Significance
Bacteria display hair-like structures called pili that enable them to adhere to host tissues, surfaces, and other bacteria. Understanding how microbes assemble and display pili is important, as these structures play a key role in host–pathogen interactions that cause disease. In this study, we have gained insight into how pathogenic Gram-positive bacteria assemble pili by visualizing the intersubunit covalent crosslink that stabilizes these fibrous structures and by investigating how pili are assembled by specialized pilus-specific sortase enzymes. We propose that pili are assembled via a conserved “latch” mechanism that is critical for driving the crosslinking of specific sites on the pilus.
Keywords: pili, sortase, Gram positive, lysine isopeptide bond, integrative structural biology
Abstract
Gram-positive bacteria assemble pili (fimbriae) on their surfaces to adhere to host tissues and to promote polymicrobial interactions. These hair-like structures, although very thin (1 to 5 nm), exhibit impressive tensile strengths because their protein components (pilins) are covalently crosslinked together via lysine–isopeptide bonds by pilus-specific sortase enzymes. While atomic structures of isolated pilins have been determined, how they are joined together by sortases and how these interpilin crosslinks stabilize pilus structure are poorly understood. Using a reconstituted pilus assembly system and hybrid structural biology methods, we elucidated the solution structure and dynamics of the crosslinked interface that is repeated to build the prototypical SpaA pilus from Corynebacterium diphtheriae. We show that sortase-catalyzed introduction of a K190-T494 isopeptide bond between adjacent SpaA pilins causes them to form a rigid interface in which the LPLTG sorting signal is inserted into a large binding groove. Cellular and quantitative kinetic measurements of the crosslinking reaction shed light onto the mechanism of pilus biogenesis. We propose that the pilus-specific sortase in C. diphtheriae uses a latch mechanism to select K190 on SpaA for crosslinking in which the sorting signal is partially transferred from the enzyme to a binding groove in SpaA in order to facilitate catalysis. This process is facilitated by a conserved loop in SpaA, which after crosslinking forms a stabilizing latch that covers the K190-T494 isopeptide bond. General features of the structure and sortase-catalyzed assembly mechanism of the SpaA pilus are likely conserved in Gram-positive bacteria.
The cellular surface of many bacteria is elaborated with thin appendages called pili (also called fimbriae) which have a range of roles including twitching motility, conjugation, immunomodulation, biofilm formation, and adherence (1, 2). These long proteinaceous fibers are key virulence factors that mediate initial host–pathogen interactions, which are subsequently strengthened by more intimate contacts from shorter pili and cell wall–attached adhesins (1–13 ). As the infection progresses, pili also facilitate biofilm formation, protecting invading microbes from host immune clearance and exogenous antibiotics (1, 5, 11, 14). Gram-positive bacteria display very thin (1 to 5 nm) (15) hair-like pili that nevertheless possess enormous tensile strength because their protein components are crosslinked together by lysine–isopeptide bonds. These crosslinked fibers are displayed by a wide range of pathogenic and commensal Gram-positive bacteria, but their structures and mechanism of assembly remain poorly understood (2, 5, 7–10).
Pili in Gram-positive bacteria are assembled by pilus-specific sortase enzymes that crosslink the pilus subunits (called pilins) together via lysine–isopeptide bonds. Our current understanding of this process has been significantly advanced by studies of the SpaA pilus in Corynebacterium diphtheriae, a pathogen that causes pharyngeal diphtheria (5, 8, 10, 16). The SpaA pilus mediates adherence to the pharyngeal epithelium and consists of three types of pilins: the pilus shaft is formed by SpaA, and the tip and base are formed by SpaC and SpaB, respectively (17). The C. diphtheriae pilus-specific sortase (CdSrtA) assembles the pilus by catalyzing a repetitive, irreversible transpeptidation reaction that covalently links pilin subunits together via an isopeptide bond. The shaft of the pilus is formed by ∼100 to 250 crosslinked SpaA pilins (16). CdSrtA-catalyzed SpaA polymerization begins when SpaA prepilin proteins containing an N-terminal signal peptide sequence are exported via the Sec pathway and retained on the extracellular surface via a C-terminal cell wall sorting signal (CWSS) (18). CdSrtA then crosslinks SpaA proteins together via a two-step process. First, an LPLTG sorting signal sequence within the CWSS is cleaved between Thr and Gly residues by the sortase, generating a thioacyl-linked CdSrtA-SpaA intermediate in which the enzyme’s active site cysteine residue is covalently linked to the carbonyl atom of the sorting signal threonine. In the second step, a lysine ε-amine group originating from another SpaA pilin attacks the thioacyl linkage in the CdSrtA-SpaA intermediate, thereby joining distinct SpaA proteins together via a K190-T494 isopeptide bond (Fig. 1A). The reactive lysine in SpaA is housed within the N-terminal domain and is part of a highly conserved WxxxVxVYPK sequence motif that is found in many pilin proteins (17). The shaft of the pilus is constructed by repeating this two-step process, and a similar CdSrtA-catalyzed reaction is used to add the SpaC tip pilin to SpaA. Pilus assembly is completed by incorporating the SpaB basal pilin, which promotes pilus attachment to the cell wall using a distinct housekeeping sortase CdSrtF (3). Pilus biogenesis is thought to occur within “pilusosomes” on the cell surface, at which pilin substrates and pilus-specific sortases colocalize to facilitate rapid polymerization (18).
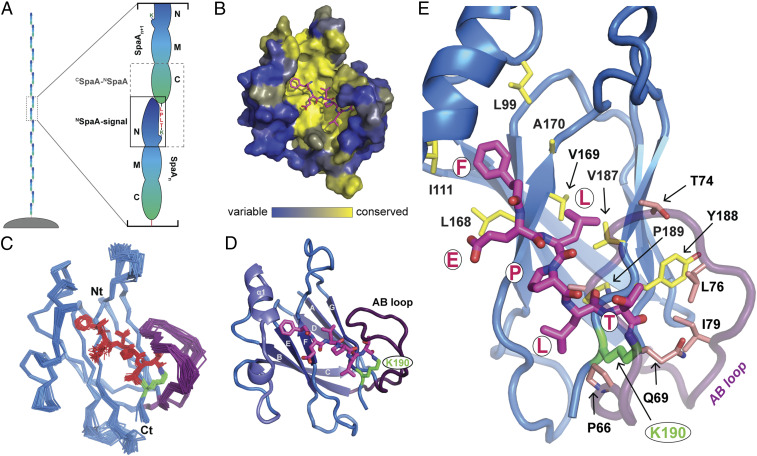
Fig. 1.
Structure of the NSpaA-signal peptide complex. (A) Schematic of pilus polymerization with full-length SpaA molecules. An expanded view of the two portions of the crosslinked SpaA polymer investigated in this study, CSpaA-NSpaA complex and NSpaA-signal, are boxed in gray dashed lines and solid black lines, respectively. (B) The NSpaA-signal peptide complex is represented in surface representation with relative conservation of each residue indicated by a color gradient ranging from highly variable positions (blue) to highly conserved residues (yellow). The peptide (magenta sticks) is docked into a highly conserved, nonpolar binding groove on SpaA. (C) A bundle of the 40 lowest energy structures of the SpaA-signal complex is displayed. The backbone of the NSpaA domain is represented by blue ribbons. The last five residues of the sorting signal peptide are depicted as red sticks, and Lys190 is shown as green sticks. (D) Secondary structural elements of the NMR structure are highlighted. (E) An expanded view of the peptide binding interface shows how the peptide is bound in the cleft of NSpaA. Residues on SpaA exhibiting intermolecular NOEs to the peptide are shown as sticks. Interacting residues in the core of the domain and within the AB loop are colored yellow and pink, respectively.
Despite their importance in bacterial physiology and pathogenesis, only structures of isolated, noncrosslinked pilins have been determined at atomic-level resolution (19). This is because it has been challenging to obtain homogenous crosslinked pili that are suitable for biophysical analyses and because Gram-positive pili are thin and flexible, making them difficult to study using cryogenic electron microscopy and X-ray crystallography. Crystal structures of isolated pilins have revealed that they contain IgG-like Cna-type domains and frequently one or more spontaneously forming intradomain isopeptide bonds that impart significant resistance to mechanical forces (19–21). Internal isopeptide bond linkages exist as either D- or E-type and are extremely stabilizing, allowing pilin domains to withstand the highest unfolding forces yet reported for a globular protein (20). Atomic-level structures of sortase crosslinked pilins have yet to be visualized, but a transmission electron microscopy study of the Streptococcus pneumoniae RrgB pilus enabled the periodicity and polarity of individual subunits within the pilus fiber to be determined (22). This work revealed that the subunits in the pilus are arranged in a head-to-tail manner, enabling sortase-catalyzed isopeptide crosslinking between the lysine and LPxTG motifs located at the N- and C-terminal ends of the pilin, respectively. In crystals, similar head-to-tail packing arrangements are observed, but whether these lattice interactions are also present in the intact pilus is not known.
In this study, we used a recently developed in vitro pilus assembly system and hybrid structural biology methods to gain insight into the structure and biogenesis mechanism of the SpaA pilus from C. diphtheriae. We first determined the NMR structure of the N-terminal domain of SpaA crosslinked to the sorting signal peptide and then used small angle X-ray scattering (SAXS), NMR, and crystallographic data to model the structure of the isopeptide-linked SpaA-SpaA building block that is repeated to construct the pilus shaft. We show that crosslinking is accompanied by a large disordered-to-ordered structural change in the SpaA pilin, which forms an interpilin interface that differs markedly from packing interactions observed in crystals of the isolated SpaA. Quantitative measurements of kinetics of the sortase-catalyzed transpeptidation reaction suggest that the enzyme uses a latch mechanism to select the appropriate lysine residue on SpaA for interpilin crosslinking.
Results
NMR Structure of the Crosslinked NSpaA-Signal Complex.
To learn how Gram-positive pili are stabilized by interpilin lysine–isopeptide bonds, we examined how these crosslinks ligate SpaA pilins together to construct the shaft of the C. diphtheriae SpaA pilus (7, 16). The SpaA shaft pilin contains three autonomously folded domains: N-terminal (NSpaA, residues 53 to 195), middle (MSpaA, residues 196 to 349), and C-terminal (CSpaA, residues 350 to 500) domains (23). SpaA pilins are joined together via interpilin crosslinks that connect the NSpaA and CSpaA domains; a lysine–isopeptide bond links the side chain ε-amine group of K190 within NSpaA to the carbonyl group of the T494 residue present in a LPLTG sorting signal sequence that immediately follows CSpaA in the primary sequence (Fig. 1A). Previously, we demonstrated that a mutationally activated CdSrtA enzyme covalently crosslinks peptides containing the LPLTG sorting signal sequence to NSpaA, a process that mimics the reaction that is repeated to build the shaft of the pilus (24–26). We first employed this enzyme to produce the NSpaA-signal complex, in which the K190 side chain in NSpaA is joined via an isopeptide bond to the threonine residue in a sorting signal peptide (KNAGFELPLT peptide that corresponds to residues K485 to T494 in CSpaA) (SI Appendix, Fig. S1). Heteronuclear multidimensional NMR spectroscopy was then used to determine the atomic structure of the complex using a total 2,070 experimental restraints, including 66 intermolecular nuclear Overhauser effect (NOE) distance restraints (SI Appendix, Table S1 and Fig. S3). The structure of the complex is well defined by the NMR data, as the backbone and heavy atom coordinates of residues T3 to Q192 and G488 to T494 in the ensemble can be superimposed to the average structure with a RMSD of 0.48 ± 0.05 and 0.81 ± 0.04 Å, respectively (Fig. 1C).
NSpaA adopts a CnaB-type fold that binds the crosslinked sorting signal via a large groove formed by residues within strands βF and βG (Fig. 1D). The bound signal contains a characteristic kink at its single proline residue, causing it to form an l-shaped structure that spans from the K190 attachment site to a wedge-shaped opening between the FG loop and helix α1 (27, 28). The conserved LPLT residues in the sorting signal form nonpolar interactions with a conserved surface on NSpaA and bury ∼630 Å2 of solvent-exposed surface area (Fig. 1B). A detailed summary of these interactions is provided in SI Appendix, Fig. S5. The C-terminal T494 residue in the signal is joined via an isopeptide linkage to the side chain of K190 located at the end of strand G and partially masked from the solvent by residues that connect strands A and B (called the AB loop) (Fig. 1D). The AB loop extends over the P492-L293-T494 portion of the sorting signal, contacting the T494 methyl group via interactions with the side chains of Q69, L76, and I79 (Fig. 1E). Interestingly, G73 in the AB loop is highly conserved among proteins that contain the pilin motif (SI Appendix, Fig. S4) and is located at the tip of the loop where the chain reverses direction and is in close contact with the bound sorting signal. The positioning of the N-terminal end of the structured portion of the signal is defined by intermolecular NOEs to the aromatic side chain of F489, which is nestled into a hydrophobic region positioned directly underneath the first turn of α1. Signal residues N-terminal to F489 are disordered and exit the binding groove via an opening between the FG loop and helix α1, which must therefore form the interpilin interface in the shaft of the SpaA pilus.
Crosslinking Triggers the Closure of a Stabilizing Latch over the Interpilin Linkage.
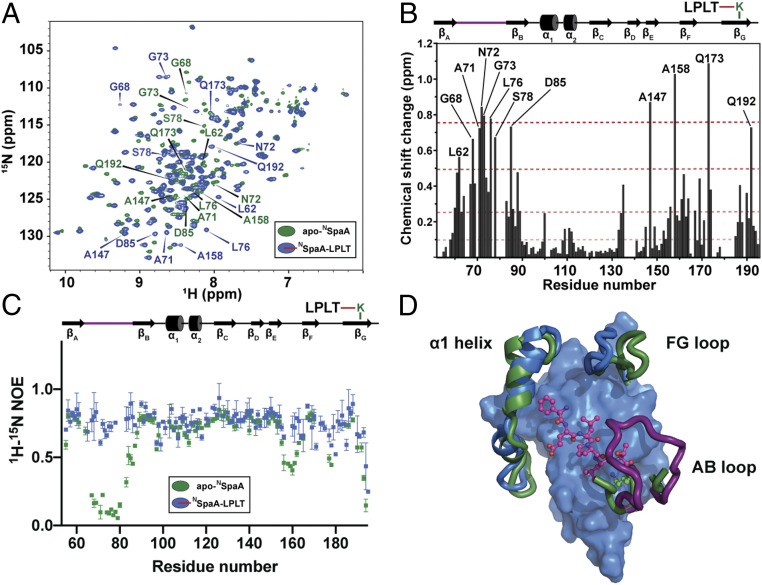
A comparison of the structure of the NSpaA-signal complex with a previously determined 1.6-Å crystal structure of unmodified SpaA protein reveals striking conformational differences (Fig. 2D). While the apo- and complexed-forms of the NSpaA domain adopt generally similar tertiary structures (their backbone coordinates can be superimposed with a RMSD of 2.3 Å), crosslinking causes a significant rearrangement in the AB loop, as well as more subtle changes in the positioning of the FG loop and α1 helix. In the complex, the AB loop rests against the body of the protein, encapsulating the K190-T494 isopeptide linkage, while in the structure of the unmodified SpaA protein, coordinates for residues Q69 to I79 in the AB loop are missing because they exhibit scant electron density (23). To determine if the AB loop undergoes a disordered-to-ordered transition upon signal attachment by sortase, we assigned the backbone chemical shifts of apo-NSpaA and acquired 1H-15N steady-state NOE relaxation data for apo-NSpaA and the NSpaA-signal complex. Consistent with the NMR structure of the complex, the largest differences in the backbone chemical shifts occur for residues that form the signal binding groove and the AB loop (Fig. 2 A and B). Interestingly, the 1H-15N steady-state NOE data reveal that signal attachment significantly retards motions in the AB loop, as residues M63 to G86 in apo-NSpaA exhibit small-magnitude steady-state NOEs indicative of high mobility, whereas in the NSpaA-signal complex, they are rigid with values around 0.8 (Fig. 2C and SI Appendix, Fig. S2). Covalent signal attachment also quenches motions on the opposite side of the binding pocket, as similar, albeit smaller trends are observed for residues in the FG loop that contact the N-terminal end of the sorting signal near the interpilin interface. Limited proteolysis experiments of apo-NSpaA and the NSpaA-signal complex indicate that the protein in the complex is ∼42% more resistant to proteolytic degradation after 24 h (SI Appendix, Fig. S6A). The NSpaA-signal complex is also slightly more thermostable based on differential scanning fluorimetry experiments (its ΔuG° increases by ∼0.9 kJ) (SI Appendix, Fig. S6 B and C) (29, 30). Taken together, these data indicate that the AB loop becomes ordered upon pilin crosslinking, forming a latch structure that shields the isopeptide linkage and stabilizes NSpaA.
Fig. 2.
The AB loop undergoes a disordered-to-ordered transition upon crosslinking. (A) 1H-15N HSQC correlation spectra of apo-NSpaA (green) and NSpaA-signal complex (blue) are overlaid. The positions of residues with differences larger than 0.5 ppm in composite chemical shift are indicated on the plot. (B) Chemical shift perturbations (CSPs) for each residue are plotted with respect to primary sequence. CSPs are binned into >0.75 ppm, 0.75 to 0.5 ppm, 0.5 to 0.25 ppm, and 0.1 to 0.25 ppm (indicated by red dotted lines of increasing transparency). (C) Heteronuclear NOE data are graphed as a function of primary sequence for apo-SpaA (green) and SpaA-signal (blue). (D) A surface rendering of the complex, with the AB loop (purple), FG loop, and α1 helix (blue) highlighted with cartoon representations and the signal peptide shown as magenta sticks. The apo-NSpaA structure is aligned to the NMR complex, and the corresponding secondary elements are shown in green to highlight conformational shifts. The backbone coordinates can be superimposed with a RMSD of 2.3 Å.
Solution Structure of the Interpilin Interface.
To gain insight into the structure and dynamics of the interpilin interface that is repeated to build the SpaA pilus, we used the activated CdSrtA enzyme to generate a crosslinked CSpaA-NSpaA dimer (M.W. 31 kDa); in the dimer, NSpaA is crosslinked via its K190 residue to the sorting signal that resides in a 11–amino acid C-terminal tail that immediately follows the CSpaA domain (Fig. 1A). NMR spectra were acquired using samples of the dimer in which either the CSpaA or NSpaA domains were selectively labeled with nitrogen-15. A comparison with the spectra of the corresponding isolated domains reveals that crosslinking causes substantial chemical shift changes, suggesting that the domains pack against one another in the dimer (SI Appendix, Fig. S7 A and B). This is substantiated by molecular correlation time (τc) measurements using NMR 15N relaxation data, as the τc of the CSpaA-NSpaA dimer is 18.0 ns, much longer than expected if the domains were simply connected by a flexible linker that enabled them to freely reorient (the τc values of the isolated NSpaA and CSpaA domains are 8.7 and 9.5 ns, respectively) (SI Appendix, Fig. S7C). SAXS data of the crosslinked dimer also indicate that it is generally inflexible as evidenced by the distance distribution (SI Appendix, Fig. S8A) and normalized Kratky plots of the data (SI Appendix, Fig. S8B). The calculated radius of gyration is smaller than expected for a freely reorienting dimer, and the Kratky plot is characterized by features which correspond to a nonflexible structure. Thus, both NMR and SAXS analyses are in agreement and indicate that the domains within the dimer are immobilized with respect to one another.
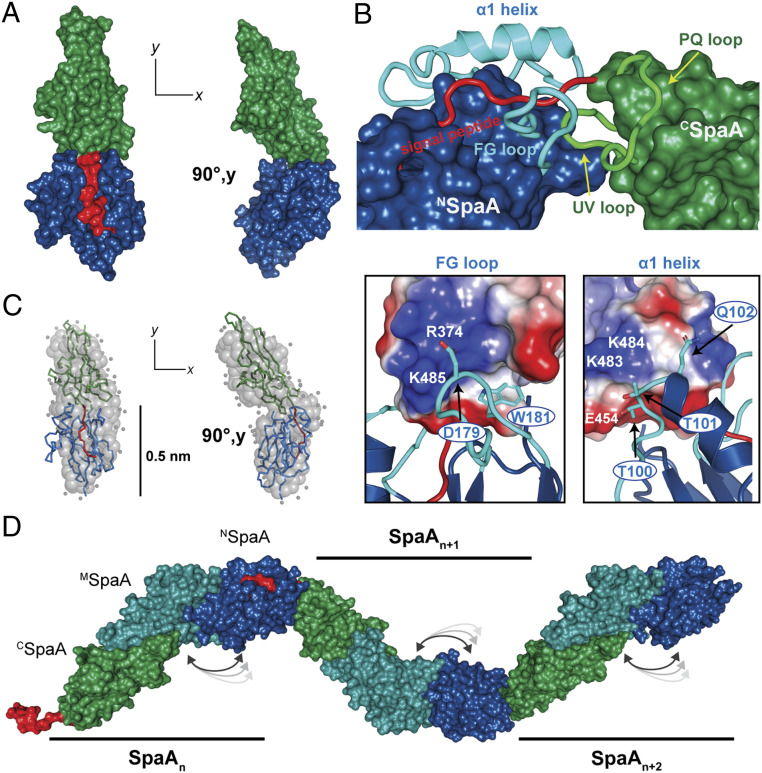
The solution structure of the crosslinked CSpaA-NSpaA dimer was modeled using an integrated approach that employed SAXS, NMR, and crystallographic data. An initial model of the CSpaA-NSpaA complex was constructed using the crystal and NMR structures of CSpaA (Protein Data Bank [PDB]: 3HR6) and NSpaA-signal (this work), respectively. SAXS data were then employed to drive multistate rigid body modeling of the complex using the MultiFOXS approach (31). To account for potential interdomain flexibility, 10,000 models were generated, and residues connecting the CSpaA and NSpaA domains (K483-A488) were allowed to move freely during the calculations (Methods). Models of the CSpaA-NSpaA dimer in which the domains adopt related orientations with respect to one another best fit the SAXS data (Fig. 3A and SI Appendix, Fig. S8 C and D). This domain orientation is further supported by lower resolution ab initio modeling of the scattering data using the program GASBOR, which yields an elongated, kinked dimeric structure that is similar to those obtained by MultiFOXS (Fig. 3C). In the MultiFOXS model of the crosslinked CSpaA-NSpaA dimer that best fits the SAXS data, the FG loop in NSpaA is inserted between the UV and PQ loops in CSpaA, thereby positioning the sorting signal following CSpaA within NSpaA’s peptide binding groove (Fig. 3 B, Top). The protein–protein interface buries 1,270 Å2 of solvent-exposed surface area (32) and is further stabilized by interactions between the N-terminal portion of helix α1 in NSpaA and UV loop in CSpaA. This packing arrangement explains why the W181 residue within the WxxxVxVYPK pilin motif is conserved, as its indole side chain presumably plays a key structural role in stabilizing the SpaA-SpaA interface by packing into a hydrophobic surface formed by residues A450 and Y453 in CSpaA’s UV loop (Fig. 3 B, Bottom Left). Stabilizing electrostatic interactions are predicted to surround this nonpolar interface. On one side, the negatively charged D179 side chain in NSpaA’s FG loop is packed against a cationic surface on CSpaA formed by residues K485 (signal peptide) and R374 (PQ loop) (Fig. 3 B, Bottom Left), while on the other side, hydrophilic interactions occur between residues at the N-terminal end of α1 helix in NSpaA (T99, T100, and Q101) and residues located in CSpaA’s UV loop (K483, K484, and E454) (Fig. 3 B, Bottom Right). Notably, the SAXS-derived solution structure of the CSpaA-NSpaA dimer presented here differs markedly from the head-to-tail packing arrangement observed in crystals of the isolated SpaA protein (23), and only the SAXS model is compatible with NMR chemical shift mapping data (described in SI Appendix, Fig. S9).
Fig. 3.
SAXS model of the crosslinked interpilin interface. (A) The best fit single-conformation rigid body model from the MultiFOXS calculation (CSpaA, green; NSpaA, blue; signal peptide, red). (B) Details of the interface formed by the solution structure of the CSpaA-NSpaA complex. (Top) The cores of the two domains and the interacting loops are shown as surface and cartoon representations, respectively. (Bottom) Expanded views of two interactions to CSpaA mediated by the FG loop and α1 helix of NSpaA. (C) GASBOR ab initio model from solution scattering, shown as gray spheres. The rigid body model calculated by MultiFOXS is aligned with the GASBOR model and depicted in ribbon representation. Left and right shapes are the same GASBOR models rotated counterclockwise by 90° around y-axis. (D) Model of a dimer of full-length SpaA molecules comprising the pilus shaft. Coordinates from the crystal structure of full-length SpaA molecules (PDB: 3HR6) were arranged in head-to-tail arrangements according to the interface determined by the MultiFOXS model of the CSpaA-NSpaA structure (color gradient blue to green from N term to C term).
The biological relevance of the SAXS-derived model of the interpilin interface is substantiated by in vitro and in vivo data. Bacteria overexpressing SpaA proteins containing L99R, P172R, D179R, or L182R single–amino acid substitutions at the predicted interface are impaired in pili display when assessed by immunoelectron microscopy or immunoblotting of fractionated cells with a specific antibody against SpaA (SI Appendix, Fig. S10). All interfacial substitutions reduce the abundance of surface-displayed pili when assessed by microscopy, with pili harboring P172R and D179R substitutions showing largest defects. The SpaA variants altering the presumed interface were further assessed for their ability to serve as a substrate in the sortase-catalyzed crosslinking reaction using an in vitro gel-based assay that monitors the production of the CSpaA-NSpaA dimer (SI Appendix, Fig. S11) (24). Consistent with formation of the interpilin interface being required for efficient transpeptidation, a densitometry analysis of the domain coupling reactions reveals that SpaA proteins harboring interfacial L99R, P172R, and L182R point substitutions produce less crosslinked product (SI Appendix, Fig. S11B). Interestingly, the integrity of the interpilin interface in the SAXS-derived model appears to be an important determinant for overall stability. This is because trypsin digestion studies reveal that the CSpaA-NSpaA dimer is significantly more resistant to proteolytic degradation than the NSpaA-signal complex, and introducing a single L99D substitution at the interface is destabilizing as it increases CSpaA-NSpaA dimer susceptibility to proteolysis (SI Appendix, Fig. S11C). In all, the results of both biochemical and cellular experiments support the SAXS-derived model of the interpilin interface and suggest that its formation is required for efficient transpeptidation and resistance to proteolytic degradation.
The Sorting Signal Must Be Partially Transferred to SpaA to Initiate Crosslinking.
Guided by the solution structures of the NSpaA-signal complex and the crosslinked CSpaA-NSpaA dimer, we employed in vitro crosslinking and cellular assays to probe the mechanism of pilus biogenesis. Initially, NSpaA proteins containing amino acid substitutions at conserved sites within the SpaA-SpaA interface were tested for their ability to serve as substrates for CdSrtA using an in vitro gel-based assay that monitors the covalent attachment of a sorting signal peptide fluorophore to NSpaA (Fig. 4A) (24). Some of the largest defects in transpeptidation activity occur when NSpaA residues that contact the LPLT sorting signal in the NSpaA-signal complex are altered (L168D and V187D) or when the K190 nucleophile and residues immediately adjacent to it are changed (H60A, Y188G, and K190A).
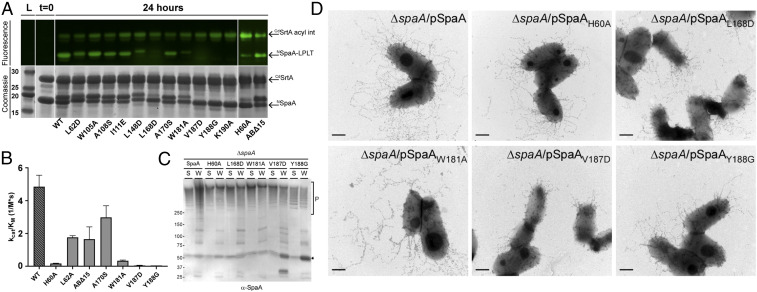
Fig. 4.
In vitro and in vivo validation of key residues on the SpaA acceptor domain. (A) Gel fluorescence assay to rapidly screen a library of NSpaA variants. The Top shows an SDS-PAGE gel visualized by FITC fluorescence, indicating the presence of CdSrtA (top band) or NSpaA (bottom band) conjugated to fluorescent peptide. The Bottom shows the same SDS-PAGE gel visualized by Coomassie staining in order to visualize the total protein composition of each lane. Peptide-labeled NSpaA variants typically have slightly lower electrophoretic mobility than the corresponding apo-NSpaA variant. (B) Bar graph comparing the catalytic efficiency (kcat/KM) of each mutant to the corresponding kinetic parameters of wild-type NSpaA. (C) Cells of the C. diphtheriae ΔspaA mutant expressing wild-type SpaA or individual SpaA mutants from a plasmid were grown to midlog phase and subjected to cell fractionation. Protein samples collected from the culture medium (S) and cell wall (W) fractions were analyzed by immunoblotting with specific antibodies against SpaA (α-SpaA). Molecular mass markers in kDa and SpaA polymers (P) and monomer (arrow) are indicated. (D) Cells of strains used in (C) were immobilized on carbon-coated nickel grids and stained with α-SpaA followed by IgG-conjugated 12 nm gold particles and 1% uranyl acetate prior to electron microscopy. (Scale bars of 0.5 μm.)
Variants exhibiting significant defects in reactivity were further evaluated using a newly developed high performance liquid chromatography (HPLC)–based assay that quantitatively measures the steady-state kinetics of crosslinking (26) (Fig. 4B and SI Appendix, Fig. S12). In the HPLC-based assay, the sorting signal peptide is held in excess, such that effects of NSpaA substitutions on the second step of transpeptidation are revealed (i.e., the rate at which the K190 lysine ε-amine group in NSpaA attacks the thioacyl-linked CdSrtA-SpaA intermediate). The V187D and Y188G variants exhibit the largest defects in transpeptidation and alter side chains that are positioned immediately proximal to the K190 nucleophile (kcat/KM values are less than 1% of wild-type protein) (Fig. 4B). Both substitutions reduce catalytic turnover, suggesting that they are needed to properly form the active site used to catalyze the isopeptide bond. The H60A variant also exhibits reduced catalytic turnover, presumably because its imidazole ring stabilizes the positioning of the Y188 sidechain through pi-stacking interactions. Interestingly, disrupting contacts between NSpaA and residues in the sorting signal that are positioned distal to the site of isopeptide formation also reduces the rate of transpeptidation by increasing the KM (A170S and W181A). This suggests that when the thioacyl CdSrtA-SpaA intermediate formed in the first step of catalysis encounters NSpaA, the sorting signal bound to the CdSrtA’s active site must move from the enzyme into the binding groove on NSpaA to form a catalytically active complex that performs the final step of transpeptidation.
Bacteria-expressing SpaA proteins containing single–amino acid substitutions in the sorting signal binding groove also show defects in pilus display. As shown in Fig. 4C, immunoblotting analysis of protein samples collected from the culture medium (S) and cell wall (W) fractions of corynebacterial cells expressing wild-type SpaA produced abundant SpaA polymers detected in the cell wall fractions, with some polymers secreted into the extracellular milieu. These protein polymers (P) have high molecular weights as they were not well separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) electrophoresis. Strains expressing SpaA with H60A, L168D, and W181A still produced SpaA polymers, albeit less abundantly as compared to wild-type SpaA. Consistent with the in vitro analysis, mutants V187D and Y188G exhibited a significant pilus assembly defect, with reduced pilus polymerization and accumulation of SpaA precursors. To corroborate the fractionation results, the same set of strains was analyzed by immunoelectron microscopy, whereby cells were stained by SpaA antibodies, followed by staining with 12 nm gold particles conjugated to IgG. Consistent with the Western blotting analysis and in vitro kinetic measurements, severe effects are observed when V187D and Y188G substitutions are introduced near the site of K190 crosslinking. These variants are unable to assemble long and abundant pili as compared to the wild-type SpaA (Fig. 4D, V187D and Y188G panels) while less severe effects are observed for the H60A mutant strain that exhibited higher activity in vitro (Fig. 4D, compare SpaA and H60A panels). While V187D and Y188G NSpaA variants are folded similar to the wild-type protein, these nonconservative alterations can be expected to significantly perturb the positioning of nearby side chains. Finally, A170S and W181A mutants designed to alter residues that contact the sorting signal but are positioned distal to the site of crosslinking also produced fewer pili as compared to the wild-type SpaA. The L168D mutant assembled short pili, and the W181A mutant appeared to be fragile, with broken pili surrounding the cells (Fig. 4D, L168D and W181A panels). Thus, both the in vitro and cellular data suggest that signal transfer from the enzyme to the sorting signal pocket on SpaA is required for efficient catalysis, as mutation of this surface impairs crosslinking. This explains previously reported findings that CdSrtA can only crosslink sorting signals to K190 when it is housed in a structurally intact NSpaA domain (24).
Discussion
Using a recently developed in vitro assembly reaction (24, 25) and integrative structural biology methods, we determined the structure and dynamics of the lysine–isopeptide bond crosslinked CSpaA-NSpaA interface that is repeated to build the shaft of the C. diphtheriae SpaA pilus (Fig. 1A). The structure of NSpaA covalently attached to the CSpaA sorting signal was first determined by NMR, and then SAXS data of the SpaA-SpaA complex was used in conjunction with the crystal structure of the CSpaA domain to model the solution structure of the interpilin interface that is formed by sortase crosslinking of the CSpaA and NSpaA domains. This work reveals that sortase crosslinking immobilizes the pilin subunits, triggering the formation of an extensive interpilin interface in which the sorting signal following CSpaA is inserted into a nonpolar groove on NSpaA (Fig. 3A). Residues within the conserved WxxxVxVYPK pilin motif line the binding groove and when altered slow transpeptidation in vitro and in cells (Fig. 4). Our results shed light onto the structure and dynamics of the shaft of the SpaA pilus, which can be modeled using our structure of the CSpaA-NSpaA complex and a previously reported structure of the intact SpaA protein (Fig. 3D) (23). The shaft is formed by SpaA pilins that are arrayed in a head-to-tail manner with successive sortase-installed interpilin isopeptide bonds positioned on opposite faces of the polymer and each crosslinked SpaA-SpaA unit forming a “S” shape because of a ∼140° kink at the CSpaA-NSpaA junction. The SpaA pilus and other Gram-positive pili are presumably flexible, as they appear as nonlinear, hair-like structures in transmission electron micrographs (Fig. 4D) (16, 22, 33). This flexibility likely originates from intrapilin motions that occur between the N-terminal and middle domains, as crystal structures of isolated pilins have revealed a small interface between the domains that allow them to adopt different positions with respect to one another (33–39). The strongest evidence comes from studies of the GG-SpaD shaft pilin from Lactobacillus rhamnosus, as its N-terminal domain adopts a range of bent conformations relative to the body of the protein (40). Some flexibility in the SpaA pilus may also originate from motions at the interpilin CSpaA-NSpaA interface, as it is primarily formed by hydrophilic interactions between the proteins (Fig. 3B). However, these motions are presumably modest, since the normalized Kratky plot of the SAXS data for the CSpaA-NSpaA complex reveals a clear bell-shaped curve at low q values with a maximum peak height of 1.32 at a peak position of 2.29, which are only slightly larger than expected for a rigid, compact structure (SI Appendix, Fig. S8) (41, 42). When adorned with its SpaC tip pilin, the SpaA pilus adheres C. diphtheriae to human pharyngeal cells, preventing disengagement of the microbe by withstanding significant pulling forces caused by coughing, sneezing, mucociliary flow, etc. (43, 44) The SpaA pilus can withstand these forces due to the optimal positioning of intra- and intermolecular lysine–isopeptide bond crosslinks (45). The model of the pilus fiber presented here (Figs. 1D and 3B) is in agreement with previous single-molecule pulling experiments, which concluded that the load bearing spine of the pilus only goes through the middle (MSpaA) and C-terminal (CSpaA) domains that contain spontaneously forming intrapilin isopeptide linkages that are capable of withstanding a pulling force of ∼525 pN (21, 23, 45).
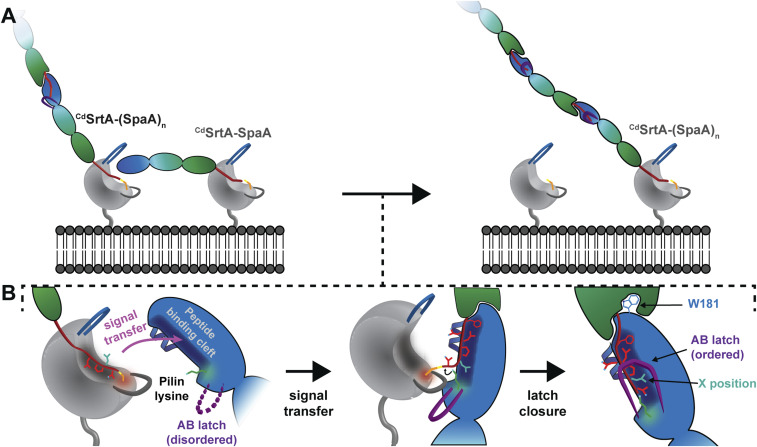
To assemble pili, CdSrtA and other pilus-specific sortases select for crosslinking a single lysine on the surface of their pilin substrates. As sortases are relatively small enzymes, how this specificity is achieved has remained unclear. Our results suggest that selectivity is achieved using a latching mechanism that relies upon tertiary structural features present in the SpaA protein. Fig. 5 shows a working model of lysine–isopeptide bond forming step catalyzed by CdSrtA that adds a single SpaA protein to the shaft of the pilus. This reaction forms a K190-T494 lysine–isopeptide bond between SpaA proteins, connecting the sorting signal (red) following the CSpaA domain (green) to the K190 amine group in NSpaA (blue) (Fig. 5A). Presumably, two thioacyl-linked enzyme–substrate intermediates mediate this interaction and are tethered to the membrane via their respective CdSrtA enzymes (7, 18). The growing (SpaA)n polymer is housed in a CdSrtA-(SpaA)n intermediate in which the carbonyl group in residue T494 of the C-terminal sorting signal on the polymer is attached via a thioacyl bond to CdSrtA’s active site cysteine. New SpaA proteins enter the reaction as similarly bonded thioacyl enzyme–substrate intermediates (CdSrtA-SpaA) after their sorting signals are nucleophilically attacked by the enzyme’s active site cysteine residue. A single protein is then added to the shaft when the reaction intermediates form a ternary complex that enables the K190 nucleophile on CdSrtA-SpaA to resolve the thioacyl bond in the CdSrtA-(SpaA)n. During this process, the growing pilus is transferred from one enzyme to another and is then poised to react with a new CdSrtA-SpaA intermediate to continue the polymerization reaction.
Fig. 5.
Latch mechanism of sortase-catalyzed pilus biogenesis. (A) The characteristic N-terminal “lid” appendage (blue) of CdSrtA occludes the catalytic cysteine residue. During the acylation step, the lid must be opened to allow substrate entry to the active site, which allows for the formation of an enzyme–SpaA acyl intermediate. The sorting signal peptide (red) is bound in the CdSrtA binding pocket (dark gray). (B) The acyl reaction intermediate encounters another SpaA molecule on the cell surface and the molecules are arranged such that the peptide docking sites and reactive cysteine and lysine of CdSrtA and SpaA, respectively, are juxtaposed. This ternary complex in which both SpaA substrates are bound to the pilin polymerase is called the attack complex. Prior to transpeptidation, the signal peptide is partially transferred to the binding groove of the SpaA acceptor substrate. After the signal peptide binds efficiently to SpaA, the pilin lysine (K190, green) nucleophilically attacks the acyl linkage, resolving the intermediate and resulting in a SpaA-SpaA isopeptide linkage. The orientation of the molecules suggests that the AB loop of the SpaA acceptor (purple) may engage in loop–loop interactions with the β7/β8 loop of CdSrtA (gray). Following transpeptidation, the previously disordered AB loop collapses into a rigid latch-like conformation over the isopeptide linkage, which dissolves the ternary complex and provides additional stability to the linkage site.
Our results suggest that the isopeptide bond–forming reaction occurs through a latch mechanism in which selectivity for K190 is achieved by first requiring that the sorting signal be transferred from the enzyme to NSpaA in order to initiate catalysis (Fig. 5B). The most parsimonious orientation of the enzyme–substrate reactants in the ternary complex aligns the sorting signal binding pockets on the enzyme and NSpaA, enabling a simple translation movement to transfer the signal between the proteins. This orientation explains why the AB loop is flexible, as it can readily be displaced outwards to allow K190 access to the enzyme’s active site. Moreover, this arrangement positions CdSrtA’s β7/β8 loop near K190, providing a rationale for why many of its residues are highly conserved and important for catalysis (25). We surmise that within the ternary complex, the sorting signal must be partially transferred to the binding groove on NSpaA in order to activate K190 for catalysis. This is because several NSpaA variants that alter contacts to sorting signal residues that are positioned distal to the site of crosslinking slow transpeptidation by increasing the enzyme’s KM for NSpaA (Fig. 4B, W181A/A170S). Partial signal transfer would act as a zipper, juxtaposing K190 and the CdSrtA-(SpaA)n thioacyl bond, potentially creating a microenvironment that deprotonates the ε-amine for nucleophilic attack on the thioacyl linkage. This would seem essential, since in the isolated NSpaA protein the AB loop is dynamic (Fig. 2C) and thus transiently exposes the side chain of K190 to solvent such that it presumably adopts a protonated, nonnucleophilic state at physiological pHs (its calculated pKa is ∼10.1) (46–48). As the isopeptide bond forms, our NMR data reveal that the AB latch closes, undergoing a disordered-to-ordered transition that shields the bond and stabilizes the protein. Latch closure may also help drive the dissolution of the ternary complex, freeing the transferred polymer for another round of catalysis. It seems likely that other sortases will build pili through a similar mechanism in which the sorting signal is first partially transferred to a binding groove on the pilin substrate to facilitate crosslinking to a specific lysine residue. While not universal, the latch may also be a conserved feature in many sortase-catalyzed pilus biogenesis reactions, as structures of several shaft pilins solved in their apo-states also contain disordered AB loops (20, 23, 36, 40, 49–53). As pili in Gram-positive bacteria are important virulence factors, the results reported here could be useful in guiding the development of antibiotics that work by inhibiting pilus assembly.
Methods
Production of the Crosslinked NSpaA-Signal Complex and CSpaA-NSpaA Dimer.
The amino-terminal domain of the SpaA protein from C. diphtheriae (NSpaA, residues E53 to S195) was produced and purified as described previously (24, 25). The NSpaA-signal complex was generated by enzymatic covalent ligation of synthetic peptide to [13C,15N] NSpaA using a previously described activated variant of CdSrtA (24, 26) (CdSrtA3M, residues N37 to Q257, containing D81G/W83G/N85A mutations). Complete modification of NSpaA to its cognate sorting signal peptide occurred after incubation of 100 μM CdSrtA3M-His6, 100 μM NSpaA, and 1 mM synthetic peptide derived from the SpaA sorting signal motif (KNAGFELPLTGGSGRI) (Peptide 2.0) in modification buffer (50 mM Tris pH 8.0, 300 mM NaCl, and 5 mM TCEP) for 24 h at room temperature. Sortase and unreacted peptide were removed from the reaction by HisPur Co2+ purification and subsequent concentration by Amicon spin filters with a 10 kDa molecular weight cut-off (MWCO). Complex formation was confirmed by matrix assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry as well as SDS-PAGE analysis. The sample was exchanged into NMR buffer (50 mM NaH2PO4 pH 6.0, 100 mM NaCl, 8% D2O, and 0.01% NaN3) and diluted to a concentration of 1.2 mM for NMR studies. Subsequently, the sample was lyophilized and redissolved into 100% deuterated NMR buffer for additional NMR studies. The crosslinked CSpaA-NSpaA dimer was prepared as described for the NSpaA-signal complex but employed 300 μM CSpaA (SpaA, residues R350 to I500) instead of peptide. To assess the relative reactivity of NSpaA variants with interfacial point mutations, the reactions were stopped at indicated timepoints and product formation was assessed by densitometry using ImageJ2 (54). For SAXS analysis, the wild-type dimer was purified from the reaction components using HisPur Co2+ immobilized metal affinity chromatography (IMAC) (Thermo Fisher Scientific) and size exclusion chromatography (Superdex Increase 10/300 GL, GE Healthcare). For NMR samples of the dimer, either NSpaA or CSpaA was uniformly labeled with nitrogen-15, and the other component was expressed in natural abundance nitrogen-14 media.
Immunoelectron Microscopy and Cell Fractionation Studies.
Cells of the C. diphtheriae ΔspaA mutant expressing wild-type SpaA or individual SpaA mutants from a plasmid were grown on Heart Infusion Broth (HIB) agar plates. A full loop of each overnight colony was collected, suspended in, and gently washed once with phosphate-buffered saline (PBS) buffer. A drop (7 μL) of bacterial suspension in PBS was immobilized on the carbon-coated nickel grids for immunogold labeling as previously reported (25, 55). Cells were stained with antibodies against SpaA (α-SpaA; 1:100 dilution), followed by IgG antibodies conjugated to 12 nm colloid gold particles. Cells were then stained with 1% uranyl acetate prior to analysis using a JEOL JEM1200 electron microscope. For fractionation studies, cells were cultured in HIB media supplemented with 30 μg/mL kanamycin and grown to mid log phase (OD600 = 0.5 to 0.6). Cells were normalized to OD600 at 1 before harvest. The harvested cells were fractionated into medium (S) and cell wall–associated (W) fractions as described previously (4). The samples were separated by 3 to 14% gradient SDS-PAGE and analyzed by immunoblotting with α-SpaA antibody.
NMR Structure Determination and Relaxation Measurements.
NMR spectra were collected at 298 K on Bruker Avance III HD 600 MHz and Avance NEO 800 MHz spectrometers equipped with triple resonance probes. NMR data were processed with NMRPipe (56), and analyzed using CARA (57) (version 1.8.4), XIPP (58) (version 1.19.6 p0), and NMRFAM-Sparky (59). 1H, 13C, and 15N protein chemical shifts were assigned using the following experiments: 15N-HSQC, 13C-HSQC, HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO, HBHA(CO)NH, HNHA, HNHB, CC(CO)NH, H(CCCO)NH, HCCH-COSY, HCCH-TOCSY, and 15N-TOCSY. Chemical shifts of the unlabeled (natural abundance) sorting signal peptide were assigned using: two-dimensional (2D) (F1,F2) 13C-filtered NOESY and 2D (F1) 13C,15N-filtered TOCSY experiments (60). Protein NOE distance restraints were acquired from 15N- and 13C-edited NOESY spectra (120 ms mixing time), and intermolecular restraints were obtained from three-dimensional (3D) (F1) 13C, 15N -filtered (F2) 13C -edited NOESY-HSQC, (F1) 13C, 15N-filtered (F2) 15N-edited NOESY-HSQC, and 2D (F1) 13C -filtered NOESY spectra. ψ and φ dihedral restraints were obtained from secondary 13C chemical shifts as calculated by TALOS-N (61) and 3JHNα measurements from the HNHA spectrum. Additional ψ angle restraints were obtained from analysis of 15N-edited NOESY spectrum. Rotamer assignments and χ1 angle restraints for β-methylene protons were obtained through analysis of 15N-TOCSY, HNHB, HN(CO)HB, and 15N -ROESY spectra.
Structures were determined using the program XPLOR-NIH (62, 63). Initially, NOE cross peaks in the 3D 15N -edited NOESY-HSQC and 13C -edited NOESY-HSQC spectra were assigned automatically using the program UNIO10 (64, 65). The NOESY data were then manually inspected using he program Xipp (58) to verify all cross peak assignments and to identify additional distance restraints. An iterative procedure was used to refine the structure of the protein–peptide complex. In the final round of calculations, 200 structures were generated, which yielded a total of 50 with no NOE, dihedral angle, or scalar coupling violations greater than 0.5 Å, 5°, or 2 Hz, respectively. The structures were sorted based on lowest overall energy, and the top 40 were selected as the ensemble to represent the structure of NSpaA-signal and have been deposited in the PDB (code 7K7F). The programs MOLMOL (66) and PyMOL (67) were used to generate figures.
The 15N relaxation data were collected using 1 mM 15N, 13C-labeled samples of the apo- and NSpaA-signal complex dissolved in H2O on a Bruker Avance 600 MHz NMR spectrometer equipped with a triple resonance cryogenic probe. Data were analyzed using SPARKY (59) and included: 15N longitudinal relaxation rates (R1), transverse relaxation rates (R2), and {1H}-15N heteronuclear NOEs. Quantifiable relaxation data could be measured for all parameters for 84 and 97 of 143 backbone amides for the apo- and NSpaA-signal complexes, respectively. For inclusion in the calculations, data from each residue must meet the following criteria: isolated 1H-15N cross peaks and {1H}-15N NOE values of >0.6. For backbone R1 (15N) and R2 (15N) measurements at 600 MHz, the same relaxation delays were used for both apo- and NSpaA-signal complex samples. R1(15N) measurements used delays of T = 1500, 1000, 500, 300 (×2), 100, and 50 ms. R2(15N) measurements used delays of 17 (×2), 34, 51, 68, 85, 119 (×2), 153, and 170 ms.
To calculate expected rotational correlation times based on molecular weight, the following relationship between hydrodynamic radius and protein molecular weight was employed:
in which M = molecular weight of the protein, ρ = the average density for proteins (1.37 g/cm3), Na = Avogadro’s number, and rw = hydration radius (1.6 to 3.2 Å) (68). After calculating the hydrodynamic radius of the protein of interest, and assuming a spherical approximation, the Stokes’ law equation was used to calculation an expected rotational correlation time:
in which η = the viscosity of the solvent, r = hydrodynamic radius (calculated above), k = Boltzmann constant, and T = acquisition temperature. Experimental values of τc for the complex and isolated domains were estimated using the ratio of T1 and T2 NMR relaxation rates (69).
Differential Scanning Fluorimetry and Protease Sensitivity Measurements.
The melting temperature and thermodynamic parameters were extracted from Differential Scanning Fluorimetry data by a method described previously (30). Briefly, NSpaA proteins were diluted to 50 μM in assay buffer (50 mM Tris pH 8.0, 300 mM NaCl), supplemented with 15x SYPRO Orange (Sigma-Aldrich) at a total volume of 20 µL. Thermal denaturation reactions were run on a CFX Connect qPCR system (Bio-Rad). A heating rate of 0.2 °C/min was employed from 4 to 95 °C, and fluorescence measurements (excitation at 525 ± 10 nm, detection at 570 ± 10 nm) were acquired after each 0.5° step increase. The melting temperature for each protein was determined by the first Derivative method after averaging the three replicate measurements. The Tm is defined as the midpoint of the transition from folded to unfolded and is identified spectroscopically as the temperature where the rate of fluorescence increases with respect to temperature is greatest. The Tm was then used to calculate the equilibrium constant of unfolding, as previously described (30). For the limited proteolysis experiments, either NSpaA, the NSpaA-signal complex, or the CSpaA-NSpaA dimer was dissolved in assay buffer (50 mM Tris pH 8.0, 300 mM NaCl) at a concentration of 20 µM in a volume of 100 µL. Trypsin protease stock solutions were created as described by the manufacturer (Sigma-Aldrich). A total of 0 to 200 ng of trypsin protease (or 400 ng for CSpaA-NSpaA dimer reactions) was added to the reactions and incubated at 37 °C. Samples from each reaction were taken after 6 and 24 h, separated by SDS-PAGE, and analyzed by densitometry.
Quantitative Transpeptidation Measurements.
The gel-based fluorescence assays were performed at room temperature as previously described (24, 26). All reactions included 25 μM CdSrtAΔ, 250 μM fluorescent sorting signal peptide (fluorescein isothiocyanate [FITC]-KNAGFELPLTGGSGRI) and 25 μM wild-type or variant NSpaA. Time points were taken after 24 h, and the resultant reaction mixture was separated by SDS-PAGE. The protein gels then washed in ddH2O and fluorescence data were acquired with a Pharos FX gel imager (Bio-Rad). FITC was detected by excitation with a 488 nm laser line and detection with a 515 to 545 nm emission filter. The same gel was then stained with Coomassie to visualize the total protein content of each lane. Quantification of the kinetic parameters of transpeptidation was carried out by separation of the reaction components by reverse-phase HPLC at various time points and substrate concentrations. Each reaction was incubated at room temperature, and proteins were dissolved in assay buffer (50 mM Tris pH 8.0, 300 mM NaCl), containing 50 μM CdSrtA Δ79–87, 1 mM sorting signal peptide (FELPLTGGSG), 5 mM DTT, and 50 to 300 μM wild-type or mutant NSpaA. Different variants were incubated either 3.5 h (wild-type, L62D, ABΔ15, and A170S) or 16 h (H60A, W181A, V187D, and Y188G) depending on reactivity. The reactions were stopped by flash freezing in liquid nitrogen after incubation. Each reaction condition was run in duplicate. A total of 25 μL of each reaction was injected onto a Water Symmetry 300 C4 HPLC column (4.6 × 150 mm, 5 μm particle size). Proteins were eluted by applying a gradient from 35 to 46% acetonitrile (with 0.1% trifluoroacetic acid) over 12 min at a flow rate of 1 mL/min. Elution of proteins was monitored by absorbance at 215 nm. Peak height of each elution in the HPLC trace was measured by integration of peak areas using Graphical Analysis (Vernier). Data were plotted as Lineweaver–Burk in order to calculate kinetic parameters for each NSpaA variant.
SAXS Analysis.
Scattering data were generated at the SIBYLS beamline (Advanced Light Source [ALS], Lawrence Berkeley National Laboratory) (70). Purified CSpaA-NSpaA complex (10 mg/mL) was dissolved in size-exclusion chromatography (SEC) buffer (50 mM NaH2PO4, pH 6.5, 100 mM NaCl, and 0.01% NaN3) and applied to a Shodex KW-802.5 SEC column for SEC-SAXS. Scattering of the buffer without protein was obtained using SAXS data from the SEC run where no protein was eluted and was subtracted from the merged data from the frames corresponding to the elution of the complex. Radius of gyration (Rg) and maximal particle dimension (Dmax) were calculated by Guinier analysis [BioXTAS RAW (71)] and GNOM (ATSAS software package), respectively. Calculated from the Guinier approximation, the radius of gyration (Rg) and forward scattering intensity (I[0]) were determined to be 25.1 Å and 51.0 Å, respectively. From the distance distribution function, the Dmax and Porod volume were calculated to be 8.7 nm and 3.72 × 104 Å3, respectively. The rod-like conformation of the dimer can be inferred from these low-resolution structural parameters, as a spherical with 287 residues would be expected to have a Rg and Dmax of 19.8 Å and 5.1 nm, respectively ( and ) (41). Inspection of the normalized Kratky plot of the SAXS data reveals a clear bell-shaped curve at low q values with a maximum peak height at 1.32 at a peak position of 2.29, which indicates a small contribution from disordered regions of the complex (idealized peak height and position are and , respectively) (41, 42). The program GASBOR (72) was used to calculate low-resolution ab initio models in which each residue of the protein is represented as dummy residues (DRs), starting from a random distribution inside a search box with long axis of diameter Dmax, followed by a simulated annealing protocol to condense the DR distribution to fit the experimental scattering data.
For rigid body modeling, we began by generating the CSpaA-NSpaA isopeptide dimer starting structure by merging the crystal and NMR coordinates of CSpaA (PDB:3HR6) and NSpaA-signal (this work), respectively, into a single coordinate file using PyMOL (67). The coordinates were energy minimized in GROMACS (73) to remove steric clashes or inappropriate geometries. Based on the lack of electron density in the crystal structure in positions C-terminal to K484 and lack of defined peptide orientation in our NMR complex N-terminal to F489, the positions in the sorting signal from K483-A488 were defined as flexible residues and generated 10,000 conformations which explored the conformational space available through rotation of those backbone dihedrals using a Rapidly Exploring Random Tree search algorithm in the Integrative Modeling Platform software package (74). A SAXS profile was then generated for each model using the FOXS method and the best scoring multistate models were enumerated. The MultiFOXS algorithm predicted two single-state models, one with a significantly better fit to the experimental SAXS data (χ2 = 0.89, 1.24). For subsequent analysis, we chose the model with the superior χ2 value. No multistate models were predicted, ruling out the possibility that the complex consists of an ensemble of conformations in solution.
Supplementary Material
Acknowledgments
We thank members of the R.T.C. and H.T.-T. laboratories for discussions and reviewing this manuscript. This research was supported by funding from the NIH under the Award Numbers AI52217 (to R.T.C.), DE025015 and DE017382 (to H.T.-T.), and T32 GM007185 and the UCLA Graduate Division Dissertation Year Fellowship (to S.A.M.). We also acknowledge NIH shared instrumentation Grants S10OD025073 and S10OD016336 and support from the US Department of Energy (DE-FC03-02ER63421). SAXS data were collected at SIBYLS, which is supported by the DOE-BER IDAT, DE-AC02-05CH11231, and National Institute of General Medical Sciences ALS-ENABLE (P30 GM124169 and S10OD018483).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019649118/-/DCSupplemental.
Data Availability
Protein structure data have been deposited in PDB (7K7F).
References
- 1.Danne C., Dramsi S., Pili of gram-positive bacteria: Roles in host colonization. Res. Microbiol. 163, 645–658 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Ramirez N. A., Das A., Ton-That H., New paradigms of pilus assembly mechanisms in gram-positive actinobacteria. Trends Microbiol. 28, 999–1009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandlik A., Das A., Ton-That H., The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 14147–14152 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang C., Mandlik A., Das A., Ton-That H., Cell surface display of minor pilin adhesins in the form of a simple heterodimeric assembly in Corynebacterium diphtheriae. Mol. Microbiol. 79, 1236–1247 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandlik A., Swierczynski A., Das A., Ton-That H., Pili in gram-positive bacteria: Assembly, involvement in colonization and biofilm development. Trends Microbiol. 16, 33–40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hospenthal M. K., Costa T. R. D., Waksman G., A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat. Rev. Microbiol. 15, 365–379 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Ton-That H., Schneewind O., Assembly of pili in Gram-positive bacteria. Trends Microbiol. 12, 228–234 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Telford J. L., Barocchi M. A., Margarit I., Rappuoli R., Grandi G., Pili in gram-positive pathogens. Nat. Rev. Microbiol. 4, 509–519 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Scott J. R., Zähner D., Pili with strong attachments: Gram-positive bacteria do it differently. Mol. Microbiol. 62, 320–330 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Kang H. J., Baker E. N., Structure and assembly of gram-positive bacterial pili: Unique covalent polymers. Curr. Opin. Struct. Biol. 22, 200–207 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Kline K. A., Dodson K. W., Caparon M. G., Hultgren S. J., A tale of two pili: Assembly and function of pili in bacteria. Trends Microbiol. 18, 224–232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Proft T., Baker E. N., Pili in Gram-negative and Gram-positive bacteria - structure, assembly and their role in disease. Cell. Mol. Life Sci. 66, 613–635 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fronzes R., Remaut H., Waksman G., Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J. 27, 2271–2280 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abee T., Kovács Á. T., Kuipers O. P., van der Veen S., Biofilm formation and dispersal in Gram-positive bacteria. Curr. Opin. Biotechnol. 22, 172–179 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Van Gerven N., Waksman G., Remaut H., “Pili and flagella: Biology, structure, and biotechnological applications” in Progress in Molecular Biology and Translational Science, Howorka S., Ed. (Elsevier B.V., 2011), 103, pp. 21–72. [DOI] [PubMed] [Google Scholar]
- 16.Ton-That H., Schneewind O., Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50, 1429–1438 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Ton-That H., Marraffini L. A., Schneewind O., Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 53, 251–261 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Guttilla I. K., et al., Acyl enzyme intermediates in sortase-catalyzed pilus morphogenesis in gram-positive bacteria. J. Bacteriol. 191, 5603–5612 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khare B., V L Narayana S., Pilus biogenesis of Gram-positive bacteria: Roles of sortases and implications for assembly. Protein Sci. 26, 1458–1473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang H. J., Coulibaly F., Clow F., Proft T., Baker E. N., Stabilizing isopeptide bonds revealed in Gram-positive bacterial pilus structure. Science 318, 1625–1628 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Kang H. J., Baker E. N., Intramolecular isopeptide bonds: Protein crosslinks built for stress? Trends Biochem. Sci. 36, 229–237 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Hilleringmann M., et al., Molecular architecture of Streptococcus pneumoniae TIGR4 pili. EMBO J. 28, 3921–3930 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang H. J., Paterson N. G., Gaspar A. H., Ton-That H., Baker E. N., The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc. Natl. Acad. Sci. U.S.A. 106, 16967–16971 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McConnell S. A. S. A., et al., Protein labeling via a specific lysine-isopeptide bond using the pilin polymerizing sortase from Corynebacterium diphtheriae. J. Am. Chem. Soc. 140, 8420–8423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang C., et al., In vitro reconstitution of sortase-catalyzed pilus polymerization reveals structural elements involved in pilin cross-linking. Proc. Natl. Acad. Sci. U.S.A. 115, E5477–E5486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sue C. K., et al., Kinetics and optimization of the lysine-isopeptide bond forming sortase enzyme from Corynebacterium diphtheriae. Bioconjug. Chem. 31, 1624–1634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan A. H., et al., Structure of the bacillus anthracis sortase a enzyme bound to its sorting signal: A flexible amino-terminal appendage modulates substrate access. J. Biol. Chem. 290, 25461–25474 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suree N., et al., The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized. J. Biol. Chem. 284, 24465–24477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baljinnyam B., Ronzetti M., Yasgar A., Simeonov A., Applications of differential scanning fluorometry and related technologies in characterization of protein-ligand interactions. Methods Mol. Biol. 2089, 47–68 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Wright T. A., Stewart J. M., Page R. C., Konkolewicz D., Extraction of thermodynamic parameters of protein unfolding using parallelized differential scanning fluorimetry. J. Phys. Chem. Lett. 8, 553–558 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Schneidman-Duhovny D., Hammel M., Tainer J. A., Sali A., FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 44, W424–W429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Spraggon G., et al., Supramolecular organization of the repetitive backbone unit of the Streptococcus pneumoniae pilus. PLoS One 5, e10919 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Budzik J. M., et al., Intramolecular amide bonds stabilize pili on the surface of bacilli. Proc. Natl. Acad. Sci. U.S.A. 106, 19992–19997 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cozzi R., et al., Structure and assembly of group B streptococcus pilus 2b backbone protein. PLoS One 10, e0125875 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang H. J., et al., A slow-forming isopeptide bond in the structure of the major pilin SpaD from Corynebacterium diphtheriae has implications for pilus assembly. Acta Crystallogr. D Biol. Crystallogr. 70, 1190–1201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishra A., et al., Two autonomous structural modules in the fimbrial shaft adhesin FimA mediate Actinomyces interactions with streptococci and host cells during oral biofilm development. Mol. Microbiol. 81, 1205–1220 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vengadesan K., Ma X., Dwivedi P., Ton-That H., Narayana S. V. L., A model for group B Streptococcus pilus type 1: The structure of a 35-kDa C-terminal fragment of the major pilin GBS80. J. Mol. Biol. 407, 731–743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nuccitelli A., et al., Structure-based approach to rationally design a chimeric protein for an effective vaccine against Group B Streptococcus infections. Proc. Natl. Acad. Sci. U.S.A. 108, 10278–10283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaurasia P., Pratap S., Palva A., von Ossowski I., Krishnan V., Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism. Commun. Biol. 1, 94 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durand D., et al., NADPH oxidase activator p67(phox) behaves in solution as a multidomain protein with semi-flexible linkers. J. Struct. Biol. 169, 45–53 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Receveur-Brechot V., Durand D., How random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci. 13, 55–75 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogers E. A., Das A., Ton-That H., Adhesion by pathogenic corynebacteria. Adv. Exp. Med. Biol. 715, 91–103 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Mandlik A., Swierczynski A., Das A., Ton-That H., Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol. Microbiol. 64, 111–124 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Echelman D. J., et al., CnaA domains in bacterial pili are efficient dissipaters of large mechanical shocks. Proc. Natl. Acad. Sci. U.S.A. 113, 2490–2495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kilambi K. P., Gray J. J., Rapid calculation of protein pKa values using Rosetta. Biophys. J. 103, 587–595 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyskov S., et al., Serverification of molecular modeling applications: The rosetta online server that includes everyone (ROSIE). PLoS One 8, e63906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagarajan R., et al., PDBparam: Online resource for computing structural parameters of proteins. Bioinform. Biol. Insights 10, 73–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Persson K., Esberg A., Claesson R., Strömberg N., The pilin protein FimP from actinomyces oris: Crystal structure and sequence analyses. PLoS One 7, e48364 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young P. G., et al., Structural conservation, variability, and immunogenicity of the T6 backbone pilin of serotype M6 Streptococcus pyogenes. Infect. Immun. 82, 2949–2957 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El Mortaji L., et al., The full-length Streptococcus pneumoniae major pilin RrgB crystallizes in a fiber-like structure, which presents the D1 isopeptide bond and provides details on the mechanism of pilus polymerization. Biochem. J. 841, 833–841 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Chaurasia P., Pratap S., von Ossowski I., Palva A., Krishnan V., New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit. Sci. Rep. 6, 28664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaik M. M., et al., A structural snapshot of type II pilus formation in Streptococcus pneumoniae. J. Biol. Chem. 290, 22581–22592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rueden C. T., et al., ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang C., Huang I. H., Hendrickx A. P. A., Ton-That H., Visualization of Gram-positive bacterial pili. Methods Mol. Biol. 966, 77–95 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Delaglio F., et al., NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 57.Keller R., The Computer Aided Resonance Assignment Tutorial (Cantina Verlag, Goldau, Switzerland, 2004). [Google Scholar]
- 58.Garrett D. S., Cai M., Clore G. M., XIPP: Multi-dimensional NMR analysis software. J. Biomol. NMR 74, 9–25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee W., Tonelli M., Markley J. L., NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clore G. M., Bax A., Gronenborn A. M., Stereospecific assignment of β-methylene protons in larger proteins using 3D 15N-separated Hartmann-Hahn and 13C-separated rotating frame Overhauser spectroscopy. J. Biomol. NMR 1, 13–22 (1991). [DOI] [PubMed] [Google Scholar]
- 61.Shen Y., Bax A., Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwieters C. D., Kuszewski J. J., Marius Clore G., Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 48, 47–62 (2006). [Google Scholar]
- 63.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M., The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Herrmann T., Güntert P., Wüthrich K., Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 (2002). [DOI] [PubMed] [Google Scholar]
- 65.Herrmann T., Güntert P., Wüthrich K., Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR 24, 171–189 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Koradi R., Billeter M., Wüthrich K., MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 (1996). [DOI] [PubMed] [Google Scholar]
- 67.DeLano W. L., The PyMOL Molecular Graphics System, Version 2.3. Schrödinger LLC www.pymol.org (2020). 10.1038/hr.2014.17. [DOI]
- 68.Cavanagh J., Protein NMR Spectroscopy : Principles and Practice (Academic Press, 2007). [Google Scholar]
- 69.Kay L. E., Torchia D. A., Bax A., Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 28, 8972–8979 (1989). [DOI] [PubMed] [Google Scholar]
- 70.Classen S., et al., Implementation and performance of SIBYLS: A dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the advanced light source. J. Appl. Cryst. 46, 1–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hopkins J. B., Gillilan R. E., Skou S., BioXTAS RAW: Improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Cryst. 50, 1545–1553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Svergun D. I., Petoukhov M. V., Koch M. H. J., Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hacker S. M., et al., Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem. 9, 1181–1190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Russel D., et al., Putting the pieces together: Integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 10, e1001244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protein structure data have been deposited in PDB (7K7F).