Abstract
Due to the beginning of vaccination against COVID-19, serological discrimination between vaccine-associated humoral response and serology-based surveillance of natural SARS-CoV-2 infections as well as breakthrough infections becomes an issue of relevance. Here, we assessed the differentiated effects of the application of an RNA vaccine using SARS-CoV-2 spike protein epitopes on the results of both anti-spike protein–based serology (EUROIMMUN) and anti-nucleocapsid-based serology (VIROTECH). A total of 80 serum samples from vaccinees acquired at different time points after vaccination was assessed. While positive or borderline serological response in the anti-spike protein assay was observed for all samples (90% both IgG and IgA, 6.3% IgA only, 3.8% borderline IgG only), only a single case of a falsely positive IgM was observed for the anti-nucleocapsid assay as expected due to this assay’s specificity. Positive anti-spike protein antibodies were already detectable in the second week after the first dose of vaccination, with higher titers after the second dose of the vaccine. In conclusion, the combined application of anti-spike protein–based serology and anti-nucleocapsid-based serology will provide a useful option for the discrimination of vaccination response and natural infection.
Keywords: SARS-CoV-2, COVID-19, serology, vaccination, surveillance, nucleocapsid, spike protein
1. Introduction
At the end of 2020, vaccination against COVID-19 was started in Germany with RNA-based vaccines using epitopes of the SARS-CoV-2 spike protein (SARS-2-S) [1] to induce protective immunity [2,3]. Vaccine-induced antibodies against the spike protein can be expected in vaccinees as indicated by the study on safety and immunogenicity prior to authorization [4]. Accordingly, vaccinated individuals without infection by SARS-CoV-2 should develop measurable antibodies in serological assays targeting the SARS-CoV-2 spike protein but not in assays targeting the nucleocapsid (SARS-2-N) protein [5] of SARS-CoV-2.
In the course of the recent months, various studies have assessed the diagnostic performance characteristics of SARS-CoV-2-specific serological assays, indicating imperfect sensitivity and specificity of the assays, with higher reliability of assays targeting IgG compared to IgM- and IgA-specific ones [6,7,8,9,10,11,12,13,14]. Specificity problems with serological assays based on whole viral antigens [15] facilitated the development of more specific assays based on chosen structures of the virus only. Although the viral nucleocapsid as well as the antigens of the viral open reading frames (ORFs) 8 and 3b were shown to elicit the strongest specific antibody responses after SARS-CoV-2-infections [16], serological assays targeting the spike protein and the nucleocapsid protein have been most frequently designed. Serological response against the spike protein has been considered as particularly interesting due to reported interaction with ACE2 (angiotensin-converting enzyme 2) binding [17]. Further, spike-protein-specific antibodies have been reported to persist longer than nucleocapsid-specific antibodies [18], which decrease within weeks to months [19]. Spike proteins offer even better sensitivity if the spike protein is applied as trimer and not as monomer in the serological assay [20]. Indeed, the trimeric spike protein was one of the first implemented as the primary antigen in various serological assays [21]. Intensity of humoral immunoreactivity toward the SARS-CoV-2 spike protein was shown to be influenced by disease severity and smoking status [22].
In spite of the shorter half-life of nucleocapsid-specific antibodies [18,19], their combination with antibodies against the receptor-binding domain of the spike protein [23] as well as their assessment alone [24] were associated with excellent specificity. In the short-term range of 2 weeks after infection, there are no differences between sensitivity of assays targeting antibodies against the nucleocapsid or the spike protein [25]. In recent comparisons of nucleocapsid- and spike-protein-based assays, no unambiguous superiority of one approach or the other could be identified [26,27,28,29]. In early infection states, sensitivity of anti-nucleocapsid antibodies can even be higher than that of anti-spike protein antibodies [24,30,31]. Other studies suggested a higher degree of inconsistency of the abundance of anti-nucleocapsid antibodies compared to anti-spike protein antibodies after SARS-CoV-2-infections [32,33]. Furthermore, spike-protein-based assays were reported to better predict the outcome of neutralization assays [34].
Considering the reported good specificity results of nucleocapsid assays [23,24], vaccination with SARS-CoV-2 spike-protein-based vaccines should elicit robust anti-spike protein antibodies as its humoral response but ought to show no cross-reaction with anti-nucleocapsid-antibody-specific assays. It should therefore be possible to differentiate between a vaccination response and at least recent natural infections with SARS-CoV-2 [18,19] on the basis of these different test strategies.
In this study, a serological assay by EUROIMMUN targeting the spike protein and an assay by VIROTECH targeting the nucleocapsid protein of SARS-CoV-2 were comparatively applied for the testing of volunteers after mRNA-based vaccination with mRNA encoding the spike protein as described [1,2,3,4]. Both the EUROIMMUN and the VIROTECH assay had already been evaluated in previous assessments [12,35,36]. By doing so, it was intended to confirm the expected differentiated response with reactivity in the spike-protein-based assay, but without reactivity in the nucleocapsid-based assay in vaccinated individuals without previous SARS-CoV-2 infection.
2. Materials and Methods
2.1. Sample Collection
Assessed samples comprised 80 sera of volunteers vaccinated with the Comirnaty COVID-19 mRNA vaccine from BioNTech (Mainz, Germany) and Pfizer (Puurs, Belgium) [36], collected after the first or second vaccination. The sera were stored in the refrigerator at 2 to 8 °C prior to assessment for a maximum of 2 days. The vaccinees were healthcare workers at the University Medical Center Göttingen, Germany.
2.2. Serological Assays
The compared serological assays comprised the spike-protein-based EUROIMMUN COVID-19 IgG/IgA assay (EUROIMMUN, Lübeck, Germany; referred to as “EUROIMMUN assay” in the following) and the nucleocapsid-based VIROTECH SARS-CoV-2 IgA/IgM/IgG ELISA (Rüsselsheim am Main, Germany; referred to as “VIROTECH assay” in the following). Both assays were performed exactly as described by the manufacturers.
2.3. Statistics
Correlation between the titers of the anti-spike protein antibodies and the time from first and second vaccination until sample acquisition as well as the vaccinees’ age at sample acquisition was calculated applying Spearman’s correlation coefficient after conducting a significant Shapiro–Wilk test for normal distribution.
Wilcoxon’s rank sum test was applied to detect statistical differences for the anti-spike protein antibody titers between males and females and between individuals who received one or two vaccinations.
2.4. Ethics
The study was ethically approved by the institutional ethics board of the University Medical Center Göttingen (identification code 21/05/20, provided on 21 May 2020).
3. Results
3.1. Vaccinees
The assessed 80 vaccinees comprised 36 (45.0%) males and 44 (55.0%) females. At the time of sample acquisition, the mean (± standard deviation SD) age in years was 39.4 (± 1.4), and the median age (interquartile range) was 37 (29, 49). At total of 53/80 (66.3%) had received the first and the second vaccine, 27/80 (33.8%) only the first vaccine. For the patients who had only received the first vaccine, the mean (± SD) number of days after this vaccine until sample acquisition was 16.5 (± 5.4), for the patients who had received both vaccines it was 4.5 (± 4.6). The raw study data on the vaccinees are shown in the Appendix A, Table A1.
3.2. Serological Test Results
In the spike-protein-specific EUROIMMUN assays, all assessed 80 vaccinees showed at least one positive (n = 77/80, 96.3%) or borderline (n = 3/80, 3.8%) result for either the immunoglobulin subclass A, G, or both. In detail, in the EUROIMMUN IgG assay 72/80 (90.0%) positive and 3/80 (3.8%) borderline results were observed. In the EUROIMMUN IgA assay, 77/80 (96.3%) showed positive results. Of note, the three cases of borderline EUROIMMUN IgG results were associated with the three negative EUROIMMUN IgA results without exemption, while positive IgA results were always associated with positive IgG results.
The three cases with negative EUROIMMUN IgA and borderline EUROIMMUN IgG comprised a 48-year-old female 19 days after her first vaccine, a 61-year-old female 1 day after her second vaccine, and a 60-year-old male 3 days after his second vaccine, respectively. The five cases with positive EUROIMMUN IgA result but negative EUROIMMUN IgG result consisted of a 21-, 51-, and a 63-year-old female and a 50-year-old male 11 days after their first vaccine, as well as a 23-year-old male 9 days after his first vaccine, respectively.
No positive results were observed for the immunoglobulin subclasses IgG and IgA of the nucleocapsid-specific VIROTECH assay. For IgM, a positive VIROTECH signal was recorded in a 51-year-old female 3 days after her second vaccine.
The raw data are provided in the Appendix A, Table A2.
3.3. Correlation of the Time in Days between the Last Vaccine and Sample Acquisition, Age, Sex and the Measured Titers
Spearman’s correlation coefficient between the time from the first and second vaccination and the titers of the anti-spike protein antibodies are indicated in Table 1. As shown there, respective correlation was weak for IgA, especially in patients who had received only one vaccine at the time of sample acquisition. It was slightly better after two vaccines, in spite of a tendency for higher titers after longer time intervals. For IgG, the correlation was better for all assessed situations.
Table 1.
Spearman’s correlation coefficient between the time from the first and second vaccination and the titers of the anti-spike protein antibodies. The p-values for the titers of the anti-spike protein antibodies and the time from first and second vaccination were 0.00003 and <0.0001, respectively.
| Population | n | Time from Vaccination | Anti-Spike Protein Antibodies | Spearman’s Correlation Coefficient with 0.95 Confidence Interval | ||
|---|---|---|---|---|---|---|
| Mean (SD) | Median (q25, q75) | Mean (SD) | Median (q25, q75) | |||
| IgA 1: All individuals | 80 | 22.53 (5.85) | 22.50 (21, 27) | 4.88 (2.73) | 4.94 (2.48, 8.03) | 0.479 [0.274, 0.676]. |
| IgA 1: Individuals with only first vaccination | 27 | 16.48 (5.38) | 19 (11, 21) | 3.87 (2.28) | 3.71 (1.73, 5.19) | 0.0075 [−0.342, 0.357 |
| IgA 2: Individuals with first and second vaccination | 53 | 4.49 (4.57) | 3 (1, 7) | 5.39 (2.82) | 5.5 (2.81, 8.52) | 0.539 [0.261, 0.760] |
| IgG 1: All individuals | 80 | 22.53 (5.85) | 22.50 (21, 27) | 4.96 (2.47) | 4.97 (3.02, 7.52) | 0.787 [0.646, 0.858] |
| IgG 1: Individuals with only first vaccination | 27 | 16.48 (5.38) | 19 (11, 21) | 2.96 (1.69) | 3.34 (1.41, 4.33) | 0.638 [0.384, 0.810] |
| IgG 2: Individuals with first and second vaccination | 53 | 4.49 (4.57) | 3 (1, 7) | 5.98 (2.18) | 6.08 (4.90, 8.08) | 0.698 [0.468, 0.839] |
1 Time from first vaccination, 2 time from second vaccination.
Table 2 indicates Spearman’s correlation coefficients between the age at the sample acquisition date and the anti-spike protein antibody titers. A non-significant trend for higher titers in young age groups was observed for both IgA and IgG antibodies.
Table 2.
Spearman’s correlation coefficients between the age at the sample acquisition date and the anti-spike protein antibody titers. The p-value for age at sample acquisition was 0.00238. The p-value for anti-spike protein IgA antibodies was 0.00008 and for IgG anti-bodies 0.00108.
| Population | n | Age in Years at Sample Acquisition | Anti-Spike Protein Antibodies | Spearman’s Correlation Coefficient with 0.95 Confidence Interval | ||
|---|---|---|---|---|---|---|
| Mean (SD) | Median (q25, q75) | Mean (SD) | Median (q25, q75) | |||
| IgA 1: All individuals | 80 | 39.4 (12.31) | 37 (29, 49) | 4.88 (2.73) | 4.94 (2.48, 8.03) | −0.153 [−0.382, 0.092] |
| IgA 1: Individuals with only first vaccination | 27 | 40.25 (13.69) | 39 (28, 50) | 3.87 (2.28) | 3.71 (1.73, 5.19) | −0.216 [−0.599, 0.187] |
| IgA 2: Individuals with first and second vaccination | 53 | 38.96 (11.66) | 36 (30, 48) | 5.39 (2.82) | 5.5 (2.81, 8.52) | −0.078 [−0.367, 0.207] |
| IgG 1: All individuals | 80 | 39.4 (12.31) | 37 (29, 49) | 4.96 (2.47) | 4.97 (3.02, 7.52) | −0.213 [−0.440, 0.044] |
| IgG 1: Individuals with only first vaccination | 27 | 40.25 (13.69) | 39 (28, 50) | 2.96 (1.69 | 3.34 (1.41, 4.33) | −0.142 [−0.569, 0.296] |
| IgG 2: Individuals with first and second vaccination | 53 | 38.96 (11.66) | 36 (30, 48) | 5.98 (2.18) | 6.08 (4.90, 8.08) | −0.211 [−0.501, 0.035] |
1 Time from first vaccination, 2 time from second vaccination.
The anti-spike protein antibody titers among males and females were compared by conducting Wilcoxon’s rank sum test and were not statistically significant assuming a level of significance of 0.05 (Table 3).
Table 3.
Comparison of spike protein antibody titers among males and females applying Wilcoxon’s rank sum test.
| Population | Males | Females | Wilcoxon Rank Sum Test, p-Value | ||||
|---|---|---|---|---|---|---|---|
| n | Mean (SD) | Median (q25, q75) | n | Mean (SD) | Median (q25, q75) | ||
| IgA 1: All individuals | 36 | 5.39 (2.63) | 5.30 (2.91, 8.42) | 44 | 4.46 (2.77) | 4.00 (1.77, 7.10) | 0.1255 |
| IgA 1: Individuals with only first vaccination | 12 | 4.59 (2.26) | 5.00 (2.86, 5.68) | 15 | 3.29 (2.19) | 3.25 (1.25, 5.19) | 0.1959 |
| IgA 2: Individuals with first and second vaccination | 24 | 5.78 (2.76) | 6.23 (2.91, 8.52) | 29 | 5.06 (2.87) | 4.90 (2.56, 8.52) | 0.4017 |
| IgG 1: All individuals | 36 | 4.87 (2.35) | 4.92 (2.92, 6.48) | 44 | 5.03 (2.60) | 5.00 (3.16, 7.69) | 0.7866 |
| IgG 1: Individuals with only first vaccination | 12 | 3.13 (1.69) | 3.07 (2.13, 4.33) | 15 | 2.83 (1.74) | 3.45 (1.03, 3.96) | 0.6256 |
| IgG 2: Individuals with first and second vaccination | 24 | 5.74 (2.16) | 5.72 (4.46, 8.09) | 29 | 6.17 (2.22) | 6.67 (4.93, 8.08) | 0.4529 |
1 Time from first vaccination, 2 time from second vaccination.
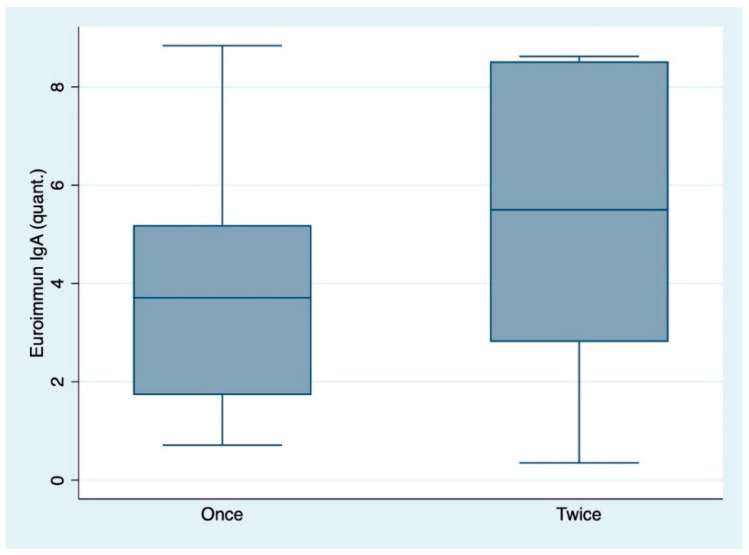
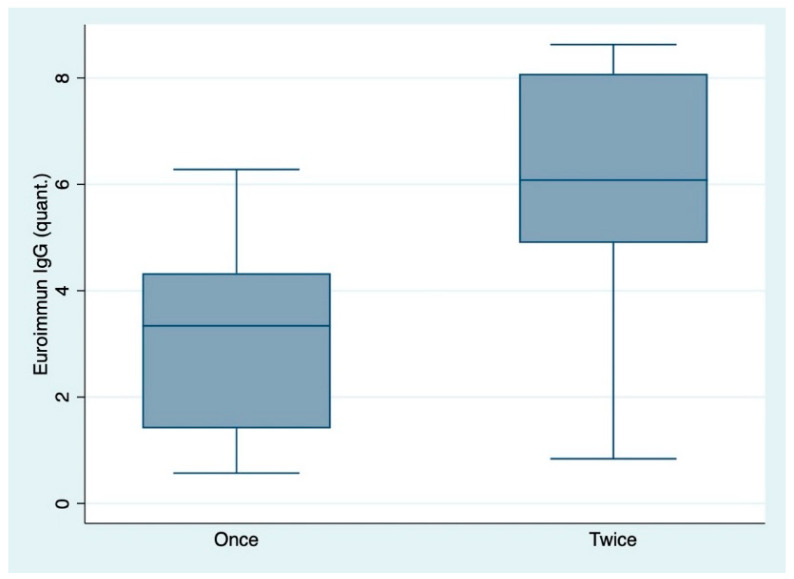
The anti-spike protein antibody titers among individuals who received one or two vaccinations were tested by conducting Wilcoxon’s rank sum test and were statistically significant assuming a level of significance of 0.05 for both subclasses IgA and IgG. The titers were higher among individuals who received both vaccinations (Table 4, Figure 1 and Figure 2).
Table 4.
Comparison of spike protein antibody titers among individuals who received one or two vaccinations applying Wilcoxon’s rank sum test.
| Population | Only First Vaccination | First and Second Vaccination | Wilcoxon Rank Sum Test, p-Value | ||||
|---|---|---|---|---|---|---|---|
| n | Mean (SD) | Median (q25, q75) | n | Mean (SD) | Median (q25, q75) | ||
| IgA: All individuals | 27 | 3.87 (2.28) | 3.71 (1.73, 5.19) | 53 | 5.39 (2.82) | 5.50 (2.81, 8.52) | 0.0217 |
| IgG: All individuals | 27 | 2.96 (1.69) | 3.34 (1.41, 4.33) | 53 | 5.98 (2.18) | 6.08 (4.90, 8.08) | <0.0001 |
Figure 1.
Box-plots indicating the difference of IgA antibody titers (as ratio of the patient sample over the extinction of the calibrator) among individuals who received one or two vaccinations. “quant.”: quantitative IgA titers. Once: one vaccination only. Twice: two vaccinations.
Figure 2.
Box-plots indicating the difference of IgG antibody titers (as ratio of the patient sample over the extinction of the calibrator) among individuals who received one or two vaccinations. “quant.”: quantitative IgG titers. Once: one vaccination only. Twice: two vaccinations.
4. Discussion
The study was conducted to differentiate the vaccination with RNA-based vaccines using epitopes of the SARS-CoV-2 spike protein (SARS-2-S) [1,2,3,4] as a proof-of-principle assessment. First of all, positive or borderline results for anti-spike protein antibodies were observed for all assessed vaccinees, irrespective of vaccination status with just one or with two vaccine doses. In contrast, anti-nucleocapsid IgA and IgG antibodies did not occur in the vaccinees. The single positive IgM result among the 80 assessed samples is as expected in line with the previously calculated specificity of the applied VIROTECH anti-nucleocapsid-IgM assay between 94% and 96.7% as indicated in a previous study [12]. Accordingly, the reported single positive IgM result can most likely be judged as a false positive. Hypothetically, a very early stage of a breakthrough infection is an alternative explanation, so follow-up assessment is recommended in case of such a serological reaction pattern. In this specific case, serological follow-up was unfeasible as the respective volunteer agreed to one blood sample acquisition only. However, in retrospect, when asked, the subject reported no symptoms of a mild upper respiratory tract infection at the relevant time.
Considering the acceptable sensitivity and specificity of the VIROTECH assay with samples from patients with COVID-19 as shown elsewhere [12], our data suggest that a combination of anti-spike protein–based serology and anti-nucleocapsid-based serology can be used to discriminate spike-protein-based vaccination-induced antibody titers from antibodies after acute SARS-CoV-2 infections as well as from potential antibodies due to breakthrough infections in spite of vaccination.
Secondly, it could be shown that anti-spike protein antibodies are already measurable early after vaccination. While 72 vaccinees showed a combination of positive IgG and IgA against the SARS-CoV-2 spike protein, five individuals with samples acquired at very early time points—9 to 11 days after the first vaccine—expressed IgA but still no IgG antibodies. This pattern was not observed at later time points of sample acquisition, suggesting its occurrence at early stages after vaccination only.
A pattern of borderline IgG anti-spike protein antibodies without measurable IgA antibodies, which was observed, compared to the study median, in older vaccinees once after the first and twice after the second vaccine dose, calls for further follow-up. Although this pattern was observed in less than 5% of the vaccinees, it raises the question of potentially insufficient humoral response toward the vaccination applied.
Thirdly, associations of antibody-titers with factors such as age, sex, and time between vaccination and serum sample acquisition were assessed. While respective correlations were generally weak, at least significance for higher anti-spike protein antibody titers in individuals after the second vaccination compared to patients with only one vaccine dose could be shown.
The study has a number of limitations. Firstly, the comparable low number of only 80 vaccinees allows a preliminary interpretation only. Secondly, the intervals between vaccination and sample acquisition were arbitrary and defined by the accessibility of the vaccinees for the sample acquisition. Thirdly, a sample acquisition on a daily basis would have been desirable to assess the seroconversion process. However, this procedure was not feasible for organizational reasons. Fourthly, neutralizing potential of the measured antibodies was not tested, as the correlation of the results of the used serological assays to the results from neutralization tests has already been demonstrated in preliminary studies [7]. Also, the scope of the work was not the confirmation of the neutralizing potential of vaccine-related antibodies but just the establishing of a ready-to-perform approach of serological discrimination of post-vaccination antibodies and wildtype virus infection–related ones in the diagnostic routine.
5. Conclusions
In a small group of recently vaccinated individuals, anti-spike protein antibodies were very consistently recorded, while anti-nucleocapsid antibodies were virtually absent. In spite of the limitations mentioned, the assessment strongly suggests combined application of anti-spike protein–based serology and anti-nucleocapsid-based serology for the discrimination of responses after spike-protein-based vaccination and natural infections. Although not directly assessed in this study, it is likely that this may also apply to breakthrough infections. Positive anti-spike protein antibody results can be expected early after vaccination, although higher titers are seen after the second vaccine. Larger studies are recommended to confirm these preliminary results. From the practical clinical point of view, a combination of serological assays for the discrimination of anti-spike protein antibodies and anti-nucleocapsid antibodies is recommended, if a recent breakthrough infection with SARS-CoV-2 is to be confirmed in vaccinees for clinical, hygiene-related, or surveillance purposes.
Acknowledgments
We are grateful to Luisa Rittmeier and Manuela Resa for excellent technical assistance.
Appendix A
Table A1.
Vaccinee-specific raw data.
| Study ID | Sex (1 = Male, 2 = Female) | Age at Sample Acquisition (Years) | Date of First Vaccine | Date of Second Vaccine | Date of sAmple Acquisition |
|---|---|---|---|---|---|
| 1 | 1 | 19 | 2021-01-21 | 2021-02-01 | |
| 2 | 2 | 21 | 2021-01-21 | 2021-02-01 | |
| 3 | 2 | 25 | 2021-01-21 | 2021-02-01 | |
| 4 | 2 | 36 | 2021-01-21 | 2021-02-01 | |
| 5 | 2 | 51 | 2021-01-21 | 2021-02-01 | |
| 6 | 1 | 50 | 2021-01-21 | 2021-02-01 | |
| 7 | 2 | 63 | 2021-01-21 | 2021-02-01 | |
| 8 | 2 | 49 | 2021-01-12 | 2021-02-02 | |
| 9 | 1 | 62 | 2021-01-15 | 2021-02-02 | |
| 10 | 2 | 48 | 2021-01-20 | 2021-02-02 | |
| 11 | 1 | 24 | 2021-01-14 | 2021-02-02 | |
| 12 | 2 | 30 | 2021-01-12 | 2021-02-02 | |
| 13 | 2 | 48 | 2021-01-14 | 2021-02-02 | |
| 14 | 1 | 44 | 2021-01-13 | 2021-02-02 | |
| 15 | 1 | 54 | 2021-01-11 | 2021-02-01 | 2021-02-02 |
| 16 | 1 | 31 | 2021-01-11 | 2021-02-01 | 2021-02-02 |
| 17 | 2 | 28 | 2021-01-11 | 2021-02-01 | 2021-02-02 |
| 18 | 1 | 31 | 2021-01-11 | 2021-02-02 | |
| 19 | 1 | 48 | 2021-01-11 | 2021-02-01 | 2021-02-02 |
| 20 | 2 | 61 | 2021-01-11 | 2021-02-01 | 2021-02-02 |
| 21 | 1 | 32 | 2021-01-12 | 2021-02-02 | |
| 22 | 2 | 45 | 2021-01-11 | 2021-02-02 | |
| 23 | 1 | 34 | 2021-01-12 | 2021-02-02 | |
| 24 | 1 | 33 | 2021-01-11 | 2021-02-02 | |
| 25 | 2 | 32 | 2021-01-11 | 2021-02-01 | 2021-02-02 |
| 26 | 1 | 40 | 2021-01-11 | 2021-02-01 | 2021-02-03 |
| 27 | 1 | 26 | 2021-01-12 | 2021-02-02 | 2021-02-03 |
| 28 | 1 | 41 | 2021-01-07 | 2021-02-01 | 2021-02-03 |
| 29 | 2 | 27 | 2021-01-13 | 2021-02-03 | |
| 30 | 1 | 43 | 2021-01-12 | 2021-02-02 | 2021-02-03 |
| 31 | 2 | 48 | 2021-01-08 | 2021-02-01 | 2021-02-04 |
| 32 | 2 | 46 | 2021-01-13 | 2021-01-03 | 2021-02-04 |
| 33 | 1 | 34 | 2021-01-11 | 2021-02-01 | 2021-02-04 |
| 34 | 2 | 50 | 2021-01-12 | 2021-02-02 | 2021-02-04 |
| 35 | 2 | 49 | 2021-01-13 | 2021-02-04 | |
| 36 | 2 | 28 | 2021-01-11 | 2021-02-01 | 2021-02-04 |
| 37 | 1 | 60 | 2021-01-11 | 2021-02-01 | 2021-02-04 |
| 38 | 2 | 28 | 2021-01-18 | 2021-02-04 | |
| 39 | 2 | 34 | 2021-01-11 | 2021-02-01 | 2021-02-04 |
| 40 | 2 | 54 | 2021-01-14 | 2021-02-04 | |
| 41 | 1 | 60 | 2021-01-20 | 2021-02-04 | |
| 42 | 1 | 39 | 2021-02-01 | 2021-02-04 | |
| 43 | 2 | 25 | 2021-01-11 | 2021-02-01 | 2021-02-04 |
| 44 | 1 | 23 | 2021-01-26 | 2021-02-04 | |
| 45 | 2 | 62 | 2021-01-15 | 2021-02-05 | |
| 46 | 2 | 25 | 2021-01-12 | 2021-02-02 | 2021-02-05 |
| 47 | 1 | 48 | 2021-01-14 | 2021-02-04 | 2021-02-05 |
| 48 | 1 | 42 | 2021-01-12 | 2021-02-02 | 2021-02-05 |
| 49 | 2 | 30 | 2021-01-14 | 2021-02-04 | 2021-02-05 |
| 50 | 2 | 41 | 2021-01-12 | 2021-02-02 | 2021-02-05 |
| 51 | 1 | 23 | 2021-01-12 | 2021-02-02 | 2021-02-05 |
| 52 | 2 | 54 | 2021-01-11 | 2021-02-04 | 2021-02-05 |
| 53 | 2 | 34 | 2021-01-12 | 2021-02-02 | 2021-02-05 |
| 54 | 2 | 39 | 2021-01-08 | 2021-02-01 | 2021-02-06 |
| 55 | 2 | 53 | 2021-01-11 | 2021-02-01 | 2021-02-08 |
| 56 | 2 | 50 | 2021-01-08 | 2021-02-01 | 2021-02-08 |
| 57 | 2 | 64 | 2021-01-08 | 2021-02-01 | 2021-02-08 |
| 58 | 2 | 38 | 2021-01-11 | 2021-02-01 | 2021-02-08 |
| 59 | 1 | 43 | 2021-01-12 | 2021-02-02 | 2021-02-08 |
| 60 | 2 | 36 | 2021-01-07 | 2021-02-01 | 2021-02-08 |
| 61 | 2 | 63 | 2021-01-11 | 2021-02-01 | 2021-02-08 |
| 62 | 1 | 42 | 2021-01-11 | 2021-02-01 | 2021-02-08 |
| 63 | 1 | 39 | 2021-01-08 | 2021-02-01 | 2021-02-08 |
| 64 | 2 | 51 | 2021-01-15 | 2021-02-05 | 2021-02-08 |
| 65 | 1 | 33 | 2021-01-12 | 2021-02-02 | 2021-02-08 |
| 66 | 2 | 31 | 2021-01-13 | 2021-02-03 | 2021-02-09 |
| 67 | 1 | 33 | 2021-01-11 | 2021-02-01 | 2021-02-09 |
| 68 | 1 | 32 | 2021-01-18 | 2021-02-08 | 2021-02-09 |
| 69 | 1 | 30 | 2021-01-11 | 2021-02-01 | 2021-02-09 |
| 70 | 2 | 58 | 2021-01-12 | 2021-02-02 | 2021-02-09 |
| 71 | 1 | 28 | 2021-01-18 | 2021-02-08 | 2021-02-09 |
| 72 | 2 | 28 | 2021-01-13 | 2021-02-03 | 2021-02-09 |
| 73 | 2 | 33 | 2021-01-12 | 2021-02-04 | 2021-02-09 |
| 74 | 1 | 31 | 2021-01-12 | 2021-02-04 | 2021-02-09 |
| 75 | 2 | 27 | 2021-01-13 | 2021-02-03 | 2021-02-09 |
| 76 | 1 | 25 | 2021-01-11 | 2021-02-01 | 2021-02-09 |
| 77 | 1 | 23 | 2021-01-12 | 2021-02-02 | 2021-02-09 |
| 78 | 2 | 57 | 2021-01-12 | 2021-02-04 | 2021-02-09 |
| 79 | 1 | 25 | 2021-01-12 | 2021-02-08 | 2021-02-09 |
| 80 | 2 | 27 | 2021-01-13 | 2021-02-03 | 2021-02-09 |
Table A2.
Raw data of the serological test results—for the VIROTECH results, only qualitative data are shown.
| Study ID | Euroimmun IgA (Qualitative, 0 = Negative, 1 = Positive) |
Euroimmun IgG (Qualitative, 0 = Negative, 1 = posItive, 3 = Borderline) |
Euroimmun IgA (Ratio of the Patient Sample over the Extinction of the Calibrator) |
Euroimmun IgG (Ratio of the Patient Sample Over the Extinction of the Calibrator) |
Virotech IgA (Qualitative, 0 = Negative, 1 = Positive) |
Virotech IgM (Qualitative, 0 = Negative, 1 = Positive) |
Virotech IgG (Qualitative, 0 = Negative, 1 = Positive) |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 8.84 | 1.41 | 0 | 0 | 0 |
| 2 | 1 | 0 | 1.73 | 0.57 | 0 | 0 | 0 |
| 3 | 1 | 1 | 4.1 | 2.20 | 0 | 0 | 0 |
| 4 | 1 | 1 | 5.2 | 3.62 | 0 | 0 | 0 |
| 5 | 1 | 0 | 3.25 | 0.63 | 0 | 0 | 0 |
| 6 | 1 | 0 | 1.99 | 0.64 | 0 | 0 | 0 |
| 7 | 1 | 0 | 1.15 | 0.62 | 0 | 0 | 0 |
| 8 | 1 | 1 | 7.86 | 4.44 | 0 | 0 | 0 |
| 9 | 1 | 1 | 7.41 | 4.26 | 0 | 0 | 0 |
| 10 | 1 | 1 | 1.32 | 1.46 | 0 | 0 | 0 |
| 11 | 1 | 1 | 3.71 | 4.37 | 0 | 0 | 0 |
| 12 | 1 | 1 | 5.19 | 5.54 | 0 | 0 | 0 |
| 13 | 0 | 3 | 0.71 | 1.03 | 0 | 0 | 0 |
| 14 | 3 | 1 | 0.95 | 3.34 | 0 | 0 | 0 |
| 15 | 1 | 1 | 1.89 | 3.03 | 0 | 0 | 0 |
| 16 | 1 | 1 | 6.26 | 5.47 | 0 | 0 | 0 |
| 17 | 1 | 1 | 1.82 | 5.02 | 0 | 0 | 0 |
| 18 | 1 | 1 | 6.18 | 4.41 | 0 | 0 | 0 |
| 19 | 1 | 1 | 1.76 | 2.48 | 0 | 0 | 0 |
| 20 | 0 | 3 | 0.35 | 0.84 | 0 | 0 | 0 |
| 21 | 1 | 1 | 3.24 | 4.33 | 0 | 0 | 0 |
| 22 | 1 | 1 | 1.25 | 3.94 | 0 | 0 | 0 |
| 23 | 1 | 1 | 4.94 | 6.28 | 0 | 0 | 0 |
| 24 | 1 | 1 | 5.07 | 2.13 | 0 | 0 | 0 |
| 25 | 1 | 1 | 3.67 | 4.93 | 0 | 0 | 0 |
| 26 | 1 | 1 | 5.41 | 5.24 | 0 | 0 | 0 |
| 27 | 1 | 1 | 3.02 | 4.90 | 0 | 0 | 0 |
| 28 | 1 | 1 | 4.94 | 5.99 | 0 | 0 | 0 |
| 29 | 1 | 1 | 1.11 | 5.44 | 0 | 0 | 0 |
| 30 | 1 | 1 | 2.55 | 4.95 | 0 | 0 | 0 |
| 31 | 1 | 1 | 3.96 | 5.62 | 0 | 0 | 0 |
| 32 | 1 | 1 | 1.61 | 2.68 | 0 | 0 | 0 |
| 33 | 1 | 1 | 2.48 | 3.68 | 0 | 0 | 0 |
| 34 | 1 | 1 | 3.32 | 3.02 | 0 | 0 | 0 |
| 35 | 1 | 1 | 4.05 | 3.96 | 0 | 0 | 0 |
| 36 | 1 | 1 | 2.26 | 4.99 | 0 | 0 | 0 |
| 37 | 0 | 3 | 0.56 | 1.09 | 0 | 0 | 0 |
| 38 | 1 | 1 | 6.57 | 3.86 | 0 | 0 | 0 |
| 39 | 1 | 1 | 4.9 | 4.47 | 0 | 0 | 0 |
| 40 | 1 | 1 | 2.33 | 1.77 | 0 | 0 | 0 |
| 41 | 1 | 1 | 5.16 | 2.77 | 0 | 0 | 0 |
| 42 | 1 | 1 | 2.48 | 2.81 | 0 | 0 | 0 |
| 43 | 1 | 1 | 6.44 | 6.67 | 0 | 0 | 0 |
| 44 | 1 | 0 | 5.19 | 0.85 | 0 | 0 | 0 |
| 45 | 1 | 1 | 3.55 | 3.45 | 0 | 0 | 0 |
| 46 | 1 | 1 | 3.31 | 6.17 | 0 | 0 | 0 |
| 47 | 1 | 1 | 8.62 | 5.63 | 0 | 0 | 0 |
| 48 | 1 | 1 | 5.51 | 2.40 | 0 | 0 | 0 |
| 49 | 1 | 1 | 4.16 | 7.57 | 0 | 0 | 0 |
| 50 | 1 | 1 | 6.96 | 6.08 | 0 | 0 | 0 |
| 51 | 1 | 1 | 2.81 | 5.37 | 0 | 0 | 0 |
| 52 | 1 | 1 | 2.56 | 2.00 | 0 | 0 | 0 |
| 53 | 1 | 1 | 1.14 | 4.32 | 0 | 0 | 0 |
| 54 | 1 | 1 | 3.13 | 7.47 | 0 | 0 | 0 |
| 55 | 1 | 1 | 5.15 | 7.82 | 0 | 0 | 0 |
| 56 | 1 | 1 | 8.59 | 8.00 | 0 | 0 | 0 |
| 57 | 1 | 1 | 8.52 | 8.50 | 0 | 0 | 0 |
| 58 | 1 | 1 | 8.52 | 8.49 | 0 | 0 | 0 |
| 59 | 1 | 1 | 8.52 | 8.12 | 0 | 0 | 0 |
| 60 | 1 | 1 | 8.52 | 8.15 | 0 | 0 | 0 |
| 61 | 1 | 1 | 8.52 | 7.88 | 0 | 0 | 0 |
| 62 | 1 | 1 | 8.32 | 8.06 | 0 | 0 | 0 |
| 63 | 1 | 1 | 8.52 | 8.26 | 0 | 0 | 0 |
| 64 | 1 | 1 | 1.73 | 3.31 | 0 | 1 | 0 |
| 65 | 1 | 1 | 8.52 | 8.23 | 0 | 0 | 0 |
| 66 | 1 | 1 | 8.33 | 8.63 | 0 | 0 | 0 |
| 67 | 1 | 1 | 8.52 | 8.24 | 0 | 0 | 0 |
| 68 | 1 | 1 | 8.21 | 5.96 | 0 | 0 | 0 |
| 69 | 1 | 1 | 8.52 | 8.13 | 0 | 0 | 0 |
| 70 | 1 | 1 | 8.52 | 8.30 | 0 | 0 | 0 |
| 71 | 1 | 1 | 3.81 | 4.03 | 0 | 0 | 0 |
| 72 | 1 | 1 | 8.52 | 8.58 | 0 | 0 | 0 |
| 73 | 1 | 1 | 7.25 | 8.08 | 0 | 0 | 0 |
| 74 | 1 | 1 | 6.2 | 6.69 | 0 | 0 | 0 |
| 75 | 1 | 1 | 1.21 | 6.80 | 0 | 0 | 0 |
| 76 | 1 | 1 | 8.52 | 8.23 | 0 | 0 | 0 |
| 77 | 1 | 1 | 8.52 | 7.99 | 0 | 0 | 0 |
| 78 | 1 | 1 | 5.5 | 6.50 | 0 | 0 | 0 |
| 79 | 1 | 1 | 6.95 | 5.81 | 0 | 0 | 0 |
| 80 | 1 | 1 | 8.52 | 8.08 | 0 | 0 | 0 |
Author Contributions
Conceptualization, A.E.Z.; methodology, A.E.Z.; software, A.E.Z.; validation, A.E.Z.; formal analysis, A.H. and J.S.; investigation, A.D., E.Y., A.H., J.S., K.M. and A.E.Z.; resources, A.H., E.Y., U.G. and A.E.Z.; data curation, A.E.Z.; writing—original draft preparation, A.D., H.F. and A.E.Z.; writing—review and editing, A.D., J.S., E.Y., H.F., K.M. and A.E.Z.; visualization, A.H., K.M. and A.E.Z.; supervision, A.E.Z.; project administration, U.G. and A.E.Z.; funding acquisition, U.G. and A.E.Z. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by the Open Access Support Program of the Deutsche Forschungsgemeinschaft and the publication fund of the Georg-August-Universität Göttingen.
Institutional Review Board Statement
The study was ethically approved by the institutional ethics board of the University Medical Center Göttingen (identification code 21/05/20, 21 May 2020).
Informed Consent Statement
Written informed consent has been obtained from the volunteers.
Data Availability Statement
All relevant data are provided in the manuscript and its tables.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Herrera N.G., Morano N.C., Celikgil A., Georgiev G.I., Malonis R.J., Lee J.H., Tong K., Vergnolle O., Massimi A.B., Yen L.Y., et al. Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis. ACS Omega. 2020;6:85–102. doi: 10.1021/acsomega.0c03512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chilamakuri R., Agarwal S. COVID-19: Characteristics and Therapeutics. Cells. 2021;10:206. doi: 10.3390/cells10020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia X. Domains and Functions of Spike Protein in Sars-Cov-2 in the Context of Vaccine Design. Viruses. 2021;13:109. doi: 10.3390/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., Mohammed A., Zhao C., Yang Y., Xie J., et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527:618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Clinical performance of different SARS-CoV-2 IgG antibody tests. J. Med. Virol. 2020;92:2243–2247. doi: 10.1002/jmv.26145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J. Clin. Virol. 2020;129:104480. doi: 10.1016/j.jcv.2020.104480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wellinghausen N., Voss M., Ivanova R., Deininger S. Evaluation of the SARS-CoV-2-IgG response in outpatients by five commercial immunoassays. GMS Infect. Dis. 2020;8:Doc22. doi: 10.3205/id000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trabaud M.A., Icard V., Milon M.P., Bal A., Lina B., Escuret V. Comparison of eight commercial, high-throughput, automated or ELISA assays detecting SARS-CoV-2 IgG or total antibody. J. Clin. Virol. 2020;132:104613. doi: 10.1016/j.jcv.2020.104613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Elslande J., Decru B., Jonckheere S., Van Wijngaerden E., Houben E., Vandecandelaere P., Indevuyst C., Depypere M., Desmet S., André E., et al. Antibody response against SARS-CoV-2 spike protein and nucleoprotein evaluated by four automated immunoassays and three ELISAs. Clin. Microbiol. Infect. 2020;26:1557.e1–1557.e7. doi: 10.1016/j.cmi.2020.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krüttgen A., Cornelissen C.G., Dreher M., Hornef M., Imöhl M., Kleines M. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J. Clin. Virol. 2020;128:104394. doi: 10.1016/j.jcv.2020.104394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dörschug A., Schwanbeck J., Hahn A., Hillebrecht A., Blaschke S., Mese K., Groß U., Dierks S., Frickmann H., Zautner A.E. Comparison of Five Serological Assays for the Detection of SARS-CoV-2 Antibodies. Diagnostics. 2021;11:78. doi: 10.3390/diagnostics11010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zilla M., Wheeler B.J., Keetch C., Mitchell G., McBreen J., Wells A., Shurin M.R., Peck-Palmer O., Wheeler S.E. Variable Performance in 6 Commercial SARS-CoV-2 Antibody Assays May Affect Convalescent Plasma and Seroprevalence Screening. Am. J. Clin. Pathol. 2020 doi: 10.1093/ajcp/aqaa228. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houlihan C.F., Beale R. The complexities of SARS-CoV-2 serology. Lancet Infect. Dis. 2020;20:1350–1351. doi: 10.1016/S1473-3099(20)30699-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meireles L.R., da Silva A.M.F., Carvalho C.A., Kesper N., Galisteo A.J., Jr., Soares C.P., Araujo D.B., Durigon E.L., Oliveira D.B.L., Morganti L., et al. Natural versus Recombinant Viral Antigens in SARS-CoV-2 Serology: Challenges in Optimizing Laboratory Diagnosis of COVID-19. Clinics. 2020;75:e2290. doi: 10.6061/clinics/2020/e2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hachim A., Kavian N., Cohen C.A., Chin A.W.H., Chu D.K.W., Mok C.K.P., Tsang O.T.Y., Yeung Y.C., Perera R.A.P.M., Poon L.L.M., et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- 17.Byrnes J.R., Zhou X.X., Lui I., Elledge S.K., Glasgow J.E., Lim S.A., Loudermilk R.P., Chiu C.Y., Wang T.T., Wilson M.R., et al. Competitive SARS-CoV-2 Serology Reveals Most Antibodies Targeting the Spike Receptor-Binding Domain Compete for ACE2 Binding. mSphere. 2020;5:e00802–e00820. doi: 10.1128/mSphere.00802-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolotin S., Tran V., Osman S., Brown K.A., Buchan S.A., Joh E., Deeks S.L., Allen V.G. SARS-CoV-2 seroprevalence survey estimates are affected by anti-nucleocapsid antibody decline. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiaa796. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lumley S.F., Wei J., O’Donnell D., Stoesser N.E., Matthews P.C., Howarth A., Hatch S.B., Marsden B.D., Cox S., James T., et al. The duration, dynamics and determinants of SARS-CoV-2 antibody responses in individual healthcare workers. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab004. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenwick C., Croxatto A., Coste A.T., Pojer F., André C., Pellaton C., Farina A., Campos J., Hacker D., Lau K., et al. Changes in SARS-CoV-2 Spike versus Nucleoprotein Antibody Responses Impact the Estimates of Infections in Population-Based Seroprevalence Studies. J. Virol. 2021;95:e01828-20. doi: 10.1128/JVI.01828-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esposito D., Mehalko J., Drew M., Snead K., Wall V., Taylor T., Frank P., Denson J.P., Hong M., Gulten G., et al. Optimizing high-yield production of SARS-CoV-2 soluble spike trimers for serology assays. Protein. Expr. Purif. 2020;174:105686. doi: 10.1016/j.pep.2020.105686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaffner A., Risch L., Aeschbacher S., Risch C., Weber M.C., Thiel S.L., Jüngert K., Pichler M., Grossmann K., Wohlwend N., et al. Characterization of a Pan-Immunoglobulin Assay Quantifying Antibodies Directed against the Receptor Binding Domain of the SARS-CoV-2 S1-Subunit of the Spike Protein: A Population-Based Study. J. Clin. Med. 2020;9:3989. doi: 10.3390/jcm9123989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mariën J., Ceulemans A., Michiels J., Heyndrickx L., Kerkhof K., Foque N., Widdowson M.A., Mortgat L., Duysburgh E., Desombere I., et al. Evaluating SARS-CoV-2 spike and nucleocapsid proteins as targets for antibody detection in severe and mild COVID-19 cases using a Luminex bead-based assay. J. Virol. Methods. 2021;288:114025. doi: 10.1016/j.jviromet.2020.114025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amrun S.N., Lee C.Y., Lee B., Fong S.W., Young B.E., Chee R.S., Yeo N.K., Torres-Ruesta A., Carissimo G., Poh C.M., et al. Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity. EBioMedicine. 2020;58:102911. doi: 10.1016/j.ebiom.2020.102911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Favresse J., Cadrobbi J., Eucher C., Elsen M., Laffineur K., Dogné J.M., Douxfils J. Clinical performance of three fully automated anti-SARS-CoV-2 immunoassays targeting the nucleocapsid or spike proteins. J. Med. Virol. 2020 doi: 10.1002/jmv.26669. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rikhtegaran Tehrani Z., Saadat S., Saleh E., Ouyang X., Constantine N., DeVico A.L., Harris A.D., Lewis G.K., Kottilil S., Sajadi M.M. Performance of nucleocapsid and spike-based SARS-CoV-2 serologic assays. PLoS ONE. 2020;15:e0237828. doi: 10.1371/journal.pone.0237828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Algaissi A., Alfaleh M.A., Hala S., Abujamel T.S., Alamri S.S., Almahboub S.A., Alluhaybi K.A., Hobani H.I., Alsulaiman R.M., AlHarbi R.H., et al. SARS-CoV-2 S1 and N-based serological assays reveal rapid seroconversion and induction of specific antibody response in COVID-19 patients. Sci. Rep. 2020;10:16561. doi: 10.1038/s41598-020-73491-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flinck H., Rauhio A., Luukinen B., Lehtimäki T., Haapala A.M., Seiskari T., Aittoniemi J. Comparison of 2 fully automated tests detecting antibodies against nucleocapsid N and spike S1/S2 proteins in COVID-19. Diagn. Microbiol. Infect. Dis. 2021;99:115197. doi: 10.1016/j.diagmicrobio.2020.115197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tehrani Z.R., Saadat S., Saleh E., Ouyang X., Constantine N., DeVico A.L., Harris A.D., Lewis G.K., Kottilil S., Sajadi M.M. Specificity and Performance of Nucleocapsid and Spike-based SARS-CoV-2 Serologic Assays. medRxiv. 2020 doi: 10.1101/2020.08.05.20168476. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., Strich J.R., Chertow D.S., Davey R.T., Jr., Cohen J.I. Detection of Nucleocapsid Antibody to SARS-CoV-2 is More Sensitive than Antibody to Spike Protein in COVID-19 Patients. medRxiv. 2020 doi: 10.1101/2020.04.20.20071423. Epub ahead of print. [DOI] [Google Scholar]
- 31.Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., Strich J.R., Chertow D.S., Davey R.T., Cohen J.I. Sensitivity in Detection of Antibodies to Nucleocapsid and Spike Proteins of Severe Acute Respiratory Syndrome Coronavirus 2 in Patients with Coronavirus Disease 2019. J. Infect. Dis. 2020;222:206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao C., Ling S., Qiu M., Deng Z., Chen L., Zhu A., Chen Y., Liu Y., Lin X., Lin F., et al. Human post-infection serological response to the spike and nucleocapsid proteins of SARS-CoV-2. Influenza Other Respir. Viruses. 2021;15:7–12. doi: 10.1111/irv.12798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McAndrews K.M., Dowlatshahi D.P., Dai J., Becker L.M., Hensel J., Snowden L.M., Leveille J.M., Brunner M.R., Holden K.W., Hopkins N.S., et al. Heterogeneous antibodies against SARS-CoV-2 spike receptor binding domain and nucleocapsid with implications for COVID-19 immunity. JCI Insight. 2020;5:e142386. doi: 10.1172/jci.insight.142386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller L., Ostermann P.N., Walker A., Wienemann T., Mertens A., Adams O., Andree M., Hauka S., Lübke N., Keitel V., et al. Sensitivity of anti-SARS-CoV-2 serological assays in a high-prevalence setting. Eur. J. Clin. Microbiol. Infect. Dis. 2021;3:1–9. doi: 10.1007/s10096-021-04169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dörschug A., Schwanbeck J., Hahn A., Hillebrecht A., Blaschke S., Groß U., Heimesaat M.M., Frickmann H., Zautner A.E. Evaluation of the Xiamen AmonMed Biotechnology rapid diagnostic test COVID-19 IgM/IgG test kit (Colloidal gold) Eur. J. Microbiol. Immunol. 2020;10:178–185. doi: 10.1556/1886.2020.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf J., Kaiser T., Pehnke S., Nickel O., Lübbert C., Kalbitz S., Arnold B., Ermisch J., Berger L., Schroth S., et al. Differences of SARS-CoV-2 serological test performance between hospitalized and outpatient COVID-19 cases. Clin. Chim. Acta. 2020;511:352–359. doi: 10.1016/j.cca.2020.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are provided in the manuscript and its tables.