Abstract
Antibiotic resistance is a significant crisis that threatens human health and safety worldwide. There is an urgent need for new strategies to control multidrug-resistant (MDR) bacterial infections. The latest breakthrough in gene-editing tools based on CRISPR/Cas9 has potential application in combating MDR bacterial infections because of their high targeting ability to specifically disrupt the drug resistance genes that microbes use for infection or to kill the pathogen directly. Despite the potential that CRISPR/Cas9 showed, its further utilization has been hampered by undesirable delivery efficiency in vivo. Nanotechnology offers an alternative way to overcome the shortcomings of traditional delivery methods of therapeutic agents. Advances in nanotechnology can improve the efficacy and safety of CRISPR/Cas9 components by using customized nanoparticle delivery systems. The combination of CRISPR/Cas9 and nanotechnology has the potential to open new avenues in the therapy of MDR bacterial infections. This review describes the recent advances related to CRISPR/Cas9 and nanoparticles for antimicrobial therapy and gene delivery, including the improvement in the packaging and localizing efficiency of the CRISPR/Cas9 components in the NP (nanoparticle)/CRISPR system. We pay particular attention to the strengths and limitations of the nanotechnology-based CRISPR/Cas9 delivery system to fight nosocomial pathogens.We highlight the need for more scientific research to explore the combinatorial efficacy of various nanoparticles and CRISPR technology to control and prevent antimicrobial resistance.
Keywords: CRISPR/Cas9, nanoparticle systems, antibiotic resistance, delivery
1. Introduction
Antibiotics have protected millions of people from bacterial infections through their remarkable ability to kill or inhibit the growth of bacterial pathogens [1,2]. Antibiotic resistance occurs when bacteria change over time and no longer respond to the available antimicrobial agents, making it harder to treat and increasing the risk of disease spread, severe illness, and death [2]. The recent increase in antibiotic resistance is dramatic and has rendered most of the available antibiotics less effective [3,4,5]. The World Health Organization (WHO, Geneva, Switzerland) has declared that antibiotic resistance is one of the top ten global public health threats facing humanity. In addition to death and disability, antibiotic has a significant social and economic impact caused by prolonged illness, extended hospitalization and healthcare, the need for more expensive medicine, which weigh most heavily on lower- and middle-income countries [6].
Misuse and overuse of antibiotics are the main driving force behind the development of multidrug resistance in bacteria [2,7]. Multidrug-resistant (MDR) bacteria succeed to acquire resistance that inactivate most of the antibiotics (including those that are considered the last resort of defense) through sequential genetic mutations and horizontal transfer of mobile genetic elements, increasing the importance for developing more effective antimicrobial agents [8]. In addition, traditional antibiotics indiscriminately kill beneficial bacteria, and deleteriously affect the commensal human microbiota [9]. Together, this highlights the need for new approaches that adopt different bactericidal mechanisms to avoid drug resistance and provide the capability to only target harmful bacteria and with minimal effect on the patient and other beneficial bacteria. The clustered regularly interspaced short palindromic repeats (CRISPR) CRISPR-associated protein (Cas) system can provide versatile and promising tools against the growing challenge of multidrug resistance prevalence [10]. Among the different types of CRISPR-Cas systems, the CRISPR-Cas9 system is the most widely applied in gene editing [11]. By designing guide RNAs, the CRISPR-Cas9 system can kill targeted bacterial species possessing specific sequences in the involved bacterial community or destroy their antibiotic resistance genes, resensitizing them to antibiotics [12]. Although the CRISPR-Cas9 system is a powerful and efficient tool to address the myriad of multidrug-resistant microbial infections, its delivery has become the major limitation for therapeutic applications [13]. Currently, both virus-based and nonviral gene delivery systems have been studied as delivery platforms for the CRISPR-Cas system [14]. However, compared to virus-based delivery, nonviral delivery systems such as nanoparticles might have more potential for future use, as it overcomes multiple disadvantages [15], such as toxicity and immunogenicity, previously reported for the use of viral vectors in gene delivery.
In this review, we aim to summarize the role of the CRISPR-Cas9 system in combating MDR bacteria, and we highlight the potential of nanoparticles to enhance the delivery of the CRISPR-Cas9 system.
2. CRISPR-Cas System
2.1. The CRISPR-Cas System Protects Bacteria from Phage Invasion
The CRISPR-Cas system, derived from the adaptive immune system of prokaryotes, has been found in approximately 50% of bacterial genomes and 87% of archaea [16]. CRISPR-Cas system was first observed in 1987 when Ishino reported a repeat sequence of unknown function in the Escherichia coli K12 genome [17]. It was not until 2002 that the repeat sequence was named CRISPR [18]. Barrangou et al. conducted phage infection experiments and reported that CRISPR and its adjacent cas gene could protect bacteria from phage invasion [19]. The CRISPR-Cas systems are composed of a genetic locus, which contains a CRISPR array of repetitive sequences (repeats) interspaced by short stretches of non-repetitive sequences (spacers), and 6–20 genes encoding CRISPR-associated (cas) proteins [20]. In addition, the leader region is adjacent to the CRISPR array and is required to guide the spacers towards the right location [21]. Sequences of leaders and repeats interact together to direct the specificity of spacer integration [22]. Spacer sequences are known as protospacers and are derived from the genetic elements of invading phages and plasmids. The cas operon, which lies upstream of the CRISPR array and determines the system′s gene-editing efficiency, plays a critical role in the CRISPR system.
The CRISPR-Cas system in bacteria degrades foreign DNA fragments in three steps: adaptation, expression, and interference [23]. In the stage of transformation or acquisition, an approximately 30 bp spacer sequence is integrated into the CRISPR array by Cas1 and Cas2. The second stage is the expression of CRISPR RNA (crRNA), in which spacers of the CRISPR locus (pre-crRNA) are transcribed and processed into crRNA [23]. These pre-crRNAs are cleaved by specific endoribonucleases to yield short mature crRNAs. The crRNA contains the spacer at the 5′ end and a repeat at the 3′ end. Ultimately, in the interference stage, crRNA recognizes and forms a base pair specific to the foreign target sequence. This hybridization leads to the sequence-specific cleavage of the crRNA-foreign sequence complex by Cas nucleases upon the second infection. It is worth noting that a short-conserved sequence known as the protospacer adjacent motif PAM (2–5 bp), located in close proximity to the sequence identical to the spacer on the foreign DNA, is indispensable for targeted DNA selection and degradation [24].
2.2. The CRISPR-Cas9 System Applied to Genome Editing
Based on the current classification put forward by Makarova, the CRISPR-Cas system is classified into two classes [25]. Class 1 CRISPR-Cas systems include types Ⅰ, Ⅲ, and Ⅳ, and Class 2 includes types Ⅱ, Ⅴ, and Ⅵ, which all contain multiple Cas proteins that function as effector proteins that are responsible for pre-crRNA processing [26]. The CRISPR-Cas type II system is an ideal choice for gene editing because all domains essential for DNA cleavage are integrated into a single protein [26,27]. Among the type II systems, the CRISPR-Cas9 system (Figure 1) has been widely applied in targeting virulence genes and specific genes that encode antibiotic resistance in bacteria [28,29]. In the CRISPR-Cas9 system, an additional small noncoding RNA, called the trans-activating crRNA (tracrRNA), is indispensable to form a unique dual RNA hybridization via base pairs complementary with the repeat sequence in the crRNA. The crRNA and tracrRNA complex is called single-guide RNA (sgRNA). sgRNA binds to Cas9 and directs it to the target site to generate double-strand breaks in chromosomal DNA. In a recent study, truncated sgRNA greatly led to a 10-fold reduction of gene knockout frequency, which are relevant for future sgRNA design approaches and studies of Cas9-DNA interactions [30]. The Cas9 endonuclease consists of two domains named the HNH and RuvC domains. The HNH domain is responsible for cutting the complementary (target) DNA strand, which complements the crRNA guide. Its active state formation and stability can be maintained in the presence of catalytic Mg2+ [31]. The RuvC domain is involved in the cleavage of a non-complementary (nontarget) DNA strand [32].
Figure 1.
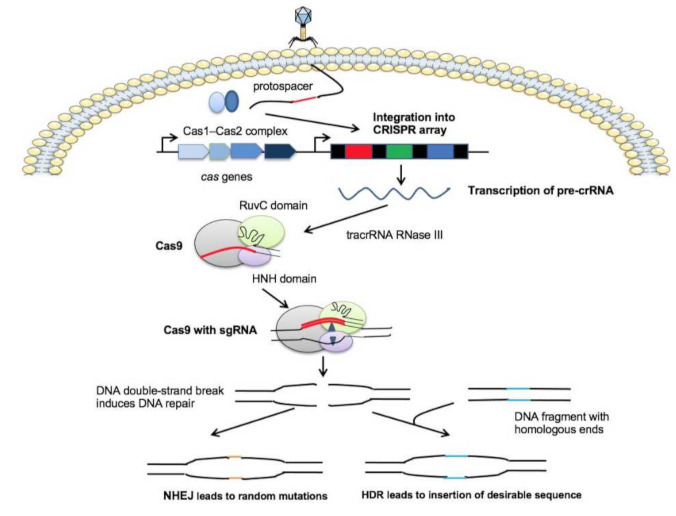
Molecular mechanism of the CRISPR-Cas 9 system. CRISPR-Cas systems are composed of a cas operon (blue arrows) and a CRISPR array which identical repeat sequences (black rectangles) that are interspersed by phage-derived spacers (colored rectangles). Upon phage infection, a sequence of the invading DNA (protospacer) is incorporated into the CRISPR array by the Cas1-Cas2 complex. The CRISPR array is then transcribed into a long precursor CRISPR RNA (pre-crRNA). In the CRISPR-Cas9 system, crRNA maturation requires tracrRNA, RNase III, and Cas9. The Cas9 protein contains two nuclease domains, the RuvC domain, and the HNH domain. Cas9 is guided by a sgRNA to induce a double-strand DNA break at a desired genomic locus. The sgRNA is composed of tracrRNA and crRNA. The tracrRNA hybridizes to the crRNA and binds to the Cas9 protein, forming the CRISPR-CAs9/sgRNA complex to edit genome sequences. DNA damage can be repaired by nonhomologous end joining (NHEJ), yielding short random insertions or deletions at the target site. Alternatively, a DNA sequence that shows partial complementarity to the target site can be inserted during homology-directed repair (HDR) for precise genome editing purposes.
Therefore, by designing multiple sgRNAs, it is facile and fast to use the CRISPR-Cas9 system to delete or insert specific sequences at the site of a genomic locus of interest in an extremely precise manner. The CRISPR-Cas9 system has received extensive attention for its extraordinary ability in genome editing and promising applications, including treating genetic diseases, genome engineering of bacteria, plants, and mammalian cells, and the reversal of antibiotic resistance as well [33]. The CRISPR-Cas system has also been successfully used in pathogenic fungi such as Candida albicans, Aspergillus, and Cryptococcus [34].
3. CRISPR-Cas System and Antibiotic Resistance
3.1. Relationships between the CRISPR-Cas System and Antibiotic Resistance
There are several studies indicate that the CRISPR-Cas system is associated with antibiotic resistance (Figure 2). For example, the Type I-F CRISPR system in E. coli was found associated with antibiotic susceptibility [35]. The CRISPR-Cas system in Francisella novicida maintains envelope integrity by regulating envelope lipoprotein expression to enhance antibiotic resistance [36]. The CRISPR-Cas system in Campylobacter jejuni was found involved in enhancing antibiotic resistance, as the deletion of the cas9 gene increased the sensitivity to antibiotics [37]. The findings of different studies reveal that the CRISPR-Cas system confers the competitive advantage over other variants for a population of bacteria that can acquire resistance genes [29]. In addition, the presence of the CRISPR-Cas system was correlated with the acquisition of antibiotic-resistant genes (ARGs), which can be positive or negative [38]: CRISPR-Cas-mediated immunity provides protection against foreign nucleic acids as well as the ARGs transfer. However, under a strong selective pressure imposed by antibiotics, the CRISPR-Cas system may be lost or devoid of function as they impede the acquisition of ARGs by horizontal gene transfer (HGT) [39].
Figure 2.
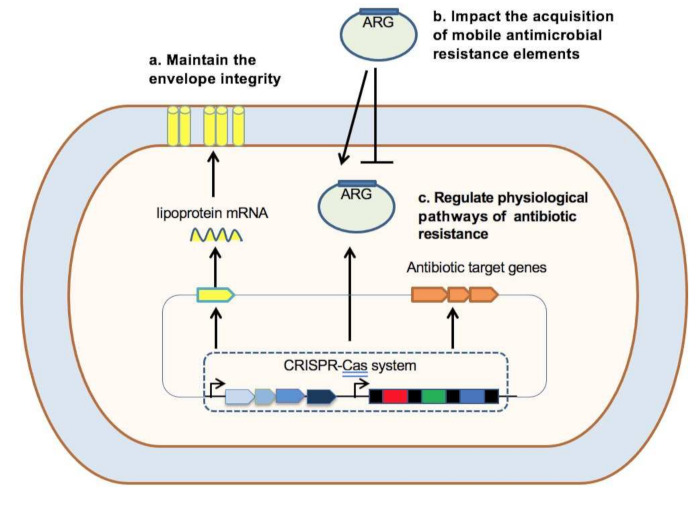
Relationships between the CRISPR-Cas system and antibiotic resistance in bacteria. a. The CRISPR-Cas system can maintain envelope integrity by regulating envelope lipoprotein expression to enhance antibiotic resistance, such as in Francisella novicida. b. The CRISPR-Cas system can impact the acquisition of mobile antimicrobial resistance elements to avoid antibiotic resistance, such as in Campylobacter jejuni and Enterococcus faecalis. c. The CRISPR-Cas system can regulate the physiological pathway involved in the antimicrobial resistance of bacteria, such as in Klebsiella pneumoniae. ARG: antibiotic resistance gene.
For example, some studies have reported that the CRISPR-Cas system can prevent the acquisition of antibiotic resistance genes in bacteria. Mackow et al. found that Klebsiella pneumoniae harboring the CRISPR-Cas system displayed a high sensitivity to carbapenems, a kind of antibiotic resistance usually caused by blaKPC plasmids [40]. In another study, Price and colleagues proved that the loss of the cas9 gene facilitated Enterococcus faecalis acquisition of resistance genes through conjugation [41]. Besides that, CRISPR-Cas expression increased the sensitivity of Mycobacterium smegmatis to the environmental stress, including acidic and oxidative stress, as well as multiple anti-tuberculosis agents, through reducing the drug-induced persistence [42]. These data support that the CRISPR-Cas system can impact the acquisition of mobile antimicrobial resistance elements or regulate the physiological pathway involved in the antimicrobial resistance of bacteria.
3.2. CRISPR-Cas System-Based Genome Editing to Combat Bacterial Infection
According to the CRISPR-Cas gene editing principle, the RNA-based spacer directs Cas proteins to target and cleave DNA that complements the spacers. In other words, guide RNAs can be designed to target virulence or antibiotic resistance genes that are specific to antimicrobial-resistant bacteria. Thus, the CRISPR-Cas9 system can be employed to neutralize antibiotic resistance genes in the targeted bacterial population without killing the beneficial bacteria in wild-type populations [43,44,45,46] (Figure 3). For example, the CRISPR-Cas9 system is being developed to restore the sensitivity to antibiotics in extend-spectrum beta-lactamase (ESBL)-producing Escherichia coli by identifying a conserved target sequence in >1000 ESBL mutants [44]. Moreover, with the high specificity of the CRISPR-Cas system, resistant strains can be selectively removed from complex bacterial populations by transforming the population with a plasmid or phage carrying a programmed CRISPR-Cas9 system targeting a unique sequence that only exists in resistant strains [43,47]. In Staphylococcus aureus, phagemid-mediated delivery of CRISPR-Cas9 was used to eliminate virulence genes and antibiotic resistance genes, thereby resensitizing bacteria to antibiotics [43]. In a recent study, Rodrigues et al. engineered conjugative plasmid pPD1 with a complete, constitutively expressed CRISPR-Cas9 targeting cassette that efficiently transfers to Enterococcus faecalis for the selective removal of ermB (encoding erythromycin resistance) and tetM (encoding tetracycline resistance) [46]. In vivo results showed that these transformants significantly reduced the prevalence of antibiotic-resistant intestinal E. faecalis and are immune to the uptake of antibiotic resistance determinants. The CRISPR-Cas9 system was also reported to cleave epidemic carbapenem-resistant plasmids, such as blaKPC-harboring IncFIIK-pKpQIL, IncN pKp58_N, and blaNDM-harboring IncX3 plasmids, through disrupting the partition gene parA in K. pneumoniae [48]. To increase the selective advantage of resensitized bacteria, Yosef et al. used temperate and lytic phages to deliver a programmed CRISPR-Cas9 system to destroy plasmids carrying beta-lactamase resistance genes blaNDM-1 and blaCTX-M-15 in E. coli [47], in which the CRISPR-Cas system targeted antimicrobial resistance genes was carried by a temperate phage. Strains transfected with this recombinant phage acquired resistance to lytic phages and thus had a selective advantage over resistant strains when treated with the same type of phage. Instead of directly killing bacteria, it aims to make bacteria sensitive to antibiotics and thereafter kill non-sensitized bacteria with lytic phages. This kind of strategy also eliminates the horizontal transfer of antibiotic resistance genes between bacteria. Apart from the CRISPR-Cas9, Kiga et al. developed CRISPR-Cas13a-based antimicrobial system in a bacteriophage capsid that is capable of effectively killing carbapenem-resistant E. coli and methicillin-resistant S. aureus targeting to sequence-specific antimicrobial resistance genes [49].
Figure 3.
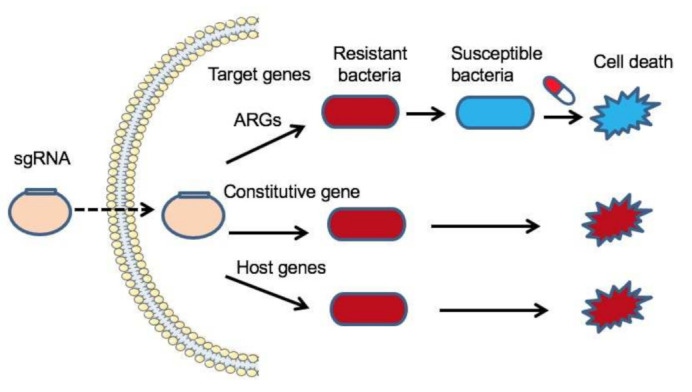
CRISPR-Cas system-based genome editing to combat bacterial infection. When the target genes are ARGs, the CRISPR-Cas system can restore the sensitivity of bacteria to antibiotics, and then the drugs will kill the susceptible bacteria. When the target genes are constitutive genes of the specific resistant bacteria, then the resistant bacteria will be killed. When the target genes are host chromosomal DNA that is cytotoxic to the resistant bacteria, the target cell will die because of the excision of the genome.
Furthermore, the CRISPR-Cas system can be used as a novel antimicrobial agent. Some reports have shown that the occasional or intentional acquisition of host chromosomal DNA by the CRISPR-Cas system is cytotoxic, which can result in cell death because of the excision of the genome [48,50]. Most of the CRISPR spacers match phages and plasmids, but some show similarity with chromosomal sequences, such as prophages, other mobile elements, and even the core genome. For example, Gomaa and colleagues reprogrammed the E. coli CRISPR-Cas type Ⅰ system with the spacer complementary to the essential ftsA gene, a critical gene involved in cell division, to kill bacteria efficiently regardless of genomic location, strand, or transcription activity [51]. Cañez et al. found that Type Ⅰ-E- and Type Ⅱ-A-based plasmids self-targeting lacZ genes were lethal in Streptococcus thermophilus survivors who had large genomic deletions during genome repair by homologous recombination [52].
The high efficiency of the CRISPR-Cas system in killing bacteria is attributed to Cas nuclease. For example, the introduction of self-targeting type Ⅰ systems in which Cas3 nuclease possesses both single-strand DNA exonuclease and 3′-5′ helicase activity induced rapid cell death and degraded the DNA away from the region of the targeted sequence region [53,54]. In another study, Hamilton et al. adopted the IncP type conjugative plasmid RK2 to deliver the CRISPR Cas9 system E. coli to Salmonella enterica with a high conjugation frequency to efficiently kill S. enterica [55]. The cis-conjugative plasmid was constructed based on pTA-Mob, a broad-host-range mobilization plasmid that is suitable for transferring large DNA to a large number of bacterial strains as donors by inserting the origin of transfer (oriT) and CRISPR system into it [56]. This plasmid had high conjugative efficiency because transconjugants became new donors for subsequent reconjugant. Therefore, the CRISPR-Cas9 system carried by this kind of plasmid kills S. enterica more efficiently. In Mycobacterium tuberculosis, an endogenous subtype III-A CRISPR-Cas system acts as an antimicrobial by introducing a recombinant phagemid carrying spacer-targeted essential genes into M. tuberculosis [57,58]. Moreover, the anti-tuberculosis bacteriophage can be delivered by inhalation devices, which implies versatile ways of drug delivery when using the CRISPR system to combat antibiotic-resistant strains [59].
These studies demonstrated that the CRISPR-Cas system can discriminate targeted strains better than antibiotics because even minor sequence differences are enough for the CRISPR-Cas system to identify. It is easy to realize that researchers can selectively eliminate closely related bacterial strains, whether in pure or mixed cultures, as long as they acquired the genome sequence information.
4. The Applications of Nanoparticles in Antibiotic Therapy
4.1. Nanoparticles Act as Antibacterial Materials
Nanoparticles (i.e., with at least one dimension between 1 nm and 100 nm) are widely used to enhance the delivery of antimicrobial agents act as novel antimicrobial material that is distinct from traditional drugs [60,61]. Nanoparticles (NPs) mainly rely on two mechanisms to act as promising antimicrobial agents against bacteria: (i) disruption of membrane potential and integrity and (ii) induction of oxidative stress via reactive oxygen species (ROS) generation catalyzed by NPs [62,63,64]. These two types of mechanisms can occur independently or simultaneously.
Inorganic nanoparticles usually possess a high surface-area-to-volume ratio and unique physical and chemical properties, which enhance their antibacterial activity. For instance, silver nanoparticles (AgNPs) have been widely used as potent antimicrobials against bacteria because of their remarkable bactericidal effect exceeding that of other metal oxides and reduced possibility of inducing resistance. Gold nanoparticles (AuNPs) are another type of inorganic nanoparticles that widely tested as antimicrobial agent because of their photothermal activity, biocompatibility, and easy modification with small antimicrobial drugs [65,66,67,68]. Furthermore, there is a particular interest in biosynthesized nanoparticles, such as copper nanoparticles prepared with Zingiber officinale, and Curcuma longa that displayed a remarkable antibacterial activity against multidrug resistant Staphylococcus aeureus [68]. Metal oxide NPs of titanium dioxide (TiO2) that adopts a similar bactericidal mechanism as AgNPs to kill both Gram-positive and Gram-negative bacteria were extensively reported [69]. Moreover, combined with other nanomaterials, such as zinc oxide (ZnO), TiO2 nanoparticles showed considerable activity against methicillin-resistant Staphylococcus aureus (MRSA) [70]. Compared with the metal oxides described above, antimicrobial ZnO is much safer and holds high biocompatibility with human cells [71,72,73,74]. ZnO NPs dispersed in ionic liquids displayed a high efficiency in killing the skin-specific bacterium Staphylococcus epidermidis through ROS production, resulting in bacterial cell lysis without toxicity to normal keratinocyte cells [72].
Besides inorganic nanoparticles, polymeric nanoparticles are used in antibacterial applications as well. Polymeric nanoparticles can directly kill microbes by interacting with bacterial cell walls, which are typically negatively charged. Polymeric nanoparticles are endowed with intrinsic antimicrobial activity by incorporating cationic and hydrophobic moieties into polymer chains, such as quaternary ammonium groups, alkyl pyridiniums, and phosphonium. Cationic groups are able to disrupt the cell membrane; meanwhile hydrophobic moieties help to penetrate and burst into the membrane. It has been reported that the antibacterial effect can be improved by increasing the density of cationic charges, which enhance the electrostatic interactions with anionic membranes [75]. Therefore, it is possible to design various polymeric nanoparticles with a positively charged surface to create different antimicrobial materials. For instance, Takahashi et al. reported a cationic amphiphilic polymeric nanoparticle that effectively killed the planktonic cariogenic bacterial Streptococcus mutants and prevented biofilm formation [76]. Another antibacterial peptide-based copolymer micelle was synthesized and exhibited potent bactericidal efficacy against both gram-negative and gram-positive bacteria. Transmission electron microscopy (TEM) results revealed that the micelles can penetrate and then destroy the bacterial cell membrane, leading to cell lysis [77]. Due to their prominent bactericidal properties, NPs were also applied in the coating of human implantable devices, wound dressings, bone cement, and dental materials [77,78,79,80,81].
4.2. Nanoparticles for Drug/Gene Delivery to Combat Bacterial Infection
Apart from directly eliminating bacteria, polymeric nanoparticles can indirectly fight bacteria by acting as drug carriers to deliver antibiotics, antimicrobial peptides, and antimicrobial agents to the target parts of the body. The main limitations of clinical antibiotic therapy are the low drug bioavailability, poor penetration to bacterial infection sites, side effects of antibiotics, and also antibiotic resistance [82]. Polymeric nanoparticles can protect antibiotics from being environmentally deactivated and improve their pharmacokinetics and distribution in the body. The most prevalent approach to produce antibiotic-loaded polymeric nanoparticles is entrapping antibiotics into polymeric particles, which enhances the solubility of hydrophobic drugs and enhances the antibacterial effect. In a study, ciprofloxacin-loaded polymeric nanoparticles were developed with a continuous moderate release rate and high antibiotic concentration at the targeted site [83]. Hasan and coworkers synthesized positively charged clindamycin-loaded polyethyleneimine nanoparticles (Cly/PPNPs) and proved that they had enhanced bactericidal efficacy against MRSA because of their strong bacterial adhesive ability [84]. In another recent study, polymeric nanoparticles loaded with the broad-spectrum antibiotic sparfloxacin and the anti-inflammatory immunosuppressant tacrolimus displayed potent antibacterial activity and the ability to precisely locate inflammatory cells to treat acute lung sepsis [85]. These results showed that polymeric nanoparticles have promising prospects in preventing or treating infectious diseases.
In addition to drug delivery, nanoparticles for gene delivery have also attracted enormous attention. Naked genetic elements cannot efficiently enter target cells because of adsorption of serum proteins, rapid clearance in blood circulation, phagocyte uptake, incapability of endosomal escape, lack of targeting ability, and toxicity induced by the immune system. To overcome these drawbacks, many types of nanoparticles have been developed as gene carriers, including polymeric nanoparticles, lipid nanoparticles, and metal nanoparticles [86]. For instance, DNA or RNA can be encapsulated in poly (lactic-coglycolic acid) nanoparticles to protect them from degradation during circulation. Among the diverse nanoparticles, lipid-based nanoparticles have been extensively explored because of their liposomal-like characteristics, which facilitate cellular entry. To improve the targeting ability, ligands such as antibodies, proteins, or peptides, can be functionalized with the gene-delivery nanoparticles to specifically bind to the receptors on the targeted cells [87,88]. Polyethyleneglycol (PEG) modification with nanoparticles is widely used to inhibit nonspecific interactions with serum proteins and increase active targeting efficiency by increasing their circulation time in the bloodstream [89,90,91].
5. Nanoparticles as CRISPR-Cas9 Delivery Systems
5.1. Delivery Strategies for CRISPR-Cas9 System
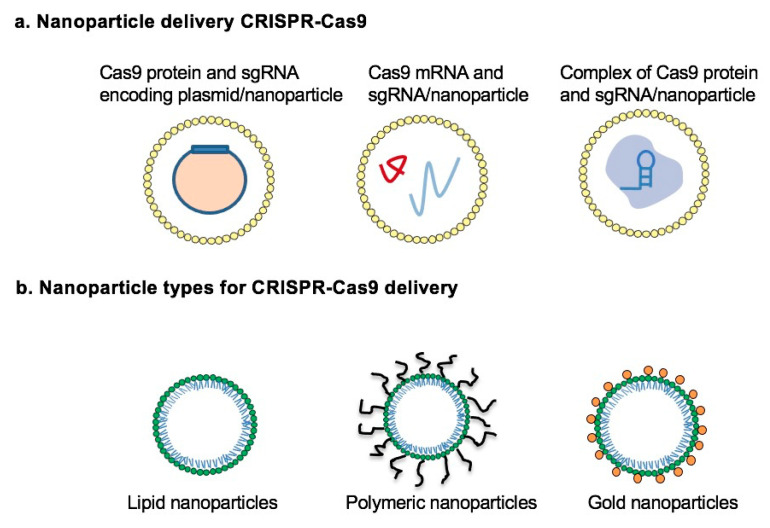
To apply CRISPR-Cas9 system gene editing, the endonuclease Cas9 and guide RNA for the CRISPR system should be included in delivery vehicles. Strategies for delivery of the CRISPR/Cas9 system (Figure 4) include: (i) a plasmid encoding Cas9 and guide RNA; (ii) two separate plasmids encoding Cas9 and guide RNA; (iii) mRNA for Cas9 and guide RNA; and (iv) a Cas9-guide RNA complex [92]. Compared with the mRNA delivery form, a plasmid encoding Cas9 and guide RNA is more stable and cost-effective than the mRNA forms of Cas9 and guide RNA. However, it is difficult for a CRISPR-delivered DNA plasmid to enter the cellular and nuclear membranes and then take effect. The large size of the plasmid, which consists of many noncoding sequences, makes it challenging to be encapsulated with nanoparticles. However, the constitutive expression of CRISPR in mammalian cells is prone to increase off-target effects and the risk of insertion mutagenesis by randomly integrating plasmid DNA into the host genome [93].
Figure 4.
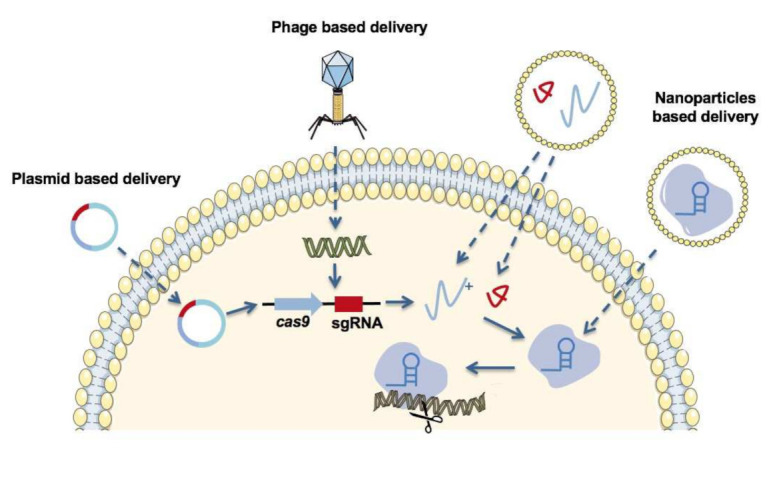
Different strategies for CRISPR-Cas9 system delivery to edit genes. Plasmid-based delivery: The plasmid-borne CRISPR-Cas system can be transferred into cells and transcribed into Cas9 mRNA and sgRNA. Cas9 mRNA is translated into the Cas9 protein, which forms a ribonucleoprotein (RNP) complex with sgRNA. Then, the RNP complex edits the target genes directed by sgRNA. Phage-based delivery: CRISPR-Cas system coding sequences are delivered by phages into cells. Nanoparticle-based delivery: Cas9 and sgRNA can be delivered either in the form of mRNA or Cas9-sgRNA ribonucleoprotein (RNP) complexes with the help of nanoparticles.
Encoded Cas9 mRNA delivery can overcome these drawbacks and be directly translated in the cytoplasm without entering the nucleus, resulting in reduced off-target effects and less risk of integration into the genome. However, mRNA is unstable and easily degraded by RNases either during the synthesis or when applied in vivo. Cas9-guide RNA ribonucleoprotein (RNP) complex delivery is the most straightforward and rapid approach for gene editing and has fewer off-target effects as well as low immunogenicity [94]. However, it is also the most challenging approach because of the large size of the Cas9 protein, the super negative charges of guide RNA, and the vulnerability to degradation and denaturation of RNPs during the entire process [95,96,97]. The development of stable and reliable nanoparticles for RNP delivery systems remains elusive. Plasmids or viral vectors, such as species-specific phages and adeno-associated viruses, are exploited for CRISPR-Cas delivery [98,99]. However, both methods are limited in practical use because of their low loading and packaging efficiency, narrow host range, risk of carcinogenesis, and immunogenicity.
5.2. Nanoparticles for CRISPR-Cas9 Delivery
To overcome the drawbacks of viral vectors, nonviral vectors for delivering the CRISPR system, mainly through nanoparticle-based deliveries such as lipid nanoparticles, polymeric nanoparticles, and gold nanoparticles (AuNPs), have attracted significant interest in researchers [100] (Figure 5). Nanoparticles with chemical modification can protect Cas9 mRNA by improving its stability. Lipid nanoparticles are one of the most extensively explored nanoparticle systems for CRISPR delivery. Several lipid-based carriers for gene therapy have been approved for clinical trials [101]. Lipid nanoparticles are amphiphilic compounds that help encapsulate negatively charged CRISPR plasmid DNA and mRNA, guiding and protect RNA from crossing the cell membrane. Liu and coworkers reported that BAMEA-O16B, a lipid nanoparticle integrated with disulfide bonds, delivered Cas9 mRNA and sgRNA simultaneously to knock out green fluorescent protein (GFP) expression in human embryonic kidney cells [102]. This gene knockout efficiency can be as high as 90%. However, when designing nanoparticles for gene delivery, it is quite challenging to selectively target specific tissues. Cheng et al. developed a strategy named selective organ targeting (SORT), which was used to modify lipid NPs with a diverse percentage of SORT molecules to precisely deliver Cas9 mRNA/sgRNA together with Cas9 ribonucleoprotein to the liver, lung, and spleen [103]. To enhance the genome editing efficiency and cell-selective ability, Tang et al. designed a cationic lipid system that includes a phenylboronic acid (PBA) group to self-assemble with CRISPR/Cas9 mRNA into nanoparticles. This kind of configuration showed an improved cellular uptake by cancer cells that overexpress surface sialic acid (SA), due to the interfacial interaction of PBA and SA [104]. This delivery system suppressed p53 mRNA significantly in Hela cancer cells with a higher efficiency than non-cancer cells. Similarly, liposome-templated hydrogel nanoparticles (LHNPs) for CRISPR/Cas9 delivery were synthesized to knock out polo-like kinase1 (PLK1) gene in a mouse flank tumor model to inhibit tumor growth with higher efficiency than commercial agent Lipofectamine 2000 [105]. Several nanoparticles delivering the CRISPR-Cas9 system are compiled in Table 1.
Figure 5.
Nanoparticles for drug/gene delivery to combat bacterial infection. (a) Nanoparticles enfold different CRISPR-Cas9 delivery forms: Cas9 protein and sgRNA encoding plasmid, Cas9 mRNA and sgRNA, Complex of Cas9 protein and sgRNA. (b) Nanoparticles for CRISPR-Cas9 delivery: lipid nanoparticles, polymeric nanoparticles, and gold nanoparticles.
Table 1.
Nanoparticle delivery systems for CRISPR-Cas9.
| Delivery System | Crispr-Cas Form | Study Objective | Target Gene | In Vitro/ Vivo |
Brief Result | Ref |
|---|---|---|---|---|---|---|
| Lipid-based NPs. | Cas9 mRNA and sgRNA | Hepatocytes, C57BL/6 mice | PCSK9 | In vivo | The lipid NPs delivered-CRISPR/Cas9 effectively knocked the protein level of PCSK9 in mouse serum down to 20%. | [99] |
| Lipid-based NPs | Cas9 mRNA and sgRNA | C57BL/6 mice | PTEN PCSK9 |
In vivo | SORT LNPs mediated effective tissue-specific genes PTEN and PCSK9 editing in the liver. | [100] |
| Lipid-based NPs | Cas9 mRNA and sgRNA | Hela cells | GFP HPV18E6 |
In vitro | treatment of HeLa cells with PBA-BADP/Cas9 mRNA/sgHPV18E6 NPs showed GFP knocked out efficiency up to 50% and resulted in 18.7% indel of HPV18E6 gene. |
[101] |
| Lipid-based NPs | Ribonucleoproteins | U87 cells Mice bearing tumor |
PLK1 | Both in vitro and in vivo | LHNPs co-encapsulated with Cas9 and minicircle sgRNA were capable of efficiently inhibiting PLK1 expression to 36.3% and inhibit tumor growth | [102] |
| Lipid-based NPs | Cas9 mRNA and sgRNA | BMDMs C57BL/6 mice |
NLRP3 | Both in vitro and in vivo | Disrupt NLRP3 of macrophages in vitro by CLANmCas9 with an efficiency rate of 70.2%, compared to the rate of 58.6% in vivo. | [106] |
| Lipid-based NPs | Cas9 mRNA and sgRNA | Splenic endothelial cells HEK293 | ICAM-2 | In vivo | LNPs can edit endothelial cells successfully, and the ideal Cas9: sgRNA ratio will depend on the relative stability of the two molecules. | [107] |
| Lipid-based NPs | Cas9 mRNA and sgRNA | HEK293, GBM 005 cells | GFP, PLK1 | In vivo | CRISPR-LNPs against PLK1 enabled up to ~70% gene editing in vivo, inhibited tumor growth by 50%, and improved survival by 30%. | [108] |
| Lipid-based NPs | Ribonucleoproteins | B16F10 cells | PD-L1 | In vitro | VLN-sgPD-L1 reduced the expression of PD-L1 to 41.3% and thus suppressed tumor growth in vivo. | [109] |
| LNP-INT01 | Cas9 mRNA and sgRNA | Cd-a-mice | TTR | In vivo | CRISPR-LNPs against TTR in the liver of mice resulted in a 97% reduction in serum protein levels that persisted for at least 12 months. | [110] |
| Lipidoid NPs | Ribonucleoproteins | Hela-DsRed cells | GFP | In vivo | LNPs-based CRISPR/Cas9 system displayed high GFP knockout efficacies ~70% with low cytotoxicities. | [58] |
| Gold/lipid NPs | Plasmid DNA | Melanoma | Plk-1 | Both in vitro and in vivo | AuNPs-based CRISPR/Cas9 system led to about 65% down-regulation of Plk-1 protein triggered by the photothermal effect. | [111] |
| Gold NPs | Ribonucleoproteins | Fragile X syndrome | mGluR5 | In vivo | CRISPR-Gold targeting the mGluR5 gene reduced the protein level by 40–50% in the mouse models that have fragile X syndrome | [112] |
| Au NPs | Ribonucleoproteins | HSPCs | CCR5 | In vitro | NPs-mediated CRISPR/Cas9 successfully penetrated into HSPCs and produced up to 17.6% total editing. | [113] |
| Au NPs | Ribonucleoproteins | Hepa 1-6 cells mice |
Pcsk9 | Both in vitro and in vivo | This Au Nps-based CRISPR/Cas9 delivery system induced significant Pcsk9 editing in vitro and reduced the LDL-C level to 30% compared with the control group by knocking out the Pcsk9 gene in mice. | [114] |
| polymeric NPs | Plasmid DNA | Chronic myeloid leukemia | CML-related BCR-ABL | Both in vitro and in vivo | CLANpCas9/gBCR-ABL disrupted the BCR-ABL gene in vitro with an efficiency rate of 46.8%, reduced the mRNA level to 41.9%, and greatly inhibited the protein expression of BCR-ABL in CML mice. | [115] |
| polymeric NPs | Plasmid DNA | HEK293T cell | dTomato | in vitro | Polymeric microcarriers for CRISPR/Cas9 displayed high gene knockout efficiency up to 70% in the transfected cells. | [116] |
| polymeric NPs | Plasmid DNA | HFD-induced T2D mice | NE | In vivo | CLANpCas9/gNE targeting the neutrophil elastase (NE) gene effectively disrupted the NE gene in the mouse have type 2 diabetes (T2D) with the gene knock-out rate of 26.4% and mitigated the insulin resistance by reducing neutrophils-related inflammation | [117] |
| polymeric NPs | Plasmid DNA | Hela cells HEK293T cell |
GFP iRFP |
In vitro | A novel reporter system involving PBAEs-CRISPR carrier for easy detection of gene knockout at one and two genomic sites | [118] |
| Polymeric NPs | Ribonucleoproteins | S. aureus | MecA | In vitro | Cr-Nanocomplex treatment resulted in a significant inhibition in MRSA growth in the presence of methicillin by disrupting the mecA gene. | [119] |
| Core-shell NPs with iron oxide core and PEI coating | Plasmid DNA | Porcine fetal fibroblasts | H11 | In vitro | Magnetic NPs carrying CRISPR/Cas9 displayed 3.5 times higher efficiency compared to the classic lipofection method | [120] |
| PEI magnetic NPs | Plasmid DNA | HEK293 cell | TLR-3 | In vitro | Magnetic NPs-CRISPR/Cas delivery system enabled site-specific incision with the combination of an inhomogeneous magnetic field. | [121] |
| PLGA NPs | Ribonucleoproteins | HSPCs | γ-globin gene | In vitro | CRISPR/Cas9-PLGA-NPs-mediated gene inaction ofγ-globin gene in HSPCs led to the increase in the HbF expression (51.7%) in a concentration-dependent manner. | [122] |
| pH-responsive polymeric NPs | Plasmid DNA | B16F10 cells | Cdk5 | Both in vitro and in vivo | The CRISPR/Cas9 encapsulated in nanoparticles specifically knock out the Cyclin-dependent kinase 5 (Cdk5) gene to significantly attenuate the expression of PD-L1 on tumor cells. | [123] |
Polymeric NPs are another important method for CRISPR delivery owing to their low immunogenicity and high biocompatibility. Polymeric nanoparticles can be conjugated with cell-penetrating peptides on the surface for quick cellular uptake and/or nuclear localization signal peptides for delivery inside cells. Carboxylated branched poly (β-amino ester) nanoparticles were reported to be an efficient way to deliver Cas9 RNPs for increased hydrogen bonding and hydrophobic effects [118]. The authors proved that this Cas9 RNP delivery polymer induced high gene editing levels both in vitro and in vivo at relatively low RNP doses. Apart from Cas9 RNPs, they also found that polymeric nanoparticles are applicable for transferring CRISPR-Cas9 plasmids [118]. Recently, Nguyen et al. reported a novel platform combining polymeric nanoparticles and a modified homology-directed repair template [124]. Polymeric NPs consisted of anionic poly-L-glutamic acid (PGA) help stabilize RNPs by shielding excess positively charged residues of the Cas9 protein, which resulted in enhancing gene editing efficiency and cell viability and reducing off-target effects. The polymeric and hybrid poly-L-arginine hydrochloride/dextran sulfate (PARG/DEXS)3 capsules that deliver CRISPR/Cas9 in the form of plasmids was reported to successfully knock out the dTomato gene in HEK293 T cells [116]. In addition, a previous research study has demonstrated the macrophage-specific gene editing using CRISPR/Cas9 components delivered through cationic lipid-assisted PEG-b-PLGA nanoparticles (CLANs), and PEGylation was proposed as an effective strategy to prevent non-specific interactions and avoid immune recognition [117].
In addition to polymeric NPs, AuNPs are considered quite suitable for CRISPR RNP complex delivery due to their unique controllable features, precise modification, and relative safety compared to lipid and polymer nanocarriers. Shahbazi and coworkers designed an AuNP-based CRISPR nanoformulation (AuNP/CRISPR) with the conjugation of the CRISPR RNP complex on the surface of AuNPs [113]. This AuNP/CRISPR delivery system successfully penetrated hard-to-transfect CD34+ hematopoietic cells (HSPCs) and edited CCR5 and γ-globin promoter gene loci without generating any adverse effects. Their results showed that other blood cell types could also be edited by AuNP/CRISPR, which provides a powerful delivery method in different cells. Moreover, another study proved that the cationic HIV-1-transactivating transcriptor (TAT) peptide-modified gold nanoclusters-carrying CRISPR/Cas9 system could be used to decrease lipoprotein cholesterol (LDL-C) level by disrupting the Pcsk9 gene in mice model, which indicates a new therapeutic approach for the treatment of cardiovascular disease [114]. Besides that, gold nanoclusters (AuNCs) were also used to deliver Streptococcus pyogenes Cas9 endonuclease (SpCas9) into the nucleus via a highly pH-dependent assembly process [125]. The SpCas9-AuNC complex is stable at higher pH and disassembles at lower pH. The authors successfully employed this delivery system to knock out the E6 oncogene responsible for cervical cancer malignant transformation. Although there are few studies on applying NP/CRISPR systems to bacteria, similar targeted strategies can be adopted to inhibit microbial infections by directly eradicating pathogens or removing antimicrobial resistance genes [15,119]. For example, Kang et al. reported a nanosized CRISPR complex that consisted of a polymer-derivatized Cas9 endonuclease and a single guide RNA targeting the major methicillin resistance gene mecA in S. aureus. The polymer-derivatized Cas9 endonuclease was produced by covalent modification with branched polyethyleneimine. This nanosized CRISPR complex can be successfully delivered into MRSA and efficiently edit the bacterial genome, resulting in reduced growth of MRSA [119].
6. Summary and Future Prospects
CRISPR-Cas systems are promising gene-editing tools for controlling the prevalence of antibiotic resistance genes among bacteria and eliminating pathogens with high precision. Off-target effects, high cost, systemic delivery, as well as delivery efficiency are major challenges in this regard. These issues may be overcome via the employment of nanomaterials as nonviral carriers for the delivery of the CRISPR/Cas9 system. Multiple innovative nanoparticles of polymers, lipids, and gold have been developed. Although tremendous progress has been made in designing these nanoparticles to optimize the effect of the CRISPR-Cas system, achieving higher efficiency and safer delivery of the system remains a challenge, and further investigations are needed.
Efficient packaging and localization of the CRISPR-Cas components are two main obstacles for NP/CRISPR application. As we discussed above, NPs can be tailored in diverse ways. PEGylation modification of the surface of the system is a common strategy to reduce reticuloendothelial system (RES)-mediated clearance and increase duration time in the blood or tissue. Other modifications, such as cell-penetrating peptides and specific cell receptors, can improve cell internalization and interaction with targeted cells. Immunogenicity and off-target editing effects are also concerns regarding in vivo applications. Nevertheless, the integration of nanoparticles and the CRISPR system is still in the early stage. There is a long way to go before the successful application of engineered nanoparticles and CRISPR-Cas systems to treat bacterial infections and control the dissemination of antimicrobial-resistant bacteria.
Author Contributions
Conceptualization, Q.L., M.S.D., and Z.R.; writing F.W.; revising M.S.D., M.G., and W.Y.; funding acquisition, Q.L. and F.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China (2017YFC1600100), National Natural Science Foundation of China (81702040, 31900021), the National Science Foundation of Zhejiang province, China (LY20H190002).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mohr K.I. History of Antibiotics Research. Curr. Top. Microbiol. Immunol. 2016;398:237–272. doi: 10.1007/82_2016_499. [DOI] [PubMed] [Google Scholar]
- 2.Dodds D.R. Antibiotic resistance: A current epilogue. Biochem. Pharmacol. 2017;134:139–146. doi: 10.1016/j.bcp.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Bassetti M., Poulakou G., Ruppe E., Bouza E., Van Hal S.J., Brink A. Antimicrobial resistance in the next 30 years, humankind, bugs and drugs: A visionary approach. Intensive Care Med. 2017;43:1464–1475. doi: 10.1007/s00134-017-4878-x. [DOI] [PubMed] [Google Scholar]
- 4.Hornischer K., Haussler S. Diagnostics and Resistance Profiling of Bacterial Pathogens. Curr. Top. Microbiol. Immunol. 2016;398:89–102. doi: 10.1007/82_2016_494. [DOI] [PubMed] [Google Scholar]
- 5.Domalaon R., Idowu T., Zhanel G.G., Schweizer F. Antibiotic Hybrids: The Next Generation of Agents and Adjuvants against Gram-Negative Pathogens? Clin. Microbiol. Rev. 2018;31 doi: 10.1128/CMR.00077-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Havenga B., Ndlovu T., Clements T., Reyneke B., Waso M., Khan W. Exploring the antimicrobial resistance profiles of WHO critical priority list bacterial strains. BMC Microbiol. 2019;19:303. doi: 10.1186/s12866-019-1687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 8.De Oliveira D.M.P., Forde B.M., Kidd T.J., Harris P.N.A., Schembri M.A., Beatson S.A., Paterson D.L., Walker M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020;33 doi: 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cieplak T., Soffer N., Sulakvelidze A., Nielsen D.S. A bacteriophage cocktail targeting Escherichia coli reduces E. coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota. Gut Microbes. 2018;9:391–399. doi: 10.1080/19490976.2018.1447291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bikard D., Barrangou R. Using CRISPR-Cas systems as antimicrobials. Curr. Opin. Microbiol. 2017;37:155–160. doi: 10.1016/j.mib.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Barman A., Deb B., Chakraborty S. A glance at genome editing with CRISPR-Cas9 technology. Curr. Genet. 2020;66:447–462. doi: 10.1007/s00294-019-01040-3. [DOI] [PubMed] [Google Scholar]
- 12.Aslam B., Rasool M., Idris A., Muzammil S., Alvi R.F., Khurshid M., Rasool M.H., Zhang D., Ma Z., Baloch Z. CRISPR-Cas system: A potential alternative tool to cope antibiotic resistance. Antimicrob. Resist. Infect. Control. 2020;9:131. doi: 10.1186/s13756-020-00795-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilbie D., Walther J., Mastrobattista E. Delivery Aspects of CRISPR/Cas for in Vivo Genome Editing. Acc. Chem. Res. 2019;52:1555–1564. doi: 10.1021/acs.accounts.9b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu M., Glass Z., Xu Q. Nonviral Nanoparticles for CRISPR-Based Genome Editing: Is It Just a Simple Adaption of What Have Been Developed for Nucleic Acid Delivery? Biomacromolecules. 2019;20:3333–3339. doi: 10.1021/acs.biomac.9b00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verma R., Sahu R., Singh D.D., Egbo T.E. A CRISPR/Cas9 based polymeric nanoparticles to treat/inhibit microbial infections. Semin. Cell Dev. Biol. 2019;96:44–52. doi: 10.1016/j.semcdb.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H., et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishino Y., Krupovic M., Forterre P. History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology. J. Bacteriol. 2018;200 doi: 10.1128/JB.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansen R., Embden J.D., Gaastra W., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 19.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 20.Shabbir M.A., Hao H., Shabbir M.Z., Hussain H.I., Iqbal Z., Ahmed S., Sattar A., Iqbal M., Li J., Yuan Z. Survival and Evolution of CRISPR-Cas System in Prokaryotes and Its Applications. Front. Immunol. 2016;7:375. doi: 10.3389/fimmu.2016.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieper S.N., Almendros C., Brouns S.J.J. Conserved motifs in the CRISPR leader sequence control spacer acquisition levels in Type I-D CRISPR-Cas systems. FEMS Microbiol. Lett. 2019;366 doi: 10.1093/femsle/fnz129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grainy J., Garrett S., Graveley B.R., Terns M.P. CRISPR repeat sequences and relative spacing specify DNA integration by Pyrococcus furiosus Cas1 and Cas2. Nucleic Acids Res. 2019;47:7518–7531. doi: 10.1093/nar/gkz548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J. CRISPR-Cas: Adapting to change. Science. 2017;356 doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 24.Globyte V., Lee S.H., Bae T., Kim J.S., Joo C. CRISPR/Cas9 searches for a protospacer adjacent motif by lateral diffusion. EMBO J. 2019;38 doi: 10.15252/embj.201899466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P., et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koonin E.V., Makarova K.S., Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adiego-Perez B., Randazzo P., Daran J.M., Verwaal R., Roubos J.A., Daran-Lapujade P., van der Oost J. Multiplex genome editing of microorganisms using CRISPR-Cas. FEMS Microbiol. Lett. 2019;366 doi: 10.1093/femsle/fnz086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bikkarolla S.K., Nordberg V., Rajer F., Muller V., Kabir M.H., Sriram K.K., Dvirnas A., Ambjornsson T., Giske C.G., Naver L., et al. Optical DNA Mapping Combined with Cas9-Targeted Resistance Gene Identification for Rapid Tracking of Resistance Plasmids in a Neonatal Intensive Care Unit Outbreak. mBio. 2019;10 doi: 10.1128/mBio.00347-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gholizadeh P., Aghazadeh M., Asgharzadeh M., Kafil H.S. Suppressing the CRISPR/Cas adaptive immune system in bacterial infections. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:2043–2051. doi: 10.1007/s10096-017-3036-2. [DOI] [PubMed] [Google Scholar]
- 30.Graf R., Li X., Chu V.T., Rajewsky K. sgRNA Sequence Motifs Blocking Efficient CRISPR/Cas9-Mediated Gene Editing. Cell Rep. 2019;26:1098–1103.e3. doi: 10.1016/j.celrep.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuo Z., Liu J. Structure and Dynamics of Cas9 HNH Domain Catalytic State. Sci. Rep. 2017;7:17271. doi: 10.1038/s41598-017-17578-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sternberg S.H., LaFrance B., Kaplan M., Doudna J.A. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Robledo J.E., Barrera M.C., Tobon G.J. CRISPR/Cas: From adaptive immune system in prokaryotes to therapeutic weapon against immune-related diseases. Int. Rev. Immunol. 2020;39:11–20. doi: 10.1080/08830185.2019.1677645. [DOI] [PubMed] [Google Scholar]
- 34.Roman E., Coman I., Prieto D., Alonso-Monge R., Pla J. Implementation of a CRISPR-Based System for Gene Regulation in Candida albicans. mSphere. 2019;4:e00001-19. doi: 10.1128/mSphere.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aydin S., Personne Y., Newire E., Laverick R., Russell O., Roberts A.P., Enne V.I. Presence of Type I-F CRISPR/Cas systems is associated with antimicrobial susceptibility in Escherichia coli. J. Antimicrob. Chemother. 2017;72:2213–2218. doi: 10.1093/jac/dkx137. [DOI] [PubMed] [Google Scholar]
- 36.Sampson T.R., Napier B.A., Schroeder M.R., Louwen R., Zhao J., Chin C.Y., Ratner H.K., Llewellyn A.C., Jones C.L., Laroui H., et al. A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proc. Natl. Acad. Sci. USA. 2014;111:11163–11168. doi: 10.1073/pnas.1323025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shabbir M.A., Wu Q., Shabbir M.Z., Sajid A., Ahmed S., Sattar A., Tang Y., Li J., Maan M.K., Hao H., et al. The CRISPR-cas system promotes antimicrobial resistance in Campylobacter jejuni. Future Microbiol. 2018;13:1757–1774. doi: 10.2217/fmb-2018-0234. [DOI] [PubMed] [Google Scholar]
- 38.Shehreen S., Chyou T.Y., Fineran P.C., Brown C.M. Genome-wide correlation analysis suggests different roles of CRISPR-Cas systems in the acquisition of antibiotic resistance genes in diverse species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019;374:20180384. doi: 10.1098/rstb.2018.0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackow N.A., Shen J., Adnan M., Khan A.S., Fries B.C., Diago-Navarro E. CRISPR-Cas influences the acquisition of antibiotic resistance in Klebsiella pneumoniae. PLoS ONE. 2019;14:e0225131. doi: 10.1371/journal.pone.0225131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price V.J., Huo W., Sharifi A., Palmer K.L. CRISPR-Cas and Restriction-Modification Act Additively against Conjugative Antibiotic Resistance Plasmid Transfer in Enterococcus faecalis. mSphere. 2016;1:e00064-16. doi: 10.1128/mSphere.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei J., Lu N., Li Z., Wu X., Jiang T., Xu L., Yang C., Guo S. The Mycobacterium tuberculosis CRISPR-Associated Cas1 Involves Persistence and Tolerance to Anti-Tubercular Drugs. Biomed. Res. Int. 2019;2019:7861695. doi: 10.1155/2019/7861695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bikard D., Euler C.W., Jiang W., Nussenzweig P.M., Goldberg G.W., Duportet X., Fischetti V.A., Marraffini L.A. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J.S., Cho D.H., Park M., Chung W.J., Shin D., Ko K.S., Kweon D.H. CRISPR/Cas9-Mediated Re-Sensitization of Antibiotic-Resistant Escherichia coli Harboring Extended-Spectrum beta-Lactamases. J. Microbiol. Biotechnol. 2016;26:394–401. doi: 10.4014/jmb.1508.08080. [DOI] [PubMed] [Google Scholar]
- 45.Price V.J., McBride S.W., Hullahalli K., Chatterjee A., Duerkop B.A., Palmer K.L. Enterococcus faecalis CRISPR-Cas Is a Robust Barrier to Conjugative Antibiotic Resistance Dissemination in the Murine Intestine. mSphere. 2019;4:e00464-19. doi: 10.1128/mSphere.00464-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodrigues M., McBride S.W., Hullahalli K., Palmer K.L., Duerkop B.A. Conjugative Delivery of CRISPR-Cas9 for the Selective Depletion of Antibiotic-Resistant Enterococci. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.01454-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yosef I., Manor M., Kiro R., Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA. 2015;112:7267–7272. doi: 10.1073/pnas.1500107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hao M., He Y., Zhang H., Liao X.P., Liu Y.H., Sun J., Du H., Kreiswirth B.N., Chen L. CRISPR-Cas9-Mediated Carbapenemase Gene and Plasmid Curing in Carbapenem-Resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00843-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiga K., Tan X.E., Ibarra-Chavez R., Watanabe S., Aiba Y., Sato’o Y., Li F.Y., Sasahara T., Cui B., Kawauchi M., et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat. Commun. 2020;11:2934. doi: 10.1038/s41467-020-16731-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levy A., Goren M.G., Yosef I., Auster O., Manor M., Amitai G., Edgar R., Qimron U., Sorek R. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gomaa A.A., Klumpe H.E., Luo M.L., Selle K., Barrangou R., Beisel C.L. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio. 2014;5:00928-13. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Canez C., Selle K., Goh Y.J., Barrangou R. Outcomes and characterization of chromosomal self-targeting by native CRISPR-Cas systems in Streptococcus thermophilus. FEMS Microbiol. Lett. 2019;366:fnz105. doi: 10.1093/femsle/fnz105. [DOI] [PubMed] [Google Scholar]
- 53.Sinkunas T., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caliando B.J., Voigt C.A. Targeted DNA degradation using a CRISPR device stably carried in the host genome. Nat. Commun. 2015;6:6989. doi: 10.1038/ncomms7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamilton T.A., Pellegrino G.M., Therrien J.A. Ham, transfer of a CRISPR nuclease for targeted bacterial killing. Nat. Commun. 2019;10:4544. doi: 10.1038/s41467-019-12448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strand T.A., Lale R., Degnes K.F., Lando M., Valla S. A new and improved host-independent plasmid system for RK2-based conjugal transfer. PLoS ONE. 2014;9:e90372. doi: 10.1371/journal.pone.0090372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X., Bai H., Yang Y., Yoon J., Wang S., Zhang X. Supramolecular Antibacterial Materials for Combatting Antibiotic Resistance. Adv. Mater. 2019;31:e1805092. doi: 10.1002/adma.201805092. [DOI] [PubMed] [Google Scholar]
- 58.Li Y., Peng N. Endogenous CRISPR-Cas System-Based Genome Editing and Antimicrobials: Review and Prospects. Front. Microbiol. 2019;10:2471. doi: 10.3389/fmicb.2019.02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carrigy N.B., Chang R.Y., Leung S.S.Y., Harrison M., Petrova Z., Pope W.H., Hatfull G.F., Britton W.J., Chan H.K., Sauvageau D., et al. Anti-Tuberculosis Bacteriophage D29 Delivery with a Vibrating Mesh Nebulizer, Jet Nebulizer, and Soft Mist Inhaler. Pharm. Res. 2017;34:2084–2096. doi: 10.1007/s11095-017-2213-4. [DOI] [PubMed] [Google Scholar]
- 60.Santos R.S., Figueiredo C., Azevedo N.F., Braeckmans K., De Smedt S.C. Nanomaterials and molecular transporters to overcome the bacterial envelope barrier: Towards advanced delivery of antibiotics. Adv. Drug Deliv. Rev. 2018;136–137:28–48. doi: 10.1016/j.addr.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 61.Naskar A., Kim K.S. Nanomaterials as Delivery Vehicles and Components of New Strategies to Combat Bacterial Infections: Advantages and Limitations. Microorganisms. 2019;16:356. doi: 10.3390/microorganisms7090356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L., Hu C., Shao L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017;12:1227–1249. doi: 10.2147/IJN.S121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang S., Zheng J. Antibacterial Activity of Silver Nanoparticles: Structural Effects. Adv. Healthc. Mater. 2018;7:e1701503. doi: 10.1002/adhm.201701503. [DOI] [PubMed] [Google Scholar]
- 64.Liao S., Zhang Y., Pan X., Zhu F., Jiang C., Liu Q., Cheng Z., Dai G., Wu G., Wang L., et al. Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant Pseudomonas aeruginosa. Int. J. Nanomed. 2019;14:1469–1487. doi: 10.2147/IJN.S191340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yougbare S., Chang T.K., Tan S.H., Kuo J.C., Hsu P.H., Su C.Y., Kuo T.R. Antimicrobial Gold Nanoclusters: Recent Developments and Future Perspectives. Int. J. Mol. Sci. 2019;20:2924. doi: 10.3390/ijms20122924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jia H., Draz M.S., Ruan Z. Functional Nanomaterials for the Detection and Control of Bacterial Infections. Curr. Top. Med. Chem. 2019;19:2449–2475. doi: 10.2174/1568026619666191023123407. [DOI] [PubMed] [Google Scholar]
- 67.Wang L., Li S., Yin J., Yang J., Li Q., Zheng W., Liu S., Jiang X. The Density of Surface Coating Can Contribute to Different Antibacterial Activities of Gold Nanoparticles. Nano Lett. 2020;20:5036–5042. doi: 10.1021/acs.nanolett.0c01196. [DOI] [PubMed] [Google Scholar]
- 68.Varghese B., Kurian M., Krishna S., Athira T.S. Biochemical synthesis of copper nanoparticles using Zingiber officinalis and Curcuma longa: Characterization and antibacterial activity study. Mater. Today Proc. 2020;25:302–306. doi: 10.1016/j.matpr.2020.01.476. [DOI] [Google Scholar]
- 69.Tahir K., Ahmad A., Li B., Nazir S., Khan A.U., Nasir T., Khan Z.U.H., Naz R., Raza M. Visible light photo catalytic inactivation of bacteria and photo degradation of methylene blue with Ag/TiO2 nanocomposite prepared by a novel method. J. Photochem. Photobiol. B. 2016;162:189–198. doi: 10.1016/j.jphotobiol.2016.06.039. [DOI] [PubMed] [Google Scholar]
- 70.Ullah K., Khan S.A., Mannan A., Khan R., Murtaza G., Yameen M.A. Enhancing the Antibacterial Activity of Erythromycin with Titanium Dioxide Nanoparticles against MRSA. Curr. Pharm. Biotechnol. 2020;21:948–954. doi: 10.2174/1389201021666200128124142. [DOI] [PubMed] [Google Scholar]
- 71.Siddiqi K.S., Ur Rahman A., Husen A. Properties of Zinc Oxide Nanoparticles and Their Activity against Microbes. Nanoscale Res. Lett. 2018;13:141. doi: 10.1186/s11671-018-2532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aditya A., Chattopadhyay S., Jha D., Gautam H.K., Maiti S., Ganguli M. Zinc Oxide Nanoparticles Dispersed in Ionic Liquids Show High Antimicrobial Efficacy to Skin-Specific Bacteria. ACS Appl. Mater. Interfaces. 2018;10:15401–15411. doi: 10.1021/acsami.8b01463. [DOI] [PubMed] [Google Scholar]
- 73.Maruthupandy M., Rajivgandhi G., Muneeswaran T., Song J.M., Manoharan N. Biologically synthesized zinc oxide nanoparticles as nanoantibiotics against ESBLs producing gram negative bacteria. Microb. Pathog. 2018;121:224–231. doi: 10.1016/j.micpath.2018.05.041. [DOI] [PubMed] [Google Scholar]
- 74.Tiwari V., Mishra N., Gadani K., Solanki P.S., Shah N.A., Tiwari M. Mechanism of Anti-bacterial Activity of Zinc Oxide Nanoparticle against Carbapenem-Resistant Acinetobacter baumannii. Front. Microbiol. 2018;9:1218. doi: 10.3389/fmicb.2018.01218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pu L., Xu J., Sun Y., Fang Z., Chan-Park M.B., Duan H. Cationic polycarbonate-grafted superparamagnetic nanoparticles with synergistic dual-modality antimicrobial activity. Biomater. Sci. 2016;4:871–879. doi: 10.1039/C5BM00545K. [DOI] [PubMed] [Google Scholar]
- 76.Takahashi H., Nadres E.T., Kuroda K. Cationic Amphiphilic Polymers with Antimicrobial Activity for Oral Care Applications: Eradication of S. mutans Biofilm. Biomacromolecules. 2017;18:257–265. doi: 10.1021/acs.biomac.6b01598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xi Y., Song T., Tang S., Wang N., Du J. Preparation and Antibacterial Mechanism Insight of Polypeptide-Based Micelles with Excellent Antibacterial Activities. Biomacromolecules. 2016;17:3922–3930. doi: 10.1021/acs.biomac.6b01285. [DOI] [PubMed] [Google Scholar]
- 78.Miola M., Fucale G., Maina G., Verne E. Antibacterial and bioactive composite bone cements containing surface silver-doped glass particles. Biomed. Mater. 2015;10:055014. doi: 10.1088/1748-6041/10/5/055014. [DOI] [PubMed] [Google Scholar]
- 79.Danti S., Azimi B., Candito M., Fusco A., Sorayani Bafqi M.S., Ricci C., Milazzo M., Cristallini C., Latifi M., Donnarumma G., et al. Lithium niobate nanoparticles as biofunctional interface material for inner ear devices. Biointerphases. 2020;15:31004. doi: 10.1116/6.0000067. [DOI] [PubMed] [Google Scholar]
- 80.Gupta A., Briffa S.M., Swingler S., Gibson H., Kannappan V., Adamus G., Kowalczuk M., Martin C., Radecka I. Synthesis of Silver Nanoparticles Using Curcumin-Cyclodextrins Loaded into Bacterial Cellulose-Based Hydrogels for Wound Dressing Applications. Biomacromolecules. 2020;21:1802–1811. doi: 10.1021/acs.biomac.9b01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ferrando-Magraner E., Bellot-Arcis C., Paredes-Gallardo V., Almerich-Silla J.M., Garcia-Sanz V., Fernandez-Alonso M., Montiel-Company J.M. Antibacterial Properties of Nanoparticles in Dental Restorative Materials: A Systematic Review and Meta-Analysis. Medicina. 2020;56:55. doi: 10.3390/medicina56020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ding X., Wang A., Tong W., Xu F.J. Biodegradable Antibacterial Polymeric Nanosystems: A New Hope to Cope with Multidrug-Resistant Bacteria. Small. 2019;15:e1900999. doi: 10.1002/smll.201900999. [DOI] [PubMed] [Google Scholar]
- 83.Gunday C., Anand S., Gencer H.B., Munafo S., Moroni L., Fusco A., Donnarumma G., Ricci C., Hatir P.C., Tureli N.G., et al. Ciprofloxacin-loaded polymeric nanoparticles incorporated electrospun fibers for drug delivery in tissue engineering applications. Drug Deliv. Transl. Res. 2020;10:706–720. doi: 10.1007/s13346-020-00736-1. [DOI] [PubMed] [Google Scholar]
- 84.Hasan N., Cao J., Lee J., Hlaing S.P., Oshi M.A., Naeem M., Ki M.H., Lee B.L., Jung Y., Yoo J.W. Bacteria-Targeted Clindamycin Loaded Polymeric Nanoparticles: Effect of Surface Charge on Nanoparticle Adhesion to MRSA, Antibacterial Activity, and Wound Healing. Pharmaceutics. 2019;11:236. doi: 10.3390/pharmaceutics11050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Y., Ding Y., Fan B., Wang Y., Mao Z., Wang W., Wu J. Inflammation-targeting polymeric nanoparticles deliver sparfloxacin and tacrolimus for combating acute lung sepsis. J. Control. Release. 2020;321:463–474. doi: 10.1016/j.jconrel.2020.02.030. [DOI] [PubMed] [Google Scholar]
- 86.Chen J., Guo Z., Tian H., Chen X. Production and clinical development of nanoparticles for gene delivery. Mol. Ther. Methods Clin. Dev. 2016;3:16023. doi: 10.1038/mtm.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin W.J., Lee W.C. Polysaccharide-modified nanoparticles with intelligent CD44 receptor targeting ability for gene delivery. Int. J. Nanomed. 2018;13:3989–4002. doi: 10.2147/IJN.S163149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ganbold T., Han S., Hasi A., Baigude H. Receptor-mediated delivery of therapeutic RNA by peptide functionalized curdlan nanoparticles. Int. J. Biol. Macromol. 2019;126:633–640. doi: 10.1016/j.ijbiomac.2018.12.152. [DOI] [PubMed] [Google Scholar]
- 89.Ge Z., Chen Q., Osada K., Liu X., Tockary T.A., Uchida S., Dirisala A., Ishii T., Nomoto T., Toh K., et al. Targeted gene delivery by polyplex micelles with crowded PEG palisade and cRGD moiety for systemic treatment of pancreatic tumors. Biomaterials. 2014;35:3416–3426. doi: 10.1016/j.biomaterials.2013.12.086. [DOI] [PubMed] [Google Scholar]
- 90.Suk J.S., Xu Q., Kim N., Hanes J., Ensign L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016;99:28–51. doi: 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hashiba K., Sato Y., Harashima H. pH-labile PEGylation of siRNA-loaded lipid nanoparticle improves active targeting and gene silencing activity in hepatocytes. J. Control. Release. 2017;262:239–246. doi: 10.1016/j.jconrel.2017.07.046. [DOI] [PubMed] [Google Scholar]
- 92.Chen F., Alphonse M., Liu Q. Strategies for nonviral nanoparticle-based delivery of CRISPR/Cas9 therapeutics. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020;12:e1609. doi: 10.1002/wnan.1609. [DOI] [PubMed] [Google Scholar]
- 93.Herai R.H. Avoiding the off-target effects of CRISPR/cas9 system is still a challenging accomplishment for genetic transformation. Gene. 2019;700:176–178. doi: 10.1016/j.gene.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 94.Liu C., Zhang L., Liu H., Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control. Release. 2017;266:17–26. doi: 10.1016/j.jconrel.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li H.Y., Kao C.Y., Lin W.H., Zheng P.X., Yan J.J., Wang M.C., Teng C.H., Tseng C.C., Wu J.J. Characterization of CRISPR-Cas Systems in Clinical Klebsiella pneumoniae Isolates Uncovers Its Potential Association with Antibiotic Susceptibility. Front. Microbiol. 2018;9:1595. doi: 10.3389/fmicb.2018.01595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lattanzi A., Meneghini V., Pavani G., Amor F., Ramadier S., Felix T., Antoniani C., Masson C., Alibeu O., Lee C., et al. Optimization of CRISPR/Cas9 Delivery to Human Hematopoietic Stem and Progenitor Cells for Therapeutic Genomic Rearrangements. Mol. Ther. 2019;27:137–150. doi: 10.1016/j.ymthe.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu C.F., Chen G.J., Luo Y.L., Zhang Y., Zhao G., Lu Z.D., Czarna A., Gu Z., Wang J. Rational designs of In Vivo CRISPR-Cas delivery systems. Adv. Drug Deliv. Rev. 2021;168:3–29. doi: 10.1016/j.addr.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 98.Zhao H., Li Y., He L., Pu W., Yu W., Li Y., Wu Y.T., Xu C., Wei Y., Ding Q., et al. In Vivo AAV-CRISPR/Cas9-Mediated Gene Editing Ameliorates Atherosclerosis in Familial Hypercholesterolemia. Circulation. 2020;141:67–79. doi: 10.1161/CIRCULATIONAHA.119.042476. [DOI] [PubMed] [Google Scholar]
- 99.Gratacap R.L., Regan T., Dehler C.E., Martin S.A.M., Boudinot P., Collet B., Houston R.D. Efficient CRISPR/Cas9 genome editing in a salmonid fish cell line using a lentivirus delivery system. BMC Biotechnol. 2020;20:35. doi: 10.1186/s12896-020-00626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Glass Z., Li Y., Xu Q. Nanoparticles for CRISPR-Cas9 delivery. Nat. Biomed. Eng. 2017;1:854–855. doi: 10.1038/s41551-017-0158-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Patel S., Ashwanikumar N., Robinson E., DuRoss A., Sun C., Murphy-Benenato K.E., Mihai C., Almarsson O., Sahay G. Boosting Intracellular Delivery of Lipid Nanoparticle-Encapsulated mRNA. Nano Lett. 2017;17:5711–5718. doi: 10.1021/acs.nanolett.7b02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu J., Chang J., Jiang Y., Meng X., Sun T., Mao L., Xu Q., Wang M. Fast and Efficient CRISPR/Cas9 Genome Editing In Vivo Enabled by Bioreducible Lipid and Messenger RNA Nanoparticles. Adv. Mater. 2019;31:e1902575. doi: 10.1002/adma.201902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tang Q., Liu J., Jiang Y., Zhang M., Mao L., Wang M. Cell-Selective Messenger RNA Delivery and CRISPR/Cas9 Genome Editing by Modulating the Interface of Phenylboronic Acid-Derived Lipid Nanoparticles and Cellular Surface Sialic Acid. ACS Appl. Mater. Interfaces. 2019;11:46585–46590. doi: 10.1021/acsami.9b17749. [DOI] [PubMed] [Google Scholar]
- 105.Chen Z., Liu F., Chen Y., Liu J., Wang X., Chen A.T., Deng G., Zhang H., Liu J., Hong Z., et al. Targeted Delivery of CRISPR/Cas9-Mediated Cancer Gene Therapy via Liposome-Templated Hydrogel Nanoparticles. Adv. Funct. Mater. 2017;27:1703036. doi: 10.1002/adfm.201703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu C., Lu Z., Luo Y., Liu Y., Cao Z., Shen S., Li H., Liu J., Chen K., Chen Z., et al. Targeting of NLRP3 inflammasome with gene editing for the amelioration of inflammatory diseases. Nat. Commun. 2018;9:4092. doi: 10.1038/s41467-018-06522-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sago C.D., Lokugamage M.P., Paunovska K., Vanover D.A., Monaco C.M., Shah N.N., Castro M.G., Anderson S.E., Rudoltz T.G., Lando G.N., et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl. Acad. Sci. USA. 2018;115:E9944–E9952. doi: 10.1073/pnas.1811276115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rosenblum D., Gutkin A., Kedmi R., Ramishetti S., Veiga N., Jacobi A.M., Schubert M.S., Friedmann-Morvinski D., Cohen Z.R., Behlke M.A., et al. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci. Adv. 2020;6:eabc9450. doi: 10.1126/sciadv.abc9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu Q., Wang C., Zheng Y., Zhao Y., Wang Y., Hao J., Zhao X., Yi K., Shi L., Kang C., et al. Virus-like nanoparticle as a co-delivery system to enhance efficacy of CRISPR/Cas9-based cancer immunotherapy. Biomaterials. 2020;258:120275. doi: 10.1016/j.biomaterials.2020.120275. [DOI] [PubMed] [Google Scholar]
- 110.Finn J.D., Smith A.R., Patel M.C., Shaw L., Youniss M.R., van Heteren J., Dirstine T., Ciullo C., Lescarbeau R., Seitzer J., et al. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 111.Wang P., Zhang L., Zheng W., Cong L., Guo Z., Xie Y., Wang L., Tang R., Feng Q., Hamada Y., et al. Thermo-triggered Release of CRISPR-Cas9 System by Lipid-Encapsulated Gold Nanoparticles for Tumor Therapy. Angew. Chem. Int. Ed. Engl. 2018;57:1491–1496. doi: 10.1002/anie.201708689. [DOI] [PubMed] [Google Scholar]
- 112.Lee B., Lee K., Panda S., Gonzales-Rojas R., Chong A., Bugay V., Park H.M., Brenner R., Murthy N., Lee H.Y. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2018;2:497–507. doi: 10.1038/s41551-018-0252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shahbazi R., Sghia-Hughes G., Reid J.L., Kubek S., Haworth K.G., Humbert O., Kiem H.P., Adair J.E. Targeted homology-directed repair in blood stem and progenitor cells with CRISPR nanoformulations. Nat. Mater. 2019;18:1124–1132. doi: 10.1038/s41563-019-0385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang L., Wang L., Xie Y., Wang P., Deng S., Qin A., Zhang J., Yu X., Zheng W., Jiang X. Triple-Targeting Delivery of CRISPR/Cas9 To Reduce the Risk of Cardiovascular Diseases. Angew. Chem. Int. Ed. Engl. 2019;58:12404–12408. doi: 10.1002/anie.201903618. [DOI] [PubMed] [Google Scholar]
- 115.Liu Y., Zhao G., Xu C.F., Luo Y.L., Lu Z.D., Wang J. Systemic delivery of CRISPR/Cas9 with PEG-PLGA nanoparticles for chronic myeloid leukemia targeted therapy. Biomater. Sci. 2018;6:1592–1603. doi: 10.1039/C8BM00263K. [DOI] [PubMed] [Google Scholar]
- 116.Timin A.S., Muslimov A.R., Lepik K.V., Epifanovskaya O.S., Shakirova A.I., Mock U., Riecken K., Okilova M.V., Sergeev V.S., Afanasyev B.V., et al. Efficient gene editing via non-viral delivery of CRISPR-Cas9 system using polymeric and hybrid microcarriers. Nanomedicine. 2018;14:97–108. doi: 10.1016/j.nano.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 117.Liu Y., Cao Z.T., Xu C.F., Lu Z.D., Luo Y.L., Wang J. Optimization of lipid-assisted nanoparticle for disturbing neutrophils-related inflammation. Biomaterials. 2018;172:92–104. doi: 10.1016/j.biomaterials.2018.04.052. [DOI] [PubMed] [Google Scholar]
- 118.Rui Y., Varanasi M., Mendes S., Yamagata H.M., Wilson D.R., Green J.J. Poly(Beta-Amino Ester) Nanoparticles Enable Nonviral Delivery of CRISPR-Cas9 Plasmids for Gene Knockout and Gene Deletion. Mol. Ther. Nucleic Acids. 2020;20:661–672. doi: 10.1016/j.omtn.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kang Y.K., Kwon K., Ryu J.S., Lee H.N., Park C., Chung H.J. Nonviral Genome Editing Based on a Polymer-Derivatized CRISPR Nanocomplex for Targeting Bacterial Pathogens and Antibiotic Resistance. Bioconjug. Chem. 2017;28:957–967. doi: 10.1021/acs.bioconjchem.6b00676. [DOI] [PubMed] [Google Scholar]
- 120.Hryhorowicz M., Grzeskowiak B., Mazurkiewicz N., Sledzinski P., Lipinski D., Slomski R. Improved Delivery of CRISPR/Cas9 System Using Magnetic Nanoparticles into Porcine Fibroblast. Mol. Biotechnol. 2019;61:173–180. doi: 10.1007/s12033-018-0145-9. [DOI] [PubMed] [Google Scholar]
- 121.Rohiwal S.S., Dvorakova N., Klima J., Vaskovicova M., Senigl F., Slouf M., Pavlova E., Stepanek P., Babuka D., Benes H., et al. Polyethylenimine based magnetic nanoparticles mediated non-viral CRISPR/Cas9 system for genome editing. Sci. Rep. 2020;10:4619. doi: 10.1038/s41598-020-61465-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cruz L.J., van Dijk T., Vepris O., Li T., Schomann T., Baldazzi F., Kurita R., Nakamura Y., Grosveld F., Philipsen S., et al. PLGA-Nanoparticles for Intracellular Delivery of the CRISPR-Complex to Elevate Fetal Globin Expression in Erythroid Cells. Biomaterials. 2020;268:120580. doi: 10.1016/j.biomaterials.2020.120580. [DOI] [PubMed] [Google Scholar]
- 123.Tu K., Deng H., Kong L., Wang Y., Yang T., Hu Q., Hu M., Yang C., Zhang Z. Reshaping Tumor Immune Microenvironment through Acidity-Responsive Nanoparticles Featured with CRISPR/Cas9-Mediated Programmed Death-Ligand 1 Attenuation and Chemotherapeutics-Induced Immunogenic Cell Death. ACS Appl. Mater. Interfaces. 2020;12:16018–16030. doi: 10.1021/acsami.9b23084. [DOI] [PubMed] [Google Scholar]
- 124.Nguyen D.N., Roth T.L., Li P.J., Chen P.A., Apathy R., Mamedov M.R., Vo L.T., Tobin V.R., Goodman D., Shifrut E., et al. Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat. Biotechnol. 2020;38:44–49. doi: 10.1038/s41587-019-0325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ju E., Li T., Ramos da Silva S., Gao S.J. Gold Nanocluster-Mediated Efficient Delivery of Cas9 Protein through pH-Induced Assembly-Disassembly for Inactivation of Virus Oncogenes. ACS Appl. Mater. Interfaces. 2019;11:34717–34724. doi: 10.1021/acsami.9b12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study.