Abstract
The maintenance of cellular homeostasis involves the participation of multiple organelles. These organelles are associated in space and time, and either cooperate or antagonize each other with regards to cell function. Crosstalk between organelles has become a significant topic in research over recent decades. We believe that signal transduction between organelles, especially the endoplasmic reticulum (ER) and mitochondria, is a factor that can influence the cell fate. As the cellular center for protein folding and modification, the endoplasmic reticulum can influence a range of physiological processes by regulating the quantity and quality of proteins. Mitochondria, as the cellular “energy factory,” are also involved in cell death processes. Some researchers regard the ER as the sensor of cellular stress and the mitochondria as an important actuator of the stress response. The scientific community now believe that bidirectional communication between the ER and the mitochondria can influence cell death. Recent studies revealed that the death signals can shuttle between the two organelles. Mitochondria-associated membranes (MAMs) play a vital role in the complex crosstalk between the ER and mitochondria. MAMs are known to play an important role in lipid synthesis, the regulation of Ca2+ homeostasis, the coordination of ER-mitochondrial function, and the transduction of death signals between the ER and the mitochondria. Clarifying the structure and function of MAMs will provide new concepts for studying the pathological mechanisms associated with neurodegenerative diseases, aging, and cancers. Here, we review the recent studies of the structure and function of MAMs and its roles involved in cell death, especially in apoptosis.
Keywords: endoplasmic reticulum, mitochondria, MAMs, Ca2+, apoptosis
1. Introduction
Apoptosis is a programmed form of cell death and can occur via three main pathways. The first pathway is the intrinsic apoptosis pathway; initiated by a range of factors, including the effects of growth factors or hormones, radiation, or cytotoxins [1]. This process involves the enhancement of pro-apoptotic signals and the weakening of anti-apoptotic signals. An imbalance in the regulation of apoptosis ultimately leads to changes in the permeability of the mitochondrial outer membrane and the release of pro-apoptotic substances from the mitochondria. These pro-apoptotic substances promote apoptosis by activating the apoptotic executive protein caspase-9, inhibiting IAPs or via the direct cleavage of DNA [2,3,4,5,6,7]. The second pathway is the extrinsic apoptosis pathway in which apoptosis is activated by the binding of specific ligands such as FasL to transmembrane receptors which contain “death domain” such as FasR. Activated death receptors then recruit adaptor proteins in the cytoplasm to assemble an apoptosis-inducing signal complex, which then activates caspase-8 to initiate apoptosis [8]. Caspase-8 can also cleave Bid to initiate the intrinsic apoptotic pathway, which plays an important role in the process of apoptotic signal amplification [9,10]. The third pathway is the perforin/granzyme pathway in which cytotoxic T cells or NK cells induce target cell apoptosis by secreting granules containing perforin or granzymes [11].
Moreover, the Bcl2 family plays a vital role in cell apoptosis. The Bcl2 family consists of 25 members, and they can be divided into pro-apoptotic proteins (Bax, Bak, etc.) and anti-apoptotic proteins (Bcl2, Bcl-XL, Mcl-1, etc.) according to their different functions. Furthermore, pro-apoptotic proteins can be divided into pro-apoptotic proteins with multiple domains and BH3-only proteins, and BH3-only proteins also can be divided into “activator” (Bim, tBid, etc.) and “sensitizer” (Bad, Bik, etc.) based on their specific mechanism of action [12]. The activated pro-apoptotic members can assemble on the outer mitochondrial membrane and change the permeability of the outer mitochondrial membrane, and promote the release of cytochrome c, AIF, Smac/Diablo and other apoptosis-inducing factors from the mitochondria [13]. Anti-apoptotic members mainly antagonize the effects of pro-apoptotic ones through protein–protein interactions to maintain the integrity of the mitochondrial outer membrane. The “activator” BH3-only members can directly activate the pro-apoptotic proteins and promote the occurrence of apoptosis, while the “sensitizer” BH3-only members can interact with the anti-apoptotic in a protein–protein interaction way to relieve the effect of the pro-apoptotic proteins [14].
As it is mentioned above, the mitochondria play a central role in the cell apoptosis, and the crosstalk between mitochondria and other organelles may impact the apoptosis process. Recent studies revealed that the communication between ER and mitochondria can influence the cell apoptosis, thus affecting the cell fate.
The ER is a key organelle that plays a crucial role in Ca2+ storage, lipid synthesis, protein folding, and assembly [15,16,17,18]. Mitochondria are the “energy factories” of eukaryotic cells, and provide energy to drive the physiological processes of cells; they also play a key role in the process of apoptosis [19,20,21]. The ER and mitochondria are independent of each other but are also closely associated in structure and function. The first spatial connection between the ER and the mitochondria was reported in the 1950s following a study of hepatocytes by transmission electron microscopy [22,23]. In 2006, an electron tomography study further confirmed the complex relationship between the ER and the mitochondria [24]. It is now believed that the mitochondrial surface juxtaposed to the ER in mammalian cells is up to 5–20% due to different cell types [25,26]. Based on this close structural connection, the ER is able to respond to a variety of stress stimuli and can transmit these stress signals to the mitochondria [27,28], thereby initiating the mitochondrial stress response. Similarly, the mitochondria can transmit signals to the ER, thus ensuring the efficient execution of compensatory responses or cell death events. Due to the special function of these precise structural associations, this biological system is usually investigated as a relatively independent sub-organelle structure referred to as a “mitochondrial-related membrane structure.”
2. The Structural Characteristics of MAMs
The structure of MAMs does not remain constant; rather, the structure of MAMs changes dynamically in response to the cell status. The width of the gap between the ER and the outer mitochondrial membrane varies from 10 to 100 nm [29,30]; the width of this gap is usually 10–15 nm at the smooth endoplasmic reticulum and 20–30 nm at the rough endoplasmic reticulum; these spatial differences may be related to the presence of ribosomes [7,31]. Different proteomic analysis of the structure of MAMs has revealed 991 [32] and 1212 [33] different proteins in MAMs [34]. Mass spectrometry analysis divided these constituent proteins into three categories: Proteins that are specifically present in MAMs; proteins that exist simultaneously in MAMs and other organelle structures; and proteins that only exist temporarily in MAMs [33]. These proteins are involved in a wide range of processes, such as structural maintenance, lipid synthesis, the regulation of Ca2+ homeostasis, mitochondrial dynamics, and apoptosis.
3. The Structure Maintenance of MAMs
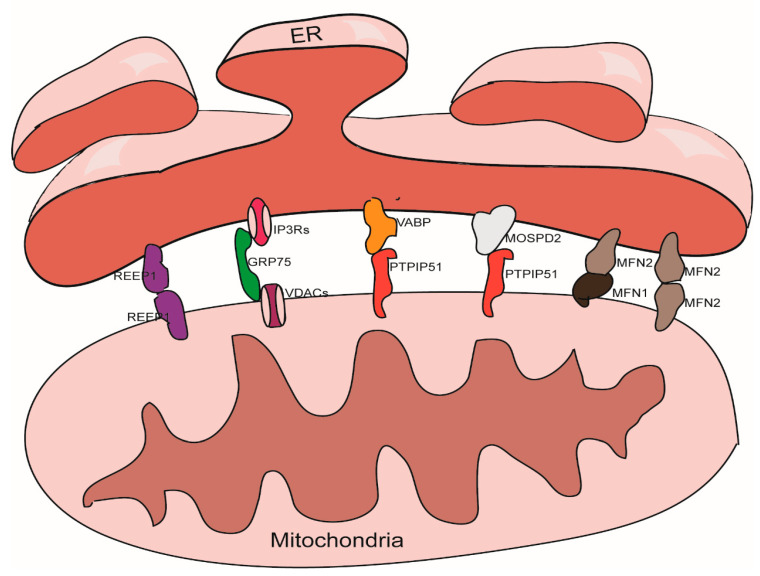
There are thousands of proteins in MAMs; the roles of these proteins are known to vary widely. Some of these proteins play a tethering role in the maintenance of MAMs [31]. According to our understanding, tethering proteins should exhibit certain characteristics. For example, tethering proteins could be (1) proteins or protein complexes that directly participate in the physical connection between the ER and the mitochondria, or (2) interfering proteins or protein complexes that can directly cause changes in the width of gap, the area of contact, or the number of contact sites between the ER and the mitochondria. These proteins and protein complexes are introduced below (Figure 1).
Figure 1.
Tethering proteins that participate in the mitochondria-associated membranes (MAMs) structure maintenance.
3.1. The IP3Rs-Grp75-VDACs Complex
IP3Rs are important Ca2+ outflow channels on the surface of the ER and mediate the release of Ca2+ from the cavity of the ER to the cytoplasm [35,36]. VDACs are ions channel located on the outer membrane of the mitochondria; these mediate the movement of a variety of ions and metabolites in and out of mitochondria, and participate in a range of cellular activities, including apoptosis, metabolism, and the regulation of Ca2+ [37]. IP3Rs and VDACs are connected by Grp75 to maintain the structure of MAMs [38]. The overexpression of VDACs is known to enhance the connection between the ER and the mitochondria and thus improve Ca2+ flux from the ER to the mitochondria [39]; while silencing VDAC1 exhibits a reduction in the connection between Grp75 and IP3R1 indicating a reduction of ER-mitochondria interactions [40]. Cells overexpressing Grp75 showed higher number of IP3R1–VDAC1 interaction sites [41]. Silencing IP3R1 or Grp75 can also reduce the connection between VDAC1 and Grp75 or IP3R1 [42].
3.2. The VAPB-PTPIP51 Complex
VAPB is located in the membrane of the ER and participates to the activation of the IRE1/XBP1 axis in the ER unfolded protein response [43,44]. VAPB can form a complex with the outer mitochondrial membrane protein PTPIP51 and help to maintain the structure of MAMs. A mutant form of VAPB, VAPBP56S, exhibits a stronger affinity for PTPIP51, thereby promoting the transfer of Ca2+ from the ER to the mitochondria; knocking out either of these two genes can reduce the transfer of Ca2+ signals [45]. Other studies have shown that knocking down either of these two proteins will reduce the level of contacts between the ER and the mitochondria [46,47].
3.3. The Mfn1/Mfn2 Complex
In addition to being located in the outer mitochondria membrane and participating to the mitochondrial fusion [48], Mfn2 can also localize on the surface of the ER. Mfn2 participates in the structural maintenance of MAMs by forming homodimers or heterodimers with Mfn1/2 on the outer membrane of the mitochondria. The function of the Mfn1/Mfn2 complex with regards to maintaining the structure of the MAMs was first discovered in 2008 [49]; this role has also been confirmed by several other studies [50,51]. However, some studies have yielded contradictory results [52,53], it is now well established that Mfn2 plays a role in the endoplasmic reticulum stress (ERS) response; the ERS induced by knockdown of Mfn2 can tighten the association between the ER and the mitochondria [54].
3.4. The MOSPD2-PTPIP51 Complex
MOSPD2, another member of VAP family, a protein that locates on the surface of the ER membrane, plays a role in connecting the ER with other membrane structures. It can also bind with proteins containing a small VAP-interacting motif, named FFAT [two phenylalanines (FF) in an acidic track (AT)] via an MSP (Major Sperm Protein domain), such as PTPIP51 on the outer membrane of the mitochondria [55].
3.5. REEP1
REEP1 is a protein that is located in the outer membrane of the ER and the mitochondria. REEP1 helps regulate the morphology of the ER. Studies have shown that REEP1 directly connects the ER and the mitochondria through oligomerization and participates in forming the structure of MAMs. In addition, through bending ER membranes, REEP1 makes it topologically possible for the ER to wrap around the mitochondria, which helps to form MAMs [56].
3.6. Other Proteins Involved in MAMs Maintance
In addition to these tethering proteins, there are some proteins that do not directly participate in the structural maintenance of MAMs. However, these proteins do affect the structure of MAMs via protein–protein interactions (Table 1). In addition to being present in the cytoplasm, α-Synuclein can also be incorporated in MAMs [57]. α-Synuclein can promote the Ca2+ transfer from ER to mitochondria by increasing the ER and mitochondria contacts; and further study showed that the C-terminal of α-Synuclein is essential to tighten the contacts [58]. Some studies revealed that the α-Synuclein existing in MAMs results in the dis-regulation of Ca2+ and lipid metabolism, which promotes substantia nigra pars compacta neurons to die, leading to the progression of PD [59]. In addition to playing an anti-apoptotic role in cells and participating in mitochondrial dynamics, DJ-1 can still exist in the MAMs, thus enhancing the connection between the ER and the mitochondria and the crosstalk between the two organelles; this effect may be related to P53 to some extent (an antagonistic relationship) [60]. Existing studies suggest that DJ-1 can bind directly to the IP3R-Grp75-VDAC complex and affect its stability. The knockout of DJ-1 resulted in the aggregation of IP3R3 in MAMs and a reduction in the formation of the IP3Rs-Grp75-VDACs complex; it is possible that this is related to the pathophysiological process of obesity [61]. Although the precise mechanism remains obscure, it has been ascertained that DJ-1 can affect the structural stability of MAMs. This also implies that MAMs may play a role in the pathogenesis of Parkinson’s syndrome. TDP-43 and FUS are proteins that are related to ALS/FTD and can activate GSK-3b by down-regulating the phosphorylation levels of serine 9 by GSK-3b. Once activated, GSK-3b can reduce the connections between VABP and PTPIP51, thereby detaching the ER from the mitochondria [47,62]. PDK4 can directly interact with the IP3Rs-Grp75-VDACs complex in MAMs and may promote the formation of this complex by regulating phosphorylation, thus increasing the area of contacts between the ER and the mitochondria [42]. In addition to participating in the post-transcriptional modification of proteins, TG2 can also be incorporated in MAMs and act directly on Grp75 to increase the number of ER-mitochondrial contacts and thus participate in the structural maintenance of MAMs [63]. The precise function of TpMs (a type of keratin binding protein that is partly located in the mitochondria) remains unclear although data indicates that this protein can negatively regulate the ER-mitochondria connections in a Mfn2-dependent manner [64]. It is generally believed that CypD, a protein located in the mitochondrial matrix, can also be incorporated in MAMs, and directly act with the IP3Rs-Grp75-VDACs complex to regulate the stability of this complex. Inhibiting the function of CypD can down-regulate the binding of Grp75 with IP3Rs and VDACs, affecting the transfer of Ca2+ between the two organelles [65]. FUNDC1 is known for maintaining the stability of IP3R2 in MAMs by direct binding, and it enhances the level of contacts and the communication of Ca2+ between the ER and the mitochondria [66]. Presenilin-2 can also promote the connection and the transfer of Ca2+ signals between the ER and the mitochondria in the presence of Mfn-2; these findings were confirmed by overexpression and knockdown experiments, which suggested that presenilin-2 works with the Mfn1/Mfn2 complex [67]. FATE1 can reduce the level of contacts between the ER and the mitochondria and downregulate the transfer of Ca2+ with an impaired sensitivity to Ca2+-related apoptosis [68]. In addition to participating in the morphological regulation of ER, NogoB can increase the gap width of MAMs and affect their function [69]. PERK, which plays an important role in ERS, can increase the level of connectivity between the ER and the mitochondria by interacting with Mfn2, and thus promote the transduction of ERS signals to the mitochondria [70,71]. Although these proteins are not considered to be directly involved in maintaining the structure of MAMs, they still attract research attention due to their specific regulatory effects on the structure of MAMs and their involvement in the pathological processes underlying many neurodegenerative diseases.
Table 1.
Proteins that play roles in MAMs structure and function regulation.
| Functions in MAMs | Proteins | Other Functions |
|---|---|---|
| α-Synuclein | Unknown | |
| CypD | Regulation of MPTP [121] | |
| DJ-1 | Antioxidant stress, chaperon activity, transcriptional regulation, degradation of proteins [122] | |
| FATE1 | Tolerance to cellular stress [123] | |
| FUNDC1 | Acts as mitophagy receptor [124] | |
| Structure maintenance | NogoB | Regulation of ER morphology [69] |
| PERK | Participating in ERS [125] | |
| PDK4 | Regulation of cellular metabolism and mitochondrial function [126] | |
| Presenilin-2 | Involved in cell adhesion, apoptosis and several cell-signaling processes [127] | |
| TDP-43 and FUS | RNA processing and transportation [128] | |
| TG2 | Modification of proteins [63] | |
| TpMs | Unknown | |
| Akt | Regulation of cell growth and differentiation [129] | |
| Bcl2 | Regulation of apoptosis [13] | |
| Bcl-xl | Regulation of apoptosis [13] | |
| Bok | Regulation of apoptosis and mitochondrial fusion/fission [130] | |
| BRCA1 | A tumor suppressor, repair of DNA damage [107,131] | |
| Caveolin-1 | Participating in the regulation of the cell cycle and cellular senescence, proliferation and invasion, cell death as well as membrane composition, lipid homeostasis and metabolism [132] | |
| ERO1α | Protein folding [133] | |
| MCL1 | Regulation of apoptosis [13] | |
| Regulating function of IP3Rs |
mTORC2 | Participating in multiple cellular processes such as proliferation, apoptosis, and differentiation [134] |
| NCS1 and WFS1 | Contributing to the maintenance of intracellular calcium homeostasis and regulation of calcium-dependent signaling pathways [135]; regulation of ER stress signaling [108] | |
| PML | Functions as a tumor suppressor and also involved in multiple cellular activities [136] | |
| PTEN | Tumor suppressor and metabolic regulator [137] | |
| Ras | Participating in proliferation, differentiation, apoptosis, senescence, and metabolism [138] | |
| Sig1-R | Regulation of ER stress, function of mitochondria, and oxidative stress, etc., [139] | |
| Tespa1 | signaling molecule in thymocyte development [140] | |
| Bcl2 | Regulation of apoptosis [13] | |
| CHOP | Participating in ER stress and apoptosis regulation [141] | |
| Regulating function of SERCAs | ERO1α and SEPN1 | Regulation of protein folding and secretion and inhibiting apoptosis, and regulates tumor progression [142]; regulation of oxidative stress [143] |
| P53 | Multiple roles in cellular activities [144] | |
| PMX1 | Participating in protein folding [116,145] | |
| S1Ts | Variants of SERCA1 [119,120] |
4. MAMs and Cell Death
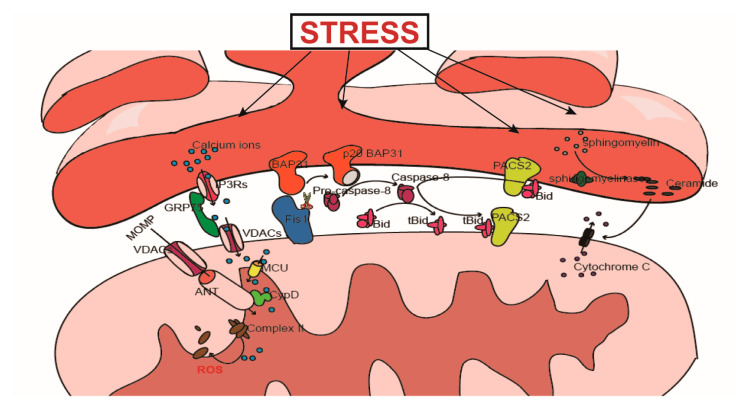
MAMs plays a crucial role in cell homeostasis as mentioned above, besides, recent studies revealed that MAMs can also influence the cell death events. The death signals in MAMs attend in multiple forms, such as the transfer of Ca2+ from the ER to the mitochondria, regulation of protein–protein interactions, the translocation of molecules or the control of lipid metabolism. These processes are described in detail in the following sections (Figure 2).
Figure 2.
Under stress conditions, endoplasmic reticulum (ER) can act as a sensor and respond immediately. Meanwhile, if the stress intensity exceeds the adaptability of the ER, it will transmit death signals to mitochondria to initiate death events. The transduction can go through some pathways as follows: 1.ER perfuse Ca2+ into mitochondria and massive Ca2+ in mitochondria works as a death signal initiating cell death; 2. translocation of PACS1 from ER to mitochondria, which is along with the activation and translocation bid; 3. ceramide synthesis and accumulation causing the mitochondrial outer membrane permeabilization (MOMP), which results in the release of cytochrome c or other pro-apoptotic substances in inter membranes space. Also, as a molecule downstream of mitochondrial apoptosis, Fis1 can transmit apoptosis signal back to ER by cleaving BAP31. The ER-mitochondria-ER amplification loop of apoptotic signals can help to coordinate the death events between the two organelles.
4.1. Ca2+-Mediated Signal Transduction and Cell Death
4.1.1. The Physiological Role of Ca2+
Ca2+ is an important vector for crosstalk between the ER and the mitochondria [72]. The ER, as the most important cellular reservoir of Ca2+, maintains proper Ca2+ level by Ca2+ pumps SERCAs [73]. SERCAs have varied isoforms, among which the ubiquitous SERCA2b shows the highest affinity to Ca2+ uptake from the cytoplasm [26]. ER releases Ca2+ into the cytoplasm via RyRs and IP3Rs. The IP3Rs are more ubiquitously expressed while RyRs mostly expressed in skeletal muscle, heart and brain [74]. IP3Rs have three isoforms and all of them can be activated by IP3, Ca2+, Ca2+-binding proteins, ATP, thiol modification and phosphorylation [75]. Although cytoplasmic Ca2+ can almost pass freely through the VDACs of the outer mitochondrial membrane, the MCU located in the inner mitochondrial membrane has a weak affinity for Ca2+ (Kd 15–20 µM), meanwhile, the concentration of Ca2+ in the cytoplasm fluctuates between 50 and 100 nM [16,76]. The mechanism responsible for the uptake of cytoplasmic Ca2+ by the mitochondria has been debated by the academic community for many years. Many models, such as Ca2+ microdomain hypothesis, was proposed to explain the Ca2+ uptake mechanism [25,77,78]. However, with the deepen understand of the MAMs, especially the IP3Rs-Grp75-VDACs complex, advancement was made with this respect. When the ER calcium channels are opened, a hotspot of calcium will be produced in microdomain between the ER and the mitochondria; this makes it possible for the mitochondria to uptake Ca2+ [16]. In this process, the IP3Rs-Grp75-VDACs complex plays a crucial role, though there exist some proteins, such as the MiCU family, MCUR1 and SLC25A23, play regulatory role in the work of MCU, which may affect the Ca2+ uptake efficiency, and the elegant mechanism can be seen in the review by Belosludtsev KN et al. [79].
The presence of Ca2+ in the mitochondrial matrix can play a variety of roles. First, it can promote the efficiency of the tricarboxylic acid cycle and the electron transport chain (ETC). Ca2+ is known to increase the activities of pyruvate dehydrogenase, ketoglutarate dehydrogenase, and isocitrate dehydrogenase, in the tricarboxylic acid cycle [30] and can directly stimulate ETC-related complexes, thus improving the activity of ATP synthase and promoting the production of ATP [80,81]. It has also been reported that Ca2+ can affect metabolism by regulating the activity of glucose transporters [82]. Second, long-term and high-level Ca2+ overload can induce cell death. Ca2+ can induce the opening of mPTP, which may result in necrosis [83], or the resultant mitochondrial swelling and outer membrane rupture in limited MPT (the open of mPTP does not involve the entire mitochondrial network, and it may appear as “flickering” mode in a small portion of mitochondria) can result in the release of pro-apoptotic substances such as cytochrome c and AIF [84,85,86]. An excessive concentration of Ca2+ in the mitochondria can bind to cardiolipin in the inner membrane of mitochondria to promote the disintegration of respiratory chain complex II, thus leading to the release of multiple subunits; this also induces the production of large amounts of ROS, thereby inducing apoptosis [87]. Like a double-edged sword, Ca2+ can both promote metabolism and induce cell apoptosis. The flux of Ca2+ determines which cellular events occur, thus highlighting the importance of MAMs structure to cells.
4.1.2. The Regulatory Effect of MAMs on Ca2+ Transfer
The IP3Rs-Grp75-VDACs complex is the basis for Ca2+ regulation in MAMs; the importance of this complex is the maintenance of physical contacts between the ER and the mitochondria, while enables the combination of IP3Rs and VDACs to overcome the MCU’s low affinity for Ca2+, thus increasing the sensitivity and efficiency of Ca2+ delivery. Also, another MAMs-presented protein, SERCAs, is fundamental to regulate the Ca2+ level in ER, whose activity can affect the Ca2+ flux pass through MAMs.
The IP3Rs-Grp75-VDACs Complex
A variety of proteins regulate the transfer of Ca2+ between the ER and the mitochondria by interacting with the IP3Rs-Grp75-VDACs complex. mTORC2 can accumulate in MAMs under the stimulation of various growth factors; and mTORC2 in MAMs can phosphorylate Akt and elevate its activity. Moreover, activated Akt can elevate the phosphorylation level of IP3Rs, thus reducing the release of Ca2+ in the ER and antagonizing cell apoptosis [88,89,90,91,92]. Antagonistically, PML can affect the phosphorylation level and activity of Akt by elevating the activity of phosphatase PP2A to regulate the function of IP3Rs [93]. As an important protein encoded by tumor suppressor genes, PTEN can counteract Akt and promote Ca2+ transfer from the ER to the mitochondria, thus increasing the sensitivity of cells to apoptotic stimuli [94].
As an important player that can affect cell fate, the functional role of the Bcl2 family may depend on the interaction between Bcl2s and MAMs, at least to a certain extent. Bcl2 and Bcl-xl can directly bind to the central regulatory domain of IP3Rs or indirectly affect the phosphorylation level of IP3Rs to inhibit the function of IP3Rs; they can also interact with VDAC1 to reduce the uptake of Ca2+ by the mitochondria [13,95,96]. In addition to directly inhibiting the activity of IP3Rs [97], Mcl-1, which is also a member of Bcl2 family, can enhance cellular metabolism by interacting with VDACs to promote the transfer of Ca2+ [98]. Bok, a pro-apoptotic member of the Bcl2 family, can interact with IP3Rs and change the ratio of IP3R1 and IP3R2 by cleavage; this process is carried out by caspase-3 and results in an increased Ca2+ transfer and increased cellular sensitivity to apoptosis [99,100]. More elegant details in this field can be seen in the review by Lewis A, et al. [101].
Sig-1R plays a role in the pathogenesis of a variety of neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s syndrome, and the lateral sclerosis of associated with muscular dystrophy [102]. Sig-1R is an important component of MAMs. A previous study showed that Sig-1R can be separated from Bip and combines with IP3Rs under conditions of mitochondrial stress, thus reducing the degradation of IP3Rs and promoting the transfer of Ca2+ to the mitochondria [103]. Caveolin-1 is known for that it helps to order the lipid bilayers organization [104]; moreover, it can directly interact with IP3Rs to promote the release of Ca2+ [105], while Ras can affect the subcellular distribution of Caveolin-1, thereby reducing the transfer of Ca2+ [106]. During the process of apoptosis, the tumor suppressor BRCA1 can be recruited to the surface of the ER in an IP3Rs-dependent manner and bind directly to IP3R1 to increase its sensitivity to IP3 and promote the transfer of Ca2+ to the mitochondria [107]. WFS1, NCS1, and IP3Rs, can form a complex to increase the connectivity between the ER and the mitochondria by enhancing Ca2+ transfer [108]. Research has shown that the mutation of WFS1 is responsible for a variety of abnormalities in the neurological and endocrine systems [109]. Tespa1 can be incorporated in MAMs and participate in the regulation of Ca2+ transfer by way of forming a complex with IP3Rs and Grp75. The knockdown of Tespa1 can down-regulate the level of Ca2+ in the mitochondria and cytoplasm [110,111]. ERO1α is also known to increase the activity of IP3Rs and promote apoptosis [112].
SERCAs
As the only channel responsible for the uptake of Ca2+ in the ER so far, SERCAs can also aggregate in MAMs. Furthermore, a variety of proteins in MAMs can regulate the function of SERCAs and affect the transfer of calcium signals. When incorporated in MAMs, P53 can change the redox state of SERCAs and promote its functional activity, thereby elevating the level of Ca2+ in the ER; this increases the flux of Ca2+ to the mitochondria, and promotes the occurrence of apoptosis [113,114]. TMX1 can negatively regulate the function of SERCA2b, and tumor cells with low expression levels of TMX1 exhibit higher levels of Ca2+ in the ER [115,116]. The anti-apoptotic protein Bcl-2 can directly bind to SERCAs to change its conformation, thereby down-regulating its functional activity and inhibiting the enrichment process of Ca2+ in the ER [117]. SEPN1 and ERO1α antagonize each other and work by regulating the redox state of SERCA2. SEPN1 can down-regulate the cysteine oxidation level of SERCA2 to maintain the stability of SERCA2 function [118].
In addition, 25 transcriptional variants of SERCA1 were detected in normal liver cells, among which 8 clones were found to be characterized by exon 11 splicing, named S1Ts. Further study revealed that S1Ts are expressed in different human tissues, such as adult pancreas, liver, kidney, lung, and placenta and fetal kidney, liver, brain, thymus [119]. S1Ts, which are localized in MAMs, can be induced by ER stress through PERK-eIF2α-ATF4 pathway and whose induction triggers Ca2+ leak from ER by forming homodimers in the ER membrane, which promotes the transfer of Ca2+ to mitochondria [119,120].
MCU
Although MCU and its regulatory proteins play crucial role in the uptake of Ca2+ by the mitochondria [79], there is no report showing specific accumulation of MCU at MAMs structure. However, MAMs can still influence the efficiency of MCU in Ca2+ uptake. Since the transport of Ca2+ by MCU is highly dependent on the “hot spot” effect of ER Ca2+ release, the MAMs proteins that can influence the spatial connection between the ER and the mitochondria (as detailed above) may influence the Ca2+ delivery efficiency by MCU indirectly (Table 1).
4.2. PACS2 Participates in the Transduction of Apoptosis Signals from the ER to the Mitochondria
PACS2 is a sorting protein located in the ER membrane which participates in the regulation of ER homeostasis, lipid synthesis, and the structural connection between the ER and the mitochondria. In addition, PACS2 mediates the process of apoptotic signal transduction from the ER to the mitochondria. High levels of ERS can cause translocation of the full-length Bid-bound PACS2 from the ER to the mitochondria; Bid is then cleaved on the surface of the mitochondria by caspase-8. Later, Bid interacts with Bax/Bak to promote permeability of the outer membrane of the mitochondria and the release of cytochrome c, thus initiating apoptosis [21].
4.3. The Role of LIPID Metabolism in the Transduction of Apoptosis Signals
MAMs plays a central role in lipid metabolism. A variety of lipids are synthesized in MAMs; some enzymes involved in the synthesis of triglyceride, ceramide, and sterol (fatty acid CoA ligase (ACS) 1/4) [146], acyl-coenzyme A: cholesterol acyltransferase-1 (ACAT1/SOAT1) [147] are known to exist only in MAMs [148]. Several lipid metabolites can affect cell fate. Of these, ceramide is the most typical. Under normal circumstances, ceramide is synthesized by the ceramide synthase pathway. However, under stress conditions (e.g., heat shock, TNF-α, Fas, chemotherapeutics, toxins, radiation and other factors) ceramide can be synthesized rapidly from sphingomyelin in the nerve sheath phospholipase [149]. The accumulation of ceramide can not only regulate and interact (directly or indirectly) with a variety of molecules involved in the transduction of apoptotic signals transduction, such as protein phosphatase 1A/2A, protein kinase C, and NF-κB, ras [150,151,152,153,154], a significant accumulation of ceramide can result in the formation of pores on the outer mitochondrial membrane, induce the release of pro-apoptotic substances, such as cytochrome c, in the intermembrane space of mitochondria [155,156], and transfer stress signals from the ER to the mitochondria. Inhibiting the activity of ACS1/4 can reduce the synthesis of ceramide and reduce the occurrence of apoptosis [157].
4.4. The Fis1-BAP31 Complex Is Involved in the Transduction of Apoptosis Signals
4.4.1. The Relationship between Fis1 and Apoptosis
As the receptor for Drp1 during mitochondrial fission, Fis1 not only plays an important role in mitochondrial fission, but also participates in the process of apoptosis. First, the downregulation of Fis1 can reduce the release of cytochrome c in apoptotic cells and can inhibit the translocation of pro-apoptotic Bcl2 family members to the mitochondria [158]. Second, the overexpression of Fis1 can induce cell apoptosis via a Bax/Bak-independent process [159,160]. Although many studies have found that mitochondrial fission is often accompanied by the release of mitochondrial apoptosis-related substances, it is currently believed that when considering the relationship between mitochondrial fission and apoptosis, mitochondrial fission is the result of cell apoptosis rather than the cause. The results of multiple time-lapse microscopy experiments have confirmed that the release of cytochrome c from mitochondria when stimulated with apoptosis-inducing drugs occurs earlier than mitochondrial fission, and that the release of cytochrome c can also occur in both reticulated and tubular mitochondria [161,162]. Therefore, Fis1 is involved in the process of apoptosis and plays a specific role that occurs downstream of mitochondrial apoptosis.
4.4.2. Fis1-BAP31 Participates in Apoptosis Signal Transduction from the Mitochondria to the ER
As a participant in the process of apoptosis, Fis1 can connect with BAP31 in MAMs to transmit mitochondrial apoptotic signals to the ER. BAP31 is an important chaperone protein on the ER membrane and is involved in the degradation of misfolded proteins and apoptosis within the ERS pathway. When Fis1 binds to BAP31, it can cleave BAP31 to produce p20 BAP31; as a pro-apoptotic protein, p20 BAP31 can convert procaspase-8 into an functional form that can truncate Bid and initiate apoptosis [21,163]. The activation of p20 BAP31 can also promote the transfer of Ca2+ from the ER to the mitochondria, thus showing that apoptosis signals can return back to the mitochondria [164,165], thus forming an amplification loop of apoptotic signals between the ER and the mitochondria that can help to coordinate the functions of these two organelles.
5. Methods of Detection
5.1. Fluorescence Microscopy
Fluorescence microscopy provides a preliminary solution for studying the structure of MAMs. Some researchers have used the co-localization of markers for the ER and mitochondria to study the connectivity between these two organelles [49,53]. However, in confocal fluorescence microscopy, the resolution of the z-axis is only 700 nm. Even though the most advanced microscopes can reach 300–400 nm [166], this is still not sufficient to measure the spatial gaps in MAMs (usually less than 100 nm). This means that results obtained via fluorescence microscopy may be questionable, although fluorescence microscopy still has certain advantages. First, fluorescence microscopy can observe living cells, making it possible to study the dynamic structure of MAMs [167]; second, with the application of a variety of fluorescent labels and proteins, the study of MAMs can be more targeted and efficient [38]. Consequently, fluorescence microscopy remains an indispensable method to study MAMs.
5.2. Transmission Electron Microscopy
Transmission electron microscopy is an irreplaceable tool for studying the structure of MAMs. The high-resolution imaging of transmission electron microscopy makes it possible to quantify the structure of MAMs [168]. At the same time, three-dimensional reconstruction technology after electron tomography can rebuild the structure of MAMs between the ER and mitochondria, making it possible to study the spatial morphology of MAMs [169,170,171]. This can provide a new perspective in the study of MAMs besides gap measurement and contact point quantification [144,172]. In addition, the combination of fluorescence microscopy and electron microscopy technology can be highly complementary and allow the study of both structure and function [159].
5.3. Gradient Centrifugation
Gradient centrifugation is a classic method for studying MAMs [173]. In combination with methods such as immunoblotting, immunofluorescence, mass spectrometry, and proteomics, the molecular composition of MAMs can be analyzed qualitatively and quantitatively [33]. In this process, gradient centrifugation is an important means to separate the structure of MAMs. However, there are also reports describing the use of biotin to separate and purify the structure and composition of MAMs and thus allow the qualitative and quantitative investigation of the composition of MAMs [174,175].
5.4. The Functional Evaluation of MAMs
The functional evaluation of MAMs can indirectly reflect the changes in the level of contact between the ER and the mitochondria. The synthesis of phospholipids and their transportation between the ER and the mitochondria, or the flux of Ca2+ transfer from the ER to the mitochondria, are usually used as indicators to reflect changes in the level of connectivity between the ER and the mitochondria [176,177].
6. Conclusions
As the most direct communicating medium between the ER and the mitochondria, MAMs play an important role in coordinating the multiple array of functions carried out by these two organelles, particularly the integration of apoptotic signals. The ER is the main site for intracellular protein modification, Ca2+ storage, and lipid synthesis. It is also evident that the ER is more susceptible to various stresses, and therefore plays a key role as a stress sensor and provides an initial response to stress. Mitochondria play an irreplaceable role in important cellular processes such as energy metabolism and apoptosis; consequently, it is logical that the mitochondria work downstream of the stress response. MAMs are located directly between these two organelles and are tightly involved in the transduction of stress signals from the ER to the mitochondria; in some cases, they transmit apoptotic signals back to the ER. This not only ensures the complementarity of the functions between the two organelles, but also amplifies apoptotic signals between the two organelles, thus promoting coordinated functional responses. Clarifying this process will provide a new perspective for the study of pathological mechanisms, such as tumors, neurodegenerative diseases, and aging.
Although some progress has been made in the study of MAMs over recent years, there are still some uncertainties that need to be addressed. First, in terms of structure, some proteins are directly involved in the maintenance of MAMs structure, while others work as regulators for structural maintenance. However, little is known about such proteins or how they can be delineated between the two organelles with respect to functionality. Second, in terms of function, especially in the regulation of Ca2+ transfer, the width of the gap between the ER and the mitochondria plays an important role. However, interfering SERCAs or IP3Rs can also affects the transfer of Ca2+. It is still not clear whether the proteins that can affect Ca2+ transfer in MAMs work by changing the tightness of the gap within MAMs or by disturbing the function of SERCA/IP3Rs. These unresolved issues will be an important direction for future research.
Acknowledgments
The authors would like to thank the Department of Pathophysiology, Jilin University for the generous support to the research that has been conducted.
Abbreviations
| ACAT1/SOAT1 | Acyl-Coenzyme A: Cholesterol Acyltransferase-1 |
| ACS | Fatty acid CoA ligase |
| AD | Alzheimer’s disease |
| AIF | Apoptosis inducing factor |
| Akt | Serine/threonine-protein kinases |
| ALS | Amyotrophic lateral sclerosis |
| ATAD3A | ATPase family AAA domain-containing protein 3A |
| ATF4 | Activating Transcription Factor 4 |
| BAP31 | B-cell receptor associated protein 31 |
| Bip | Immunoglobulin heavy chain binding protein |
| BRCA1 | Breast cancer susceptibility gene |
| CypD | Cyclophilin D |
| IRE1 | Inositol-requiring enzyme 1 |
| eIF2α | eukaryotic Initiation Factor 2α |
| ER | Endoplasmic reticulum |
| ERO1α | Endoplasmic reticulum oxidoreductase 1-α |
| ERS | Endoplasmic reticulum stress |
| ETC | Electron transport chain |
| FADD | Fas-associating protein with a novel death domain |
| FasL | Fas ligand |
| FATE1 | Fetal and adult testis expressed 1 |
| Fis1 | Mitochondrial fission 1 |
| FTD | Frontotemporal dementia |
| FUNDC1 | FUN14 domain containing 1 |
| FUS | Fused in sarcoma |
| Grp75 | 75 KDa Glucose-regulated protein |
| GSK-3b | Glycogen synthase kinase 3 beta |
| IP3Rs | Inositol1,4,5-trisphosphatereceptors |
| MAMs | Mitochondria associated membranes |
| MCU | Mitochondrial Ca(2+) uniporter |
| Mfn1/2 | Mitofusins 1/2 |
| MOSPD2 | Motile sperm domain-containing protein 2 |
| mPTP | Mitochondrial permeability transition pore |
| mTORC2 | Mammalian target of rapamycin complex2 |
| NCS1 | Nucleobase cation symporter-1 |
| NF-κB | Nuclear factor kappa B |
| PACS2 | Phosphoacidic cluster sorting protein 2 |
| PD | Parkinson’s disease |
| PDK4 | Pyruvate dehydrogenase kinase 4 |
| PERK | Protein kinase R (PKR)-like endoplasmic reticulum kinase |
| PTPIP51 | Protein tyrosine phosphatase interacting protein 51 |
| REEP1 | Receptor expression-enhancing protein 1 |
| ROS | Reactive oxygen species |
| RyRs | Ryanodine receptors |
| SEPN1 | Selenoprotein N |
| SERCAs | Sarco(endo)plasmic reticulum calcium-ATPases |
| Sig-R | ECF sigma factor sigma(R) |
| S1Ts | Exon 4 and/or exon 11 spliced SERCA1 splice variants |
| TDP-43 | TAR DNA binding protein 43 |
| TG2 | Transglutaminase 2 |
| TMX1 | Transmembrane thioredoxin-related protein 1 |
| TNFR1 | Tumor necrosis factor receptor 1 |
| TpMs | Trichoplein keratin filament binding protein |
| TRADD | TNF receptor-associated death domain |
| VABP | Vitamin A binding protein |
| VDAC | Voltage-dependent anion channel |
| WASF3 | Wiskott-Aldridge syndrome family 3 |
| WFS1 | Wolfram syndrome type 1 |
| XBP1 | X-box binding protein 1 |
Author Contributions
N.W.: writing the original draft; C.W.: drawing figures; H.Z.: literature searching; Y.H.: review and editing; B.L.: review and editing; L.S.: review, study design and funding acquisition; Y.G.: conceptualization, project administration, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by: Department of Science and Technology of Jilin Province (20180101136JC), Department of Finance of Jilin Province (2018SCZ030), the Education Department of Jilin Province (JJKH20190005KJ), Development and Reform Commission Engineering Laboratory Project of Jilin Province (2019C031), the Lateral Research Funds of Jilin University (2015377), National Natural Science Foundation of China (81772794, 81472419).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Elmore S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santucci R., Sinibaldi F., Cozza P., Polticelli F., Fiorucci L. Cytochrome c: An extreme multifunctional protein with a key role in cell fate. Int. J. Biol. Macromol. 2019;136:1237–1246. doi: 10.1016/j.ijbiomac.2019.06.180. [DOI] [PubMed] [Google Scholar]
- 3.Korga A., Korobowicz E., Dudka J. Role of mitochondrial protein Smac/Diablo in regulation of apoptotic pathways. Pol. Merkur. Lekarski. 2006;20:573–576. [PubMed] [Google Scholar]
- 4.Vande Walle L., Lamkanfi M., Vandenabeele P. The mitochondrial serine protease HtrA2/Omi: An overview. Cell Death Differ. 2008;15:453–460. doi: 10.1038/sj.cdd.4402291. [DOI] [PubMed] [Google Scholar]
- 5.Bano D., Prehn J.H.M. Apoptosis-Inducing Factor (AIF) in Physiology and Disease: The Tale of a Repented Natural Born Killer. EBioMedicine. 2018;30:29–37. doi: 10.1016/j.ebiom.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Low R.L. Mitochondrial Endonuclease G function in apoptosis and mtDNA metabolism: A historical perspective. Mitochondrion. 2003;2:225–236. doi: 10.1016/S1567-7249(02)00104-6. [DOI] [PubMed] [Google Scholar]
- 7.Larsen B.D., Sorensen C.S. The caspase-activated DNase: Apoptosis and beyond. FEBS J. 2017;284:1160–1170. doi: 10.1111/febs.13970. [DOI] [PubMed] [Google Scholar]
- 8.D’Arcy M.S. Cell death: A review of the major forms of apoptosis, necrosis and autophagy. Cell Biol. Int. 2019;43:582–592. doi: 10.1002/cbin.11137. [DOI] [PubMed] [Google Scholar]
- 9.Kantari C., Walczak H. Caspase-8 and bid: Caught in the act between death receptors and mitochondria. Biochim. Biophys. Acta. 2011;1813:558–563. doi: 10.1016/j.bbamcr.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 10.Huang K., Zhang J., O’Neill K.L., Gurumurthy C.B., Quadros R.M., Tu Y., Luo X. Cleavage by Caspase 8 and Mitochondrial Membrane Association Activate the BH3-only Protein Bid during TRAIL-induced Apoptosis. J. Biol. Chem. 2016;291:11843–11851. doi: 10.1074/jbc.M115.711051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voskoboinik I., Whisstock J.C., Trapani J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015;15:388–400. doi: 10.1038/nri3839. [DOI] [PubMed] [Google Scholar]
- 12.Hassan M., Watari H., AbuAlmaaty A., Ohba Y., Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed. Res. Int. 2014;2014:150845. doi: 10.1155/2014/150845. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Siddiqui W.A., Ahad A., Ahsan H. The mystery of BCL2 family: Bcl-2 proteins and apoptosis: An update. Arch. Toxicol. 2015;89:289–317. doi: 10.1007/s00204-014-1448-7. [DOI] [PubMed] [Google Scholar]
- 14.Kim H., Rafiuddin-Shah M., Tu H.C., Jeffers J.R., Zambetti G.P., Hsieh J.J., Cheng E.H. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 2006;8:1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 15.Schwarz D.S., Blower M.D. The endoplasmic reticulum: Structure, function and response to cellular signaling. Cell. Mol. Life Sci. 2016;73:79–94. doi: 10.1007/s00018-015-2052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braakman I., Hebert D.N. Protein folding in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2013;5:a013201. doi: 10.1101/cshperspect.a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quon E., Sere Y.Y., Chauhan N., Johansen J., Sullivan D.P., Dittman J.S., Rice W.J., Chan R.B., Di Paolo G., Beh C.T., et al. Endoplasmic reticulum-plasma membrane contact sites integrate sterol and phospholipid regulation. PLoS Biol. 2018;16:e2003864. doi: 10.1371/journal.pbio.2003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Islam M.S. Calcium Signaling: From Basic to Bedside. Adv. Exp. Med. Biol. 2020;1131:1–6. doi: 10.1007/978-3-030-12457-1_1. [DOI] [PubMed] [Google Scholar]
- 19.Annesley S.J., Fisher P.R. Mitochondria in Health and Disease. Cells. 2019;8:680. doi: 10.3390/cells8070680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desagher S., Martinou J.C. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 2000;10:369–377. doi: 10.1016/S0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- 21.Galluzzi L., Kepp O., Trojel-Hansen C., Kroemer G. Mitochondrial control of cellular life, stress, and death. Circ. Res. 2012;111:1198–1207. doi: 10.1161/CIRCRESAHA.112.268946. [DOI] [PubMed] [Google Scholar]
- 22.Doghman-Bouguerra M., Lalli E. ER-mitochondria interactions: Both strength and weakness within cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:650–662. doi: 10.1016/j.bbamcr.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Bernhard W., Rouiller C. Close topographical relationship between mitochondria and ergastoplasm of liver cells in a definite phase of cellular activity. J. Biophys. Biochem. Cytol. 1956;2:73–78. doi: 10.1083/jcb.2.4.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mannella C.A. The relevance of mitochondrial membrane topology to mitochondrial function. Biochim. Biophys. Acta. 2006;1762:140–147. doi: 10.1016/j.bbadis.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Rizzuto R., Pinton P., Carrington W., Fay F.S., Fogarty K.E., Lifshitz L.M., Tuft R.A., Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 26.Marchi S., Patergnani S., Missiroli S., Morciano G., Rimessi A., Wieckowski M.R., Giorgi C., Pinton P. Mitochondrial and endoplasmic reticulum calcium homeostasis and cell death. Cell Calcium. 2018;69:62–72. doi: 10.1016/j.ceca.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Iurlaro R., Munoz-Pinedo C. Cell death induced by endoplasmic reticulum stress. FEBS J. 2016;283:2640–2652. doi: 10.1111/febs.13598. [DOI] [PubMed] [Google Scholar]
- 28.Gardner B.M., Pincus D., Gotthardt K., Gallagher C.M., Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb. Perspect. Biol. 2013;5:a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giacomello M., Pellegrini L. The coming of age of the mitochondria-ER contact: A matter of thickness. Cell Death Differ. 2016;23:1417–1427. doi: 10.1038/cdd.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filadi R., Theurey P., Pizzo P. The endoplasmic reticulum-mitochondria coupling in health and disease: Molecules, functions and significance. Cell Calcium. 2017;62:1–15. doi: 10.1016/j.ceca.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Csordas G., Renken C., Varnai P., Walter L., Weaver D., Buttle K.F., Balla T., Mannella C.A., Hajnoczky G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang A., Williamson C.D., Wong D.S., Bullough M.D., Brown K.J., Hathout Y., Colberg-Poley A.M. Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection. Mol. Cell. Proteom. 2011;10:M111-009936. doi: 10.1074/mcp.M111.009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poston C.N., Krishnan S.C., Bazemore-Walker C.R. In-depth proteomic analysis of mammalian mitochondria-associated membranes (MAM) J. Proteom. 2013;79:219–230. doi: 10.1016/j.jprot.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 34.Janikiewicz J., Szymanski J., Malinska D., Patalas-Krawczyk P., Michalska B., Duszynski J., Giorgi C., Bonora M., Dobrzyn A., Wieckowski M.R. Mitochondria-associated membranes in aging and senescence: Structure, function, and dynamics. Cell Death Dis. 2018;9:332. doi: 10.1038/s41419-017-0105-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parys J.B., Vervliet T. New Insights in the IP3 Receptor and Its Regulation. Adv. Exp. Med. Biol. 2020;1131:243–270. doi: 10.1007/978-3-030-12457-1_10. [DOI] [PubMed] [Google Scholar]
- 36.Tada M., Nishizawa M., Onodera O. Roles of inositol 1,4,5-trisphosphate receptors in spinocerebellar ataxias. Neurochem. Int. 2016;94:1–8. doi: 10.1016/j.neuint.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 37.Mazure N.M. VDAC in cancer. Biochim. Biophys. Acta Bioenerg. 2017;1858:665–673. doi: 10.1016/j.bbabio.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Szabadkai G., Bianchi K., Varnai P., De Stefani D., Wieckowski M.R., Cavagna D., Nagy A.I., Balla T., Rizzuto R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 2006;175:901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rapizzi E., Pinton P., Szabadkai G., Wieckowski M.R., Vandecasteele G., Baird G., Tuft R.A., Fogarty K.E., Rizzuto R. Recombinant expression of the voltage-dependent anion channel enhances the transfer of Ca2+ microdomains to mitochondria. J. Cell Biol. 2002;159:613–624. doi: 10.1083/jcb.200205091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tubbs E., Theurey P., Vial G., Bendridi N., Bravard A., Chauvin M.A., Ji-Cao J., Zoulim F., Bartosch B., Ovize M., et al. Mitochondria-associated endoplasmic reticulum membrane (MAM) integrity is required for insulin signaling and is implicated in hepatic insulin resistance. Diabetes. 2014;63:3279–3294. doi: 10.2337/db13-1751. [DOI] [PubMed] [Google Scholar]
- 41.Honrath B., Metz I., Bendridi N., Rieusset J., Culmsee C., Dolga A.M. Glucose-regulated protein 75 determines ER-mitochondrial coupling and sensitivity to oxidative stress in neuronal cells. Cell Death Discov. 2017;3:17076. doi: 10.1038/cddiscovery.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thoudam T., Ha C.M., Leem J., Chanda D., Park J.S., Kim H.J., Jeon J.H., Choi Y.K., Liangpunsakul S., Huh Y.H., et al. PDK4 Augments ER-Mitochondria Contact to Dampen Skeletal Muscle Insulin Signaling during Obesity. Diabetes. 2019;68:571–586. doi: 10.2337/db18-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vinay Kumar C., Kumar K.M., Swetha R., Ramaiah S., Anbarasu A. Protein aggregation due to nsSNP resulting in P56S VABP protein is associated with amyotrophic lateral sclerosis. J. Theor. Biol. 2014;354:72–80. doi: 10.1016/j.jtbi.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 44.Kanekura K., Nishimoto I., Aiso S., Matsuoka M. Characterization of amyotrophic lateral sclerosis-linked P56S mutation of vesicle-associated membrane protein-associated protein B (VAPB/ALS8) J. Biol. Chem. 2006;281:30223–30233. doi: 10.1074/jbc.M605049200. [DOI] [PubMed] [Google Scholar]
- 45.De Vos K.J., Morotz G.M., Stoica R., Tudor E.L., Lau K.F., Ackerley S., Warley A., Shaw C.E., Miller C.C. VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum. Mol. Genet. 2012;21:1299–1311. doi: 10.1093/hmg/ddr559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoica R., De Vos K.J., Paillusson S., Mueller S., Sancho R.M., Lau K.F., Vizcay-Barrena G., Lin W.L., Xu Y.F., Lewis J., et al. ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat. Commun. 2014;5:3996. doi: 10.1038/ncomms4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiao X., Jia S., Ye J., Fang X., Zhang C., Cao Y., Xu C., Zhao L., Zhu Y., Wang L., et al. PTPIP51 regulates mouse cardiac ischemia/reperfusion through mediating the mitochondria-SR junction. Sci. Rep. 2017;7:45379. doi: 10.1038/srep45379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Formosa L.E., Ryan M.T. Mitochondrial fusion: Reaching the end of mitofusin’s tether. J. Cell Biol. 2016;215:597–598. doi: 10.1083/jcb.201611048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Brito O.M., Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 50.Alford S.C., Ding Y., Simmen T., Campbell R.E. Dimerization-dependent green and yellow fluorescent proteins. ACS Synth. Biol. 2012;1:569–575. doi: 10.1021/sb300050j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naon D., Zaninello M., Giacomello M., Varanita T., Grespi F., Lakshminaranayan S., Serafini A., Semenzato M., Herkenne S., Hernandez-Alvarez M.I., et al. Critical reappraisal confirms that Mitofusin 2 is an endoplasmic reticulum-mitochondria tether. Proc. Natl. Acad. Sci. USA. 2016;113:11249–11254. doi: 10.1073/pnas.1606786113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cosson P., Marchetti A., Ravazzola M., Orci L. Mitofusin-2 independent juxtaposition of endoplasmic reticulum and mitochondria: An ultrastructural study. PLoS ONE. 2012;7:e46293. doi: 10.1371/journal.pone.0046293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Filadi R., Greotti E., Turacchio G., Luini A., Pozzan T., Pizzo P. Mitofusin 2 ablation increases endoplasmic reticulum-mitochondria coupling. Proc. Natl. Acad. Sci. USA. 2015;112:E2174–E2181. doi: 10.1073/pnas.1504880112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herrera-Cruz M.S., Simmen T. Cancer: Untethering Mitochondria from the Endoplasmic Reticulum? Front. Oncol. 2017;7:105. doi: 10.3389/fonc.2017.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Mattia T., Wilhelm L.P., Ikhlef S., Wendling C., Spehner D., Nomine Y., Giordano F., Mathelin C., Drin G., Tomasetto C., et al. Identification of MOSPD2, a novel scaffold for endoplasmic reticulum membrane contact sites. EMBO Rep. 2018;19 doi: 10.15252/embr.201745453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lim Y., Cho I.T., Schoel L.J., Cho G., Golden J.A. Hereditary spastic paraplegia-linked REEP1 modulates endoplasmic reticulum/mitochondria contacts. Ann. Neurol. 2015;78:679–696. doi: 10.1002/ana.24488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guardia-Laguarta C., Area-Gomez E., Rub C., Liu Y., Magrane J., Becker D., Voos W., Schon E.A., Przedborski S. alpha-Synuclein is localized to mitochondria-associated ER membranes. J. Neurosci. 2014;34:249–259. doi: 10.1523/JNEUROSCI.2507-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cali T., Ottolini D., Negro A., Brini M. alpha-Synuclein controls mitochondrial calcium homeostasis by enhancing endoplasmic reticulum-mitochondria interactions. J. Biol. Chem. 2012;287:17914–17929. doi: 10.1074/jbc.M111.302794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gomez-Suaga P., Bravo-San Pedro J.M., Gonzalez-Polo R.A., Fuentes J.M., Niso-Santano M. ER-mitochondria signaling in Parkinson’s disease. Cell Death Dis. 2018;9:337. doi: 10.1038/s41419-017-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ottolini D., Cali T., Negro A., Brini M. The Parkinson disease-related protein DJ-1 counteracts mitochondrial impairment induced by the tumour suppressor protein p53 by enhancing endoplasmic reticulum-mitochondria tethering. Hum. Mol. Genet. 2013;22:2152–2168. doi: 10.1093/hmg/ddt068. [DOI] [PubMed] [Google Scholar]
- 61.Liu Y., Ma X., Fujioka H., Liu J., Chen S., Zhu X. DJ-1 regulates the integrity and function of ER-mitochondria association through interaction with IP3R3-Grp75-VDAC1. Proc. Natl. Acad. Sci. USA. 2019;116:25322–25328. doi: 10.1073/pnas.1906565116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stoica R., Paillusson S., Gomez-Suaga P., Mitchell J.C., Lau D.H., Gray E.H., Sancho R.M., Vizcay-Barrena G., De Vos K.J., Shaw C.E., et al. ALS/FTD-associated FUS activates GSK-3beta to disrupt the VAPB-PTPIP51 interaction and ER-mitochondria associations. EMBO Rep. 2016;17:1326–1342. doi: 10.15252/embr.201541726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D’Eletto M., Rossin F., Occhigrossi L., Farrace M.G., Faccenda D., Desai R., Marchi S., Refolo G., Falasca L., Antonioli M., et al. Transglutaminase Type 2 Regulates ER-Mitochondria Contact Sites by Interacting with GRP75. Cell Rep. 2018;25:3573–3581 e3574. doi: 10.1016/j.celrep.2018.11.094. [DOI] [PubMed] [Google Scholar]
- 64.Cerqua C., Anesti V., Pyakurel A., Liu D., Naon D., Wiche G., Baffa R., Dimmer K.S., Scorrano L. Trichoplein/mitostatin regulates endoplasmic reticulum-mitochondria juxtaposition. EMBO Rep. 2010;11:854–860. doi: 10.1038/embor.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paillard M., Tubbs E., Thiebaut P.A., Gomez L., Fauconnier J., Da Silva C.C., Teixeira G., Mewton N., Belaidi E., Durand A., et al. Depressing mitochondria-reticulum interactions protects cardiomyocytes from lethal hypoxia-reoxygenation injury. Circulation. 2013;128:1555–1565. doi: 10.1161/CIRCULATIONAHA.113.001225. [DOI] [PubMed] [Google Scholar]
- 66.Wu S., Lu Q., Wang Q., Ding Y., Ma Z., Mao X., Huang K., Xie Z., Zou M.H. Binding of FUN14 Domain Containing 1 With Inositol 1,4,5-Trisphosphate Receptor in Mitochondria-Associated Endoplasmic Reticulum Membranes Maintains Mitochondrial Dynamics and Function in Hearts In Vivo. Circulation. 2017;136:2248–2266. doi: 10.1161/CIRCULATIONAHA.117.030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Filadi R., Greotti E., Turacchio G., Luini A., Pozzan T., Pizzo P. Presenilin 2 Modulates Endoplasmic Reticulum-Mitochondria Coupling by Tuning the Antagonistic Effect of Mitofusin 2. Cell Rep. 2016;15:2226–2238. doi: 10.1016/j.celrep.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 68.Doghman-Bouguerra M., Granatiero V., Sbiera S., Sbiera I., Lacas-Gervais S., Brau F., Fassnacht M., Rizzuto R., Lalli E. FATE1 antagonizes calcium- and drug-induced apoptosis by uncoupling ER and mitochondria. EMBO Rep. 2016;17:1264–1280. doi: 10.15252/embr.201541504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sutendra G., Dromparis P., Wright P., Bonnet S., Haromy A., Hao Z., McMurtry M.S., Michalak M., Vance J.E., Sessa W.C., et al. The role of Nogo and the mitochondria-endoplasmic reticulum unit in pulmonary hypertension. Sci. Transl. Med. 2011;3:88ra55. doi: 10.1126/scitranslmed.3002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Munoz J.P., Ivanova S., Sanchez-Wandelmer J., Martinez-Cristobal P., Noguera E., Sancho A., Diaz-Ramos A., Hernandez-Alvarez M.I., Sebastian D., Mauvezin C., et al. Mfn2 modulates the UPR and mitochondrial function via repression of PERK. EMBO J. 2013;32:2348–2361. doi: 10.1038/emboj.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Vliet A.R., Agostinis P. When under pressure, get closer: PERKing up membrane contact sites during ER stress. Biochem. Soc. Trans. 2016;44:499–504. doi: 10.1042/BST20150272. [DOI] [PubMed] [Google Scholar]
- 72.Veeresh P., Kaur H., Sarmah D., Mounica L., Verma G., Kotian V., Kesharwani R., Kalia K., Borah A., Wang X., et al. Endoplasmic reticulum-mitochondria crosstalk: From junction to function across neurological disorders. Ann. N. Y. Acad. Sci. 2019;1457:41–60. doi: 10.1111/nyas.14212. [DOI] [PubMed] [Google Scholar]
- 73.Bravo-Sagua R., Rodriguez A.E., Kuzmicic J., Gutierrez T., Lopez-Crisosto C., Quiroga C., Diaz-Elizondo J., Chiong M., Gillette T.G., Rothermel B.A., et al. Cell death and survival through the endoplasmic reticulum-mitochondrial axis. Curr. Mol. Med. 2013;13:317–329. doi: 10.2174/156652413804810781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chami M., Checler F. Alterations of the Endoplasmic Reticulum (ER) Calcium Signaling Molecular Components in Alzheimer’s Disease. Cells. 2020;9:2577. doi: 10.3390/cells9122577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ivanova H., Vervliet T., Missiaen L., Parys J.B., De Smedt H., Bultynck G. Inositol 1,4,5-trisphosphate receptor-isoform diversity in cell death and survival. Biochim. Biophys. Acta. 2014;1843:2164–2183. doi: 10.1016/j.bbamcr.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 76.Rizzuto R., Simpson A.W., Brini M., Pozzan T. Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature. 1992;358:325–327. doi: 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- 77.Giacomello M., Drago I., Bortolozzi M., Scorzeto M., Gianelle A., Pizzo P., Pozzan T. Ca2+ hot spots on the mitochondrial surface are generated by Ca2+ mobilization from stores, but not by activation of store-operated Ca2+ channels. Mol. Cell. 2010;38:280–290. doi: 10.1016/j.molcel.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 78.Brasen J.C., Olsen L.F., Hallett M.B. Cell surface topology creates high Ca2+ signalling microdomains. Cell Calcium. 2010;47:339–349. doi: 10.1016/j.ceca.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Belosludtsev K.N., Dubinin M.V., Belosludtseva N.V., Mironova G.D. Mitochondrial Ca2+ Transport: Mechanisms, Molecular Structures, and Role in Cells. Biochemistry. 2019;84:593–607. doi: 10.1134/S0006297919060026. [DOI] [PubMed] [Google Scholar]
- 80.Territo P.R., Mootha V.K., French S.A., Balaban R.S. Ca2+ activation of heart mitochondrial oxidative phosphorylation: Role of the F0/F1-ATPase. Am. J. Physiol. Cell Physiol. 2000;278:C423–C435. doi: 10.1152/ajpcell.2000.278.2.C423. [DOI] [PubMed] [Google Scholar]
- 81.Glancy B., Willis W.T., Chess D.J., Balaban R.S. Effect of calcium on the oxidative phosphorylation cascade in skeletal muscle mitochondria. Biochemistry. 2013;52:2793–2809. doi: 10.1021/bi3015983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mitani Y., Behrooz A., Dubyak G.R., Ismail-Beigi F. Stimulation of GLUT-1 glucose transporter expression in response to exposure to calcium ionophore A-23187. Am. J. Physiol. 1995;269:C1228–C1234. doi: 10.1152/ajpcell.1995.269.5.C1228. [DOI] [PubMed] [Google Scholar]
- 83.Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., Alnemri E.S., Altucci L., Amelio I., Andrews D.W., et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giorgi C., Baldassari F., Bononi A., Bonora M., De Marchi E., Marchi S., Missiroli S., Patergnani S., Rimessi A., Suski J.M., et al. Mitochondrial Ca2+ and apoptosis. Cell Calcium. 2012;52:36–43. doi: 10.1016/j.ceca.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pinton P., Ferrari D., Rapizzi E., Di Virgilio F., Pozzan T., Rizzuto R. The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: Significance for the molecular mechanism of Bcl-2 action. EMBO J. 2001;20:2690–2701. doi: 10.1093/emboj/20.11.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carraro M., Jones K., Sartori G., Schiavone M., Antonucci S., Kucharczyk R., di Rago J.P., Franchin C., Arrigoni G., Forte M., et al. The Unique Cysteine of F-ATP Synthase OSCP Subunit Participates in Modulation of the Permeability Transition Pore. Cell Rep. 2020;32:108095. doi: 10.1016/j.celrep.2020.108095. [DOI] [PubMed] [Google Scholar]
- 87.Hwang M.S., Schwall C.T., Pazarentzos E., Datler C., Alder N.N., Grimm S. Mitochondrial Ca2+ influx targets cardiolipin to disintegrate respiratory chain complex II for cell death induction. Cell Death Differ. 2014;21:1733–1745. doi: 10.1038/cdd.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marchi S., Marinello M., Bononi A., Bonora M., Giorgi C., Rimessi A., Pinton P. Selective modulation of subtype III IP3R by Akt regulates ER Ca2+ release and apoptosis. Cell Death Dis. 2012;3:e304. doi: 10.1038/cddis.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Szado T., Vanderheyden V., Parys J.B., De Smedt H., Rietdorf K., Kotelevets L., Chastre E., Khan F., Landegren U., Soderberg O., et al. Phosphorylation of inositol 1,4,5-trisphosphate receptors by protein kinase B/Akt inhibits Ca2+ release and apoptosis. Proc. Natl. Acad. Sci. USA. 2008;105:2427–2432. doi: 10.1073/pnas.0711324105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marchi S., Rimessi A., Giorgi C., Baldini C., Ferroni L., Rizzuto R., Pinton P. Akt kinase reducing endoplasmic reticulum Ca2+ release protects cells from Ca2+-dependent apoptotic stimuli. Biochem. Biophys. Res. Commun. 2008;375:501–505. doi: 10.1016/j.bbrc.2008.07.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Betz C., Stracka D., Prescianotto-Baschong C., Frieden M., Demaurex N., Hall M.N. Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc. Natl. Acad. Sci. USA. 2013;110:12526–12534. doi: 10.1073/pnas.1302455110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hagiwara A., Cornu M., Cybulski N., Polak P., Betz C., Trapani F., Terracciano L., Heim M.H., Ruegg M.A., Hall M.N. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 93.Giorgi C., Ito K., Lin H.K., Santangelo C., Wieckowski M.R., Lebiedzinska M., Bononi A., Bonora M., Duszynski J., Bernardi R., et al. PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science. 2010;330:1247–1251. doi: 10.1126/science.1189157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bononi A., Bonora M., Marchi S., Missiroli S., Poletti F., Giorgi C., Pandolfi P.P., Pinton P. Identification of PTEN at the ER and MAMs and its regulation of Ca2+ signaling and apoptosis in a protein phosphatase-dependent manner. Cell Death Differ. 2013;20:1631–1643. doi: 10.1038/cdd.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Monaco G., Decrock E., Akl H., Ponsaerts R., Vervliet T., Luyten T., De Maeyer M., Missiaen L., Distelhorst C.W., De Smedt H., et al. Selective regulation of IP3-receptor-mediated Ca2+ signaling and apoptosis by the BH4 domain of Bcl-2 versus Bcl-Xl. Cell Death Differ. 2012;19:295–309. doi: 10.1038/cdd.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arbel N., Shoshan-Barmatz V. Voltage-dependent anion channel 1-based peptides interact with Bcl-2 to prevent antiapoptotic activity. J. Biol. Chem. 2010;285:6053–6062. doi: 10.1074/jbc.M109.082990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eckenrode E.F., Yang J., Velmurugan G.V., Foskett J.K., White C. Apoptosis protection by Mcl-1 and Bcl-2 modulation of inositol 1,4,5-trisphosphate receptor-dependent Ca2+ signaling. J. Biol. Chem. 2010;285:13678–13684. doi: 10.1074/jbc.M109.096040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang H., Shah K., Bradbury N.A., Li C., White C. Mcl-1 promotes lung cancer cell migration by directly interacting with VDAC to increase mitochondrial Ca2+ uptake and reactive oxygen species generation. Cell Death Dis. 2014;5:e1482. doi: 10.1038/cddis.2014.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schulman J.J., Wright F.A., Kaufmann T., Wojcikiewicz R.J. The Bcl-2 protein family member Bok binds to the coupling domain of inositol 1,4,5-trisphosphate receptors and protects them from proteolytic cleavage. J. Biol. Chem. 2013;288:25340–25349. doi: 10.1074/jbc.M113.496570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Assefa Z., Bultynck G., Szlufcik K., Nadif Kasri N., Vermassen E., Goris J., Missiaen L., Callewaert G., Parys J.B., De Smedt H. Caspase-3-induced truncation of type 1 inositol trisphosphate receptor accelerates apoptotic cell death and induces inositol trisphosphate-independent calcium release during apoptosis. J. Biol. Chem. 2004;279:43227–43236. doi: 10.1074/jbc.M403872200. [DOI] [PubMed] [Google Scholar]
- 101.Lewis A., Hayashi T., Su T.P., Betenbaugh M.J. Bcl-2 family in inter-organelle modulation of calcium signaling; roles in bioenergetics and cell survival. J. Bioenerg. Biomembr. 2014;46:1–15. doi: 10.1007/s10863-013-9527-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cai Y., Yang L., Niu F., Liao K., Buch S. Role of Sigma-1 Receptor in Cocaine Abuse and Neurodegenerative Disease. Adv. Exp. Med. Biol. 2017;964:163–175. doi: 10.1007/978-3-319-50174-1_12. [DOI] [PubMed] [Google Scholar]
- 103.Hayashi T., Su T.P. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell. 2007;131:596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 104.Raggi C., Diociaiuti M., Caracciolo G., Fratini F., Fantozzi L., Piccaro G., Fecchi K., Pizzi E., Marano G., Ciaffoni F., et al. Caveolin-1 Endows Order in Cholesterol-Rich Detergent Resistant Membranes. Biomolecules. 2019;9:287. doi: 10.3390/biom9070287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sundivakkam P.C., Kwiatek A.M., Sharma T.T., Minshall R.D., Malik A.B., Tiruppathi C. Caveolin-1 scaffold domain interacts with TRPC1 and IP3R3 to regulate Ca2+ store release-induced Ca2+ entry in endothelial cells. Am. J. Physiol. Cell Physiol. 2009;296:C403–C413. doi: 10.1152/ajpcell.00470.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rimessi A., Marchi S., Patergnani S., Pinton P. H-Ras-driven tumoral maintenance is sustained through caveolin-1-dependent alterations in calcium signaling. Oncogene. 2014;33:2329–2340. doi: 10.1038/onc.2013.192. [DOI] [PubMed] [Google Scholar]
- 107.Hedgepeth S.C., Garcia M.I., Wagner L.E., 2nd, Rodriguez A.M., Chintapalli S.V., Snyder R.R., Hankins G.D., Henderson B.R., Brodie K.M., Yule D.I., et al. The BRCA1 tumor suppressor binds to inositol 1,4,5-trisphosphate receptors to stimulate apoptotic calcium release. J. Biol. Chem. 2015;290:7304–7313. doi: 10.1074/jbc.M114.611186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Angebault C., Fauconnier J., Patergnani S., Rieusset J., Danese A., Affortit C.A., Jagodzinska J., Megy C., Quiles M., Cazevieille C., et al. ER-mitochondria cross-talk is regulated by the Ca2+ sensor NCS1 and is impaired in Wolfram syndrome. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aaq1380. [DOI] [PubMed] [Google Scholar]
- 109.Delprat B., Maurice T., Delettre C. Wolfram syndrome: MAMs’ connection? Cell Death Dis. 2018;9:364. doi: 10.1038/s41419-018-0406-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matsuzaki H., Fujimoto T., Ota T., Ogawa M., Tsunoda T., Doi K., Hamabashiri M., Tanaka M., Shirasawa S. Tespa1 is a novel inositol 1,4,5-trisphosphate receptor binding protein in T and B lymphocytes. FEBS Open Bio. 2012;2:255–259. doi: 10.1016/j.fob.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Matsuzaki H., Fujimoto T., Tanaka M., Shirasawa S. Tespa1 is a novel component of mitochondria-associated endoplasmic reticulum membranes and affects mitochondrial calcium flux. Biochem. Biophys. Res. Commun. 2013;433:322–326. doi: 10.1016/j.bbrc.2013.02.099. [DOI] [PubMed] [Google Scholar]
- 112.Li G., Mongillo M., Chin K.T., Harding H., Ron D., Marks A.R., Tabas I. Role of ERO1-alpha-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. J. Cell Biol. 2009;186:783–792. doi: 10.1083/jcb.200904060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Giorgi C., Bonora M., Sorrentino G., Missiroli S., Poletti F., Suski J.M., Galindo Ramirez F., Rizzuto R., Di Virgilio F., Zito E., et al. p53 at the endoplasmic reticulum regulates apoptosis in a Ca2+-dependent manner. Proc. Natl. Acad. Sci. USA. 2015;112:1779–1784. doi: 10.1073/pnas.1410723112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Giorgi C., Bonora M., Missiroli S., Poletti F., Ramirez F.G., Morciano G., Morganti C., Pandolfi P.P., Mammano F., Pinton P. Intravital imaging reveals p53-dependent cancer cell death induced by phototherapy via calcium signaling. Oncotarget. 2015;6:1435–1445. doi: 10.18632/oncotarget.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Krols M., Bultynck G., Janssens S. ER-Mitochondria contact sites: A new regulator of cellular calcium flux comes into play. J. Cell Biol. 2016;214:367–370. doi: 10.1083/jcb.201607124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Raturi A., Gutierrez T., Ortiz-Sandoval C., Ruangkittisakul A., Herrera-Cruz M.S., Rockley J.P., Gesson K., Ourdev D., Lou P.H., Lucchinetti E., et al. TMX1 determines cancer cell metabolism as a thiol-based modulator of ER-mitochondria Ca2+ flux. J. Cell Biol. 2016;214:433–444. doi: 10.1083/jcb.201512077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dremina E.S., Sharov V.S., Kumar K., Zaidi A., Michaelis E.K., Schoneich C. Anti-apoptotic protein Bcl-2 interacts with and destabilizes the sarcoplasmic/endoplasmic reticulum Ca2+-ATPase (SERCA) Biochem. J. 2004;383:361–370. doi: 10.1042/BJ20040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marino M., Stoilova T., Giorgi C., Bachi A., Cattaneo A., Auricchio A., Pinton P., Zito E. SEPN1, an endoplasmic reticulum-localized selenoprotein linked to skeletal muscle pathology, counteracts hyperoxidation by means of redox-regulating SERCA2 pump activity. Hum. Mol. Genet. 2015;24:1843–1855. doi: 10.1093/hmg/ddu602. [DOI] [PubMed] [Google Scholar]
- 119.Chami M., Gozuacik D., Lagorce D., Brini M., Falson P., Peaucellier G., Pinton P., Lecoeur H., Gougeon M.L., le Maire M., et al. SERCA1 truncated proteins unable to pump calcium reduce the endoplasmic reticulum calcium concentration and induce apoptosis. J. Cell Biol. 2001;153:1301–1314. doi: 10.1083/jcb.153.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chami M., Oules B., Szabadkai G., Tacine R., Rizzuto R., Paterlini-Brechot P. Role of SERCA1 truncated isoform in the proapoptotic calcium transfer from ER to mitochondria during ER stress. Mol. Cell. 2008;32:641–651. doi: 10.1016/j.molcel.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fayaz S.M., Raj Y.V., Krishnamurthy R.G. CypD: The Key to the Death Door. CNS Neurol. Disord. Drug Targets. 2015;14:654–663. doi: 10.2174/1871527314666150429113239. [DOI] [PubMed] [Google Scholar]
- 122.Hijioka M., Inden M., Yanagisawa D., Kitamura Y. DJ-1/PARK7: A New Therapeutic Target for Neurodegenerative Disorders. Biol. Pharm. Bull. 2017;40:548–552. doi: 10.1248/bpb.b16-01006. [DOI] [PubMed] [Google Scholar]
- 123.Maxfield K.E., Taus P.J., Corcoran K., Wooten J., Macion J., Zhou Y., Borromeo M., Kollipara R.K., Yan J., Xie Y., et al. Comprehensive functional characterization of cancer-testis antigens defines obligate participation in multiple hallmarks of cancer. Nat. Commun. 2015;6:8840. doi: 10.1038/ncomms9840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen M., Chen Z., Wang Y., Tan Z., Zhu C., Li Y., Han Z., Chen L., Gao R., Liu L., et al. Mitophagy receptor FUNDC1 regulates mitochondrial dynamics and mitophagy. Autophagy. 2016;12:689–702. doi: 10.1080/15548627.2016.1151580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ohno M. PERK as a hub of multiple pathogenic pathways leading to memory deficits and neurodegeneration in Alzheimer’s disease. Brain Res. Bull. 2018;141:72–78. doi: 10.1016/j.brainresbull.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 126.Leem J., Lee I.K. Mechanisms of Vascular Calcification: The Pivotal Role of Pyruvate Dehydrogenase Kinase 4. Endocrinol. Metab. 2016;31:52–61. doi: 10.3803/EnM.2016.31.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen Q., Schubert D. Presenilin-interacting proteins. Expert Rev. Mol. Med. 2002;4:1–18. doi: 10.1017/S1462399402005008. [DOI] [PubMed] [Google Scholar]
- 128.Ratti A., Buratti E. Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J. Neurochem. 2016;138(Suppl. 1):95–111. doi: 10.1111/jnc.13625. [DOI] [PubMed] [Google Scholar]
- 129.Revathidevi S., Munirajan A.K. Akt in cancer: Mediator and more. Semin. Cancer Biol. 2019;59:80–91. doi: 10.1016/j.semcancer.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 130.Schulman J.J., Szczesniak L.M., Bunker E.N., Nelson H.A., Roe M.W., Wagner L.E., 2nd, Yule D.I., Wojcikiewicz R.J.H. Bok regulates mitochondrial fusion and morphology. Cell Death Differ. 2019;26:2682–2694. doi: 10.1038/s41418-019-0327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Clark S.L., Rodriguez A.M., Snyder R.R., Hankins G.D., Boehning D. Structure-Function Of The Tumor Suppressor BRCA1. Comput. Struct. Biotechnol. J. 2012;1 doi: 10.5936/csbj.201204005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ketteler J., Klein D. Caveolin-1, cancer and therapy resistance. Int. J. Cancer. 2018;143:2092–2104. doi: 10.1002/ijc.31369. [DOI] [PubMed] [Google Scholar]
- 133.Shergalis A.G., Hu S., Bankhead A., 3rd, Neamati N. Role of the ERO1-PDI interaction in oxidative protein folding and disease. Pharmacol. Ther. 2020;210:107525. doi: 10.1016/j.pharmthera.2020.107525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yuan T., Lupse B., Maedler K., Ardestani A. mTORC2 Signaling: A Path for Pancreatic beta Cell’s Growth and Function. J. Mol. Biol. 2018;430:904–918. doi: 10.1016/j.jmb.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 135.Boeckel G.R., Ehrlich B.E. NCS-1 is a regulator of calcium signaling in health and disease. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:1660–1667. doi: 10.1016/j.bbamcr.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hsu K.S., Kao H.Y. PML: Regulation and multifaceted function beyond tumor suppression. Cell Biosci. 2018;8:5. doi: 10.1186/s13578-018-0204-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen C.Y., Chen J., He L., Stiles B.L. PTEN: Tumor Suppressor and Metabolic Regulator. Front. Endocrinol. 2018;9:338. doi: 10.3389/fendo.2018.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Slack C. Ras signaling in aging and metabolic regulation. Nutr. Healthy Aging. 2017;4:195–205. doi: 10.3233/NHA-160021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Penke B., Fulop L., Szucs M., Frecska E. The Role of Sigma-1 Receptor, an Intracellular Chaperone in Neurodegenerative Diseases. Curr. Neuropharmacol. 2018;16:97–116. doi: 10.2174/1570159X15666170529104323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang D., Zheng M., Lei L., Ji J., Yao Y., Qiu Y., Ma L., Lou J., Ouyang C., Zhang X., et al. Tespa1 is involved in late thymocyte development through the regulation of TCR-mediated signaling. Nat. Immunol. 2012;13:560–568. doi: 10.1038/ni.2301. [DOI] [PubMed] [Google Scholar]
- 141.Li Y., Guo Y., Tang J., Jiang J., Chen Z. New insights into the roles of CHOP-induced apoptosis in ER stress. Acta Biochim. Biophys. Sin. 2014;46:629–640. doi: 10.1093/abbs/gmu048. [DOI] [PubMed] [Google Scholar]
- 142.Hu J., Jin J., Qu Y., Liu W., Ma Z., Zhang J., Chen F. ERO1alpha inhibits cell apoptosis and regulates steroidogenesis in mouse granulosa cells. Mol. Cell. Endocrinol. 2020;511:110842. doi: 10.1016/j.mce.2020.110842. [DOI] [PubMed] [Google Scholar]
- 143.Pozzer D., Varone E., Chernorudskiy A., Schiarea S., Missiroli S., Giorgi C., Pinton P., Canato M., Germinario E., Nogara L., et al. A maladaptive ER stress response triggers dysfunction in highly active muscles of mice with SELENON loss. Redox Biol. 2019;20:354–366. doi: 10.1016/j.redox.2018.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Laptenko O., Prives C. p53: Master of life, death, and the epigenome. Genes Dev. 2017;31:955–956. doi: 10.1101/gad.302364.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Matsuo Y., Hirota K. Transmembrane thioredoxin-related protein TMX1 is reversibly oxidized in response to protein accumulation in the endoplasmic reticulum. FEBS Open Bio. 2017;7:1768–1777. doi: 10.1002/2211-5463.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lewin T.M., Kim J.H., Granger D.A., Vance J.E., Coleman R.A. Acyl-CoA synthetase isoforms 1, 4, and 5 are present in different subcellular membranes in rat liver and can be inhibited independently. J. Biol. Chem. 2001;276:24674–24679. doi: 10.1074/jbc.M102036200. [DOI] [PubMed] [Google Scholar]
- 147.Rusinol A.E., Cui Z., Chen M.H., Vance J.E. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 1994;269:27494–27502. doi: 10.1016/S0021-9258(18)47012-3. [DOI] [PubMed] [Google Scholar]
- 148.Simoes I.C.M., Morciano G., Lebiedzinska-Arciszewska M., Aguiari G., Pinton P., Potes Y., Wieckowski M.R. The mystery of mitochondria-ER contact sites in physiology and pathology: A cancer perspective. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866:165834. doi: 10.1016/j.bbadis.2020.165834. [DOI] [PubMed] [Google Scholar]
- 149.Mullen T.D., Obeid L.M. Ceramide and apoptosis: Exploring the enigmatic connections between sphingolipid metabolism and programmed cell death. Anticancer Agents Med. Chem. 2012;12:340–363. doi: 10.2174/187152012800228661. [DOI] [PubMed] [Google Scholar]
- 150.Chalfant C.E., Szulc Z., Roddy P., Bielawska A., Hannun Y.A. The structural requirements for ceramide activation of serine-threonine protein phosphatases. J. Lipid Res. 2004;45:496–506. doi: 10.1194/jlr.M300347-JLR200. [DOI] [PubMed] [Google Scholar]
- 151.Lozano J., Berra E., Municio M.M., Diaz-Meco M.T., Dominguez I., Sanz L., Moscat J. Protein kinase C zeta isoform is critical for kappa B-dependent promoter activation by sphingomyelinase. J. Biol. Chem. 1994;269:19200–19202. doi: 10.1016/S0021-9258(17)32152-X. [DOI] [PubMed] [Google Scholar]
- 152.Zhang Y., Yao B., Delikat S., Bayoumy S., Lin X.H., Basu S., McGinley M., Chan-Hui P.Y., Lichenstein H., Kolesnick R. Kinase suppressor of Ras is ceramide-activated protein kinase. Cell. 1997;89:63–72. doi: 10.1016/S0092-8674(00)80183-X. [DOI] [PubMed] [Google Scholar]
- 153.Saddoughi S.A., Ogretmen B. Diverse functions of ceramide in cancer cell death and proliferation. Adv. Cancer Res. 2013;117:37–58. doi: 10.1016/B978-0-12-394274-6.00002-9. [DOI] [PubMed] [Google Scholar]
- 154.Huang C., Freter C. Lipid metabolism, apoptosis and cancer therapy. Int. J. Mol. Sci. 2015;16:924–949. doi: 10.3390/ijms16010924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Stiban J., Caputo L., Colombini M. Ceramide synthesis in the endoplasmic reticulum can permeabilize mitochondria to proapoptotic proteins. J. Lipid Res. 2008;49:625–634. doi: 10.1194/jlr.M700480-JLR200. [DOI] [PubMed] [Google Scholar]
- 156.Siskind L.J., Kolesnick R.N., Colombini M. Ceramide channels increase the permeability of the mitochondrial outer membrane to small proteins. J. Biol. Chem. 2002;277:26796–26803. doi: 10.1074/jbc.M200754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Coleman R.A., Lewin T.M., Van Horn C.G., Gonzalez-Baro M.R. Do long-chain acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J. Nutr. 2002;132:2123–2126. doi: 10.1093/jn/132.8.2123. [DOI] [PubMed] [Google Scholar]
- 158.Lee Y.J., Jeong S.Y., Karbowski M., Smith C.L., Youle R.J. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol. Biol. Cell. 2004;15:5001–5011. doi: 10.1091/mbc.e04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Alirol E., James D., Huber D., Marchetto A., Vergani L., Martinou J.C., Scorrano L. The mitochondrial fission protein hFis1 requires the endoplasmic reticulum gateway to induce apoptosis. Mol. Biol. Cell. 2006;17:4593–4605. doi: 10.1091/mbc.e06-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yu T., Fox R.J., Burwell L.S., Yoon Y. Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1. J. Cell Sci. 2005;118:4141–4151. doi: 10.1242/jcs.02537. [DOI] [PubMed] [Google Scholar]
- 161.Arnoult D., Grodet A., Lee Y.J., Estaquier J., Blackstone C. Release of OPA1 during apoptosis participates in the rapid and complete release of cytochrome c and subsequent mitochondrial fragmentation. J. Biol. Chem. 2005;280:35742–35750. doi: 10.1074/jbc.M505970200. [DOI] [PubMed] [Google Scholar]
- 162.Gao W., Pu Y., Luo K.Q., Chang D.C. Temporal relationship between cytochrome c release and mitochondrial swelling during UV-induced apoptosis in living HeLa cells. J. Cell Sci. 2001;114:2855–2862. doi: 10.1242/jcs.114.15.2855. [DOI] [PubMed] [Google Scholar]
- 163.Iwasawa R., Mahul-Mellier A.L., Datler C., Pazarentzos E., Grimm S. Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 2011;30:556–568. doi: 10.1038/emboj.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Area-Gomez E., Schon E.A. On the Pathogenesis of Alzheimer’s Disease: The MAM Hypothesis. FASEB J. 2017;31:864–867. doi: 10.1096/fj.201601309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang B., Nguyen M., Chang N.C., Shore G.C. Fis1, Bap31 and the kiss of death between mitochondria and endoplasmic reticulum. EMBO J. 2011;30:451–452. doi: 10.1038/emboj.2010.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sivaguru M., Urban M.A., Fried G., Wesseln C.J., Mander L., Punyasena S.W. Comparative performance of airyscan and structured illumination superresolution microscopy in the study of the surface texture and 3D shape of pollen. Microsc. Res. Tech. 2018;81:101–114. doi: 10.1002/jemt.22732. [DOI] [PubMed] [Google Scholar]
- 167.Elgass K.D., Smith E.A., LeGros M.A., Larabell C.A., Ryan M.T. Analysis of ER-mitochondria contacts using correlative fluorescence microscopy and soft X-ray tomography of mammalian cells. J. Cell Sci. 2015;128:2795–2804. doi: 10.1242/jcs.169136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Csordas G., Weaver D., Hajnoczky G. Endoplasmic Reticulum-Mitochondrial Contactology: Structure and Signaling Functions. Trends Cell Biol. 2018;28:523–540. doi: 10.1016/j.tcb.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., Voeltz G.K. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Murley A., Lackner L.L., Osman C., West M., Voeltz G.K., Walter P., Nunnari J. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wu Y., Whiteus C., Xu C.S., Hayworth K.J., Weinberg R.J., Hess H.F., De Camilli P. Contacts between the endoplasmic reticulum and other membranes in neurons. Proc. Natl. Acad. Sci. USA. 2017;114:E4859–E4867. doi: 10.1073/pnas.1701078114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhao Y.G., Chen Y., Miao G., Zhao H., Qu W., Li D., Wang Z., Liu N., Li L., Chen S., et al. The ER-Localized Transmembrane Protein EPG-3/VMP1 Regulates SERCA Activity to Control ER-Isolation Membrane Contacts for Autophagosome Formation. Mol. Cell. 2017;67:974–989.e6. doi: 10.1016/j.molcel.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 173.Williamson C.D., Wong D.S., Bozidis P., Zhang A., Colberg-Poley A.M. Isolation of Endoplasmic Reticulum, Mitochondria, and Mitochondria-Associated Membrane and Detergent Resistant Membrane Fractions from Transfected Cells and from Human Cytomegalovirus-Infected Primary Fibroblasts. Curr. Protoc. Cell Biol. 2015;68:3.27.1–3.27.33. doi: 10.1002/0471143030.cb0327s68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cho I.T., Adelmant G., Lim Y., Marto J.A., Cho G., Golden J.A. Ascorbate peroxidase proximity labeling coupled with biochemical fractionation identifies promoters of endoplasmic reticulum-mitochondrial contacts. J. Biol. Chem. 2017;292:16382–16392. doi: 10.1074/jbc.M117.795286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hung V., Lam S.S., Udeshi N.D., Svinkina T., Guzman G., Mootha V.K., Carr S.A., Ting A.Y. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife. 2017;6 doi: 10.7554/eLife.24463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Herrera-Cruz M.S., Simmen T. Over Six Decades of Discovery and Characterization of the Architecture at Mitochondria-Associated Membranes (MAMs) Adv. Exp. Med. Biol. 2017;997:13–31. doi: 10.1007/978-981-10-4567-7_2. [DOI] [PubMed] [Google Scholar]
- 177.Szymanski J., Janikiewicz J., Michalska B., Patalas-Krawczyk P., Perrone M., Ziolkowski W., Duszynski J., Pinton P., Dobrzyn A., Wieckowski M.R. Interaction of Mitochondria with the Endoplasmic Reticulum and Plasma Membrane in Calcium Homeostasis, Lipid Trafficking and Mitochondrial Structure. Int. J. Mol. Sci. 2017;18:1576. doi: 10.3390/ijms18071576. [DOI] [PMC free article] [PubMed] [Google Scholar]