Significance
Genetic markers in TRANK1 gene and its vicinity have been weakly associated with bipolar disorder and schizophrenia (10% increased risk). We found that the same polymorphisms are associated with Kleine-Levin syndrome (50% increased risk), a rare sleep disorder characterized by recurrent episodes of severe hypersomnia and cognitive abnormalities. Response to lithium treatment is suggestive of a pathophysiological overlap between KLS and bipolar disorder. The study also shows that variants in the TRANK1 gene region may predispose to KLS when patients have had a difficult birth, suggesting that TRANK1 gene region modulates newborns’ response to brain injury, with consequences for mental and neurological health in adulthood. Another possibility may be that the polymorphism impacts birth and KLS.
Keywords: Kleine-Levin syndrome, hypersomnia, bipolar disorder, birth difficulties, GWAS
Abstract
Kleine-Levin syndrome (KLS) is a rare disorder characterized by severe episodic hypersomnia, with cognitive impairment accompanied by apathy or disinhibition. Pathophysiology is unknown, although imaging studies indicate decreased activity in hypothalamic/thalamic areas during episodes. Familial occurrence is increased, and risk is associated with reports of a difficult birth. We conducted a worldwide case−control genome-wide association study in 673 KLS cases collected over 14 y, and ethnically matched 15,341 control individuals. We found a strong genome-wide significant association (rs71947865, Odds Ratio [OR] = 1.48, P = 8.6 × 10−9) within the 3′region of TRANK1 gene locus, previously associated with bipolar disorder and schizophrenia. Strikingly, KLS cases with rs71947865 variant had significantly increased reports of a difficult birth. As perinatal outcomes have dramatically improved over the last 40 y, we further stratified our sample by birth years and found that recent cases had a significantly reduced rs71947865 association. While the rs71947865 association did not replicate in the entire follow-up sample of 171 KLS cases, rs71947865 was significantly associated with KLS in the subset follow-up sample of 59 KLS cases who reported birth difficulties (OR = 1.54, P = 0.01). Genetic liability of KLS as explained by polygenic risk scores was increased (pseudo R2 = 0.15; P < 2.0 × 10−22 at P = 0.5 threshold) in the follow-up sample. Pathway analysis of genetic associations identified enrichment of circadian regulation pathway genes in KLS cases. Our results suggest links between KLS, circadian regulation, and bipolar disorder, and indicate that the TRANK1 polymorphisms in conjunction with reported birth difficulties may predispose to KLS.
Kleine-Levin syndrome (KLS) was identified as a unique disease entity almost a century ago (1, 2). It affects primarily male teenagers and is characterized by unpredictable periods (median length of ∼10 d) of intense hypersomnia (≥18 h of sleep per day), occasionally associated with megaphagia and sexual disinhibition. More recent descriptions in large case series have confirmed a stereotypic appearance of abrupt onset episodes in mostly male (66 to 75%) patients and association of severe hypersomniac episodes with cognitive and behavioral disturbances (mental slowness, confusion, apathy, derealization, altered perception, and disinhibited behavior). Remarkably, there is a complete absence of the symptoms between episodes, and a generally favorable evolution, with spontaneous disappearance of episodes after one to two decades (3–6). KLS episodes are dramatic in appearance, and patients are fully incapacitated. Electroencephalogram (EEG) studies occasionally reveal nonspecific EEG slowing during episodes. Further, lithium has been shown to reduce episodes in about one-third of cases (4, 7). Episodes may be associated with flu-like symptoms at the onset, with seasonal winter occurrence suggested by some (8).
Although most cases of KLS are sporadic, familial occurrence (3 to 8% of relatives) has been reported in studies (5, 9), and there are reports of concordant twin pairs (9, 10) which, considering the rarity of the condition (a few cases per million individuals) (11), suggests genetic or shared environmental effects. Early studies had suggested a human leukocyte antigen (HLA) association suggestive of an autoimmune mechanism (12), but this was not confirmed in larger samples (3, 8, 13). Further, karyotyping was normal in 112 patients, except for one patient with sporadic KLS who had a duplication in Xp22.31 (13).
This study reports genetic associations in a discovery cohort of 673 KLS cases with attempted replication in 171 cases across the world. Additionally, targeted exome sequencing in 32 cases derived from 16 multiplex families (9) was conducted. A KLS genome-wide association study (GWAS) found a strong signal in the TRANK1 region (rs71947865), similar to that reported in bipolar disorder and schizophrenia. In these latter disorders, the TRANK1 association variably maps to rs9834970 in Caucasians or to rs3732386 in multiethnic populations, both single-nucleotide polymorphisms (SNPs) located immediately 3′ of the TRANK1 gene (14–16). Strikingly, the TRANK1 signal was mostly present in KLS patients reporting birth difficulties and decreased over time (defined by birth year), paralleling well-established improved perinatal care. Birth difficulties (and maternal infections) have also long been associated with increased risk of bipolar disorder and schizophrenia (17–29). These results indicate possible gene × environment interactions in KLS, with possibly similar effects in other TRANK1-associated disorders such as bipolar disorder and schizophrenia.
Results
GWAS of KLS.
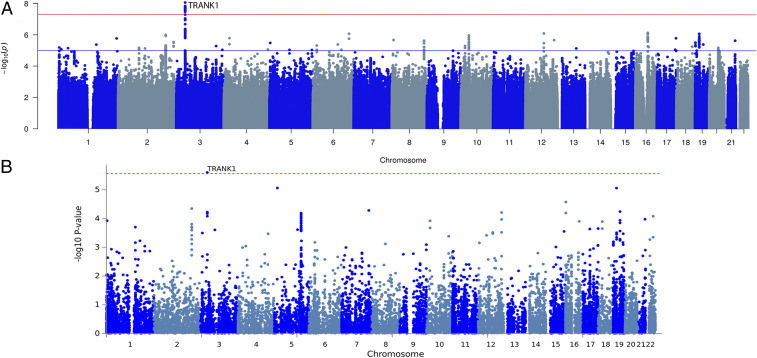
A worldwide multiethnic cohort of 673 KLS cases, the largest sample of KLS cases ever to be analyzed, was collected between the years of 2003 and 2017 and compared to 15,341 matched controls (the “discovery cohort”; see Dataset S1 for sample construction, and see SI Appendix, Fig. S1 for corresponding principal component analysis [PCA] plots). Genetic association analyses (genomic inflation λGC = 1.06) found a genome-wide significant effect (Fig. 1) with no heterogeneity across ethnic groups, with more than 20 tightly linked SNPs encompassing a 35-kb region of human chromosome 3 in the 3′ region of the tetratricopeptide repeat and ankyrin repeat-containing protein (TRANK1) gene. The TRANK1 gene was observed to have the highest expression in the fallopian tube, closely followed by the cerebellum (GTEx v8) (see Table 1 and Fig. 2 for zoom plot, and see Dataset S2, where all variants in this locus are listed). The most significant association was observed at rs71947865 (OR = 1.47, P = 8.6 × 10−9, presence of G versus GGGAGCCA), although the highest OR was observed at rs6789885 (OR = 1.53, P = 8.9 × 10−8, T versus C; Dataset S2). This TRANK1 region SNP rs71947865 is in absolute linkage disequilibrium (LD) with rs3732386 (r2 = 1), an SNP associated with bipolar disorder and schizophrenia in multiethnic samples (Dataset S3).
Fig. 1.
Multiethnic GWAS in 673 KLS cases and 15,431 control individuals. (A) Manhattan plot of the metaanalysis association (discovery cohort) results from SNP-based GWAS study. The x axis shows genomic location by chromosome, and the y axis shows −log10 P values. Red horizontal line indicates genome-wide significant P value threshold of 5 × 10−8. Individual loci are annotated. (B) Manhattan plot of gene-based association test association results. SNPs were mapped to 18,247 protein coding genes (distance 0). Genome-wide significance (red dashed line) was defined at P = 0.05/18,247 = 2.74 × 10−6.
Table 1.
Top variants in TRANK1 region from chromosome 3 associated with KLS in worldwide discovery cohort GWAS metaanalysis in 673 KLS cases and 15,341 control individuals and in a follow-up replication cohort of 171 KLS cases and 1,956 controls
| SNP | BP | A/B | GWAS metaanalysis, n = 673 | Follow-up samples, n = 171 | Combined, n = 844 | ||||
| Freq. B | OR | P value | OR | P value | OR | P value | |||
| rs71947865 | 36877798 | GGGAGCCA/G | 0.29 | 1.48 | 8.61×10−09 | 1.17 | 0.19 | 1.40 | 3.51×10−8 |
| rs3732385 | 36872193 | C/A | 0.30 | 1.47 | 1.44×10−8 | 1.17 | 0.18 | 1.39 | 5.00×10−8 |
| rs3732384 | 36872224 | T/C | 0.30 | 1.47 | 1.44×10−8 | 1.17 | 0.18 | 1.39 | 5.00×10−8 |
| rs7652637 | 36871507 | T/C | 0.30 | 1.47 | 1.47×10−8 | 1.18 | 0.18 | 1.39 | 5.06×10−8 |
| rs9821223 | 36883369 | T/C | 0.29 | 1.47 | 1.5×10−8 | 1.11 | 0.39 | 1.38 | 1.67×10−7 |
| rs9811916 | 36878086 | A/G | 0.30 | 1.47 | 1.63×10−8 | 1.17 | 0.19 | 1.39 | 5.78×10−8 |
| rs4678909 | 36878563 | A/G | 0.30 | 1.47 | 1.73×10−8 | 1.16 | 0.21 | 1.39 | 7.42×10−8 |
| rs6771655 | 36884370 | C/A | 0.29 | 1.47 | 1.74×10−8 | 1.11 | 0.39 | 1.38 | 1.91×10−7 |
| rs9882911 | 36885392 | T/C | 0.29 | 1.47 | 1.78×10−8 | 1.11 | 0.39 | 1.38 | 1.96×10−7 |
| rs9863798 | 36885960 | C/T | 0.29 | 1.47 | 1.99×10−8 | 1.11 | 0.39 | 1.37 | 2.16×10−7 |
| rs4678910 | 36887973 | C/G | 0.29 | 1.47 | 2.03×10−8 | 1.11 | 0.41 | 1.37 | 2.38×10−7 |
| rs4678911 | 36888064 | G/T | 0.29 | 1.47 | 2.09×10−8 | 1.11 | 0.40 | 1.37 | 2.29×10−7 |
| rs9819304 | 36888172 | T/G | 0.29 | 1.47 | 2.1×10−8 | 1.11 | 0.40 | 1.37 | 2.30×10−7 |
| rs17807744 | 36894048 | C/T | 0.29 | 1.46 | 2.18×10−8 | 1.10 | 0.45 | 1.37 | 2.92×10−7 |
| rs4234258 | 36878970 | A/G | 0.30 | 1.46 | 2.26×10−8 | 1.16 | 0.23 | 1.38 | 9.95×10−8 |
| rs3732386 | 36871993 | C/T | 0.30 | 1.46 | 2.26×10−8 | 1.17 | 0.18 | 1.39 | 7.16×10−8 |
| rs6550435 | 36864489 | T/G | 0.29 | 1.47 | 2.37×10−8 | 1.17 | 0.20 | 1.39 | 8.62×10−8 |
| rs17195098 | 36893965 | C/T | 0.29 | 1.46 | 2.83×10−8 | 1.10 | 0.45 | 1.36 | 3.63×10−7 |
| rs9883001 | 36894579 | C/T | 0.29 | 1.46 | 2.9×10−8 | 1.10 | 0.45 | 1.36 | 3.70×10−7 |
| rs4678912 | 36894646 | A/T | 0.29 | 1.46 | 2.91×10−8 | 1.10 | 0.45 | 1.36 | 3.71×10−7 |
| rs1532965 | 36861196 | T/G | 0.30 | 1.46 | 2.92×10−8 | 1.16 | 0.22 | 1.38 | 1.20×10−7 |
| rs4678913 | 36895741 | C/G | 0.29 | 1.46 | 3.05×10−8 | 1.10 | 0.45 | 1.36 | 3.93×10−7 |
| rs11706780 | 36896235 | C/T | 0.29 | 1.46 | 3.12×10−8 | 1.09 | 0.46 | 1.36 | 4.04×10−7 |
| rs4624519 | 36862980 | C/T | 0.30 | 1.45 | 3.39×10−8 | 1.16 | 0.22 | 1.38 | 1.36×10−7 |
Effect allele is B; genomic coordinates are in hg19/GRCh37; Freq. B is the frequency of the minor allele B.
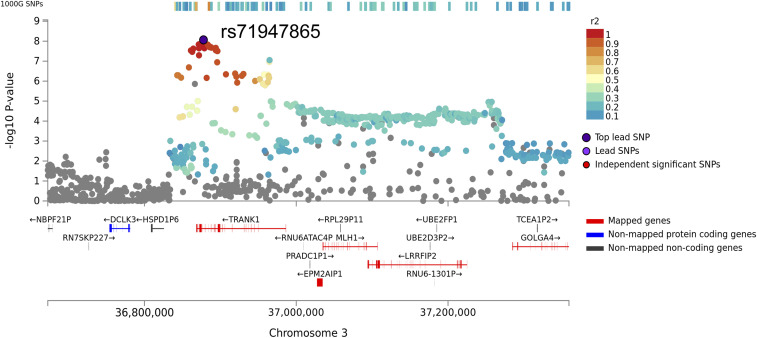
Fig. 2.
Regional association plot of the GWA significance of chromosome 3 flanked by rs76652637 and rs11706780 in the 3′ region of TRANK1. Highest significance was observed at rs71947865 (OR = 1.47, P = 8.6 × 10−9, presence of G versus GGGAGCCA). The variants are color coded by the magnitude of LD with the rs71947865.
The LD block contains several eQTL (expression quantitative trait loci) effects affecting numerous genes such as TRANK1, LRRFIP2, MLH1, GOLGA4, DCLK3, STAC, and long noncoding RNA RP11-640L9.2. The SNP rs11129735 (KLS discovery GWAS P = 9.9 × 10−6; LD r2 = 0.4 with our top GWAS significant variants) had the highest coloc (CLPP) probability (>0.95 causal probability) using eCAVIAR, with the expression of LRRFIP2 in the thyroid, DCLK3 in skin and brain frontal cortex, C3orf35 in testis, MLH1 in breast tissue, TRANK1 in the spinal cord, and EPM2AIP1 in the hypothalamus (GTEx v7). Querying the GTEx v8 database (30), we found our lead SNP rs71947865 variant to be a strong eQTL for LRRFIP2 gene expression in the thyroid (P = 8.2 × 10−20) and TRANK1 expression in breast tissue and whole blood. The rs71947865 also had multiple effects on LRRFIP2 expression in various tissues (P < 10−6, GTEx portal v8). LRRFIP2 together with MLH1 and GOLGA4 are highly expressed in placenta and differentially expressed in placentae from complicated compared to normal pregnancies (31, 32). The tissue expression for most of these genes is widespread in the brain and poorly characterized. For the reason of convenience, we further called this effect “TRANK1-region polymorphisms” or “TRANK1 effect,” although data to date do not allow genetic attribution of the observed effects to secondary changes in TRANK1 gene expression.
Association of Symptoms with Polymorphisms.
Clinical data collected from 592 of 673 KLS patients from the discovery cohort were next scrutinized for genotype effects. Our a priori hypothesis for testing was to explore whether TRANK1 polymorphisms were associated with more depressive symptoms, considering lithium responsiveness in the syndrome, and/or with a differential effect on birth difficulties, a robust finding in our prior study (3). Other variables were compared as exploratory analyses. TRANK1 rs71947865 (G) genotype was most significantly associated with reports of birth difficulties (OR = 1.65; P < 0.005 Fisher’s exact test of allelic counts) and developmental delays. In addition, patients with TRANK1 rs71947865 (G) have more severe cognitive symptoms, a longer duration of first episode, a more frequent family history of KLS, an older age of onset, and more behavioral disinhibition and altered speech during episodes, generally suggesting more severe disease (Table 2; full list of variables tested are provided in Dataset S4). Types of birth difficulties are summarized in Dataset S5. In a further analysis of the association of clinical variables with the genotype dose of the TRANK1 deletion, we adjusted for reports of birth difficulties. We noted that cognitive signs, age of onset, and longer first episode duration were significantly (P < 0.05) increased in KLS cases with TRANK1 deletion allele, while there was a trend to association of TRANK1 deletion allele with current depression in KLS cases (Table 3).
Table 2.
TRANK1 deletion (rs71947865) and its univariate association with KLS clinical variables
| Clinical variable | Model1 | Model2 | Fisher’s | BB (n = 81) | AB (n = 276) | AA (n = 235) | |||
| P | beta | P | Beta | P | Allelic OR | ||||
| Personal history of birth difficulties | 0.0021 | 0.50 | 0.0147 | 0.41 | 0.0021 | 1.66 | 32.4% (22/68) | 21.9% (48/219) | 14.8% (27/183) |
| Personal history of birth difficulties or developmental delay | 0.0024 | 0.46 | 0.0148 | 0.38 | 0.0026 | 1.58 | 38.2% (26/68) | 25.7% (58/226) | 19.1% (36/188) |
| Severe cognitive symptoms during episodes | 0.011 | 0.82 | 0.0505 | 0.65 | 0.0089 | 2.24 | 97.1% (67/69) | 96.3% (233/242) | 90.4% (179/198) |
| First episode duration, d | 0.0271 | 3.46 | 0.0512 | 3.13 | — | — | 17.64 ± 3.76 (72) | 16.13 ± 1.87 (241) | 11.51 ± 0.86 (204) |
| Age of KLS onset, y | 0.0347 | 0.62 | 0.0476 | 0.60 | — | — | 15.99 ± 0.56 (72) | 15.6 ± 0.29 (249) | 14.83 ± 0.32 (210) |
| Family history of KLS | 0.0427 | 0.27 | 0.614 | −0.07 | 0.0484 | 1.31 | 53.1% (34/64) | 61.7% (145/235) | 46.6% (97/208) |
| Behavioral disinhibition | 0.0453 | 0.28 | 0.287 | 0.15 | 0.0475 | 1.34 | 69.6% (48/69) | 72.4% (163/225) | 60.7% (116/191) |
| Altered speech during episodes | 0.0464 | 0.45 | 0.227 | 0.30 | 0.0476 | 1.55 | 91.8% (56/61) | 90.8% (207/228) | 84.6% (165/195) |
| Family history of Depression | 0.0585 | 0.26 | 0.542 | 0.09 | 0.0605 | 1.31 | 63.8% (44/69) | 73.5% (169/230) | 58.2% (113/194) |
| Abnormal behavior | 0.0707 | 0.36 | 0.978 | 0.006 | 0.0859 | 1.43 | 88.4% (61/69) | 89.7% (217/242) | 82.6% (166/201) |
| Sleep time during episodes (h) | 0.0831 | 0.37 | 0.107 | 0.348 | — | — | 18.74 ± 0.34 (68) | 18.43 ± 0.23 (216) | 18.01 ± 0.23 (170) |
| Attention deficit hyperactivity disorder | 0.0922 | 0.68 | 0.137 | 0.62 | 0.103 | 1.95 | 4.3% (3/69) | 3.4% (8/238) | 1.1% (2/189) |
We coded the minor allele additively and used linear regression for continuous variables and logistic regression for categorical variables: in model1, the minor allele genotype was fit as a function of the clinical variable; model2 was similar to model1 except for including ethnicity, gender, and missing data proportion as three covariates. A Fisher’s exact test of allelic counts was further performed for categorical variables. In these analyses, 592 KLS cases were included (frequency % | mean ± SEM was computed). BB, allele G/G; AB, GGGAGCCA/G; AA, GGGAGCCA/GGGAGCCA; Minor Allele G (deletion) was the effect allele.
Table 3.
Association of clinical variables with genotype dose of TRANK1 deletion (rs71947865) after adjusting for medical history of birth difficulties, gender, referrer/race, and missing data proportion in 592 KLS cases (linear regression for continuous variables and logistic regression for categorical variables)
| Clinical variable | P value | beta | BB (n = 81) | AB (n = 276) | AA (n = 235) |
| Severe cognitive symptoms during episodes | 0.011 | 0.99 | 97.1% (67/69) | 96.3% (233/242) | 90.4% (179/198) |
| Age at KLS onset, y | 0.026 | 0.76 | 15.99 ± 0.56 (72) | 15.6 ± 0.29 (249) | 14.83 ± 0.32 (210) |
| First episode duration, d | 0.046 | 3.63 | 17.64 ± 3.76 (72) | 16.13 ± 1.87 (241) | 11.51 ± 0.86 (204) |
| Current depression | 0.097 | 0.50 | 7.2% (5/69) | 7.1% (17/238) | 2.7% (5/188) |
Frequency % | mean ± SEM was computed. *Minor Allele G (deletion) was the effect allele.
TRANK1 genotyping was conducted in 195 controls with birth difficulty data available. These controls were from the KLS case control study reported by Arnulf et al. (3), with further addition of controls (friend or spouse of KLS patient) over more recent years. In the control sample, 22 subjects reported birth difficulties and, unlike in the KLS cases, we found no significant association of birth difficulties with TRANK1 rs71947865 genotype (P = 0.5, OR = 1.05). To formally test for an interaction effect of TRANK1 genotype on birth difficulties in KLS versus controls, we used the combined KLS cases−control samples matched by PCA. Whereas both birth difficulties and TRANK1 rs71947865 had a significant effect on KLS risk (P < 0.05), the interaction term of TRANK1 rs71947865 and presence of birth difficulties did not reach statistical significance. Note that the sample of controls with clinical (birth difficulty) data available is small and, although it was also used in the GWAS as a control, most of the controls used for GWAS were drawn from the Genetic Epidemiology Research on Aging (GERA) (33) cohort and/or other controls obtained through other studies from the same countries, which come from similar countries and collaborators.
The TRANK1 Association Is Birth Year Dependent.
As indicated above, our analysis revealed a significant association of the TRANK1 variant with birth difficulties in KLS patients but not controls. As perinatal care has improved over recent decades (34–39) (https://www.cdc.gov/nchs/data/nvsr/nvsr56/nvsr56_10.pdf; https://mchb.hrsa.gov/sites/default/files/mchb/Data/Chartbooks/child-health-2014.pdf), most notably, in the 1990–2000s, we asked whether the year of birth was an essential factor correlated to KLS in conjunction with TRANK1 rs71947865 variant. A possible birth year-dependent TRANK1 interaction is also supported by the recent largest bipolar disorder GWAS ever published, where it was noteworthy that the strong TRANK1 SNP association found in the discovery sample in bipolar disorder cases did not replicate in a new, presumably more recent, bipolar GWAS replication set (15), a caveat being that birth year data were not made available in this study (15) for all but one replication cohort where the birth year range was 1981–2005. Further, later reported TRANK1 ORs for bipolar disorder decreased in size from earlier studies in Brazil (40) and other countries (41–43) to the most recent metanalysis (15), even independently of the nonsignificant replication sample of this most recent metanalysis (15). This is illustrated in Dataset S3, where published TRANK1 association results are reported, with the caveat that the same samples are typically used cumulatively across studies.
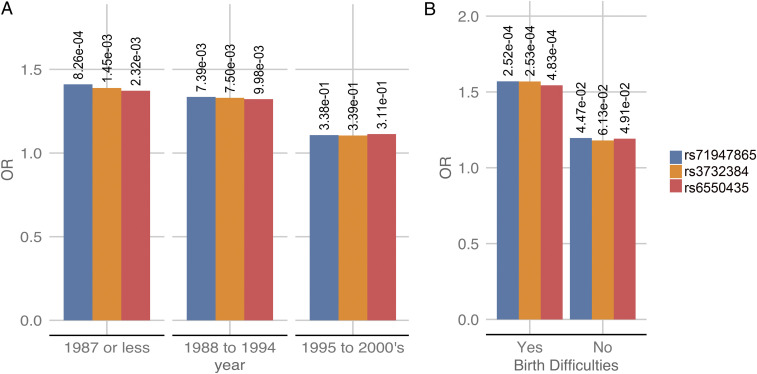
In a subset of the full cohort (discovery and replication sets) when a valid birth year was available (n = 650), we performed TRANK1 SNP association by birth year stratified into three approximately equal-sized groups; 1987 or earlier (n = 220, median year of birth: 1978), 1988–1994 (n = 202, median year of birth: 1992), and 1995–2000s (n = 228, median year of birth: 1999). Using genetic principle component matched controls for each stratum mainly derived from the GERA cohort (birth year range ∼1928–1989), we observed a decrease in TRANK1 SNP rs71947865 association with time (Fig. 3A and Dataset S6A). Notably, while individuals born in 1987 or earlier had the strongest association TRANK1 association (P = 8.3 × 10−4; OR = 1.41), this effect decreased in KLS cases born 1988–1994 (P = 7.48.3 × 10−3; OR = 1.34) to finally disappear in the 1995–2000s group (P = 0.31; OR = 1.11). In addition, within KLS cases, a Fisher’s test of allelic counts was performed, and birth year bin 1987 or earlier was compared to 1988–1994 and 1995–2000s in a 2 × 2 contingency table. We observed the highest association of TRANK1 SNP rs71947865 (P = 0.002) in year bin 1987 or earlier when compared to the minor allele count of TRANK1 SNP rs71947865 in the birth year bin 1995–2000s (Dataset S6B).
Fig. 3.
(A) The TRANK1-region SNP associations in KLS cases stratified by birth years; the y axis represents the odds ratio of the effect allele, while the x axis is the birth year. P value of the association is labeled. (B) The TRANK1-region SNP associations in KLS cases stratified by birth difficulties; the y axis represents the odds ratio of the effect allele, while the x axis is the presence of birth difficulties.
GWAS Analysis of Follow-up KLS Cases with and without Reports of Birth Difficulties.
To replicate these findings, we collected and genotyped 171 new KLS cases collected between 2017 and 2019, collecting information on birth difficulties whenever possible. In the resulting sample set, birth difficulties data were available for 151 of the 171 cases. In the overall replication sample (n = 171), generally born more recently (median year of birth: 1998; range: 1960–2015), TRANK1 rs71947865 was not significantly associated with KLS (P = 0.1, OR = 1.1) (Table 1 and Dataset S2). Strikingly, however, when we stratified this sample into cases with versus without birth difficulties, then a significant TRANK1 rs71947865 association was seen only in the first group (P = 0.01; OR = 1.54 in n = 59 cases reporting birth difficulties; Dataset S7). In KLS cases with no birth difficulties, no TRANK1 rs71947865 effect was observed (P = 0.4; OR = 1.12 in n = 88 cases). No birth information was available for n = 24 patients. A combined TRANK1 SNP metaanalysis (OR = 1.4;P = 3.5 × 10−8) of both discovery and replication cohorts is presented in Table 1.
Our prior analyses of the discovery cohort had revealed significant association of TRANK1 variant rs71947865 with birth difficulties; we therefore determined the association of TRANK1 rs71947865 variants in KLS cases with birth difficulties when compared to controls individuals in both discovery and replication cohorts. We observed that TRANK1 rs71947865 variants were significantly associated with birth difficulties in KLS cases from both the discovery cohort (P = 0.002; OR = 1.61; see Dataset S8A) and the follow-up sample of KLS cases (P = 0.03; OR = 1.53). A combined analysis of KLS cases with and without birth difficulties from combining both the discovery and replication cohorts compared to genetic PCA matched controls also revealed significantly increased TRANK1 variant rs71947865 frequencies in KLS cases with birth difficulties (Fisher’s test, OR = 1.54; P = 4.83 × 10−4; Fig. 3B and Dataset S8B), while, in KLS cases without birth difficulties, the effect was weaker (Fisher’s test, OR = 1.20; P = 0.04).
Fine Mapping of the KLS Signal across Ethnicity.
GWAS studies in schizophrenia and bipolar disorders have mapped the TRANK1 signal at SNP rs9834970 (r2 = 0.74 with KLS SNP rs71947865) in Caucasians (15, 42–44), while, when a mixed ethnic sample that included Asians and Caucasian was used, the top SNP is rs3732386 (16, 45–47), a signal entirely linked (r2 = 1) with the KLS-associated SNP rs71947865 (Dataset S3 and SI Appendix, Fig. S2 A–C).
Since our KLS sample is derived from multiple ethnicities, as an additional validation, we sought to determine TRANK1 loci association KLS cases of European/Caucasian origin only. In the overall European KLS metaanalysis, we observed that the TRANK1 region SNP rs71947865 (OR = 1.28; P = 1.9 × 10−4) was retained as the top association (Dataset S9 and SI Appendix, Fig. S2 A–C), with no evidence of heterogeneity. Our KLS European/Caucasians-only analysis (discovery plus replication cohort) revealed that rs9834970 was among the top associations in the loci (SI Appendix, Fig. S2D and Dataset S9). The rs3732386 and rs71947865 were the top associations in the TRANK1 region and were dependent on the existence of associated birth difficulties (SI Appendix, Fig. S2). Further, on comparing the associations in the TRANK1 loci in discovery KLS versus combined schizophrenia and bipolar disorder GWAS, there is remarkable overlap of signals, with only the magnitude of ORs being 1.3× increased in KLS (SI Appendix, Fig. S2D).
Polygenic Risk Score of KLS.
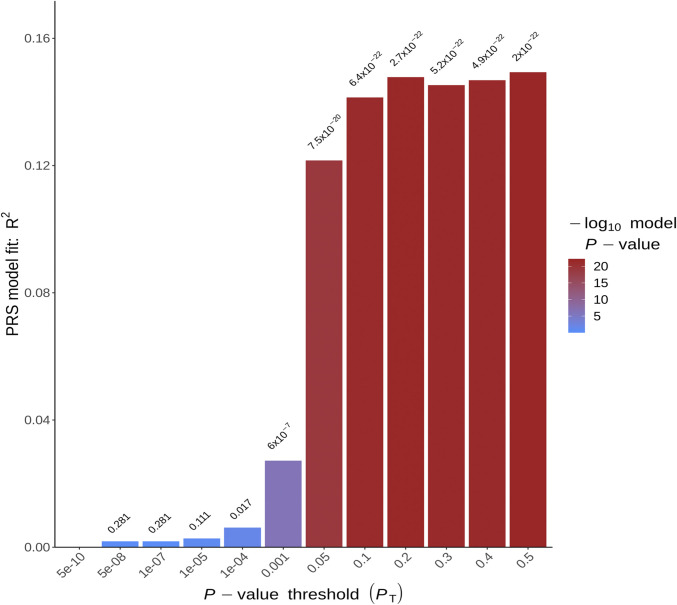
We found significant evidence that the KLS discovery cohort polygenic risk scores (PRSs) were indicative of increased genetic liability to KLS phenotype in a follow-up replication cohort (n = 171) under various P value thresholds using PRSice2 (48). The most predictive P value threshold was 0.5 (pseudo R2 P = 2 × 10−22; Fig. 4) using 81,570 variants. P value thresholds ranging from 0.05 to 0.4 were also indicative of increased genetic liability to KLS (P < 1 × 10−20, mean pseudo R2 = 0.14; see Dataset S10). Further, as an additional control, we also used only Caucasian KLS sample discovery metaanalysis (n = 299) to build the PRS with validation only in Caucasian KLS replication sample (n = 160); the PRSs were indicative again of increased genetic liability to KLS (pseudo R2 = 0.094), with the best score at P = 0.4 threshold (SI Appendix, Fig. S3).
Fig. 4.
Phenotypic variance explained by PRSs (pseudo R2) built on the KLS discovery cohort and tested on the KLS replication cohort; the y axis is the pseudo R2, while the x axis represents the P value thresholds. The color gradient over each bar is representative of the P value association between the PRS and the KLS trait in replication cohort.
Considering the results above, we built PRSs using GWAS summary statistics from bipolar disorder (15), schizophrenia (49), circadian chronotype (50), sleep duration (51), and excessive daytime sleepiness (52), using previously published GWAS studies. These PRSs were used to score our KLS follow up cohort. Notably, shared KLS genetic liability (pseudo R2 = 0.06) was found using bipolar disorder and schizophrenia GWAS summary statistics as the training set (SI Appendix, Fig. S3 and Dataset S10). LD score regression analysis estimated the total observed scale heritability h2 = 0.09 (intercept = 1.044) and also revealed significant genetic correlation of KLS with bipolar disorder (rg genetic correlation = 0.41; P < 0.05) and depressive symptoms (rg = 0.32; P < 0.05; Dataset S11).
Gene Pathway Analysis.
Biological processes pathway enrichment analysis was next conducted using MAGMA (53). The MAGMA module gene-based association analysis was first performed in the combined discovery−replication sample summary statistics mapping SNPs to genes with a 1-kb window upstream and downstream. This first step entails collapsing mean individual SNP statistics into a per gene-level statistic. Then a gene P value is obtained by using a known approximation of the sampling distribution as detailed in MAGMA (53). The gene-based P values converted to z scores are then used to conduct gene set enrichment analyses on gene sets derived from MsigDBv6.1 (5,346 curated gene sets). At 5% FDR (false discovery rate), pathways “Reactome- RORA activates circadian expression” (54) and “Reactome-Circadian Repression of Expression By Rev ERBA” (55) were observed as significantly enriched in KLS genetic association analyses (Table 4). Notably, these two pathways shared identical gene members except for a single gene. Gene-based association tests within these enriched pathways are presented in Dataset S12. Both ROR Alpha (retinoic acid receptor-related alpha) and Rev ERBA are critical transcriptional regulators of periodic expression of the BMAl1 gene, directly regulating circadian pathways (56–59). Other circadian pathways of interest included “Reactome-Bmal1 Clock NPAS2 Activates Circadian Expression” but did not reach 5% FDR threshold.
Table 4.
A gene set enrichment analysis was carried out on the combined discovery and follow-up cohort of KLS cases using the MAGMA module
| Gene set name | Ngenes | Beta | P value | FDR P value |
| Reactome_Rora_Activates_Circadian_Expression | 22 | 1.03 | 1.29×10−7 | 0.00069 |
| Reactome_Circadian_Repression_Of_Expression_By_Rev_Erba | 21 | 0.97 | 2.17×10−6 | 0.00579 |
| Go_Regulation_Of_Tumor_Necrosis_Factor_Superfamily_Cytokine_Production | 88 | 0.29 | 0.00050 | 0.61043 |
| Go_Positive_Regulation_Of_Multicellular_Organismal_Metabolic_Process | 17 | 0.66 | 0.00051 | 0.61043 |
| Reactome_Bmal1_Clock_Npas2_Activates_Circadian_Expression | 33 | 0.51 | 0.00057 | 0.61043 |
A total of 5,346 sets including curated gene sets, KEGG pathways, Reactome pathways, and GO biological processes derived from msigdb6.1 were used (5% FDR P value was computed). Beta is the direction of association of the pathway/gene set with the KLS case control status. Ngenes is the number of genes.
KLS Exome Sequencing Analysis.
Targeted exome sequencing was performed in 32 individuals with KLS, derived from 16 multiplex families (Dataset S13A). The variant frequencies and variant by variant analyses compared to 286 European controls from 1000 Genomes phase 3 were largely unremarkable. Damaging or probably damaging variants in each gene were collapsed, and mutational burden in KLS probands and/or shared by at least three families was identified and presented below.
LMOD3 Gene Variants Are Not Associated with KLS.
Following up on a recent study suggesting increased mutational load in the LMOD3 gene in familial KLS cases (60), we analyzed the LMOD3 burden in our exome sequencing cohort of KLS probands and sporadic cases (n = 17) compared to controls (n = 286). We observed no significant differences in mutational burden in the LMOD3 gene between cases and controls (P = 0.13; see Dataset S13B). Variant level analyses in KLS probands (n = 17) revealed a previously reported single site rs35740823 (chr3:69171290; p.R83H) to be enriched in KLS cases (allelic OR = 3.8; P = 0.06) as compared to controls (60) (Dataset S13B). We therefore imputed this LMOD3 variant rs35740823 and analyzed for associations in 151 KLS cases and 478 controls derived from replication cohort and predominantly of European origin, but did not find any remarkable differences (OR = 1.1; P = 0.77; Dataset S13B).
Gene Mutational Burden Test in KLS.
We also compared the overall frequency of modifier variants tagged using SnpEff (61) as moderate or high effects in all genes in all familial KLS cases versus 1000 Genome controls (n = 286) running gene-based mutational burden analyses (Dataset S14A), matched for ethnicity and SNP principal components. Although significant effects were found, none were found in genes within the TRANK1 loci. However, we observed increased mutational burden in the rare alleles in genes NRXN2, GABRG2, CHORDC1, AATK, and CASP7 (FDR P < 1 × 10−4) among probands (n = 17) relative to 1000 Genomes reference samples (Dataset S14A). Genes with excessive rare alleles across each KLS family were next analyzed. Notably, GABRG2, CASP7, and ANKRD31, among other genes, were shared across at least three KLS families (Dataset S14B).
Further, pathway burden test from the list of 21 genes belonging to the circadian regulation pathway identified through the MAGMA analysis revealed significantly increased minor allele burden (Dataset S15A) in the whole KLS cohort (n = 32) compared to European origin controls. We also used pathway sets from msigDB6.1 to profile mutational burden in each pathway in KLS probands (n = 14). Notably, we found increased mutational burden in GO term GAMMA AMINOBUTYRIC ACID SIGNALING PATHWAY (FDR P = 0.007) in KLS probands among other pathways that included many neuronal pathways (Dataset S15B). This is in agreement with results of the gene burden test where we observed significantly increased burden in GABRG2 in KLS probands.
Discussion
Our study explores the interaction of genetic and environmental contributions to the pathophysiology of KLS. Results of GWAS revealed that TRANK1 gene variants were predisposing and associated with birth difficulties in KLS. PRS constructed on discovery GWAS analysis found strongly replicable PRS that overlapped with bipolar disorder and schizophrenia. Pathway analysis also revealed the involvement of circadian regulation genes.
This large study did not confirm findings made in other smaller genetic studies of KLS. For example, a 20-y-old study had suggested an HLA-DQ2 association (12) in KLS, but this was not confirmed either in other studies (3, 8, 13) or in our current analyses. A recent study (60) focused on a large Saudi pedigree KLS-affected family where an rs111848977 p.E142D mutation in LMOD3 was observed to cosegregate. This study also reported an increased burden of damaging mutations, including p.R23H in LMOD3 in KLS, a gene involved in nemaline myopathy 10 (62). In our sample of 32 familial cases, we could not find any overall increase in the frequency of LMOD3 mutations, and, although p.R83H was nominally increased in familial cases, we did not find this association in a larger cohort of 151 KLS cases (Dataset S13B).
Although the phenomenology of KLS is quite different from that of bipolar disorder or schizophrenia, these syndromes have some clinical similarities. Most notably, in both KLS and bipolar disorder, patients are asymptomatic between episodes. Similarly, exacerbations are usually episodic in some schizophrenia. In KLS, however, onset/offset of episodes is rapid (typically within 1 d), while it is generally slower in most bipolar patients, except for “rapid cyclers” (63). Further, although not a dominant feature, depression, anxiety, or hallucinations/delusions can occur during KLS episodes (3, 4, 6, 64), and, at the end of episodes, some patients may have brief (1 d to 2 d) period of elation, insomnia, and logorrhea which can resemble a manic miniepisode.
Most interestingly, hypersomnia is a common feature of depressive episodes in bipolar type 1 disorder (65–67), unlike in other depressions, where patients most often exhibit insomnia. Like KLS, bipolardisorder is also associated with cognitive deficiencies, although these are milder (68, 69) as compared to KLS patients. This, together with the fact that lithium is the only treatment reported to have an effect in preventing KLS episodes (7), suggests possible shared pathophysiology with bipolar disorder. Pathophysiological overlap among KLS, bipolar disorder, and schizophrenia is also in line with genetic studies in the UK Biobank cohort that have shown that the genetic architecture of long self-reported sleep duration overlaps with that of bipolar disorder, in contrast with the genetic architecture of insomnia that shares a similarity with anxiety and restless leg syndrome (70). Most strikingly, strong correlations between PRS scores of long sleep duration and bipolar disorder type 1 in the UK Biobank sample were found (71). This suggests that hypersomnia, periodic or not, could share some commonality with bipolar disorder and schizophrenia.
One possible hypothesis is that KLS, like bipolar disorder and other psychosis, is characterized by periodic instability of specific brain networks involving, in the case of KLS, reduced metabolism in cortical and subcortical areas, as reported by imaging studies (72). In this syndrome, cognitive and derealization symptoms may correlate with decreased activity in cortical areas, whereas decreased activity in thalamus and hypothalamus may be involved in the functional hypersomnia observed in disorder (72–74). Involvement of circadian or behavioral rhythmicity genes may be essential for maintaining hypersomnia episodes once an episode is initiated, for example, if subjects are suddenly spending much time in bed, unexposed to light, a behavior which could result in difficulties entraining in genetically susceptible individuals (75). This hypothesis could also explain why patients with KLS can exhibit sleep onset REM periods during Multiple Sleep Latency Test (MSLT; the circadian clock strongly drives occurrence of REM sleep) (76, 77), a feature also associated with shift work (78). Other investigators have found abnormal circadian entrainment during KLS episodes using actigraphy (79). A role for circadian abnormalities has also been suggested in bipolar disorder type 1 (67, 80, 81). Subjects with bipolar type 1 also reported increased eveningness tendencies (71), although this was not found in patients with KLS (3).
The most striking finding of this study is the observation of a strong genome-wide significant association of KLS with TRANK1-region variants (OR of ∼1.50) that are strongly associated with bipolar disorder (OR of ∼1.12) (43, 82, 83) and schizophrenia (OR of ∼1.08) (46, 84–86), but not mood instability (87). Although it remains possible this effect could be due to a rarer variant linked with this haplotype in KLS, this is more likely mediated by a similar effect of these common variants across these diseases. Of notable importance is the fact the TRANK1 effect size in this study is four to five orders of magnitude higher (OR of ∼1.5) than in studies in these other pathologies (OR of ∼1.1), explaining detection of association in a small sample of less than 1,000 KLS patients. This association with KLS is thus unlikely to be explained by contamination of our current KLS cohort with bipolar cases, although the opposite, contamination of bipolar or schizophrenia samples with KLS patients, could explain weaker associations in these other pathologies.
The reason for the weaker than expected TRANK1 replication in the KLS replication cohort became evident after stratifying our discovery sample by birth decade, finding that the TRANK1 association was highest in subjects born before the 1980s and decreased thereafter. Further, KLS subjects with TRANK1 polymorphism have reported increased birth difficulties (summarized in Dataset S5). We hypothesize that improved perinatal outcomes that have occurred in the past 50 y may explain this tendency (34–39) (https://www.cdc.gov/nchs/data/nvsr/nvsr56/nvsr56_10.pdf; https://mchb.hrsa.gov/sites/default/files/mchb/Data/Chartbooks/child-health-2014.pdf). The TRANK1 birth difficulties relationship was also confirmed in the independent replication sample, where we found that only the subset of KLS patient reporting birth difficulties had a significant TRANK1 association. Overall, patients who reported birth difficulties had the highest association (Fisher’s test, OR = 1.57; P = 2.5 × 10−4) in both the discovery and replication cohorts (Dataset S9). TRANK1 association also increases disease severity during episodes independently of birth difficulty effects (Table 3).
Interestingly, although the association with TRANK1 was the highest genome-significant finding in bipolar disorder in a recent metanalysis of 32 cohorts from 14 countries in Europe, North America, and Australia, totaling 20,352 cases and 31,358 controls of European descent (rs9834970; OR = 1.10, P = 5.5 × 10−14) (15), as in our current KLS sample, replication in follow-up bipolar disorder samples totaling 9,412 cases and 137,760 controls of European ancestry did not replicate (OR = 1.02, P = 0.15). This finding deviated more than expected by chance (P = 0.01, although not significant if Bonferroni corrected for all loci detected in the study). We hypothesize that a similar time-dependent TRANK1 birth difficulty interaction may play a role in bipolar disorder and schizophrenia. Indeed, these disorders are known to be strongly associated with early-life complications (17–28, 88, 89), and data in the UK Biobank cohort indicate a dose-dependent increased prevalence of major depressive disorders and bipolar disorders in association with reported low birth weight (90), even when controlled for a history of the physical disorder known to be associated with low birth weight.
Two possibilities, which are not necessarily mutually exclusive, could explain these data. First, TRANK1-region genes could affect placental physiology, modulating the risk of birth difficulties and long-term fetal damage that could manifest as KLS or other disorders. Ursini et al. (31) found strong significant genetic interaction between the PRS of schizophrenia and the occurrence of early-life complications, in multiple case−control samples. Further, MLH1, LRRFIP2, and GOLGA4 associated with these overall genetic components were highly expressed in the placenta and modulated by pathological placental processes such as preeclampsia and intrauterine growth restriction, but not in other disease processes (31). Problematically, however, recent papers did not observe that the KLS-associated SNPs identified in this study had substantial effects on placental expression of these genes (31, 32, 91, 92). Further, TRANK1-region polymorphisms are not known maternal or fetal genetic factors associated with low birth weight (93–95), prematurity (96), or birth complications (91, 97) in large samples, and were not associated with birth difficulties in a small sample of 198 controls ascertained similarly to our KLS subjects for birth difficulties. Finally, this hypothesis would not explain the time dependency/cohort effect of the TRANK1 association we observed with KLS, which is likely due to improved or changes in perinatal care with time.
Another possible explanation, which we favor as it better fits our results and the lack of effects of TRANK1 on birth difficulties in previously published studies, is that newborns carrying TRANK1-region polymorphisms are more susceptible to injury in the presence of birth difficulties (notably hypoxia) in a way that produces behavioral instability such as KLS or other psychiatric conditions such as bipolar disorder in adulthood. This would be a true gene × environment interaction, although our study lacked sufficient control to formally test this hypothesis statistically. The mechanism underlying this phenomenon, which could have evolutionary advantages in times of stress (98), remains to be investigated. Interestingly, rs9834970, an SNP linked with the TRANK1-region susceptibility haplotype, is associated with decreased expression of TRANK1 in induced pluripotent stem cells (iPSCs) and neural progenitor cells (99); such an effect has not been reported in the adult brain, but, as discussed, TRANK1-associated polymorphism reduces expression in various dividing tissues such as blood, esophageal mucosa, and B cells. The iPSC/neuronal precursor cell effect was rescued by valproic acid but not lithium, and the TRANK1 gene knockdown was found to modulate the expression of many genes, suggesting developmental effects (99).
Because the susceptibility haplotype modulates expression of many other genes in a tissue-specific manner, however, we believe the study of other genes in this region across tissues and development with the susceptibility haplotype will be necessary before concluding that TRANK1 is the gene behind this genetic association, which is a limitation of our analyses. Additional studies aiming at studying more precisely which types of birth difficulties are associated with TRANK1 in these disorders are also needed. Our results offer clues on the developmental window in which the TRANK1-susceptibility haplotype could affect risk for KLS and mental disorders in adulthood and mandate a more careful analysis of what and how the reported birth difficulties interact with specific TRANK1 genetic variants.
Our data support a genetic predisposition in the presence of TRANK1 polymorphisms and link KLS, circadian regulation, and bipolar disorder. PRSs derived from discovery KLS samples are strongly predictive of the KLS phenotype in the replication sample. Further, PRSs support a genetic overlap with bipolar disorder and schizophrenia, with a caveat being that these associations in various P thresholds were unadjusted for multiple comparisons and likely are underpowered, since only the replication follow-up KLS sample was used. Strikingly, the temporal effect of TRANK1 polymorphism was observed in KLS cases in relation to the year of birth. In the presence of birth difficulties, the TRANK1 polymorphism may predispose to pathophysiological consequences such as KLS, bipolar disorder, and schizophrenia.
Materials and Methods
Inclusion Criteria for KLS and Origin of Cases.
Patients with KLS were included if they met International Classification of Sleep Disorders (ICSD3) criteria for the disorder, including the presence of recurrent episodes (≥2) of severe hypersomnia (>18/24 h) lasting days to weeks, associated with cognitive disturbances (confusion, amnesia, and derealization) and behavioral abnormalities (major apathy, megaphagia, and hypersexuality). These episodes had to be interspersed with long periods of normal sleep, cognition, behavior, and mood. A sleep-trained physician diagnosed all patients. They were also asked to answer a detailed questionnaire, or a systematic interview as described (3, 6). Informed consent, in accordance with governing institutions, was obtained from all subjects. The research protocol was approved by Institutional Review Board (IRB) panels on Medical Human Subjects at Stanford University, and by respective IRB panels in each country providing samples for the study.
KLS Cases.
The discovery sample of 673 KLS cases includes 261 cases recruited in the United States, with the help of the Kleine-Levin Syndrome Foundation (KLSF). US patients were diagnosed by an outside physician or referred by the KLSF (N.F.), and the diagnosis was confirmed at Stanford, where the questionnaire and blood sample were collected. Other major recruitment sites include Pitié-Salpêtrière, Paris, France (I.A., n = 140); University of Peking, Beijing, People’s Republic of China, (F.H., n = 62); Taiwan University, Taipei, Republic of China, Taiwan (Y.-S.H., n = 58); Guy de Chauliac Hospital, Montpellier, France (Y.D., n = 35); Safra Children’s Hospital, Sheba Medical Center, Tel Aviv University, Tel Aviv, Israel (Y.S., n = 34); Hepata Clinic, Germany (G.M., n = 24); Uppsala University, Sweden (A.M.L., n = 15); Hospital Robert Debre, Paris, France (M.L., n = 7); Tokyo Metropolitan Institute of Medical Science Tokyo, Japan (M.H., n = 7); University of Bologna, Italy (F.P. and G.P., n = 6); Maynei Hayeshua Medical Center, Bnei Barak, and The Sackler Faculty of Medicine, Tel-Aviv University (N.G., n = 6); Akita University, Akita, Japan (T.K., n = 2); St. Vincent's Hospital, The Catholic University of Korea (S.C.H., n = 2); 1st Faculty of Medicine and General Teaching Hospital, Prague, Czech Republic (S.N., n = 2); Bambino Gesù Children Hospital, Rome, Italy (Marina Macchialio, n = 2); and other centers who contributed single sample (n = 10).
The replication cohort included 171 additional KLS cases. These included cases from Pitié-Salpêtrière, Paris, France (I.A., S.L.-S., and P.D., n = 66); Uppsala University, Sweden (A.M.L., n = 26); KLSF (n = 23); University of Bologna, Italy (F.P. and G.P., n = 13); London Bridge Hospital, London, United Kingdom (G.D.L., n = 11); Guy de Chauliac Hospital, Montpellier, France (Y.D., n = 9); Emory University, Atlanta (D.R., n = 6); Hepata Clinic, Germany (G.M., n = 5); Stanford University, (E.J.-M.M., n = 5); Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan (M.H., n = 4); and other centers (n = 3).
GWAS Patients and Matched Controls.
Because the samples were collected over a long time frame (2003−2019), genotyping platforms evolved, and hence each platform required its cohort of cases and controls (Dataset S1). As a result, the discovery sample (n = 673, 2003–2017) was subdivided into seven subcohorts of cases and controls based on genotyping platform/ethnicity differences. Cases and ethnically matched controls (n = 15,339) were genotyped at University of California, San Francisco genomics core with Affymetrix Axiom Genome-Wide LAT, EUR, CHB, Axiom 55066 arrays, and Axiom world arrays, while multiethnic Axiom PMRA arrays were genotyped at Akesogen Inc. These include three European ancestry cohorts, an Asian cohort, a Latino cohort, and Ashkenazi and non-Ashkenazi Jewish cohorts. The replication sample (n = 171, collected between the years 2017 and 2019), subcohort 8, was treated as a single sample and genotyped solely on Axiom PMRA arrays. DNA was extracted using Qiagen DNeasy kit, and samples were stored at −20 °C until genotyping; a minimum of ∼200 ng was used for genotyping.
Genotype Analysis and Imputation.
Raw CEL files were imported into Affymetrix Genotyping console, and Affymetrix best practices workflow was followed. Genotype calls were exported as plink ped/map files (100). For CEL files generated from Axiom world arrays, primarily AffyPipe was used to call the genotypes and exported as plink ped/map files (101). Stringent quality control (QC) was applied, variants and individuals with >0.05 missingness were excluded, related individuals with identity by descent pihat > 0.2 were excluded, and markers with Hardy−Weinberg equilibrium P < 1e−6 in controls were excluded. HapMap and 1000 Genomes phase 3 reference population genotypes (102) were used to guide ethnicity matching in our sample subcohorts. Genetic Prinicipal Components (PCs) were extracted using plink. PC plots were visually inspected for unexpected deviations and outliers, after which Euclidean distance was used to accurately and automatically match cases and controls. Post QC, genotypes were phased one chromosome at a time with shapeit (103, 104) using the build37 coordinate system of the human genome. Phased haplotypes were then subject to imputation to 1000 Genomes phase 1 in 1-mb chunks for each chromosome, using impute2 (105).
Association Testing and Variant Prioritization.
Frequentist association tests were then performed on the imputed genotypes by including genetic principal components (PCA) and missing data proportion using snptest (106). Metaanalysis was subsequently performed using cohort-specific β estimates using a fixed effects model (106). The genome-wide significance threshold was set at P < 5e-8 based on Bonferroni adjustment. Gene-based association tests, including pathway analysis, were performed with MAGMA modules (107). Manhattan plots were constructed with R package qqman (108). Additional eQTL annotation was carried out using SNIPA, webserver (109), and GTEx v8 (30). eCAVIAR was used to perform eQTL colocalization (110). LD hub was used to calculate LD score regression and find shared SNP heritability across phenotypes (111). PRSs were computed using PRSice2 (48), briefly imputed genotypes in BGEN format from the replication dataset were quality controlled (minor allele frequency > 0.05 and impute info score ≥ 0.9) and clumped for LD (250-kb window; r2 = 0.1) and used as a target dataset along with the first five genetic PCAs. The number of common variants remaining after clumping from the base and target dataset were 104626. Apart from using summary statistics from the KLS discovery, we also used summary statistics from various other phenotypes. PRSs at various P value thresholds (5 × 10−10, 5 × 10−8, 1 × 10−7, 1 × 10−5, 0.0001, 0.001, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, and 1) were derived and validated in KLS replication cohort (n = 171).
Exome Sequencing Sample and Matched Controls.
Agilent Sure Select V4 system workflow was used to capture regions of interest; the resulting fragments were subject to Illumina paired-end sequencing run with an average depth of 130× and average read length of 100 in the target regions for the KLS family DNA samples. The KLS family DNA consisted of 32 individuals derived from 16 multiplex families (Dataset S13A). On the other hand, for controls, exome FASTQ files from 286 individuals part of 1000 Genomes (predominantly CEU, TSI, GBR, IBS, and FIN) were used. The FASTQ files were aligned to UCSC hg19 reference FASTA using BWA (112) and read groups inserted for downstream variant quality score recalibration. GATK best practices workflow (https://gatk.broadinstitute.org/hc/en-us/sections/360007226651) was implemented, and joint genotyping was performed on both cases and controls simultaneously. Raw variant call sets were subject to variant quality score recalibration, and variants passing the filters were then imported into plinkseq (https://www.atgu.mgh.harvard.edu/resources/software/) and used in downstream association analysis. Annovar and SnpEff were used to annotate variants (61, 113), following which moderate and high modifier variants were collapsed, and gene-based burden tests in various subcohorts were carried out using plinkseq.
A gene burden test was carried out within each family, probands only and all 32 individuals. Each analysis used the 1000 Genomes European dataset matched by principle components as a control cohort. The burden analyses were focused only in rare variants (minor allele counts 2 to 20) and restricted to predicted to be moderate or high modifiers as tagged by Annovar and SnpEff. Further, genes that were part of the rhythmic behavior pathway were collapsed, and SKAT-based (sequence kernel association test) gene burden tests were performed (114) with resulting P values adjusted using a 5% FDR.
Taqman Genotyping.
To validate the genotype calls from SNP genotyping arrays, we conducted Taqman assays for TRANK1 SNPs rs71947865 and rs6550435 on a random subset of KLS cases and controls typed across multiple platforms (n = 122). Allele concordance for rs71947865 was 100%, while, for rs6550435, it was 97% in 122 subjects.
Clinical Associations.
Demographic and various clinical features within the KLS cohort (n = 592) were analyzed for associations with TRANK1 rs71947865 variant (deletion). Three kinds of analyses were performed: In model 1, the minor allele genotype (additively coded) was fit as a function of the clinical variable; model 2 was similar to model 1 except for including ethnicity, gender, and missing data proportion as three covariates. A Fisher’s exact test of allelic counts was further performed for categorical variables.
Birth Year Associations.
Two analysis strategies were pursued. First, when birth year was available for KLS cases (n = 650), we stratified the KLS cases into three approximately equal-sized groups: 1987 or before (n = 220, median year of birth: 1978), 1988–1994 (n = 202, median year of birth: 1992), and 1995–2000s (n = 228, median year of birth: 1999). Within each group, we matched control samples based on genetic principle components. Fisher's exact and χ2 tests on TRANK1 rs71947865 allelic counts were performed to find significant difference in allele frequencies. A Fisher’s test of TRANK1 SNPs allelic counts was performed within KLS cases stratified by the three birth year bins; the minor allele counts in birth year bin 1987 or earlier was compared to minor allele counts 1988–1994 and 1995–2000s in 2 × 2 contingency table.
Supplementary Material
Acknowledgments
We thank countless staff members, clinicians, patients, and patient advocates who are not listed here and have provided material and logistic support over decades for these 15-y-long studies. The study was primarily funded by the KLSF (2003−2020) and by NIH Grant 1R01MH080957 (2007−2013) to E.J.-M.M. The French Kleine-Levin Syndrome research program is financed by a grant to I.A. (Grant PHRC 070138) from the French Ministry of Health. H.M.O. has been supported by the Academy of Finland (Grant 309643), Yrjö Jahnsson Foundation, and Oskar Öflund Foundation. Computing for this project was performed on the Stanford Genomics cluster supported by NIH Grant 1S10OD023452-01. We thank the Stanford Research Computing Center for providing computational resources and support that contributed to these research results.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005753118/-/DCSupplemental.
Data Availability
All the analyses presented in the manuscript is from data in Datasets S1 to S15 which are available. Complete summary statistics of the genetic associations are available at https://doi.org/10.6084/m9.figshare.14128475.v2.To protect the privacy of the study participants, the individual genetic data generated and analyzed during this study are available from the corresponding authors on reasonable request.
References
- 1.Kleine D. W., Periodische Schlafsucht. Eur. Neurol. 57, 305–320 (1925). [Google Scholar]
- 2.Levin M., Periodic somnolence and morbid hunger: A new syndrome. Brain 59, 494–504 (1936). [Google Scholar]
- 3.Arnulf I., et al., Kleine-Levin syndrome: A systematic study of 108 patients. Ann. Neurol. 63, 482–493 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Arnulf I., Rico T. J., Mignot E., Diagnosis, disease course, and management of patients with Kleine-Levin syndrome. Lancet Neurol. 11, 918–928 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Billiard M., Jaussent I., Dauvilliers Y., Besset A., Recurrent hypersomnia: A review of 339 cases. Sleep Med. Rev. 15, 247–257 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Arnulf I., Zeitzer J. M., File J., Farber N., Mignot E., Kleine-Levin syndrome: A systematic review of 186 cases in the literature. Brain 128, 2763–2776 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Leu-Semenescu S., et al., Lithium therapy in Kleine-Levin syndrome: An open-label, controlled study in 130 patients. Neurology 85, 1655–1662 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Huang Y. S., et al., Relationship between Kleine-Levin syndrome and upper respiratory infection in Taiwan. Sleep (Basel) 35, 123–129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen Q. T., et al., Familial Kleine-Levin syndrome: A specific entity? Sleep (Basel) 39, 1535–1542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peraita-Adrados R., Vicario J. L., Tafti M., García de León M., Billiard M., Monozygotic twins affected with Kleine-Levin syndrome. Sleep (Basel) 35, 595–596 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Habra O., Heinzer R., Haba-Rubio J., Rossetti A. O., Prevalence and mimics of Kleine-Levin syndrome: A survey in French-speaking Switzerland. J. Clin. Sleep Med. 12, 1083–1087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dauvilliers Y., et al., Kleine-Levin syndrome: An autoimmune hypothesis based on clinical and genetic analyses. Neurology 59, 1739–1745 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Lavault S., et al., Kleine-Levin syndrome in 120 patients: Differential diagnosis and long episodes. Ann. Neurol. 77, 529–540 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Lam M.et al.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Indonesia Schizophrenia Consortium; Genetic REsearch on schizophreniA neTwork-China and the Netherlands (GREAT-CN) , Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 51, 1670–1678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stahl E. A.et al.; eQTLGen Consortium; BIOS Consortium; Bipolar Disorder Working Group of the Psychiatric Genomics Consortium , Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda M., et al., Genome-wide association study detected novel susceptibility genes for schizophrenia and shared trans-populations/diseases genetic effect. Schizophr. Bull. 45, 824–834 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stilo S. A., Murray R. M., Non-genetic factors in schizophrenia. Curr. Psychiatry Rep. 21, 100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Söderlund J., Wicks S., Jörgensen L., Dalman C., Comparing cohort incidence of schizophrenia with that of bipolar disorder and affective psychosis in individuals born in Stockholm County 1955-1967. Psychol. Med. 45, 3433–3439 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Chudal R., et al., Perinatal factors and the risk of bipolar disorder in Finland. J. Affect. Disord. 155, 75–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haukvik U. K., et al., Pre- and perinatal hypoxia associated with hippocampus/amygdala volume in bipolar disorder. Psychol. Med. 44, 975–985 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nosarti C., et al., Preterm birth and psychiatric disorders in young adult life. Arch. Gen. Psychiatry 69, E1–E8 (2012). [DOI] [PubMed] [Google Scholar]
- 22.McGrath J., Scott J., Urban birth and risk of schizophrenia: A worrying example of epidemiology where the data are stronger than the hypotheses. Epidemiol. Psichiatr. Soc. 15, 243–246 (2006). [PubMed] [Google Scholar]
- 23.Cannon M., Jones P. B., Murray R. M., Obstetric complications and schizophrenia: Historical and meta-analytic review. Am. J. Psychiatry 159, 1080–1092 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Kunugi H., Nanko S., Murray R. M., Obstetric complications and schizophrenia: Prenatal underdevelopment and subsequent neurodevelopmental impairment. Br. J. Psychiatry Suppl. 40, s25–s29 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Kinney D. K., Yurgelun-Todd D. A., Tohen M., Tramer S., Pre- and perinatal complications and risk for bipolar disorder: A retrospective study. J. Affect. Disord. 50, 117–124 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Geddes J. R., Lawrie S. M., Obstetric complications and schizophrenia: A meta-analysis. Br. J. Psychiatry 167, 786–793 (1995). [DOI] [PubMed] [Google Scholar]
- 27.McNeil T. F., Perinatal risk factors and schizophrenia: Selective review and methodological concerns. Epidemiol. Rev. 17, 107–112 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Brixey S. N., Gallagher B. J. 3rd, McFalls J. A. Jr., Parmelee L. F., Gestational and neonatal factors in the etiology of schizophrenia. J. Clin. Psychol. 49, 447–456 (1993). [DOI] [PubMed] [Google Scholar]
- 29.Shimizu H., Hirose A., Tatsuno T., Nakamura M., Katsube J., Pharmacological properties of SM-3997: A new anxioselective anxiolytic candidate. Jpn. J. Pharmacol. 45, 493–500 (1987). [DOI] [PubMed] [Google Scholar]
- 30.GTEx Consortium , The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ursini G., et al., Convergence of placenta biology and genetic risk for schizophrenia. Nat. Med. 24, 792–801 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Peng S., et al., Expression quantitative trait loci (eQTLs) in human placentas suggest developmental origins of complex diseases. Hum. Mol. Genet. 26, 3432–3441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann T. J., et al., Next generation genome-wide association tool: Design and coverage of a high-throughput European-optimized SNP array. Genomics 98, 79–89 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sander A., Wauer R., From single-case analysis of neonatal deaths toward a further reduction of the neonatal mortality rate. J. Perinat. Med. 47, 125–133 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Sugai M. K., Gilmour S., Ota E., Shibuya K., Trends in perinatal mortality and its risk factors in Japan: Analysis of vital registration data, 1979-2010. Sci. Rep. 7, 46681 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider E. B., Fetal health stagnation: Have health conditions in utero improved in the United States and Western and Northern Europe over the past 150 years? Soc. Sci. Med. 179, 18–26 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Verstraete E., et al., Healthcare-associated bloodstream infections in a neonatal intensive care unit over a 20-year period (1992-2011): Trends in incidence, pathogens, and mortality. Infect. Control Hosp. Epidemiol. 35, 511–518 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Glinianaia S. V., et al., Temporal changes in key maternal and fetal factors affecting birth outcomes: A 32-year population-based study in an industrial city. BMC Pregnancy Childbirth 8, 39 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rush D., Alvir J. M., Kenny D. A., Johnson S. S., Horvitz D. G., The National WIC evaluation: Evaluation of the special supplemental food program for women, infants, and children. III. Historical study of pregnancy outcomes. Am. J. Clin. Nutr. 48, 412–428 (1988). [DOI] [PubMed] [Google Scholar]
- 40.Secolin R., et al., Family-based association study for bipolar affective disorder. Psychiatr. Genet. 20, 126–129 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Charney A. W., et al., Evidence for genetic heterogeneity between clinical subtypes of bipolar disorder. Transl. Psychiatry 7, e993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou L., et al., Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum. Mol. Genet. 25, 3383–3394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mühleisen T. W., et al., Genome-wide association study reveals two new risk loci for bipolar disorder. Nat. Commun. 5, 3339 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Ruderfer D. M.et al.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Bipolar Disorder Working Group of the Psychiatric Genomics Consortium; Cross-Disorder Working Group of the Psychiatric Genomics Consortium , Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol. Psychiatry 19, 1017–1024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z., et al., Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat. Genet. 49, 1576–1583 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Schizophrenia Working Group of the Psychiatric Genomics Consortium , Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen D. T.et al.; BiGS , Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol. Psychiatry 18, 195–205 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Choi S. W., O’Reilly P. F., PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience 8, giz082 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruderfer D. M., et al., Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell 173, 1705–1715.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones S. E., et al., Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat. Commun. 10, 343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dashti H. S., et al., Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat. Commun. 10, 1100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H., et al., Genome-wide association analysis of self-reported daytime sleepiness identifies 42 loci that suggest biological subtypes. Nat. Commun. 10, 3503 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Leeuw C. A., Mooij J. M., Heskes T., Posthuma D., MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giguère V., et al., Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. 8, 538–553 (1994). [DOI] [PubMed] [Google Scholar]
- 55.Griffin P., et al., Circadian clock protein Rev-erbα regulates neuroinflammation. Proc. Natl. Acad. Sci. U.S.A. 116, 5102–5107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y., et al., GENE REGULATION. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348, 1488–1492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Solt L. A., Kojetin D. J., Burris T. P., The REV-ERBs and RORs: Molecular links between circadian rhythms and lipid homeostasis. Future Med. Chem. 3, 623–638 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guillaumond F., Dardente H., Giguère V., Cermakian N., Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms 20, 391–403 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Sato T. K., et al., A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43, 527–537 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Al Shareef S. M., et al., Kleine-Levin syndrome is associated with LMOD3 variants. J. Sleep Res. 28, e12718 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Cingolani P., et al., A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schatz U. A., et al., Evidence of mild founder LMOD3 mutations causing nemaline myopathy 10 in Germany and Austria. Neurology 91, e1690–e1694 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Tillman R., Geller B., Definitions of rapid, ultrarapid, and ultradian cycling and of episode duration in pediatric and adult bipolar disorders: A proposal to distinguish episodes from cycles. J. Child Adolesc. Psychopharmacol. 13, 267–271 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Groos E., et al., Emerging psychiatric disorders in Kleine-Levin syndrome. J. Sleep Res. 27, e12690 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Kaplan K. A., et al., Hypersomnia subtypes, sleep and relapse in bipolar disorder. Psychol. Med. 45, 1751–1763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grigolon R. B., et al., Hypersomnia and bipolar disorder: A systematic review and meta-analysis of proportion. J. Affect. Disord. 246, 659–666 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Pagani L., et al., Genetic contributions to circadian activity rhythm and sleep pattern phenotypes in pedigrees segregating for severe bipolar disorder. Proc. Natl. Acad. Sci. U.S.A. 113, E754–E761 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bora E., Neurocognitive features in clinical subgroups of bipolar disorder: A meta-analysis. J. Affect. Disord. 229, 125–134 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Lima I. M. M., Peckham A. D., Johnson S. L., Cognitive deficits in bipolar disorders: Implications for emotion. Clin. Psychol. Rev. 59, 126–136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lane J. M., et al., Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat. Genet. 49, 274–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lewis K. J. S., et al., Comparison of genetic liability for sleep traits among individuals with bipolar disorder I or II and control participants. JAMA Psychiatry 77, 303–310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Engström M., Latini F., Landtblom A. M., Neuroimaging in the Kleine-Levin syndrome. Curr. Neurol. Neurosci. Rep. 18, 58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hong S. B., Neuroimaging of narcolepsy and Kleine-Levin syndrome. Sleep Med. Clin. 12, 359–368 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Kas A., Lavault S., Habert M. O., Arnulf I., Feeling unreal: A functional imaging study in patients with Kleine-Levin syndrome. Brain 137, 2077–2087 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Blume C., Garbazza C., Spitschan M., Effects of light on human circadian rhythms, sleep and mood. Somnologie (Berl.) 23, 147–156 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Czeisler C. A., Zimmerman J. C., Ronda J. M., Moore-Ede M. C., Weitzman E. D., Timing of REM sleep is coupled to the circadian rhythm of body temperature in man. Sleep 2, 329–346 (1980). [PubMed] [Google Scholar]
- 77.Carskadon M. A., Dement W. C., Sleep studies on a 90-minute day. Electroencephalogr. Clin. Neurophysiol. 39, 145–155 (1975). [DOI] [PubMed] [Google Scholar]
- 78.Goldbart A., et al., Narcolepsy and predictors of positive MSLTs in the Wisconsin sleep cohort. Sleep (Basel) 37, 1043–1051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen L., Huang Y.-S., “Actigraphy study and different hypersomnia disorders” in 2nd Asian Narcolepsy & Hypersomnolence Society Meeting (2019), vol. 2, p. 39. [Google Scholar]
- 80.Gold A. K., Kinrys G., Treating circadian rhythm disruption in bipolar disorder. Curr. Psychiatry Rep. 21, 14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vreeker A., et al., Genetic analysis of activity, brain and behavioral associations in extended families with heavy genetic loading for bipolar disorder. Pyschol. Med., 1–9, 10.1017/S0033291719003416 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Ikeda M., et al., A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol. Psychiatry 23, 639–647 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Psychiatric GWAS Consortium Bipolar Disorder Working Group , Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 43, 977–983 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Forstner A. J., et al., Identification of shared risk loci and pathways for bipolar disorder and schizophrenia. PLoS One 12, e0171595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ripke S.et al.; Multicenter Genetic Studies of Schizophrenia Consortium; Psychosis Endophenotypes International Consortium; Wellcome Trust Case Control Consortium 2 , Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 45, 1150–1159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium , Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 43, 969–976 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ward J., et al., Genome-wide analysis in UK Biobank identifies four loci associated with mood instability and genetic correlation with major depressive disorder, anxiety disorder and schizophrenia. Transl. Psychiatry 7, 1264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weber G., Investigation of metal-species (Ca, Mg, Zn, Fe, Cu, Pb, Cd, Sn) in urine by HPLC-AAS. J. Trace Elem. Electrolytes Health Dis. 2, 61–65 (1988). [PubMed] [Google Scholar]
- 89.Tsuchiya K. J., Byrne M., Mortensen P. B., Risk factors in relation to an emergence of bipolar disorder: A systematic review. Bipolar Disord. 5, 231–242 (2003). [DOI] [PubMed] [Google Scholar]
- 90.Lyall D. M., et al., Low birth weight and features of neuroticism and mood disorder in 83 545 participants of the UK Biobank cohort. BJPsych Open 2, 38–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kikas T., Rull K., Beaumont R. N., Freathy R. M., Laan M., The effect of genetic variation on the placental transcriptome in humans. Front. Genet. 10, 550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brockenshire A., The “mini-mental state”: A handy tool. Perspectives 11, 7–8 (1987). [PubMed] [Google Scholar]
- 93.Horikoshi M.et al.; CHARGE Consortium Hematology Working Group; Early Growth Genetics (EGG) Consortium , Genome-wide associations for birth weight and correlations with adult disease. Nature 538, 248–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Warrington N. M.et al.; EGG Consortium , Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat. Genet. 51, 804–814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Beaumont R. N.et al.; Early Growth Genetics (EGG) Consortium , Genome-wide association study of offspring birth weight in 86 577 women identifies five novel loci and highlights maternal genetic effects that are independent of fetal genetics. Hum. Mol. Genet. 27, 742–756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang G., et al., Genetic associations with gestational duration and spontaneous preterm birth. N. Engl. J. Med. 377, 1156–1167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Srinivasan L.et al.; Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network , Genome-wide association study of sepsis in extremely premature infants. Arch. Dis. Child. Fetal Neonatal Ed. 102, F439–F445 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pasqualini J. R., Mercat P., Giambiagi N., Histone acetylation decreased by estradiol in the MCF-7 human mammary cancer cell line. Breast Cancer Res. Treat. 14, 101–105 (1989). [DOI] [PubMed] [Google Scholar]
- 99.Jiang X., et al., Sodium valproate rescues expression of TRANK1 in iPSC-derived neural cells that carry a genetic variant associated with serious mental illness. Mol. Psychiatry 24, 613–624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Purcell S., et al., PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nicolazzi E. L., Iamartino D., Williams J. L., AffyPipe: An open-source pipeline for Affymetrix Axiom genotyping workflow. Bioinformatics 30, 3118–3119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Auton A.et al.; 1000 Genomes Project Consortium , A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Delaneau O., Marchini J., Zagury J. F., A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2011). [DOI] [PubMed] [Google Scholar]
- 104.Delaneau O., Marchini J.; 1000 Genomes Project Consortium; 1000 Genomes Project Consortium , Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun. 5, 3934 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Howie B. N., Donnelly P., Marchini J., A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Marchini J., Howie B., Myers S., McVean G., Donnelly P., A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007). [DOI] [PubMed] [Google Scholar]
- 107.Watanabe K., Taskesen E., van Bochoven A., Posthuma D., Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Turner S. D., qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv [Preprint] (2014). 10.1101/005165 (Accessed 20 March 2019). [DOI]
- 109.Arnold M., Raffler J., Pfeufer A., Suhre K., Kastenmüller G., SNiPA: An interactive, genetic variant-centered annotation browser. Bioinformatics 31, 1334–1336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hormozdiari F., et al., Colocalization of GWAS and eQTL signals detects target genes. Am. J. Hum. Genet. 99, 1245–1260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zheng J.et al.; Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium , LD hub: A centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang K., Li M., Hakonarson H., ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee S., Abecasis G. R., Boehnke M., Lin X., Rare-variant association analysis: Study designs and statistical tests. Am. J. Hum. Genet. 95, 5–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the analyses presented in the manuscript is from data in Datasets S1 to S15 which are available. Complete summary statistics of the genetic associations are available at https://doi.org/10.6084/m9.figshare.14128475.v2.To protect the privacy of the study participants, the individual genetic data generated and analyzed during this study are available from the corresponding authors on reasonable request.