Abstract
In the past decade, gene- and cell-based therapies have been at the forefront of the biomedical revolution. Synthetic biology, the engineering discipline of building sophisticated ‘genetic software’ to enable precise regulation of gene activities in living cells, has been a decisive success factor of these new therapies. Here, we discuss the core technologies and treatment strategies that have already gained approval for therapeutic applications in humans. We also review promising preclinical work that could either enhance the efficacy of existing treatment strategies or pave the way for new precision medicines to treat currently intractable human conditions.
Keywords: translational medicine, synthetic biology, cell-based therapies, gene therapy, ATMP
1. Introduction
Unlike small-molecular drugs or antibodies, cell-based therapies have the potential to sense various input signals and respond by initiating context-dependent treatment actions (1, 2). Although gene- and cell-based therapies have been seen as offering tremendous promise since the early days of recombinant DNA and virus technologies, they have only begun taking center stage in the pharmaceutical industry over the past decade (3–5). Currently, regulatory approval of such therapies is accelerating the technological revolution in biotechnology and medicine (6), and these changes have the potential to produce tectonic shifts in the global economy and in society. For example, Glybera was released in the European market in 2012 as a gene therapy treatment designed to reverse lipoprotein lipase deficiency, but treatment costs of >1 million USD per patient forced its withdrawal from the market a few years later (5). On the other hand, although official price tags for the recently approved chimeric antigen receptor T cell (CAR-T) anticancer therapies Kymriah (tisagenlecleucel) and Yescarta (axicabtagene ciloleucel) remain in a similar range, the National Health Service (NHS) in the UK has already made these drugs available to patients (7), and Japan has also recently made Kymriah available under their national health insurance system (7). Thus, these breakthrough technologies are becoming clinical realities.
At present, a clean technical distinction between the terms ‘gene therapy’ and ‘cell-based therapy’ remains elusive. Whereas the US Food and Drug Administration (FDA) defines gene therapy as an attempt to ‘modify or manipulate the expression of a gene or to alter the biological properties of living cells for therapeutic use’, the European Medicines Agency (EMA) describes it as ‘medicinal products administered as nucleic acids, lipid complexes, viruses or genetically engineered micro-organisms for a therapeutic, prophylactic or diagnostic effect’ (3). Cell therapies on the other hand, are based on patient-derived cells that are genetically modified ex vivo and finally returned to the patient (3). However, in view of the increasing complexity of potential therapies that are already in (pre)clinical stages, we believe that the term ‘advanced therapy medicinal products (ATMPs)’ introduced by EMA might be a more suitable description for all classes of ‘gene therapies’, ‘cell therapies’ and ‘cell-based gene therapies’ that have gained clinical consideration or approval (Supplementary Table S1) and would also cover most next-generation precision medicines that will shape the pharmaceutical landscape in the future (Supplementary Table S2). Therefore, we use this term in the present review.
1.1 Treatment approaches and molecular targets of current ATMPs
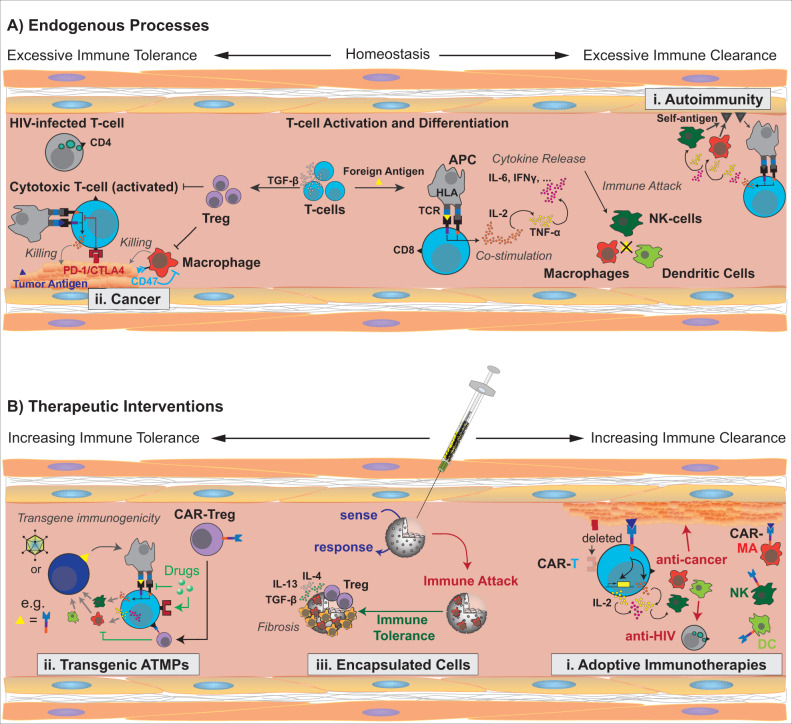
In principle, any ATMP therapy works by strategic manipulation of a patient’s immune tolerance, but an unbalanced intervention may result in severe adverse effects (Figure 1). Autoimmune diseases represent a chronic state of compromised immune (self)-tolerance caused by premature T-cell activation against auto-antigens (Figure 1A-i), while cancers result from excessive immune tolerance that has allowed tumor cells to evade timely elimination (Figure 1A-ii) (8). Thus, therapies based on adoptive transfer of cytotoxic T lymphocytes (e.g. CAR-T cells) essentially focus on site-specific reduction of (self)-tolerance to cancer cells; specifically, activation of T-cell-mediated killing is engineered to no longer depend on the binding of native T-cell receptors (TCRs) to human leukocyte antigens (HLA) on antigen-presenting cells but can be directly activated by tailored tumor-specific antigens (Figure 1B-i) (9). In addition, some tumor cells evade leukocyte-mediated clearance by expressing immune checkpoint inhibitors [e.g. programmed cell death protein 1 (PD-1) or cytotoxic T-lymphocyte-associated protein 4 (CTLA-4)] that block (co)stimulation of TCRs (Figure 1A-ii). Thus, antibodies that selectively bind to PD-1 or CTLA-4 and block their binding to their cognate receptors on the T cell have shown great clinical success in the treatment of many cancers (10, 11). Paradoxically, many ATMPs involve allogeneic and xenogeneic components that could trigger transgene immunogenicity upon implantation or infusion (12). Stimulation of immune tolerance for the transplant occurs through antagonism of very same molecular targets used in adoptive T-cell therapies, such as PD-1/CTLA-4 activation, TCR inhibition or secretion of immunomodulatory cytokines (e.g. TGF-β, IL-12, CXCL12 or CCL22) that trigger regulatory T-cell (Treg) differentiation (Figure 1B-ii) (8). Therefore, the safety and efficacy profile of every ATMP depends directly on how selectively each therapy component suppresses or stimulates the various targets involved in the regulation of immune tolerance.
Figure 1.
Treatment strategies and molecular targets of ATMPs. (A) Endogenous (im)balances of immune tolerance exemplified by (i) autoimmune diseases and (ii) cancer progression. (B) Consequences of different therapeutic interventions for immune tolerance, including (i) cellular adoptive immunotherapies, (ii) transgenic ATMPs and (iii) treatments based on implantation of encapsulated cells. Left: molecular mechanisms stimulating immune tolerance (avoiding immune clearance). Right: molecular mechanisms stimulating immune clearance (suppressing immune tolerance).
Similarly, ATMP therapies involving implantation of foreign materials (e.g. medical devices or encapsulated therapeutic cells) also need to overcome rejection mechanisms associated with immune clearance. Implanted biomaterials often trigger the host immune system to initiate a foreign body reaction, a ‘diverted’ wound-healing process that ultimately forms a fibrotic capsule around the implanted device (Figure 1B-iii) (13). Proinflammatory cytokines are secreted during the early phase of the foreign body reaction. The elevated cytokine level at the implantation site recruits leukocytes to the implantation site, activates macrophages and attracts fibroblasts, which deposit collagen. The eventual formation of the fibrotic tissue triggers secretion of anti-inflammatory cytokines (e.g. IL-4, IL-10, IL-13 and TGF-β), angiogenesis and the induction of immune (self)-tolerance through Tregs (14). Finally, the foreign body is tolerated by the host immune system as ‘self’; however, the fibrotic capsule reduces the permeability of the cell chamber and often compromises oxygen supply to and/or protein secretion from encapsulated cells (15–17) (Figure 1B-iii). This determines the lifetime of therapeutic implants in vivo. Antifibrotic strategies to extend this window mostly involve (i) co-delivery of immunosuppressants during the entire implantation period or (ii) hydrophilic treatments of the implant surface to hinder immune cells from docking on the foreign material (18, 19).
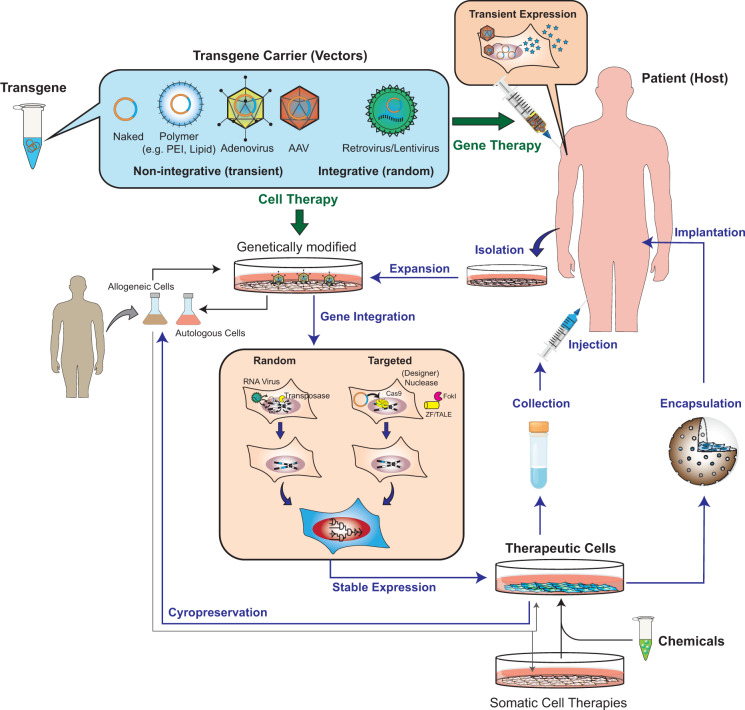
Technically, every ATMP can be described as the choice of an appropriate integration technology for ectopic overexpression of one or multiple therapeutic transgenes in a corresponding host cell. The transgene product determines the eventual mechanism of action of the treatment. The location of this gene integration process defines the ATMP approach; treatments of cells ex vivo prior to implantation are designated as conventional cell therapy approaches, whereas gene integration processes that occur directly in a patient’s living tissue in vivo are classed as gene therapy (Figure 2). Therefore, ATMPs can be sufficiently characterized by the gene integration technology (i.e. viral vectors, non-viral polymer shells or direct electroporation of the transgenic material), the type of host cell and site of gene integration (i.e. gene therapy or cell therapy) and the delivery strategy in vivo (local or systemic) (Supplementary Table S1) (3, 12, 20).
Figure 2.
Cell therapy and gene therapy products using ATMPs. Cell and gene therapy approaches either use non-viral materials (naked plasmids, oligonucleotides or proteins or materials formulated in cationic polymer shells or lipid particles) or viral transgene carriers (non-integrative DNA viruses such as adenoviruses or AAV or integrative RNA viruses such as lentivirus or retrovirus) to integrate one or multiple therapeutic transgenes into host cells. In gene therapy approaches, this integration occurs directly in the patient following injection of the appropriate transgene carrier into the tissue. In cell therapy approaches, this integration occurs ex vivo in cells isolated from the patient (autologous) or derived from a donor (allogeneic). Therapeutic cells (genetically modified cells or somatic cell therapies) can be directly injected into the patient or encapsulated into a semipermeable biocompatible device to provide immune-isolation and nutrient exchange. While retroviruses and transposases trigger random transgene integration (into multiple and different genomic loci), designer nucleases such as ZFN/TALEN/Cas9 enable targeted transgene integration (into known genomic loci). For safety reasons, gene therapy approaches should prefer the use of non-integrative transgene carriers. To facilitate commercialization, therapeutically active batches of cell therapy products should be compatible with cryopreservation methods.
2. Part 1: ATMP therapies under clinical investigation
2.1 Cell therapy approaches (ex vivo cell production)
Medicinal products containing genetically modified cells
To create long-lasting therapeutic effects, the encoding transgenes must be stably integrated into the host cell prior to implantation or infusion (12). According to current EMA regulations, this kind of medicine may belong to the class of ‘medicinal products containing genetically modified cells’ during clinical application (EMA/CAT/GTWP/671639/2008). Currently, retroviral vectors are the predominant technology for stable transgene integration (Supplementary Tables S1 and S2). Retroviruses are single-stranded RNA viruses capable of accommodating up to 8 kb of transgene cargo, which randomly integrates into the genome of the dividing host cell upon transduction (21). Lentiviruses belong to the family of retroviruses but have the capability to infect both dividing and nondividing cells, thus providing a broader target range (20). In contrast, transposase systems such as Sleeping Beauty or PiggyBac are efficient non-viral alternatives for random gene integration with higher packaging capacities (<20 kb) (22, 23). Although random gene integration processes bear substantial safety risks (24–26), these technologies can achieve high transgene expression levels (1). In theory, current advances in omics and gene-editing technologies could be applied to identify and correct erroneous gene disruption resulting from the integration process in individual cell clones.
Cellular adoptive immunotherapies
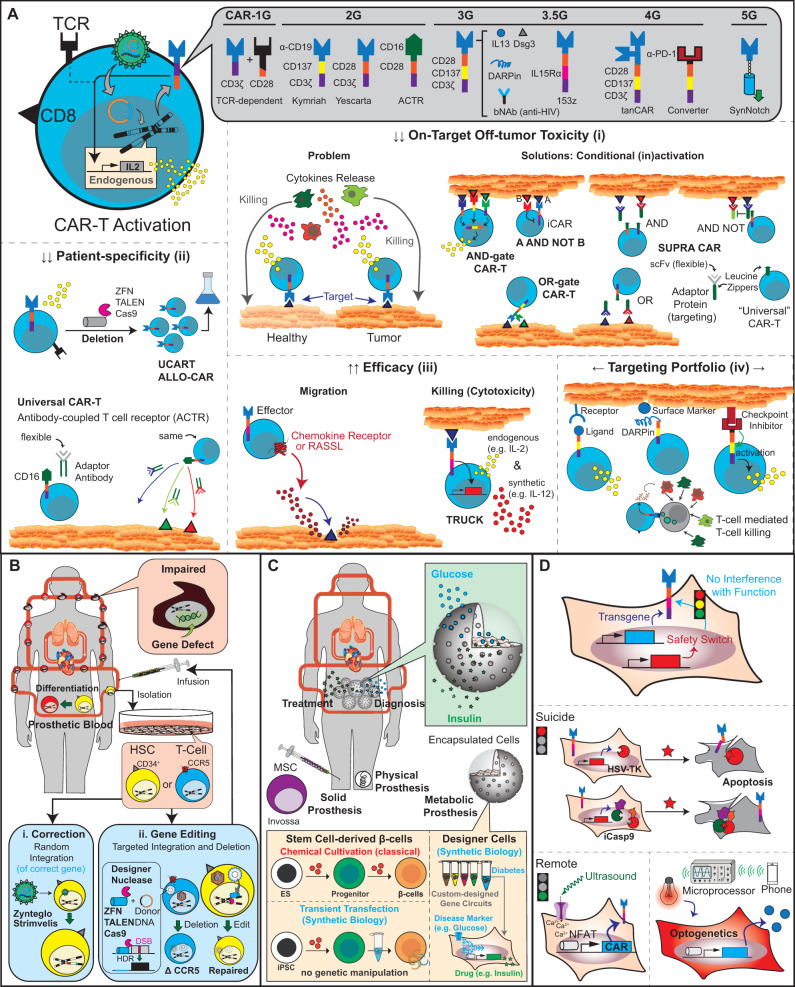
CAR-T cell therapies illustrate successful clinical applications of retroviral vectors. This type of immunotherapy is based on the collection of a patient’s T cells via leukapheresis, followed by an ex vivo manufacturing process that includes T-cell expansion and transduction of a chimeric antigen receptor (CAR) before reinfusion of the engineered cells into the patient (27). The process from cell harvest to patient administration is highly dependent on the cell source and takes ∼2 weeks. Particularly for autologous T cells, these resource-intensive procedures account for the high cost of such therapies (28–30). The CAR is an artificial TCR in which the extracellular TCR domain is substituted by a single-chain variable fragment (scFv), which allows antigen-specific and HLA-independent activation of intracellular signaling (Figure 3A) (11, 31). For example, the CAR of the FDA-approved product Kymriah (tisagenlecleucel) for the treatment of acute B-cell lymphoblastic leukemia (ALL) is composed of an scFv against the B-cell lineage antigen CD19. Anti-CD19 is fused to the native TCR signaling domain CD137 (4-1BB) and the CD3ζ costimulatory domain (Supplementary Table S1) (32). Upon infusion of T cells stably expressing this lentivirus-transduced CART19 (anti-CD19-CD137-CD3ζ), the genetically modified cells autonomously migrate through the body and bind to CD19-expressing tumors, where they trigger cell killing (Figure 1B-i) (33–35). Similarly, the CAR of Yescarta (axicabtagene ciloleucel) and Tecartus (brexucabtagene autoleucel) also contains an scFv against CD19, but the CD3ζ is fused to a different costimulatory domain (CD28) (Supplementary Table S1) (36, 37). Retroviral transduction of this 19-28z CAR (anti-CD19-CD28-CD3ζ) generates CAR-T cells for the treatment of adult patients with refractory large B-cell lymphoma and mantle cell lymphoma, respectively (Supplementary Table S1) (38–41). All these clinically approved CAR-T products (i.e. Kymriah, Yescarta and Tecartus) employ so-called ‘second-generation’ CAR designs, where only one costimulatory domain (CD28 or CD137) precedes CD3ζ (42). In many clinical trials of ‘second-generation’ CARs, suboptimal efficiency of T-cell activation must be compensated with elevated cell doses and this can cause severe adverse effects such as cytokine release syndrome (CRS) (Figure 3A-i) (43–47). For instance, when the target-specific scFv domain was changed to B-cell maturation antigen for the treatment of multiple myeloma (Supplementary Table S1) (48), only 16 out of 33 patients showed a response and >75% of the patients developed CRS (49). Similar problems have been observed for a variety of other treatments, such as in the use of HER2-specific CARs for the treatment of breast cancer (Supplementary Table S1) (50, 51). ‘Third-generation’ CARs contain multiple costimulatory domains alongside CD3ζ (Figure 3A) (52). Nevertheless, the possible safety risks, which have actually occurred in the clinic, remain similar to those of second-generation CARs (Supplementary Table S1) (53, 54). Interestingly, an improved third-generation CAR engineered with an intracellular IL-15Rα domain instead of CD137 (153z; scFv-CD28-IL15Rα-CD3ζ) has decreased CRS incidences in patients (Supplementary Table S1) (55).
Figure 3.
Clinically eligible cell therapy approaches. (A) Evolution of CARs. Most CAR-transgenic T-cell lines (CAR-T) were produced by retro/lentiviral transduction. First-generation CARs (1G) consist of an scFv-antibody fused to the native TCR signaling domain CD3ζ, and therefore require costimulatory domains such as CD28, CD137 (4-1BB) or IL15Rα on other receptors for full activation. Second-generation CARs (2G) integrate a costimulatory domain into the receptor architecture, while third-generation CARs comprise two costimulatory domains. The scFv-domain can also be replaced with antigens such as CD16 (to design universal CAR-Ts), ligands such as IL13 (to target cognate cell-surface receptors), DARPins (to target almost any protein of interest) or PD-1-receptors (to create converter CARs that transform the inhibitory effect of PD-1 into activating signals). Using the synNotch concept, scFv binding can be synchronized with expression of any therapeutic transgene (see Figure 4C). Current CAR-T research primarily focuses on (i) reduction of adverse effects related to on-target off-tumor toxicity, (ii) creation of allogeneic and/or universal CAR-T products to avoid patient- and/or tumor-specific re-engineering of new CAR-T cells, (iii) increasing CAR-T efficacy in terms of killing capacity and migration efficiency and (iv) broadening of the portfolio and types of possible CAR-T targets. (B) Blood replacement therapies (prosthetic blood). Defective blood cells caused by inherited genetic diseases can be ‘washed away’ by infusions of ex vivo-corrected CD34+ HSCs that can differentiate into the appropriate cell type in vivo. (i) Currently approved gene correction strategies use retroviral vectors to restore the expression of missing genes. (ii) In the future, gene-editing technologies based on designer nucleases could directly correct genomic sequences through site-specific DNA repair. (C) Cell-based prostheses. Prostheses complement or restore defective body functions in a seamless and automated manner. Physical prostheses are implanted at the appropriate body site to restore mechanical processes. Solid prostheses are based on the injection of cells (such as MSC) at body sites where specific cell–cell contact is missing. By constantly coordinating diagnosis (detection of disease markers) with treatment (synchronized production and secretion of therapeutic proteins), encapsulated cells also provide prosthetic functions by restoring metabolic balances of nutrients and hormones. For the treatment of diabetes, impaired metabolic function of glucose-dependent insulin production can be restored with stem-cell-derived β cells or β-cell-mimetic designer cells. Because designer cells use gene circuits to program any type of sense-and-respond behavior, this strategy can be applied in principle to any metabolic disease. (D) Safety switches in ATMP therapies and synthetic biology. Safety switches allow orthogonal control compounds to abort or resume the activity of transgenic cells at any point in time and operate in parallel with the therapeutic core program. Early generations of safety switches are based on drug-induced apoptosis to eliminate genetically modified cells in vivo on demand. Current and future generations of safety switches are based on trigger-inducible cell activity using soluble compounds (such as antibody mixes in SUPRA CAR; see A) or traceless remote signals such as ultrasound and light. Optogenetics involves the regulation of cell activities with light.
In contrast to viral transduction technologies, gene correction of cells ex vivo using designer nucleases allows precision editing of any target DNA sequence at the single base-pair level (12). Designer nucleases comprise zinc-finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN) or CRISPR-associated (Cas) proteins capable of targeting any genomic sequence of interest by triggering site-specific double strand breaks (DSBs). Upon co-delivery of an appropriate donor template that contains a custom-designed DNA sequence (the therapeutic transgene), the endogenous homology-directed repair (HDR) machinery can assist in fixing the DSB and simultaneously incorporating the therapeutic transgene into the targeted locus. This process enables site-specific correction, deletion or insertion of any genetic sequence (12). Such strategies were applied to knock-out specific gene products of existing ‘off-the-shelf’ CAR-T cells, such as disruption of PD-1 using CRISPR/Cas9 to achieve resistance to immune checkpoint inhibition (56). This strategy was also used to engineer allogeneic CAR-T cells (also known as ALLO-CAR and UCART products), either by eliminating endogenous TCR components with ZFN (57, 58), TALEN (59) or CRISPR/Cas9 (60), or by deleting HLA-markers with ZFN (61) (Figure 3A-ii). Allogenic cell therapies have higher compliance and lower manufacturing costs, since both leukapheresis and patient-specific therapy designs can be avoided. These therapies are also extremely valuable for patients for whom leukapheresis would be technically too demanding or risky (e.g. with infants or severely ill people) (3).
Similar to allogeneic CAR-T, ‘universal’ CAR-T allows the same batches of CAR-T cells to target different tumor antigens, thereby avoiding time- and resource-intensive reengineering of new CAR constructs when targeting a different antigen (62). To achieve this goal, an antibody-coupled T-cell receptor (ACTR) system was engineered by replacing the scFv domain of a second-generation CAR with the extracellular domain of CD16 (Supplementary Table S1) (63). Since CD16 binds to the constant fragment of antibodies with high affinity, the same ACTR-expressing T cells can be used to target different tumors by using different antibodies specific for different cell surface markers (Figure 3A-ii). The activity of universal CAR-T cells is tunable and makes the dosing of CAR-T cells much safer (64). Alternatively, tandem CARs (tanCAR) feature multiple scFvs on a single CAR moiety, increasing the number of antigens that can be recognized by the same CAR-T cell (Figure 3A-i; Supplementary Table S1) (65–68). Apart from the engineering of antigen-specific receptors and cells, killing efficacy and specificity have been other important parameters for CAR-T development. To amplify the local cytotoxic cell response in the tumor microenvironment, CAR-T cells were engineered to secrete immunostimulatory cytokines such as IL-12 either in a constitutive (69) or NFAT-dependent manner (70), based on the so-called TRUCK concept (T cells redirected for universal cytokine killing) (Figure 3A-iii; Supplementary Table S1). To increase the target specificity of CAR-T cell therapies, various ‘logic gate’ principles have been designed that allow only a pre-defined combination of input signals to activate T-cell signaling (Figure 3A-i). For example, multiple CAR (multiCAR) systems use two (dual-CAR; (71, 72)) or three (tri-CAR; (73)) different CAR molecules to program AND-type expression logics. Intracellular TCR domains (e.g. CD3ζ) and costimulatory domains (e.g. CD28 or CD137) are either expressed alone or are separately linked to different scFv domains. Thus, activation of multiCAR occurs only when all scFvs interact with their specific antigens at the same time (Figure 3A-i). Also, co-expression of synthetic receptors for immune checkpoint inhibitors (e.g. PD-1 or CTLA4) alongside the CAR yields inhibitory CAR systems following an A AND NOT B logic (74) (Figure 3A-i; Supplementary Tables S1 and S2).
To explore cell-specific advantages of other leukocytes in adoptive tumor targeting and killing, some CAR systems have been tested in natural killer (NK) cells (75, 76), dendritic cells (DC) (77) and macrophages (78,79) (Figure 1B-i; Supplementary Table S1). As compared with T cells, the isolation and engineering of NK cells are technically more demanding. However, CAR-NK cells can offer higher tumor-killing efficacies and greater safety in vivo (76). Monocyte-derived macrophages, on the other hand, are highly mobile and can penetrate deep tissues. Thus, CAR macrophages are an excellent option for solid tumor clearance (79). Importantly, cellular adoptive immunotherapies using genetically modified leukocytes are not limited to the treatment of cancer. For example, HIV-specific CARs have been clinically investigated; CAR-T cells were engineered to target infected CD4+ T cells through broadly neutralizing antibodies (bNAbs) and are intended to attenuate HIV survival by induction of T-cell-mediated T-cell killing (80–82) (Figure 3A-iv; Supplementary Table S1). Also, immunosuppressive Tregs were engineered to express CARs that trigger site-specific immune tolerance to alloantigens or foreign transplants (83–85) (Figure 1A-i).
Prosthetic blood
While the engineering of patient-specific leukocytes has focused mainly on cancer immunotherapy, cell therapy approaches using hematopoietic stem cells (HSCs) as the host cells are promising for the treatment of various inherited blood disorders. For example, β-hemoglobinopathies are characterized by impaired blood cells resulting from inborn gene deficiencies. The gene deficiencies manifest in abnormal cell phenotypes, and the blood cells are unable to fully carry out their intended function. Since HSCs can repopulate an entire hematopoietic system, expansion and permanent genetic modification of isolated CD34+ HSCs enable mass production of gene-corrected progenitor cells ex vivo, which differentiate into the appropriate cell type in vivo upon infusion (Figure 3B). For example, the ATMP therapy Zynteglo is based on autologous CD34+ cells transduced with a lentiviral vector harboring the βA-T87Q-globin gene and is approved for patients with transfusion-dependent β-thalassemia (86, 87) (Supplementary Table S1). Similarly, Strimvelis consists of autologous CD34+ cells transduced with a retroviral vector encoding a human adenosine deaminase (ADA) transgene (Supplementary Table S1). It is approved for the treatment of severe combined immunodeficiency (SCID) due to ADA shortage (ADA-SCID) and was the first commercially available ATMP based on corrective ex vivo cell therapy (88, 89). Gene correction of HSCs through random integration using viral vectors to achieve ectopic overexpression of the correct gene is reminiscent of manufacturing blood ‘prostheses’ (i.e. direct replacement of a defective organ with a functional one) (Figure 3B-i). In contrast, targeted gene editing (also known as ‘genomic surgery’) using designer nucleases allows high-fidelity knock-in and knock-out of any set of (trans)genes almost at will (90) (Figure 3B-ii). Blood prostheses created with such ‘gene-editing’ technologies may therefore be substantially safer for patients than their virally transduced counterparts. Driven by the clinical success of Zynteglo and Strimvelis, many blood prosthesis therapies are currently under clinical trial, including treatments intended for sickle cell disease and β-thalassaemia (91–96), X-linked severe combined immunodeficiency (97), Wiskott-Aldrich syndrome (98) and chronic granuloma (99) (Supplementary Table S1). In addition, blood prosthesis therapies have played a major role in the treatment of HIV-infected patients. In the clinic, ZFN-based deletion of the CCR5 and/or CXCR4 genes in patient-specific T cells has resulted in markedly reduced virus proliferation rates in vivo upon transfusion (100–103) (Supplementary Table S1). Blood prostheses can also effectively assist CAR-T therapies. In fact, on-target off-tumor adverse effects remain a critical risk factor in CAR-mediated cell therapies. In an elegant treatment approach for myeloid leukemia, CAR-T cells targeting the lineage-specific CD33 were co-administered with gene-edited HSCs in which CD33 had been deleted with the CRISPR/Cas9 system (Supplementary Table S2) (104). Even if anti-CD33 CAR-T cells could kill healthy CD33-expressing myeloid cells (on-target, off-tumor; Figure 3A-i), new myeloid cells would repopulate from the gene-edited HSCs that were engineered to resist elimination by the CAR-T cells.
Solid prostheses
Apart from the engineering of blood prostheses, gene editing can also overcome various limitations of other cell therapies. In fact, ex vivo modification in somatic cells can similarly create ‘solid’ prostheses (Figure 3C). By definition, prostheses are devices that can complement or restore defective body functions in a seamless and automated manner. While physical prostheses (e.g. artificial organs or joints) are implanted at an appropriate body site to restore mechanical functions, solid biological prostheses are based on injection of cells at body sites where specific cell-cell contacts have been missing. For example, Invossa consists of human allogeneic chondrocytes transduced with TGF-β and is approved in Korea for the treatment of knee osteoarthritis (Supplementary Table S1) (105, 106).
Somatic cell therapies (without genetic intervention)
Traditional organ and cell transplantations were the first kinds of solid prostheses developed in biomedicine. The majority of these approaches, including stem cell therapies, do not involve (any) genetic modification. Instead, the isolated host cells are expanded ex vivo to allow production of sufficient cell numbers for (re)-implantation, and these cells may optionally be treated with specific chemicals and/or growth factors to trigger epigenetic changes, such as differentiation. According to EMA guidelines, cell therapies of this kind that do not intentionally change the host cell’s genomic integrity should be registered as ‘somatic cell therapy medicinal products’ (EMA/CAT/600280/2010). For example, Alofisel (darvadstrocel) is simply based on the expansion of human allogeneic adipose-derived mesenchymal stem cells (ADSC) and was approved in the EU for the treatment of complex perianal fistulas in patients with Crohn’s disease; single intralesional injections of 120 million ADSCs provided clinical remission rates of >50% at 1 year follow-up (107) (Supplementary Table S1). Mesenchymal stem cell (MSC) treatments are generally accepted as safe and have enormous therapeutic potential, but clarification of the specific molecular mechanism accounting for the proposed treatment effect remains a major challenge during clinical investigations (108). In general, major mechanisms of actions of MSCs include (i) tissue regeneration through self-sufficient differentiation into specific cell types, (ii) immunomodulation through tissue migration and local secretion of cytokines and exosomes or (iii) cell-type-specific activation through direct cell contacts (109). Stem cells are particularly attractive because these multipotent cells have the potential to differentiate into various cell types to aid in tissue repair (110). For stem-cell-based medicines, the ideal therapeutic approach would comprise complete differentiation and purification of the therapeutically active cell type ex vivo prior to implantation or injection into the patient (6). However, the main limiting factors are the technical challenge in identifying and isolating proper stem/progenitor cells, the scarcity of robust and scalable cultivation protocols and the shortage of quality-assurance technologies capable of characterizing differentiation efficiency (111). Therefore, various treatment strategies currently under clinical testing employ a semi-rational approach, which is based on implantation of progenitor cells into the patient in the expectation that full differentiation and maturation will eventually occur in vivo. For the treatment of glaucoma and other optic neuropathies, for example, implantation-ready progenitor cells can be extracted from bone marrow (112, 113), or generated ex vivo by treatment with specific inhibitors (114) (Supplementary Table S1).
Metabolic prostheses
Cell-based treatment strategies also play a central role in the treatment of type-1 diabetes, an autoimmune disorder characterized chronic hyperglycemia due to the loss of pancreatic β cells (115). Although direct transplantation of allogeneic pancreatic islets remains the gold standard in terms of therapeutic efficacy (116), this approach either critically depends on the timely availability of donors or requires life-long immunosuppression (117, 118). To overcome these issues, transplantation of allogenic islets can be achieved through CRISPR/Cas9-mediated knock-out of endogenous retrovirus genomes in porcine islet cells (119) and/or encapsulation of islets into biocompatible and semipermeable cell chambers (macroencapsulation; (120–122)) or alginate beads (microencapsulation; (123–125)) that afford xenograft tolerance (Figure 1B-iii; Supplementary Table S1). Encapsulation of β-like cells produced by differentiation of human embryonic stem cells (ES; 126) or induced pluripotent stem cells ex vivo (iPSC; 127) also has potential for diabetes treatment (128). Encapsulated islets or β-like cells can be seen as ‘metabolic prostheses’, which are capable of restoring the function of a defective organ even if the implant itself is placed at a distant location in the body (Figure 3C). However, stem cell-based approaches have similar types of advantages and limitations as with allogenic islet transplantation. To achieve optimal glycemic control and xenograft lifetime, it is essential that encapsulated β-like cells already show a glucose-stimulated insulin secretion profile similar to that of clinical-grade human islets prior to implantation. For example, based on an earlier observation that pancreatic progenitor (PP) cells could differentiate more efficiently into glucose-responsive β-like cells in vivo than in vitro (129), a clinical trial driven by ViaCyte Inc. used an semi-rational strategy of implanting macroencapsulated PPs inside a cell chamber that was optimized for nutrient exchange and oxygen supply (130, 131) (Supplementary Table S1). However, differentiation in vivo might be much slower than the fibrotic reaction triggered by the implant itself, which probably accounts for the limited therapeutic efficacy, in spite of good safety results (116) (Figure 1B-iii). In contrast, ES-derived β-like cells microencapsulated in alginate beads 1.5 mm in diameter could restore self-sufficient glycemic control over 174 days upon implantation into mice, and this is currently the most promising preclinical study of stem cell-based diabetes treatments (132) (Figure 3C;Supplementary Table S2).
Safety switches
To avoid hyperactivity of therapeutic cells in vivo, one alternative to cell number titration is the engineering of safety switches. Safety switches allow orthogonal control compounds to block the activity of transgenic cells on demand, while operating in parallel with the therapeutic core program (Figure 3D). Safety switches under clinical testing include the herpes simplex virus-thymidine kinase (HSV-TK) suicide gene system (133) and the inducible caspase 9 (iCasp9) system (134). Both systems involve transgenes that encode drug-controlled initiators of apoptosis (Figure 3D). Constitutively expressed HSV-TK only triggers controlled cell death when ganciclovir is administered exogenously; the iCasp9 suicide switch consists of two inactive caspase 9 monomers, which are activated by dimerization in the presence of a bio-inert rapamycin analogue, AP1903. In the clinic, the iCasp9 system has been incorporated into a GD-2-specific CAR construct for the treatment of neuroblastoma (Supplementary Table S1) (135, 136).
2.2 Gene therapy approaches (in vivo gene delivery)
Non-integrative transgene delivery
Oncolytic DNA viruses
Non-integrative gene delivery is analogous to transient transfection experiments in cell culture. Transgenes are carried into the host cell to allow constant expression until the episomal vector (i.e. non-chromosomal gene carrier) is eventually ‘out-diluted’ through cell division (20) (Figure 2). To achieve high-level expression, non-integrating DNA viruses such as adenovirus and the related AAV system (adenovirus-associated virus) are the most widely used transgene carriers (3, 21). While adenoviral vectors allow cargo sizes of up to 8.5 kb, AAVs are generally restricted to the delivery of shorter transgenes of < 5.0 kb (137). In comparison to adenoviral vectors, AAVs are safer due to their lower immunogenic and oncogenic potentials. In fact, some gene-editing-based cell therapy approaches use non-replicative adenoviral and AAV vectors to more efficiently deliver the donor DNA required for HDR-based repair (Figure 3B-ii) (138–142). For gene therapy approaches, non-replicative adenovirus and AAV strains are very effective to transiently deliver therapeutic transgenes into the living tissue of patients (20). For example, Gendicine, arguably the first commercial gene therapy drug that was approved for cancer treatment, is based on a replication-deficient adenovirus encoding for transgenic p53 under the control of the Rous sarcoma virus promoter (Supplementary Table S1) (143). Local injection of the purified virus into the tumor triggers tumor-specific p53 expression and apoptosis (Figure 4A;Supplementary Table S1) (144). Thus, Gendicine can be classified either as a conventional gene delivery approach based on ectopic transgene overexpression, or as an oncolytic virus (3). According to a strict definition, however, an oncolytic virus must have the ability not only to infect and kill, but also to specifically replicate within the tumor (145). Based on a selectively replicating adenovirus vector ONYX-015 (146), Oncocrine (H101) was therefore designed as an oncolytic virus for the treatment of nasopharyngeal carcinoma (Supplementary Table S1) (147). Both Gendicine and Oncocrine have been approved in China for more than a decade but were not approved by the US FDA due to a lack of sufficient information about the two therapies (20). Because wild-type adenovirus replication lyses its host cell before transmission, engineering oncolytic adenoviruses for tumor-specific expression of the replication factor E1A is a good strategy to program tumor-specific apoptosis (Figure 4B). This approach has been tested in many clinical trials, including for the treatment of bladder cancer (148) and neuroendocrine tumors (149) (Supplementary Table S1).
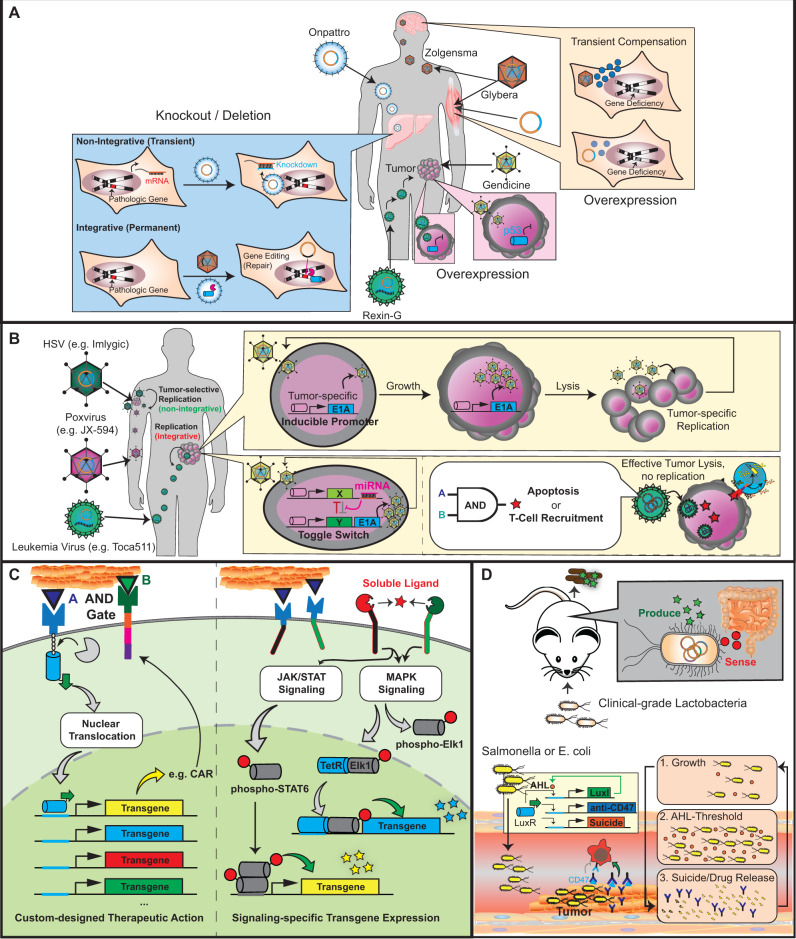
Figure 4.
Gene therapies and future prospects for ATMPs. (A) Gene correction. Inherited genetic disorders in somatic tissues can be corrected by ectopic overexpression of the missing gene or by transient knock-down of pathologic genes through RNAi or antisense mRNAs. Non-integrative overexpression is based on episomal expression from viral (adenovirus or AAV; stronger) or non-viral transgene carriers (plasmid DNA; weaker), which transiently override defective or deficient genomic expression. Integrative approaches enabling genome correction using designer nucleases are also possible but must overcome off-target editing events. (B) Oncolytic virotherapy. Adenoviruses lyse the infected host cell upon replication. Engineering oncolytic adenoviruses for tumor-specific replication (through E1A expression) enables tumor-selective re-infection and killing. Oncolytic HSV and poxviruses are DNA viruses with higher packaging capacities for therapeutic transgenes. Poxviruses and the RNA virus Toca511 are suitable for systemic administration. Lentiviruses can also be used for the local delivery of oncolytic transgenes into tumors. (C) Receptor-mediated transcription. Activation of SynNotch receptors triggers proteolytic cleavage of the transmembrane domain, resulting in the nuclear translocation of synthetic transcription factors and the initiation of transgene expression from cognate-specific promoters. Transcription of therapeutic transgenes can also be synchronized with endogenous signaling pathways such as JAK/STAT and MAPK. Stimulation of these pathways by custom-designed cell-surface receptor actions activates synthetic promoters engineered to contain signaling-specific response elements. (D) Therapeutic bacteria. Lactobacteria can be engineered to sense infection markers in the gastrointestinal system. Upon oral ingestion, genetically engineered bacteria self-sufficiently migrate to the gut and produce specific reporter signals that can be measured in feces. Some bacterial strains such as E. coli or Salmonella preferentially colonize and proliferate in hypoxic and immune-privileged tumor microenvironments upon systemic administration. Engineering these bacteria to carry gene circuits that trigger population-density-dependent cell lysis and drug release is a promising alternative for cancer therapy.
In addition to adenoviruses, herpes simplex viruses (HSV) and poxviruses (e.g. vaccinia virus) are employed for the engineering of oncolytic viruses. Both HSV and vaccinia virus are non-integrative DNA viruses with readily designable replication-specificity but have higher transgene packaging capacities (>30 kb) than adenoviruses (21). HSV can efficiently infect almost every type of host cell upon local injection (150). In contrast, a major advantage of oncolytic poxviruses is their ability to travel systemically through the blood and migrate self-sufficiently to a specific tumor site (145) (Figure 4B). The oncolytic HSV Imlygic (talimogene laherparepvec), engineered for tumor-specific replication and expression of the immune stimulatory protein GM-CSF, was approved by the US FDA for the treatment of melanoma in 2006 (151) (Supplementary Table S1). In the following year, JX-594 (pexastimogene devacirepvec), an oncolytic poxvirus also capable of tumor-specific GM-CSF expression (152), received orphan drug designation from the US FDA and EMA for the treatment of hepatocellular carcinoma (Supplementary Table S1).
Gene correction by overexpression
Currently, recombinant AAVs are exclusively used for ‘encapsidation’ to deliver DNA into specific cell types and tissues, enabling ectopic (over)expression of therapeutic transgenes (153) (Figure 4A). In the clinics, AAV-based gene therapy plays an important role in the correction of gene defects within particular tissues and organs (3). For instance, patients with familial lipoprotein lipase deficiency (LPLD) have elevated levels of serum triglycerides, which may cause recurrent and life-threatening pancreatitis (154). Glybera (alipogene tiparvovec) consisted of an AAV1 vector encoding for LPL and is injected intramuscularly, leading to the transient expression and subsequent secretion of the lipase in the bloodstream (155) (Figure 4A;Supplementary Table S1). Similarly, Luxturna (voretigene neparvovec-rzyl) is an AAV2 vector genetically engineered to express the human retinal pigment epithelial-specific protein 65 kDa (RPE65) and is approved for the treatment of mutation-associated retinal dystrophy (156) (Figure 4A;Supplementary Table S1). Intravenous administration of Zolgensma (onasemnogene abeparvovec-xioi), an AAV9 vector containing a transgene encoding the human survival motor neuron (SMN) protein, can effectively target spinal motor neurons, neuronal and glial cells of the brain (157) (Figure 4A;Supplementary Table S1). Despite questionable efficiency results due to lack of an available control group, Zolgensma is currently approved for the treatment of spinal muscular atrophy in infants (157). In the not-too-distant future, AAV-based gene therapy for the treatment of hemophilia is also expected to enter the market (158, 159).
AAV and adenoviruses generally provide high transgene expression levels owing to their very high delivery efficiencies (20). However, if weak constitutive levels are needed for a specific therapeutic goal, non-viral vectors such as plasmid DNA and oligonucleotides are preferred (3). For example, direct injection or electroporation of plasmid DNA into the target tissue provides transient gene expression in a similar manner to AAV-based gene correction (Figure 4A). Currently approved plasmid therapies include Neovasculgen (cambiogenplasmid) for the treatment of peripheral vascular disease through constitutive VEGF165 expression (160), as well as Collategene (a constitutive hepatocyte growth factor expression vector) for the treatment of critical limb ischemia (161) (Supplementary Table S1). To achieve transient transgene overexpression with non-viral vectors, mRNA encoding the desired protein or purified protein can also be directly administered into the host cell by electroporation. In contrast to ectopic overexpression of a transgene to temporarily compensate for a gene deficiency, gene correction can also be achieved by transient knock-down of specific pathologic genes using custom-designed oligonucleotides (Figure 4A). For example, the first therapies based on RNA interference (RNAi) (162) and antisense mRNA technology (163–167) have recently entered the market.
Integrative gene delivery in vivo
Gene editing
Intuitively, targeted gene-editing technologies would seem to be preferable for gene correction. Indeed, many preclinical studies are currently testing the feasibility of using ZFN (168), mega-nucleases (169) and CRISPR/Cas9 (170–176), aiming to emulate the treatment efficacies of AAV-based gene correction therapies. For example, one study involving intravenous delivery of the CRISPR/Cas9 system formulated in a lipid nanoparticle carrier resulted in >80% editing of the Pcsk9 gene in mouse liver (Figure 4A;Supplementary Table S2) (177). However, since this type of gene-editing results in permanent modification of the host cell genome, this editing efficiency remains too low for clinical consideration. Although optimized Cas9 variants can reduce off-target indel rates to <0.1%, even an error rate of this order is still immense in the context of the size of the human genome—and the absence of evidence in editing mistakes is not equivalent to evidence of absence (178). Therefore, even though a recent study has performed a thorough risk analysis for CRISPR/Cas9-based therapeutic genome editing while developing a rational strategy for guide RNA selection (179), we believe that gene correction with non-integrative methods such as AAV, adenoviruses, plasmid DNA and oligonucleotides (Figure 4A) remains preferable for safety reasons.
Oncolytic RNA viruses
Apart from gene correction, integrative gene delivery in vivo is clinically acceptable if the therapeutic goal is to ultimately kill the targeted host cell. For example, Rexin-G (Mx-dnG1) is a systemically injected RNA virus approved for the treatment of metastatic cancers (180). Rexin-G is a non-replicative retrovirus selectively targeting tumor-specific signature (SIG) proteins through the cryptic collagen-binding motif in the extracellular matrix and encodes a dominant-negative mutant of human cyclin G1 (181) (Supplementary Table S1). Mechanistically, it can be defined as a systemic variant of Gendicine, which kills cancer cells by tumor-specific induction of cell cycle arrest instead of selective replication (Figure 4A). In contrast, Toca511 is a replicative retrovirus engineered on the basis of amphotrophic murine leukemia viruses (182). Upon systemic administration, this oncolytic RNA virus selectively infects tumor cells in the brain and expresses the yeast enzyme cytosine deaminase, which converts the prodrug 5-fluorocytosine into a potent anticancer drug, 5-fluorouracil (183) (Supplementary Table S2). Toca511 is currently in a Phase 2/3 clinical trial for malignant glioma and has shown promising interim results (184). One shortcoming of systemically delivered oncolytic viruses is the strong transgene immunogenicity triggered by these particles. In reality, clinical strains of oncolytic viruses are indeed considered ‘too’ safe as they are efficiently cleared from the circulation by the immune system, which severely limits treatment efficacy (185). Therefore, the current view in the field of oncolytic virotherapy is that the oncolytic virus cannot (yet) be considered as a stand-alone therapy for cancer but can be highly efficacious when used in combination with other more established therapeutic interventions (186). As an interesting alternative to circumvent the host immune barrier, oncolytic viruses can be encapsulated prior to systemic injection, either within cationic liposomes or polymers as an immuno-isolating shell (187, 188), or encoded into autologous cells to create immuno-compatible virus carriers (185). Cell-based virus carriers are often described a ‘Trojan horse’ strategy that uses human cells for tumor-specific migration and site-specific stimulation of virus production and secretion (185, 186). Various cell types are currently being investigated as oncolytic virus carriers in (pre)clinical tests, including MSC s (189, 190), neural stem cells (191), T cells (192, 193) and cytokine-induced killer cells (194) (Supplementary Tables S1 and S2). Hence, oncolytic viruses delivered by cells to their target location in vivo may be classified as a cell therapy (Figure 2).
3. Part 2: future perspectives, opportunities and lessons
ATMP therapies have already provided impressive treatment results. The current century has witnessed ∼50 ATMP approvals, not including a variety of mysterious therapies that are not scientifically proven but have nevertheless notoriously been commercialized in some parts of the world (5). In this context, Google has recently announced a new Healthcare and Medicines policy that prohibits digital advertisements of unproven ATMPs with no clear scientific basis (7), and this may help to minimize both the risk to patients and the likelihood of concomitant reputational damage to properly established therapies. It seems clear that genuine ATMP therapies are at the forefront of a new technological revolution in biomedicine, and the scope for engineered precision medicines in the coming decades is truly enormous (6). Of course, issues remain to be overcome. For example, the spatiotemporal control capacity of ATMP mechanisms in vivo must be dramatically increased to further enhance treatment safety and efficacy. In CAR-T cell therapies, the need for this is illustrated in classical approaches for dose-efficacy titration, which was purely based on an empirical testing of different cell numbers for injection. Such testing in patients has resulted in serious adverse effects, with overdoses of constitutively active CAR-T cells in the bloodstream causing severe CRS, neurotoxicity and, in some cases, even death (43–47, 54). In current gene therapy approaches, transgene expression strengths in vivo are typically determined by the choice of the vector system. Delivery to the target tissue and the expression level of the very same transgene vary greatly, depending upon whether a viral transduction or a naked plasmid transfection is chosen (3) (Figure 4A). Among ATMP therapies already in the clinic (Supplementary Table S1), oncolytic viruses might have made the largest step toward achieving programmable and context-dependent therapeutic activity. Key features, including target specificity, replication and transgene expression, have all been subjected to application-specific bioengineering, providing an excellent illustration of the huge impact that this class of precision medicines is expected to have (148,149, 152, 181,182, 190) (Figure 4B).
3.1 Current research focuses
CAR-T therapy
Synthetic biology, by focusing on the design of genetic circuits to control human cell functions with high spatiotemporal precision, is a key driver of the progress of ATMP therapies (195, 196). In essence, synthetic biology comprises any type of genetic intervention in any biological system that aims to improve the functionality of cells, tissues or organisms. Therefore, synthetic biology can advance the development of ATMP products in almost every respect. For example, the engineering of CARs in which the scFv domain is replaced by membrane-tethered ligands (197) or designed ankyrin repeat proteins (DARPin; 198) has broadened the range of possible targets for CTL-mediated killing (Figure 3A-iv). Indeed, chimeric autoantigen receptor (CAAR)-T cells using the autoantigen desmoglein 3 (Dsg3) as an extracellular targeting domain were recently engineered to target and eliminate autoreactive B lymphocytes in pemphigus vulgaris (199). While the CAR moiety is responsible for TCR-dependent effector functions, co-expression of TCR-independent cell membrane receptors can facilitate the migration of CAR-T cells to specific sites (31). Not surprisingly, if CAR-T cells cannot access their target cells, it is very unlikely that they can effectively control tumor growth (200). Thus, examples for co-receptors guiding the trafficking of CAR-T cells to their assigned destinations in vivo include natural chemokine receptors (201–203) and synthetic receptors activated solely by a synthetic ligand engineered to be specific for the small molecule clozapine-N-oxide (204) (Figure 3A-iii). In the tumor microenvironment, CAR-T-cell-mediated killing inherently involves the secretion of proinflammatory cytokines, such as IL-2 and IL-10 (205) (Figure 1B-i). To create customized therapeutic programs, classical TRUCK approaches co-express effector proteins to add another killing program in parallel, such as IL-12 (69, 70), anticancer proteins (206) or ion channels (207) (Figure 3A-iii; Supplementary Table S2). ‘Converter CARs’ containing an extracellular binding domain for immunosuppressive ligands (e.g. IL-4 or PD-1) fused to a TCR-activating intracellular domain were also engineered and co-expressed in CAR-T cells to redirect immune tolerance (208,209) (Figure 3A-iv). To achieve full control over T-cell activity, back-to-back publications from the same laboratory presented a novel CAR architecture designed on the basis of the Notch receptor (205, 210,211) (Figure 3A). With such synthetic Notch (synNotch) receptors, T-cell activation through scFv is no longer (exclusively) synchronized with endogenous TCR signaling but can directly initiate transcription of any user-defined therapeutic transgene from synthetic receptor-specific promoters (205). Importantly, this design could substantially improve the dynamics of conditional CAR-T-cell activation (Figure 4C). For example, whereas conventional multiCAR systems require the omnipresence of different CAR parts to achieve AND gate logics (Figure 3A-i), synNotch receptors can spatiotemporally program CAR-T-cell activation by only expressing the CAR construct when specific cell contacts are matched (210,211) (Figure 4C). As an alternative to synNotch receptors, scFvs can also be fused to intracellular domains of interleukin receptors to synchronize antigen-sensing with user-defined, JAK-STAT-mediated transgene transcription (Figure 4C) (212). To improve the sensitivity to soluble molecules, JAK-STAT-mediated transgene expression was synchronized with various dimerization-based EpoR activation strategies (213) (Figure 4C).
Safety switches
To achieve tight control over transgene expression, toggle switches based on mutually repressive transcription units are important tools in synthetic biology (214–216). Withdrawal of one repressive module results in activation of the other module and thus, the dynamic control is superior to that achievable with inducible ON/OFF promoters (Figure 4B) (196). For the treatment of hepatocellular carcinoma, a toggle switch was incorporated into an oncolytic adenovirus, allowing the tumor-specific α-fetoprotein promoter to control toggle switch expression and a specific miRNA signature to trigger activation of adenovirus replication (through E1A expression) and immunomodulation (through co-expression of IL-2, GM-CSF and anti-PD-1 scFvs) (217) (Figure 4B). Replicating oncolytic adenoviruses containing surface modifications were also engineered for the treatment of pancreatic cancer with enhanced tumor specificity (218). To achieve maximum spatiotemporal control of self-sufficient ATMP activity in vivo, the canonical approach would be the rational choice of most appropriate combinations of tissue-specific markers to create conditionally activated transgene responses (Figures 3A-i and 4C). In clinical settings, however, patient safety would be substantially increased if the physician could use specific control signals to intervene in the therapy at any point in time and thus react to emergent situations. Current versions of such safety switches tested in the clinic are based on trigger-controlled cell death using small-molecular drugs to abort the treatment (133–136) (Figure 3D). However, future safety switches could use inducible transgene expression systems controlled by traceless, orthogonal and biocompatible trigger compounds, which would prevent the irreversible elimination of ‘expensive’ ATMP products in vivo (219). For example, intracellular TCR signaling has been engineered to depend on drug-inducible protein dimerization in split CARs, which permit T-cell activation only if an exogenous drug is present (220). In addition, expanding the ‘universal’ CAR-T-cell principle to accept multiple input signals renders CAR-T-cell activation dependent on different combinations of exogenously applied adaptor molecules (62). In this ‘split, universal and programmable’ (SUPRA) CAR system, the extracellular domain of CAR consists of a leucine zipper, which acts as a protein tether. Activation of SUPRA CAR depends on bi-specific adaptors simultaneously binding a target antigen through scFv and the tether through leucine zipper interactions (62) (Figure 3A-i). By using different combinations of adaptor molecules, which are either competing for the same SUPRA CAR or simultaneously activating multiple SUPRA CARs in parallel, the same CAR-T cells can be pre-programmed to respond to different user-defined therapeutic contexts following specific antigen recognition logics (62) (Figure 3A-i). By synchronizing CAR expression with activation of the mechanosensitive piezo channel through intracellular calcium signaling, CAR-T activity may be remote-controlled with traceless and noninvasive ultrasound waves (221) (Figure 3D).
Metabolic prostheses
An ideal ATMP therapy must be safe, efficient, easy to manufacture and easy to administer (10, 111). Scientifically, implantation of encapsulated stem cell-derived β cells is considered the optimal solution for diabetes treatment (115) (Figure 3C). However, the clinical success of this strategy is primarily constrained by practical issues. First, β-cell differentiation using chemical cultivation protocols requires embryonic stem cells as the source material (126, 132), which not only raises ethical and safety concerns when applied to humans but also complicates production on a commercial scale (222). iPSCs produced from somatic tissues can also be used as source materials but show much lower differentiation efficiency (127). To improve the differentiation efficiency, a synthetic biology approach based on transient transfection of lineage-specific gene circuits was introduced and afforded unprecedented production yields of iPSCs-derived β-like cells. This success was achieved by the implementation of time- and context-dependent transcription factors, which regulate the expression of proteins involved in endogenous cell fate (223). However, a further hurdle facing stem-cell-based diabetes therapies is the high cost of production and quality assurance (111). Therapeutically active batches of β cells cannot be cryopreserved for re-use. In addition, technologies that can measure and standardize the quality of individual differentiation-based production runs are not yet available (224, 225). Consequently, manufacture of stem cell-derived cell therapies is time- and resource-demanding; typically, several months are required to produce sufficient cell numbers for implantation (222). Therefore, functional mimetics of β cells that pragmatically combine therapeutic efficacy and commercial applicability are attractive alternatives for cell-based diabetes therapy (226). For example, by using synthetic gene circuits mediating depolarization-dependent calcium entry and calcium-specific gene expression, any glycolytic human cell can be reprogrammed for glucose-dependent insulin secretion (227). These engineered cells, also known as β-cell-mimetic designer cells, have the potential to be cost-effectively produced, expanded and freeze-stored according to GMP regulations for biopharmaceutical manufacturing; these are critical features for commercialization (Figure 3C).
By using different synthetic gene circuits encoding for other sense-and-respond therapeutic programs, metabolic prostheses can be principally engineered for self-sufficient and closed-loop control of any metabolic disorder, such as insulin resistance (228), obesity (229), gouty arthritis (230), hypertension (231), Grave’s disease (232), bacterial infections (233) or psoriasis (234) (Figure 3C). Furthermore, open-loop systems that allow exogenous control compounds to potentially act as safety switches have also been engineered (235–237). For example, one study used optogenetics to provide communication between electronic devices and biological systems (238) (Figure 3D). Specifically, human cells engineered for far-red-light-dependent transgene expression were controlled by an electronic microprocessor that coordinated LED illumination upon reception of specific wireless signals. This allows portable electronics, such as smartphones, to regulate gene expression, representing the ultimate level of traceless, noninvasive and long-distance remote control of cell activities.
Therapeutic bacteria
Bacteria are also a powerful host cell type for the design of next-generation ATMPs. For example, engineered probiotic strains, such as Lactococcus lactis, can be programmed to attach to the mammalian gastrointestinal tract and trigger site-specific attenuation of Vibrio cholerae infections upon oral ingestion (239) (Figure 4D). In contrast, several bacterial species, such as Escherichia coli (240–242) or Salmonella (243–245), preferentially colonize and proliferate in hypoxic and immune-privileged tumor microenvironments upon systemic administration (Figure 4D). Therefore, the engineering of tumor-trophic bacterial strains to contain tumor-specific killing programs offers a novel and promising alternative for cancer therapies. In essence, ATMP therapies based on CAR-transgenic leukocytes (Figure 3A), oncolytic viruses (Figure 4B) and tumor-trophic bacteria (Figure 4D) rely on pre-programmed targeting of cancer cells in vivo through unique cell-type- and species-specific properties, while encoding for similar therapeutic effects. For example, most cancer cells overexpress the cell surface marker CD47, which binds to and inactivates macrophages in the tumor microenvironment (1) (Figure 1A-ii). To overcome this survival strategy of cancer cells, the CD47 targets of macrophages can be deleted by CRISPR/Cas9-based gene editing (246). Such deletion of CD47 targets may enhance the tumor clearance capabilities of CAR-carrying macrophages (79). Alternatively, bacteria can also be programmed for tumor-specific CD47 inhibition (240). For instance, nonpathogenic E. coli were engineered to self-sufficiently invade tumors and locally mediate density-dependent control of cell lysis and drug release. During tumor-specific proliferation, these bacteria constantly produce a quorum signal AHL and an anti-CD47 nanobody. Upon exceeding a critical cell density, AHL triggers the expression of bacterial suicide genes, resulting in pulse-like induction and amplification of macrophage-mediated immune responses while attenuating CD47-mediated immune tolerance (Figure 4D) (240).
3.2. Conclusions
In conclusion, advances in synthetic biology have the potential not only to improve existing ATMP therapies but also to create novel therapeutic opportunities. It is not going too far to say that cell-based immunotherapies, oncolytic viruses and therapeutic bacteria might form the next three pillars of cancer therapy. Each strategy allows disease-specific programming of sophisticated sense-and-respond functions almost at will, but may differ in the ability to penetrate deep tissues due to differences in cell and particle sizes. Therefore, an ideal treatment strategy might not exist by default, as each therapy would require a rational choice of the most suitable combination of drug targets, therapeutic transgenes, vectors and administration routes (Supplementary Table S2) according to disease-specific conditions. In this context, enormous progress might be achieved by using oncolytic viruses in combination with gene circuit therapies—a novel class of medicines using synthetic interconnected gene switches to compute the malignancy of individual cell populations (247, 248). At present, the clinical utility of such ‘therapeutic biocomputers’ is limited by the delivery efficiency of large-scale gene circuits into specific cells in vivo. Recently, a lentiviral transduction strategy has shown promising preclinical results (249) (Figure 4B). Novel vector systems enabling the delivery of therapeutic transgenes with designable target specificities, such as phages, are also under active development (250).
When translating innovative synthetic biology-inspired ATMP therapies to the clinic, the complexity of therapeutic transgenes may present the greatest uncertainty. Gene- and cell therapies engineered to contain large amounts of foreign molecules potentially increase the level of transgene immunogenicity in the patient (Figure 1B-ii). Therefore, selecting human-derived components when designing complicated gene circuits is advantageous, as was demonstrated in a recent report on designer-cell-based therapy for diabetes and muscle atrophy (235). Apart from the therapeutic transgene, the gene delivery vectors and administration routes used by next-generation ATMP products (Supplementary Table S2) have remained largely identical to those already submitted for regulatory approval and/or approved for clinical use (Supplementary Table S1). Therefore, currently approved ATMP products provide an extensive pool of clinical data and experience that should be helpful to successfully translate novel therapies to the clinic. For example, designer-cell-based therapies for diabetes essentially use the same cell encapsulation techniques as used for allogenic islets and stem-cell-derived β cells (Figure 3C). Alginate beads have also been optimized for the encapsulation of bacteria, showcasing the possibility of preventing cell leakage when implanted into humans (251). However, therapies based on encapsulated mammalian cells must overcome issues related to foreign body responses and fibrosis, which will determine the lifetime of each implantation therapy. For clinical applications, the therapeutic transgenes of designer cells can be effectively integrated into MSCs using the Sleeping Beauty transposase system (227, 228, 252). Both MSC-derived transplants (e.g. Alofisel) and the Sleeping Beauty technology (253) are already used in clinical applications. Most recently, another designer cell-based therapy showed promising safety and efficacy results in non-human primates upon implantation of 1.5 × 108 microencapsulated cells (237), providing clues to clinically applicable cell doses (20, 109, 179). Thus, all the technical cornerstones of designer cell-based therapies have been indirectly proven safe through various independent studies, paving the way for first-in-class clinical trials in humans. In theory, the majority of synthetic biology-inspired ATMPs are already eligible for initiation of clinical trials. However, regulatory approval for registration of a clinical trial does not automatically flow from scientifically valid studies (254); for example animal models cannot always sufficiently predict all clinically relevant information on safety and efficacy (12). Thus, careful design of clinical trials is essential to address the regulatory and ethical challenges of novel ATMP therapies (179). Constant (self-)education through exploration of the academic literature, compliance with current regulatory and ethical guidelines and careful monitoring during commercialization are all imperative to anticipate potential adverse events that may be encountered by patients, in addition to post-marketing monitoring to ensure that, if unexpected events do occur, they are picked up as early as possible (20, 109, 179).
Supplementary data
Supplementary data are available at SYNBIO Online.
Supplementary Material
Acknowledgments
We thank Hui Wang for critical discussions on this manuscript. This review is dedicated to Professor Angelika Viviani, an exemplary teacher, fine mentor and visionary scientist.
Funding
Research in the laboratory of M.X. is supported by the Westlake Education Foundation. Research in the laboratory of M.F. is supported by the National Center of Competence in Research on Molecular Systems Engineering and in part by the European Research Council advanced grant (ElectroGene, no. 785800).
Conflict of interest statement. None declared.
References
- 1. Bailey S.R., Maus M.V. (2019) Gene editing for immune cell therapies. Nat. Biotechnol., 37, 1425–1434. [DOI] [PubMed] [Google Scholar]
- 2. Lim W.A., June C.H. (2017) The principles of engineering immune cells to treat cancer. Cell, 168, 724–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ma C.C., Wang Z.L., Xu T., He Z.Y., Wei Y.Q. (2019) The approved gene therapy drugs worldwide: from 1998 to 2019. Biotechnol. Adv., 107502. [DOI] [PubMed] [Google Scholar]
- 4. Naldini L. (2015) Gene therapy returns to centre stage. Nature, 526, 351–360. [DOI] [PubMed] [Google Scholar]
- 5. Yla-Herttuala S. (2019) Gene and cell therapy: success stories and future challenges. Mol. Ther., 27, 891–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mount N.M., Ward S.J., Kefalas P., Hyllner J. (2015) Cell-based therapy technology classifications and translational challenges. Philos. Trans. R. Soc. London Ser. B: Biol. Sci., 370, 20150017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ilic D., Liovic M., Noli L. (2019). Industry updates from the field of stem cell research and regenerative medicine in September 2019. Regen Med. [Google Scholar]
- 8. Luo X., Miller S.D., Shea L.D. (2016) Immune tolerance for autoimmune disease and cell transplantation. Annu. Rev. Biomed. Eng., 18, 181–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fesnak A.D., June C.H., Levine B.L. (2016) Engineered T cells: the promise and challenges of cancer immunotherapy. Nat. Rev. Cancer, 16, 566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brenner M.J., Cho J.H., Wong N.M.L., Wong W.W. (2018) Synthetic biology: immunotherapy by design. Annu. Rev. Biomed. Eng., 20, 95–118. [DOI] [PubMed] [Google Scholar]
- 11. Cassetta L., Kitamura T. (2018) Macrophage targeting: opening new possibilities for cancer immunotherapy. Immunology, 155, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Porteus M.H. (2019) A new class of medicines through DNA editing. N. Engl. J. Med, 380, 947–959. [DOI] [PubMed] [Google Scholar]
- 13. Kastellorizios M., Tipnis N., Burgess D.J. (2015) Foreign body reaction to subcutaneous implants. Adv. Exp. Med. Biol., 865, 93–108. [DOI] [PubMed] [Google Scholar]
- 14. Wick G., Grundtman C., Mayerl C., Wimpissinger T.F., Feichtinger J., Zelger B., Sgonc R., Wolfram D. (2013) The immunology of fibrosis. Annu. Rev. Immunol., 31, 107–135. [DOI] [PubMed] [Google Scholar]
- 15. Evron Y., Colton C.K., Ludwig B., Weir G.C., Zimermann B., Maimon S., Neufeld T., Shalev N., Goldman T., Leon A.. et al. (2018) Long-term viability and function of transplanted islets macroencapsulated at high density are achieved by enhanced oxygen supply. Sci. Rep., 8, 6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Faleo G., Russ H.A., Wisel S., Parent A.V., Nguyen V., Nair G.G., Freise J.E., Villanueva K.E., Szot G.L., Hebrok M.. et al. (2017) Mitigating ischemic injury of stem cell-derived insulin-producing cells after transplant. Stem Cell Rep., 9, 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ludwig B., Rotem A., Schmid J., Weir G.C., Colton C.K., Brendel M.D., Neufeld T., Block N.L., Yavriyants K., Steffen A.. et al. (2012) Improvement of islet function in a bioartificial pancreas by enhanced oxygen supply and growth hormone releasing hormone agonist. Proc. Natl. Acad. Sci. USA, 109, 5022–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vegas A.J., Veiseh O., Doloff J.C., Ma M., Tam H.H., Bratlie K., Li J., Bader A.R., Langan E., Olejnik K.. et al. (2016. a) Combinatorial hydrogel library enables identification of materials that mitigate the foreign body response in primates. Nat. Biotechnol., 34, 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Veiseh O., Doloff J.C., Ma M., Vegas A.J., Tam H.H., Bader A.R., Li J., Langan E., Wyckoff J., Loo W.S.. et al. (2015) Size- and shape-dependent foreign body immune response to materials implanted in rodents and non-human primates. Nat. Mater., 14, 643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sheridan C. (2011) Gene therapy finds its niche. Nat. Biotechnol., 29, 121–128. [DOI] [PubMed] [Google Scholar]
- 21. Lundstrom K. (2018) Viral vectors in gene therapy. Diseases, 6, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grabundzija I., Irgang M., Mates L., Belay E., Matrai J., Gogol-Doring A., Kawakami K., Chen W., Ruiz P., Chuah M.K.. et al. (2010) Comparative analysis of transposable element vector systems in human cells. Mol. Ther., 18, 1200–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holstein M., Mesa-Nunez C., Miskey C., Almarza E., Poletti V., Schmeer M., Grueso E., Ordonez Flores J.C., Kobelt D., Walther W.. et al. (2018) Efficient non-viral gene delivery into human hematopoietic stem cells by minicircle sleeping beauty transposon vectors. Mol. Ther., 26, 1137–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hacein-Bey-Abina S., Garrigue A., Wang G.P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K.. et al. (2008) Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin Invest., 118, 3132–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marcucci K.T., Jadlowsky J.K., Hwang W.T., Suhoski-Davis M., Gonzalez V.E., Kulikovskaya I., Gupta M., Lacey S.F., Plesa G., Chew A.. et al. (2018) Retroviral and lentiviral safety analysis of gene-modified T cell products and infused HIV and oncology patients. Mol. Ther., 26, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCormack M.P., Rabbitts T.H. (2004) Activation of the T-cell oncogene LMO2 after gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med., 350, 913–922. [DOI] [PubMed] [Google Scholar]
- 27. Yip A., Webster R.M. (2018) The market for chimeric antigen receptor T cell therapies. Nat. Rev. Drug Discov., 17, 161–162. [DOI] [PubMed] [Google Scholar]
- 28. Locke F.L., Neelapu S.S., Bartlett N.L., Siddiqi T., Chavez J.C., Hosing C.M., Ghobadi A., Budde L.E., Bot A., Rossi J.M.. et al. (2017) Phase 1 results of ZUMA-1: a multicenter study of KTE-C19 anti-CD19 CAR T cell therapy in refractory aggressive lymphoma. Mol. Ther., 25, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Valton J., Guyot V., Boldajipour B., Sommer C., Pertel T., Juillerat A., Duclert A., Sasu B.J., Duchateau P., Poirot L. (2018) A versatile safeguard for chimeric antigen receptor T-cell immunotherapies. Sci. Rep., 8, 8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao J., Lin Q., Song Y., Liu D. (2018) Universal CARs, universal T cells, and universal CAR T cells. J. Hematol. Oncol., 11, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chakravarti D., Wong W.W. (2015) Synthetic biology in cell-based cancer immunotherapy. Trends Biotechnol., 33, 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vairy S., Garcia J.L., Teira P., Bittencourt H. (2018) CTL019 (tisagenlecleucel): CAR-T therapy for relapsed and refractory B-cell acute lymphoblastic leukemia. Drug Des. Dev. Ther., 12, 3885–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maude S.L., Laetsch T.W., Buechner J., Rives S., Boyer M., Bittencourt H., Bader P., Verneris M.R., Stefanski H.E., Myers G.D.. et al. (2018) Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl. J. Med., 378, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prasad V. (2018) Immunotherapy: tisagenlecleucel - the first approved CAR-T-cell therapy: implications for payers and policy makers. Nat. Rev. Clin. Oncol., 15, 11–12. [DOI] [PubMed] [Google Scholar]
- 35. Schuster S.J., Bishop M.R., Tam C.S., Waller E.K., Borchmann P., McGuirk J.P., Jager U., Jaglowski S., Andreadis C., Westin J.R.. et al. (2019) Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N. Engl. J. Med., 380, 45–56. [DOI] [PubMed] [Google Scholar]
- 36. Brentjens R.J., Davila M.L., Riviere I., Park J., Wang X., Cowell L.G., Bartido S., Stefanski J., Taylor C., Olszewska M.. et al. (2013) CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl. Med., 5, 177ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang M., Munoz J., Goy A., Locke F.L., Jacobson C.A., Hill B.T., Timmerman J.M., Holmes H., Jaglowski S., Flinn I.W.. et al. (2020) KTE-X19 CAR T-cell therapy in relapsed or refractory mantle-cell lymphoma. N. Engl. J. Med., 382, 1331–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Locke F.L., Ghobadi A., Jacobson C.A., Miklos D.B., Lekakis L.J., Oluwole O.O., Lin Y., Braunschweig I., Hill B.T., Timmerman J.M.. et al. (2019) Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1-2 trial. Lancet Oncol., 20, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neelapu S.S., Locke F.L., Bartlett N.L., Lekakis L.J., Miklos D.B., Jacobson C.A., Braunschweig I., Oluwole O.O., Siddiqi T., Lin Y.. et al. (2017) Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med., 377, 2531–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Park J.H., Riviere I., Gonen M., Wang X., Senechal B., Curran K.J., Sauter C., Wang Y., Santomasso B., Mead E.. et al. (2018) Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med., 378, 449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roberts Z.J., Better M., Bot A., Roberts M.R., Ribas A. (2018) Axicabtagene ciloleucel, a first-in-class CAR T cell therapy for aggressive NHL. Leuk. Lymphoma, 59, 1785–1796. [DOI] [PubMed] [Google Scholar]
- 42. Kowolik C.M., Topp M.S., Gonzalez S., Pfeiffer T., Olivares S., Gonzalez N., Smith D.D., Forman S.J., Jensen M.C., Cooper L.J. (2006) CD28 costimulation provided through a CD19-specific chimeric antigen receptor enhances in vivo persistence and antitumor efficacy of adoptively transferred T cells. Cancer Res., 66, 10995–11004. [DOI] [PubMed] [Google Scholar]
- 43. Grupp S.A., Kalos M., Barrett D., Aplenc R., Porter D.L., Rheingold S.R., Teachey D.T., Chew A., Hauck B., Wright J.F.. et al. (2013) Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med., 368, 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kochenderfer J.N., Dudley M.E., Kassim S.H., Somerville R.P., Carpenter R.O., Stetler-Stevenson M., Yang J.C., Phan G.Q., Hughes M.S., Sherry R.M.. et al. (2015) Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J. Clin. Oncol., 33, 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee D.W., Kochenderfer J.N., Stetler-Stevenson M., Cui Y.K., Delbrook C., Feldman S.A., Fry T.J., Orentas R., Sabatino M., Shah N.N.. et al. (2015) T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet, 385, 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maude S.L., Frey N., Shaw P.A., Aplenc R., Barrett D.M., Bunin N.J., Chew A., Gonzalez V.E., Zheng Z., Lacey S.F.. et al. (2014) Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med., 371, 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Porter D.L., Levine B.L., Kalos M., Bagg A., June C.H. (2011) Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med., 365, 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Friedman K.M., Garrett T.E., Evans J.W., Horton H.M., Latimer H.J., Seidel S.L., Horvath C.J., Morgan R.A. (2018) Effective targeting of multiple B-cell maturation antigen-expressing hematological malignances by anti-B-cell maturation antigen chimeric antigen receptor T cells. Hum. Gene Ther., 29, 585–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Raje N., Berdeja J., Lin Y., Siegel D., Jagannath S., Madduri D., Liedtke M., Rosenblatt J., Maus M.V., Turka A.. et al. (2019) Anti-BCMA CAR T-cell therapy bb2121 in relapsed or refractory multiple myeloma. N. Engl. J. Med., 380, 1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu X., Zhang N., Shi H. (2017) Driving better and safer HER2-specific CARs for cancer therapy. Oncotarget, 8, 62730–62741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sun M., Shi H., Liu C., Liu J., Liu X., Sun Y. (2014) Construction and evaluation of a novel humanized HER2-specific chimeric receptor. Breast Cancer Res., 16, R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang J., Jensen M., Lin Y., Sui X., Chen E., Lindgren C.G., Till B., Raubitschek A., Forman S.J., Qian X.. et al. (2007) Optimizing adoptive polyclonal T cell immunotherapy of lymphomas, using a chimeric T cell receptor possessing CD28 and CD137 costimulatory domains. Hum. Gene Ther., 18, 712–725. [DOI] [PubMed] [Google Scholar]
- 53. Karlsson H., Svensson E., Gigg C., Jarvius M., Olsson-Stromberg U., Savoldo B., Dotti G., Loskog A. (2015) Evaluation of intracellular signaling downstream chimeric antigen receptors. PLoS One, 10, e0144787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Morgan R.A., Yang J.C., Kitano M., Dudley M.E., Laurencot C.M., Rosenberg S.A. (2010) Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol. Ther., 18, 843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nair S., Wang J.B., Tsao S.T., Liu Y., Zhu W., Slayton W.B., Moreb J.S., Dong L., Chang L.J. (2019) Functional improvement of chimeric antigen receptor through intrinsic interleukin-15Ralpha signaling. Curr. Gene Ther., 19, 40–53. [DOI] [PubMed] [Google Scholar]
- 56. Rupp L.J., Schumann K., Roybal K.T., Gate R.E., Ye C.J., Lim W.A., Marson A. (2017) CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci. Rep., 7, 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Provasi E., Genovese P., Lombardo A., Magnani Z., Liu P.Q., Reik A., Chu V., Paschon D.E., Zhang L., Kuball J.. et al. (2012) Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat. Med., 18, 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Torikai H., Reik A., Liu P.-Q., Zhou Y., Zhang L., Maiti S., Huls H., Miller J.C., Kebriaei P., Rabinovitch B.. et al. (2012) A foundation for universal T-cell based immunotherapy: t cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood, 119, 5697–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Qasim,W., Zhan,H., Samarasinghe,S., Adams,S., Amrolia,P., Stafford,S., Butler,K., Rivat,C., Wright,G., Somana,K., et al. (2017) Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Science Translational Medicine, 9, eaaj2013. [DOI] [PubMed] [Google Scholar]
- 60. Ren J., Liu X., Fang C., Jiang S., June C.H., Zhao Y. (2017) Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin. Cancer Res., 23, 2255–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Torikai H., Reik A., Soldner F., Warren E.H., Yuen C., Zhou Y., Crossland D.L., Huls H., Littman N., Zhang Z.. et al. (2013) Toward eliminating HLA class I expression to generate universal cells from allogeneic donors. Blood, 122, 1341–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cho J.H., Collins J.J., Wong W.W. (2018) Universal chimeric antigen receptors for multiplexed and logical control of T cell responses. Cell, 173, 1426–1438 e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kudo K., Imai C., Lorenzini P., Kamiya T., Kono K., Davidoff A.M., Chng W.J., Campana D. (2014) T lymphocytes expressing a CD16 signaling receptor exert antibody-dependent cancer cell killing. Cancer Res., 74, 93–103. [DOI] [PubMed] [Google Scholar]
- 64. Si W., Li C., Wei P. (2018) Synthetic immunology: T-cell engineering and adoptive immunotherapy. Synth. Syst. Biotechnol/, 3, 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Grada Z., Hegde M., Byrd T., Shaffer D.R., Ghazi A., Brawley V.S., Corder A., Schonfeld K., Koch J., Dotti G.. et al. (2013) TanCAR: a novel bispecific chimeric antigen receptor for cancer immunotherapy. Mol. Ther. Nucleic Acids, 2, e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hegde M., Mukherjee M., Grada Z., Pignata A., Landi D., Navai S.A., Wakefield A., Fousek K., Bielamowicz K., Chow K.K.. et al. (2019) Tandem CAR T cells targeting HER2 and IL13Ralpha2 mitigate tumor antigen escape. J. Clin. Invest., 129, 3464–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Qin H., Ramakrishna S., Nguyen S., Fountaine T.J., Ponduri A., Stetler-Stevenson M., Yuan C.M., Haso W., Shern J.F., Shah N.N.. et al. (2018) Preclinical development of bivalent chimeric antigen receptors targeting both CD19 and CD22. Mol. Ther. Oncolytics, 11, 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zah E., Lin M.Y., Silva-Benedict A., Jensen M.C., Chen Y.Y. (2016) T cells expressing CD19/CD20 bispecific chimeric antigen receptors prevent antigen escape by malignant B cells. Cancer Immunol. Res., 4, 498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pegram H.J., Lee J.C., Hayman E.G., Imperato G.H., Tedder T.F., Sadelain M., Brentjens R.J. (2012) Tumor-targeted T cells modified to secrete IL-12 eradicate systemic tumors without need for prior conditioning. Blood, 119, 4133–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang L., Morgan R.A., Beane J.D., Zheng Z., Dudley M.E., Kassim S.H., Nahvi A.V., Ngo L.T., Sherry R.M., Phan G.Q.. et al. (2015) Tumor-infiltrating lymphocytes genetically engineered with an inducible gene encoding interleukin-12 for the immunotherapy of metastatic melanoma. Clin. Cancer Res., 21, 2278–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kloss C.C., Condomines M., Cartellieri M., Bachmann M., Sadelain M. (2013) Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat. Biotechnol., 31, 71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wilkie S., van Schalkwyk M.C., Hobbs S., Davies D.M., van der Stegen S.J., Pereira A.C., Burbridge S.E., Box C., Eccles S.A., Maher J. (2012) Dual targeting of ErbB2 and MUC1 in breast cancer using chimeric antigen receptors engineered to provide complementary signaling. J. Clin. Immunol., 32, 1059–1070. [DOI] [PubMed] [Google Scholar]
- 73. Bielamowicz K., Fousek K., Byrd T.T., Samaha H., Mukherjee M., Aware N., Wu M.F., Orange J.S., Sumazin P., Man T.K.. et al. (2018) Trivalent CAR T cells overcome interpatient antigenic variability in glioblastoma. Neuro Oncol., 20, 506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fedorov V.D., Themeli M., Sadelain M. (2013) PD-1- and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci. Transl. Med., 5, 215ra172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xia N., Haopeng P., Gong J.U., Lu J., Chen Z., Zheng Y., Wang Z., Sun Y.U., Yang Z., Hoffman R.M.. et al. (2019) Robo1-specific CAR-NK immunotherapy enhances efficacy of (125)I seed brachytherapy in an orthotopic mouse model of human pancreatic carcinoma. Anticancer Res., 39, 5919–5925. [DOI] [PubMed] [Google Scholar]
- 76. Xu X., Huang W., Heczey A., Liu D., Guo L., Wood M., Jin J., Courtney A.N., Liu B., Di Pierro E.J.. et al. (2019. b) NKT cells coexpressing a GD2-specific chimeric antigen receptor and IL15 show enhanced in vivo persistence and antitumor activity against neuroblastoma. Clin. Cancer Res., 25, 7126–7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Narayanan P., Lapteva N., Seethammagari M., Levitt J.M., Slawin K.M., Spencer D.M. (2011) A composite MyD88/CD40 switch synergistically activates mouse and human dendritic cells for enhanced antitumor efficacy. J. Clin. Invest., 121, 1524–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hung C.F., Xu X., Li L., Ma Y., Jin Q., Viley A., Allen C., Natarajan P., Shivakumar R., Peshwa M.V.. et al. (2018) Development of anti-human mesothelin-targeted chimeric antigen receptor messenger RNA-transfected peripheral blood lymphocytes for ovarian cancer therapy. Hum. Gene Ther., 29, 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Moyes K.W., Lieberman N.A., Kreuser S.A., Chinn H., Winter C., Deutsch G., Hoglund V., Watson R., Crane C.A. (2017) Genetically engineered macrophages: a potential platform for cancer immunotherapy. Hum. Gene Ther., 28, 200–215. [DOI] [PubMed] [Google Scholar]
- 80. Ali A., Kitchen S.G., Chen I.S.Y., Ng H.L., Zack J.A., Yang O.O. (2016) HIV-1-specific chimeric antigen receptors based on broadly neutralizing antibodies. J. Virol., 90, 6999–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hale M., Mesojednik T., Romano Ibarra G.S., Sahni J., Bernard A., Sommer K., Scharenberg A.M., Rawlings D.J., Wagner T.A. (2017) Engineering HIV-resistant, anti-HIV chimeric antigen receptor T cells. Mol. Ther., 25, 570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu B., Zou F., Lu L., Chen C., He D., Zhang X., Tang X., Liu C., Li L., Zhang H. (2016. a) Chimeric antigen receptor T cells guided by the single-chain Fv of a broadly neutralizing antibody specifically and effectively eradicate virus reactivated from latency in CD4+ T lymphocytes isolated from HIV-1-infected individuals receiving suppressive combined antiretroviral therapy. J. Virol., 90, 9712–9724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Elinav E., Adam N., Waks T., Eshhar Z. (2009) Amelioration of colitis by genetically engineered murine regulatory T cells redirected by antigen-specific chimeric receptor. Gastroenterology, 136, 1721–1731. [DOI] [PubMed] [Google Scholar]
- 84. MacDonald K.G., Hoeppli R.E., Huang Q., Gillies J., Luciani D.S., Orban P.C., Broady R., Levings M.K. (2016) Alloantigen-specific regulatory T cells generated with a chimeric antigen receptor. J. Clin. Invest., 126, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Taylor P.A., Lees C.J., Blazar B.R. (2002) The infusion of ex vivo activated and expanded CD4(+)CD25(+) immune regulatory cells inhibits graft-versus-host disease lethality. Blood, 99, 3493–3499. [DOI] [PubMed] [Google Scholar]
- 86. Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K.. et al. (2010) Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature, 467, 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Negre O., Eggimann A.V., Beuzard Y., Ribeil J.A., Bourget P., Borwornpinyo S., Hongeng S., Hacein-Bey S., Cavazzana M., Leboulch P.. et al. (2016) Gene therapy of the beta-hemoglobinopathies by lentiviral transfer of the beta(A(T87Q))-globin gene. Hum. Gene Ther., 27, 148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Schimmer J., Breazzano S. (2016) Investor outlook: rising from the ashes; GSK's European approval of strimvelis for ADA-SCID. Hum. Gene Ther. Clin. Dev., 27, 57–61. [DOI] [PubMed] [Google Scholar]
- 89. Stirnadel-Farrant H., Kudari M., Garman N., Imrie J., Chopra B., Giannelli S., Gabaldo M., Corti A., Zancan S., Aiuti A.. et al. (2018) Gene therapy in rare diseases: the benefits and challenges of developing a patient-centric registry for Strimvelis in ADA-SCID. Orphanet J. Rare Dis., 13, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bolukbasi M.F., Gupta A., Wolfe S.A. (2016) Creating and evaluating accurate CRISPR-Cas9 scalpels for genomic surgery. Nat. Methods, 13, 41–50. [DOI] [PubMed] [Google Scholar]
- 91. Canver M.C., Smith E.C., Sher F., Pinello L., Sanjana N.E., Shalem O., Chen D.D., Schupp P.G., Vinjamur D.S., Garcia S.P.. et al. (2015) BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature, 527, 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. DeWitt M.A., Magis W., Bray N.L., Wang T., Berman J.R., Urbinati F., Heo S.J., Mitros T., Munoz D.P., Boffelli D.. et al. (2016) Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med., 8, 360ra134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Traxler E.A., Yao Y., Wang Y.D., Woodard K.J., Kurita R., Nakamura Y., Hughes J.R., Hardison R.C., Blobel G.A., Li C.. et al. (2016) A genome-editing strategy to treat beta-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat. Med., 22, 987–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wu Y., Zeng J., Roscoe B.P., Liu P., Yao Q., Lazzarotto C.R., Clement K., Cole M.A., Luk K., Baricordi C.. et al. (2019) Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat. Med., 25, 776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Xu S., Luk K., Yao Q., Shen A.H., Zeng J., Wu Y., Luo H.Y., Brendel C., Pinello L., Chui D.H.K.. et al. (2019. a) Editing aberrant splice sites efficiently restores beta-globin expression in beta-thalassemia. Blood, 133, 2255–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ye L., Wang J., Tan Y., Beyer A.I., Xie F., Muench M.O., Kan Y.W. (2016) Genome editing using CRISPR-Cas9 to create the HPFH genotype in HSPCs: an approach for treating sickle cell disease and beta-thalassemia. Proc. Natl. Acad. Sci. USA, 113, 10661–10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Schiroli,G., Ferrari,S., Conway,A., Jacob,A., Capo,V., Albano,L., Plati,T., Castiello,M. C., Sanvito,F., Gennery,A. R., et al. (2017) Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Science Translational Medicine, 9, eaan0820. [DOI] [PubMed] [Google Scholar]
- 98. Ferrua F., Cicalese M.P., Galimberti S., Giannelli S., Dionisio F., Barzaghi F., Migliavacca M., Bernardo M.E., Calbi V., Assanelli A.A.. et al. (2019) Lentiviral haemopoietic stem/progenitor cell gene therapy for treatment of Wiskott-Aldrich syndrome: interim results of a non-randomised, open-label, phase 1/2 clinical study. Lancet Haematol., 6, e239–e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. De Ravin,S. S., Li,L., Wu,X., Choi,U., Allen,C., Koontz,S., Lee,J., Theobald-Whiting,N., Chu,J., Garofalo,M., et al. (2017) CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Science Translational Medicine, 9, eaah3480. [DOI] [PubMed] [Google Scholar]
- 100. DiGiusto D.L., Cannon P.M., Holmes M.C., Li L., Rao A., Wang J., Lee G., Gregory P.D., Kim K.A., Hayward S.B.. et al. (2016) Preclinical development and qualification of ZFN-mediated CCR5 disruption in human hematopoietic stem/progenitor cells. Mol. Ther. Methods Clin. Dev., 3, 16067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Perez E.E., Wang J., Miller J.C., Jouvenot Y., Kim K.A., Liu O., Wang N., Lee G., Bartsevich V.V., Lee Y.L.. et al. (2008) Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol., 26, 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G.. et al. (2014) Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med., 370, 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Voit R.A., McMahon M.A., Sawyer S.L., Porteus M.H. (2013) Generation of an HIV resistant T-cell line by targeted “stacking” of restriction factors. Mol. Ther., 21, 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kim M.Y., Yu K.R., Kenderian S.S., Ruella M., Chen S., Shin T.H., Aljanahi A.A., Schreeder D., Klichinsky M., Shestova O.. et al. (2018. b) Genetic inactivation of CD33 in hematopoietic stem cells to enable CAR T cell immunotherapy for acute myeloid leukemia. Cell, 173, 1439–1453 e1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Evans C.H., Ghivizzani S.C., Robbins P.D. (2018) Arthritis gene therapy is becoming a reality. Nat. Rev. Rheumatol., 14, 381–382. [DOI] [PubMed] [Google Scholar]
- 106. Kim M.K., Ha C.W., In Y., Cho S.D., Choi E.S., Ha J.K., Lee J.H., Yoo J.D., Bin S.I., Choi C.H.. et al. (2018. a) A multicenter, double-blind, phase III clinical trial to evaluate the efficacy and safety of a cell and gene therapy in knee osteoarthritis patients. Hum. Gene Ther. Clin. Dev., 29, 48–59. [DOI] [PubMed] [Google Scholar]
- 107. Scott L.J. (2018) Darvadstrocel: a review in treatment-refractory complex perianal fistulas in Crohn's disease. BioDrugs, 32, 627–634. [DOI] [PubMed] [Google Scholar]
- 108. Galipeau J., Sensebe L. (2018) Mesenchymal stromal cells: clinical challenges and therapeutic opportunities. Cell Stem Cell, 22, 824–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kabat M., Bobkov I., Kumar S., Grumet M. (2020) Trends in mesenchymal stem cell clinical trials 2004-2018: is efficacy optimal in a narrow dose range? Stem Cells Transl. Med., 9, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Graf T., Enver T. (2009) Forcing cells to change lineages. Nature, 462, 587–594. [DOI] [PubMed] [Google Scholar]
- 111. Lipsitz Y.Y., Bedford P., Davies A.H., Timmins N.E., Zandstra P.W. (2017) Achieving efficient manufacturing and quality assurance through synthetic cell therapy design. Cell Stem Cell, 20, 13–17. [DOI] [PubMed] [Google Scholar]
- 112. Park S.S., Bauer G., Abedi M., Pontow S., Panorgias A., Jonnal R., Zawadzki R.J., Werner J.S., Nolta J. (2014. b) Intravitreal autologous bone marrow CD34+ cell therapy for ischemic and degenerative retinal disorders: preliminary phase 1 clinical trial findings. Invest. Ophthalmol. Vis. Sci., 56, 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Tsai R.J., Li L.M., Chen J.K. (2000) Reconstruction of damaged corneas by transplantation of autologous limbal epithelial cells. N. Engl. J. Med., 343, 86–93. [DOI] [PubMed] [Google Scholar]
- 114. Kinoshita S., Koizumi N., Ueno M., Okumura N., Imai K., Tanaka H., Yamamoto Y., Nakamura T., Inatomi T., Bush J.. et al. (2018) Injection of cultured cells with a ROCK inhibitor for bullous keratopathy. N. Engl. J. Med., 378, 995–1003. [DOI] [PubMed] [Google Scholar]
- 115. Sneddon J.B., Tang Q., Stock P., Bluestone J.A., Roy S., Desai T., Hebrok M. (2018) Stem cell therapies for treating diabetes: progress and remaining challenges. Cell Stem Cell, 22, 810–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Latres E., Finan D.A., Greenstein J.L., Kowalski A., Kieffer T.J. (2019) Navigating two roads to glucose normalization in diabetes: automated insulin delivery devices and cell therapy. Cell Metab., 29, 545–563. [DOI] [PubMed] [Google Scholar]
- 117. Bertuzzi F., Colussi G., Lauterio A., De Carlis L. (2018) Intramuscular islet allotransplantation in type 1 diabetes mellitus. Eur. Rev. Med. Pharmacol. Sci., 22, 1731–1736. [DOI] [PubMed] [Google Scholar]
- 118. Ricordi C., Goldstein J.S., Balamurugan A.N., Szot G.L., Kin T., Liu C., Czarniecki C.W., Barbaro B., Bridges N.D., Cano J.. et al. (2016) National institutes of health-sponsored clinical islet transplantation consortium phase 3 trial: manufacture of a complex cellular product at eight processing facilities. Diabetes, 65, 3418–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Niu D., Wei H.J., Lin L., George H., Wang T., Lee I.H., Zhao H.Y., Wang Y., Kan Y., Shrock E.. et al. (2017) Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science, 357, 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Barkai U., Weir G.C., Colton C.K., Ludwig B., Bornstein S.R., Brendel M.D., Neufeld T., Bremer C., Leon A., Evron Y.. et al. (2013) Enhanced oxygen supply improves islet viability in a new bioartificial pancreas. Cell Transpl., 22, 1463–1476. [DOI] [PubMed] [Google Scholar]
- 121. Ludwig B., Reichel A., Steffen A., Zimerman B., Schally A.V., Block N.L., Colton C.K., Ludwig S., Kersting S., Bonifacio E.. et al. (2013) Transplantation of human islets without immunosuppression. Proc. Natl. Acad. Sci. USA, 110, 19054–19058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Pepper A.R., Pawlick R., Gala-Lopez B., MacGillivary A., Mazzuca D.M., White D.J., Toleikis P.M., Shapiro A.M. (2015) Diabetes is reversed in a murine model by marginal mass syngeneic islet transplantation using a subcutaneous cell pouch device. Transplantation, 99, 2294–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Bochenek M.A., Veiseh O., Vegas A.J., McGarrigle J.J., Qi M., Marchese E., Omami M., Doloff J.C., Mendoza-Elias J., Nourmohammadzadeh M.. et al. (2018) Alginate encapsulation as long-term immune protection of allogeneic pancreatic islet cells transplanted into the omental bursa of macaques. Nat. Biomed. Eng., 2, 810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Duvivier-Kali V.F., Omer A., Parent R.J., O'Neil J.J., Weir G.C. (2001) Complete protection of islets against allorejection and autoimmunity by a simple barium-alginate membrane. Diabetes, 50, 1698–1705. [DOI] [PubMed] [Google Scholar]
- 125. Jacobs-Tulleneers-Thevissen D., Chintinne M., Ling Z., Gillard P., Schoonjans L., Delvaux G., Strand B.L., Gorus F., Keymeulen B., Pipeleers D; on behalf of the Beta Cell Therapy Consortium EU-FP7 (2013) Sustained function of alginate-encapsulated human islet cell implants in the peritoneal cavity of mice leading to a pilot study in a type 1 diabetic patient. Diabetologia, 56, 1605–1614. [DOI] [PubMed] [Google Scholar]
- 126. Pagliuca F.W., Millman J.R., Gurtler M., Segel M., Van Dervort A., Ryu J.H., Peterson Q.P., Greiner D., Melton D.A. (2014) Generation of functional human pancreatic beta cells in vitro. Cell, 159, 428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Rezania A., Bruin J.E., Arora P., Rubin A., Batushansky I., Asadi A., O'Dwyer S., Quiskamp N., Mojibian M., Albrecht T.. et al. (2014) Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol., 32, 1121–1133. [DOI] [PubMed] [Google Scholar]
- 128. Millman J.R., Pagliuca F.W. (2017) Autologous pluripotent stem cell-derived beta-like cells for diabetes cellular therapy. Diabetes, 66, 1111–1120. [DOI] [PubMed] [Google Scholar]
- 129. Agulnick A.D., Ambruzs D.M., Moorman M.A., Bhoumik A., Cesario R.M., Payne J.K., Kelly J.R., Haakmeester C., Srijemac R., Wilson A.Z.. et al. (2015) Insulin-producing endocrine cells differentiated in vitro from human embryonic stem cells function in macroencapsulation devices in vivo. Stem Cells Transl. Med., 4, 1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kumagai-Braesch M., Jacobson S., Mori H., Jia X., Takahashi T., Wernerson A., Flodstrom-Tullberg M., Tibell A. (2013) The TheraCyte device protects against islet allograft rejection in immunized hosts. Cell Transpl., 22, 1137–1146. [DOI] [PubMed] [Google Scholar]
- 131. Robert T., De Mesmaeker I., Stange G.M., Suenens K.G., Ling Z., Kroon E.J., Pipeleers D.G. (2018) Functional beta cell mass from device-encapsulated hESC-derived pancreatic endoderm achieving metabolic control. Stem Cell Rep., 10, 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Vegas A.J., Veiseh O., Gurtler M., Millman J.R., Pagliuca F.W., Bader A.R., Doloff J.C., Li J., Chen M., Olejnik K.. et al. (2016. b) Long-term glycemic control using polymer-encapsulated human stem cell-derived beta cells in immune-competent mice. Nat. Med., 22, 306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Greco R., Oliveira G., Stanghellini M.T., Vago L., Bondanza A., Peccatori J., Cieri N., Marktel S., Mastaglio S., Bordignon C.. et al. (2015) Improving the safety of cell therapy with the TK-suicide gene. Front. Pharmacol., 6, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Straathof K.C., Pule M.A., Yotnda P., Dotti G., Vanin E.F., Brenner M.K., Heslop H.E., Spencer D.M., Rooney C.M. (2005) An inducible caspase 9 safety switch for T-cell therapy. Blood, 105, 4247–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Chen Y., Sun C., Landoni E., Metelitsa L., Dotti G., Savoldo B. (2019) Eradication of neuroblastoma by T cells redirected with an optimized GD2-specific chimeric antigen receptor and interleukin-15. Clin. Cancer Res., 25, 2915–2924. [DOI] [PubMed] [Google Scholar]
- 136. Louis C.U., Savoldo B., Dotti G., Pule M., Yvon E., Myers G.D., Rossig C., Russell H.V., Diouf O., Liu E.. et al. (2011) Antitumor activity and long-term fate of chimeric antigen receptor-positive T cells in patients with neuroblastoma. Blood, 118, 6050–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Wang D., Tai P.W.L., Gao G. (2019) Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov., 18, 358–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. De Ravin S.S., Reik A., Liu P.Q., Li L., Wu X., Su L., Raley C., Theobald N., Choi U., Song A.H.. et al. (2016) Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease. Na. Biotechnol., 34, 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S.. et al. (2016) CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature, 539, 384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Hubbard N., Hagin D., Sommer K., Song Y., Khan I., Clough C., Ochs H.D., Rawlings D.J., Scharenberg A.M., Torgerson T.R. (2016) Targeted gene editing restores regulated CD40L function in X-linked hyper-IgM syndrome. Blood, 127, 2513–2522. [DOI] [PubMed] [Google Scholar]
- 141. MacLeod D.T., Antony J., Martin A.J., Moser R.J., Hekele A., Wetzel K.J., Brown A.E., Triggiano M.A., Hux J.A., Pham C.D.. et al. (2017) Integration of a CD19 CAR into the TCR alpha chain locus streamlines production of allogeneic gene-edited CAR T cells. Mol. Ther., 25, 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Sather B.D., Romano Ibarra G.S., Sommer K., Curinga G., Hale M., Khan I.F., Singh S., Song Y., Gwiazda K., Sahni J.. et al. (2015) Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Transl. Med., 7, 307ra156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Guo W., Song H. (2018) Development of gene therapeutics for head and neck cancer in China: from bench to bedside. Hum. Gene Ther., 29, 180–187. [DOI] [PubMed] [Google Scholar]
- 144. Peng Z. (2005) Current status of gendicine in China: recombinant human Ad-p53 agent for treatment of cancers. Hum. Gene Ther., 16, 1016–1027. [DOI] [PubMed] [Google Scholar]
- 145. Kirn D.H., Thorne S.H. (2009) Targeted and armed oncolytic poxviruses: a novel multi-mechanistic therapeutic class for cancer. Nat. Rev. Cancer, 9, 64–71. [DOI] [PubMed] [Google Scholar]
- 146. Kirn D., Hermiston T., McCormick F. (1998) ONYX-015: clinical data are encouraging. Nat. Med., 4, 1341–1342. [DOI] [PubMed] [Google Scholar]
- 147. Liang M. (2018) Oncorine, the world first oncolytic virus medicine and its update in China. Curr. Cancer Drug Targets, 18, 171–176. [DOI] [PubMed] [Google Scholar]
- 148. Ramesh N., Ge Y., Ennist D.L., Zhu M., Mina M., Ganesh S., Reddy P.S., Yu D.C. (2006) CG0070, a conditionally replicating granulocyte macrophage colony-stimulating factor–armed oncolytic adenovirus for the treatment of bladder cancer. Clin. Cancer Res., 12, 305–313. [DOI] [PubMed] [Google Scholar]
- 149. Leja J., Yu D., Nilsson B., Gedda L., Zieba A., Hakkarainen T., Åkerström G., Öberg K., Giandomenico V., Essand M. (2011) Oncolytic adenovirus modified with somatostatin motifs for selective infection of neuroendocrine tumor cells. Gene Ther., 18, 1052–1062. [DOI] [PubMed] [Google Scholar]
- 150. Watanabe D., Goshima F. (2018) Oncolytic virotherapy by HSV. Adv. Exp. Med. Biol., 1045, 63–84. [DOI] [PubMed] [Google Scholar]
- 151. Raman S.S., Hecht J.R., Chan E. (2019) Talimogene laherparepvec: review of its mechanism of action and clinical efficacy and safety. Immunotherapy, 11, 705–723. [DOI] [PubMed] [Google Scholar]
- 152. Heo J., Reid T., Ruo L., Breitbach C.J., Rose S., Bloomston M., Cho M., Lim H.Y., Chung H.C., Kim C.W.. et al. (2013) Randomized dose-finding clinical trial of oncolytic immunotherapeutic vaccinia JX-594 in liver cancer. Nat. Med., 19, 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Weinmann J., Grimm D. (2017) Next-generation AAV vectors for clinical use: an ever-accelerating race. Virus Genes, 53, 707–713. [DOI] [PubMed] [Google Scholar]
- 154. Bryant L.M., Christopher D.M., Giles A.R., Hinderer C., Rodriguez J.L., Smith J.B., Traxler E.A., Tycko J., Wojno A.P., Wilson J.M. (2013) Lessons learned from the clinical development and market authorization of Glybera. Hum. Gene Ther. Clin. Dev., 24, 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Scott L.J. (2015) Alipogene tiparvovec: a review of its use in adults with familial lipoprotein lipase deficiency. Drugs, 75, 175–182. [DOI] [PubMed] [Google Scholar]
- 156. Ameri H. (2018) Prospect of retinal gene therapy following commercialization of voretigene neparvovec-rzyl for retinal dystrophy mediated by RPE65 mutation. J. Curr. Ophthalmol., 30, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Hoy S.M. (2019) Onasemnogene abeparvovec: first global approval. Drugs, 79, 1255–1262. [DOI] [PubMed] [Google Scholar]
- 158. Chapin J.C., Monahan P.E. (2018) Gene therapy for hemophilia: progress to date. BioDrugs, 32, 9–25. [DOI] [PubMed] [Google Scholar]
- 159. Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D.. et al. (2014) Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med., 371, 1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Deev R., Plaksa I., Bozo I., Mzhavanadze N., Suchkov I., Chervyakov Y., Staroverov I., Kalinin R., Isaev A. (2018) Results of 5-year follow-up study in patients with peripheral artery disease treated with PL-VEGF165 for intermittent claudication. Ther. Adv. Cardiovasc. Dis., 12, 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Suda H., Murakami A., Kaga T., Tomioka H., Morishita R. (2014) Beperminogene perplasmid for the treatment of critical limb ischemia. Expert Rev. Cardiovasc. Ther/, 12, 1145–1156. [DOI] [PubMed] [Google Scholar]
- 162. Hoy S.M. (2018) Patisiran: first global approval. Drugs, 78, 1625–1631. [DOI] [PubMed] [Google Scholar]
- 163. Keam S.J. (2018) Inotersen: first global approval. Drugs, 78, 1371–1376. [DOI] [PubMed] [Google Scholar]
- 164. Nandakumar R., Matveyenko A., Thomas T., Pavlyha M., Ngai C., Holleran S., Ramakrishnan R., Ginsberg H.N., Karmally W., Marcovina S.M.. et al. (2018) Effects of mipomersen, an apolipoprotein B100 antisense, on lipoprotein (a) metabolism in healthy subjects. J. Lipid Res., 59, 2397–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Ottesen E.W. (2017) ISS-N1 makes the first FDA-approved drug for spinal muscular atrophy. Transl. Neurosci., 8, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Paik J., Duggan S. (2019) Volanesorsen: first global approval. Drugs, 79, 1349–1354. [DOI] [PubMed] [Google Scholar]
- 167. Syed Y.Y. (2016) Eteplirsen: first global approval. Drugs, 76, 1699–1704. [DOI] [PubMed] [Google Scholar]
- 168. Sharma R., Anguela X.M., Doyon Y., Wechsler T., DeKelver R.C., Sproul S., Paschon D.E., Miller J.C., Davidson R.J., Shivak D.. et al. (2015) In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood, 126, 1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Wang L., Smith J., Breton C., Clark P., Zhang J., Ying L., Che Y., Lape J., Bell P., Calcedo R.. et al. (2018. b) Meganuclease targeting of PCSK9 in macaque liver leads to stable reduction in serum cholesterol. Nat. Biotechnol., 36, 717–725. [DOI] [PubMed] [Google Scholar]
- 170. Ehrke-Schulz E., Zhang W., Gao J., Ehrhardt A. (2017) Recent advances in preclinical developments using adenovirus hybrid vectors. Hum. Gene Ther., 28, 833–841. [DOI] [PubMed] [Google Scholar]
- 171. Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. (2016) Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science, 351, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Rivera R.M.C., Madhavan S., Pan X., Ran F.A., Yan W.X.. et al. (2016) In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science, 351, 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S.. et al. (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature, 520, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Tabebordbar M., Zhu K., Cheng J.K.W., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A.. et al. (2016) In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science, 351, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Yang Y., Wang L., Bell P., McMenamin D., He Z., White J., Yu H., Xu C., Morizono H., Musunuru K.. et al. (2016) A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol., 34, 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A.. et al. (2016) Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol., 34, 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Yin H., Song C.Q., Suresh S., Wu Q., Walsh S., Rhym L.H., Mintzer E., Bolukbasi M.F., Zhu L.J., Kauffman K.. et al. (2017) Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol., 35, 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Davidovich C., Cech T.R. (2015) The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA, 21, 2007–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Scott D.A., Zhang F. (2017) Implications of human genetic variation in CRISPR-based therapeutic genome editing. Nat. Med., 23, 1095–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Gordon E.M., Hall F.L. (2009) The ‘timely’ development of Rexin-G: first targeted injectable gene vector (review). Int. J. Oncol., 35, 229–238. [PubMed] [Google Scholar]
- 181. Chawla S.P., Bruckner H., Morse M.A., Assudani N., Hall F.L., Gordon E.M. (2019) A phase I-II study using rexin-G tumor-targeted retrovector encoding a dominant-negative cyclin G1 inhibitor for advanced pancreatic cancer. Mol. Ther. Oncolytics, 12, 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Perez O.D., Logg C.R., Hiraoka K., Diago O., Burnett R., Inagaki A., Jolson D., Amundson K., Buckley T., Lohse D.. et al. (2012) Design and selection of Toca 511 for clinical use: modified retroviral replicating vector with improved stability and gene expression. Mol. Ther., 20, 1689–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Huang T.T., Parab S., Burnett R., Diago O., Ostertag D., Hofman F.M., Espinoza F.L., Martin B., Ibanez C.E., Kasahara N.. et al. (2015) Intravenous administration of retroviral replicating vector, Toca 511, demonstrates therapeutic efficacy in orthotopic immune-competent mouse glioma model. Hum. Gene Ther., 26, 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Lawler S.E., Speranza M.C., Cho C.F., Chiocca E.A. (2017) Oncolytic viruses in cancer treatment: a review. JAMA Oncol., 3, 841–849. [DOI] [PubMed] [Google Scholar]
- 185. Willmon C., Harrington K., Kottke T., Prestwich R., Melcher A., Vile R. (2009) Cell carriers for oncolytic viruses: fed Ex for cancer therapy. Mol. Ther., 17, 1667–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Bell J., McFadden G. (2014) Viruses for tumor therapy. Cell Host Microbe, 15, 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Lundstrom K. (2005) Biology and application of alphaviruses in gene therapy. Gene Ther., 12, S92–97. [DOI] [PubMed] [Google Scholar]
- 188. Naito T., Kaneko Y., Kozbor D. (2007) Oral vaccination with modified vaccinia virus Ankara attached covalently to TMPEG-modified cationic liposomes overcomes pre-existing poxvirus immunity from recombinant vaccinia immunization. J. Gen. Virol., 88, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Duebgen M., Martinez-Quintanilla J., Tamura K., Hingtgen S., Redjal N., Wakimoto H., Shah K. (2014) Stem cells loaded with multimechanistic oncolytic herpes simplex virus variants for brain tumor therapy. J. Natl. Cancer Inst., 106, dju090. [DOI] [PubMed] [Google Scholar]
- 190. Mader E.K., Butler G., Dowdy S.C., Mariani A., Knutson K.L., Federspiel M.J., Russell S.J., Galanis E., Dietz A.B., Peng K.W. (2013) Optimizing patient derived mesenchymal stem cells as virus carriers for a phase I clinical trial in ovarian cancer. J. Transl. Med., 11, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Tobias A.L., Thaci B., Auffinger B., Rincon E., Balyasnikova I.V., Kim C.K., Han Y., Zhang L., Aboody K.S., Ahmed A.U.. et al. (2013) The timing of neural stem cell-based virotherapy is critical for optimal therapeutic efficacy when applied with radiation and chemotherapy for the treatment of glioblastoma. Stem Cells Transl. Med., 2, 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Cole C., Qiao J., Kottke T., Diaz R.M., Ahmed A., Sanchez-Perez L., Brunn G., Thompson J., Chester J., Vile R.G. (2005) Tumor-targeted, systemic delivery of therapeutic viral vectors using hitchhiking on antigen-specific T cells. Nat. Med., 11, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 193. Ilett E.J., Prestwich R.J., Kottke T., Errington F., Thompson J.M., Harrington K.J., Pandha H.S., Coffey M., Selby P.J., Vile R.G.. et al. (2009) Dendritic cells and T cells deliver oncolytic reovirus for tumour killing despite pre-existing anti-viral immunity. Gene Ther., 16, 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Thorne S.H., Negrin R.S., Contag C.H. (2006) Synergistic antitumor effects of immune cell-viral biotherapy. Science, 311, 1780–1784. [DOI] [PubMed] [Google Scholar]
- 195. Esensten J.H., Bluestone J.A., Lim W.A. (2017) Engineering therapeutic T cells: from synthetic biology to clinical trials. Annu. Rev. Pathol., 12, 305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Xie M., Fussenegger M. (2018) Designing cell function: assembly of synthetic gene circuits for cell biology applications. Nat. Rev. Mol. Cell Biol., 19, 507–525. [DOI] [PubMed] [Google Scholar]
- 197. Brown C.E., Aguilar B., Starr R., Yang X., Chang W.C., Weng L., Chang B., Sarkissian A., Brito A., Sanchez J.F.. et al. (2018) Optimization of IL13Ralpha2-targeted chimeric antigen receptor T cells for improved anti-tumor efficacy against glioblastoma. Mol. Ther., 26, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Hammill J.A., VanSeggelen H., Helsen C.W., Denisova G.F., Evelegh C., Tantalo D.G., Bassett J.D., Bramson J.L. (2015) Designed ankyrin repeat proteins are effective targeting elements for chimeric antigen receptors. J. Immunother. Cancer, 3, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Ellebrecht C.T., Bhoj V.G., Nace A., Choi E.J., Mao X., Cho M.J., Di Zenzo G., Lanzavecchia A., Seykora J.T., Cotsarelis G.. et al. (2016) Reengineering chimeric antigen receptor T cells for targeted therapy of autoimmune disease. Science, 353, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Tokarew N., Ogonek J., Endres S., von Bergwelt-Baildon M., Kobold S. (2019) Teaching an old dog new tricks: next-generation CAR T cells. Br. J. Cancer, 120, 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Craddock J.A., Lu A., Bear A., Pule M., Brenner M.K., Rooney C.M., Foster A.E. (2010) Enhanced tumor trafficking of GD2 chimeric antigen receptor T cells by expression of the chemokine receptor CCR2b. J. Immunother., 33, 780–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Hong M., Puaux A.L., Huang C., Loumagne L., Tow C., Mackay C., Kato M., Prevost-Blondel A., Avril M.F., Nardin A.. et al. (2011) Chemotherapy induces intratumoral expression of chemokines in cutaneous melanoma, favoring T-cell infiltration and tumor control. Cancer Res., 71, 6997–7009. [DOI] [PubMed] [Google Scholar]
- 203. Moon E.K., Carpenito C., Sun J., Wang L.C., Kapoor V., Predina J., Powell D.J. Jr., Riley J.L., June C.H., Albelda S.M. (2011) Expression of a functional CCR2 receptor enhances tumor localization and tumor eradication by retargeted human T cells expressing a mesothelin-specific chimeric antibody receptor. Clin. Cancer Res., 17, 4719–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Park J.S., Rhau B., Hermann A., McNally K.A., Zhou C., Gong D., Weiner O.D., Conklin B.R., Onuffer J., Lim W.A. (2014. a) Synthetic control of mammalian-cell motility by engineering chemotaxis to an orthogonal bioinert chemical signal. Proc. Natl. Acad. Sci. USA, 111, 5896–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Roybal K.T., Williams J.Z., Morsut L., Rupp L.J., Kolinko I., Choe J.H., Walker W.J., McNally K.A., Lim W.A. (2016. b) Engineering T cells with customized therapeutic response programs using synthetic notch receptors. Cell, 167, 419–432 e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Boice M., Salloum D., Mourcin F., Sanghvi V., Amin R., Oricchio E., Jiang M., Mottok A., Denis-Lagache N., Ciriello G.. et al. (2016) Loss of the HVEM tumor suppressor in lymphoma and restoration by modified CAR-T cells. Cell, 167, 405–418 e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Eil R., Vodnala S.K., Clever D., Klebanoff C.A., Sukumar M., Pan J.H., Palmer D.C., Gros A., Yamamoto T.N., Patel S.J.. et al. (2016) Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature, 537, 539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Leen A.M., Sukumaran S., Watanabe N., Mohammed S., Keirnan J., Yanagisawa R., Anurathapan U., Rendon D., Heslop H.E., Rooney C.M.. et al. (2014) Reversal of tumor immune inhibition using a chimeric cytokine receptor. Mol. Ther., 22, 1211–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Liu X., Ranganathan R., Jiang S., Fang C., Sun J., Kim S., Newick K., Lo A., June C.H., Zhao Y.. et al. ,. (2016. b) A chimeric switch-receptor targeting PD1 augments the efficacy of second-generation CAR T cells in advanced solid tumors. Cancer Res., 76, 1578–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Morsut L., Roybal K.T., Xiong X., Gordley R.M., Coyle S.M., Thomson M., Lim W.A. (2016) Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell, 164, 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Roybal K.T., Rupp L.J., Morsut L., Walker W.J., McNally K.A., Park J.S., Lim W.A. (2016. a) Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell, 164, 770–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Kojima R., Scheller L., Fussenegger M. (2018) Nonimmune cells equipped with T-cell-receptor-like signaling for cancer cell ablation. Nat. Chem. Biol., 14, 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Scheller L., Strittmatter T., Fuchs D., Bojar D., Fussenegger M. (2018) Generalized extracellular molecule sensor platform for programming cellular behavior. Nat. Chem. Biol., 14, 723–729. [DOI] [PubMed] [Google Scholar]
- 214. Gardner T.S., Cantor C.R., Collins J.J. (2000) Construction of a genetic toggle switch in Escherichia coli. Nature, 403, 339–342. [DOI] [PubMed] [Google Scholar]
- 215. Kramer B.P., Viretta A.U., Daoud-El-Baba M., Aubel D., Weber W., Fussenegger M. (2004) An engineered epigenetic transgene switch in mammalian cells. Nat. Biotechnol., 22, 867–870. [DOI] [PubMed] [Google Scholar]
- 216. Muller K., Engesser R., Metzger S., Schulz S., Kampf M.M., Busacker M., Steinberg T., Tomakidi P., Ehrbar M., Nagy F.. et al. (2013) A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res., 41, e77–e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Huang H., Liu Y., Liao W., Cao Y., Liu Q., Guo Y., Lu Y., Xie Z. (2019) Oncolytic adenovirus programmed by synthetic gene circuit for cancer immunotherapy. Nat. Commun., 10, 4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Yamamoto Y., Nagasato M., Rin Y., Henmi M., Ino Y., Yachida S., Ohki R., Hiraoka N., Tagawa M., Aoki K. (2017) Strong antitumor efficacy of a pancreatic tumor-targeting oncolytic adenovirus for neuroendocrine tumors. Cancer Med., 6, 2385–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Caliendo F., Dukhinova M., Siciliano V. (2019) Engineered cell-based therapeutics: synthetic biology meets immunology. Front. Bioeng. Biotechnol., 7, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Wu C.Y., Roybal K.T., Puchner E.M., Onuffer J., Lim W.A. (2015) Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science, 350, aab4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Pan Y., Yoon S., Sun J., Huang Z., Lee C., Allen M., Wu Y., Chang Y.J., Sadelain M., Shung K.K.. et al. (2018) Mechanogenetics for the remote and noninvasive control of cancer immunotherapy. Proc. Natl. Acad. Sci. USA, 115, 992–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Rostovskaya M., Bredenkamp N., Smith A. (2015) Towards consistent generation of pancreatic lineage progenitors from human pluripotent stem cells. Philos. Trans. R. Soc. London Ser. B: Biol. Sci., 370, 20140365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Saxena P., Heng B.C., Bai P., Folcher M., Zulewski H., Fussenegger M. (2016. b) A programmable synthetic lineage-control network that differentiates human IPSCs into glucose-sensitive insulin-secreting beta-like cells. Nat. Commun., 7, 11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Carcamo-Orive I., Hoffman G.E., Cundiff P., Beckmann N.D., D'Souza S.L., Knowles J.W., Patel A., Papatsenko D., Abbasi F., Reaven G.M.. et al. (2016) Analysis of transcriptional variability in a large human iPSC library reveals genetic and non-genetic determinants of heterogeneity. Cell Stem Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Tapia N., Scholer H.R. (2016) Molecular obstacles to clinical translation of iPSCs. Cell Stem Cell, 19, 298–309. [DOI] [PubMed] [Google Scholar]
- 226. Strzyz P. (2017) Synthetic biology: designer cells tackle diabetes. Nat. Rev. Mol. Cell Biol., 18, 69. [DOI] [PubMed] [Google Scholar]
- 227. Xie M., Ye H., Wang H., Charpin-El Hamri G., Lormeau C., Saxena P., Stelling J., Fussenegger M. (2016) beta-cell-mimetic designer cells provide closed-loop glycemic control. Science, 354, 1296–1301. [DOI] [PubMed] [Google Scholar]
- 228. Ye H., Xie M., Xue S., Charpin-El Hamri G., Yin J., Zulewski H., Fussenegger M. (2017) Self-adjusting synthetic gene circuit for correcting insulin resistance. Nat. Biomed. Eng., 1, 0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Rossger K., Charpin-El-Hamri G., Fussenegger M. (2013. b) A closed-loop synthetic gene circuit for the treatment of diet-induced obesity in mice. Nat. Commun., 4, 2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Kemmer C., Gitzinger M., Daoud-El Baba M., Djonov V., Stelling J., Fussenegger M. (2010) Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat. Biotechnol., 28, 355–360. [DOI] [PubMed] [Google Scholar]
- 231. Rossger K., Charpin-El Hamri G., Fussenegger M. (2013. a) Reward-based hypertension control by a synthetic brain-dopamine interface. Proc. Natl. Acad. Sci. USA, 110, 18150–18155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Saxena P., Charpin-El Hamri G., Folcher M., Zulewski H., Fussenegger M. (2016. a) Synthetic gene network restoring endogenous pituitary-thyroid feedback control in experimental Graves' disease. Proc. Natl. Acad. Sci. USA, 113, 1244–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Liu Y., Bai P., Woischnig A.K., Charpin-El Hamri G., Ye H., Folcher M., Xie M., Khanna N., Fussenegger M. (2018) Immunomimetic designer cells protect mice from MRSA infection. Cell, 174, 259–270 e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Schukur L., Geering B., Charpin-El Hamri G., Fussenegger M. (2015) Implantable synthetic cytokine converter cells with AND-gate logic treat experimental psoriasis. Sci. Transl. Med., 7, 318ra201. [DOI] [PubMed] [Google Scholar]
- 235. Bai P., Liu Y., Xue S., Hamri G.C., Saxena P., Ye H., Xie M., Fussenegger M. (2019) A fully human transgene switch to regulate therapeutic protein production by cooling sensation. Nat. Med., 25, 1266–1273. [DOI] [PubMed] [Google Scholar]
- 236. Wang H., Ye H., Xie M., Daoud El-Baba M., Fussenegger M. (2015) Cosmetics-triggered percutaneous remote control of transgene expression in mice. Nucleic Acids Res., 43, e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Yin J., Yang L., Mou L., Dong K., Jiang J., Xue S., Xu Y., Wang X., Lu Y., Ye H. (2019) A green tea-triggered genetic control system for treating diabetes in mice and monkeys. Sci. Transl. Med., 11, [DOI] [PubMed] [Google Scholar]
- 238. Shao,J., Xue,S., Yu,G., Yu,Y., Yang,X., Bai,Y., Zhu,S., Yang,L., Yin,J., Wang,Y., et al. (2017) Smartphone-controlled optogenetically engineered cells enable semiautomatic glucose homeostasis in diabetic mice. Science Translational Medicine, 9, eaal2298. [DOI] [PubMed] [Google Scholar]
- 239. Mao,N. Cubillos-Ruiz,A. Cameron,D. E. and Collins,J. J. (2018) Probiotic strains detect and suppress cholera in mice. Science Translational Medicine, 10, eaao2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Chowdhury S., Castro S., Coker C., Hinchliffe T.E., Arpaia N., Danino T. (2019) Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat. Med., 25, 1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Geller L.T., Barzily-Rokni M., Danino T., Jonas O.H., Shental N., Nejman D., Gavert N., Zwang Y., Cooper Z.A., Shee K.. et al. (2017) Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science, 357, 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Ho C.L., Tan H.Q., Chua K.J., Kang A., Lim K.H., Ling K.L., Yew W.S., Lee Y.S., Thiery J.P., Chang M.W. (2018) Engineered commensal microbes for diet-mediated colorectal-cancer chemoprevention. Nat. Biomed. Eng., 2, 27–37. [DOI] [PubMed] [Google Scholar]
- 243. Din M.O., Danino T., Prindle A., Skalak M., Selimkhanov J., Allen K., Julio E., Atolia E., Tsimring L.S., Bhatia S.N.. et al. (2016) Synchronized cycles of bacterial lysis for in vivo delivery. Nature, 536, 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Swofford C.A., Van Dessel N., Forbes N.S. (2015) Quorum-sensing Salmonella selectively trigger protein expression within tumors. Proc. Natl. Acad. Sci. USA, 112, 3457–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Xu X., Hegazy W.A., Guo L., Gao X., Courtney A.N., Kurbanov S., Liu D., Tian G., Manuel E.R., Diamond D.J.. et al. (2014) Effective cancer vaccine platform based on attenuated salmonella and a type III secretion system. Cancer Res., 74, 6260–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Ray M., Lee Y.-W., Hardie J., Mout R., Yeşilbag Tonga G., Farkas M.E., Rotello V.M. (2018) CRISPRed macrophages for cell-based cancer immunotherapy. Bioconjug. Chem., 29, 445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Liu Y., Zeng Y., Liu L., Zhuang C., Fu X., Huang W., Cai Z. (2014) Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nat. Commun., 5, 5393. [DOI] [PubMed] [Google Scholar]
- 248. Xie Z., Wroblewska L., Prochazka L., Weiss R., Benenson Y. (2011) Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science, 333, 1307–1311. [DOI] [PubMed] [Google Scholar]
- 249. Nissim L., Wu M.R., Pery E., Binder-Nissim A., Suzuki H.I., Stupp D., Wehrspaun C., Tabach Y., Sharp P.A., Lu T.K. (2017) Synthetic RNA-based immunomodulatory gene circuits for cancer immunotherapy. Cell, 171, 1138–1150.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Lemire S., Yehl K.M., Lu T.K. (2018) Phage-based applications in synthetic biology. Annu. Rev. Virol., 5, 453–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Li P., Muller M., Chang M.W., Frettloh M., Schonherr H. (2017) Encapsulation of autoinducer sensing reporter bacteria in reinforced alginate-based microbeads. ACS Appl. Mater. Interfaces, 9, 22321–22331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Wang H., Xie M., Charpin-El Hamri G., Ye H., Fussenegger M. (2018. a) Treatment of chronic pain by designer cells controlled by spearmint aromatherapy. Nat. Biomed. Eng., 2, 114–123. [DOI] [PubMed] [Google Scholar]
- 253. Kebriaei P., Singh H., Huls M.H., Figliola M.J., Bassett R., Olivares S., Jena B., Dawson M.J., Kumaresan P.R., Su S.. et al. (2016) Phase I trials using Sleeping Beauty to generate CD19-specific CAR T cells. J. Clin. Invest., 126, 3363–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Chari S., Nguyen A., Saxe J. (2018) Stem cells in the clinic. Cell Stem Cell, 22, 781–782. [DOI] [PubMed] [Google Scholar]
- 255. Hurton L.V., Singh H., Najjar A.M., Switzer K.C., Mi T., Maiti S., Olivares S., Rabinovich B., Huls H., Forget M.A.. et al. (2016) Tethered IL-15 augments antitumor activity and promotes a stem-cell memory subset in tumor-specific T cells. Proc. Natl. Acad. Sci. USA, 113, E7788–E7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Zhou R., Yazdanifar M., Roy L.D., Whilding L.M., Gavrill A., Maher J., Mukherjee P. (2019) CAR T cells targeting the tumor MUC1 glycoprotein reduce triple-negative breast cancer growth. Front. Immunol., 10, 1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.