Significance
Plant miRNAs can move from cell to cell through cytoplasmic channels called plasmodesmata, transmitting positional information that is decoded to make developmental decisions; one well-studied example of this is xylem patterning in the root of the model plant Arabidopsis thaliana, which depends on the formation of opposing gradients of two miRNAs and their target transcripts. Here, we show that two plasma membrane- and plasmodesmata-localized proteins, BAM1 and BAM2, are required for maintaining appropriate levels and distribution of the miRNA target transcripts, and hence for proper root xylem patterning, uncovering additional players in the regulation of this developmental process and adding one more function, direct or indirect, to the ever-growing portfolio of BAM1 and BAM2.
Keywords: receptor-like kinase, BAM1/2, xylem patterning, C4, miRNA
Abstract
Xylem patterning in the root is established through the creation of opposing gradients of miRNAs and their targets, transcripts of the HD-ZIP III family of transcriptions factors, enabled by the cell-to-cell spread of the former. The miRNAs regulating xylem patterning, miR165/6, move through plasmodesmata, but how their trafficking is regulated remains elusive. Here, we describe that simultaneous mutation of the plasma membrane- and plasmodesmata-localized receptor-like kinases (RLKs) BARELY ANY MERISTEM (BAM) 1 and 2 or expression of the geminivirus-encoded BAM1/2-interactor C4 results in higher accumulation and broader distribution of the HD-ZIP III transcripts despite normal total accumulation of miR165/6, and ultimately causes defects in xylem patterning, which depend on the function of the aforementioned miRNA targets. Taken together, our results show that BAM1 and BAM2 are redundantly required for proper xylem patterning in the Arabidopsis root, by ensuring the proper distribution and accumulation of miR165/6-targeted transcripts.
Tissue patterning in plant organ development depends primarily on positional information, which must be communicated between cells. Different mobile molecules can mediate cell-to-cell communication, including phytohormones, transcription factors, or peptides. In the past decade, multiple studies have uncovered the relevance of small noncoding RNAs (sRNAs) as mobile molecules capable of acting as morphogens in plant development, determining leaf polarity, root vascular patterning, embryo meristem formation, female gametogenesis, and maintenance of the shoot apical meristem, regulating the acquisition of cell fate in a dose-dependent fashion (reviewed in refs. 1–3). Interestingly, it has been recently shown that the cell-to-cell movement of microRNAs (miRNAs) is directional (4), indicating that this process must be precisely regulated.
An elegant example of how sRNAs can determine pattern formation is provided by the study of xylem patterning in the root of Arabidopsis thaliana (hereafter referred to as Arabidopsis). Xylem patterning is established by a robust regulatory pathway comprising the action of miRNAs 165 and 166 (miR165/6) and the transcription factors SHORT ROOT (SHR) and SCARECROW (SCR) (5). Xylem precursors differentiate into two types of xylem vessels: metaxylem cells, with pitted secondary cell walls, in the center of the vascular cylinder, and protoxylem cells, distinguishable by their spiral wall thickenings, in a peripheral position (Fig. 1A). SHR is produced in the stele, and moves from cell to cell to the endodermis, where it induces SCR expression and, together with the latter, that of MIR165a and MIR166b. The resulting miR165 and miR166 move into the stele, restricting the accumulation of the class III HOMEODOMAIN-LEUCIN ZIPPER (HD-ZIP III) mRNA, particularly that of PHABULOSA (PHB), to the center of the stele, which results in correct xylem patterning with formation of both metaxylem (high HD-ZIP III activity) and protoxylem (low HD-ZIP III activity) (5, 6). Although miR165/6 have been shown to move symplastically through plasmodesmata (7), how their trafficking is regulated remains elusive.
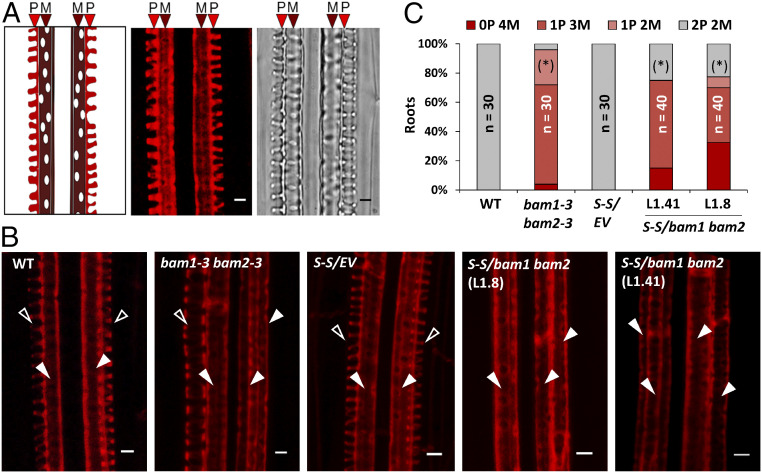
Fig. 1.
BAM1 and BAM2 play a redundant role in xylem patterning. (A) Typical structure of protoxylem (P) and metaxylem (M) in a longitudinal section of the root; on the right, confocal image of a longitudinal section of the root showing basic fuchsin-stained protoxylem (P) and metaxylem (M), and brightfield image of the same region. (Scale bar, 3 µm.) (B) Confocal micrographs of basic fuchsin-stained xylem in the roots of 6-d-old control and bam1 bam2 seedlings. Empty arrowheads indicate protoxylem; filled arrowheads indicate metaxylem. WT: wild type (Ler); S-S: SUC:SUL; EV: empty vector. (Scale bar, 3 µm.) (C) Quantification of the number of protoxylem (P) and metaxylem files (M) in the roots of listed genotypes. Phenotypic categories are displayed in vertical sequence. Statistical differences in the distribution of % of xylem phenotypes (normal patterning = 2P vs. abnormal patterning <2P) between each genotype and its respective control (WT Ler or Col-0 S-S/EV) were assessed by applying Fisher’s exact test; asterisks in parentheses indicate significant differences at P < 0.05. n = number of roots.
The plasma membrane- and plasmodesmata-localized receptor-like kinases (RLKs) BARELY ANY MERISTEM (BAM) 1 and 2 have been recently described as required, directly or indirectly, for the cell-to-cell spread of RNA silencing in the reporter SUC-SUL plants (8), in which production of mobile small interfering RNA (siRNA) against the endogenous At4g18480 gene (encoding the CHLI subunit of the magnesium chelatase) in phloem companion cells causes non–cell-autonomous silencing observable as a chlorotic phenotype around the leaf veins (9). The underlying molecular mechanism of this potential function of BAM1/2, and whether they might also play a role in the cell-to-cell movement of other sRNAs, such as miRNAs, is yet to be determined.
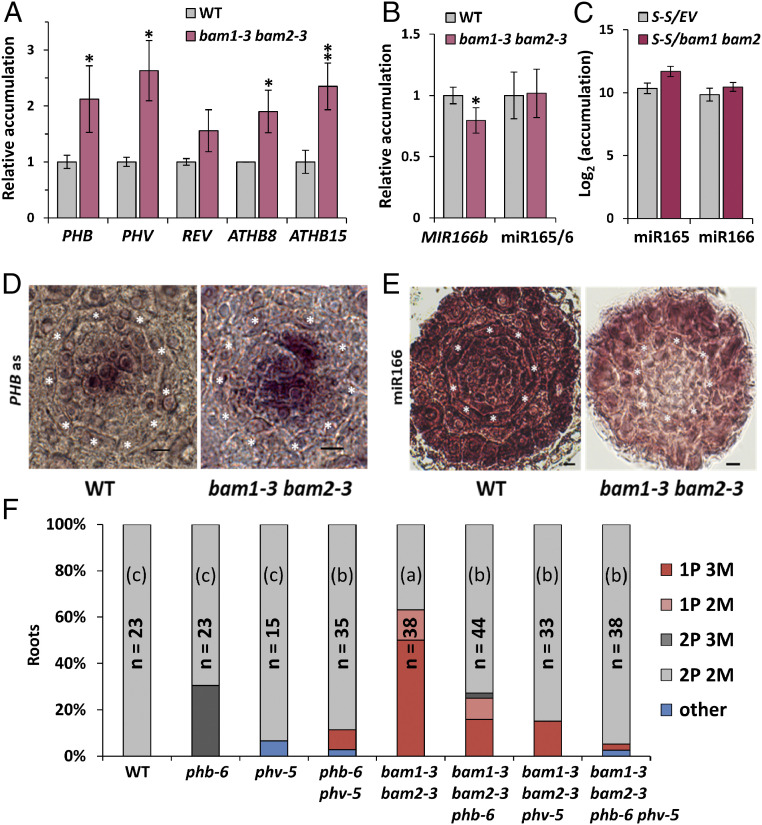
In Arabidopsis roots, BAM1 is strongly expressed in the stele (SI Appendix, Fig. S1) (10). We reasoned that, considering this expression pattern, if BAM1 regulates movement of miRNAs, it could mediate the cell-to-cell spread of miR165/6, hence acting as a regulator of xylem patterning. In order to determine whether BAM1/2 are required for correct xylem formation in the root, we observed xylem patterning in bam1/2 mutants (9, 11). Interestingly, bam1 bam2 double mutants, but not bam1 or bam2 single mutants, display shorter roots (SI Appendix, Fig. S2) and show discontinuous xylem defects consistent with a malfunction of miR165/6-mediated patterning, namely absence of protoxylem files and differentiation of metaxylem in place of protoxylem (Fig. 1 and SI Appendix, Fig. S3). This phenotype can also be observed in a bam1 bam2 bam3 mutant (12) (SI Appendix, Fig. S3). At the molecular level, bam1 bam2 mutants display increased accumulation of HD-ZIP III transcripts, targets of miR165/6; however, these mutants are not affected in regulatory processes upstream of the action of the miRNAs, namely expression of MIR166b, SHR, or SCR, accumulation of miR165/6 (Fig. 2 A–C and SI Appendix, Fig. S4), and a triple bam1 bam2 bam3 mutant is not affected in SHR trafficking (10). The HD-ZIP III family member PHB is a major regulator of xylem patterning; miRNA-resistant mutant versions of PHB (phb-1d, phb-7d) display a xylem patterning phenotype reminiscent of that observed in bam1 bam2 mutants (5). In situ hybridization shows that the spatial distribution of the PHB transcript is less restricted in the root stele in the absence of BAM1/2 (Fig. 2D; negative hybridization control in SI Appendix, Fig. S5), while lower levels of miR166 seem to be detected in this area (Fig. 2E). Confirming that the altered xylem patterning in bam1 bam2 results from the increased accumulation of HD-ZIP III transcription factors, mutations in either PHB and/or PHV are sufficient to partially restore normal xylem patterning (Fig. 2F and SI Appendix, Fig. S6). Considering that in the bam1 bam2 mutant there is a wild-type (WT)-like accumulation of miR165/6 (Fig. 2C) but a wider spatial distribution of its target transcript PHB, our results suggest that it is the action of the miRNA that is affected by the lack of BAM1/2, either cell-autonomously or non–cell-autonomously. Transgenic plants overexpressing BAM1 have normal xylem and WT-like accumulation of the HD-ZIP III transcripts (SI Appendix, Fig. S7, Right), suggesting that either BAM1 is not rate-limiting in this process, or an additional element, e.g., a ligand peptide, is required.
Fig. 2.
miR165/6 activity, but not accumulation, is impaired in bam1 bam2 mutants, leading to increased levels of HD-ZIPIII factors and altered xylem patterning. (A and B) Accumulation of transcripts of the HD-ZIPIII family genes (A), transcripts of the MIR166b gene, and mature miR165/6 (B) in the roots of 6-d-old WT and bam1-3 bam2-3 double mutant seedlings as measured by qRT-PCR. Results are the average of three biological replicates. Error bars indicate SD. Statistical comparisons of means relative to the control group (WT) were assessed by applying Student’s t test; asterisks indicate significant differences with P < 0.05 (*) and P < 0.01 (**). (C) miR165/6 accumulation in SUC-SUL/bam1 bam2 double mutants and SUC-SUL/EV control as measured by sRNA-seq. S-S: SUC:SUL; PHB: PHABULOSA; MIR: pri-miRNA species; miR: mature miRNA species. (D) In situ hybridization with a PHB mRNA specific probe on cross-sections of WT and bam1-3 bam2-3 roots. Seven roots were checked per genotype; all roots showed a similar PHB mRNA spatial distribution. (E) In situ hybridization with a miRNA166-specific LNA probe on cross-sections of WT, bam1-3 bam2-3 roots. Ten roots were checked per genotype; five bam1-3 bam2-3 roots showed the miR166 distribution pattern displayed in this figure. (Scale bar, 6 µm.) Asterisks indicate the position of the endodermis. (F) Quantification of the number of protoxylem (P) and metaxylem (M) files in fuchsin-stained roots of bam1-3 bam2-3 and phb-6 phv-5 double mutants, bam1-3 bam2-3 phb-6 and bam1-3 bam2-3 phv-5 triple mutants, and the bam1-3 bam2-3 phb-6 phv-5 quadruple mutant. The graph shows the aggregate data obtained in two independent replicates, each of them with similar results. WT: wild type (Ler); n: number of roots analyzed; “other” includes rare (low represented) xylem patterning phenotypes such as 3P 2M and 2P 1M. Statistical differences in the distribution of % of xylem phenotypes (normal patterning = 2P vs. abnormal patterning <2P) between different genotypes were assessed by applying Fisher’s exact test; letters in parentheses indicate significant differences at P < 0.0018 (Bonferroni’s adjusted level for multiple comparisons). n = number of roots.
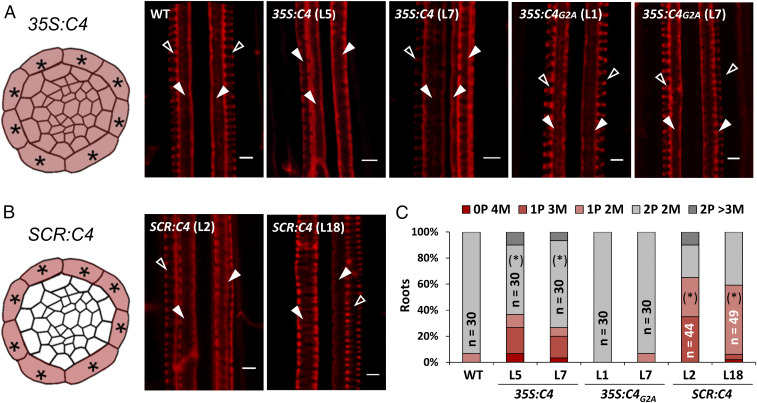
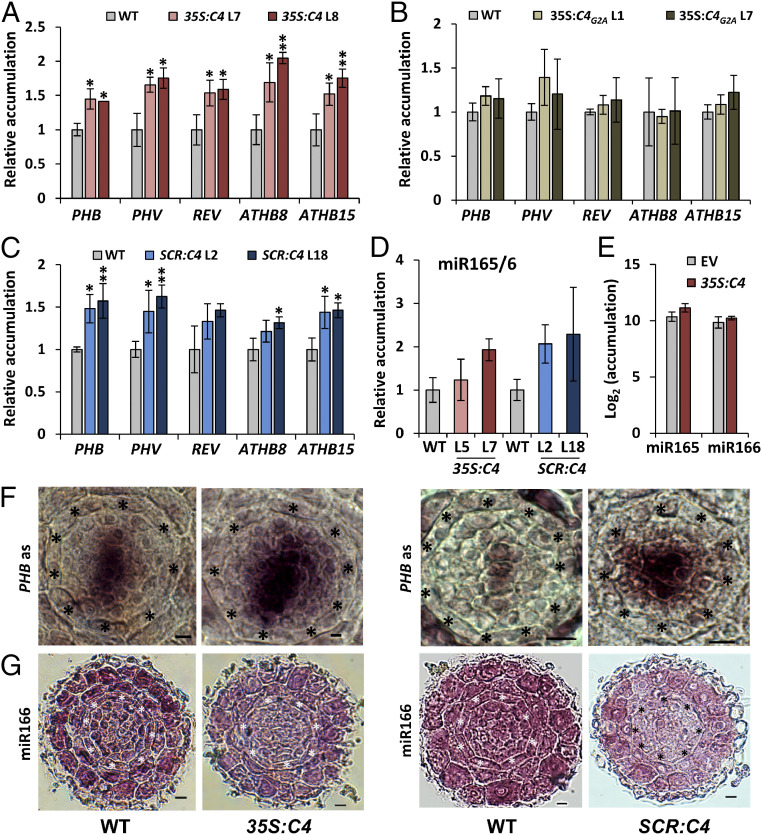
The C4 protein from the geminivirus Tomato yellow leaf curl virus (TYLCV) interacts with the intracellular domain of BAM1/2 at the plasma membrane and has a negative impact on the cell-to-cell spread of RNA silencing (9). In order to test whether the activity of C4 can have an effect on xylem patterning, we observed the xylem in roots of transgenic Arabidopsis plants expressing C4 under the control of the constitutive 35S promoter (9). Strikingly, expression of C4 led to defects in xylem patterning similar to those observed in bam1 bam2 mutants (Fig. 3 A and C). Plasma membrane localization of C4 is essential for this phenotype, since plants expressing the mutated version C4G2A, which loses its membrane association and localizes to chloroplasts (9, 13), have WT-like xylem (Fig. 3A). Transgenic plants expressing C4, but not those expressing C4G2A, show increased accumulation of HD-ZIP III transcripts (Fig. 4 A and B). However, similarly to the lack of BAM1 and BAM2, C4 does not negatively affect the expression of MIR166b, SHR, or SCR, or the accumulation of miR165/6 (Fig. 4 D and E and SI Appendix, Fig. S8 A and C). As observed for bam1 bam2 mutants, the distribution of the PHB transcript in the stele is broader in the C4-expressing plants (Fig. 4F), and lower levels of miR166 seem to be detected in this part of the root (Fig. 4G). Taken together, these results support the idea that C4 affects xylem patterning through its interaction with BAM1/2. Importantly, root length in transgenic 35S:C4 seedlings is comparable to that of WT seedlings (SI Appendix, Fig. S9), ruling out a general effect on root development.
Fig. 3.
Ubiquitous or endodermis-specific expression of the viral BAM1/2 interactor C4 interferes with xylem patterning. (A) Schematic representation of the 35S:C4 expression pattern in a cross-section of the root stele (Left) and confocal micrographs of basic fuchsin-stained xylem in the roots of 5-d-old WT, 35S:C4, and 35S:C4G2A seedlings (Right). (B) Schematic representation of the SCR:C4 expression pattern in a cross-section of the root stele (Left) and confocal micrographs of basic fuchsin-stained xylem in the roots of 5-d-old SCR:C4 seedlings (Right). Empty arrowheads indicate protoxylem; filled arrowheads indicate metaxylem. (Scale bar, 4 μm.) (C) Quantification of the number of protoxylem (P) and metaxylem files (M) in the roots of listed genotypes. Asterisks indicate the position of endodermis. Empty arrowheads indicate protoxylem; filled arrowheads indicate metaxylem. Phenotypic categories are displayed in vertical sequence. WT: wild type (Col-0). Statistical differences in the distribution of % of xylem phenotypes (normal patterning = 2P vs. abnormal patterning <2P) between each genotype and the WT control (Col-0) were assessed by applying Fisher’s exact test; asterisks in parentheses indicate significant differences at P < 0.05. n = number of roots.
Fig. 4.
miR165/6 cell-to-cell movement, but not accumulation, is impaired in C4-expressing plants. (A–D) Accumulation of transcripts of the HD-ZIP III family genes (A, B, and C), and mature miR165/6 (D) in the roots of 11-d-old WT, 35S:C4 (A and D), or 35S:C4G2A (B) seedlings and in the roots of 5-d-old WT or SCR:C4 (C and D) seedlings as measured by qRT-PCR. Results are the average of three biological replicates. Error bars indicate SD. Statistical comparisons of means relative to the control group (WT) were assessed by applying Dunnett’s test; asterisks indicate significant differences with P < 0.05 (*) and P < 0.01 (**). (E) miR165/6 accumulation in SUC-SUL/35S:C4 and SUC-SUL/EV control as measured by sRNA-seq. (F) In situ hybridization with a PHB mRNA specific probe on cross-sections of WT, 35S:C4, and SCR:C4 roots. Five 35S:C4 roots and eight SCR:C4 roots were checked, together with the same number of WT roots; all roots showed a similar PHB mRNA spatial distribution. (G) In situ hybridization with a miRNA166-specific LNA probe on cross-sections of WT, 35S:C4, and SCR:C4 roots. Thirteen 35S:C4 roots and 17 SCR:C4 roots were checked; 5 35S:C4 roots and 7 SCR:C4 roots showed the miR166 distribution pattern displayed in this figure. (Scale bar, 6 µm.) Asterisks indicate the position of endodermis. WT: wild type (Col-0); PHB: PHABULOSA; MIR: pri-miRNA species; miR: miRNA species.
Since miR165/6 are produced in the endodermis, and from here traffic inwards into the stele establishing a gradient that determines HD-ZIP III dosage (5), we reasoned that if C4 is exerting its effect on xylem patterning through the interference with the cell-to-cell movement of miRNAs, then expressing C4 in the endodermal layer should have a non–cell-autonomous effect and be sufficient to cause the observed phenotype. Indeed, transgenic plants expressing C4 under the control of the endodermis-specific SCR promoter display xylem patterning and related molecular phenotypes similar to those previously described for bam1 bam2 and 35S:C4 transgenic lines (Figs. 3 B and C and 4 C and D), including a broader PHB domain and lower miR166 in the root stele (Fig. 4 F and G). Moreover, despite its localization to plasmodesmata (9), C4 does not disturb the movement of SHR-GFP (SI Appendix, Fig. S10); all other upstream regulatory steps analyzed are similarly unaffected (SI Appendix, Fig. S8 B and D). This is consistent with the finding that SHR trafficking is not altered in a bam1 bam2 bam3 mutant (10). Of note, transgenic plants expressing C4 under the SCR promoter display wild-type-like rosettes, but abnormal floral stems (SI Appendix, Fig. S11); as previously described for the 35S:C4 transgenic lines, root length is not altered in SCR:C4 seedlings (SI Appendix, Fig. S9).
Transgenic 35S:C4 or SCR:C4 plants exhibit normal promoter activity of the xylem markers AHP6, ATHB8, and TMO5 (14–16) (SI Appendix, Fig. S12) as well as of MIR165a (5) (SI Appendix, Fig. S13); reconstructions of the cross-view of roots from z-stack images from Col-0, Ler, bam1-3 bam2-3, 35S:C4 (L7), and SCR:C4 (L18) are available as Movies S1–S5. qRT-PCR analyses of the transcript accumulation of these xylem markers show an increase in 35S:C4 plants, and a tendency toward higher accumulation in SCR:C4 and bam1-3 bam2-3 plants, when compared to their WT controls (SI Appendix, Fig. S14). Thus, cell type specification in the C4-expressing lines and the bam1 bam2 mutant seems to be largely unaffected, despite the altered xylem patterning, in agreement with a role of HD-ZIP III transcription factors in xylem differentiation (17–19). These observations are in contrast with results obtained with the gain-of-function mutants phb-1d and phb-7d, as well as with the shr mutant, where increased PHB transcript levels correlated with the activation of cytokinin signaling and inhibition of the expression of AHP6 (5). It is worth noting that both C4-expressing plants and bam1 bam2 mutants show an overall weaker xylem phenotype compared with phb-1d and phb-7d (5). Nevertheless, the bam1 bam2 mutants present additional developmental phenotypes similar to those observed concomitant to the broadening of the PHB domain (20), namely a decrease in the number of cells in the stele and alterations in the endodermis/cortex architecture (SI Appendix, Fig. S15). C4-expressing plants also exhibit abnormalities in the endodermis layout, but a clear trend in the number of cells in the stele could not be detected (SI Appendix, Fig. S15).
Recently, BAM1 was shown to act as a receptor for the CLE9/10 peptides to regulate periclinal cell division of xylem precursor cells, but not xylem identity (21); BAM-CLE signaling has also been demonstrated to mediate formative cell divisions in the Arabidopsis root (10). Importantly, the cle9 cle10 mutant displays WT-like xylem patterning, and transgenic plants expressing C4 under the 35S or the SCR promoters respond normally to CLE9/10 in root growth inhibition assays (SI Appendix, Fig. S16). The results presented here unveil an additional, redundant role of BAM1 and BAM2 in the regulation of xylem cell fate in the root stele; whether the activity of BAM1/2 in this pathway also depends on the presence of a peptide ligand, as previously shown for the regulation of cell division in xylem precursor cells (21) or cortex/endodermis initials (10), remains to be determined. bam1 bam2 double mutants display defects in xylem patterning, which are mimicked cell-autonomously and non–cell-autonomously by the expression of the viral BAM1/2-interactor C4; however, all regulatory steps occurring upstream of the action of miR165/6 appear to be unaltered in the absence of BAM1/2 or in the presence of C4. Despite normal accumulation of miR165/6, the activity of these miRNAs on their target PHB is compromised in the bam1 bam2 mutant or C4 transgenic lines, which correlates with an apparently reduced distribution of miR166 in the root stele, underpinning the observed defective xylem patterning. Importantly, although the bam1 bam2 double mutant has shorter roots than the wild type (SI Appendix, Fig. S2), root length is not severely and consistently affected in plants expressing C4 between days 6 and 11 (SI Appendix, Fig. S9), despite these plants displaying defects in xylem patterning comparable to those observed in the mutant. These results indicate that the two developmental phenotypes can be uncoupled, and suggest that the altered root length in the bam1 bam2 mutant may result from other functions of these RLKs; this would be in agreement with the observation that not all BAM1-mediated responses are hampered by C4 (9).
Taken together, our results uncover the plasma membrane- and plasmodesmata-localized BAM1 and BAM2 as novel regulators of root xylem patterning in Arabidopsis, adding one more function to the ever-growing portfolio of these RLKs.
Methods
Plant Materials and Growth Conditions.
Mutants and transgenic plants used in this study are summarized in SI Appendix, Table S1. Seedlings used for qRT-PCR, xylem phenotype analysis, CLE9/10 treatment, and confocal imaging were grown on half strength Murashige and Skoog (1/2 MS) medium containing 1% sucrose and 1% agar. Plates were placed vertically in a growth chamber with a photoperiod of 16 h light/8 h dark at 22 °C. SCR:C4 plants used for phenotyping were grown in soil under the same environmental conditions described above. For CLE9/10 peptide treatments, SCR:C4 line 2 (L2) and line 18 (L18), 35S:C4 line 3 (L3) and line 7 (L7), and WT Col-0 seeds were sterilized and sown in 1% agar 1% sucrose 1/2 MS medium, and grown for 3 d in vertical plates in a chamber with a photoperiod of 16 h light/8 h dark at 22 °C. After 3 d, the seedlings were transferred to new 1/2 MS medium supplemented with 0, 100 nM, or 1 µM of CLE9/10 peptide solution, and grown for 3 more days in vertical plates. Finally, 6-d-old seedlings were imaged and root length was quantified by using ImageJ software. The average and SD of at least 6 to 10 seedlings was calculated per genotype and treatment. This experiment was repeated three times with similar results.
qRT-PCR.
For qRT-PCR, total RNA was extracted using Plant RNA Kit (Omega) and reverse transcribed by First Chain cDNA Synthesis Kit (TonkBio, China). qPCR was performed using C1000 Touch Thermal Cycler (Bio-Rad); 20 μL of PCR mixture contained 10 μL of SYBR Green mix (Bio-Rad), 1 μL of primer mix (10 μM), 1 μL reverse-transcribed product, and 8 μL of water. ACTIN (ACT2) was used as normalizer. Data were analyzed using the 2ΔΔCT method. To quantify the accumulation of miR165/6 mature forms, stem-loop qPCR was conducted as previously described (22). The primers used in these experiments amplify all the mature forms of both miRNA165 and miR166 (miR165/6). To quantify the accumulation of the pri-miRNA166b (MIR166b), primers annealing specifically in the noncommon regions of this transcript were designed. All primers used for qRT-PCR are listed in SI Appendix, Table S2.
Generation of Transgenic and Mutant Lines.
To generate the pSCR:C4 construct, the coding sequence of C4 was cloned into pENTR/D-TOPO (Invitrogen) and subsequently Gateway cloned into the pSCR:GW vector (23) through an LR reaction (Invitrogen). A. thaliana plants were transformed using the floral dipping method (24). To generate multiple mutants containing phb-6 and phv-5 (the triple mutants bam1-3 bam2-3 phb-6 and bam1-3 bam2-3 phv-5 and the quadruple mutant bam1-3 bam2-3 phb-6 phv-5), parental lines phb-6 phv-5 and bam1-3 bam2-3+/− were crossed, and rounds of genotyping and selection were carried out until reaching F3, when the isolation of homozygous bam1-3 bam2-3 phb-6 phv-5 seedlings was possible. To introduce the phb-13 mutation into CRISPR-Cas9-derived bam1 bam2 mutants, parental lines phb-13 er-2 and S-S/CRISPR-Cas9 bam1 bam2+/− (L1.41) were crossed; rounds of genotyping and selection were performed until reaching F3 in order to segregate S-S (SUC2:SUL cassette) and Cas9, obtaining the bam1 bam2 double mutant and bam1 bam2 phb-13 triple mutant. The primers used for genotyping are listed in SI Appendix, Table S2. To analyze xylem precursor cells in C4-expressing backgrounds, the marker lines pAHP6 > GR > mTurquoise2, pATHB8 SAND line, and pTMO5:n3GFP (14–16) were crossed with Col-0, 35S:C4 L7, and SCR:C4 L18, and fluorescence was analyzed under the confocal microscope in the F1 generation. To induce the expression of mTurquoise2 in the pAHP6 > GR > mTurquoise2 line, transgenic seedlings were grown in 1/2 MS supplemented with dexamethasone (DEX) 50 µM (in dimethyl sulfoxide, DMSO). The different genotypes used in this study are listed in SI Appendix, Table S1. To generate pSHR:SHR-GFP/SCR:C4 lines, the marker line pSHR:SHR-GFP (25) was crossed with Col-0, SCR:C4 L2, and SCR:C4 L18. To generate MIR165a:GFP/35S:C4 and MIR165a:GFP/SCR:C4, the marker line pMIR165a:GFP (5) was crossed with 35S:C4 L7 and SCR:C4 L18.
In Situ Hybridization.
In situ hybridization was performed as previously described (26, 27). The probe for PHB detection (or its negative hybridization control) was cloned into the pGEM-T Easy vector (Promega), using the primers listed in SI Appendix, Table S2. For microRNA in situ hybridization, a specific miR166 locked nucleic acid (LNA) probe (QIAGEN) was used. A total of 100 ng probe were used per slide. The hybridization temperature was 52 °C for PHB detection, and 58 °C for miR166 detection. Note that miR165 and miR166 mature forms only differ in one nucleotide (nt 17), so cross-reactivity may occur (5).
sRNA Sequencing.
sRNA data analyses were performed using a pipeline previously described (28). Briefly, raw reads were trimmed using trim_galore v0.4.0 (https://www. bioinformatics.babraham.ac.uk/projects/trim_galore/) to remove the adapter sequences and bases that have a quality score lower than 10. Reads that could not be aligned to structural RNA sequences (rRNA, tRNA, snoRNA, snRNA, etc.) were aligned to the TAIR10 genome using the Burrows–Wheeler aligner by allowing one mismatch per read (28). The Tair10 genome was divided into nonoverlapping 200-bp bins. The number of sRNA reads (with different lengths) in each 200-bp bin or specific genes were summarized and normalized to the structural RNA-removed library size (reads per 10 million) using bedtools v2.26.0 (https://bedtools.readthedocs.io/en/latest/). Results from two independent transgenic lines per construct were pooled.
Confocal Imaging.
Confocal images were acquired using a Leica TCS SP8 point scanning confocal microscope or a Leica TCS SMD FLCS single molecule detection system. For basic fuchsin staining, 5- or 6-d-old seedlings were first treated with 1 M KOH solution for 6 h at 37 °C. Seedlings were then stained with 0.01% basic fuchsin solution in water for 5 min and subsequently destained in 70% ethanol for 10 min. To check BAM1 expression pattern and SHR-GFP movement in the root tip, 5-d-old seedlings were imaged after propidium iodide (PI) staining. Marker lines for AHP6, ATHB8, TMO5, and MIR165 (5, 14–16) were stained with FM4-64 (1 μM FM4-64 solution, 15 min) before imaging. The settings used for the laser scanning are as follows: excitation (Ex):561 nm, emission (Em):600 to 700 nm for basic fuchsin staining; Ex:488 nm, Em:500 to 550 nm for GFP; Ex:514 nm, Em:525 to 570 nm for YFP; Ex:561 nm, Em:630 to 680 nm for PI staining; and Ex:561 nm, Em:650 to 695 nm for FM4-64 staining. ImageJ and IMARIS 7.7.0 were used for the imaging processing. For root cross-view reconstruction, modified pseudo Schiff propidium iodide (mPS-PI) staining was performed in the roots of 5- or 6-d-old seedlings (29), and the Z-stack images were captured using the Andor Spinning Disk Revolution WD system with a 561-nm laser. Root cross-view reconstructions were made in Fiji. To quantify stele and endodermal cells, 5- or 6-d-old seedlings were fixed with 4% paraformaldehyde, then treated with ClearSee solution as previously described (30), and finally stained with calcofluor-white and analyzed in a Spinning Disk microscope with a 561-nm laser. Root cross-view reconstructions were made in Fiji.
Statistical Analysis.
Statistical comparisons between pairs of means were performed by applying Student’s t test (with equal variances) or Welch’s t test (assuming unequal variances). Statistical multiple comparisons of means were made by one-way ANOVA followed by Dunnett’s test (comparisons of multiple groups to one reference group) or Scheffé’s multiple range test (comparisons between multiple groups). Statistical analyses of xylem and endodermal phenotypes (%) relative to the control plants were performed by applying Fisher’s exact test. For xylem phenotypes, multiple comparisons (%) between genotypes were done by applying Fisher’s exact test with the Bonferroni’s correction of significance level. Statistical analyses were performed with IBM SPSS Statistics v.20 software.
Supplementary Material
Acknowledgments
The authors thank Steven Clark, Zachary Nimchuk, Philip N. Benfey, Ykä Helariutta, Jijie Chai, and Tatsuo Kakimoto for kindly sharing materials; Wenjie Zeng, Xinyu Jian, Aurora Luque, Yujing (Ada) Liu, and the staff at the Cell Biology Core Facility in the Shanghai Center for Plant Stress Biology for technical assistance; and all members of R.L.-D.’s and Alberto Macho’s groups for stimulating discussions and helpful suggestions. This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences, Grant XDB27040206, and by the National Natural Science Foundation of China (NSFC) (Grants 31671994 and 31870250). E.A. is the recipient of a Young Investigator Grant from the NSFC (Grant 31950410534) and a Marie Skłodowska-Curie Grant from the European Union’s Horizon 2020 Research and Innovation Programme (Grant 896910-GeminiDECODER). Research in R.L.-D.’s laboratory is funded by the Shanghai Center for Plant Stress Biology of the Chinese Academy of Sciences and the 100 Talent Program of the Chinese Academy of Sciences.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. Y.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022547118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Furuta K., Lichtenberger R., Helariutta Y., The role of mobile small RNA species during root growth and development. Curr. Opin. Cell Biol. 24, 211–216 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Hisanaga T., Miyashima S., Nakajima K., Small RNAs as positional signal for pattern formation. Curr. Opin. Plant Biol. 21, 37–42 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Skopelitis D. S., Husbands A. Y., Timmermans M. C., Plant small RNAs as morphogens. Curr. Opin. Cell Biol. 24, 217–224 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Skopelitis D. S., et al., Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nat. Commun. 9, 3107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlsbecker A., et al., Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyashima S., Koi S., Hashimoto T., Nakajima K., Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138, 2303–2313 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Vatén A., et al., Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 21, 1144–1155 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Himber C., Dunoyer P., Moissiard G., Ritzenthaler C., Voinnet O., Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22, 4523–4533 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosas-Diaz T., et al., A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi. Proc. Natl. Acad. Sci. U.S.A. 115, 1388–1393 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crook A. D., et al., BAM1/2 receptor kinase signaling drives CLE peptide-mediated formative cell divisions in Arabidopsis roots. Proc. Natl. Acad. Sci. U.S.A. 117, 32750–32756 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeYoung B. J., et al., The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45, 1–16 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Nimchuk Z. L., Zhou Y., Tarr P. T., Peterson B. A., Meyerowitz E. M., Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142, 1043–1049 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medina-Puche L., et al., A defense pathway linking plasma membrane and chloroplasts and Co-opted by pathogens. Cell 182, 1109–1124.e25 (2020). [DOI] [PubMed] [Google Scholar]
- 14.De Rybel B., et al., A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev. Cell 24, 426–437 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Marques-Bueno M. D. M., et al., A versatile multisite gateway-compatible promoter and transgenic line collection for cell type-specific functional genomics in Arabidopsis. Plant J. 85, 320–333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schürholz A. K., et al., A comprehensive toolkit for inducible, cell type-specific gene expression in Arabidopsis. Plant Physiol. 178, 40–53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Q., et al., Activation of miR165b represses AtHB15 expression and induces pith secondary wall development in Arabidopsis. Plant J. 83, 388–400 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Endo H., et al., Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56, 242–254 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Taylor-Teeples M., et al., An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sebastian J., et al., PHABULOSA controls the quiescent center-independent root meristem activities in Arabidopsis thaliana. PLoS Genet. 11, e1004973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian P., et al., The CLE9/10 secretory peptide regulates stomatal and vascular development through distinct receptors. Nat. Plants 4, 1071–1081 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Varkonyi-Gasic E., Wu R., Wood M., Walton E. F., Hellens R. P., Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3, 12 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michniewicz M., Frick E. M., Strader L. C., Gateway-compatible tissue-specific vectors for plant transformation. BMC Res. Notes 8, 63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X., Henriques R., Lin S. S., Niu Q. W., Chua N. H., Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Nakajima K., Sena G., Nawy T., Benfey P. N., Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307–311 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Li H., et al., The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell 17, 2157–2171 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao X., Huang H., Xu L., In situ detection of mature miRNAs in plants using LNA-modified DNA probes. Methods Mol. Biol. 883, 143–154 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truernit E., et al., High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20, 1494–1503 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ursache R., Andersen T. G., Marhavy P., Geldner N., A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 93, 399–412 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.