Abstract
Three years after a prospective study on wound infections in a rural hospital in Ghana revealed no emergence of carbapenem-resistant bacteria we initiated a new study to assess the prevalence of multidrug-resistant pathogens. Three hundred and one samples of patients with wound infections were analysed for the presence of resistant bacteria in the period August 2017 till March 2018. Carbapenem-resistant Acinetobacter (A.) baumannii were further characterized by resistance gene sequencing, PCR-based bacterial strain typing, pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST “Oxford scheme”). A. baumanni was detected in wound infections of 45 patients (15%); 22 isolates were carbapenem-resistant. Carbapenemases NDM-1 and/or OXA-23 were detected in all isolates; two isolates harboured additionally OXA-420. PFGE and MLST analyses confirmed the presence of one A. baumannii strain in 17 patients that was assigned to the worldwide spread sequence type ST231 and carried NDM-1 and OXA-23. Furthermore, two new A. baumannii STs (ST2145 and ST2146) were detected in two and three patients, respectively. Within three years the prevalence of carbapenem-resistant A. baumannii increased dramatically in the hospital. The early detection of multidrug-resistant bacteria and prevention of their further spread are only possible if continuous surveillance and molecular typing will be implemented.
Keywords: Acinetobacter baumannii, carbapenem resistance, OXA-23, OXA-420, NDM-1, wound infections, Ghana, rural
1. Introduction
Antibiotics are essential medicines whose use in human or veterinary medicine, no matter how prudent, is inevitably associated with accelerated development of antimicrobial resistance (AMR). β-lactams are still considered the most successful antibiotic classes. With a proportion of almost two thirds of all antibiotic prescriptions they are even the most widely used antibacterial agents against infectious diseases [1]. The carbapenems have the broadest spectrum of activity against various bacteria and are widely regarded as the class of last resort for treatment of infections with multidrug-resistant pathogens [2]. However, resistance to carbapenems has increased dramatically worldwide [3]. One reason for this resistance is the production of different carbapenem-hydrolyzing enzymes, the carbapenemases. In the last 20 years, the number of newly detected carbapenemases has increased continuously. The most prevalent carbapenemases in Enterobacterales are OXA-48, KPC, VIM and NDM; in Pseudomonas (P.) aeruginosa VIM and IMP have been frequently detected, and different OXA-enzymes (OXA-23, OXA-58, OXA-40-like) and NDM have been found mainly in Acinetobacter (A.) baumannii [3,4,5,6]. The location of carbapenemase genes within transposons and/or conjugative plasmids enables the emergence in various bacterial species and facilitates their worldwide spread [7,8].
Treatment of infections caused by carbapenem-resistant pathogens is quite a big challenge in industrialized countries and almost impossible for health systems with limited resources [9,10,11]. Of great concern are carbapenemase-producing A. baumannii, P. aeruginosa and Enterobacterales [12,13,14]. Remaining treatment option for Enterobacterales producing serine carbapenemases, e.g., KPC or OXA-48, might be ceftazidime-avibactam, meropenem-vaborbactam, imipenem-cilastatin-relebactam, or cefiderocol; for Enterobacterales producing metallo-beta-lactamases, e.g., NDM, aztreonam plus ceftazidim-avibactam or cefiderocol. Alternatively, colistin (plus meropenem or tigecycline or eravacycline) could be an option. Therapeutic options for carbapenem-resistant A. baumannii might be cefiderocol or colistin (plus a carbapenem, minocycline, tigecycline, or rifampicin); for carbapenem-resistant P. aeruginosa ceftolozane-tazobactam, imipenem-cilastatin-relebactam, cefiderocol or colistin plus meropenem [15]. However, their availability especially in low- and middle-income countries is rather unlikely [16].
A. baumannii is a non-fermenting bacterium of importance in both humid and temperate climates [17]. Colonization of the human skin may lead to community-acquired or nosocomial traumatic or surgical wound infections [18,19]. However, the distinction between true infection and colonization is often difficult to decide [20]. Wound infections are a common disease entity that is frequently not given the necessary attention, including lack of microbiological investigation, leading to chronicity and disability [21]. Biofilms are present in most chronic wounds and may contribute to delayed healing and persistent inflammation. The environment of the biofilm facilitates the horizontal spread of antibiotic resistance genes and virulence factors between embedded pathogenic bacteria [22]. Moreover, bacteria are protected from external threats creating additional bacterial tolerance to antimicrobial agents [23]. Microbiological culture of wound specimens performed through deep swabbing techniques and processed within two hours frequently detects a polymicrobial flora. Deciding which of these different pathogens should be treated is always a big challenge for the clinician.
Prevention and control of multidrug-resistant bacteria [24] and availability of up to date recommendations for antimicrobial therapy [25] require continuous surveillance of antimicrobial resistance which is still not established in most Sub-Saharan African countries [26]. A pilot study by our study group in a rural hospital in Eikwe, Western Region of Ghana, found a high prevalence of Enterobacterales with combined resistance to third-generation cephalosporins and fluoroquinolones but no carbapenem-resistant bacteria at all in wound infections [27]. The results of this pilot study in 2014 prompted the project partners in Eikwe and Göttingen to conduct another study. Therefore, a more comprehensive microbiological analysis of wound infections in Ghana was carried out from August 2017 to March 2018. All detected carbapenem-resistant A. baumannii were characterized in the present study.
2. Materials and Methods
2.1. Ethics
The study was approved by the Ghana Health Service Ethics Review Committee, Research & Development Division, Ghana Health Service, Accra, Ghana (GHS-ERC: 04/06/17, 21 July 2017), and the Ethics Committee of the University Medical Center in Göttingen, Germany (5/6/17, 20 June 2017), respectively. All patients provided written informed consent before inclusion in the study.
2.2. Clinical Diagnosis and Sample Collection
Wound infection was diagnosed clinically using the classic signs of inflammation. Before taking the sample, wounds were cleaned with sterile cotton swabs moistened with sterile sodium chloride solution (0.9%). The sample was taken with an eSwabTM (Copan, Brescia, Italy) from the wound bed and edge and then immediately brought to the bacteriology laboratory for further processing.
2.3. Microbiological Culture and Susceptibility Testing
Basic biochemical identification of the bacterial isolates and antibiotic susceptibility testing were initially carried out in the bacteriology laboratory of the hospital in Eikwe adapted to the locally available resources. The anti-infective treatment of wound infections was adjusted according to the results of the microbiological analysis as described before [27]. In brief, samples were inoculated on 5% blood agar (Merck, Darmstadt, Germany) and on McConkey agar (Merck, Darmstadt, Germany), incubated aerobically at 37 °C, and read after 24 h and 48 h. Gram-negative bacteria were identified through: (i) hydrogen sulphide production, indole production and motility in semi-solid SIM (sulphide, indole, motility) medium (Merck, Darmstadt, Germany) vertically in tubes; (ii) sucrose fermentation in Hugh Leifson medium (Merck, Darmstadt, Germany); and (iii) Oxidase test, respectively. With this local approach, identification down to the genus level was possible. The microscopical examination of a Gram-stained smear was done in order to ensure wound specimen quality and to check for presence of bacteria, neutrophils, and epithelial cells. Furthermore, disc diffusion tests for Gram-negative bacteria were performed using the following paper antibiotic discs: ampicillin 10 µg; ampicillin-sulbactam 10 µg–10 µg; cefotaxime 5 µg; ciprofloxacin 5 µg; gentamicin 10 µg; and trimethoprim-sulfamethoxazole 1.25–23.75 µg, respectively [28]. There was no routine screening for carbapenem susceptibility.
All bacterial isolates, independent of their antibiotic resistance, were stored at −20 °C in microbank systems, and were reanalysed at the Institute for Medical Microbiology of the University Medical Center in Göttingen, Germany, and at the Robert Koch Institute in Wernigerode, Germany. Species identification was done with MALDI Biotyper 3.0 (Bruker Daltonics, Bremen, Germany). Antibiotic susceptibility testing for A. baumannii isolates was performed by broth microdilution and VITEK 2 (bioMérieux, Marcy-l’Étoile, France) using card AST-N248 (bioMérieux, Nuertingen, Germany) with interpretation of results according to EUCAST criteria (EUCAST v10.0).
2.4. Molecular Analyses
For all A. baumannii isolates with resistance to meropenem and/or imipenem the presence of intrinsic blaOXA-51-like genes and the presence of different carbapenemase and other β-lactamase genes (ISAba1+blaOXA-51-like, blaOXA-23-like, blaOXA-24-like, blaOXA-58-like, blaIMP-like, blaVIM-like, blaNDM-like, blaGES-like, blaPER-like, blaVEB-like) was investigated by PCR and Sanger sequencing, as described previously [29,30,31]. Furthermore, all carbapenem-resistant A. baumannii were tested for assignment to the important international clones 1–3 (formerly named European clones I–III) by multiplex-PCR [32]. Bacterial strain typing was done by Apal-macrorestriction and subsequent pulsed-field gel electrophoresis (PFGE) and results were interpreted according to the criteria of Tenover et al. [33]. Finally, multilocus sequence typing (MLST) was performed for selected isolates (PFGE types A-1, A-2, A-3) according to the “Oxford” scheme (https://pubmlst.org/organisms/acinetobacter-baumannii, accessed on 3 August 2020).
3. Results
3.1. Wound Infection Classification
Overall, wound swabs from 301 patients with wound infections were analysed. Wound infection (WI) (duration of infection ≤ three month), chronic wound infection (CWI) (duration of infection > three month), and surgical site infection (SSI) was diagnosed in 144 (48%), 70 (23%), and 74 (25%) patients, respectively. No information was available in 13 (4%) patients.
3.2. Identification and Susceptibility of Detected Bacterial Pathogens
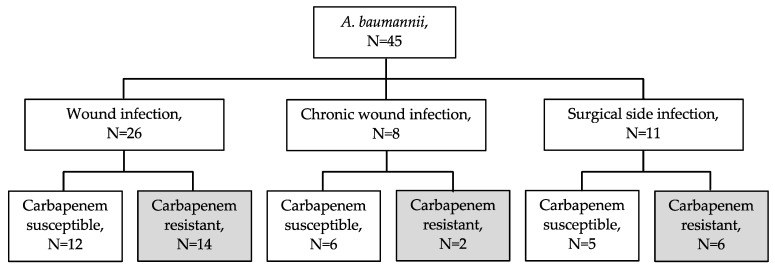
A. baumannii was isolated in wound swabs of 45 patients. Carbapenem resistance was detected in 22 (49%) of these 45 A. baumannii isolates, Figure 1. Further bacterial pathogens which were detected beside these carbapenem-resistant A. baumannii in wound swabs are listed in Table 1. However, no other carbapenem-resistant bacteria except three Stenotrophomonas maltophilia and one P. aeruginosa without transmissible carbapenemase genes were found. The 22 carbapenem-resistant A. baumannii isolates were additionally resistant to ciprofloxacin, trimethoprim-sulfamethoxazole, gentamicin and amikacin (20 of 22 isolates) but remained susceptible to colistin (Table 2).
Figure 1.
Distribution of Acinetobacter baumannii isolates according to wound infection type and carbapenem susceptibility.
Table 1.
List of bacterial pathogens detected beside carbapenem-resistant Acinetobacter baumannii in wound infections (n = 22).
| Co-Detected Bacterial Species in Wound Infections (n = Number of Wound Infections) | |
|---|---|
| Gram-Positive Pathogens | Gram-Negative Pathogens |
| Staphylococcus aureus, n = 6 | Proteus mirabilis, n = 6 |
| Corynebacterium striatum, n = 3 | Escherichia coli, n = 4 |
| Bacillus cereus, n = 2 | Stenotrophomonas maltophilia, n = 3 |
| Corynebacterium amycolatum, n = 1 | Enterobacter cloacae, n = 2 |
| Enterococcus avium, n = 1 | Klebsiella pneumoniae, n = 2 |
| Enterococcus casseliflavus, n = 1 | Pseudomonas aeruginosa, n = 2 |
| Enterococcus faecalis, n = 1 | Achromobacter spanius, n = 1 |
| Providencia stuartii, n = 1 | |
Between 0 and 5 further bacterial pathogens beside carbapenem-resistant A. baumannii were detected in the individual wound infection. The average number of pathogens per wound infection was 2.7. Coagulase-negative staphylococci (n = 7) other than Staphylococcus lugdunensis or Staphylococcus schleiferi subsp. schleiferi were present but not further analysed.
Table 2.
Characteristics of 22 carbapenem-resistant Acinetobacter baumannii isolates from wound infections, Ghana, 2017/2018.
| Isolate No. | Sampling Date |
Species |
A. baumannii Specific blaOXA-51-like1 |
Carbapenemase Genes |
A. baumannii Typing PCR (clones EI-III) 2 |
PFGE-Type | Sequence Type (ST)OX |
Resistances 3 |
|---|---|---|---|---|---|---|---|---|
| 5B | 21.08.2017 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 36B | 22.09.2017 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 39B | 26.09.2017 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 43A | 27.09.2017 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1a | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 67A | 12.10.2017 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1a | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 105A | 06.11.2017 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 109C | 08.11.2017 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 116F | 10.11.2017 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 146C | 27.11.2017 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 165B | 06.12.2017 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 168A | 11.12.2017 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 178A | 15.12.2017 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 190E | 22.12.2017 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 229C | 15.01.2018 | A. baumannii | bla OXA-378 | blaOXA-23, ISAba1-blaOXA-378 | n.t. | A-2 | ST2145 | IPM, MEM, GEN, CIP, SXT |
| 236A | 17.01.2018 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 246-1A | 24.01.2018 | A. baumannii | bla OXA-378 | blaNDM-1, blaOXA-420 | n.t. | A-3 | ST2146 | IPM, MEM, GEN, AMK, CIP, SXT |
| 258A | 05.02.2018 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1b | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 267B | 06.02.2018 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
| 274A | 14.02.2018 | A. baumannii | bla OXA-378 | blaOXA-23, ISAba1-blaOXA-378 | n.t. | A-2 | ST2145 | IPM, MEM, GEN, CIP, SXT |
| 278D | 19.02.2018 | A. baumannii | bla OXA-378 | blaNDM-1, blaOXA-420 | n.t. | A-3 | ST2146 | IPM, MEM, GEN, AMK, CIP, SXT |
| 280C | 19.02.2018 | A. baumannii | bla OXA-378 | blaOXA-23, ISAba1-blaOXA-378, blaNDM-1 | n.t. | A-2a | ST2145 | IPM, MEM, GEN, AMK, CIP, SXT |
| 288A | 24.02.2018 | A. baumannii | bla OXA-69 | blaNDM-1, blaOXA-23 | EU-I (IC 1) | A-1 | ST231 | IPM, MEM, GEN, AMK, CIP, SXT |
1 PCR Turton et al. 2006 [31]. 2 PCR Turton et al. 2007 [32]. n.t. non-typeable. 3 All carbapenem-resistant A. baumannii isolates were susceptible to colistin. STOX, sequence type according multilocus sequence typing (MLST “Oxford” scheme (https://pubmlst.org/organisms/acinetobacter-baumannii, accessed on 3 August 2020). IPM, imipenem; MEM, meropenem; GEN, gentamicin; AMK; amikacin; CIP, ciprofloxacin; SXT, trimethoprim-sulfamethoxazole.
3.3. Molecular Characteristics of A. baumannii
The majority of the 22 carbapenem-resistant A. baumannii isolates (n = 17, 77%) harboured the two carbapenemase genes blaOXA-23 and blaNDM-1. In two isolates the combination blaNDM-1 and blaOXA-420 was detected. The three remaining isolates carried blaOXA-23, and the insertion sequence ISAba1 was present upstream of the intrinsic gene blaOXA-378 (Table 2).
By PCR-based typing the 17 A. baumannii isolates with carbapenemase gene combination blaOXA-23/blaNDM-1 were assigned to international clone 1 (IC 1). All 17 isolates harboured blaOXA-69, a variant of the intrinsic, A. baumannii specific blaOXA-51 gene. The five remaining isolates were non-typeable by this multiplex-PCR and carried the intrinsic gene variant blaOXA-378.
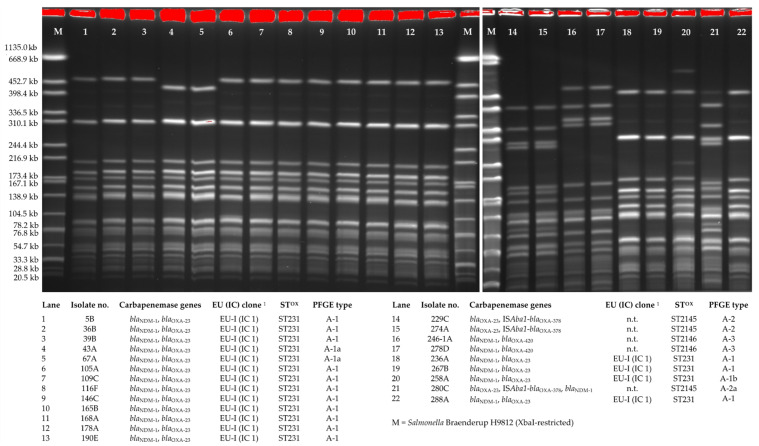
PFGE-typing of the 22 carbapenemase-producing isolates revealed three distinctly distinguishable macrorestriction patterns designated as A. baumannii PFGE-types A-1, A-2 and A-3 (Figure 2). The 17 A. baumannii isolates of IC 1 were assigned to PFGE-type A-1 and showed highly related macrorestriction patterns that differed in 0–2 bands. PFGE-type A-2 isolates (n = 3) differed in six bands from PFGE-type A-3 isolates (n = 2) but all five isolates carried the intrinsic gene blaOXA-378.
Figure 2.
ApaI-macrorestriction and pulsed-field gel electrophoresis of 22 carbapenem-resistant A. baumannii isolates from wound infections, Ghana, 2017/2018. 1 PCR Turton et al. 2007 [32]. STOX, sequence type assigned by multilocus sequence typing (MLST Oxford scheme https://pubmlst.org/organisms/acinetobacter-baumannii, assessed on 3 August 2020); n.t. non-typeable.
Using MLST (“Oxford” scheme) the 17 A. baumanni isolates of IC 1 were assigned to sequence type ST231 (Table 2). The five other isolates were designated as new sequence types ST2145 and ST2146, respectively.
4. Discussion
The molecular analyses of the 22 detected carbapenem-resistant A. baumannii isolates in this study confirmed the presence of an NDM-1 and OXA-23 carbapenemase-producing strain of sequence type ST231 in wounds of 17 patients. According to the MLST database ST231 (Table 2) has been detected for clinical isolates worldwide, e.g., Brasilia, Libya, the USA, the Netherlands and Germany [29]. The emergence and spread of this carbapenemase-producing strain of ST231 within the hospital in Eikwe is of concern and needs continuous surveillance.
Furthermore, three and two patients carried A. baumannii of the novel sequence types ST2145 and ST2146, respectively. Both STs differed in only one allele (gpi) to ST1452 and ST1459, that were detected previously for several isolates with blaOXA-378 from white stork nestlings in Poland, 2015 [34]. The strain of ST2145 carried carbapenemase gene blaOXA-23 and insertion sequence ISAba1 upstream of blaOXA-378 that is known to provide a strong promoter for blaOXA-51-like genes in A. baumannii resulting in increased gene expression and resistance to carbapenems [31]. For the strain of ST2146 carbapenemase genes blaNDM-1 and blaOXA-420 were detected. OXA-420 has been reported first in carbapenem-resistant A. baumannii isolates from Nepal in 2014 and it shows one amino acid substitution (A256D) compared to the worldwide prevalent carbapenemase OXA-58 [35]. Furthermore, the blaOXA-420 gene sequence has been submitted to the NCBI database from A. baumannii isolates in the Netherlands (CP038646.1, April 2019); and the genome analysis of 36 A. baumannii from three hospitals in Ghana (2016–2017) revealed one ST107 isolate from a sputum sample with this rare enzyme variant [36]. Since carbapenemase genes are often located on plasmids a transfer between different A. baumannii strains might be possible but this was not analyzed further in the present study.
Our pilot study in Eikwe, Western Region of Ghana, 2014 [27], and a study from another rural district hospital in Asante Akim North Municipality of Ghana, 2016 [37] found no carbapenem-resistant A. baumannii. However, three and a half years later, the present study has shown that almost half of the A. baumannii isolates from wound infections are carbapenem-resistant. Several circumstances might have been contributed. Exposure to carbapenems is a necessary but not sufficient prerequisite. Carbapenems are available in Ghana, but their clinical use seems to be very limited and most likely restricted to very selected patients in private or university hospitals [38]. The increasing mobility of local people nationally and even internationally for private and/or professional reasons could lead to the acquisition of carbapenem-resistant A. baumannii isolates [39,40,41]. In addition, colonized patients admitted to multiple health facilities might contribute to the further spread of opportunistic bacteria. Once carbapenem-resistant A. baumannii is introduced into a local health facility and there is no continuous microbiological surveillance to detect the phenomenon of carbapenem resistance, together with inadequate infection control measures the gateway to further transmission is open.
The treatment of patients with carbapenem-resistant A. baumannii wound infection is difficult. There is no oral treatment option available. Colistin might be therapeutic options but in resource-limited countries such as Sub-Saharan Africa, these drugs are simply not available or too expensive [42]. Ultimately, only wound debridement, use of antiseptics which favour the formation of granulation tissue and appropriate wound care with careful adherence to hygiene standards are realistic options. Therefore, investment in and continuous training of horizontal infection control measures like appropriate hand hygiene, e.g., WHO 5 moments of hand hygiene, need to be promoted and trained continuously [43].
5. Conclusions
In conclusion, the emergence and nosocomial spread of carbapenem-resistant A. baumannii in this follow up investigation in Ghana demonstrates that continuous surveillance of antimicrobial resistance of clinical isolates is of paramount importance. Health systems with limited resources need to be enabled to perform nationwide basic microbiological investigations on a routine basis. However, there is also need to get access to more sophisticated technologies like molecular typing methods to analyse emergence and spread of multidrug-resistant bacteria and to take immediate and appropriate preventive hygienic measures. Finally, access to effective anti-infective drugs according to the local resistance situation is urgently needed.
Acknowledgments
We would like to thank the patients in Ghana for their participation in the study. We also would like to thank the staff from St. Martin de Porres Hospital in Eikwe, Ghana, and the staff from the Institute for Medical Microbiology of the University Medical Center Göttingen, Germany, for their commitment to perform the study. Finally, we thank Sybille Müller-Bertling and Kirstin Ganske from the Robert Koch Institute for excellent technical assistance.
Author Contributions
Conceptualization, M.M., P.C., U.G. and M.H.S.; methodology, M.M., P.C., U.G. and M.H.S.; software, M.M., Y.P. and M.H.S.; validation, M.M., Y.P. and M.H.S.; formal analysis, M.M., Y.P. and M.H.S.; investigation, M.M., H.K.A., Y.P. and M.H.S.; resources, M.M., P.C., H.K.A., T.P., U.G., Y.P. and M.H.S.; data curation, M.M., Y.P. and M.H.S.; writing—original draft preparation, M.M., Y.P. and M.H.S.; writing—review and editing, M.M., P.C., H.K.A., T.P., U.G., Y.P. and M.H.S.; visualization, M.M., Y.P. and M.H.S.; supervision, P.C., T.P., U.G. and M.H.S.; project administration, M.M., P.C., U.G. and M.H.S.; funding acquisition, U.G., Y.P. and M.H.S. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by the Open Access Publication Funds of the Georg August University of Göttingen and the University Medical Center Göttingen.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ghana Health Service Ethics Review Committee, Research & Development Division, Ghana Health Service, Accra, Ghana (GHS-ERC: 04/06/17, 21 July 2017) and the Ethics Committee of the University Medical Center in Göttingen, Germany (5/6/17, 20 June 2017), respectively.
Informed Consent Statement
Informed consent was obtained from all persons involved in the study.
Data Availability Statement
The information on two A. baumanni isolates with novel sequence types ST2145 (no. 71-20, ID5021) and ST2146 (no. 73-20, ID5022) was submitted to the MLST database (https://pubmlst.org/bigsdb?db=pubmlst_abaumannii_isolates, assessed on 3 August 2020).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vrancianu C.O., Gheorghe I., Dobre E.G., Barbu I.C., Cristian R.E., Popa M., Lee S.H., Limban C., Vlad I.M., Chifiriuc M.C. Emerging Strategies to Combat beta-Lactamase Producing ESKAPE Pathogens. Int. J. Mol. Sci. 2020;21:8527. doi: 10.3390/ijms21228527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shallcross L.J., Howard S.J., Fowler T., Davies S.C. Tackling the threat of antimicrobial resistance: From policy to sustainable action. Philos. Trans. R. Soc. B. 2015;370:20140082. doi: 10.1098/rstb.2014.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen G.T. Continuous Evolution: Perspective on the Epidemiology of Carbapenemase Resistance Among Enterobacterales and Other Gram-Negative Bacteria. Infect. Dis. Ther. 2021 doi: 10.1007/s40121-020-00395-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halat D.H., Moubareck C.A. The Current Burden of Carbapenemases: Review of Significant Properties and Dissemination among Gram-Negative Bacteria. Antibiotics. 2020;9:186. doi: 10.3390/antibiotics9040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Livermore D.M., Woodford N. The beta-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 2006;14:413–420. doi: 10.1016/j.tim.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Livermore D.M. Carbapenemases. J. Antimicrob. Chemother. 1992;29:609–613. doi: 10.1093/jac/29.6.609. [DOI] [PubMed] [Google Scholar]
- 7.Bush K., Bradford P.A. Epidemiology of beta-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020;33 doi: 10.1128/CMR.00047-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber R.E., Pietsch M., Fruhauf A., Pfeifer Y., Martin M., Luft D., Gatermann S., Pfennigwerth N., Kaase M., Werner G., et al. IS26-Mediated Transfer of bla NDM-1 as the Main Route of Resistance Transmission During a Polyclonal, Multispecies Outbreak in a German Hospital. Front. Microbiol. 2019;10:2817. doi: 10.3389/fmicb.2019.02817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labi A.K., Nielsen K.L., Marvig R.L., Bjerrum S., Enweronu-Laryea C., Bennedbaek M., Newman M.J., Ayibor P.K., Andersen L.P., Kurtzhals J.A.L. Oxacillinase-181 Carbapenemase-Producing Klebsiella pneumoniae in Neonatal Intensive Care Unit, Ghana, 2017–2019. Emerg. Infect. Dis. 2020;26:2235–2238. doi: 10.3201/eid2609.200562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strydom K.A., Chen L., Kock M.M., Stoltz A.C., Peirano G., Nobrega D.B., Lowe M., Ehlers M.M., Mbelle N.M., Kreiswirth B.N., et al. Klebsiella pneumoniae ST307 with OXA-181: Threat of a high-risk clone and promiscuous plasmid in a resource-constrained healthcare setting. J. Antimicrob. Chemother. 2020;75:896–902. doi: 10.1093/jac/dkz550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leski T.A., Bangura U., Jimmy D.H., Ansumana R., Lizewski S.E., Li R.W., Stenger D.A., Taitt C.R., Vora G.J. Identification of blaOXA-(5)(1)-like, blaOXA-(5)(8), blaDIM-(1), and blaVIM carbapenemase genes in hospital Enterobacteriaceae isolates from Sierra Leone. J. Clin. Microbiol. 2013;51:2435–2438. doi: 10.1128/JCM.00832-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonomo R.A., Burd E.M., Conly J., Limbago B.M., Poirel L., Segre J.A., Westblade L.F. Carbapenemase-Producing Organisms: A Global Scourge. Clin. Infect. Dis. 2018;66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manenzhe R.I., Zar H.J., Nicol M.P., Kaba M. The spread of carbapenemase-producing bacteria in Africa: A systematic review. J. Antimicrob. Chemother. 2015;70:23–40. doi: 10.1093/jac/dku356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semret M., Haraoui L.P. Antimicrobial Resistance in the Tropics. Infect. Dis. Clin. N. Am. 2019;33:231–245. doi: 10.1016/j.idc.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Doi Y. Treatment Options for Carbapenem-resistant Gram-negative Bacterial Infections. Clin. Infect. Dis. 2019;69:S565–S575. doi: 10.1093/cid/ciz830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laxminarayan R., Matsoso P., Pant S., Brower C., Rottingen J.A., Klugman K., Davies S. Access to effective antimicrobials: A worldwide challenge. Lancet. 2016;387:168–175. doi: 10.1016/S0140-6736(15)00474-2. [DOI] [PubMed] [Google Scholar]
- 17.Munoz-Price L.S., Weinstein R.A. Acinetobacter infection. N. Engl. J. Med. 2008;358:1271–1281. doi: 10.1056/NEJMra070741. [DOI] [PubMed] [Google Scholar]
- 18.Sebeny P.J., Riddle M.S., Petersen K. Acinetobacter baumannii skin and soft-tissue infection associated with war trauma. Clin. Infect. Dis. 2008;47:444–449. doi: 10.1086/590568. [DOI] [PubMed] [Google Scholar]
- 19.Davis K.A., Moran K.A., McAllister C.K., Gray P.J. Multidrug-resistant Acinetobacter extremity infections in soldiers. Emerg. Infect. Dis. 2005;11:1218–1224. doi: 10.3201/1108.050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petersen K., Cannegieter S.C., van der Reijden T.J., van Strijen B., You D.M., Babel B.S., Philip A.I., Dijkshoorn L. Diversity and clinical impact of Acinetobacter baumannii colonization and infection at a military medical center. J. Clin. Microbiol. 2011;49:159–166. doi: 10.1128/JCM.00766-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leaper D., Assadian O., Edmiston C.E. Approach to chronic wound infections. Br. J. Dermatol. 2015;173:351–358. doi: 10.1111/bjd.13677. [DOI] [PubMed] [Google Scholar]
- 22.Savage V.J., Chopra I., O’Neill A.J. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob. Agents Chemother. 2013;57:1968–1970. doi: 10.1128/AAC.02008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowler P.G. Antibiotic resistance and biofilm tolerance: A combined threat in the treatment of chronic infections. J. Wound Care. 2018;27:273–277. doi: 10.12968/jowc.2018.27.5.273. [DOI] [PubMed] [Google Scholar]
- 24.Singh N., Manchanda V. Control of multidrug-resistant Gram-negative bacteria in low- and middle-income countries-high impact interventions without much resources. Clin. Microbiol. Infect. 2017;23:216–218. doi: 10.1016/j.cmi.2017.02.034. [DOI] [PubMed] [Google Scholar]
- 25.Cox J.A., Vlieghe E., Mendelson M., Wertheim H., Ndegwa L., Villegas M.V., Gould I., Hara G.L. Antibiotic stewardship in low- and middle-income countries: The same but different? Clin. Microbiol. Infect. 2017;23:812–818. doi: 10.1016/j.cmi.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Nordmann P., Poirel L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019;69:S521–S528. doi: 10.1093/cid/ciz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janssen H., Janssen I., Cooper P., Kainyah C., Pellio T., Quintel M., Monnheimer M., Gross U., Schulze M.H. Antimicrobial-Resistant Bacteria in Infected Wounds, Ghana, 2014. Emerg. Infect. Dis. 2018;24:916–919. doi: 10.3201/eid2405.171506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheesbrough M. District Laboratory Practice in Tropical Countries. 2nd ed. Part 2 Cambridge University Press; Cambridge, UK: 2006. [Google Scholar]
- 29.Rieber H., Frontzek A., Pfeifer Y. Molecular Investigation of Carbapenem-Resistant Acinetobacter spp. from Hospitals in North Rhine-Westphalia, Germany. Microb. Drug Resist. 2017;23:25–31. doi: 10.1089/mdr.2016.0035. [DOI] [PubMed] [Google Scholar]
- 30.Pfeifer Y., Wilharm G., Zander E., Wichelhaus T.A., Gottig S., Hunfeld K.P., Seifert H., Witte W., Higgins P.G. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 2011;66:1998–2001. doi: 10.1093/jac/dkr256. [DOI] [PubMed] [Google Scholar]
- 31.Turton J.F., Ward M.E., Woodford N., Kaufmann M.E., Pike R., Livermore D.M., Pitt T.L. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 2006;258:72–77. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 32.Turton J.F., Gabriel S.N., Valderrey C., Kaufmann M.E., Pitt T.L. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 2007;13:807–815. doi: 10.1111/j.1469-0691.2007.01759.x. [DOI] [PubMed] [Google Scholar]
- 33.Tenover F.C., Arbeit R.D., Goering R.V., Mickelsen P.A., Murray B.E., Persing D.H., Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995;33:2233–2239. doi: 10.1128/JCM.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilharm G., Skiebe E., Higgins P.G., Poppel M.T., Blaschke U., Leser S., Heider C., Heindorf M., Brauner P., Jackel U., et al. Relatedness of wildlife and livestock avian isolates of the nosocomial pathogen Acinetobacter baumannii to lineages spread in hospitals worldwide. Environ. Microbiol. 2017;19:4349–4364. doi: 10.1111/1462-2920.13931. [DOI] [PubMed] [Google Scholar]
- 35.Shrestha S., Tada T., Miyoshi-Akiyama T., Ohara H., Shimada K., Satou K., Teruya K., Nakano K., Shiroma A., Sherchand J.B., et al. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii isolates in a university hospital in Nepal reveals the emergence of a novel epidemic clonal lineage. Int. J. Antimicrob. Agents. 2015;46:526–531. doi: 10.1016/j.ijantimicag.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 36.Ayibieke A., Kobayashi A., Suzuki M., Sato W., Mahazu S., Prah I., Mizoguchi M., Moriya K., Hayashi T., Suzuki T., et al. Prevalence and Characterization of Carbapenem-Hydrolyzing Class D beta-Lactamase-Producing Acinetobacter Isolates From Ghana. Front. Microbiol. 2020;11:587398. doi: 10.3389/fmicb.2020.587398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krumkamp R., Oppong K., Hogan B., Strauss R., Frickmann H., Wiafe-Akenten C., Boahen K.G., Rickerts V., McCormick Smith I., Gross U., et al. Spectrum of antibiotic resistant bacteria and fungi isolated from chronically infected wounds in a rural district hospital in Ghana. PLoS ONE. 2020;15:e0237263. doi: 10.1371/journal.pone.0237263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ingelbeen B., Koirala K.D., Verdonck K., Barbe B., Mukendi D., Thong P., El Safi S., Van Duffel L., Bottieau E., van der Sande M.A.B., et al. Antibiotic use prior to seeking medical care in patients with persistent fever: A cross-sectional study in four low- and middle-income countries. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Peirano G., Ahmed-Bentley J., Fuller J., Rubin J.E., Pitout J.D. Travel-related carbapenemase-producing Gram-negative bacteria in Alberta, Canada: The first 3 years. J. Clin. Microbiol. 2014;52:1575–1581. doi: 10.1128/JCM.00162-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van der Bij A.K., Pitout J.D. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J. Antimicrob. Chemother. 2012;67:2090–2100. doi: 10.1093/jac/dks214. [DOI] [PubMed] [Google Scholar]
- 41.Rogers B.A., Aminzadeh Z., Hayashi Y., Paterson D.L. Country-to-country transfer of patients and the risk of multi-resistant bacterial infection. Clin. Infect. Dis. 2011;53:49–56. doi: 10.1093/cid/cir273. [DOI] [PubMed] [Google Scholar]
- 42.Kallberg C., Ardal C., Blix H.S., Klein E., Martinez E., Lindbaek M., Outterson K., Rottingen J.A., Laxminarayan R. Introduction and geographic availability of new antibiotics approved between 1999 and 2014. PLoS ONE. 2018;13:e0205166. doi: 10.1371/journal.pone.0205166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gould D.J., Moralejo D., Drey N., Chudleigh J.H., Taljaard M. Interventions to improve hand hygiene compliance in patient care. Cochrane Database Syst. Rev. 2017;9:CD005186. doi: 10.1002/14651858.CD005186.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The information on two A. baumanni isolates with novel sequence types ST2145 (no. 71-20, ID5021) and ST2146 (no. 73-20, ID5022) was submitted to the MLST database (https://pubmlst.org/bigsdb?db=pubmlst_abaumannii_isolates, assessed on 3 August 2020).