14-7R, Spain; Spanish Ministry of Economy and Competitiveness PI15/01983, PI18/01777 and CIBERCV. We want to particularly acknowledge the patients and the Biobank of the Aragon Health System (PT17/0015/0039) integrated in the Spanish National Biobanks Network for their collaboration.
Author contributions
A.M.B.: Data acquisition, methodology, writing—original draft. E.F.-M.: Data acquisition. V.M.-B., E.J. and I.G.-R.: Data acquisition and methodology. A.C.: Conceptualization, Formal analysis, Writing—original draft, Supervision. F.C.: Conceptualization, Funding acquisition, Project administration, writing—original draft, Supervision. I.L.-M.: Data acquisition and methodology, formal analysis and writing—original draft. All authors have reviewed and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Conklin D, et al. Identification of a mammalian angiopoietin-related protein expressed specifically in liver. Genomics. 1999;62:477–482. doi: 10.1006/geno.1999.6041. [DOI] [PubMed] [Google Scholar]
- 2.Shimizugawa T, et al. ANGPTL3 decreases very low density lipoprotein triglyceride clearance by inhibition of lipoprotein lipase. J. Biol. Chem. 2002;277:33742–33748. doi: 10.1074/jbc.M203215200. [DOI] [PubMed] [Google Scholar]
- 3.Shimamura M, et al. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler. Thromb. Vasc. Biol. 2007;27:366–372. doi: 10.1161/01.ATV.0000252827.51626.89. [DOI] [PubMed] [Google Scholar]
- 4.Dewey FE, et al. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. N. Engl. J. Med. 2017;377:211–221. doi: 10.1056/NEJMoa1612790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ono M, et al. Protein region important for regulation of lipid metabolism in angiopoietin-like 3 (ANGPTL3): ANGPTL3 is cleaved and activated in vivo. J. Biol. Chem. 2003;278:41804–41809. doi: 10.1074/jbc.M302861200. [DOI] [PubMed] [Google Scholar]
- 6.Korstanje R, et al. Locating Ath8, a locus for murine atherosclerosis susceptibility and testing several of its candidate genes in mice and humans. Atherosclerosis. 2004;177:443–450. doi: 10.1016/j.atherosclerosis.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Stitziel NO, et al. ANGPTL3 deficiency and protection against coronary artery disease. J. Am. Coll. Cardiol. 2017;69:2054–2063. doi: 10.1016/j.jacc.2017.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatsuda S, et al. Association between plasma angiopoietin-like protein 3 and arterial wall thickness in healthy subjects. J. Vasc. Res. 2007;44:61–66. doi: 10.1159/000098153. [DOI] [PubMed] [Google Scholar]
- 9.Ando Y, et al. A decreased expression of angiopoietin-like 3 is protective against atherosclerosis in apoE-deficient mice. J. Lipid Res. 2003;44:1216–1223. doi: 10.1194/jlr.M300031-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein JL, Schrott HG, Hazzard WR, Bierman EL, Motulsky AG. Hyperlipidemia in coronary heart disease. II. Genetic analysis of lipid levels in 176 families and delineation of a new inherited disorder, combined hyperlipidemia. J. Clin. Invest. 1973;52:1544–1568. doi: 10.1172/JCI107332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaddi A, Galetti C, Pauciullo P, Arca M. Familial combined hyperlipoproteinemia: experts panel position on diagnostic criteria for clinical practice. Committee of experts of the Atherosclerosis and Dysmetabolic Disorders Study Group. Nutr. Metab. Cardiovasc. Dis. NMCD. 1999;9:304–311. [PubMed] [Google Scholar]
- 12.Babirak SP, Brown BG, Brunzell JD. Familial combined hyperlipidemia and abnormal lipoprotein lipase. Arterioscler. Thromb. 1992;12(10):1176–1183. doi: 10.1161/01.atv.12.10.1176. [DOI] [PubMed] [Google Scholar]
- 13.Brouwers MCGJ, van Greevenbroek MMJ, Stehouwer CDA, de Graaf J, Stalenhoef AFH. The genetics of familial combined hyperlipidaemia. Nat. Rev. Endocrinol. 2012;8:352–362. doi: 10.1038/nrendo.2012.15. [DOI] [PubMed] [Google Scholar]
- 14.Veerkamp MJ, de Graaf J, Stalenhoef AFH. Role of insulin resistance in familial combined hyperlipidemia. Arterioscler. Thromb. Vasc. Biol. 2005;25:1026–1031. doi: 10.1161/01.ATV.0000160612.18065.29. [DOI] [PubMed] [Google Scholar]
- 15.Yang WS, Nevin DN, Peng R, Brunzell JD, Deeb SS. A mutation in the promoter of the lipoprotein lipase (LPL) gene in a patient with familial combined hyperlipidemia and low LPL activity. Proc. Natl. Acad. Sci. 1995;92:4462–4466. doi: 10.1073/pnas.92.10.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Civeira F, et al. Frequency of low-density lipoprotein receptor gene mutations in patients with a clinical diagnosis of familial combined hyperlipidemia in a clinical setting. J. Am. Coll. Cardiol. 2008;52:1546–1553. doi: 10.1016/j.jacc.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 17.Minicocci I, et al. Contribution of mutations in low density lipoprotein receptor (LDLR) and lipoprotein lipase (LPL) genes to familial combined hyperlipidemia (FCHL): A reappraisal by using a resequencing approach. Atherosclerosis. 2015;242:618–624. doi: 10.1016/j.atherosclerosis.2015.06.036. [DOI] [PubMed] [Google Scholar]
- 18.Austin MA, et al. Cardiovascular disease mortality in familial forms of hypertriglyceridemia: A 20-year prospective study. Circulation. 2000;101:2777–2782. doi: 10.1161/01.CIR.101.24.2777. [DOI] [PubMed] [Google Scholar]
- 19.Di Costanzo A, et al. Clinical and biochemical characteristics of individuals with low cholesterol syndromes: A comparison between familial hypobetalipoproteinemia and familial combined hypolipidemia. J. Clin. Lipidol. 2017;11:1234–1242. doi: 10.1016/j.jacl.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Arca M, D’Erasmo L, Minicocci I. Familial combined hypolipidemia: Angiopoietin-like protein-3 deficiency. Curr. Opin. Lipidol. 2020;31:41–48. doi: 10.1097/MOL.0000000000000668. [DOI] [PubMed] [Google Scholar]
- 21.De Castro-Orós I, et al. Common genetic variants contribute to primary hypertriglyceridemia without differences between familial combined hyperlipidemia and isolated hypertriglyceridemia. Circ. Cardiovasc. Genet. 2014;7:814–821. doi: 10.1161/CIRCGENETICS.114.000522. [DOI] [PubMed] [Google Scholar]
- 22.Marcovina SM, et al. Use of a reference material proposed by the International Federation of Clinical Chemistry and Laboratory Medicine to evaluate analytical methods for the determination of plasma lipoprotein(a) Clin. Chem. 2000;46:1956–1967. doi: 10.1093/clinchem/46.12.1956. [DOI] [PubMed] [Google Scholar]
- 23.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 1972;18:499–502. doi: 10.1093/clinchem/18.6.499. [DOI] [PubMed] [Google Scholar]
- 24.Bea AM, et al. Lipid-lowering response in subjects with the p.(Leu167del) mutation in the APOE gene. Atherosclerosis. 2019;282:143–147. doi: 10.1016/j.atherosclerosis.2019.01.024. [DOI] [PubMed] [Google Scholar]
- 25.Palacios L, et al. Molecular characterization of familial hypercholesterolemia in Spain. Atherosclerosis. 2012;221:137–142. doi: 10.1016/j.atherosclerosis.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bendl J, et al. PredictSNP2: A unified platform for accurately evaluating SNP effects by exploiting the different characteristics of variants in distinct genomic regions. PLoS Comput. Biol. 2016;12:e1004962. doi: 10.1371/journal.pcbi.1004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J. Comput. Biol. J. Comput. Mol. Cell Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 29.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karczewski, K. J. et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. http://biorxiv.org/lookup/. 10.1101/531210 (2019).
- 31.ClinVar. https://www.ncbi.nlm.nih.gov/clinvar/.
- 32.Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42:D86–D91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Team, R. C. R Foundation for Statistical Computing; Vienna, Austria: 2014. R Lang. Environ. Stat. Comput. 2013 (2018).
- 34.Solanas-Barca M, et al. Apolipoprotein E gene mutations in subjects with mixed hyperlipidemia and a clinical diagnosis of familial combined hyperlipidemia. Atherosclerosis. 2012;222:449–455. doi: 10.1016/j.atherosclerosis.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 35.Abifadel M, et al. A PCSK9 variant and familial combined hyperlipidaemia. J. Med. Genet. 2008;45:780–786. doi: 10.1136/jmg.2008.059980. [DOI] [PubMed] [Google Scholar]
- 36.van der Vleuten GM, et al. Haplotype analyses of the APOA5 gene in patients with familial combined hyperlipidemia. Biochim. Biophys. Acta. 2007;1772:81–88. doi: 10.1016/j.bbadis.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 37.Di Taranto MD, et al. Association of USF1 and APOA5 polymorphisms with familial combined hyperlipidemia in an Italian population. Mol. Cell. Probes. 2015;29:19–24. doi: 10.1016/j.mcp.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Baila-Rueda L, et al. Cholesterol oversynthesis markers define familial combined hyperlipidemia versus other genetic hypercholesterolemias independently of body weight. J. Nutr. Biochem. 2018;53:48–57. doi: 10.1016/j.jnutbio.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 39.van Himbergen TM, et al. Familial combined hyperlipidemia is associated with alterations in the cholesterol synthesis pathway. Arterioscler. Thromb. Vasc. Biol. 2010;30:113–120. doi: 10.1161/ATVBAHA.109.196550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hess AL, et al. Analysis of circulating angiopoietin-like protein 3 and genetic variants in lipid metabolism and liver health: The DiOGenes study. Genes Nutr. 2018;13:7. doi: 10.1186/s12263-018-0597-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ripatti P, et al. The contribution of GWAS Loci in Familial Dyslipidemias. PLOS Genet. 2016;12:e1006078. doi: 10.1371/journal.pgen.1006078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Natarajan P, et al. Deep-coverage whole genome sequences and blood lipids among 16,324 individuals. Nat. Commun. 2018;9:3391. doi: 10.1038/s41467-018-05747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Surakka I, et al. The impact of low-frequency and rare variants on lipid levels. Nat. Genet. 2015;47:589–597. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.UK10K consortium The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]