Abstract
Microbiological strategies are currently being considered as methods for reducing the ethanol content of wine. Fermentations started with a multistarter of three non-Saccharomyces yeasts (Metschnikowia pulcherrima (Mp), Torulaspora delbrueckii (Td) and Zygosaccharomyces bailii (Zb)) at different inoculum concentrations. S. cerevisiae (Sc) was inoculated into fermentations at 0 h (coinoculation), 48 h or 72 h (sequential fermentations). The microbial populations were analyzed by a culture-dependent approach (Wallerstein Laboratory Nutrient (WLN) culture medium) and a culture-independent method (PMA-qPCR). The results showed that among these three non-Saccharomyces yeasts, Td became the dominant non-Saccharomyces yeast in all fermentations, and Mp was the minority yeast. Sc was able to grow in all fermentations where it was involved, being the dominant yeast at the end of fermentation. We obtained a significant ethanol reduction of 0.48 to 0.77% (v/v) in sequential fermentations, with increased concentrations of lactic and acetic acids. The highest reduction was achieved when the inoculum concentration of non-Saccharomyces yeast was 10 times higher (107 cells/mL) than that of S. cerevisiae. However, this reduction was lower than that obtained when these strains were used as single non-Saccharomyces species in the starter, indicating that interactions between them affected their performance. Therefore, more combinations of yeast species should be tested to achieve greater ethanol reductions.
Keywords: wine, PMA-qPCR, Metschnikowia pulcherrima, Torulaspora delbrueckii, Zygosaccharomyces bailii, mixed fermentation, coinoculation, sequential fermentation
1. Introduction
In recent years, the average ethanol concentration in wine has increased, mainly due to climate change and consumer preference for wine styles [1,2,3]. Different strategies have been applied to reduce ethanol production in wine, such as decreasing the leaf area to lower the sugar content in grape berries, reducing the maturity of grapes, removing sugar from the grape must, developing or screening low-alcohol wine yeasts and removing alcohol from wine (reviewed by [4]). Compared with microbiological strategies, other strategies might have negative effects on wine, such as delaying maturity, reducing the yield of berries, causing a significant reduction in anthocyanins, soluble solids and volatile compounds, and decreasing the wine color [5,6,7,8]. Therefore, microbiological strategies were considered to be effective and accompanied by smaller impacts on the wine sensory profile and quality. In particular, the use of non-Saccharomyces yeast strains for reducing the alcohol content of wines has been proven to improve the wine aroma complexity and has become a consistent proposal [9,10,11].
Non-Saccharomyces yeasts have been applied in fermentations to reduce ethanol using different inoculation strategies. For example, several studies have shown that single-culture fermentations with Hanseniaspora uvarum, Lachancea thermotolerans, Metschnikowia pulcherrima, Starmerella bombicola, Starmerella bacillaris, Zygosaccharomyces bailii, Zygosaccharomyces bisporus, and Zygosaccharomyces sapae species are able to reduce the ethanol content in wine [10,12,13,14,15,16,17,18,19]. However, most of these single yeast fermentations became stuck. To solve this problem, mixed fermentations with non-Saccharomyces yeasts and Saccharomyces cerevisiae have been proposed, demonstrating that simultaneous inoculation of S. cerevisiae with L. thermotolerans, S. bacillaris or M. pulcherrima species produced a reduction in ethanol [10,12,14,15,20]. In addition, to achieve this purpose, most researchers preferred to use non-Saccharomyces yeasts (Hanseniaspora osmophila, H. uvarum, L. thermotolerans, M. pulcherrima, S. bombicola, Torulaspora delbrueckii and Z. bailii species) in sequential fermentations, with S. cerevisiae inoculated at 48 h, 72 h, or when 50% of the sugar from grape must was consumed [11,12,17,21,22,23,24].
Due to the growing interest in mixed fermentations, it is necessary to understand the behavior and interactions of strains throughout the fermentation process. Thus, the population dynamics of yeasts have become a key factor in the study of yeast interactions. The traditional microbial counting method usually uses different solid media, such as Wallerstein Laboratory Nutrient (WLN) and lysine agar, to distinguish or isolate different yeast species based on their dissimilar morphological characteristics or selective growth [25,26]. However, this is not an effective method of discrimination when two or more non-Saccharomyces yeast species are simultaneously inoculated in mixed fermentations because some of them show similar morphological profiles. In addition, for an accurate evaluation of the population dynamics of different yeast species, we have to consider the existence of viable but nonculturable (VBNC) cells caused by the different metabolic statuses of cells. In these cases, the use of the plating method would not be appropriate because those VBNC cells would not be detectable [27].
Compared with the classical method, researchers have confirmed that quantitative real-time polymerase chain reaction (qPCR) is a more sensitive and specific technique for the detection of nucleic acids, and it shows a wide detection range for different cell concentrations, usually from 10 to 108 cells/mL [28,29,30]. This sensitive and low-detection limit method provides the possibility to detect species present in low quantities in the mixed fermentation process. However, the population of yeasts is often overestimated from qPCR because the method does not discriminate the DNA of living and dead cells in fermenting must [31,32]. To distinguish the population of living cells from that of dead cells, DNA-binding dyes, such as ethidium monoazide (EMA) and propidium monoazide (PMA), are applied to must samples as a pretreatment before DNA extraction. The qPCR analysis of these samples quantifies only living cells because the living cell membrane is impermeable to DNA-binding dye, and therefore, it will only bind to free DNA from dead cells, avoiding its amplification. Nocker et al. [33] showed that PMA is more effective than EMA, which is the reason why PMA has been favored by researchers in the counting of living cells in recent years [34,35,36,37,38].
Most articles have focused on evaluating the effects of single strains or mixed starters composed of one S. cerevisiae strain and one non-Saccharomyces species. However, this work aims to test a multistarter culture consisting of several non-Saccharomyces species previously selected for their ability to reduce ethanol [39] and a commercial S. cerevisiae strain. The use of multiple selected species as the inoculum may improve ethanol reduction, as well as the overall complexity of wines [32].
The aim of the present work was to analyze the effect of the mixed inoculation of S. cerevisiae and several non-Saccharomyces yeasts on ethanol reduction and population dynamics. Three non-Saccharomyces strains belonging to M. pulcherrima, T. delbrueckii and Z. bailii species, which have been demonstrated to have the ability to reduce ethanol in single fermentations [39], were simultaneously inoculated into the must. To determine the best inoculation protocol to achieve ethanol reduction, different mixed inoculation conditions with S. cerevisiae were evaluated (sequential or coinoculation at different inoculation ratios). The population dynamics of the different yeasts were assessed by WLN counting and PMA-qPCR analysis during the fermentation process, and the concentrations of the main organic compounds were analyzed by HPLC in the resulting wines.
2. Materials and Methods
2.1. Yeast Strains
Four different yeast strains were used in this study: the commercial wine yeast S. cerevisiae (Lalvin QA23®, referred to as Sc) from Lallemand Inc. (Montreal, QC, Canada), M. pulcherrima 51 (Mp) and T. delbrueckii Priorat (Td), selected from the Priorat Appellation of Origin (URV collection, Tarragona, Spain) [9], and Z. bailii CECT 11043 (Zb), obtained from the Spanish Type Culture Collection. These strains were previously selected for their ability to produce low ethanol yield [39] when several strains of each species were tested.
The strains were stored at −80 °C in YPD liquid medium (2% (w/v) glucose, 2% (w/v) yeast extract, and 1% (w/v) peptone, Cultimed, Barcelona, Spain) with 40% (v/v) glycerol. Before starting the fermentations, the yeasts were grown at 28 °C in YPD agar (YPD liquid with 1.7% (w/v) agar) and Wallerstein laboratory nutrient (WLN) agar (Becton, Dickinson and Company, Isère, France). The species identification of the four strains was confirmed by PCR-RFLP analysis of 5.8S-ITS rDNA according to Esteve-Zarzoso et al. [40].
2.2. Natural Must and Starter Cultures
Natural must (NM) was obtained from Muscat grapes harvested from Finca Experimental Mas dels Frares of Rovira i Virgili University (Constantí, Spain) during the 2019 vintage (after treatment with 50 mg/L SO2 and settling, the must had 220 g/L sugars, 4.49 g/L total acidity (as tartaric acid), 77.8 mg/L assimilable nitrogen and a pH of 3.27). The concentration of assimilable nitrogen was determined in a Miura autoanalyzer (EE-MIURAONE Rev., I.S.E. S.r.l., Italy) using an Ammonia and α-Aminic Nitrogen Enzymatic KIT (Tecnología Difusión Ibérica, S.L., Barcelona, Spain) and corrected with diammonium phosphate (Panreac Quimica SA, E.U.) until reaching a final concentration of 250 mg N/L. Before the start of the fermentations, dimethyl dicarbonate (0.2 mL/L) (ChemCruz, Santa Cruz Biotechnology, America) was added to NM and kept at 4 °C for 24 h to eliminate endogenous microorganisms. The absence of microorganisms after this treatment was confirmed by plating must samples with no dilution and 10-fold dilution on YPD agar.
The starter cultures were performed by transferring a single colony from YPD agar to YPD liquid medium and incubating it for 24 h (S. cerevisiae) or 48 h (non-Saccharomyces) at 28 °C with a stirring rate of 120 rpm in an orbital shaker. After incubation, the cells were counted in a Neubauer chamber (Leica Microsystems GMS QmbH, Leica, Germany) and inoculated at the indicated concentrations into the NM.
2.3. Fermentation Trials in Natural Must
Eight different mixed fermentations were carried out in this study (Table 1). Initially, all fermentations were simultaneously inoculated with three non-Saccharomyces strains at two different concentrations: 106 cells/mL (fermentations named 1CN) or 107 cells/mL (fermentations named 10CN). Additionally, Sc was inoculated into these two mixed non-Saccharomyces starters at the same moment (coinoculated fermentations were named 1CA and 10CA) and at 48 or 72 h later (sequential fermentations named 1S48, 1S72, 10S48 and 10S72, respectively), always at the same concentration (106 cells/mL). A single fermentation with Sc (106 cells/mL) was used as the control fermentation (C).
Table 1.
Fermentation conditions in 230 mL Muscat sterile must. Non-Saccharomyces (Mp, Td and Zb) and S. cerevisiae (Sc) strains were inoculated at the listed cell concentrations (cells/mL) and times.
| Fermentations | Inoculum Procedures | Inoculum Ratios (Mp:Td:Zb:Sc) |
Mp | Td | Zb | Sc |
|---|---|---|---|---|---|---|
| 1CA | Co-inoculation of all strains | 1:1:1:1 | 1 × 106 | 1 × 106 | 1 × 106 | 1 × 106 |
| 1S48 | Sequential inoculation of Sc at 48 h | 1:1:1:1 | 1 × 106 | 1 × 106 | 1 × 106 | 1 × 106 |
| 1S72 | Sequential inoculation of Sc at 72 h | 1:1:1:1 | 1 × 106 | 1 × 106 | 1 × 106 | 1 × 106 |
| 1CN | Co-inoculation of non-Saccharomyces strains | 1:1:1:0 | 1 × 106 | 1 × 106 | 1 × 106 | |
| 10CA | Co-inoculation of all strains | 10:10:10:1 | 1 × 107 | 1 × 107 | 1 × 107 | 1 × 106 |
| 10S48 | Sequential inoculation of Sc at 48 h | 10:10:10:1 | 1 × 107 | 1 × 107 | 1 × 107 | 1 × 106 |
| 10S72 | Sequential inoculation of Sc at 72 h | 10:10:10:1 | 1 × 107 | 1 × 107 | 1 × 107 | 1 × 106 |
| 10CN | Co-inoculation of non- Saccharomyces strains | 10:10:10:0 | 1 × 107 | 1 × 107 | 1 × 107 | |
| C | Single inoculation of Sc | 1 × 106 |
Triplicate fermentations were conducted in 250 mL glass bottles with 230 mL of NM and incubated at 22 °C with stirring at 120 rpm. The bottle cap had two ports—one connected to a 0.22 μm filter (Dominique Dutscher, Brumath, France) for gas flow and the other clamped by an iron clip for sampling. Before inoculation with Sc in sequential fermentations, the concentration of assimilable nitrogen in the fermented must was determined and supplemented, if needed, to 100 mg/L. Fermentations were monitored by measuring the must density with an electronic densitometer (Densito 30PX Portable Density Meter, Mettler Toledo, Hospitalet de Llobregat, Spain). Fermentations were considered finished when the residual sugars were below 2 g/L, checked by D-glucose/D-fructose enzymatic assays (Biosystems S.A., Barcelona, Spain) in the Miura autoanalyzer, or arrested when the must density did not decrease for more than two days. Samples from the end of fermentation were centrifuged at 7800 rpm for 5 min, and supernatants were frozen at 20 °C until chemical analysis.
2.4. Colony Counting
The yeast population during fermentation was analyzed by colony growth on WLN plates according to the different colony morphologies of Saccharomyces and non-Saccharomyces strains [35]. Briefly, samples were serially diluted in sterilized Milli-Q water from a Milli-Q purification system (Millipore S.A.S., Molsheim, France). The number of colony-forming units per milliliter (CFU/mL) was determined by plating 100 μL of three appropriately chosen dilutions on WLN agar. The plates were incubated at 28 °C for 2 or 3 days.
2.5. PMAxx Treatment
To obtain only the DNA of living cells, PMAxxTM viability dye (Biotium Inc., Fremont, CA, USA) was added to must samples as previously described by Navarro et al. [38]. Briefly, selected samples of must (1 mL) were centrifuged, and the pellets were washed with sterilized distilled water and treated with 25 μM PMAxx. After incubation for 10 min in darkness, PMAxx was permanently linked to the DNA of dead cells by subjecting samples twice to light for 30 s, with an interval of 1 min on ice. Pellets were recovered by centrifugation and frozen until DNA extraction.
2.6. DNA Extraction and qPCR Analysis
DNA was extracted using the DNeasy Plant Mini Kit (QIAGEN GmbH, Hilden, Germany) following the instructions of the manufacturer. Cell quantification of the different yeast species was conducted by species-specific qPCR using the primers CESP-F/SCER-R for S. cerevisiae [31], MP2-F/MP2-R for M. pulcherrima [41], Tods L2/Tods R2 for T. delbrueckii [42] and ZBF1/ZBR1 for Z. bailii [29] (all primer sequences are shown in Table A1). For all samples, qPCR was performed in triplicate using TB GreenTM Premix Ex TaqTM II (2×) (Takara Bio Inc., Kusatsu, Japan) with a final volume of 20 μL (2 μL of DNA, 0.8 μL of each primer, 0.08 μL of ROX Reference Dye (50×), 10 μL TB Green Premix Ex Taq II (2×) and 6.32 μL of sterilized Milli-Q water) on a QuantStudioTM 5 real-time PCR instrument (Applied Biosystems by Thermo Fisher Scientific, Waltham, MA, USA). A 96-well nonskirted PCR plate (4titude® Ltd., Wotton, UK) was used for the reaction. The amplification reaction was one cycle of 95 °C for 1 min and 40 cycles of 95 °C for 5 s and 60 °C for 35 s, followed by a dissociation step. Milli-Q water was used as a negative control. Ct (the cycle threshold) was determined using Thermo Fisher Scientific software (Waltham, MA, USA).
Standard curves were calculated for each species with and without PMAxx treatment, as described by Navarro et al. [38] but slightly modified. After incubating the yeast colony in YPD liquid medium at 28 °C for 24 h, samples with 108 cells/mL were collected in triplicate and centrifuged at 10,000 rpm for 2 min. The pellet was washed once with 1 mL sterilized distilled water and was subjected to PMAxx treatment as previously described. The pellet was stored at −20 °C until DNA extraction. Standard curves were created by plotting the average Ct values of a tenfold serial dilution of DNA from 108 to 10 cells/mL against the log of cells/mL.
2.7. Chemical Analysis
Residual sugars of samples at the end of fermentation were quantified by D-glucose/D-fructose enzymatic assays. Ethanol, glycerol and organic acids (citric acid, malic acid, tartaric acid, acetic acid, lactic acid and succinic acid) in the samples were determined by high-performance liquid chromatography (HPLC) using an Agilent 1100 (Agilent Technologies, Waldbronn, Germany) as previously described by Quirós et al. [43] and Zhu et al. [39].
2.8. Statistical Analysis
All graphs were generated using GraphPad Prism® version 8 (GraphPad Software, San Diego, CA, USA). The results are expressed as the mean ± standard deviation (SD). Statistically significant differences (one-way ANOVA) were analyzed by IBM SPSS Statistics version 23.0 (IBM, NY, USA). The ethanol yield was calculated with the Equation (1).
| Ethanol yield (g/g) = ethanol production (g/L)/sugar consumption (g/L) | (1) |
3. Results
3.1. Fermentation Kinetics
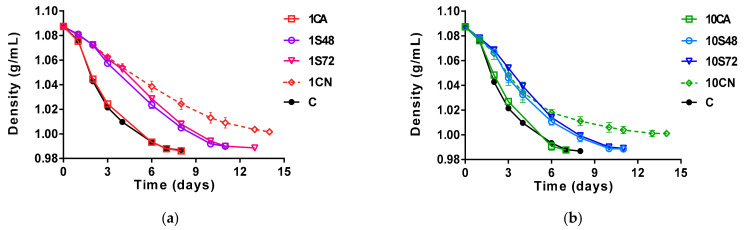
The fermentation kinetic profiles obtained under the different inoculation conditions, measured as must density reduction, are shown in Figure 1. The fastest fermentations were those with single inoculation with Sc and with the coinoculation of all the strains at the same time (1CA and 10CA), which showed similar fermentation profiles and completed fermentations in 7–8 days (10CA completed the fermentation 1 day earlier than that of Sc alone (C)). The fermentation kinetics were less affected by the inoculum concentration of non-Saccharomyces yeasts but were influenced by the inoculation time of Sc. The sequential inoculations, in which only non-Saccharomyces strains were fermented for 48–72 h, resulted in slower fermentations, finishing all the sugars in 11 (1S48, 10S48, 10S72) or 13 days (1S72) (Figure 1). These fermentations were slightly slower when Sc was inoculated later, at 72 h, mainly in the lower non-Saccharomyces inoculum (1S72). Finally, fermentations with only non-Saccharomyces species (1CN and 10CN) showed the slowest fermentation kinetics, getting stuck on the 13th day, at densities of 1.002 and 1.001 g/mL, respectively (Figure 1a,b). In general, the fermentations with non-Saccharomyces strains inoculated at 107 cells/mL showed slightly faster fermentation kinetics than did those inoculated at 106 cells/mL (Figure 1a,b).
Figure 1.
Density of single and mixed fermentations of S. cerevisiae and non-Saccharomyces yeasts with different inoculation conditions (see Table 1). The initial non-Saccharomyces inoculum size was 106 cells/mL (a) and 107 cells/mL (b).
3.2. Yeast Population Dynamics
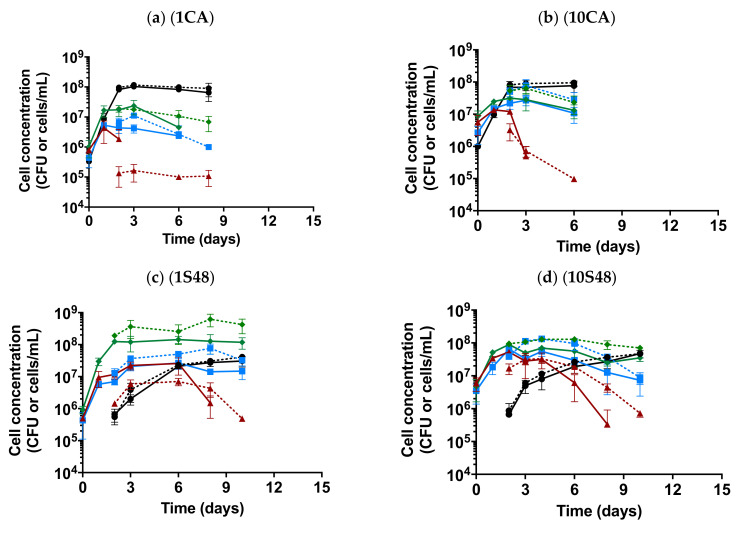
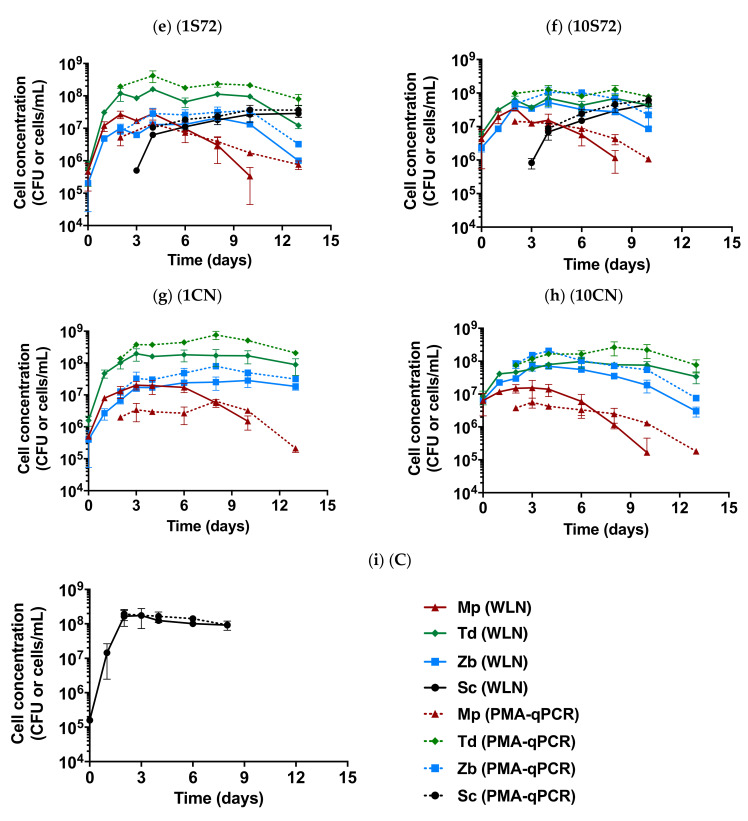
To study the yeast population dynamics during single or mixed fermentations, the growth of different species was obtained by different methodologies (Figure 2). On one hand, viability was determined during the fermentation process by plating the samples on WLN agar. Due to the different colony morphologies of the four yeast species on WLN agar [35], the colonies of these four strains could be counted on the same plate. On the other hand, PMA-qPCR analysis was performed to quantify the living yeast population using specific primers for each species. We applied PMA-qPCR from the second day of fermentation because the adaptation of the cells to the medium may produce an underestimation of the population by qPCR [38].
Figure 2.
Yeast population analysis based on Wallerstein Laboratory Nutrient (WLN) plates (solid lines  ) and qPCR with PMAxx treatment (dotted lines
) and qPCR with PMAxx treatment (dotted lines  ), determined in the different fermentative conditions: non-Saccharomyces multistarter coinoculated with (a,b) and without (g,h) S. cerevisaie (Sc); non-Saccharomyces multistarter with sequential inoculation of Sc at 48 h (c,d) or 72 h (e,f) and the single Sc fermentations (i). The initial non-Saccharomyces inoculum size was 106 cells/mL (a,c,e,g) and 107 cells/mL (b,d,f,h). Each species is shown in different colors (Mp in red, Td in green, Zb in blue and Sc in black).
), determined in the different fermentative conditions: non-Saccharomyces multistarter coinoculated with (a,b) and without (g,h) S. cerevisaie (Sc); non-Saccharomyces multistarter with sequential inoculation of Sc at 48 h (c,d) or 72 h (e,f) and the single Sc fermentations (i). The initial non-Saccharomyces inoculum size was 106 cells/mL (a,c,e,g) and 107 cells/mL (b,d,f,h). Each species is shown in different colors (Mp in red, Td in green, Zb in blue and Sc in black).
First, the standard curves were calculated by plotting the Ct values (<30) of DNA from 103 to 108 cells/mL (M. pulcherrima was from 104 to 108 cells/mL) against the log input cells/mL (Table A2), with efficiencies between 87.92% and 98.83%.
After yeast inoculation, all strains were able to grow in fermentation media and showed different population dynamics, as seen in the results obtained from WLN plates. In fermentations in which each non-Saccharomyces species was inoculated at 106 cells/mL, the population of non-Saccharomyces yeasts grew rapidly on the first day (Figure 2a,c,e,g). Td quickly increased the population, reaching up to 2 × 108 CFU/mL on the third day. The Td population was then stable until the end of fermentation, becoming the dominant yeast in all mixed inoculations, except in the coinoculated fermentations with Sc (1CA and 10CA, Figure 2a,b), in which Sc significantly impaired growth, with the fermentation reaching a maximum Td population of only 107 CFU/mL under 1CA conditions. Mp reached a maximum population of 2.7 × 107 CFU/mL in the sequential or non-Saccharomyces coinoculations, with densities higher than that of Zb within the first days but decreasing after the sixth day under most conditions (the Mp decrease occurred much earlier in the coinoculation with Sc, 1CA). In the case of Zb, the maximum growth was reached in the late fermentation stage, with a population of 2.8 × 107 CFU/mL, and was maintained rather stably until the end of fermentation.
The population dynamics in fermentations inoculated with 107 cells/mL of each non-Saccharomyces yeast differed from those inoculated at 106 cells/mL (Figure 2b,d,f,h). In those fermentations, Td and Zb presented similar population dynamics, and although Td was again the yeast with the highest population among all non-Saccharomyces yeasts, the difference relative to Zb was not as large (Td reached 9.7 × 107 CFU/mL and Zb 7.0 × 107 CFU/mL). Mp achieved the highest population (5.0 × 107 CFU/mL) on the second day and became the minority strain until the end of fermentation.
Regarding the S. cerevisiae dynamics, the dominance was only achieved in both coinoculated fermentations (1CA and 10CA), reaching a similar concentration to that of the Sc single inoculation on the third day of fermentation (1.8 × 108 CFU/mL, Figure 2a,b). However, the growth of Sc was restricted in sequential fermentations, where its population increased gradually until the end of fermentation, achieving up to 4.7 × 107 CFU/mL (Figure 2c–f). In these sequential fermentations, Sc together with Td were the majority strains at the end of the fermentation. This loss of the full imposition was not due to the lack of assimilable nitrogen, since supplementation to 100 mg/L was carried out in the deficient fermentations (1S72, 10S48 and 10S72) before the inoculation of Sc (as detailed in Table A3).
As already mentioned, during the fermentation process, Td and Zb colonies were observed until the end of fermentation (except in fermentation 1CA, where Td and Zb were absent at the end point), while Mp was absent on WLN agar after mid-fermentation. However, PMA-qPCR analysis allowed the detection of Mp until the end of fermentation, with the population ranging from 1 × 105 to 1.1 × 106 cells/mL, as well as that of Td and Zb at the end of fermentation in 1CA (Figure 2a). After the second day of fermentation, the trends of the yeast populations of Td, Zb and Sc obtained by PMA-qPCR analysis were similar to those of the WLN counting, although the cell concentrations obtained by PMA-qPCR were higher, reaching a difference of one order of magnitude for most non-Saccharomyces yeasts (Figure 2). Specifically, for the Td, Zb and Sc strains, the maximum populations obtained by PMA-qPCR from mixed fermentations reached 7.6 × 108, 1.7 × 108 and 1.2 × 108 cells/mL, respectively. In contrast, the population of Mp obtained by PMA-qPCR analysis before mid-fermentations was lower than that obtained by WLN counting in the first 6 days for fermentations inoculated with 106 cells/mL and in the first 3 days for fermentations inoculated with 107 cells/mL.
3.3. Main Fermentation Byproducts
To detect the ethanol reduction of the different inoculated fermentations and the metabolic characteristics of yeasts under the different conditions, the residual sugar, ethanol production and main byproducts at the end of fermentation were analyzed, as shown in Table 2.
Table 2.
Concentrations of sugars, ethanol, glycerol and organic acids at the end of fermentation.
| Compounds | C | 1CA | 1S48 | 1S72 | 1CN | 10CA | 10S48 | 10S72 | 10CN |
|---|---|---|---|---|---|---|---|---|---|
| Residual sugars (g/L) | 0.13 ± 0.17 | 0.39 ± 0.29 | 1.22 ± 0.62 | 0.42 ± 0.57 | 29.61 ± 5.65 * | 0.42 ± 0.41 | 0.37 ± 0.22 | 0.88 ± 1.03 | 28.30 ± 6.29 * |
| Ethanol production (%, v/v) | 13.06 ± 0.03 | 12.93 ± 0.06 | 12.58 ± 0.10 * | 12.57 ± 0.08 * | 11.04 ± 0.13 * | 12.77 ± 0.05 * | 12.41 ± 0.11 * | 12.29 ± 0.06 * | 10.91 ± 0.06 * |
| Ethanol yield (g/g) | 0.47 ± 0 | 0.46 ± 0 | 0.45 ± 0 * | 0.45 ± 0 * | 0.46 ± 0.01 | 0.46 ± 0 | 0.45 ± 0 * | 0.44 ± 0 * | 0.45 ± 0.01 * |
| Ethanol reduction (%, v/v) | 0 | 0.13 ± 0.06 | 0.48 ± 0.10 * | 0.49 ± 0.08 * | NC | 0.29 ± 0.05 * | 0.65 ± 0.11 * | 0.77 ± 0.06 * | NC |
| Glycerol (g/L) | 6.57 ± 0.08 | 6.57 ± 0.22 | 5.41 ± 0.75 * | 5.74 ± 0.93 | 4.96 ± 0.41 * | 6.23 ± 0.11 | 6.70 ± 0.48 | 6.38 ± 0.18 | 5.44 ± 0.68 * |
| Citric acid (g/L) | 0.13 ± 0 | 0.13 ± 0 | 0.11 ± 0.01 * | 0.12 ± 0.01 * | 0.12 ± 0 * | 0.12 ± 0.01 * | 0.12 ± 0.01 * | 0.11 ± 0 * | 0.11 ± 0 * |
| Tartaric acid (g/L) | 2.79 ± 0.07 | 3.10 ± 0.14 | 3.41 ± 0.38 | 3.30 ± 0.31 | 3.38 ± 0.34 | 2.47 ± 1.10 | 3.23 ± 0.25 | 3.44 ± 0.14 | 3.38 ± 0.28 |
| Malic acid (g/L) | 0.81 ± 0.13 | 0.63 ± 0.05 * | 0.50 ± 0.03 * | 0.45 ± 0.03 * | 0.39 ± 0.03 * | 0.61 ± 0.02 * | 0.54 ± 0.03 * | 0.51 ± 0.01 * | 0.44 ± 0.02 * |
| Succinic acid (g/L) | 0.44 ± 0 | 0.43 ± 0.05 | 0.40 ± 0.05 | 0.36 ± 0.05 | 0.37 ± 0.02 | 0.44 ± 0.06 | 0.52 ± 0.03 | 0.52 ± 0.02 | 0.67 ± 0.05 * |
| Lactic acid (g/L) | 0.20 ± 0.03 | 0.24 ± 0.02 | 0.24 ± 0.01 | 0.30 ± 0.10 * | 0.29 ± 0.01 * | 0.23 ± 0.05 | 0.30 ± 0 * | 0.33 ± 0.04 * | 0.25 ± 0.03 |
| Acetic acid (g/L) | 0.10 ± 0.01 | 0.14 ± 0.03 | 0.29 ± 0.08 * | 0.36 ± 0.05 * | 0.32 ± 0.06 * | 0.22 ± 0.05 * | 0.23 ± 0.05 * | 0.22 ± 0.08 * | 0.21 ± 0.04 * |
Values are mean ± standard deviation of three independent replicates; The initial sugar concentration of synthetic must was 220 g/L; * means statistically significant differences from the control sample of C on the same row (LSD test, p < 0.05); NC means no calculate ethanol reduction due to stuck fermentation.
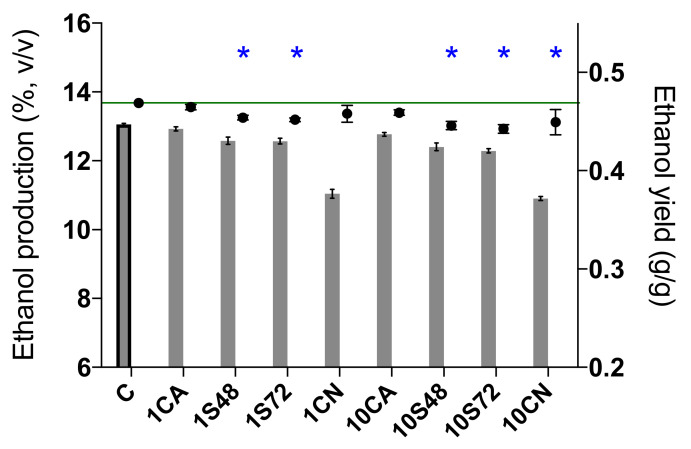
All fermentations with a mixed inoculum of non-Saccharomyces and Sc were completed, with less than 2 g/L residual sugars. However, the fermentations with only non-Saccharomyces strains, 1CN and 10CN, presented higher concentrations of residual sugars due to the stagnation of these two conditions (Table 2). Among all fermentations, the control fermentation (C) had the highest ethanol production (13.06%, v/v) and ethanol yield (0.47 g/g) (Figure 3). Mixed fermentations decreased ethanol production by 0.13 to 0.77% (v/v) compared to single fermentation by Sc (C), and this decrease was significant in 5 fermentations (1S48, 1S72, 10CA, 10S48 and 10S72) (Table 2). Fermentation 10S72 showed the highest ethanol reduction with the lowest ethanol yield (0.44 g/g), followed by 10S48, 1S72, 1S48 and 10CA (Table 2, Figure 3). Among these sequential fermentations, our results show a trend in relation to the inoculum concentration of non-Saccharomyces yeast, since a higher ethanol reduction was achieved when a higher inoculum of non-Saccharomyces was used (0.65 ± 0.11 and 0.77 ± 0.06 with 107 cells/mL vs. 0.48 ± 0.10 and 0.49 ± 0.08 with 106 cells/mL).
Figure 3.
Ethanol production (%, v/v) and ethanol yield (g/g) at the end of single and mixed fermentations. * indicates statistically significant differences from the control sample (C) (LSD (least significant difference) test, p < 0.05). The value of the green line is 0.47 g/g (ethanol yield of C). 1CN and 10CN were stuck fermentations.
Regarding the other fermentation byproducts, the concentration of glycerol in the mixed fermentations was similar to that of single fermentation with Sc (C), except in the 1S48, 1CN and 10CN fermentations, which had lower glycerol contents, although the latter two conditions had residual sugars left. On the other hand, in all fermentations, the concentrations of lactic and acetic acids were higher in the mixed fermentations, although they remained below 0.33 g/L and 0.36 g/L, respectively (Table 2). Surprisingly, more acetic acid was detected at a lower inoculum concentration of non-Saccharomyces yeasts (106 cells/mL).
4. Discussion
In recent years, non-Saccharomyces yeasts have been proposed for use as starters in alcoholic fermentation to reduce the ethanol content in wine [13,15,18,22,39,44,45,46]. Most studies have focused on evaluating the effects of single or mixed starters composed of one S. cerevisiae strain and one non-Saccharomyces species. Previous studies showed that as single starters, strains of M. pulcherrima and Z. bailii were not able to complete fermentation or had a low fermentation capacity [21,44], while T. delbrueckii has been reported to be a strong fermenter and able to finish fermentations [22,47]. However, fermentations inoculated with several non-Saccharomyces species as a multistarter have been poorly studied. In the current study, we evaluated the use of a multistarter of three non-Saccharomyces strains in Muscat grape fermentation. These strains were selected in a previous study [39], in which several strains of these three species and other non-Saccharomyces species were screened for their ability to reduce ethanol. When this multistarter of non-Saccharomyces strains was used, we observed stagnation of fermentation, even though Td was the dominant strain throughout the fermentation process and has shown its ability to complete fermentation when used as a single inoculum [47,48,49]. This lower performance of Td when used in multistarter fermentation could be due to interspecific microbial interactions, such as competition for nutrients [50,51,52], or the production of inhibitory compounds, such as killer toxins or other antimicrobial peptides or vesicles [53,54,55,56], as part of cell-cell interaction mechanisms. The higher population size of the non-Saccharomyces inoculum sped the consumption of sugars until mid-fermentation, although this was not enough to complete the fermentation, and similar final populations and residual sugars were obtained under both conditions. Mp was the minority strain under all conditions, and its population decreased significantly throughout the fermentation process, which agrees with previous studies [26,57,58].
As most non-Saccharomyces yeasts are incapable of completing alcoholic fermentation, S. cerevisiae is usually added, either simultaneously as a coinoculum or sequentially at 24–72 h after non-Saccharomyces yeast inoculation. These inoculation strategies reduce the risk of a stuck fermentation [59,60]. Indeed, in the current study, only the fermentations involving S. cerevisiae were complete. When all strains were coinoculated, Sc was the dominant strain and became the most abundant yeast at the end of fermentation, regardless of the inoculum concentration of non-Saccharomyces yeasts. The dominance of Sc during the fermentation process could be explained by its high ability to tolerate different stresses, such as high ethanol levels, especially relevant in the last stage of fermentation [61]. Additionally, different mechanisms, such as cell-to-cell contact, nutrient competition, secretion of toxic compounds or changes in media [26,62,63,64] could also be responsible of the dominance. The effect of metabolite production and changes in the fermentative medium produced by S. cerevisiae has been proven to reduce the competition and persistence of M. pulcherrima [65], which agrees with the drastic reduction in the Mp population observed in fermentations when Sc was coinoculated. As described above, the fermentation kinetics were mainly affected by the inoculation time of Sc. Specifically, coinoculated fermentations were faster and finished earlier compared to sequential fermentations, which agrees with previous studies [64,66].
Focusing on competition for nutrients, a key factor in wine fermentation is the availability of assimilable nitrogen. On one hand, in pure-culture fermentations, some non-Saccharomyces yeasts (H. uvarum, H. vineae, S. bacillaris and T. delbrueckii) consume less assimilable nitrogen than does S. cerevisiae [47,67,68]. On the other hand, studies have shown that in sequential fermentations, some non-Saccharomyces species, such as T. delbrueckii, L. thermotolerans and S. bacillaris, are capable of consuming almost all assimilable nitrogen within 48 or 72 h before S. cerevisiae is inoculated, which could result in incomplete fermentation due to low growth of S. cerevisiae [47,69]. In the current study, to avoid stuck fermentations due to nitrogen limitation and to promote the growth of Sc, assimilable nitrogen was restored before Sc inoculation, as non-Saccharomyces yeasts had been consuming up to 80% of assimilable nitrogen present in the must. As expected, this addition produced a quick increase in the population of Sc. This conclusion is supported by Medina et al. [68], who also observed an increase in the percentage of the Saccharomyces strain after assimilable nitrogen addition.
A critical aspect for studying yeast interactions during fermentation is to evaluate the yeast population, especially living cells. The traditional method used for determining the yeast living cells during alcoholic fermentation is based on colony counting in different culture media, which takes 2 or 3 days and might cause some deviations [27,70]. In mixed or spontaneous fermentations, to differentiate and count the different yeast species, we need to use selective or differential media [27,41]. One of these media is WLN agar, which is able to distinguish different yeast species according to their colony morphology [25,38,71]. In our work, the four yeast species had different colony morphologies on this medium, which allowed their differential counting. In comparison, qPCR analysis is a relatively fast and sensitive technique to simultaneously identify and quantify different targeted yeast species [29,41,72]. However, as described above, this method cannot distinguish the populations of viable and dead cells some DNA-binding dyes, such as PMA, are used and combined with qPCR, allowing the quantification of only the viable cell population [34,38,73].
In the current study, cell counting on WLN plates and PMA-qPCR were used to monitor the viable populations of the different yeast species during the fermentation process. Specific primers for qPCR were available for all species used in the current work [34,38], which presented a high efficiency and good quantification limit. As described above, even though yeast population profiles were similar between WLN counting and PMA-qPCR analysis, they still showed some differences. First, in the coinoculated fermentations with all strains (1CA and 10CA), Sc was the dominant species at the end of fermentation, without detection of these species in the WLN counting in 1CA. Indeed, WLN medium has been described to be useful for quantifying species with similar log populations but is unable to detect species with a population 1 or 2 logs lower than the main species [38], which would explain the inability to detect some species, especially Mp, when its population started decreasing. In contrast, PMA-qPCR sensitively detected the populations of all non-Saccharomyces strains until the end of fermentation, even at low concentrations, as observed in previous studies [32,38,41,74]. Surprisingly, the viable cell population of Mp obtained from PMA-qPCR analysis was lower than that from WLN counting before mid-fermentation. This could be explained by the different membrane compositions and permeabilities of this species. Vázquez et al. [75] showed that the cellular lipid composition of M. pulcherrima during grape must fermentation contains a high percentage of polyunsaturated fatty acids, which results in more fluid membranes. Moreover, the permeability of the cell membrane is known to be increased upon contact with grape must or ethanol [76]. Thus, this higher permeability could facilitate the penetration of PMAxx into viable Mp cells, resulting in lower quantification by PMA-qPCR analysis. Navarro et al. [38] described this effect in different strains in the early stage of fermentation.
In recent years, M. pulcherrima, T. delbrueckii and Z. bailii have been proven to be species able to reduce ethanol content, mainly in mixed fermentations with S. cerevisiae. For example, M. pulcherrima was able to reduce 0.7–1.5% (v/v) ethanol in sequential fermentations in grape must or defined medium [11,15,39,44], while T. delbrueckii and Z. bailii achieved ethanol reductions of 0.6–1.0% (v/v) and 0.7–1.8% (v/v), respectively, in sequential fermentations with S. cerevisiae compared to single S. cerevisiae fermentation [3,11,39]. All these ethanol reductions were achieved with fermentations initiated by a single non-Saccharomyces strain; however, this reduction changed when a multistarter inoculation was used. For example, according to the results from Varela et al. [23], in sequential fermentations with M. pulcherrima or S. uvarum, the ethanol content decreased by 1.09 and 1.01% (v/v), respectively, while when those species were simultaneously inoculated as mixed starters with S. cerevisiae, the ethanol content decreased by 1.85% (v/v), showing an additive effect of coinoculation. In the current study, we used a mixed starter of three non-Saccharomyces species selected based on their ability to reduce ethanol. Our previous studies showed that the selected Mp, Td and Zb strains were able to reduce the ethanol content by 1.39, 0.84 and 1.02% (v/v), respectively, in sequential fermentations with Sc (inoculated at 48 h), using the same natural must [39]. In this research, the mixed inoculation of these three species in sequential fermentations with Sc resulted in a lower ethanol reduction (less than 0.77% (v/v)) compared to sequential fermentation with a single non-Saccharomyces strain. Thus, we did not observe an additive effect, as did Contreras et al. [77] or Varela et al. [23]. Instead, those species showed a dissipative impact on reducing ethanol production when used together. Similar results were also observed in the study of Contreras et al. [77], in which sequential fermentations by the single starter of M. pulcherrima achieved an ethanol reduction of 1.16–1.76% (v/v), while sequential fermentations initiated by mixed starters of M. pulcherrima, H. uvarum, P. kluyveri and T. delbrueckii reduced the ethanol content by only 0.38% (v/v). Therefore, Contreras et al. [77] demonstrated that ethanol reduction is very dependent on the yeast combination used in the starter, since a yeast strain with a high ability to reduce ethanol, such as M. pulcherrima, was affected by the presence of other yeasts, and as a result, its ability to reduce ethanol could be weakened or strengthened. Moreover, the relative proportion between yeast species during the fermentation could affect the metabolite production. Indeed, comparisons between pure cultures and sequentially inoculated cultures revealed changes in the distribution of carbon fluxes during fermentation [78]. Finally, the initial must composition and nutrient availability can also have an impact in the metabolite production, as previously demonstrated by Seguinot et al. [78].
In addition, our results showed that the inoculum size of non-Saccharomyces yeasts also has an impact on ethanol production, with a lower ethanol content in wines inoculated with higher populations of non-Saccharomyces yeasts. This agrees with the results obtained by Maturano et al. [79], in which a higher inoculum size of non-Saccharomyces yeasts (5 × 106 cells/mL vs. 1 × 106 cells/mL) produced wines with a lower ethanol content. Thus, more combinations of yeast species in terms of both the number and diversity of strains and the amount of each strain inoculated should be tested to find the best combination for ethanol reduction.
Non-Saccharomyces yeasts usually differ from S. cerevisiae in the distribution of their metabolic flux during fermentation [46,67,78,80]. Indeed, several non-Saccharomyces species are able to aerobically respire sugar, which results in altered formation of the main metabolites produced during fermentation, including ethanol, glycerol and organic acids [3,20,45,46]. However, although ethanol production can be decreased when providing non-Saccharomyces yeasts with oxygen, this sometimes has undesirable side effects, such as higher acetic acid or ethyl acetate levels [3,81]. In this study, a significant decrease in ethanol levels was observed in mixed fermentations, even without the addition of oxygen and without a detrimental increase in acetic acid. Indeed, higher levels of acetic and lactic acids were obtained in the presence of non-Saccharomyces yeast, mainly in sequential inoculations, but the levels were kept below 0.33 g/L for lactic acid and 0.36 g/L for acetic acid, with nondetrimental concentrations for the final wines. On the other hand, the concentration of glycerol did not increase in our reduced-ethanol wines, which implied that the production of glycerol was not the main pathway of ethanol reduction in mixed culture fermentations, as occurred in other studies [39,82]. Nevertheless, the final amounts of acetic and lactic acids are not enough to counterbalance the decrease in ethanol and glycerol. Previous studies had also observed that the reduction of the ethanol yield by some non-Saccharomyces strains, could not be fully explained by the overproduction of glycerol or organic acids, suggesting that respiration would be responsible, at least in part, of the poor ethanol yield observed for these strains [83,84]. However, the analysis of the volatile composition of wines (higher alcohols, volatile fatty acids, esters, carbonyl compounds) would help the understanding of the effect of this type of inoculation on the overall complexity of wines [11,85].
In summary, our results confirm that PMA-qPCR analysis is a fast and sensitive method for monitoring the viable cell population dynamics in mixed fermentations of T. delbrueckii, M. pulcherrima, Z. bailii and S. cerevisiae. T. delbruecckii was the dominant non-Saccharomyces species under all conditions, and M. pulcherrima was the minority species, being detected by PMA-qPCR throughout fermentation but at lower and decreasing concentrations. The use of a multistarter culture consisting of several non-Saccharomyces species previously selected for their ability to reduce ethanol and a S. cerevisiae strain resulted in reduced-alcohol wines, even if no aeration was applied. The sequential fermentations obtained ethanol reductions from 0.48–0.77% (v/v), which were accompanied by increases in the lactic acid and acetic acid contents. Among all fermentations, the highest reduction was obtained in the sequential inoculation with a higher inoculum size (107 cells/mL), when S. cerevisiae was added at 72 h. Nevertheless, the ethanol reduction obtained when using a multistarter of non-Saccharomyces species was lower than that when using each non-Saccharomyces species separately [39], which indicates no additive effect among them but a lower efficiency to deliver this outcome due to microbial interactions. Although not as efficient in reducing ethanol, a multistarter inoculation strategy could have additional benefits, such as improving the aroma profile and overall complexity of wines [23,32]. Nevertheless, the use of several species can also be challenged by winemaking conditions and the initial yeast population; therefore, special care has to be taken in the winery for this kind of procedure to be applied [32,77].
Acknowledgments
The authors would like to thank Braulio Esteve-Zarzoso for support on the qPCR design.
Appendix A
Table A1.
Primer sequences used for quantitative PCR analysis.
| Target Species | Primer Name | Primer Sequence 5′-3′ | References |
|---|---|---|---|
| S. cerevisiae | CESP-F | ATCGAATTTTTGAACGCACATTG | Hierro et al. (2007) [28] |
| SCER-R | CGCAGAGAAACCTCTCTTTGGA | ||
| M. pulcherrima | MP2-F | AGACACTTAACTGGGCCAGC | García et al. (2017a) [38] |
| MP2-R | GGGGTGGTGTGGAAGTAAGG | ||
| T. delbrueckii | Tods L2 | CAAAGTCATCCAAGCCAGC | Zott et al. (2010) [39] |
| Tods R2 | TTCTCAAACAATCATGTTTGGTAG | ||
| Z. bailii | ZBF1 | CATGGTGTTTTGCGCC | Rawsthorne and Phister. (2006) [26] |
| ZBR1 | CGTCCGCCACGAAGTGGTAGA |
Table A2.
Slopes, Y-Intersections, correlation coefficients (R2), efficiencies (%), limits of quantification (LoQ) and limits of detection (LoD) of standard curves obtained from serially diluted DNA of S. cerevisiae, M. pulcherrima, T. delbrueckii and Z. bailii with PMAxx treatment. Efficiency was estimated by the formula E = (10−1/slope) − 1.
| Slope | Y-Intersection | R2 | Efficiency (%) | LoQ | LoD | |
|---|---|---|---|---|---|---|
| S. cerevisiae | −3.45 ± 0.04 | 40.38 ± 0.20 | 0.9985 | 94.80 | 103 | 10 |
| M. pulcherrima | −3.40 ± 0.03 | 43.51 ± 0.19 | 0.9991 | 97.01 | 104 | 102 |
| T. delbrueckii | −3.35 ± 0.04 | 40.02 ± 0.25 | 0.9978 | 98.83 | 103 | 10 |
| Z. bailii | −3.65 ± 0.03 | 41.81 ± 0.17 | 0.9986 | 87.92 | 103 | 10 |
Table A3.
The concentration of assimilable nitrogen and the added concentration on the 2nd and 3rd day of fermentation.
| Days | Fermentation | Nitrogen (mg/L) | Added Nitrogen (mg/L) |
|---|---|---|---|
| 2nd day | 1S48 | 102 ± 5.66 | 0 |
| 10S48 | 79.67 ± 9.02 | 20 ± 5.50 | |
| 3rd day | 1S72 | 70.67 ± 13.05 | 31 ± 8.54 |
| 10S72 | 48 ± 5.29 | 53.33 ± 5.77 |
Author Contributions
Conceptualization, X.Z., Y.N., M.-J.T., G.B. and A.M.; methodology, X.Z., Y.N. and M.-J.T.; formal analysis, X.Z., Y.N.; investigation, X.Z, Y.N.; resources, M.-J.T., G.B. and A.M.; data curation, X.Z., Y.N.; writing-original draft preparation, X.Z., Y.N.; writing-review and editing, M.-J.T., G.B. and A.M.; visualization, X.Z., Y.N.; supervision, M.-J.T., G.B. and A.M.; project administration, G.B. and A.M.; funding acquisition, G.B. and A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science, Innovation and Universities, Spain (Project CoolWine, PCI2018-092962), under the call ERA-NET ERA COBIOTECH. Y.N. is a postdoctoral fellow of this project.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Godden P. Persistent Wine Instability Issues. Aust. Grapegrow. Winemak. 2000;443:10–14. [Google Scholar]
- 2.Jones G.V., White M.A., Cooper O.R., Storchmann K. Climate Change and Global Wine Quality. Clim. Chang. 2005;73:319–343. doi: 10.1007/s10584-005-4704-2. [DOI] [Google Scholar]
- 3.Contreras A., Hidalgo C., Schmidt S., Henschke P., Curtin C., Varela C. The application of non-Saccharomyces yeast in fermentations with limited aeration as a strategy for the production of wine with reduced alcohol content. Int. J. Food Microbiol. 2015;205:7–15. doi: 10.1016/j.ijfoodmicro.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 4.Varela C., Dry P., Kutyna D., Francis I., Henschke P., Curtin C., Chambers P. Strategies for reducing alcohol concentration in wine. Aust. J. Grape Wine Res. 2015;21:670–679. doi: 10.1111/ajgw.12187. [DOI] [Google Scholar]
- 5.García-Martín N., Perez-Magariño S., Ortega-Heras M., González-Huerta C., Mihnea M., González-Sanjosé M.L., Palacio L., Prádanos P., Hernández A. Sugar reduction in musts with nanofiltration membranes to obtain low alcohol-content wines. Sep. Purif. Technol. 2010;76:158–170. doi: 10.1016/j.seppur.2010.10.002. [DOI] [Google Scholar]
- 6.Bindon K., Varela C., Kennedy J., Holt H., Herderich M. Relationships between harvest time and wine composition in Vitis vinifera L. cv. Cabernet Sauvignon 1. Grape and wine chemistry. Food Chem. 2013;138:1696–1705. doi: 10.1016/j.foodchem.2012.09.146. [DOI] [PubMed] [Google Scholar]
- 7.De Toda F.M., Sancha J., Balda P. Reducing the Sugar and pH of the Grape (Vitis vinifera L. cvs. ‘Grenache’ and ‘Tempranillo’) Through a Single Shoot Trimming. S. Afr. J. Enol. Vitic. 2016;34:246–251. doi: 10.21548/34-2-1101. [DOI] [Google Scholar]
- 8.Longo R., Blackman J.W., Torley P.J., Rogiers S.Y., Schmidtke L.M. Changes in volatile composition and sensory attributes of wines during alcohol content reduction. J. Sci. Food Agric. 2016;97:8–16. doi: 10.1002/jsfa.7757. [DOI] [PubMed] [Google Scholar]
- 9.Padilla B., García-Fernández D., González B., Izidoro I., Esteve-Zarzoso B., Beltran G., Mas A. Yeast Biodiversity from DOQ Priorat Uninoculated Fermentations. Front. Microbiol. 2016;7:930. doi: 10.3389/fmicb.2016.00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varela C., Barker A., Tran T., Borneman A., Curtin C. Sensory profile and volatile aroma composition of reduced alcohol Merlot wines fermented with Metschnikowia pulcherrima and Saccharomyces uvarum. Int. J. Food Microbiol. 2017;252:1–9. doi: 10.1016/j.ijfoodmicro.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Canonico L., Solomon M., Comitini F., Ciani M., Varela C. Volatile profile of reduced alcohol wines fermented with selected non-Saccharomyces yeasts under different aeration conditions. Food Microbiol. 2019;84:103247. doi: 10.1016/j.fm.2019.103247. [DOI] [PubMed] [Google Scholar]
- 12.Gobbi M., Comitini F., Domizio P., Romani C., Lencioni L., Mannazzu I., Ciani M. Lachancea thermotolerans and Saccharomyces cerevisiae in simultaneous and sequential co-fermentation: A strategy to enhance acidity and improve the overall quality of wine. Food Microbiol. 2013;33:271–281. doi: 10.1016/j.fm.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Gobbi M., De Vero L., Solieri L., Comitini F., Oro L., Giudici P., Ciani M. Fermentative aptitude of non-Saccharomyces wine yeast for reduction in the ethanol content in wine. Eur. Food Res. Technol. 2014;239:41–48. doi: 10.1007/s00217-014-2187-y. [DOI] [Google Scholar]
- 14.Englezos V., Rantsiou K., Cravero F., Torchio F., Ortiz-Julien A., Gerbi V., Rolle L., Cocolin L. Starmerella bacillaris and Saccharomyces cerevisiae mixed fermentations to reduce ethanol content in wine. Appl. Microbiol. Biotechnol. 2016;100:5515–5526. doi: 10.1007/s00253-016-7413-z. [DOI] [PubMed] [Google Scholar]
- 15.Hranilovic A., Gambetta J.M., Jeffery D.W., Grbin P.R., Jiranek V. Lower-alcohol wines produced by Metschnikowia pulcherrima and Saccharomyces cerevisiae co-fermentations: The effect of sequential inoculation timing. Int. J. Food Microbiol. 2020;329:108651. doi: 10.1016/j.ijfoodmicro.2020.108651. [DOI] [PubMed] [Google Scholar]
- 16.Castrillo D., Rabuñal E., Neira N., Blanco P. Oenological potential of non-Saccharomyces yeasts to mitigate effects of climate change in winemaking: Impact on aroma and sensory profiles of Treixadura wines. FEMS Yeast Res. 2019;19:foz065. doi: 10.1093/femsyr/foz065. [DOI] [PubMed] [Google Scholar]
- 17.Binati R.L., Junior W.J.L., Luzzini G., Slaghenaufi D., Ugliano M., Torriani S. Contribution of non-Saccharomyces yeasts to wine volatile and sensory diversity: A study on Lachancea thermotolerans, Metschnikowia spp. and Starmerella bacillaris strains isolated in Italy. Int. J. Food Microbiol. 2020;318:108470. doi: 10.1016/j.ijfoodmicro.2019.108470. [DOI] [PubMed] [Google Scholar]
- 18.Junior W.J.F.L., Nadai C., Crepalde L.T., de Oliveira V.S., de Matos A.D., Giacomini A., Corich V. Potential use of Starmerella bacillaris as fermentation starter for the production of low-alcohol beverages obtained from unripe grapes. Int. J. Food Microbiol. 2019;303:1–8. doi: 10.1016/j.ijfoodmicro.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Furlani M.V.M., Maturano Y.P., Combina M., Mercado L.A., Toro M.E., Vazquez F. Selection of non-Saccharomyces yeasts to be used in grape musts with high alcoholic potential: A strategy to obtain wines with reduced ethanol content. FEMS Yeast Res. 2017;17:1–11. doi: 10.1093/femsyr/fox010. [DOI] [PubMed] [Google Scholar]
- 20.Morales P., Rojas V., Quirós M., Gonzalez R. The impact of oxygen on the final alcohol content of wine fermented by a mixed starter culture. Appl. Microbiol. Biotechnol. 2015;99:3993–4003. doi: 10.1007/s00253-014-6321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comitini F., Gobbi M., Domizio P., Romani C., Lencioni L., Mannazzu I., Ciani M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011;28:873–882. doi: 10.1016/j.fm.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Ecanonico L., Ecomitini F., Eoro L., Eciani M. Sequential Fermentation with Selected Immobilized Non-Saccharomyces Yeast for Reduction of Ethanol Content in Wine. Front. Microbiol. 2016;7:278. doi: 10.3389/fmicb.2016.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varela C., Sengler F., Solomon M., Curtin C. Volatile flavour profile of reduced alcohol wines fermented with the non-conventional yeast species Metschnikowia pulcherrima and Saccharomyces uvarum. Food Chem. 2016;209:57–64. doi: 10.1016/j.foodchem.2016.04.024. [DOI] [PubMed] [Google Scholar]
- 24.Canonico L., Comitini F., Ciani M. Metschnikowia pulcherrima Selected Strain for Ethanol Reduction in Wine: Influence of Cell Immobilization and Aeration Condition. Foods. 2019;8:378. doi: 10.3390/foods8090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallmann C.L., Brown J.A., Olineka T.L., Cocolin L., Mills D.A., Bisson L.F. Use of WL medium to profile native flora fermentations. Am. J. Enol. Vitic. 2001 52:198–203. [Google Scholar]
- 26.Wang C., Mas A., Esteve-Zarzoso B. The Interaction between Saccharomyces cerevisiae and Non-Saccharomyces Yeast during Alcoholic Fermentation Is Species and Strain Specific. Front. Microbiol. 2016;7:502. doi: 10.3389/fmicb.2016.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Díaz C., Molina A.M., Nähring J., Fischer R. Characterization and Dynamic Behavior of Wild Yeast during Spontaneous Wine Fermentation in Steel Tanks and Amphorae. BioMed Res. Int. 2013;2013:540465. doi: 10.1155/2013/540465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phister T.G., Mills D.A. Real-Time PCR Assay for Detection and Enumeration of Dekkera bruxellensis in Wine. Appl. Environ. Microbiol. 2003;69:7430–7434. doi: 10.1128/AEM.69.12.7430-7434.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawsthorne H., Phister T.G. A real-time PCR assay for the enumeration and detection of Zygosaccharomyces bailii from wine and fruit juices. Int. J. Food Microbiol. 2006;112:1–7. doi: 10.1016/j.ijfoodmicro.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Forootan A., Sjöback R., Björkman J., Sjögreen B., Linz L., Kubista M. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR) Biomol. Detect. Quantif. 2017;12:1–6. doi: 10.1016/j.bdq.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hierro N., Esteve-Zarzoso B., Mas A., Guillamón J.M. Monitoring of Saccharomyces and Hanseniaspora populations during alcoholic fermentation by real-time quantitative PCR. FEMS Yeast Res. 2007;7:1340–1349. doi: 10.1111/j.1567-1364.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 32.Padilla B., Zulian L., Ferreres À, Pastor R., Esteve-Zarzoso B., Beltran G., Mas A. Sequential Inoculation of Native Non-Saccharomyces and Saccharomyces cerevisiae Strains for Wine Making. Front. Microbiol. 2017;8:1293. doi: 10.3389/fmicb.2017.01293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nocker A., Cheung C.-Y., Camper A.K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods. 2006;67:310–320. doi: 10.1016/j.mimet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 34.Andorrà I., Esteve-Zarzoso B., Guillamon J.M., Mas A. Determination of viable wine yeast using DNA binding dyes and quantitative PCR. Int. J. Food Microbiol. 2010;144:257–262. doi: 10.1016/j.ijfoodmicro.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Fittipaldi M., Nocker A., Codony F. Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods. 2012;91:276–289. doi: 10.1016/j.mimet.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Vendrame M., Iacumin L., Manzano M., Comi G. Use of propidium monoazide for the enumeration of viable Oenococcus oeni in must and wine by quantitative PCR. Food Microbiol. 2013;35:49–57. doi: 10.1016/j.fm.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Li Y., Yang L., Fu J., Yan M., Chen D., Zhang L. The novel loop-mediated isothermal amplification based confirmation methodology on the bacteria in Viable but Non-Culturable (VBNC) state. Microb. Pathog. 2017;111:280–284. doi: 10.1016/j.micpath.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Navarro Y., Torija M.-J., Mas A., Beltran G. Viability-PCR Allows Monitoring Yeast Population Dynamics in Mixed Fermentations Including Viable but Non-Culturable Yeasts. Foods. 2020;9:1373. doi: 10.3390/foods9101373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu X., Navarro Y., Mas A., Torija M.-J., Beltran G. A Rapid Method for Selecting Non-Saccharomyces Strains with a Low Ethanol Yield. Microorg. 2020;8:658. doi: 10.3390/microorganisms8050658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esteve-Zarzoso B., Belloch C., Uruburu F., Querol A. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Evol. Microbiol. 1999;49:329–337. doi: 10.1099/00207713-49-1-329. [DOI] [PubMed] [Google Scholar]
- 41.García M., Esteve-Zarzoso B., Crespo J., Cabellos J.M., Arroyo T. Yeast Monitoring of Wine Mixed or Sequential Fermentations Made by Native Strains from D.O. “Vinos de Madrid” Using Real-Time Quantitative PCR. Front. Microbiol. 2017;8:2520. doi: 10.3389/fmicb.2017.02520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zott K., Claisse O., Lucas P., Coulon J., Lonvaud-Funel A., Masneuf-Pomarede I. Characterization of the yeast ecosystem in grape must and wine using real-time PCR. Food Microbiol. 2010;27:559–567. doi: 10.1016/j.fm.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Quirós M., Gonzalez-Ramos D., Tabera L., Gonzalez R. A new methodology to obtain wine yeast strains overproducing mannoproteins. Int. J. Food Microbiol. 2010;139:9–14. doi: 10.1016/j.ijfoodmicro.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 44.Contreras A., Hidalgo C., Henschke P.A., Chambers P.J., Curtin C., Varela C. Evaluation of Non-Saccharomyces Yeasts for the Reduction of Alcohol Content in Wine. Appl. Environ. Microbiol. 2014;80:1670–1678. doi: 10.1128/AEM.03780-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quirós M., Rojas V., Gonzalez R., Morales P. Selection of non-Saccharomyces yeast strains for reducing alcohol levels in wine by sugar respiration. Int. J. Food Microbiol. 2014;181:85–91. doi: 10.1016/j.ijfoodmicro.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 46.Ciani M., Morales P., Comitini F., Tronchoni J., Canonico L., Curiel J.A., Oro L., Rodrigues A.J., Gonzalez R. Non-conventional Yeast Species for Lowering Ethanol Content of Wines. Front. Microbiol. 2016;7:642. doi: 10.3389/fmicb.2016.00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roca-Mesa H., Sendra S., Mas A., Beltran G., Torija M.-J. Nitrogen Preferences during Alcoholic Fermentation of Different Non-Saccharomyces Yeasts of Oenological Interest. Microorganisms. 2020;8:157. doi: 10.3390/microorganisms8020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen D., Liu S.Q. Impact of simultaneous and sequential fermentation with Torulaspora delbrueckii and Saccharomyces cerevisiae on non-volatiles and volatiles of lychee wines. LWT. 2016;65:53–61. doi: 10.1016/j.lwt.2015.07.050. [DOI] [Google Scholar]
- 49.Canonico L., Comitini F., Ciani M. Torulaspora delbrueckii contribution in mixed brewing fermentations with different Saccharomyces cerevisiae strains. Int. J. Food Microbiol. 2017;259:7–13. doi: 10.1016/j.ijfoodmicro.2017.07.017. [DOI] [PubMed] [Google Scholar]
- 50.Curiel J.A., Morales P., Gonzalez R., Tronchoni J. Different Non-Saccharomyces Yeast Species Stimulate Nutrient Consumption in S. cerevisiae Mixed Cultures. Front. Microbiol. 2017;8:2121. doi: 10.3389/fmicb.2017.02121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tronchoni J., Curiel J.A., Morales P., Torres-Pérez R., Gonzalez R. Early transcriptional response to biotic stress in mixed starter fermentations involving Saccharomyces cerevisiae and Torulaspora delbrueckii. Int. J. Food Microbiol. 2017;241:60–68. doi: 10.1016/j.ijfoodmicro.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 52.Rollero S., Bloem A., Ortiz-Julien A., Camarasa C., Divol B. Altered Fermentation Performances, Growth, and Metabolic Footprints Reveal Competition for Nutrients between Yeast Species Inoculated in Synthetic Grape Juice-Like Medium. Front. Microbiol. 2018;9:196. doi: 10.3389/fmicb.2018.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yap N., Lopes M.D.B., Langridge P., Henschke P. The incidence of killer activity of non-Saccharomycesyeasts towards indigenous yeast species of grape must: Potential application in wine fermentation. J. Appl. Microbiol. 2000;89:381–389. doi: 10.1046/j.1365-2672.2000.01124.x. [DOI] [PubMed] [Google Scholar]
- 54.Branco P., Francisco D., Chambon C., Hébraud M., Arneborg N., Almeida M.G., Caldeira J., Albergaria H. Identification of novel GAPDH-derived antimicrobial peptides secreted by Saccharomyces cerevisiae and involved in wine microbial interactions. Appl. Microbiol. Biotechnol. 2014;98:843–853. doi: 10.1007/s00253-013-5411-y. [DOI] [PubMed] [Google Scholar]
- 55.Evelázquez R., Ezamora E., Álvarez M.L., Hernández L.M., Eramírez M. Effects of new Torulaspora delbrueckii killer yeasts on the must fermentation kinetics and aroma compounds of white table wine. Front. Microbiol. 2015;6:1222. doi: 10.3389/fmicb.2015.01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mencher A., Morales P., Valero E., Tronchoni J., Patil K.R., Gonzalez R. Proteomic characterization of extracellular vesicles produced by several wine yeast species. Microb. Biotechnol. 2020;13:1581–1596. doi: 10.1111/1751-7915.13614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sadoudi M., Tourdot-Maréchal R., Rousseaux S., Steyer D., Gallardo-Chacón J.-J., Ballester J., Vichi S., Guérin-Schneider R., Caixach J., Alexandre H. Yeast–yeast interactions revealed by aromatic profile analysis of Sauvignon Blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol. 2012;32:243–253. doi: 10.1016/j.fm.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 58.Escribano-Viana R., González-Arenzana L., Portu J., Garijo P., López-Alfaro I., López R., Santamaría P., Gutiérrez A.R. Wine aroma evolution throughout alcoholic fermentation sequentially inoculated with non- Saccharomyces/Saccharomyces yeasts. Food Res. Int. 2018;112:17–24. doi: 10.1016/j.foodres.2018.06.018. [DOI] [PubMed] [Google Scholar]
- 59.Zironi R., Romano P., Suzzi G., Battistutta F., Comi G. Volatile metabolites produced in wine by mixed and sequential cultures of Hanseniaspora guilliermondii or Kloeckera apiculata and Saccharomyces cerevisiae. Biotechnol. Lett. 1993;15:235–238. doi: 10.1007/BF00128311. [DOI] [Google Scholar]
- 60.Soden A., Francis I., Oakey H., Henschke P. Effects of co-fermentation with Candida stellata and Saccharomyces cerevisiae on the aroma and composition of Chardonnay wine. Aust. J. Grape Wine Res. 2000;6:21–30. doi: 10.1111/j.1755-0238.2000.tb00158.x. [DOI] [Google Scholar]
- 61.Querol A., Fernández-Espinar M.T., Del Olmo M., Barrio E. Adaptive evolution of wine yeast. Int. J. Food Microbiol. 2003;86:3–10. doi: 10.1016/S0168-1605(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 62.Goddard M.R. Quantifying the complexities ofsaccharomyces cerevisiae’s ecosystem engineering via fermentation. Ecology. 2008;89:2077–2082. doi: 10.1890/07-2060.1. [DOI] [PubMed] [Google Scholar]
- 63.Wang C., Mas A., Esteve-Zarzoso B. Interaction between Hanseniaspora uvarum and Saccharomyces cerevisiae during alcoholic fermentation. Int. J. Food Microbiol. 2015;206:67–74. doi: 10.1016/j.ijfoodmicro.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 64.Lleixà J., Manzano M., Mas A., Portillo M.D.C. Saccharomyces and non-Saccharomyces Competition during Microvinification under Different Sugar and Nitrogen Conditions. Front. Microbiol. 2016;7:1959. doi: 10.3389/fmicb.2016.01959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.García M., Arroyo T., Crespo J., Cabellos J.M., Esteve-Zarzoso B. Use of native non-Saccharomyces strain: A new strategy in D.O. Eur. J. Food Sci. Technol. 2017;5:1–31. [Google Scholar]
- 66.Loira I., Morata A., Comuzzo P., Callejo M.J., González C., Calderón F., Suárez-Lepe J.A. Use of Schizosaccharomyces pombe and Torulaspora delbrueckii strains in mixed and sequential fermentations to improve red wine sensory quality. Food Res. Int. 2015;76:325–333. doi: 10.1016/j.foodres.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 67.Ciani M., Beco L., Comitini F. Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int. J. Food Microbiol. 2006;108:239–245. doi: 10.1016/j.ijfoodmicro.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 68.Medina K., Boido E., Dellacassa E., Carrau F. Growth of non-Saccharomyces yeasts affects nutrient availability for Saccharomyces cerevisiae during wine fermentation. Int. J. Food Microbiol. 2012;157:245–250. doi: 10.1016/j.ijfoodmicro.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 69.Taillandier P., Lai Q.P., Julien-Ortiz A., Brandam C. Interactions between Torulaspora delbrueckii and Saccharomyces cerevisiae in wine fermentation: Influence of inoculation and nitrogen content. World J. Microbiol. Biotechnol. 2014;30:1959–1967. doi: 10.1007/s11274-014-1618-z. [DOI] [PubMed] [Google Scholar]
- 70.Chambers P., Borneman A., Varela C., Cordente A., Bellon J., Tran T., Henschke P., Curtin C. Ongoing domestication of wine yeast: Past, present and future. Aust. J. Grape Wine Res. 2015;21:642–650. doi: 10.1111/ajgw.12190. [DOI] [Google Scholar]
- 71.Cavazza A., Poznanski E., Guzzon R. Must treatments and wild yeast growth before and during alcoholic fermentation. Ann. Microbiol. 2010;61:41–48. doi: 10.1007/s13213-010-0132-2. [DOI] [Google Scholar]
- 72.Andorrà I., Landi S., Mas A., Esteve-Zarzoso B., Guillamon J.M. Effect of fermentation temperature on microbial population evolution using culture-independent and dependent techniques. Food Res. Int. 2010;43:773–779. doi: 10.1016/j.foodres.2009.11.014. [DOI] [Google Scholar]
- 73.Vendrame M., Manzano M., Comi G., Bertrand J., Iacumin L. Use of propidium monoazide for the enumeration of viable Brettanomyces bruxellensis in wine and beer by quantitative PCR. Food Microbiol. 2014;42:196–204. doi: 10.1016/j.fm.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 74.Andorrà I., Monteiro M., Esteve-Zarzoso B., Albergaria H., Mas A. Analysis and direct quantification of Saccharomyces cerevisiae and Hanseniaspora guilliermondii populations during alcoholic fermentation by fluorescence in situ hybridization, flow cytometry and quantitative PCR. Food Microbiol. 2011;28:1483–1491. doi: 10.1016/j.fm.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 75.Vázquez J., Grillitsch K., Daum G., Mas A., Beltran G., Torija M.J. The role of the membrane lipid composition in the oxidative stress tolerance of different wine yeasts. Food Microbiol. 2019;78:143–154. doi: 10.1016/j.fm.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 76.Pérez-Torrado R., Carrasco P., Aranda A., Gimeno-Alcañiz J., Pérez-Ortín J.E., Matallana E., Del Olmo M.L. Study of the First Hours of Microvinification by the Use of Osmotic Stress-response Genes as Probes. Syst. Appl. Microbiol. 2002;25:153–161. doi: 10.1078/0723-2020-00087. [DOI] [PubMed] [Google Scholar]
- 77.Contreras A., Curtin C., Varela C. Yeast population dynamics reveal a potential ‘collaboration’ between Metschnikowia pulcherrima and Saccharomyces uvarum for the production of reduced alcohol wines during Shiraz fermentation. Appl. Microbiol. Biotechnol. 2014;99:1885–1895. doi: 10.1007/s00253-014-6193-6. [DOI] [PubMed] [Google Scholar]
- 78.Seguinot P., Ortiz-Julien A., Camarasa C. Impact of Nutrient Availability on the Fermentation and Production of Aroma Compounds Under Sequential Inoculation With M. pulcherrima and S. cerevisiae. Front. Microbiol. 2020;11:305. doi: 10.3389/fmicb.2020.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maturano Y.P., Mestre M.V., Kuchen B., Toro M.E., Mercado L.A., Vazquez F., Combina M. Optimization of fermentation-relevant factors: A strategy to reduce ethanol in red wine by sequential culture of native yeasts. Int. J. Food Microbiol. 2019;289:40–48. doi: 10.1016/j.ijfoodmicro.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 80.Minebois R., Pérez-Torrado R., Querol A. A time course metabolism comparison among Saccharomyces cerevisiae, S. uvarum and S. kudriavzevii species in wine fermentation. Food Microbiol. 2020;90:103484. doi: 10.1016/j.fm.2020.103484. [DOI] [PubMed] [Google Scholar]
- 81.Röcker J., Strub S., Ebert K., Grossmann M. Usage of different aerobic non-Saccharomyces yeasts and experimental conditions as a tool for reducing the potential ethanol content in wines. Eur. Food Res. Technol. 2016;242:2051–2070. doi: 10.1007/s00217-016-2703-3. [DOI] [Google Scholar]
- 82.Rodrigues A.J., Raimbourg T., Gonzalez R., Morales P. Environmental factors influencing the efficacy of different yeast strains for alcohol level reduction in wine by respiration. LWT. 2016;65:1038–1043. doi: 10.1016/j.lwt.2015.09.046. [DOI] [Google Scholar]
- 83.Gonzalez R., Quirós M., Morales P. Yeast respiration of sugars by non-Saccharomyces yeast species: A promising and barely explored approach to lowering alcohol content of wines. Trends Food Sci. Technol. 2013;29:55–61. doi: 10.1016/j.tifs.2012.06.015. [DOI] [Google Scholar]
- 84.Magyar I., Tóth T. Comparative evaluation of some oenological properties in wine strains of Candida stellata, Candida zemplinina, Saccharomyces uvarum and Saccharomyces cerevisiae. Food Microbiol. 2011;28:94–100. doi: 10.1016/j.fm.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 85.Tronchoni J., Curiel J.A., Sáenz-Navajas M.P., Morales P., Blanco A.D.L.F., Fernández-Zurbano P., Ferreira V., Gonzalez R. Aroma profiling of an aerated fermentation of natural grape must with selected yeast strains at pilot scale. Food Microbiol. 2018;70:214–223. doi: 10.1016/j.fm.2017.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.