Abstract
Simple Summary
Recent genomic studies have identified chromatin-spliceosome (CS)-acute myeloid leukemia (AML) as a new subgroup of AML. CS-AML is defined by several mutations that perturb epigenetic regulation, such as those affecting splicing factors, cohesin components, transcription factors, and chromatin modifiers, which are also frequently mutated in other myeloid malignancies, such as myelodysplastic syndrome and secondary AML. Thus, these mutations identify myeloid neoplasms that lie on the boundaries of conventional differential diagnosis. CS-AML shares several clinical characteristics with secondary AML. Therefore, the presence of CS-mutations may help to better classify and manage patients with AML and related disorders. The aim of this review is to discuss the genetic and clinical characteristics of CS-AML and roles of driver mutations defining this unique genomic subgroup of AML.
Abstract
Recent genetic studies on large patient cohorts with acute myeloid leukemia (AML) have cataloged a comprehensive list of driver mutations, resulting in the classification of AML into distinct genomic subgroups. Among these subgroups, chromatin-spliceosome (CS)-AML is characterized by mutations in the spliceosome, cohesin complex, transcription factors, and chromatin modifiers. Class-defining mutations of CS-AML are also frequently identified in myelodysplastic syndrome (MDS) and secondary AML, indicating the molecular similarity among these diseases. CS-AML is associated with myelodysplasia-related changes in hematopoietic cells and poor prognosis, and, thus, can be treated using novel therapeutic strategies and allogeneic stem cell transplantation. Functional studies of CS-mutations in mice have revealed that CS-mutations typically cause MDS-like phenotypes by altering the epigenetic regulation of target genes. Moreover, multiple CS-mutations often synergistically induce more severe phenotypes, such as the development of lethal MDS/AML, suggesting that the accumulation of many CS-mutations plays a crucial role in the progression of MDS/AML. Indeed, the presence of multiple CS-mutations is a stronger indicator of CS-AML than a single mutation. This review summarizes the current understanding of the genetic and clinical features of CS-AML and the functional roles of driver mutations characterizing this unique category of AML.
Keywords: acute myeloid leukemia, myelodysplastic syndrome, splicing factor, cohesin, chromatin, epigenetic regulation, hematopoiesis
1. Introduction
Acute myeloid leukemia (AML) is a group of myeloid malignancies characterized by expanded, undifferentiated myeloid precursor cells and impaired hematopoiesis, however with highly variable clinical manifestations. Despite considerable improvement in the response to initial remission induction chemotherapy, many patients develop refractory disease or experience relapse even after achieving complete remission (CR), which underlines the unmet need for better management and novel therapeutic strategies [1,2]. Although AML was initially classified based on morphological features of leukemic cells, the discovery of recurrent cytogenetic abnormalities and driver mutations has substantially promoted our understanding of AML pathogenesis. Particularly, driver mutations are now known to play a central role in the evolution of AML and have been incorporated into the definition of different AML subtypes according to the World Health Organization (WHO) classification [3,4] of myeloid neoplasms and acute leukemia.
Recent genetic studies using next-generation sequencing (NGS) technologies have almost completely cataloged driver mutations in AML [4,5]. Through the analysis of these mutations in a large cohort of patients, a novel genomic classification of AML was proposed, which comprises 11 unique subgroups with distinct molecular and prognostic features [6]. Among these subgroups, a novel category was defined as “AML with mutations in genes encoding chromatin and/or RNA splicing regulators” (hereafter called chromatin-spliceosome (CS)-AML), which is yet to be fully characterized. Mutations defining CS-AML (hereafter referred to as CS-mutations) consist of RUNX1, ASXL1, BCOR, STAG2, EZH2, SRSF2, SF3B1, U2AF1, ZRSR2, and partial tandem duplication (PTD) of KMT2A (MLL) gene (MLL-PTD) [6]. Importantly, these mutations affecting epigenetic regulators are also frequently found in myelodysplastic syndrome (MDS) and secondary AML (sAML) (or AML with myelodysplasia-related changes), a subtype of AML that develops after antecedent hematological diseases (Table 1) [7,8,9,10,11,12], suggesting that these mutations define a group of mutually overlapping myeloid neoplasms with unique pathophysiology distinct from typical de novo AML. Clinically, CS-AML correlates with progressive myelodysplasia-related changes in hematopoietic cells and poor prognosis [6].
Table 1.
Mutations defining chromatin-spliceosome acute myeloid leukemia (CS-AML).
| Driver Mutations | Pathway/Functions | Approximate Frequency (%) | ||
|---|---|---|---|---|
| de novo AML | sAML | MDS | ||
| SRSF2 | Spliceosome | 2–7 | 12–20 * | 12–17 † |
| SF3B1 | Spliceosome | 2–10 | 7–11 * | 13–33 † |
| U2AF1 | Spliceosome | 1–4 | 11–16 * | 5–11 † |
| ZRSR2 | Spliceosome | 0–1 | 3–8 * | 3–8 † |
| STAG2 | Cohesin | 2–7 | 10–14 * | 3–8 † |
| RUNX1 | Transcription factor | 5–20 | 20–31 * | 6–14 |
| EZH2 | Chromatin modification | 2–4 | 5–9 * | 4–15 † |
| BCOR | Chromatin modification | 2–3 | 7–8 * | 2–6 † |
| ASXL1 | Chromatin modification | 5–15 | 19–32 * | 10–23 † |
| MLL-PTD | Chromatin modification | 5–8 | 14 | 4–5 |
In terms of the functional implications of CS-mutations, recent studies have demonstrated that these mutations cause abnormal hematopoiesis and leukemogenesis by altering the epigenetic regulation of target genes [13,14,15,16,17,18,19,20,21]. Typically, in vivo mouse models with single CS-mutation show relatively mild abnormalities in hematopoiesis. Meanwhile, multiple CS-mutations often synergistically cause more prominent phenotypes in mice, such as the development of lethal MDS/AML, underscoring the significance of accumulation of multiple CS-mutations in the pathogenesis of MDS/AML [22,23,24].
In this review, we summarise and discuss the current understanding of the genetic and clinical characteristics of CS-AML and the functional roles of the relevant mutations in the pathogenesis of this unique category of AML.
2. Genetics of Chromatin-Spliceosome Acute Myeloid Leukemia (CS-AML)
2.1. Genetic Classification of AML
The molecular pathogenesis of AML, initially studied by cytogenetic analysis, is currently assessed by examining recurrent chromosomal abnormalities, such as t(15;17) and t(8;21), which are efficient diagnostic and prognostic markers for AML [1]. However, nearly half of all AML patients show a normal karyotype without apparent chromosomal structural abnormalities [25]. Given this background, The Cancer Genome Atlas (TCGA), a landmark cancer genomics program, analyzed the genomes of 200 patients with AML using whole-genome sequencing (WGS) (50 cases) and whole-exome sequencing (WES) (150 cases), together with RNA, microRNA, and DNA methylation analysis, to comprehensively identify genetic and epigenetic abnormalities in AML [5]. The number of coding mutations, including single nucleotide variations and insertions/deletions, was 13 per patient, on average. This project also identified 23 significantly mutated genes, including previously known driver genes, such as DNMT3A, FLT3, NPM1, IDH1, IDH2, and CEBPA, as well as genes recently implicated in AML leukemogenesis, such as U2AF1, SRSF2, EZH2, SMC1A, and STAG2. Almost all samples had at least one nonsynonymous mutation in one of nine functional categories of genes relevant to AML pathogenesis, including transcription factor fusions (e.g., t(15;17), t(8;21), and inv(16)/t(16;16)) (18%), signaling genes (e.g., FLT3, KIT) (59%), DNA methylation-related genes (e.g., DNMT3A and TET2) (44%), chromatin-modifying genes (e.g., ASXL1 and EZH2) (30%), the gene encoding nucleophosmin (NPM1) (27%), myeloid transcription factor genes (e.g., RUNX1 and CEBPA) (22%), tumor suppressor genes (e.g., TP53 and WT1) (16%), spliceosome genes (e.g., SRSF2 and U2AF1) (14%), and cohesin complex genes (e.g., STAG2 and SMC1A) (13%). Thus, driver events have been identified in most AML cases using integrated NGS and cytogenetic analysis in recent years.
A landmark study analyzed a large cohort of 1540 patients with AML using targeted-capture sequencing of 111 driver genes to understand how genetic diversity defines AML pathogenesis and informs clinical practice [6]. This identified over 5000 driver mutations across 76 genes or genomic regions, with mutations in two or more drivers discovered in 86% of patients. Intriguingly, the patterns of co-occurrence and mutual exclusivity of genetic alterations stratified the cohort into 11 categories with distinct diagnostic features and prognosis. In addition to previously defined AML subgroups, such as AML with mutated NPM1 or biallelic mutated CEBPA, three new heterogeneous genomic categories emerged in this study. These included CS-AML (18%), AML with TP53 mutations and/or chromosomal aneuploidies (13%), and, AML with IDH2R172 mutations (without other class-defining abnormalities; 1%). This classification strategy uniquely defined approximately 80% of AML into a single group, depending solely on the presence of genetic abnormalities. CS-AML was associated with old age and poor clinical outcome, with 20% of CS-AML patients having a pre-existing myeloid disorder or dysplasia. Due to limited available data on this new category, CS-AML cases have not been assigned a specific risk group by the European Leukemia Net (ELN) guideline, which defines risk stratification by genetics [2].
Another recent study analyzed 672 samples collected from a cohort of 562 AML patients (Beat AML program) using WES, RNA-sequencing, and assaying ex vivo drug sensitivity for 122 small-molecule inhibitors [26]. The Beat AML and TCGA studies, both of which analyzed AML mutations by WES or WGS, generally showed similar frequencies of driver mutations. Interestingly, the strength of drug responses correlated with mutational status, thus implying that the classification of AML patients based on mutational profiles may prove useful for risk prediction and application of precision medicine.
In summary, recent large genomic studies have helped catalog a list of driver mutations in AML, most of which occur with the acquisition of at least one genetic abnormality. The molecular findings of these studies inform disease classification, risk prediction, and facilitate better clinical management, including the application of novel molecular therapies.
2.2. Definition of CS-AML
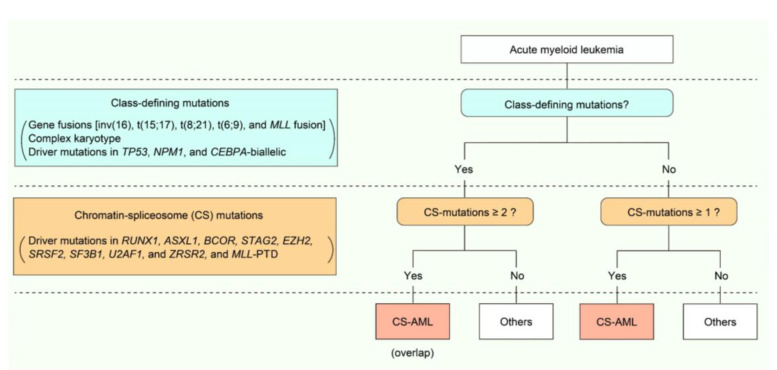
CS-AML is defined by one or more mutations in genes encoding proteins involved in chromatin regulation or the splicing machinery pathway. These CS-mutations include those in RUNX1, ASXL1, BCOR, STAG2, EZH2, SRSF2, SF3B1, U2AF1, ZRSR2, and MLL-PTD [6]. In combination with other class-defining mutations, such as t(15;17), t(8;21), inv(16), t(6;9), KMT2A fusion genes, complex karyotype, or driver mutations in NPM1, TP53, or CEBPA biallelic mutations, two or more of the CS-mutations are required to define CS-AML (Figure 1). Thus, CS-AML is a subtype of AML defined solely by genetic profiles, and not by a single mutation. In the following sections, we describe the functional roles of driver mutations in CS-AML pathogenesis as well as associated genetic and clinical features in detail.
Figure 1.
Definition and genetic features of chromatin-spliceosome acute myeloid leukemia (CS-AML). Scheme demonstrating the definition of CS-AML.
3. Driver Genes Mutated in CS-AML
3.1. Splicing Factors (SF)
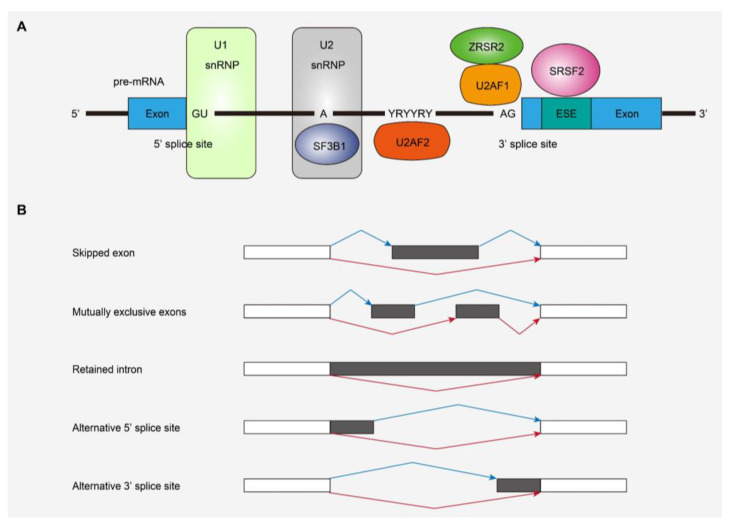
SF mutations were first reported in MDS and other myeloid neoplasms, as well as chronic lymphocytic leukemia, and represent a novel class of driver mutations in cancers [27,28]. As described above, SF mutations are one of the major driver mutations in CS-AML [6,7,8,9,10], and have been found in various myeloid neoplasms, such as MDS, AML, and myeloproliferative neoplasms (MPNs) [7,8,9,10,11]. Frequently affected SF genes include SF3B1, SRSF2, U2AF1, and ZRSR2, all of which are included in class-defining mutations for CS-AML (Figure 2A) [29]. In the former three genes, mutations change specific amino acid residues, suggesting the gain-of-function nature of those mutations [27]. In contrast, most mutations in ZRSR2, located on the X chromosome, are nonsense or frameshift mutations, suggesting inactivation of the gene function [27]. SF mutations are almost always heterozygous (or hemizygous) and seen in a mutually exclusive manner, suggesting that multiple abnormalities in the vital splicing pathway are not compatible with the survival and clonal selection of leukemia cells [27].
Figure 2.
Splicing factors (SFs) and alternative splicing affected by mutations. (A) SFs frequently affected in myeloid malignancies and their functions in RNA splicing. (B) Types of alternative splicing events detected in SF-mutated leukemias. snRNP, small nuclear ribonucleoprotein; ESE, exonic splicing enhancer.
Previous studies have revealed that SF mutations generally induce widespread RNA splicing alterations, the patterns of which depend on the mutated SFs (Figure 2B). Specific splicing alterations induced by each SF mutation have been vigorously studied using in vitro and in vivo models of SF-mutated leukemia as well as transcriptome studies of human leukemia samples with SF mutations. Mutant SF3B1 induces the usage of alternative branch points and causes an alternative 3’-splice site [30,31]. SRSF2 and U2AF1 mutations have been shown to cause alternative exon usage [13,32,33,34]. The ZRSR2 mutation involves retention of minor (U12-type) introns that make up less than 1% of introns in humans, while splicing of major (U2-type) introns is not significantly affected [35]. These specific patterns of alternative splicing due to SF mutations are also observed in primary human leukemia samples [13,14,15,36]. SF mutations lead to mis-splicing of key hematopoietic regulators, such as EZH2 and INTS3 in SRSF2-mutated MDS and ERFE, BRD9, and SF3B1 in SF3B1-mutated MDS, which may partially explain the disease phenotypes caused by SF mutations [13,37,38,39,40]. Thus, SF mutations included in CS-mutations exert differential effects on global RNA splicing, indicating that not all SF mutations cause leukemogenesis in the same way. In this regard, there may be an overlapping mechanism involved in the development of leukemia caused by SF mutations. An intriguing report demonstrated that SF mutations in SRSF2 and U2AF1 induce cell growth defects through elevated levels of R-loops, replication stress, and ATR-Chk1 activation [41]. As enhanced R-loops correlated with compromised proliferation of blood progenitors derived from the bone marrow, which was rescued by forced RNase H expression, they may contribute to aberrant hematopoiesis and leukemogenesis [41].
In mouse models, SF3B1, SRSF2, and U2AF1 mutations cause not only splicing changes but also aberrant hematopoiesis and/or MDS-like phenotypes, confirming the functional importance of these mutations in hematopoiesis and leukemogenesis [13,14,15]. A recent report also demonstrated that SRSF2 and the isocitrate dehydrogenase gene, IDH2, mutations frequently co-exist in human AML cases, and that co-expression of mutant SRSF2 and IDH2 caused lethal MDS/MPN-like diseases with myelodysplasia and proliferative features in mice, possibly through the synergistic effects of these mutations on the epigenome and RNA splicing [39]. Thus, SFs contribute to leukemogenesis by affecting epigenetic regulation and alternative splicing.
3.2. Cohesin Complex
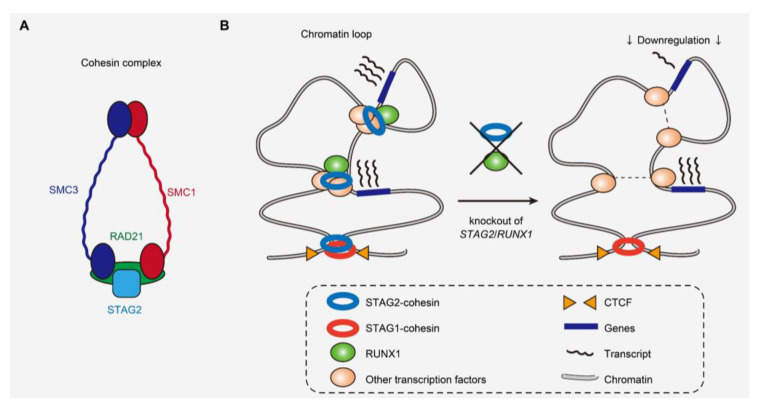
Cohesin mutations also represent a novel class of driver mutations in cancers (Figure 3A) [5,42]. Comprised of STAG1 or STAG2, RAD21, SMC1, and SMC3, the cohesin complex is involved in multiple different cellular functions, such as maintaining sister chromatid cohesion during cell division, and DNA repair [43]. Moreover, cohesin has recently also been implicated in the maintenance of the 3D genome architecture to regulate gene transcription [44]. Cohesin mutations are found in ~10–15% of AML, MDS, and MPN cases [42,45,46]. Interestingly, cohesin mutations are particularly frequent in Down syndrome-related acute megakaryoblastic leukemia, in which together with CTCF mutations, cohesin mutations were found in >50% of the cases, although rarely found in transient myeloid disease [47]. A member of the cohesin complex, STAG2 is also mutated in bladder cancers [48,49,50]. Mutations have been reported to affect all cohesin components, such as STAG2, RAD21, SMC1, and SMC3 in a mutually exclusive manner [51]. Among cohesin mutations, STAG2 mutation is most frequent and is one of the class-defining mutations in CS-AML [6,42]. Most STAG2 mutations are nonsense or frameshift, predicted to cause protein truncation and loss-of-function [42].
Figure 3.
Cohesin complex and the 3D genome structure affected by cohesin and RUNX1 mutations. (A) Cohesin complex and its components frequently mutated in myeloid malignancies. (B) Scheme demonstrating the perturbation of short-range loops and deregulation of a subset of genes caused by loss of STAG2 and RUNX1 [22].
Several studies have illustrated that cohesin mutations cause enhanced self-renewal and aberrant differentiation of hematopoietic stem and progenitor cells (HSPCs) in both in vitro and in vivo models [16,17,22,52,53,54]. Cohesin mutations particularly alter transcriptional regulation of several transcription factors, such as RUNX1, GATA2, and ERG in hematopoietic cells [16,17,22,52,53]. Moreover, recent research showed that STAG2 loss in mice preferentially disrupted short-range 3D chromatin interactions [22,53]. Genes showing high basal levels of transcriptional pausing or those regulated by super-enhancers were particularly prone to downregulation, indicating that disruption of chromosomal interactions does not necessarily result in global downregulation of gene expression (Figure 3B) [22]. The genes downregulated by cohesin deficiency included those involved in the interferon response pathway, downregulation of which is also observed in human leukemias with cohesin mutations [22,55]. In the background of STAG2 absence, additional STAG1 loss abrogated hematopoiesis, consistent with the reported synthetic lethality between STAG1 and STAG2 in leukemia cell lines and the redundancy in STAG1 and STAG2 function in chromatid segregation [53,56,57].
STAG2 mutations are almost always accompanied by other driver mutations, such as in RUNX1, SRSF2, and ASXL1 [6,8,9,22], but how cohesin mutations induce myeloid neoplasms in conjunction with other driver mutations remains to be determined. A recent study showed that combined loss of STAG2 and RUNX1, that colocalize at enhancer regions, synergistically attenuated enhancer–promoter loops and caused lethal MDS-like phenotypes in mice (Figure 3B) [22]. Another report demonstrated the interaction of cohesin with the chromatin modifying protein ASXL1, which also supports the functional relationship of cohesin with other driver genes frequently co-mutated with cohesin [58]. These studies underscore the importance of understanding cohesin function in the context of co-mutations in human leukemias to further elucidate the functional relationship between different driver genes that could synergistically affect common molecular processes.
3.3. Transcription Factors
Transcription factors regulate gene expression by binding to specific DNA sequences, and are frequent targets of genetic alterations in cancers, including AML. Representative mutational targets include RUNX1, CEBPA, and GATA2, of which RUNX1 is included in genes defining CS-AML. RUNX1 mutations have frequently been identified in a variety of hematological malignancies, including AML, MDS, MPN, and acute lymphoblastic leukemia (ALL) [4,5,6,7,8,9,10,59,60,61,62,63]. RUNX1 mutations are also found in the germline and cause familial platelet disorder with a predisposition to AML [64,65]. AML accompanied with mutated RUNX1 is assigned to the adverse prognosis group based on the ELN guidelines [2]. RUNX1 is also affected by more than 50 chromosomal translocations [66], such as t(8;21) and t(3;21), that generate RUNX1-RUNX1T1 and RUNX1-MECOM, respectively and are commonly involved in AML [66,67,68].
RUNX1 is a master transcription factor of hematopoietic cells that plays crucial roles in embryogenesis, and both definitive and adult hematopoiesis in vertebrates [18,19,20,69,70,71,72,73]. Functional studies have shown that RUNX1 regulates the transcription of key genes in hematopoiesis, such as KIT, together with other hematopoietic transcription factors, such as GATA proteins and SPI1, and mutated RUNX1 or RUNX1 fusion proteins deregulate target genes to cause aberrant hematopoiesis [59,74,75,76]. In several mouse models, the loss of RUNX1 alone does not seem to cause phenotypes of hematological malignancies [18,19,20]. However, in bone marrow transplantation models, the RUNX1 mutant causes MDS in collaboration with EVI1 [77,78]. Moreover, recent studies have shown the interaction of RUNX1 with other frequently co-mutated drivers, such as STAG2 and ASXL1, thus promoting the advancement of myeloid neoplasms [22,23]. In the Beat AML study describing drug sensitivity assays performed ex vivo on primary leukemia cells, RUNX1-mutated AML showed higher sensitivity to PIK3C and mTOR inhibitors and to the multi-kinase vascular endothelial growth factor receptor (VEGFR) inhibitor, suggesting the possibility of targeted therapy for mutated RUNX1; however, this remains to be validated in vivo [26].
3.4. Chromatin Modifiers
Genes related to the Polycomb group (PcG) of proteins are frequently affected in myeloid neoplasms. In mammals, there are two major PcG complexes, namely polycomb repressive complex 1 (PRC1) and 2 (PRC2), which regulate ubiquitination and methylation of histone modifications, respectively [79,80,81,82]. EZH2, which encodes a component of PRC2 complex, is mutated in hematological neoplasms, such as AML, MDS, MPN, and B-cell lymphoma [5,6,8,10,83,84,85]. Loss-of-function mutations of EZH2 reduce global H3K27me3 levels to deregulate expression of target genes, and result in the development of MDS, MDS/MPN, and T-cell acute lymphoblastic leukemia [24,86]. In a mouse model with a JAK2 mutation that developed MPN, additional loss of PRC2 increased sensitivity to bromodomain inhibition [87]. In the background of EZH2 deficiency, EZH1 becomes essential for maintaining hematopoiesis, indicating the synthetic lethality between EZH1 and EZH2 [86,88].
BCOR and BCORL1 proteins function as components of PRC1.1, a noncanonical PRC1, and are frequent targets of somatic mutations in AML, sAML, MDS, and chronic myelomonocytic leukemia (CMML) [6,7,8,9,10,89,90]. BCOR loss in mice promotes myeloid cell proliferation and differentiation along with an upregulation of HoxA genes [91,92]. Combined loss of BCOR and TET2 causes lethal MDS in mice [92]. The Beat AML program, a functional genomics study of AML, suggests that patients with BCOR mutations show higher sensitivity to JAK inhibitors compared to those with BCOR wild-type, which remains to be confirmed in future studies [26]. Interestingly, patients carrying both BCOR and RUNX1 mutations, both of which are members of CS-mutations, were particularly sensitive to JAK inhibitors, indicating that cooperation between CS-AML drivers may influence therapeutic strategies.
ASXL1, an epigenetic modulator, interacts with EZH2 [93]. Mutations in ASXL1 are frequently found in a variety of myeloid neoplasms, such as MDS, AML, MPNs, and CMML, as well as age-related clonal hematopoiesis in healthy individuals [5,6,8,9,10,94,95,96,97,98]. AML with mutated ASXL1 is assigned to the adverse prognosis group by the ELN genetic risk stratification guidelines [2]. Studies investigating ASXL1 function have suggested that ASXL1 mutations alter the pattern of histone modifications, such as H3K4me3, H3K27me3, and H2AK119Ub, and impair hematopoietic function [93,96,99,100,101]. Perturbed expression of ASXL1 induces MDS- or MPN-like phenotypes in several in vivo models [21,102,103,104]. Moreover, mutations in ASXL1 and SETBP1 induce leukemic transformation and MDS in a mouse model [105]. The combination of SETBP1 and ASXL1 mutations is seen in patients with germline GATA2 mutation-related MDS, in whom monosomy 7 occurred as an early somatic event followed by the acquisition of both mutations [106]. Thus, combined mutations of SETBP1 and ASXL1 seem to synergistically induce myelodysplasia. Molecules with therapeutic properties, including BRD4 and HDAC inhibitors, that reverse the action of mutant ASXL1 have been investigated, which suggests the potential treatment strategy against ASXL1-mutated leukemia [26,103,107].
MLL-PTD abnormality is found in a subset of MDS and AML, which typically shows normal cytogenetics or trisomy 11 [6,108,109,110,111,112,113,114]. Particularly, patients with acute erythroid leukemia, a subtype of AML characterized by proliferation of erythroid and myeloid blast cells in the bone marrow, carry this abnormality more frequently compared to other non-erythroid AML [115]. Functional studies have shown that MLL-PTD mutation results in aberrant chromatin remodeling and alters the transcription of target genes, and causes expansion of HSPCs and enhanced colony-formation [109,116,117,118,119]. However, MLL-PTD abnormality alone is not sufficient to develop leukemia in mice [116,117], suggesting that additional mutations together with MLL-PTD are necessary for leukemic transformation. In this regard, considerable co-occurrence of MLL-PTD and STAG2 mutations in acute erythroid leukemia [115], both of which are CS-mutations, are interesting avenues to be explored in future functional studies.
4. Clinical Features of CS-AML
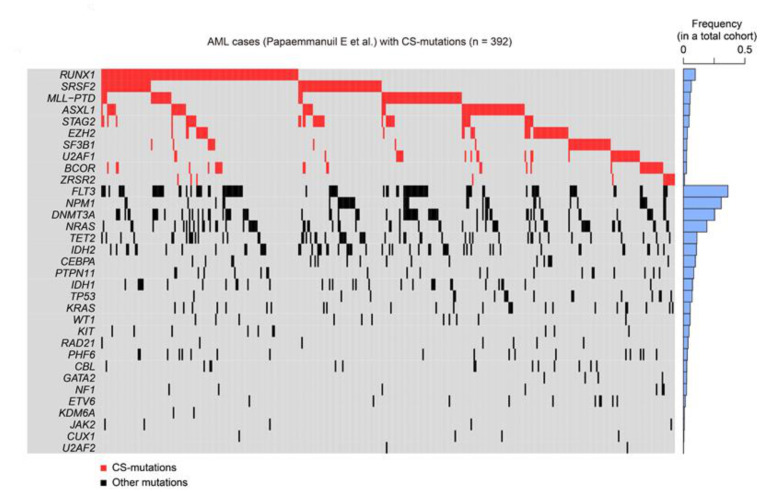
CS-AML is defined by mutations in several genes implicated in epigenetic regulation. To fully understand the molecular features of CS-AML, it is worth noting that CS-mutations are also frequently found in other myeloid tumors, such as sAML and MDS (Table 1) [7,8,9,10,11]. Another interesting feature of CS-mutations is the frequent co-occurrence of these mutations. For instance, all cases with mutated STAG2 had another CS-mutation in the original report defining CS-AML [6], suggesting that co-existence of multiple driver mutations plays a crucial role in the development of this type of leukemia (Figure 4).
Figure 4.
CS-mutations in AML. Heatmap representing CS-mutations found in a previous study [6]. Blue bars on the right show the frequency of each mutation in a large cohort of patients with AML (1540 cases). Not all cases demonstrated here satisfy the definition of CS-AML, as a subset of AML with CS-mutations is excluded from CS-AML due to presence of other class-defining mutations, as shown in Figure 1.
A previous study analyzed mutations in 194 patients with sAML or therapy-related AML and 105 de novo AML cases [7]. This study demonstrated that the presence of one or more mutations in SRSF2, SF3B1, U2AF1, ZRSR2, ASXL1, EZH2, BCOR, or STAG2, called “secondary-type” mutations, was highly specific to sAML. Notably, all secondary-type mutations are included in CS-mutations. These secondary-type mutations define a distinct genetic subgroup that has poor prognosis in elderly patients with de novo AML, suggesting that secondary-type mutations identify sAML-like disease within de novo AML [7]. Another study investigating gene expression and DNA methylation profiles in leukemia revealed that SF-mutant MDS and SF-mutant AML were clinically, cytologically, and molecularly similar [12]. This suggests that SF-mutant MDS/AML may be considered as myeloid disorders lying on the boundaries of MDS and AML. Collectively, CS-AML shares similar molecular features with MDS and sAML.
According to the report that defined CS-AML, CS-AML accounts for approximately one-fifth (18%) of AML, the second largest among the 11 subgroups [6]. This was corroborated by a recent study from the Northern Italy Leukemia Group (NILG), which analyzed a prospective cohort of 413 patients with de novo AML enrolled in a randomized trial and reported a similar frequency of CS-AML in those patients (17.6%) [120]. This study also revealed that CS-AML shared clinical characteristics with sAML (older age, lower white blood cell counts, and higher rate of multilineage dysplasia), and was associated with adverse prognosis compared to other de novo AML (overall survival, 30% in CS-AML and 17% in sAML vs. 61% in other de novo AML). Importantly, allogeneic stem cell transplantation after the first CR improved survival in both de novo AML within CS-AML category and sAML. This study emphasizes the clinical significance of diagnosing CS-AML for improving prognosis by application of therapeutic strategies such as allogeneic stem cell transplantation.
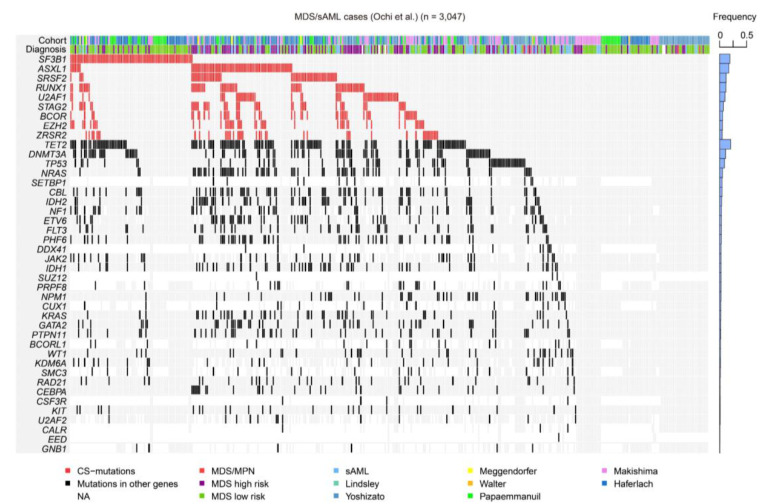
Despite being among the major driver mutations in both MDS and de novo AML, CS-mutations are more frequently seen in the former than in the latter (Table 1) [8,9,10]. In a meta-analysis of 3047 patients with MDS, related myeloid disorders, and sAML, approximately half of them carried at least one CS-mutation, suggesting that these mutations play a central role in MDS and AML pathogenesis (Figure 5) [22]. Furthermore, this study demonstrated that CS-mutations in four representative genes (STAG2, RUNX1, SRSF2, and ASXL1, called “SRSA” genes) were conspicuously enriched and co-occurred in both MDS and sAML [22]. At least one of these genes was found mutated in 31.8% of patients, and 46.8% of those carried mutations in two or more SRSA genes. More SRSA mutations were found in MDS patients that, importantly, increased the likelihood of transformation to sAML in a linear manner; 5.1%, 11.1%, and 32.9% of patients with MDS carrying 0, 1, or ≥2 SRSA mutations, respectively, experienced transformation to sAML. Consistently, patients with more than one SRSA mutation had a poorer overall survival than those with only one, which also negatively affected survival. The variant allele frequencies of the SRSA mutations suggested that SRSF2 mutations were acquired earlier than other mutations, followed by RUNX1, STAG2, and ASXL1 mutations [22]. The co-existence of SRSA mutations was also confirmed within CS-AML cases in an AML cohort at a similar frequency as that reported above [6,22]. This study revealed similar patterns of co-occurrence of particular mutations in a subset of MDS and AML, and identified those that induced leukemic transformation of MDS. Importantly, the presence of two or more CS-mutations is a stronger indicator to define CS-AML than a single mutation when other category-defining genetic events exist, such as transcription factor fusions (Figure 1) [6].
Figure 5.
CS-mutations in MDS/sAML. A heatmap representing the mutations found in a large cohort of MDS, related myeloid disorders, and sAML, as shown in our previous study [22]. Blue bars on the right show frequencies of mutations in all cases. MPN, myeloproliferative neoplasm; NA, not available.
In summary, CS-AML is a genomic subgroup of AML defined by several gene mutations, which are also frequently identified in MDS and sAML. These mutations may identify myeloid neoplasms lying on the boundaries of conventional categorization of AML. CS-AML exhibits clinical characteristics similar to those of sAML, and the unambiguous presence of CS-mutations may help clinicians to better classify and manage patients with AML, in the absence of detailed clinical history, and apparent morphological or cytogenetic abnormalities. The accumulation of multiple CS-mutations also seems to be important for the development and progression of AML, as multiple CS-mutations, such as in STAG2, RUNX1, SRSF2, and ASXL1, preferentially co-occur in patients with high-risk MDS who later develop sAML.
5. Conclusions and Future Perspectives
CS-AML is a novel form of AML and is characterized by mutations in several genes, such as those encoding the spliceosome, cohesin complex, transcription factors, and chromatin modifiers, all of which are implicated in various epigenetic pathways. Class-defining mutations in CS-AML are also frequently found in MDS and sAML, indicating a molecular similarity among a subset of CS-AML, MDS, and sAML. Clinically, CS-AML correlates with myelodysplasia-related changes in hematopoietic cells and poor patient prognosis, and has been successfully treated by allogeneic stem cell transplantation, with the potential of using novel therapeutics. Notably, the mutational status in patients with AML is associated with sensitivity to various anticancer drugs, which suggests the need to adjust treatment based on the mutational status [26].
Although the functional role of each CS-mutation in hematopoiesis and leukemogenesis is not completely homogenous, at least some aspects are shared between mutations. Functional studies using mouse models have revealed that CS-mutations typically perturb epigenetic regulation and transcription, and impair hematopoietic functions, causing aberrant hematopoiesis and/or MDS-like phenotypes. Moreover, multiple CS-mutations in mice often synergize to induce more severe phenotypes in the hematopoietic system, such as the development of lethal MDS/AML, suggesting that the accumulation of abnormalities in genes involved in epigenetic pathways may play a critical role in the development and progression of a subset of MDS/AML.
Several questions remain to be addressed to better understand CS-AML. For instance, the distinct diagnostic boundaries between CS-AML, MDS, and sAML become obscure when considering the similarity in the molecular signatures of these diseases. This ambiguity hinders the development of optimal diagnosis and therapeutic strategies for patients carrying CS-mutations. Future studies on pan-myeloid tumors should identify ways to integrate the conventional morphological and clinical diagnosis with the mutational profiles to optimize patient diagnosis and disease management. Furthermore, the molecular mechanism by which distinct CS-mutations induce comparable phenotypes and define the same disorder remains nebulous. As CS-mutations may alter epigenetic regulation, integrative analysis of genomic and epigenomic alterations in AML may prove the common molecular pathogenesis of CS-AML. Recent work suggests applying precision medicine that targets CS-mutations, which, although promising and potentially effective, warrants validation in multiple systems before translating to human trails. Given that CS-AML and MDS share common molecular signatures, patients with CS-AML may be treated using the hypomethylating agents azacitidine and decitabine that have been used in MDS treatment. Future studies should comprehensively identify the landscape of genetic and epigenetic features that lead to the pathogenesis of CS-AML, which can be used to develop novel therapeutic strategies for this unique category of AML.
Acknowledgments
The super-computing resource was provided by Human Genome Center, the Institute of Medical Science, the University of Tokyo.
Author Contributions
Conceptualization, Y.O. and S.O.; writing—original draft preparation, Y.O.; writing—review and editing, S.O.; visualization, Y.O.; supervision, S.O.; project administration, S.O.; funding acquisition, Y.O. and S.O. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Grant-in-Aid for JSPS KAKENHI (JP19H05656: S.O., 20K22809: Y.O.) and MEXT (hp190158: S.O.), Grant-in-Aid for Scientific Research on Innovative Areas (15H05909: S.O), and grants from the Japan Agency for Medical Research and Development, AMED (JP19cm0106501h0004: S.O.).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Döhner H., Weisdorf D.J., Bloomfield C.D. Acute Myeloid Leukemia. N. Engl. J. Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2.Döhner H., Estey E., Grimwade D., Amadori S., Appelbaum F.R., Büchner T., Dombret H., Ebert B.L., Fenaux P., Larson R.A., et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 4.Bullinger L., Döhner K., Döhner H. Genomics of Acute Myeloid Leukemia Diagnosis and Pathways. J. Clin. Oncol. 2017;35:934–946. doi: 10.1200/JCO.2016.71.2208. [DOI] [PubMed] [Google Scholar]
- 5.Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J., Jr., Laird P.W., Baty J.D., et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papaemmanuil E., Gerstung M., Bullinger L., Gaidzik V.I., Paschka P., Roberts N.D., Potter N.E., Heuser M., Thol F., Bolli N., et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsley R.C., Mar B.G., Mazzola E., Grauman P.V., Shareef S., Allen S.L., Pigneux A., Wetzler M., Stuart R.K., Erba H.P., et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–1376. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa S. Genetics of MDS. Blood. 2019;133:1049–1059. doi: 10.1182/blood-2018-10-844621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haferlach T., Nagata Y., Grossmann V., Okuno Y., Bacher U., Nagae G., Schnittger S., Sanada M., Kon A., Alpermann T., et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papaemmanuil E., Gerstung M., Malcovati L., Tauro S., Gundem G., Van Loo P., Yoon C.J., Ellis P., Wedge D.C., Pellagatti A., et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makishima H., Yoshizato T., Yoshida K., Sekeres M.A., Radivoyevitch T., Suzuki H., Przychodzen B., Nagata Y., Meggendorfer M., Sanada M., et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat. Genet. 2017;49:204–212. doi: 10.1038/ng.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taskesen E., Havermans M., van Lom K., Sanders M.A., van Norden Y., Bindels E., Hoogenboezem R., Reinders M.J.T., Figueroa M.E., Valk P.J.M., et al. Two splice-factor mutant leukemia subgroups uncovered at the boundaries of MDS and AML using combined gene expression and DNA-methylation profiling. Blood. 2014;123:3327–3335. doi: 10.1182/blood-2013-07-512855. [DOI] [PubMed] [Google Scholar]
- 13.Kim E., Ilagan J.O., Liang Y., Daubner G.M., Lee S.C., Ramakrishnan A., Li Y., Chung Y.R., Micol J.B., Murphy M.E., et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell. 2015;27:617–630. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirai C.L., Ley J.N., White B.S., Kim S., Tibbitts J., Shao J., Ndonwi M., Wadugu B., Duncavage E.J., Okeyo-Owuor T., et al. Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo. Cancer Cell. 2015;27:631–643. doi: 10.1016/j.ccell.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obeng E.A., Chappell R.J., Seiler M., Chen M.C., Campagna D.R., Schmidt P.J., Schneider R.K., Lord A.M., Wang L., Gambe R.G., et al. Physiologic Expression of Sf3b1(K700E) Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer Cell. 2016;30:404–417. doi: 10.1016/j.ccell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullenders J., Aranda-Orgilles B., Lhoumaud P., Keller M., Pae J., Wang K., Kayembe C., Rocha P.P., Raviram R., Gong Y., et al. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J. Exp. Med. 2015;212:1833–1850. doi: 10.1084/jem.20151323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viny A.D., Ott C.J., Spitzer B., Rivas M., Meydan C., Papalexi E., Yelin D., Shank K., Reyes J., Chiu A., et al. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J. Exp. Med. 2015;212:1819–1832. doi: 10.1084/jem.20151317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ichikawa M., Asai T., Saito T., Seo S., Yamazaki I., Yamagata T., Mitani K., Chiba S., Ogawa S., Kurokawa M., et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 19.Sun W., Downing J.R. Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood. 2004;104:3565–3572. doi: 10.1182/blood-2003-12-4349. [DOI] [PubMed] [Google Scholar]
- 20.Growney J.D., Shigematsu H., Li Z., Lee B.H., Adelsperger J., Rowan R., Curley D.P., Kutok J.L., Akashi K., Williams I.R., et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdel-Wahab O., Gao J., Adli M., Dey A., Trimarchi T., Chung Y.R., Kuscu C., Hricik T., Ndiaye-Lobry D., Lafave L.M., et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J. Exp. Med. 2013;210:2641–2659. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochi Y., Kon A., Sakata T., Nakagawa M.M., Nakazawa N., Kakuta M., Kataoka K., Koseki H., Nakayama M., Morishita D., et al. Combined Cohesin-RUNX1 Deficiency Synergistically Perturbs Chromatin Looping and Causes Myelodysplastic Syndromes. Cancer Discov. 2020;10:836–853. doi: 10.1158/2159-8290.CD-19-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bera R., Chiu M.C., Huang Y.J., Lin T.H., Kuo M.C., Shih L.Y. RUNX1 mutations promote leukemogenesis of myeloid malignancies in ASXL1-mutated leukemia. J. Hematol. Oncol. 2019;12:104. doi: 10.1186/s13045-019-0789-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sashida G., Harada H., Matsui H., Oshima M., Yui M., Harada Y., Tanaka S., Mochizuki-Kashio M., Wang C., Saraya A., et al. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat. Commun. 2014;5:4177. doi: 10.1038/ncomms5177. [DOI] [PubMed] [Google Scholar]
- 25.Rowley J.D. Chromosomal translocations: Revisited yet again. Blood. 2008;112:2183–2189. doi: 10.1182/blood-2008-04-097931. [DOI] [PubMed] [Google Scholar]
- 26.Tyner J.W., Tognon C.E., Bottomly D., Wilmot B., Kurtz S.E., Savage S.L., Long N., Schultz A.R., Traer E., Abel M., et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M., et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida K., Ogawa S. Splicing factor mutations and cancer. Wiley Interdiscip. Rev. RNA. 2014;5:445–459. doi: 10.1002/wrna.1222. [DOI] [PubMed] [Google Scholar]
- 29.Wahl M.C., Will C.L., Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Darman R.B., Seiler M., Agrawal A.A., Lim K.H., Peng S., Aird D., Bailey S.L., Bhavsar E.B., Chan B., Colla S., et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3’ Splice Site Selection through Use of a Different Branch Point. Cell Rep. 2015;13:1033–1045. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 31.Alsafadi S., Houy A., Battistella A., Popova T., Wassef M., Henry E., Tirode F., Constantinou A., Piperno-Neumann S., Roman-Roman S., et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 2016;7:10615. doi: 10.1038/ncomms10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ilagan J.O., Ramakrishnan A., Hayes B., Murphy M.E., Zebari A.S., Bradley P., Bradley R.K. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015;25:14–26. doi: 10.1101/gr.181016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao C., Yang B., Wu T., Huang J., Tang P., Zhou Y., Zhou J., Qiu J., Jiang L., Li H., et al. Mechanisms for U2AF to define 3’ splice sites and regulate alternative splicing in the human genome. Nat. Struct. Mol. Biol. 2014;21:997–1005. doi: 10.1038/nsmb.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J., Lieu Y.K., Ali A.M., Penson A., Reggio K.S., Rabadan R., Raza A., Mukherjee S., Manley J.L. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E4726–E4734. doi: 10.1073/pnas.1514105112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madan V., Kanojia D., Li J., Okamoto R., Sato-Otsubo A., Kohlmann A., Sanada M., Grossmann V., Sundaresan J., Shiraishi Y., et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun. 2015;14:6042. doi: 10.1038/ncomms7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiozawa Y., Malcovati L., Gallì A., Sato-Otsubo A., Kataoka K., Sato Y., Watatani Y., Suzuki H., Yoshizato T., Yoshida K., et al. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat. Commun. 2018;9:3649. doi: 10.1038/s41467-018-06063-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergot T., Lippert E., Douet-Guilbert N., Commet S., Corcos L., Bernard D.G. Human Cancer-Associated Mutations of SF3B1 Lead to a Splicing Modification of Its Own RNA. Cancers. 2020;12:652. doi: 10.3390/cancers12030652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bondu S., Alary A.S., Lefèvre C., Houy A., Jung G., Lefebvre T., Rombaut D., Boussaid I., Bousta A., Guillonneau F., et al. A variant erythroferrone disrupts iron homeostasis in SF3B1-mutated myelodysplastic syndrome. Sci. Transl. Med. 2019;11:652. doi: 10.1126/scitranslmed.aav5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshimi A., Lin K.-T., Wiseman D.H., Rahman M.A., Pastore A., Wang B., Lee S.C.-W., Micol J.-B., Zhang X.J., de Botton S., et al. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature. 2019;574:273–277. doi: 10.1038/s41586-019-1618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoue D., Chew G.-L., Liu B., Michel B.C., Pangallo J., D’Avino A.R., Hitchman T., North K., Lee S.C.-W., Bitner L., et al. Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature. 2019;574:432–436. doi: 10.1038/s41586-019-1646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L., Chen J.-Y., Huang Y.-J., Gu Y., Qiu J., Qian H., Shao C., Zhang X., Hu J., Li H., et al. The Augmented R-Loop Is a Unifying Mechanism for Myelodysplastic Syndromes Induced by High-Risk Splicing Factor Mutations. Mol. Cell. 2018;69:412–425. doi: 10.1016/j.molcel.2017.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kon A., Shih L.Y., Minamino M., Sanada M., Shiraishi Y., Nagata Y., Yoshida K., Okuno Y., Bando M., Nakato R., et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat. Genet. 2013;45:1232–1237. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 43.Losada A. Cohesin in cancer: Chromosome segregation and beyond. Nat. Rev. Cancer. 2014;14:389–393. doi: 10.1038/nrc3743. [DOI] [PubMed] [Google Scholar]
- 44.Rowley M.J., Corces V.G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018;19:789–800. doi: 10.1038/s41576-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thol F., Bollin R., Gehlhaar M., Walter C., Dugas M., Suchanek K.J., Kirchner A., Huang L., Chaturvedi A., Wichmann M., et al. Mutations in the cohesin complex in acute myeloid leukemia: Clinical and prognostic implications. Blood. 2014;123:914–920. doi: 10.1182/blood-2013-07-518746. [DOI] [PubMed] [Google Scholar]
- 46.Thota S., Viny A.D., Makishima H., Spitzer B., Radivoyevitch T., Przychodzen B., Sekeres M.A., Levine R.L., Maciejewski J.P. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood. 2014;124:1790–1798. doi: 10.1182/blood-2014-04-567057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida K., Toki T., Okuno Y., Kanezaki R., Shiraishi Y., Sato-Otsubo A., Sanada M., Park M.J., Terui K., Suzuki H., et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat. Genet. 2013;45:1293–1299. doi: 10.1038/ng.2759. [DOI] [PubMed] [Google Scholar]
- 48.Solomon D.A., Kim T., Diaz-Martinez L.A., Fair J., Elkahloun A.G., Harris B.T., Toretsky J.A., Rosenberg S.A., Shukla N., Ladanyi M., et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balbás-Martínez C., Sagrera A., Carrillo-de-Santa-Pau E., Earl J., Márquez M., Vazquez M., Lapi E., Castro-Giner F., Beltran S., Bayés M., et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat. Genet. 2013;45:1464–1469. doi: 10.1038/ng.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor C.F., Platt F.M., Hurst C.D., Thygesen H.H., Knowles M.A. Frequent inactivating mutations of STAG2 in bladder cancer are associated with low tumour grade and stage and inversely related to chromosomal copy number changes. Hum. Mol. Genet. 2014;23:1964–1974. doi: 10.1093/hmg/ddt589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merkenschlager M., Odom D.T. CTCF and cohesin: Linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 52.Mazumdar C., Shen Y., Xavy S., Zhao F., Reinisch A., Li R., Corces M.R., Flynn R.A., Buenrostro J.D., Chan S.M., et al. Leukemia-Associated Cohesin Mutants Dominantly Enforce Stem Cell Programs and Impair Human Hematopoietic Progenitor Differentiation. Cell Stem Cell. 2015;17:675–688. doi: 10.1016/j.stem.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Viny A.D., Bowman R.L., Liu Y., Lavallée V.P., Eisman S.E., Xiao W., Durham B.H., Navitski A., Park J., Braunstein S., et al. Cohesin Members Stag1 and Stag2 Display Distinct Roles in Chromatin Accessibility and Topological Control of HSC Self-Renewal and Differentiation. Cell Stem Cell. 2019;25:682–696.e8. doi: 10.1016/j.stem.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galeev R., Baudet A., Kumar P., Rundberg Nilsson A., Nilsson B., Soneji S., Torngren T., Borg A., Kvist A., Larsson J. Genome-wide RNAi Screen Identifies Cohesin Genes as Modifiers of Renewal and Differentiation in Human HSCs. Cell Rep. 2016;14:2988–3000. doi: 10.1016/j.celrep.2016.02.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jojic V., Shay T., Sylvia K., Zuk O., Sun X., Kang J., Regev A., Koller D., Best A.J., Knell J., et al. Identification of transcriptional regulators in the mouse immune system. Nat. Immunol. 2013;14:633–643. doi: 10.1038/ni.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benedetti L., Cereda M., Monteverde L., Desai N., Ciccarelli F.D. Synthetic lethal interaction between the tumour suppressor STAG2 and its paralog STAG1. Oncotarget. 2017;8:37619–37632. doi: 10.18632/oncotarget.16838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van der Lelij P., Lieb S., Jude J., Wutz G., Santos C.P., Falkenberg K., Schlattl A., Ban J., Schwentner R., Hoffmann T., et al. Synthetic lethality between the cohesin subunits STAG1 and STAG2 in diverse cancer contexts. eLife. 2017;6:e26980. doi: 10.7554/eLife.26980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z., Zhang P., Yan A., Guo Z., Ban Y., Li J., Chen S., Yang H., He Y., Li J., et al. ASXL1 interacts with the cohesin complex to maintain chromatid separation and gene expression for normal hematopoiesis. Sci. Adv. 2017;3:e1601602. doi: 10.1126/sciadv.1601602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sood R., Kamikubo Y., Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129:2070–2082. doi: 10.1182/blood-2016-10-687830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaidzik V.I., Bullinger L., Schlenk R.F., Zimmermann A.S., Röck J., Paschka P., Corbacioglu A., Krauter J., Schlegelberger B., Ganser A., et al. RUNX1 mutations in acute myeloid leukemia: Results from a comprehensive genetic and clinical analysis from the AML study group. J. Clin. Oncol. 2011;29:1364–1372. doi: 10.1200/JCO.2010.30.7926. [DOI] [PubMed] [Google Scholar]
- 61.Tang J.L., Hou H.A., Chen C.Y., Liu C.Y., Chou W.C., Tseng M.H., Huang C.F., Lee F.Y., Liu M.C., Yao M., et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: Prognostic implication and interaction with other gene alterations. Blood. 2009;114:5352–5361. doi: 10.1182/blood-2009-05-223784. [DOI] [PubMed] [Google Scholar]
- 62.Mendler J.H., Maharry K., Radmacher M.D., Mrózek K., Becker H., Metzeler K.H., Schwind S., Whitman S.P., Khalife J., Kohlschmidt J., et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J. Clin. Oncol. 2012;30:3109–3118. doi: 10.1200/JCO.2011.40.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaidzik V.I., Teleanu V., Papaemmanuil E., Weber D., Paschka P., Hahn J., Wallrabenstein T., Kolbinger B., Köhne C.H., Horst H.A., et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia. 2016;30:2160–2168. doi: 10.1038/leu.2016.126. [DOI] [PubMed] [Google Scholar]
- 64.Song W.J., Sullivan M.G., Legare R.D., Hutchings S., Tan X., Kufrin D., Ratajczak J., Resende I.C., Haworth C., Hock R., et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 65.Michaud J., Wu F., Osato M., Cottles G.M., Yanagida M., Asou N., Shigesada K., Ito Y., Benson K.F., Raskind W.H., et al. In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: Implications for mechanisms of pathogenesis. Blood. 2002;99:1364–1372. doi: 10.1182/blood.V99.4.1364. [DOI] [PubMed] [Google Scholar]
- 66.De Braekeleer E., Douet-Guilbert N., Morel F., Le Bris M.J., Férec C., De Braekeleer M. RUNX1 translocations and fusion genes in malignant hemopathies. Future Oncol. 2011;7:77–91. doi: 10.2217/fon.10.158. [DOI] [PubMed] [Google Scholar]
- 67.Nucifora G., Begy C.R., Kobayashi H., Roulston D., Claxton D., Pedersen-Bjergaard J., Parganas E., Ihle J.N., Rowley J.D. Consistent intergenic splicing and production of multiple transcripts between AML1 at 21q22 and unrelated genes at 3q26 in (3;21)(q26;q22) translocations. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4004–4008. doi: 10.1073/pnas.91.9.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Christen F., Hoyer K., Yoshida K., Hou H.A., Waldhueter N., Heuser M., Hills R.K., Chan W., Hablesreiter R., Blau O., et al. Genomic landscape and clonal evolution of acute myeloid leukemia with t(8;21): An international study on 331 patients. Blood. 2019;133:1140–1151. doi: 10.1182/blood-2018-05-852822. [DOI] [PubMed] [Google Scholar]
- 69.De Bruijn M., Dzierzak E. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood. 2017;129:2061–2069. doi: 10.1182/blood-2016-12-689109. [DOI] [PubMed] [Google Scholar]
- 70.Behrens K., Triviai I., Schwieger M., Tekin N., Alawi M., Spohn M., Indenbirken D., Ziegler M., Müller U., Alexander W.S., et al. Runx1 downregulates stem cell and megakaryocytic transcription programs that support niche interactions. Blood. 2016;127:3369–3381. doi: 10.1182/blood-2015-09-668129. [DOI] [PubMed] [Google Scholar]
- 71.Okuda T., van Deursen J., Hiebert S.W., Grosveld G., Downing J.R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/S0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 72.Speck N.A., Gilliland D.G. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 73.Ichikawa M., Goyama S., Asai T., Kawazu M., Nakagawa M., Takeshita M., Chiba S., Ogawa S., Kurokawa M. AML1/Runx1 negatively regulates quiescent hematopoietic stem cells in adult hematopoiesis. J. Immunol. 2008;180:4402–4408. doi: 10.4049/jimmunol.180.7.4402. [DOI] [PubMed] [Google Scholar]
- 74.Wilson N.K., Foster S.D., Wang X., Knezevic K., Schütte J., Kaimakis P., Chilarska P.M., Kinston S., Ouwehand W.H., Dzierzak E., et al. Combinatorial transcriptional control in blood stem/progenitor cells: Genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 75.Debaize L., Jakobczyk H., Avner S., Gaudichon J., Rio A.G., Sérandour A.A., Dorsheimer L., Chalmel F., Carroll J.S., Zörnig M., et al. Interplay between transcription regulators RUNX1 and FUBP1 activates an enhancer of the oncogene c-KIT and amplifies cell proliferation. Nucleic Acids Res. 2018;46:11214–11228. doi: 10.1093/nar/gky756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loke J., Assi S.A., Imperato M.R., Ptasinska A., Cauchy P., Grabovska Y., Soria N.M., Raghavan M., Delwel H.R., Cockerill P.N., et al. RUNX1-ETO and RUNX1-EVI1 Differentially Reprogram the Chromatin Landscape in t(8;21) and t(3;21) AML. Cell Reports. 2017;19:1654–1668. doi: 10.1016/j.celrep.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harada Y., Inoue D., Ding Y., Imagawa J., Doki N., Matsui H., Yahata T., Matsushita H., Ando K., Sashida G., et al. RUNX1/AML1 mutant collaborates with BMI1 overexpression in the development of human and murine myelodysplastic syndromes. Blood. 2013;121:3434–3446. doi: 10.1182/blood-2012-06-434423. [DOI] [PubMed] [Google Scholar]
- 78.Watanabe-Okochi N., Kitaura J., Ono R., Harada H., Harada Y., Komeno Y., Nakajima H., Nosaka T., Inaba T., Kitamura T. AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood. 2008;111:4297–4308. doi: 10.1182/blood-2007-01-068346. [DOI] [PubMed] [Google Scholar]
- 79.Sauvageau M., Sauvageau G. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blackledge N.P., Rose N.R., Klose R.J. Targeting Polycomb systems to regulate gene expression: Modifications to a complex story. Nat. Rev. Mol. Cell Biol. 2015;16:643–649. doi: 10.1038/nrm4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sashida G., Oshima M., Iwama A. Deregulated Polycomb functions in myeloproliferative neoplasms. Int. J. Hematol. 2019;110:170–178. doi: 10.1007/s12185-019-02600-6. [DOI] [PubMed] [Google Scholar]
- 82.Sparmann A., Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 83.Nikoloski G., Langemeijer S.M.C., Kuiper R.P., Knops R., Massop M., Tönnissen E.R.L.T.M., van der Heijden A., Scheele T.N., Vandenberghe P., de Witte T., et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 84.Sneeringer C.J., Scott M.P., Kuntz K.W., Knutson S.K., Pollock R.M., Richon V.M., Copeland R.A. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ernst T., Chase A.J., Score J., Hidalgo-Curtis C.E., Bryant C., Jones A.V., Waghorn K., Zoi K., Ross F.M., Reiter A., et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 86.Mochizuki-Kashio M., Aoyama K., Sashida G., Oshima M., Tomioka T., Muto T., Wang C., Iwama A. Ezh2 loss in hematopoietic stem cells predisposes mice to develop heterogeneous malignancies in an Ezh1-dependent manner. Blood. 2015;126:1172–1183. doi: 10.1182/blood-2015-03-634428. [DOI] [PubMed] [Google Scholar]
- 87.Sashida G., Wang C., Tomioka T., Oshima M., Aoyama K., Kanai A., Mochizuki-Kashio M., Harada H., Shimoda K., Iwama A. The loss of Ezh2 drives the pathogenesis of myelofibrosis and sensitizes tumor-initiating cells to bromodomain inhibition. J. Exp. Med. 2016;213:1459–1477. doi: 10.1084/jem.20151121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gu Z., Liu Y., Cai F., Patrick M., Zmajkovic J., Cao H., Zhang Y., Tasdogan A., Chen M., Qi L., et al. Loss of EZH2 Reprograms BCAA Metabolism to Drive Leukemic Transformation. Cancer Discov. 2019;9:1228–1247. doi: 10.1158/2159-8290.CD-19-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grossmann V., Tiacci E., Holmes A.B., Kohlmann A., Martelli M.P., Kern W., Spanhol-Rosseto A., Klein H.U., Dugas M., Schindela S., et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood. 2011;118:6153–6163. doi: 10.1182/blood-2011-07-365320. [DOI] [PubMed] [Google Scholar]
- 90.Damm F., Chesnais V., Nagata Y., Yoshida K., Scourzic L., Okuno Y., Itzykson R., Sanada M., Shiraishi Y., Gelsi-Boyer V., et al. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood. 2013;122:3169–3177. doi: 10.1182/blood-2012-11-469619. [DOI] [PubMed] [Google Scholar]
- 91.Cao Q., Gearhart M.D., Gery S., Shojaee S., Yang H., Sun H., Lin D.c., Bai J.w., Mead M., Zhao Z., et al. BCOR regulates myeloid cell proliferation and differentiation. Leukemia. 2016;30:1155–1165. doi: 10.1038/leu.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tara S., Isshiki Y., Nakajima-Takagi Y., Oshima M., Aoyama K., Tanaka T., Shinoda D., Koide S., Saraya A., Miyagi S., et al. Bcor insufficiency promotes initiation and progression of myelodysplastic syndrome. Blood. 2018;132:2470–2483. doi: 10.1182/blood-2018-01-827964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abdel-Wahab O., Adli M., LaFave L.M., Gao J., Hricik T., Shih A.H., Pandey S., Patel J.P., Chung Y.R., Koche R., et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Asada S., Fujino T., Goyama S., Kitamura T. The role of ASXL1 in hematopoiesis and myeloid malignancies. Cell. Mol. Life Sci. 2019;76:2511–2523. doi: 10.1007/s00018-019-03084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carbuccia N., Murati A., Trouplin V., Brecqueville M., Adélaïde J., Rey J., Vainchenker W., Bernard O.A., Chaffanet M., Vey N., et al. Mutations of ASXL1 gene in myeloproliferative neoplasms. Leukemia. 2009;23:2183–2186. doi: 10.1038/leu.2009.141. [DOI] [PubMed] [Google Scholar]
- 96.Fujino T., Kitamura T. ASXL1 mutation in clonal hematopoiesis. Exp. Hematol. 2020;83:74–84. doi: 10.1016/j.exphem.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 97.Genovese G., Kähler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M., et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A., et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scheuermann J.C., de Ayala Alonso A.G., Oktaba K., Ly-Hartig N., McGinty R.K., Fraterman S., Wilm M., Muir T.W., Müller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang J., Scribner K.T. Parentage and sibship inference from markers in polyploids. Mol. Ecol. Resour. 2014;14:541–553. doi: 10.1111/1755-0998.12210. [DOI] [PubMed] [Google Scholar]
- 101.Inoue D., Fujino T., Sheridan P., Zhang Y.-z., Nagase R., Horikawa S., Li Z., Matsui H., Kanai A., Saika M., et al. A novel ASXL1–OGT axis plays roles in H3K4 methylation and tumor suppression in myeloid malignancies. Leukemia. 2018;32:1327–1337. doi: 10.1038/s41375-018-0083-3. [DOI] [PubMed] [Google Scholar]
- 102.Wang J., Li Z., He Y., Pan F., Chen S., Rhodes S., Nguyen L., Yuan J., Jiang L., Yang X., et al. Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood. 2014;123:541–553. doi: 10.1182/blood-2013-05-500272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang H., Kurtenbach S., Guo Y., Lohse I., Durante M.A., Li J., Li Z., Al-Ali H., Li L., Chen Z., et al. Gain of function of ASXL1 truncating protein in the pathogenesis of myeloid malignancies. Blood. 2018;131:328–341. doi: 10.1182/blood-2017-06-789669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Uni M., Masamoto Y., Sato T., Kamikubo Y., Arai S., Hara E., Kurokawa M. Modeling ASXL1 mutation revealed impaired hematopoiesis caused by derepression of p16Ink4a through aberrant PRC1-mediated histone modification. Leukemia. 2019;33:191–204. doi: 10.1038/s41375-018-0198-6. [DOI] [PubMed] [Google Scholar]
- 105.Inoue D., Kitaura J., Matsui H., Hou H.A., Chou W.C., Nagamachi A., Kawabata K.C., Togami K., Nagase R., Horikawa S., et al. SETBP1 mutations drive leukemic transformation in ASXL1-mutated MDS. Leukemia. 2015;29:847–857. doi: 10.1038/leu.2014.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wlodarski M.W., Collin M., Horwitz M.S. GATA2 deficiency and related myeloid neoplasms. Semin. Hematol. 2017;54:81–86. doi: 10.1053/j.seminhematol.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Saika M., Inoue D., Nagase R., Sato N., Tsuchiya A., Yabushita T., Kitamura T., Goyama S. ASXL1 and SETBP1 mutations promote leukaemogenesis by repressing TGFβ pathway genes through histone deacetylation. Sci. Rep. 2018;8:15873. doi: 10.1038/s41598-018-33881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Steudel C., Wermke M., Schaich M., Schäkel U., Illmer T., Ehninger G., Thiede C. Comparative analysis of MLL partial tandem duplication and FLT3 internal tandem duplication mutations in 956 adult patients with acute myeloid leukemia. Genes Chromosomes Cancer. 2003;37:237–251. doi: 10.1002/gcc.10219. [DOI] [PubMed] [Google Scholar]
- 109.Whitman S.P., Liu S., Vukosavljevic T., Rush L.J., Yu L., Liu C., Klisovic M.I., Maharry K., Guimond M., Strout M.P., et al. The MLL partial tandem duplication: Evidence for recessive gain-of-function in acute myeloid leukemia identifies a novel patient subgroup for molecular-targeted therapy. Blood. 2005;106:345–352. doi: 10.1182/blood-2005-01-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Döhner K., Tobis K., Ulrich R., Fröhling S., Benner A., Schlenk R.F., Döhner H. Prognostic significance of partial tandem duplications of the MLL gene in adult patients 16 to 60 years old with acute myeloid leukemia and normal cytogenetics: A study of the Acute Myeloid Leukemia Study Group Ulm. J. Clin. Oncol. 2002;20:3254–3261. doi: 10.1200/JCO.2002.09.088. [DOI] [PubMed] [Google Scholar]
- 111.Strout M.P., Marcucci G., Bloomfield C.D., Caligiuri M.A. The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2390–2395. doi: 10.1073/pnas.95.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schnittger S., Kinkelin U., Schoch C., Heinecke A., Haase D., Haferlach T., Büchner T., Wörmann B., Hiddemann W., Griesinger F. Screening for MLL tandem duplication in 387 unselected patients with AML identify a prognostically unfavorable subset of AML. Leukemia. 2000;14:796–804. doi: 10.1038/sj.leu.2401773. [DOI] [PubMed] [Google Scholar]
- 113.Sarah M.C., Rajan D., Patrick W.B., Lina S. Partial tandem duplication of KMT2A (MLL) may predict a subset of myelodysplastic syndrome with unique characteristics and poor outcome. Haematologica. 2018;103:e131–e134. doi: 10.3324/haematol.2017.185249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dicker F., Haferlach C., Sundermann J., Wendland N., Weiss T., Kern W., Haferlach T., Schnittger S. Mutation analysis for RUNX1, MLL-PTD, FLT3-ITD, NPM1 and NRAS in 269 patients with MDS or secondary AML. Leukemia. 2010;24:1528–1532. doi: 10.1038/leu.2010.124. [DOI] [PubMed] [Google Scholar]
- 115.Iacobucci I., Wen J., Meggendorfer M., Choi J.K., Shi L., Pounds S.B., Carmichael C.L., Masih K.E., Morris S.M., Lindsley R.C., et al. Genomic subtyping and therapeutic targeting of acute erythroleukemia. Nat. Genet. 2019;51:694–704. doi: 10.1038/s41588-019-0375-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dorrance A.M., Liu S., Chong A., Pulley B., Nemer D., Guimond M., Yuan W., Chang D., Whitman S.P., Marcucci G., et al. The Mll partial tandem duplication: Differential, tissue-specific activity in the presence or absence of the wild-type allele. Blood. 2008;112:2508–2511. doi: 10.1182/blood-2008-01-134338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dorrance A.M., Liu S., Yuan W., Becknell B., Arnoczky K.J., Guimond M., Strout M.P., Feng L., Nakamura T., Yu L., et al. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J. Clin. Investig. 2006;116:2707–2716. doi: 10.1172/JCI25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Whitman S.P., Hackanson B., Liyanarachchi S., Liu S., Rush L.J., Maharry K., Margeson D., Davuluri R., Wen J., Witte T., et al. DNA hypermethylation and epigenetic silencing of the tumor suppressor gene, SLC5A8, in acute myeloid leukemia with the MLL partial tandem duplication. Blood. 2008;112:2013–2016. doi: 10.1182/blood-2008-01-128595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang Y., Yan X., Sashida G., Zhao X., Rao Y., Goyama S., Whitman S.P., Zorko N., Bernot K., Conway R.M., et al. Stress hematopoiesis reveals abnormal control of self-renewal, lineage bias, and myeloid differentiation in Mll partial tandem duplication (Mll-PTD) hematopoietic stem/progenitor cells. Blood. 2012;120:1118–1129. doi: 10.1182/blood-2012-02-412379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Caprioli C., Lussana F., Salmoiraghi S., Cavagna R., Buklijas K., Elidi L., Zanghi P., Michelato A., Delaini F., Oldani E., et al. Clinical significance of chromatin-spliceosome acute myeloid leukemia: A report from the Northern Italy Leukemia Group (NILG) randomized trial 02/06. Haematologica. 2020 doi: 10.3324/haematol.2020.252825. [DOI] [PMC free article] [PubMed] [Google Scholar]