Abstract
Background
In a hospital setting, there is a need for rapid detection of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) to guide isolation measures and targeted admission.
Aim
To evaluate the diagnostic performance of five SARS-CoV-2 rapid nucleocapsid protein antigen detection (RAD) assays (Biosynex, Biotical, Orient Gene, Panbio and SD Biosensor), and describe the performance and impact of implementation of the SD Biosensor assay in an emergency department.
Methods
Sensitivity and specificity of the five RAD assays were analysed on 100 respiratory samples: 60 real-time reverse transcriptase polymerase chain reaction (rRT-PCR)-confirmed SARS-CoV-2-positive samples, 24 SARS-CoV-2 RNA-negative samples and 16 samples positive for other respiratory pathogens. The manufacturer's protocol was adapted to validate the antigen tests on transport media used for rRT-PCR in the authors' routine practice. The SD Biosensor RAD assay was implemented as a screening method for rapid diagnosis and targeted admission.
Findings
Sensitivity of the five RAD assays ranged from 88.9% to 100% for samples with cycle threshold values <26, and specificity ranged from 46.2% to 100%. During the implementation period, 4195 RAD tests were performed. Due to the rapid RAD result, 157 patients were transferred directly to the coronavirus disease 2019 (COVID-19) cohort ward instead of the regular ward (N=47) or the temporary COVID-19 ward (N=110).
Conclusion
The SD Biosensor, Biotical and Panbio SARS-CoV-2 antigen tests showed acceptable overall performance, and identified the majority of contagious patients. In the context of high prevalence of SARS-CoV-2, RAD tests can be used as a rapid screening tool to guide infection prevention measures and aid targeted admission.
Keywords: SARS-CoV-2, Rapid antigen detection, rRT-PCR, Infection prevention, Isolation precautions
Introduction
Since the start of the coronavirus disease 2019 (COVID-19) pandemic in December 2019, the gold standard for the diagnosis of COVID-19 has been the detection of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) viral RNA in respiratory tract samples by real-time reverse transcriptase polymerase chain reaction (rRT-PCR) [1,2]. Despite excellent sensitivity and specificity, rRT-PCR is a complex analysis and has a long turnaround time (5–17 h) [3]. Several new rapid antigen detection (RAD) tests for qualitative detection of SARS-CoV-2 antigen on nasopharyngeal swab samples have now been introduced. They are easy to perform and results are available after 10–30 min. Specificity is consistently reported to be high. Sensitivity of the RAD tests compared with rRT-PCR is highly variable (0–94%) and is related to the viral load, with higher sensitivity reported for samples with higher viral loads [[4], [5], [6], [7]]. Overall, RAD assays are less sensitive than rRT-PCR. The World Health Organization (WHO) recommends that RAD tests should have a minimum performance of ≥80% sensitivity and ≥97% specificity compared with rRT-PCR [4].
In a hospital setting, taking the appropriate measures to prevent transmission of SARS-CoV-2 is a key priority to protect patients and healthcare workers (HCWs) [8]. Assuming that RAD assays have high specificity, patients with positive RAD tests can be diagnosed with COVID-19 and transferred directly to COVID-19 wards where appropriate infection prevention precautions are applied. Due to the higher limit of detection of RAD tests compared with rRT-PCR, RAD assays cannot exclude COVID-19, but they can be used to aid infection prevention and control precautions as they are expected to detect the most contagious patients and HCWs.
At the study institution, the COVID-19 pandemic led to the intensification of standard infection prevention precautions with strict application of hand and respiratory hygiene guidelines and social distancing. Additionally, HCWs wear a surgical mask at all times, and patients and visitors use comfort masks. Strict isolation precautions are applied to patients with suspected or confirmed COVID-19. Table S1 shows a local epidemiological view (see online supplementary material).
All patients admitted to the study hospital are tested for SARS-CoV-2 using an in-house rRT-PCR [2]. Patients with COVID-19 symptoms on admission are admitted to the pre-cohort area (suspected COVID-19 ward) in a single isolation room while waiting for the rRT-PCR result. If their COVID-19 test is positive, they are transferred to the cohort area (confirmed COVID-19 ward). If COVID-19 is excluded (based on rRT-PCR in combination with clinical presentation and radiology), they are transferred to a regular ward. Due to the broad clinical presentation of COVID-19, many patients meet the definition of a possible case at initial presentation [9], and are admitted to the pre-cohort ward, delaying specialized care and investigations for their actual medical problem.
Patients who are not suspected to have COVID-19 on admission are hospitalized on a specific hospital ward in a single or double room. Notwithstanding the recommendation to wear a mask and to keep the curtain closed between the beds while awaiting the rRT-PCR result, this involves a risk of SARS-CoV-2 transmission in the case of highly infectious patients (pre-/asymptomatic or atypical). For HCWs, COVID-19 should be excluded even in the case of mild symptoms. Using the in-house rRT-PCR test, this takes at least 5 h.
Implementation of a RAD assay could help overcome some of the current issues by allowing rapid identification of (highly) contagious patients/HCWs.
This study evaluated the analytical performance of five new commercially available SARS-CoV-2 RAD assays in comparison with rRT-PCR. In addition, the performance and effect of implementation of a RAD assay as a first-line screening test for patients in an emergency department who needed to be hospitalized were analysed. It was hypothesized that the RAD test has potential as a rapid diagnostic tool for decision-making concerning isolation measures and targeted admission for patients admitted to hospital in an emergency setting.
Methods
Samples/patients
Part 1: validation
Sensitivity was analysed prospectively from 27th October to 1st November 2020 at the OLV Hospital Aalst, Belgium on 60 SARS-CoV-2-positive nasopharyngeal swabs, confirmed by in-house rRT-PCR (N1 gene) [2] (stored at room temperature for a maximum of 12 h). Nasopharyngeal samples were collected in various transport media used routinely in the hospital laboratory [ESwab Amies, Copan, Brescia, Italy; Sigma Transwab Liquid Amies, Medical Wire and Equipment (MWE), Corsham, UK; UTM, Copan; Sigma Virocult, MWE; viral stabilization tube, Vacuette Greiner Bio-One, Vilvoorde, Belgium; virus sampling tube, Lingen, Shanghai, China).
The selection of SARS-CoV-2-positive samples was based on their cycle threshold (Ct) value, regardless of the time since exposure or symptom onset. Samples from confirmed cases of COVID-19 were grouped into four categories according to their Ct value: Ct<20 (N=16); 20≤Ct<26 (N=18); 26≤Ct<30 (N=18); and 30≤Ct<36 (N=8).
Specificity was evaluated using 40 samples: 24 nasopharyngeal SARS-CoV-2 RNA-negative swabs (collected between 27th October and 1st November 2020) and a cross-reactivity panel consisting of 16 samples (nasopharyngeal swabs and aspirates) positive for one or multiple other respiratory pathogens (obtained prior to the COVID-19 outbreak and stored at -80°C). These samples contained human metapneumovirus (N=3), human coronavirus HKU (N=3), human coronavirus OC43 (N=2), human coronavirus NL63 (N=2), human coronavirus 229E (N=2), influenza virus A/B (N=6), bocavirus (N=1), respiratory syncytial virus (N=4), rhinovirus (N=2), adenovirus (N=3), enterovirus (N=1), herpes simplex virus (N=1) and Streptococcus pneumoniae (N=7) (Table S2, see online supplementary material).
Part 2: implementation
On 5th November 2020, the SD Biosensor RAD assay was introduced in the emergency room as a screening method for SARS-CoV-2 on nasopharyngeal swabs for all patients with an indication for hospital admission for whom no recent positive SARS-CoV-2 rRT-PCR result was available (<8 weeks). All RAD tests were followed by an in-house rRT-PCR (from 07:00 to 22.00 h). The prevalence of COVID-19 was calculated based on these rRT-PCR results. Sensitivity, specificity and positive predictive value (PPV) of the RAD test were determined in comparison with the in-house method.
During the study period, variants of concern (VOCs) began to circulate in Belgium. From 26th January 2021, variant analysis using targeted rRT-PCR was performed on all SARS-CoV-2-positive samples. In this way, it was possible to identify the 20I/501Y.V1 (British), 20H/501Y.V2 (South African) and 20J/501Y.V3 (Brazilian) variants. As the mutations present in these variants do not target the nucleocapsid protein, it is unlikely that the analytical performance of the RAD test would be affected. This was confirmed by retrospective analysis of some samples containing VOCs. Sensitivity of the RAD test was also determined in VOC-containing samples.
SARS-CoV-2 testing
Part 1: validation
After detailed market research considering performance data, rapid and sustained availability and price, five SARS-CoV-2 RAD tests were selected for evaluation: COVID-19 ag BSS (Biosynex, Fribourg, Switzerland), SARS-CoV-2 Ag card (Biotical health, Madrid, Spain), Coronavirus AG Rapid test cassette (Zhejiang Orient Gene Biotech Co., Zhejiang, China), Panbio COVID-19 Ag Rapid Test Device (Abbott, Ludwigshafen, Germany) and SARS-CoV-2 Rapid Antigen test (SD Biosensor, Gyeonggi-do, Korea). All five RAD tests work on the basis of detection of viral nucleocapsid protein SARS-CoV-2 antigen (Table S3, see online supplementary material). The manufacturers' instructions recommend performing the tests directly on the nasopharyngeal swabs included in the test kits. The protocol was adapted for this study as the authors wanted to validate the antigen tests on the transport media used for rRT-PCR in routine practice in order to avoid additional sampling (limiting discomfort, risk of complications and workload). In order to compare all five RAD tests on the same sample, the sample and buffer volumes were adapted. A one-on-one dilution of sample extraction buffer was used for all assays. Depending on the test, results were read manually after 10–15 min. In the presence of a control line, any signal was interpreted as positive. Tests were read by two independent persons who were blinded to the rRT-PCR results.
Part 2: implementation
On 5th November 2020, the SD Biosensor RAD assay was implemented in the authors' laboratory. This test was selected based on good sensitivity and specificity, in combination with the price and ease of use. The test was performed according to the manufacturer's instructions with a sample volume of 350 μL, but using Amies transport medium instead of the swabs included in the test kit. Every sample tested with a RAD test was confirmed by in-house rRT-PCR. In case of discordant results (i.e. a RAD-positive/rRT-PCR-negative or RAD-negative/rRT-PCR-positive result with Ct value <26), the RAD test cassette was checked retrospectively by a second reader. The RAD test and rRT-PCR were repeated on the same sample. If no explanation was found from these additional interventions, a new sample was requested to repeat both the RAD test and rRT-PCR.
Clinical data
To gain insight into the effect of implementation of the RAD test, clinical data were obtained retrospectively from RAD-positive patients from 5th November 2020 to 14th March 2021.
RAD-positive patients were divided into two groups based on their clinical presentation at the emergency ward, in accordance with the case definition of Sciensano, the national public health institute of Belgium [9]: ‘COVID-19-suspected cases’, who would have been admitted to the pre-cohort ward; and ‘non-COVID-19-suspected cases’, who would have been admitted to a regular ward.
Ethical approval
This study was approved by the Ethics Committee of OLV Hospital Aalst (Ref. 2020/123).
Statistical analysis
Sensitivity and specificity were calculated using Excel version 16.0 (Microsoft Corp., Redmond, WA, USA). Sensitivity was evaluated for all samples, as well as for the four subgroups based on the Ct values. PPV was calculated at the beginning and end of the study period, and a projection was made to other prevalences.
Statistically significant differences of the Ct values were verified using MEDCALC Statistical Software version 17.1 (MedCalc Software Ltd, Ostend, Belgium). D'Agostino-Pearson test was used to test normality. In the case of a normal distribution, unpaired Student's t-test was applied; otherwise, Mann–Whitney U-test for independent samples was used. P<0.05 was considered to indicate significance.
Results
Part 1: validation
Sixty rRT-PCR-positive and 40 rRT-PCR-negative samples were selected (demographics, Table S4, see online supplementary material). The first two samples collected in virus sampling tubes (Lingen) gave invalid results (no control line) for the SD Biosensor assay and were excluded for all assays.
The specificity of the RAD assays ranged from 46.2% to 100%. The overall sensitivity ranged from 67.2% to 89.7%. More importantly, in the subgroup analyses based on the Ct values, all five RAD assays showed excellent sensitivity for samples with Ct values <20, with sensitivity reducing to <50% for samples with Ct values ≥30. Detailed results are presented in Table I .
Table I.
Sensitivity and specificity of the included rapid antigen detection tests on validation, and the SD Biosensor assay on implementation
| Biosynex | Biotical | Orient Gene (validation) | Panbio | SD Biosensor | SD Biosensor (implementation) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Ct value | Median Ct | Range | Median Ct | Range | ||||||
| <20 | 17.7 | 14.6–19.2 | 14/14 (100%) |
14/14 (100%) |
14/14 (100%) |
13/13a (100%) |
14/14 (100%) |
18.4 | 11.7–19.9 | 61/61 (100%) |
|
| 20–<26 | 21.7 | 20.3–25.4 | 18/18 (100%) |
16/18 (88.9%) |
18/18 (100%) |
17/18 (94.4%) |
18/18 (100%) |
23.1 | 20.0–25.9 | 100/108 (92.6%) |
|
| 26–<30 | 28.5 | 26.2–29.9 | 16/18 (88.9%) |
8/18 (44.4%) |
13/18 (72.2%) |
13/18 (72.2%) |
13/18 (72.2%) |
28.2 | 26.1–29.9 | 35/64 (54.7%) |
|
| 30–<36 | 31.3 | 30.8–35.5 | 4/8 (50.0%) |
1/8 (12.5%) |
3/8 (37.5%) |
2/8 (25.0%) |
3/8 (37.5%) |
33.2 | 30.0–35.9 | 4/84 (4.8%) |
|
| >36 | 37.6 | 36.1–42.0 | 0/52 (0.0%) |
||||||||
| Overall | 52/58 (89.7%) |
39/58 (67.2%) |
48/58 (82.8%) |
45/57 (78.9%) |
48/58 (82.8%) |
200/369 (54.2%) |
|||||
| Specificity | Negative samples | 7/23a (69.6%) |
0/24 (100%) |
2/24 (91.7%) |
0/24 (100%) |
0/24 (100%) |
|||||
| Cross-reactivity samples | 14/16 (12.5%) |
0/16 (100%) |
1/16 (93.8%) |
0/16 (100%) |
0/16 (100%) |
||||||
| Overall | 21/39 (46.2%) |
0/40 (100%) |
3/40 (92.5%) |
0/40 (100%) |
0/40 (100%) |
3814/3826 (99.7%) |
Ct, cycle threshold.
One sample with insufficient volume to perform the test.
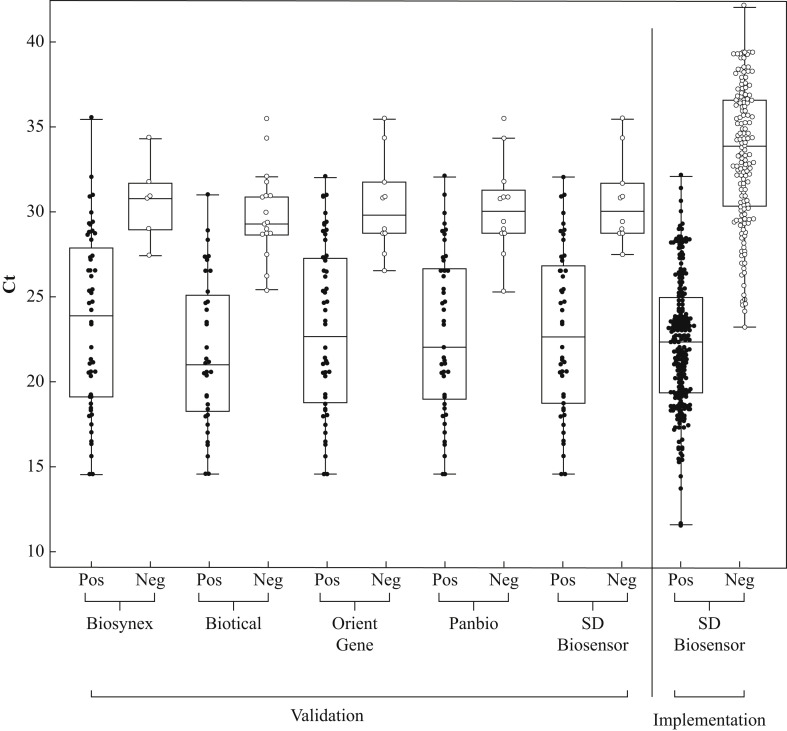
A significant (P<0.001) difference was observed in Ct values of positive RAD tests compared with negative RAD tests (Figure 1 ), resulting in sensitivity of 100% for all studied RAD assays for samples with rRT-PCR Ct values between 25 and 27.
Figure 1.
Real-time reverse transcriptase polymerase chain reaction cycle threshold (Ct) values in positive vs negative test results by rapid antigen detection (RAD) assay: validation and implementation. The average Ct value was significantly lower in all positive RAD tests compared with negative RAD tests (P<0.001). In the box-and-whisker plot, the central box represents the interquartile range, with the central line being the median. The whiskers represent the range, excluding outlier values.
Part 2: implementation
From 5th November to 14th March 2021, 4195 SD Biosensor RAD tests were performed (Table S4, see online supplementary material). The median rRT-PCR Ct value was 22.4 for the RAD-positive samples and 33.9 for the RAD-negative/rRT-PCR-positive samples (P<0.0001) (Figure 1). Two hundred and twelve of 4195 (5.1%) RAD tests were positive, compared with 369/4195 (8.8%) rRT-PCR tests. In the observed time period, the SD Biosensor RAD test showed overall sensitivity of 54.2% and specificity of 99.7% compared with rRT-PCR. For Ct values <20 and 20–<26, the SD Biosensor RAD test showed sensitivity of 100% and 92.6%, respectively (Table I and Figure S1, see online supplementary material). There were 12 false-positive results. Review of the test cassette, in combination with repeat RAD testing, revealed incorrect reading of three of 12 false-positive results. There were nine false-positive results which remained unexplained.
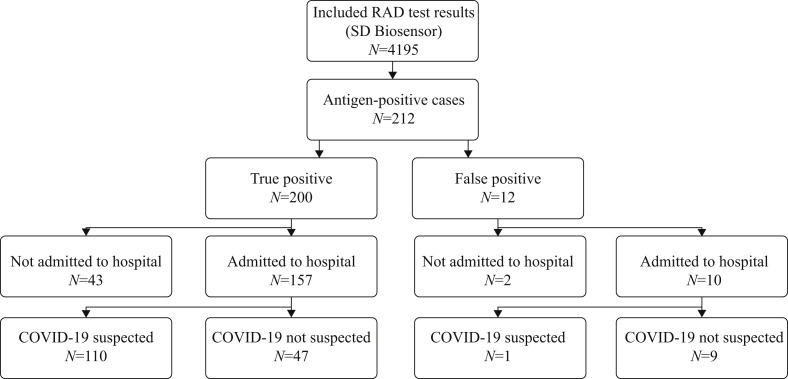
Of the 212 patients who tested positive using the RAD test, as mentioned above, 200 were true positives and 12 were false positives. One hundred and fifty-seven of 200 (78.5%) true-positive patients were admitted to hospital. Of these, 47/157 (29.9%) patients had symptoms that were not typical of COVID-19. They were transferred immediately to the cohort ward instead of the regular ward. One hundred and ten patients (70.1%) were suspected to have COVID-19 and were transferred directly to the cohort ward instead of the pre-cohort ward (Figure 2 ).
Figure 2.
Flowchart describing the impact of positive rapid antigen detection (RAD) test results for guiding isolation decisions.
Ten patients with a false-positive result were admitted to hospital and transferred to a cohort ward. Only one of these 10 patients had symptoms suggestive of COVID-19. Eight patients were transferred to single rooms on the cohort ward. Two patients were hospitalized in a multi-person room. After the negative rRT-PCR result was known, these patients were transferred immediately to single rooms on regular wards. One of the patients who stayed in a multi-person room was tested on day 4 and day 8 after his admission, and remained SARS-CoV-2 negative. Unfortunately, the second patient hospitalized in a multi-person room tested positive on day 4 after his admission. He was transferred to the cohort ward. Two of the 12 patients with a false-positive result were not admitted to hospital. One of them had signs of fever and dyspnoea. After investigation, he was discharged home from the emergency department. He was readmitted after 48 h and rRT-PCR was COVID-19 negative. Indeed cardial investigation showed he suffered from infective endocarditis. The second patient suffered from epigastric pain, caused by a duodenal ulcer. This patient was not retested.
Over the study period, 32 SD Biosensor RAD tests were performed on samples containing a VOC. There were 25 samples with 20I/501Y.V1, five samples with 20H/501Y.V2 and two samples with 20J/501Y.V3. For samples containing 20I/501Y.V1, the SD Biosensor RAD test showed overall sensitivity of 76%, and sensitivity of 100% and 87.5% for samples with Ct values <20 and 20–<26, respectively. Except for one sample with a Ct value of 27 containing 20H/501Y.V2, all samples containing 20H/501Y.V2 and 20J/501Y.V3 tested positive using the RAD test.
At the beginning of the study period, the prevalence of COVID-19 was 25%. At that time, PPV for the RAD test was 98.4%. During the study period, prevalence reduced to 4%, resulting in a marked decrease in PPV to 88.3%. The impact of the prevalence of COVID-19 on PPV is shown in Figure S2 (see online supplementary material).
Discussion
In a hospital setting with ongoing spread of COVID-19, rapid and easy-to-perform diagnostic tests are very important for fast diagnosis and optimization of infection control measures in order to limit further nosocomial spread.
In this study, the analytical performance of five rapid SARS-CoV-2 antigen tests was determined. In addition, the performance of the SD Biosensor RAD test in routine practice was analysed, and the added value of this RAD test for guiding isolation decisions and optimal patient care in a hospital setting was evaluated.
The SD Biosensor RAD test was the only assay which met the WHO performance criteria in the validation study but not in the implementation study (overall sensitivity 54.2%) [4]. In the subgroups with a Ct value <26, all assays met the WHO sensitivity criterion during the validation period and the SD Biosensor met the criterion during the implementation period. As the Biotical and Panbio assays also showed excellent specificity, they seem to be appropriate for use in settings similar to the study setting. This is in line with recently published literature on the SD Biosensor RAD test and the Panbio RAD test [7,[10], [11], [12], [13], [14], [15]]. In general, comparison of sensitivity between studies remains difficult as various Ct value categories are used. Moreover, Ct values between different rRT-PCR platforms cannot be compared directly.
Specificity in this validation study was 100% for the Biotical, Panbio and SD Biosensor assays. In the literature, specificity of the SD Biosensor and Panbio assays varies between 92% and 100%, and between 95% and 100% respectively [7,[10], [11], [12], [13], [14], [15]]. However, only one study included a cross-reactivity panel, in common with the present study [10]. The Orient Gene and Biosynex assays showed specificity of 92.5% and 46.2%, respectively. These suboptimal to poor specificity results are very different from the data provided in the package inserts. This can probably be attributed to the use of transport medium instead of the swab provided in the test kit. One transport medium was not compatible with the SD Biosensor assay. Additionally, Biotical has declared partial incompatibility of this test with Amies media for high Ct samples. This highlights the importance of every laboratory verifying the performance of a RAD assay as it will be used in practice.
In the implementation period, lower specificity of the SD Biosensor RAD test was observed in comparison with the validation period (99.7% vs. 100%). This can probably be explained by the larger data set included. In three of 12 samples, the false-positive results could not be confirmed by repeat RAD testing, emphasizing the risk of sample mix-up and subjective interpretation of the test result. The use of a reader would help to standardize the interpretation of test results. In addition, 10 of 12 false-positive results were reported in the first 6 weeks of the implementation period. As such, it is believed that specificity improves with the learning curve of the staff.
For all RAD tests (validation and implementation), the rRT-PCR Ct values of RAD-positive samples were significantly lower than the Ct values of the RAD-negative samples. This confirms the lower sensitivity of RAD assays compared with rRT-PCR, and almost exclusively concerns samples of patients with relatively low viral loads (Ct values >26). The cut-offs mentioned in the literature for infectivity are between >25 and >34 based on Ct values, and between <100,000 and 6.63 log10 RNA copies/mL based on viral load, probably depending on the test characteristics and the type of sample used [7,[16], [17], [18], [19], [20]]. For the in-house rRT-PCR, a Ct value of 26 correlates with approximately 105 RNA copies/mL. This implies that the SD Biosensor RAD assay, with sensitivity of 95.3% for samples with Ct values <26, can probably differentiate between contagious and non-contagious individuals [17].
Early identification of patients infected with SARS-CoV-2 is essential to isolate confirmed cases on cohort wards to prevent nosocomial transmission of SARS-CoV-2 to other patients or HCWs [21]. The authors implemented the SD Biosensor assay as a rapid screening test in the emergency department at the study hospital. As the Biotical and Panbio RAD tests also had acceptable analytical performance, the decision to use the SD Biosensor RAD test was guided by ease of use, price, and rapid and sustained availability.
During the implementation period of approximately 4 months, more than half of the rRT-PCR-positive patients presenting at the emergency department tested positive with the RAD test. Due to the rapid positive result of the antigen test, 47 non-COVID-19-suspected cases were transferred immediately to the COVID-19 ward instead of a regular ward, limiting the risk of nosocomial transmission and outbreaks. In the study institution, the results from the RAD test were available approximately 30 min after the nasopharyngeal swab was sent to the laboratory. Due to the user-friendly procedure, it was possible to perform the RAD test 24 h/day on 7 days/week. In contrast, the rRT-PCR test results were available, at best, 5 h after the sample was sent to the laboratory. For samples taken after 19:00 h, the result was not known until 12:00 h the next day as no runs were performed at night. With reports of nosocomial transmission of SARS-CoV-2 in patients hospitalized in the same room as early as day 1 [22,23], the fast results of the RAD test hold the potential to reduce transmission in the hospital environment.
One hundred and ten COVID-19-suspected cases were admitted immediately to the cohort ward instead of the pre-cohort ward, reducing pressure on the single-room hospital beds in the pre-cohort ward and limiting extra transfers of infectious patients between the pre-cohort and cohort wards. All but eight of the remaining 157 (RAD-negative) rRT-PCR positive patients had a Ct value >26, corresponding to limited to no contagiousness. Unfortunately, 10 COVID-19-negative patients were admitted to the cohort ward due to a false-positive RAD test, exposing them to the potential risk of nosocomial transmission of COVID-19. One patient became infected. This highlights the importance of confirmation of each RAD test with rRT-PCR.
When evaluating the performance of an antigen test, it is crucial to perform the evaluation in the context the test will be used. It was calculated that in a situation with low prevalence of COVID-19 in the target population, for example 0.5%, the PPV for the RAD test is reduced to less than 50%. In this context, the disadvantages outweigh the benefits. The decision was made to stop using the RAD test as a screening tool when the prevalence of SARS-CoV-2 falls below 3% in the targeted population. This is in accordance with the recommendations of WHO and earlier studies [4,24].
Meanwhile, the RAD test is also used in the study hospital as a screening tool for hospital staff with mild symptoms when they start their shift. A RAD test is always followed by a rRT-PCR test, but with a negative RAD test, the person can go to work while waiting for his/her rRT-PCR result. This implies an enormous advantage in terms of work planning and absenteeism. The RAD test is not currently used as a pre-operative screening method at the study hospital, as endotracheal intubation is an aerosol-generating procedure which has been linked to increased risk of transmission [25,26]. A negative RAD test on an upper respiratory sample is felt to be insufficient in this context, so rapid molecular tests are reserved for unplanned/urgent surgical procedures at the study hospital.
One of the strengths of this study is the head-to-head comparison of five RAD tests, three of which have not been described previously in the literature (Biosynex, Biotical and Orient Gene). Furthermore, to investigate specificity, a cross-reactivity panel consisting of respiratory samples positive for other respiratory viruses was included. With the exception of Mak et al., none of the previous studies on the SD Biosensor or Panbio assays included such a panel [10]. All RAD assays and rRT-PCR were performed in parallel on the same samples, so between-sample variations due to pre-analytical issues, such as the sample collection technique, are excluded. RAD assays were performed in the same time frame as rRT-PCR without cooling or freezing.
One of the main limitations of this study is that the RAD tests were not performed directly on the nasopharyngeal swabs supplied in the kits, as described in the manufacturers' instructions. A second limitation is that the subgroup analyses were performed on small sample cohorts. Furthermore, Ct values obtained using the in-house rRT-PCR were used as a proxy for viral load and infectivity in the absence of viral culture. Clinical data used to study the effect of the implementation of the RAD assay were retrieved retrospectively, and only for the RAD-positive patients. Unfortunately, the incidence of nosocomial SARS-CoV-2 infection before and after implementation of the RAD tests was not recorded.
During the study period, VOCs were first identified and became present in Belgium. VOCs are now present in approximately 77.2% [27] of positive samples in Belgium. As the mutations included in these variants do not target the nucleocapsid protein, it is unlikely that the analytical performance of the RAD test would be affected. The limited data show equal performance of the RAD test in VOC-containing samples compared with all samples. The overall sensitivity of the RAD test for 20I/501Y.V1-containing samples was 76%. Sensitivity met the WHO sensitivity criterion for samples with a Ct value <20 (100% for 20I/501Y.V1; no samples for 20H/501Y.V2; 100% for 20J/501Y.V3) and 20–<26 (87.5% for 20I/501Y.V1; 100% for 20H/501Y.V2; 100% for 20J/501Y.V3). Similar findings have been published for the British variant (20I/501Y.V1) [28].
In conclusion, the Biotical, Panbio and SD Biosensor RAD tests demonstrated acceptable performance in this study. All RAD assays are less sensitive than the current gold standard, rRT-PCR. However, from a clinical point of view, they seem to be able to detect the majority of infectious patients. Exclusive use of a RAD test cannot replace rRT-PCR in a hospital setting because a negative RAD test result cannot rule out a diagnosis of COVID-19. In the context of high prevalence of SARS-CoV-2 in the community, RAD tests can be used as a rapid screening tool complementary to rRT-PCR to guide infection prevention measures and aid targeted admission.
Acknowledgements
The authors wish to thank all laboratory staff at OLVZ Hospital, Aalst, Belgium for their contribution towards the survey.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhin.2021.03.021.
Conflict of interest statement
None declared.
Funding source
The research group kindly received the following test assays for validation: Biosynex (Life, Belgium), Biotical (BMD, Belgium), Orient Gene (Aqua Nano, Belgium) and Panbio (Abbott, Germany).
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Smithgall M.C., Dowlatshahi M., Spitalnik S.L., Hod E.A., Rai A.J. Types of assays for SARS-CoV-2 testing: a review. Lab Med. 2020;51:e59–e65. doi: 10.1093/labmed/lmaa039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. pii=2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lippi G., Simundic A.-M., Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin Chem Lab Med. 2020;58:1070–1076. doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization . WHO; Geneva: 2020. Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays.https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays Available at: [last accessed November 2020] [Google Scholar]
- 5.Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C.S., Lam E.T., et al. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol. 2020;129:104500. doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinnes J., Deeks J.J., Adriano A., Berhane S., Davenport C., Dittrich S., et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2020;8:CD013705. doi: 10.1002/14651858.CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albert E., Torres I., Bueno F., Huntley D., Molla E., Fernández-Fuentes M.Á., et al. Field evaluation of a rapid antigen test (PanbioTM COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centres. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2020;3 doi: 10.1016/j.cmi.2020.11.004. P472.E7-472.E1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.European Centre for Disease Prevention and Control . ECDC; Stockholm: 2020. Infection prevention and control and preparedness for COVID-19 in healthcare settings.https://www.ecdc.europa.eu/sites/default/files/documents/Infection-prevention-and-control-in-healthcare-settings-COVID-19_5th_update.pdf Fifth update. Available at: [last accessed November 2020] [Google Scholar]
- 9.Sciensano . Sciensano; Brussels: 2020. Gevalsdefinitie, indicaties voor testen en verplichte melding van COVID-19 versie 23 November 2020.https://covid-19.sciensano.be/sites/default/files/Covid19/COVID-19_Case definition_Testing_NL.pdf Available at: [last accessed December 2020] [Google Scholar]
- 10.Mak G.C., Lau S.S., Wong K.K., Chow N.L., Lau C.S., Lam E.T., et al. Analytical sensitivity and clinical sensitivity of the three rapid antigen detection kits for detection of SARS-CoV-2 virus. J Clin Virol. 2020;133:104684. doi: 10.1016/j.jcv.2020.104684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krüttgen A., Cornelissen C.G., Dreher M., Hornef M.W., Imöhl M., Kleines M. Comparison of the SARS-CoV-2 rapid antigen test to the real star Sars-CoV-2 RT PCR kit. J Virol Methods. 2020;288:114024. doi: 10.1016/j.jviromet.2020.114024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerutti F., Burdino E., Milia M.G., Allice T., Gregori G., Bruzzone B., et al. Urgent need of rapid tests for SARS CoV-2 antigen detection: evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J Clin Virol. 2020;132:104654. doi: 10.1016/j.jcv.2020.104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nalumansi A., Lutalo T., Kayiwa J., Watera C., Balinandi S., Kiconco J., et al. Field evaluation of the performance of a SARS-CoV-2 antigen rapid diagnostic test in Uganda using nasopharyngeal samples. Int J Infect Dis. 2021;104:282–286. doi: 10.1016/j.ijid.2020.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenollar F., Bouam A., Ballouche M., Fuster L., Prudent E., Colson P., et al. Evaluation of the Panbio Covid-19 rapid antigen detection test device for the screening of patients with Covid-19. J Clin Microbiol. 2021;59 doi: 10.1128/JCM.02589-20. e02589-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linares M., Pérez-Tanoira R., Carrero A., Romanyk J., Pérez-García F., Gómez-Herruz P., et al. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol. 2020;133:104659. doi: 10.1016/j.jcv.2020.104659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 17.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020;71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Kampen J.J.A., van de Vijver D.A.M.C., Fraaij P.L.A., Haagmans B.L., Lamers M.M., Okba N., et al. Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (COVID-19): duration and key determinants. MedRxiv. 2020 doi: 10.1038/s41467-020-20568-4. 2020.06.08.20125310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korean Centers for Disease Control and Prevention . KCDC; Cheongju: 2020. Findings from investigation and analysis of re-positive cases.https://is.cdc.go.kr/upload_comm/syview/doc.html?fn=159118745823700.pdf&rs=/upload_comm/docu/0030/ Available at: [last accessed December 2020] [Google Scholar]
- 21.Cheng V.C.C., Wong S.-C., Chen J.H.K., Yip C.C.Y., Chuang V.W.M., Tsang O.T.Y., et al. Escalating infection control response to the rapidly evolving epidemiology of the coronavirus disease 2019 (COVID-19) due to SARS-CoV-2 in Hong Kong. Infect Control Hosp Epidemiol. 2020;41:493–498. doi: 10.1017/ice.2020.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rickman H.M., Rampling T., Shaw K., Martinez-Garcia G., Hail L., Coen P., et al. Nosocomial transmission of coronavirus disease 2019: a retrospective study of 66 hospital-acquired cases in a London teaching hospital. Clin Infect Dis. 2021;72:690–693. doi: 10.1093/cid/ciaa816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanhems P., Saadatian-Elahi M., Chuzeville M., Marion E., Favrelle L., Hilliquin D., et al. Rapid nosocomial spread of SARS-CoV-2 in a French geriatric unit. Infect Control Hosp Epidemiol. 2020;41:866–867. doi: 10.1017/ice.2020.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaimayo C., Kaewnaphan B., Tanlieng N., Athipanyasilp N., Sirijatuphat R., Chayakulkeeree M., et al. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol J. 2020;17:177. doi: 10.1186/s12985-020-01452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization . WHO; Geneva: 2020. Infection prevention and control during health care when coronavirus disease (COVID-19) is suspected or confirmed: interim guidance, 29 June 2020.https://apps.who.int/iris/bitstream/handle/10665/332879/WHO-2019-nCoV-IPC-2020.4-eng.pdf?sequence=1&isAllowed=y Available at: [last accessed November 2020] [Google Scholar]
- 26.Tran K., Cimon K., Severn M., Pessoa-Silva C.L., Conly J. Aerosol generating procedures and risk of transmission of acute respiratory infections to healthcare workers: a systematic review. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Reference Laboratory Belgium (UZ Leuven & KU Leuven) National Reference Laboratory Belgium; Leuven: 2021. Genomic surveillance of SARS-CoV-2 in Belgium.https://www.uzleuven.be/nl/laboratoriumgeneeskunde/genomic-surveillance-sars-cov-2-belgium Report of the National Reference Laboratory (UZ Leuven & KU Leuven) situation update – 16th March 2021 (report 2021_18) Available at: [last accessed March 2021] [Google Scholar]
- 28.Public Health England . PHE; London: 2020. Guidance SARS-CoV-2 lateral flow antigen tests: evaluation of VUI-202012/01.https://www.gov.uk/government/publications/sars-cov-2-lateral-flow-antigen-tests-evaluation-of-vui-20201201/sars-cov-2-lateral-flow-antigen-tests-evaluation-of-vui-20201201 Available at: [last accessed March 2021] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.