Significance
Understanding when Neanderthals disappeared is a hotly debated topic. When radiocarbon dating placed the Spy Neanderthals amongst the latest surviving in Northwest Europe, questions were raised regarding the reliability of the dates. Using a procedure more efficient in removing contamination and ancient genomic analysis, we show that previous dates produced on Neanderthal specimens from Spy are too young by up to 10,000 y. Our direct radiocarbon dates on the Neanderthals from Spy and those from Engis and Fonds-de-Forêt show a reduction of the uncertainty for the time window corresponding to Neanderthal disappearance in Northwest Europe. This population disappeared at 44,200 to 40,600 cal B.P. (at 95.4% probability). This is also earlier than previous suggestions based on dates on bulk collagen.
Keywords: Neanderthal disappearance, Belgium, compound-specific radiocarbon dating, ancient genomic analysis
Abstract
Elucidating when Neanderthal populations disappeared from Eurasia is a key question in paleoanthropology, and Belgium is one of the key regions for studying the Middle to Upper Paleolithic transition. Previous radiocarbon dating placed the Spy Neanderthals among the latest surviving Neanderthals in Northwest Europe with reported dates as young as 23,880 ± 240 B.P. (OxA-8912). Questions were raised, however, regarding the reliability of these dates. Soil contamination and carbon-based conservation products are known to cause problems during the radiocarbon dating of bulk collagen samples. Employing a compound-specific approach that is today the most efficient in removing contamination and ancient genomic analysis, we demonstrate here that previous dates produced on Neanderthal specimens from Spy were inaccurately young by up to 10,000 y due to the presence of unremoved contamination. Our compound-specific radiocarbon dates on the Neanderthals from Spy and those from Engis and Fonds-de-Forêt demonstrate that they disappeared from Northwest Europe at 44,200 to 40,600 cal B.P. (at 95.4% probability), much earlier than previously suggested. Our data contribute significantly to refining models for Neanderthal disappearance in Europe and, more broadly, show that chronometric models regarding the appearance or disappearance of animal or hominin groups should be based only on radiocarbon dates obtained using robust pretreatment methods.
Determining when Neanderthal populations disappeared from Eurasia and anatomically modern humans (AMH) arrived is a key question in paleoanthropology (1). Recent work at more than 40 archaeological sites has established that stone tool industries linked with the Neanderthals in Europe end by ∼41,000 to 39,000 calibrated years (cal) B.P. (at 95.4% probability) (2). Belgium is one of the key regions for studying the Middle to Upper Paleolithic transition. Chronological and stratigraphic evidence from several sites in this region has hinted at a gap of around 4,000 y (ca. 42 to 38 ky cal B.P.) between the Late Mousterian (Neanderthal) and the earliest dated Aurignacian (AMH) settlements (3, 4). Direct dating of Neanderthal remains, however, has produced a series of results that span this gap (5), suggesting a late-surviving Neanderthal population that could be associated with the Lincombian–Ranisian–Jerzmanowician (LRJ) (6), a so-called “transitional industry” (7). A reliable chronology is therefore crucial to test whether these Neanderthals survived as late as the dating evidence suggests. In this paper, we redate key Neanderthal specimens from Belgium using an advanced pretreatment method producing uncontaminated and reliable radiocarbon dates, allowing us to reevaluate the latest presence of Neanderthals in Northwest Europe.
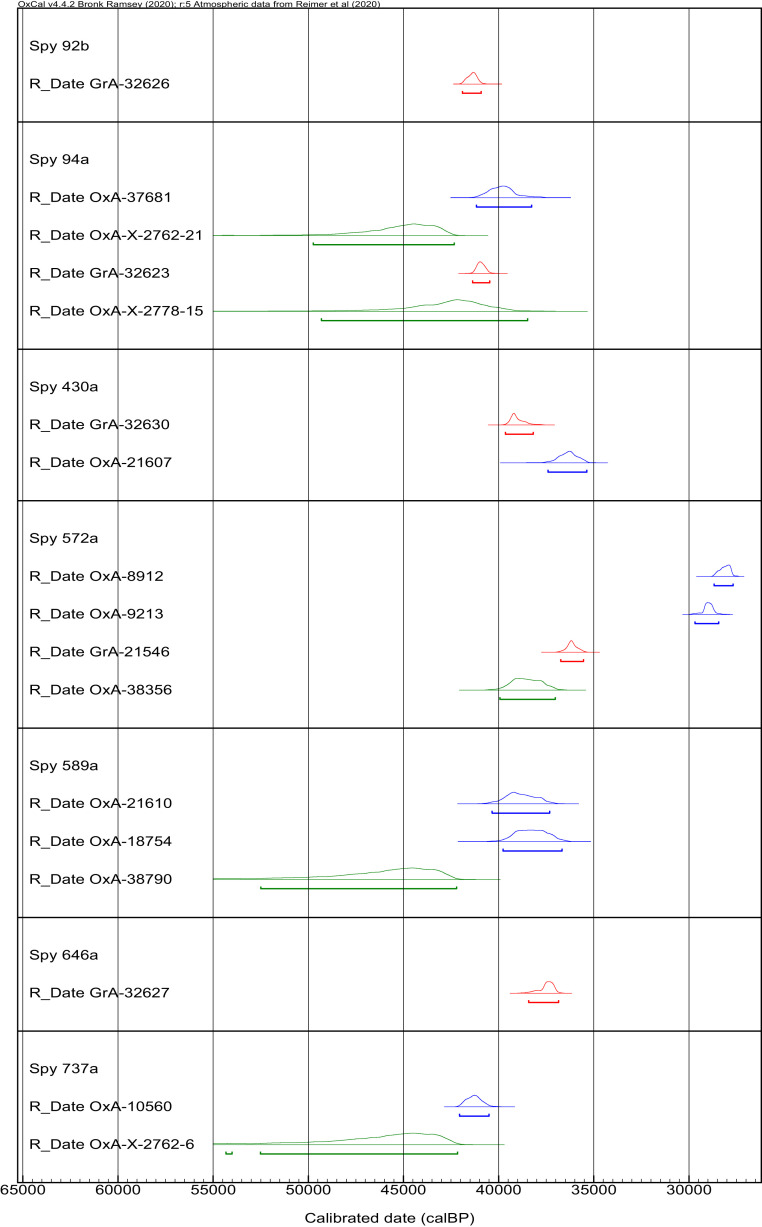
Neanderthal remains have been recovered from nine caves in Belgium: La Naulette Cave, Spy Cave, Première Caverne du Bay Bonnet (Fonds-de-Forêt), Schmerling Cave (Engis), Scladina Cave, Walou Cave, Troisième Caverne de Goyet, Trou de l’Abîme, and Trou Magrite (SI Appendix, Fig. S1 and Table S1). Among these, Spy Cave is of particular interest because of the large number of human remains that have come from this site (description of the site in SI Appendix). Recent work has focused on reassessing the collection of human and faunal material excavated from the site over the last century. The original collection (excavated from 1885 to 1886) comprised 89 hominin bone fragments related to two Neanderthal individuals. Further investigations have identified more than 1,800 hominin remains, including 24 associated with the Spy Neanderthals (Spy I, Spy II, and the newly identified juvenile Spy VI) (8, 9). Seven remains of these Neanderthals have previously been directly radiocarbon dated at the Oxford Radiocarbon Accelerator Unit (ORAU) and/or at the Centre for Isotope Research at the University of Groningen following the protocols as described in Brock et al. (10) and Dee et al. (11), respectively. Twelve accelerator mass spectrometry (AMS) dates have been obtained on collagen (SI Appendix, Table S2). They span a wide range from ∼42,000 to 28,000 cal B.P. (5, 6, 9, 12). The ages for some of these specimens, therefore, fall within and after the temporal gap believed to separate the Late Mousterian and the Early Upper Paleolithic in this region (Fig. 1). Some of these specimens, dated multiple times, produced different ages, and we observed that several of these radiocarbon dates have high C:N atomic ratios (6). Caution is therefore required, because of the high probability that the radiocarbon determinations were affected by unremoved contamination.
Fig. 1.
Calibrated age ranges of the Neanderthal specimen from Spy Cave. Dates obtained on collagen in Groningen are reported in red. Dates obtained on collagen and hydroxyproline at the ORAU are reported in blue and green, respectively. The calibrated ages were obtained using OxCal 4.5 (87) and the IntCal20 calibration curve (88).
In addition to the contamination from the sedimentary environment, ancient human bones are often treated and consolidated for better preservation, but such treatments were not systematically recorded. It is sometimes difficult to fully remove all the contamination using routine radiocarbon pretreatments, particularly when the contaminants are cross-linked to the collagen. We have optimized a more robust and chemically reliable method, which uses preparative liquid chromatography to isolate a single amino acid from the bone collagen for AMS dating (13, 14). The amino acid hydroxyproline (HYP) is the key target, since it is almost exclusively found in mammalian collagen. Extracting and dating HYP provides a level of reliability unmatched by the other methods. When redating Paleolithic sites using this so-called “compound-specific radiocarbon analysis” (CSRA) approach, we have observed that many of the previous radiocarbon dates obtained after less robust pretreatments are inaccurate. This can in turn lead to erroneous interpretations of human and faunal dispersal and rates of change in the archaeological, climatic, and evolutionary record (15–21).
Results
Four of the Neanderthal remains from the Spy collection were redated using the CSRA method, and five dates were obtained (Figs. 1 and 2 and SI Appendix, Table S2). The collagen of the vertebra fragment (Spy 737a) was previously dated at 36,250 ± 500 B.P. (OxA-10560). Our HYP date is 41,600 ± 2,400 B.P. (OxA-X-2762-6). Although the age uncertainty is larger, this date is significantly older than the previous one. This specimen was found outside of the cave and was never preserved with conservation treatments. Specimens Spy 589a and Spy 94a were discovered in the faunal material during the reassessment of the collection (22). Like Spy 737a, they have not been treated with any conservation material. Spy 589a was first dated to 33,950 ± 550 B.P. (OxA-21610) and 33,450 ± 600 B.P. (OxA-18754). The HYP measurement on the same bone produced an age of 41,700 ± 2,300 B.P. (OxA-38790). We obtained two dates for specimen Spy 94a: one on the third upper molar and one on the maxilla attached to the molar. The maxilla fragment was previously dated in Groningen and produced an age of 35,810 +260/−240 B.P. (GrA-32623) (6). We obtained a HYP date that is statistically identical by virtue of its high measurement uncertainty: 37,700 ± 2,200 B.P. (OxA-X-2778-15). The CSRA date obtained on the tooth (41,500 ± 1,800 B.P. [OxA-X-2762-21]) is, however, significantly older than the date that was previously obtained on the collagen extracted from the same tooth (34,650 ± 600 B.P. [OxA-37681]). The two HYP dates obtained on the tooth (OxA-X-2762-21) and the maxilla (OxA-X-2778-15) are statistically identical. The fourth reanalyzed specimen from Spy Cave is the scapula (Spy 572a) from the original collection of human remains. It was previously dated twice in Oxford, producing ages of 23,880 ± 240 B.P. (OxA-8912) and 24,730 ± 240 B.P. (OxA-9213). It was also dated at 31,810 ± 250 B.P. (GrA-21546) in Groningen. These three dates appear extremely young for Neanderthals, so unremoved contamination was suspected (2, 6). We redated the same scapula using our CSRA approach and obtained an age of 33,700 ± 550 B.P. (OxA-38356). This date is older than all previous ones obtained on this specimen, but it still appears surprisingly young compared with other dates obtained on Neanderthal specimens in Belgium and more generally in western Europe. We suspected that collagen-containing animal glue might have been used to consolidate the bone, since this preservation method was common for bones excavated in the 19th century (23, 24). The chemical composition of animal glue would be identical to endogenous collagen and therefore remain undetected during the radiocarbon dating. To explore this, we extracted DNA from Spy 572a (25), constructed libraries (26), and performed hybridization capture (27) to test for the presence of ancient hominin and other mammalian mitochondrial DNA (mtDNA). Even though we did not find evidence for the preservation of ancient hominin DNA (SI Appendix, Tables S4–S8), we found that between 9.7 and 12.8% of the isolated mtDNA fragments from Spy 572a (between 158,384 and 240,213, SI Appendix, Table S6) originated from Bovidae (28), whereas the remaining ones were assigned to modern human. The frequency of deamination-induced cytosine (C) to thymine (T) substitutions, which are typically elevated in authentic ancient DNA (29), were very low at the ends of both the human and bovid fragments (consistently less than 2.7%, SI Appendix, Tables S5 and S7). Moreover, we did not detect DNA from Bovidae in the associated extraction and library negative controls (SI Appendix, Table S6). These results indicate that the Spy 572a scapula is heavily contaminated with bovid DNA of relatively young age, pointing to preservation with glue prepared from cattle bones. We therefore conclude that the relatively young radiocarbon age of Spy 572a is an artifact of contamination with collagen from cattle, which cannot be separated from endogenous collagen prior to the radiocarbon measurements. This implies that OxA-38256 is an underestimate of the real age of the specimen and should be set to one side.
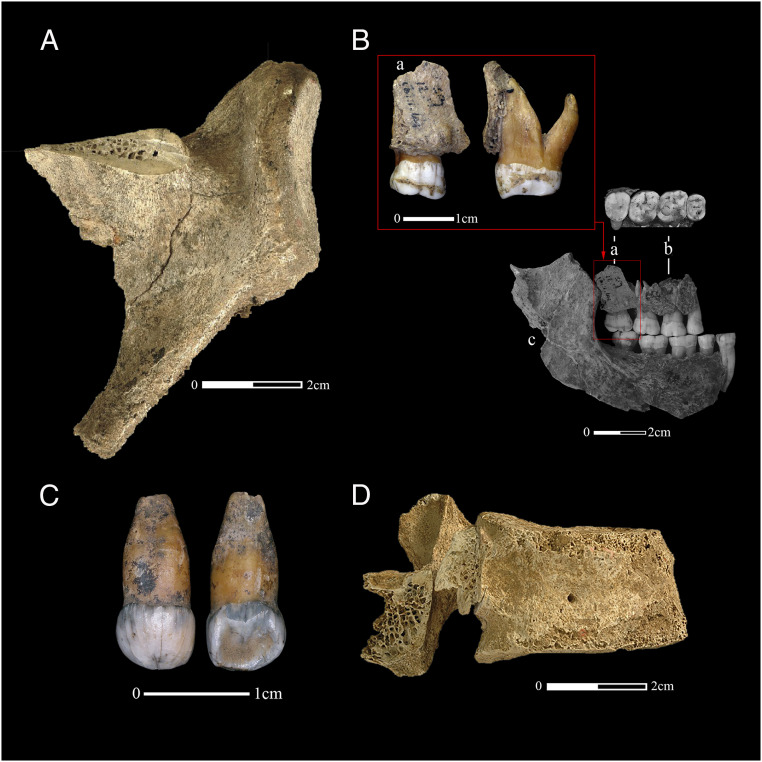
Fig. 2.
The four Neanderthal specimens from Spy Cave radiocarbon dated in this study: (A) right scapula (Spy 572a, https://www.morphosource.org/concern/biological_specimens/000S32268); (B, a) maxillary fragment and upper third molar (Spy 94a) associated with a maxillary fragment (B, b) (Spy 11a) and a mandibular fragment (B, c) (Spy 12a); (C) a first upper right deciduous incisor (Spy 589a); and (D) the lumbar vertebra (Spy 737a). Image by G. Abrams after pictures from M. Toussaint (A, D) and E. De Wamme (B, C; Copyright RBINS).
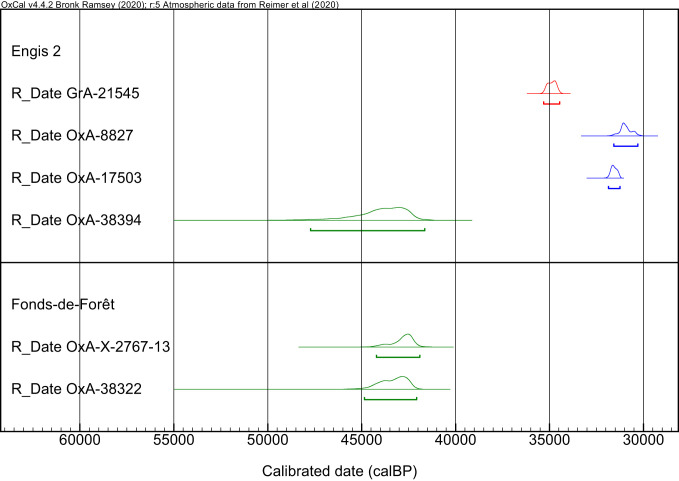
We also dated a Neanderthal lower right deciduous (or milk) second molar (LRdm2) from Engis (Fig. 3B) and the adult Neanderthal femur from Fonds-de-Forêt (Fig. 3C) using the CSRA approach (see SI Appendix for site information). We dated the tooth to 39,900 ± 1,700 B.P. (OxA-38394). This is significantly older than the dates previously obtained on the parietal bone from the same specimen: 26,820 ± 340 B.P. (OxA-8827), 27,670 ± 140 B.P. (OxA-17503), and 30,460 ± 210 B.P. (GrA-21545) (Fig. 4 and SI Appendix, Table S3). Since the femur from Fonds-de-Forêt has a very shiny surface (Fig. 3), we investigated the presence of conservation materials. Several analytical techniques can be applied to explore whether contaminants are present in bone specimens, such as Fourier transform infrared (FTIR) spectroscopy (30, 31) and pyrolysis combined with gas chromatography and mass spectrometry (Py-GC/MS) (21, 32). FTIR is rapid, but the detection of residual contamination is constrained given the detection limits of the method. Pyrolysis, however, provides information at a molecular level and can therefore be used to identify trace contaminants from the burial environment and/or derived from the conservation of the specimens. Py-GC/MS analyses revealed the presence of paraffin characterized by a series of alkanes as well as a copolymer of methyl methacrylate and butyl methacrylate (SI Appendix, Fig. S2). This shows two different and hitherto undocumented events for the conservation of the specimen. Such heavily contaminated bones can sometimes be difficult to clean with routine radiocarbon pretreatments (31, 33). The sample was prepared twice with our CSRA method and produced two statistically identical dates: 38,800 ± 900 B.P. (OxA-X-2767-13) and 39,500 ± 1,100 B.P. (OxA-38322) (Fig. 4 and SI Appendix, Table S3). The specimens from Engis and Fonds-de-Forêt both produced C:N ratios within the acceptable range for the hydroxyproline molecule, indicating that any exogenous carbon from soil contamination and conservation materials had been removed before AMS measurement. In contrast to Spy 572a, DNA analysis of the Fonds-de-Forêt femur yielded more than 152,069 hominin mtDNA fragments (SI Appendix, Table S4), 94.66% of which could be assigned to Neanderthals (SI Appendix, Table S8), but no DNA from animals (SI Appendix, Table S6). Thus, we found no indication for the presence of bone glue, which could confound compound-specific radiocarbon dating. Moreover, we reconstructed a complete mtDNA genome from the Fonds-de-Forêt femur based on 135-fold coverage in deaminated DNA fragments. This genome falls within the variation of late Neanderthals (SI Appendix, Fig. S3) and differs by only a single base pair (bp) from the mitochondrial genomes of Goyet Q57-1, Feldhofer 1, and Vindija 33.25.
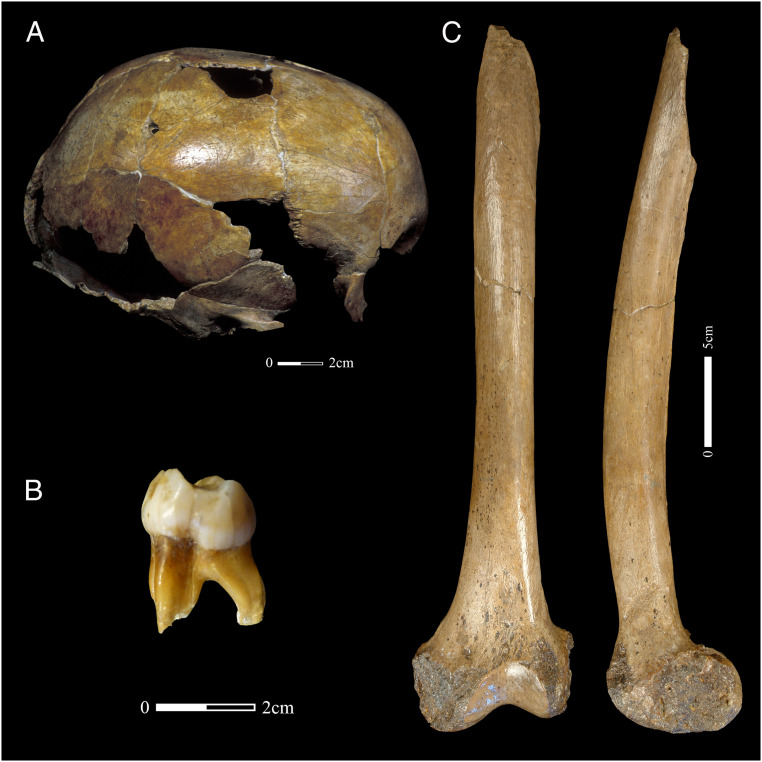
Fig. 3.
The Neanderthal Child Engis 2. (A) Skullcap and (B) second lower right deciduous molar (LRdm2) radiocarbon dated in this study. Image by G. Abrams after pictures from G. Focant (A, Copyright SPW) and A. Le Cabec (B). (C) The Neanderthal left femur from Fonds-de-Forêt radiocarbon dated in this study. Image by G. Abrams modified after pictures from E. De Wamme (Copyright RBINS).
Fig. 4.
Calibrated age ranges of the Neanderthal specimens from Engis and Fonds-de-Forêt caves. The calibrated ages were obtained using OxCal 4.5 (87) and the IntCal20 calibration curve (88). Dates obtained on collagen in Groningen are reported in red. Dates obtained on collagen and hydroxyproline at the ORAU are reported in blue and green, respectively.
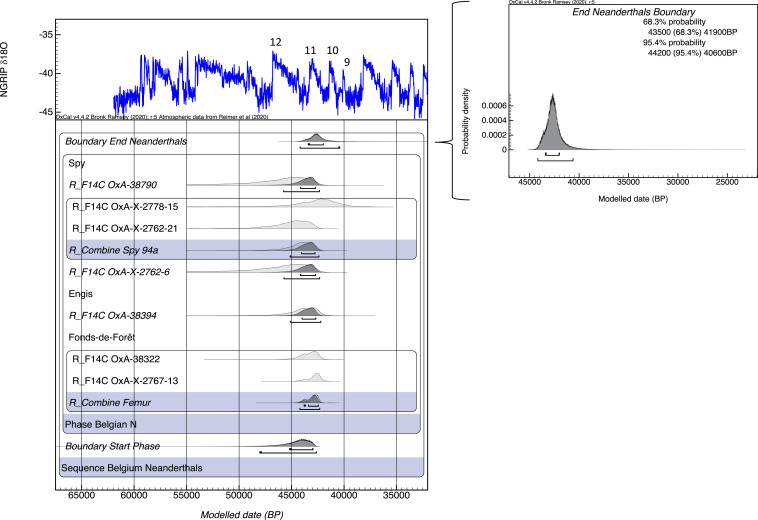
We then used the CSRA dates to build a Bayesian model to assess probabilistically the latest Neanderthal occupation in Belgium. We built a simple single-phase model using OxCal 4.5 and the IntCal20 calibration curve to generate a probability density function (PDF) for the end boundary of the HYP determinations from Spy, Engis, and Fonds-de-Forêt Neanderthals (Fig. 5). We did not include the other six Belgian sites that produced Neanderthal specimens in the PDF because they were obviously too old, or they have not yet been directly dated, or the dates were not produced using CSRA pretreatments (see additional text in SI Appendix). By comparing this end boundary for Belgian Neanderthals with the previous Spy end boundary (2), we observe a substantial shift of the PDF toward earlier ages: 44,200 to 40,600 cal B.P. (at 95.4% probability) instead of 41,210 to 37,830 cal B.P. (at 95.4% probability).
Fig. 5.
Bayesian age model for Spy, Engis, and Fonds-de-Forêt caves using compound-specific determinations. The data are modeled in a single-phase model and compared tentatively against the North Greenland Ice Core Project dataset from Greenland (89). Numbers indicate interstadials in Greenland.
Discussion
Our dating shows that Neanderthals disappeared from Northwest Europe much earlier than previously suggested, at 44,200 to 40,600 cal B.P. (at 95.4% probability). The potential causes of this extinction could be linked to climate change, competition, and inbreeding (34), but these are beyond the scope of this article.
It was hypothesized that the Spy Neanderthals, with three dates around 36,000 B.P., may be associated with the LRJ, a transitional technocomplex defined in northern Europe, and also discovered at this site (6, 35). This association is now less likely (at Spy Cave) because the late Neanderthal dates have been proven unreliable. It will, however, be necessary to redate the LRJ using the same CSRA approach in order to be able to discern whether Neanderthals or AMH produced the LRJ. In other regions, there is evidence that Neanderthals survived later than they did in Belgium. The Châtelperronian transitional industry from France and northern Spain, which has been linked with Neanderthals, for example, is estimated to end at 41,200 to 38,300 cal B.P. (2). Similarly, the Peștera cu Oase AMH in Romania, dated to 41,000 to 39,000 cal B.P., has recent Neanderthal introgression four to six generations prior to him (36). The earliest evidence for AMH in Europe predates 40,000 cal B.P. significantly ∼45,000 cal B.P. at the Italian site of Grotta del Cavallo (37), and perhaps as early as 47,000 cal B.P. at Bacho Kiro, Bulgaria (38).
At present, a long period of overlap with AMH seems apparent. The possibility, however, remains that, because of the pervasive and influential effects of contamination on the reliability of bone dates in this period, underestimated ages are still present in the wider corpus of dates we have available to interpret. Next to sedimentary contamination, human remains are particularly prone to contamination from conservation materials, and therefore we recommend that all Paleolithic human remains should be (re)dated using CSRA approaches combined with DNA analyses to guarantee reliable results, as demonstrated here with Spy. Chronometric models regarding the appearance or disappearance of hominin groups should be based only on dates obtained using robust pretreatment methods. The crucial dates from Oase, for example, which reflect a terminus ante quem for Neanderthals in Europe based on the dates and genetic evidence, were not produced using a CSRA approach. There is a possibility that they are a minimum age. These, and other outlying dates, need to be tested first before we can be wholly confident in the wider picture of the latest Neanderthals and their spatiotemporal distribution.
Materials and Methods
Materials.
Spy Cave.
In the current study, four specimens were dated and are described below (Fig. 2). They are listed after the nomenclature proposed by H. Rougier and colleagues during the reassessment of the collections (8, 22).
Spy 572a.
This fragment of a right scapula (Spy 572a, Fig. 2A) belongs to the original collection and was mentioned in the first publication of the human remains and was associated with the most complete individual (39) and should now be associated with Spy II (22). The scapula presents a morphology compatible with Neanderthals (40), although part of its morphological traits can be found in a minority of human remains from the Upper Paleolithic. However, none of Spy's successive excavators have discovered human remains associated with lithic tools from the Upper Paleolithic, and no radiocarbon date of human bones support such attribution (41). This specimen is part of the collections of the Department of Geology of the University of Liège. Analyses presented in this article revealed that the scapula was treated with a consolidant made of animal-derived collagen, as is probably the case with the other remains from the original collection.
Spy 94a.
During the reassessment of the collections, 24 new Neanderthal remains were discovered among the faunal material, mainly in the collections of the Royal Belgian Institute of Natural Sciences (RBINS) gathered during F. Twiesselmann’s excavations in the 1950s (42). These excavations were carried out on the slope leading from the cave to the Orneau River, which consisted mainly of an accumulation of backfill from earlier excavations (43). From this collection, Spy 94a and Spy 589a (see below) were dated in this study. Spy 94a (Fig. 2 B, a) is an upper right third molar still attached to a small alveolar fragment of maxilla that refits with two fragments of the original collection: Spy 11a (a fragment of maxilla; Fig. 2 B, b) and Spy 12a (a fragment of mandible; Fig. 2 B, c) which were attributed first to the second individual and specifically mentioned as missing in the publication by Fraipont and Lohest (39). These remains are now attributed to Spy I (5).
Spy 589a.
Spy 589a (Fig. 2C) belongs to the same collection as Spy 94a described above. It is a deciduous upper first right incisor, which, together with three other deciduous teeth (Spy 645a, Spy 592a, and Spy 594a) and a fragment of mandible (Spy 646a), is attributed to Spy VI, a third Neanderthal (immature) individual (9).
Spy 737a.
A fragmentary Neanderthal lumbar vertebra (Spy 737a, Fig. 2D) was discovered in the early 2000s on the slope below Spy Cave (12). The body of the vertebra is almost complete, and the vertebral arch is well preserved. However, both transverse processes are missing, and the spinous process is very incomplete except for a small part of its root. So far, it was not possible to attribute it to a specific individual, and even the Neanderthal attribution is made difficult by the absence of derived features. The vertebra was partially destroyed to produce the ultrafiltration date OxA-10560 (12), which was compatible with other dates made on Spy Neanderthals (6). The remaining vertebral fragment is stored in the private collection of Ph. Pirson, who found the specimen.
Even though these four specimens originate from the same site, they were discovered separately at different times. They have different taphonomic histories that can affect the collagen preservation and, therefore, the quality and reliability of the results. With the exception of the scapula fragment (Spy 572a) from the original collection (also known as De Puydt and Lohest collection), all the other remains dated here were found among the faunal material during the collection reassessment (Spy 94a and Spy 589a) or discovered by chance on the slope in front of the site (Spy 737a). The recently discovered human remains that refit with the original collection are of particular interest, since they are untreated, which helps with analyses such as radiocarbon dating and DNA analyses (6).
Schmerling Cave.
The Engis 2 individual has been stored in the collections of the Department of Geology of the University of Liège since its discovery during the winter 1829 to 1830. It is composed of an almost complete calotte (skullcap, Fig. 3A), isolated teeth and a fragmentary maxilla of an infant, which is illustrated in the original publication (44). The remains of Engis 2 were located at the bottom of the cave, near an elephantidae tooth (45). The skull fell apart when Schmerling removed it from the sediments (45). However, Engis 2 is quite well preserved considering its young age [ca. 3 y old at the time of death (46)]. The calotte was first described in detail and attributed to Neanderthals by C. Fraipont, more than a century after its discovery (47), primarily using La Quina H18 as comparison. This interpretation, as well as the attribution of the isolated teeth, were later confirmed by A. M. Tillier, who had access to a larger number of comparative individuals (48). The Neanderthal attribution was also confirmed by genetic analyses (49).
The calotte was previously dated to 26,820 ± 340 B.P. (OxA-8827) and 30,460 ± 210 B.P. (GrA-21545) (12), but these dates were interpreted as too recent considering its Neanderthal attribution.
There is a real possibility, considering the antiquity of the discovery, the fragmentation that occurred during the excavation, and the shiny surface of the bone that the calotte was treated using consolidants made of animal-derived collagen. Moreover, the high porosity and thinness of these bones make the extraction of uncontaminated material difficult. To avoid the risk of extracting mixed collagen (Neanderthals and modern animals), which may lead to an erroneous date, it was decided that the second lower right deciduous molar (LRdm2), named Engis2_N°4, would be used for this study (Fig. 3B). This molar was still complete and is associated with the Neanderthal calotte (48).
Première Caverne du Bay Bonnet (Fonds-de-Forêt).
Two human bones were recovered from the Première Caverne du Bay Bonnet (Fonds-de-Forêt) by F. Tihon in 1895 (50): an upper molar, which is now lost (51), and an incomplete left femur missing its proximal part (Fig. 3C). Based on its similarities to the femora from Spy, F. Tihon attributed the femur to Neanderthals. This interpretation was later confirmed by F. Twiesselmann, who was the first to study the adult femur in detail (52). Tihon also proposed that the femur was broken intentionally, raising the question of cannibalism (50). However, the breakage pattern of the proximal part, testifying that it occurred when the bone was still fresh, in combination with tooth marks on the distal extremity suggest carnivore activity rather than cannibalism (53). The femur was part of the collection of the Royal Museums of Art and History before its permanent deposition in the collections of the RBINS. The shiny surface of the bone is suggestive of the presence of some conservation treatment, as is usually the case for specimens excavated in the 19th century. The femur was then sampled and sequenced for the presence of exogenous mtDNA of nonhuman origin and analyzed by pyrolysis to discern the presence of consolidants before being radiocarbon dated. These methods are described below, and the results discussed in the article.
Methods
Radiocarbon Dating.
We (re)dated the Spy, Engis, and Fonds-de-Forêt Neanderthal specimens using the single amino acid radiocarbon dating method optimized at the ORAU (13). This method involves separation of the underivatized amino acids from hydrolyzed bone or tooth collagen samples using preparative Liquid Chromatography (Prep-LC). The HYP is isolated, combusted, graphitized, and AMS dated. This pretreatment approach (Coded “HYP” in the ORAU) is the most efficient technique to remove contaminants, including, but not limited to, conservation materials (unless collagen-based glue has been applied). The percent of C, percent of N, and atomic C:N ratio were measured using an automated carbon and nitrogen elemental analyzer (Carlo Erba EA1108) coupled with a continuous-flow isotope monitoring mass spectrometer (Europa Geo 20/20).
Py-GC/MS.
The Py-GC/MS analysis was performed on 450 µg of bone powder from the Fonds-de-Forêt femur and 250µg of an uncontaminated mammoth bone sample from Latton (UK), which is used as a background standard at the ORAU. The samples were successively placed in a stainless-steel cup and inserted in the microfurnace, where they were pyrolysed at 600 °C for 20 s. The analyses were performed on an EGA/PY-3030D Micro Furnace Pyrolyzer (Frontier Lab, Japan) connected to a 6890 gas chromatograph equipped with a split/splitless injector. An HP 5MS column (30 m × 0.25 mm, film thickness 0.25 µm, Agilent Technologies) coupled with a deactivated silica precolumn (2 m × 0.32 mm, Agilent Technologies, USA) was used for the chromatographic separation. The program of the oven temperature used for the chromatographic separation was 40 °C isothermal for 6 min followed by a linear ramp at 20 °C/min up to 310 °C. Analyses were performed under a constant flow of helium at 1 mL/min, with a split ratio 1:20. The gas chromatograph was coupled with a 5973 Mass spectrometer (MS) (Agilent Technologies, USA). The temperature of the transfer line to the MS was 300 °C. The MS was operated in electron ionization positive mode (70 eV, scanning m/z 50 to 600). The MS ion source was kept at 230 °C and the MS quadrupole at 150 °C.
DNA Analyses.
Sampling.
We removed powder samples from the Spy scapula (Spy 572a) and the femur from Fonds-de-Forêt in a clean room facility at the RBINS in Brussels using a sterile dentistry drill. We removed a thin layer of surface amounting to 15.2 mg (sample A) from an area of Spy 572a that was adjacent to where a sample was previously taken for HYP dating. An additional sample of 24.3 mg (sample B) of bone powder was taken by drilling into the cortical portion of the scapula directly underneath. A second thin layer of surface amounting to 14.7 mg (sample C) was removed from close to sampling area B. Sampling of the Fonds-de-Forêt femur was performed by extending the hole from which a sample for hydroxyproline dating had previously been taken, amounting to 26.7 mg of bone powder.
DNA extraction, library preparation, and mtDNA capture.
All subsequent steps were performed at the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany. DNA was extracted on an automated liquid handling platform (Bravo NGS workstation, Agilent Technologies) and as described in ref. 25. DNA extracts were converted into single-stranded DNA libraries (26) and the efficiency of library preparation was assessed using a control spike-in oligonucleotide and qPCR assays as described in (54). Libraries were amplified (55, 56) and labeled with two unique indices (57). An aliquot of each amplified library was further enriched for human mtDNA using a probe set designed based on the revised Cambridge Reference Sequence (rCRS, GenBank accession NC_01290) of the human mtDNA genome synthesized on an oligonucleotide array (27) and two consecutive rounds of on-bead hybridization capture (58). Extraction and library negative controls were carried alongside the samples throughout all steps. Enriched libraries were pooled and sequenced on an Illumina Miseq in a double index configuration (2 × 75 cycles) (57). Base calling was done using Bustard (Illumina).
Sequencing, data processing, and ancient DNA damage profiling.
Adapters were trimmed and overlapping paired-end reads were merged into single sequences using leeHom (59). We used Burrows–Wheeler Aligner (60) with ancient DNA (aDNA) parameters [“-n 0.01 -o 2 -l 16500” (61)] to align sequences to the rCRS. Only sequences with perfect matches to the expected index combinations were retained for subsequent analyses and PCR duplicates were removed using bam-rmdup (https://github.com/mpieva/biohazard-tools). Sequences shorter than 35 bp and with a mapping quality lower than 25 (MQ < 25) were discarded from all downstream analyses.
To investigate the presence of nonhominin mammalian DNA, we additionally aligned sequences to a set of 242 mammalian mtDNA genomes (62) and processed the alignments with a pipeline originally developed to assign mtDNA sequences from sediment samples to mammalian families (63). In brief, this pipeline uses Basic Local Alignment Search Tool (BLAST) (64) for realignments of sequences to a database of mammalian mtDNA genomes and the “lowest common ancestor” algorithm implemented in Metagenome Analyzer (MEGAN) for taxonomic assignments (65). Sequences were then evaluated separately by the family they were assigned to for the presence of elevated frequencies of terminal C to T substitutions at their ends to determine if they derived from authentic aDNA (29).
Phylogenetic inferences.
To distinguish between Neanderthal, modern human, Denisovan, or Sima de los Huesos mtDNA being present in Spy 572a and the femur from Fonds-de-Forêt, we investigated the state of DNA fragments overlapping phylogenetically informative (or “diagnostic”) positions (i.e., positions where the mitochondrial genomes of one of these groups differ from those of all others) (66).
We reconstructed the mitochondrial genome of the femur from Fonds-de-Forêt using mtDNA fragments longer than 35 bp that produced alignments with a mapping quality of at least 25 and that showed a C-to-T difference to the reference genome within the first and/or last three positions [deaminated fragments (67)]. We called a consensus base at each position of the mtDNA that was covered by at least three DNA fragments showing a base quality score of 20 or higher and in which at least two thirds of fragments carried an identical base (67). We converted Ts appearing in the first and last three positions of each DNA fragment in the orientation as sequenced into Ns to hinder deamination-induced substitutions affecting the calling of a consensus base.
The reconstructed mtDNA genome of the femur from Fonds-de-Forêt was aligned to the mtDNA genomes of 54 present-day humans (68), 24 Neanderthals (69–79), 4 Denisovans (28, 80, 81), a Sima de los Huesos individual (67), and a chimpanzee (82) using multiple alignment using fast Fourier transform (MAFFT) [version 7.271 (83)]. We used jModelTest2 (84) to determine the best-fitting substitution model (TrN+I+G) and generated a maximum likelihood tree using PhyML (85) with 200 bootstrap replications. All positions containing gaps and missing data were excluded from this analysis. The number of pairwise differences among the genomes was calculated using Molecular Evolutionary Genetics Analysis (MEGA) [version 7] (86).
Supplementary Material
Acknowledgments
We thank Svante Pääbo (Max Planck Institute for Evolutionary Anthropology, Germany), Michael Dee (University of Groningen, Netherlands), Erika Ribechini (University of Pisa, Italy), Marco Matonai (University of Pisa, Italy), Elena Essel (Max Planck Institute for Evolutionary Anthropology, Germany), Sarah Nagel (Max Planck Institute for Evolutionary Anthropology, Germany), Julia Richter (Max Planck Institute for Evolutionary Anthropology, Germany), and Dominique Bonjean (Scladina Cave Archaeological Centre) for their support and helpful discussions. Funding was provided by the European Research Council through Grant 324139 (PalaeoChron) to T.H. It was also provided by a research mobility grant from the Royal Society of Chemistry (Grant Agreement RM1802-6433) and a research fellowship from the University of Pisa awarded to T.D. We would like to acknowledge support from the Max Planck Society and the UK Natural Environment Research Council (NERC) for the Oxford node of the national NERC Radiocarbon facility.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022466118/-/DCSupplemental.
Data Availability
All the radiocarbon data generated at the ORAU are archived internally and are also available on the laboratory’s website, along with a link to the paper. The mitochondrial genome from Fonds-de-Forêt is deposited in GenBank with the accession number PRJEB39136. All other study data are included in the article and/or SI Appendix.
References
- 1.Hublin J.-J., The last Neanderthal. Proc. Natl. Acad. Sci. U.S.A. 114, 10520–10522 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Higham T., et al., The timing and spatiotemporal patterning of Neanderthal disappearance. Nature 512, 306–309 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Di Modica K., Toussaint M., Abrams G., Pirson S., The Middle Palaeolithic from Belgium: Chronostratigraphy, territorial management and culture on a mosaic of contrasting environments. Quat. Int. 411, 77–106 (2016). [Google Scholar]
- 4.Pirson S., et al., Chronostratigraphic context of the Middle to Upper Palaeolithic transition: Recent data from Belgium. Quat. Int. 259, 78–94 (2012). [Google Scholar]
- 5.Semal P., et al., “Radiocarbon dating of human remains and associated archaeological material” in Spy Cave. 125 Years of Multidisciplinary Research at the Betche aux Rotches (Jemeppe-sur-Sambre, Province of Namur, Belgium), Rougier H., Semal P., Eds. (Bull. Soc. Roy. Belge Anthrop. Préhist., 2013), vol. 123/2012, pp. 331–356. [Google Scholar]
- 6.Semal P., et al., New data on the late Neandertals: Direct dating of the Belgian Spy fossils. Am. J. Phys. Anthropol. 138, 421–428 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Flas D., The Middle to Upper Paleolithic transition in Northern Europe: The Lincombian-Ranisian-Jerzmanowician and the issue of acculturation of the last Neanderthals. World Archaeol. 43, 605–627 (2011). [Google Scholar]
- 8.Rougier H., et al., Collections de la Grotte de Spy: (re)découvertes et inventaire anthropologique. Notae Praehistoricae 24, 181–190 (2004). [Google Scholar]
- 9.Crevecoeur I., et al., The Spy VI child: A newly discovered Neandertal infant. J. Hum. Evol. 59, 641–656 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Brock F., Higham T., Ditchfield P., Bronk Ramsey C., Current pretreatment methods for AMS radiocarbon dating at the Oxford radiocarbon accelerator unit (ORAU). Radiocarbon 52, 103–112 (2010). [Google Scholar]
- 11.Dee M. W., et al., Radiocarbon dating at Groningen: New and updated chemical pretreatment procedures. Radiocarbon 62, 63–74 (2020). [Google Scholar]
- 12.Toussaint M., Pirson S., Neandertal studies in Belgium: 2000–2005. Period. Biol. 108, 373–387 (2006). [Google Scholar]
- 13.Devièse T., Comeskey D., McCullagh J., Bronk Ramsey C., Higham T., New protocol for compound-specific radiocarbon analysis of archaeological bones. Rapid Commun. Mass Spectrom. 32, 373–379 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Tripp J. A., Devièse T., McCullagh J. S. O., Preparative HPLC separation of underivatized amino acids for isotopic analysis. Methods Mol. Biol. 2030, 69–83 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Devièse T., et al., Direct dating of Neanderthal remains from the site of Vindija cave and implications for the Middle to Upper Paleolithic transition. Proc. Natl. Acad. Sci. U.S.A. 114, 10606–10611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourrillon R., et al., A new Aurignacian engraving from Abri Blanchard, France: Implications for understanding Aurignacian graphic expression in Western and Central Europe. Quat. Int. 491, 46–64 (2018). [Google Scholar]
- 17.Becerra-Valdivia L., et al., Reassessing the chronology of the archaeological site of Anzick. Proc. Natl. Acad. Sci. U.S.A. 115, 7000–7003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devièse T., et al., Increasing accuracy for the radiocarbon dating of sites occupied by the first Americans. Quat. Sci. Rev. 198, 171–180 (2018). [Google Scholar]
- 19.Dinnis R., et al., New data for the early Upper Paleolithic of Kostenki (Russia). J. Hum. Evol. 127, 21–40 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Kosintsev P., et al., Evolution and extinction of the giant rhinoceros Elasmotherium sibiricum sheds light on late Quaternary megafaunal extinctions. Nat. Ecol. Evol. 3, 31–38 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Devièse T., et al., Compound-specific radiocarbon dating and mitochondrial DNA analysis of the Pleistocene hominin from Salkhit Mongolia. Nat. Commun. 10, 274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rougier H., Semal P., Eds., Spy Cave: 125 Years of Multidisciplinary Research at the Betche Aux Rotches (Jemeppe-sur-Sambre, Province of Namur, Belgium), (Bull. Soc. Roy. Belge Anthrop. Préhist., Brussels, 2013), vol. 123/2012. [Google Scholar]
- 23.Schellmann N. C., Animal glues: A review of their key properties relevant to conservation. Stud. Conserv. 52, 55–66 (2007). [Google Scholar]
- 24.Masschelein-Kleiner L., Ancient Binding Media, Varnishes and Adhesives (A & J Servizi Grafici Editoriali, Rome, Italy, ed. 2, 1995). [Google Scholar]
- 25.Rohland N., Glocke I., Aximu-Petri A., Meyer M., Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing. Nat. Protoc. 13, 2447–2461 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Gansauge M.-T., Aximu-Petri A., Nagel S., Meyer M., Manual and automated preparation of single-stranded DNA libraries for the sequencing of DNA from ancient biological remains and other sources of highly degraded DNA. Nat. Protoc. 15, 2279–2300 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Fu Q., et al., DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl. Acad. Sci. U.S.A. 110, 2223–2227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slon V., et al., A fourth Denisovan individual. Sci. Adv. 3, e1700186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briggs A. W., et al., Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. U.S.A. 104, 14616–14621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gianfrate G., et al., Qualitative application based on IR spectroscopy for bone sample quality control in radiocarbon dating. Nucl. Instrum. Methods Phys. Res. B 259, 316–319 (2007). [Google Scholar]
- 31.Brock F., et al., Testing the effectiveness of protocols for removal of common conservation treatments for radiocarbon dating. Radiocarbon 60, 35–50 (2018). [Google Scholar]
- 32.Devièse T., Ribechini E., Querci D., Higham T., Assessing the efficiency of supercritical fluid extraction for the decontamination of archaeological bones prior to radiocarbon dating. Analyst 144, 6128–6135 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Higham T. F. G., Removing contaminants: A restatement of the value of isolating single compounds for AMS dating. Antiquity 93, 1072–1075 (2019). [Google Scholar]
- 34.Timmermann A., Quantifying the potential causes of Neanderthal extinction: Abrupt climate change versus competition and interbreeding. Quat. Sci. Rev. 238, 106331 (2020). [Google Scholar]
- 35.Flas D., “Jerzmanowice points from Spy and the issue of the Lincombian-Ranisian-Jerzmanowician” in Spy Cave. 125 Years of Multidisciplinary Research at the Betche aux Rotches (Jemeppe-sur-Sambre, Province of Namur, Belgium), Rougier H., Semal P., Eds. (Bull. Soc. Roy. Belge Anthrop. Préhist., 2013), vol. 123/2012, pp. 217–230. [Google Scholar]
- 36.Fu Q., et al., An early modern human from Romania with a recent Neanderthal ancestor. Nature 524, 216–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benazzi S., et al., Early dispersal of modern humans in Europe and implications for Neanderthal behaviour. Nature 479, 525–528 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Hublin J.-J., et al., Initial Upper Palaeolithic Homo sapiens from Bacho Kiro cave, Bulgaria. Nature 581, 299–302 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Fraipont J., Lohest M., La race humaine de Néanderthal ou de Canstadt en Belgique: Recherches ethnographiques sur des ossements humains découverts dans les dépôts quaternaires d’une grotte à Spy et détermination de leur âge géologique. Arch. Biol. (Liege) 7, 586–757 (1887). [Google Scholar]
- 40.Trinkaus E., Modern human versus Neandertal evolutionary distinctiveness. Curr. Anthropol. 47, 597–620 (2006). [Google Scholar]
- 41.Trinkaus E., European early modern humans and the fate of the Neandertals. Proc. Natl. Acad. Sci. U.S.A. 104, 7367–7372 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Semal P., et al., “History of excavations, discoveries and collections” in Spy Cave. 125 Years of Multidisciplinary Research at the Betche aux Rotches (Jemeppe-sur-Sambre, Province of Namur, Belgium), Rougier H., Semal P., Eds. (Bull. Soc. Roy. Belge Anthrop. Préhist., 2013), vol. 123/2012, pp. 13–39. [Google Scholar]
- 43.Pirson S., et al., “The stratigraphy of Spy Cave. A review of the available lithostratigraphic and archaeostratigraphic information” in Spy Cave. 125 Years of Multidisciplinary Research at the Betche aux Rotches (Jemeppe-sur-Sambre, Province of Namur, Belgium), Rougier H., Semal P., Eds. (Bull. Soc. Roy. Belge Anthrop. Préhist., 2013), vol. 123/2012, pp. 91–131. [Google Scholar]
- 44.Schmerling P.-C., Recherches Sur les Ossemens Fossiles Découverts Dans les Cavernes de la Province de Liége (Collardin, Liège, 1834), vol. 2. [Google Scholar]
- 45.Schmerling P.-C., Recherches Sur les Ossemens Fossiles Découverts Dans les Cavernes de la Province de Liège (Collardin, Liège, 1833), vol. 1. [Google Scholar]
- 46.Smith T. M., et al., Dental evidence for ontogenetic differences between modern humans and Neanderthals. Proc. Natl. Acad. Sci. U.S.A. 107, 20923–20928 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fraipont C., Les Hommes Fossiles d’Engis (Archi. Inst. Paléonto. Hum, Paris, France, 1936), vol. 16. [Google Scholar]
- 48.Tillier A. M., Le crâne d’enfant d’Engis 2: Un exemple de distribution des caractères juvéniles, primitifs et néanderthaliens. Bull. Soc. Roy. Bel. Anthrop. Préhist 94, 51–75 (1983). [Google Scholar]
- 49.Serre D., et al., No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol. 2, E57 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tihon F., Les cavernes préhistoriques de la vallée de la Vesdre. Fouilles à Fond-de-Forêt (2ème article). Ann. Soci. Archéo. Bruxelles. 12, 145–173 (1898). [Google Scholar]
- 51.Toussaint M., Semal P., Pirson S., “Les néandertaliens du bassin mosan belge: Bilan 2006-2011” in Le Paléolithique Moyen en Belgique. Mélanges Marguerite Ulrix-Closset, Toussaint M., Di Modica K., Pirson S., Eds. (Etu. Rech. Archéo. Univ., Liège, Liège, 2011), vol. 128, pp. 149–196. [Google Scholar]
- 52.Twiesselmann F., Le Fémur Néanderthalien de Fond-de-Forêt (Province de Liège) (Mém. Inst. Roy. Sci. Nat. Bel, Brussels, 1961), vol. 148. [Google Scholar]
- 53.Camarós E., et al., Hunted or scavenged Neanderthals? Taphonomic approach to hominin fossils with carnivore damage. Int. J. Osteoarchaeol. 27, 606–620 (2017). [Google Scholar]
- 54.Glocke I., Meyer M., Extending the spectrum of DNA sequences retrieved from ancient bones and teeth. Genome Res. 27, 1230–1237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dabney J., Meyer M., Length and GC-biases during sequencing library amplification: A comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries. Biotechniques 52, 87–94 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Korlević P., et al., Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. Biotechniques 59, 87–93 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Kircher M., Sawyer S., Meyer M., Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maricic T., Whitten M., Pääbo S., Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One 5, e14004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Renaud G., Stenzel U., Kelso J., leeHom: Adaptor trimming and merging for Illumina sequencing reads. Nucleic Acids Res. 42, e141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyer M., et al., A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Slon V., et al., Mammalian mitochondrial capture, a tool for rapid screening of DNA preservation in faunal and undiagnostic remains, and its application to Middle Pleistocene specimens from Qesem Cave (Israel). Quat. Int. 398, 210–218 (2016). [Google Scholar]
- 63.Slon V., et al., Neandertal and denisovan DNA from Pleistocene sediments. Science 356, 605–608 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 65.Huson D. H., Auch A. F., Qi J., Schuster S. C., MEGAN analysis of metagenomic data. Genome Res. 17, 377–386 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meyer M., et al., Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature 531, 504–507 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Meyer M., et al., A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature 505, 403–406 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Green R. E., et al., A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell 134, 416–426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Briggs A. W., et al., Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science 325, 318–321 (2009). [DOI] [PubMed] [Google Scholar]
- 70.Gansauge M.-T., Meyer M., Selective enrichment of damaged DNA molecules for ancient genome sequencing. Genome Res. 24, 1543–1549 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Green R. E., et al., A draft sequence of the Neandertal genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hajdinjak M., et al., Reconstructing the genetic history of late Neanderthals. Nature 555, 652–656 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Posth C., et al., Deeply divergent archaic mitochondrial genome provides lower time boundary for African gene flow into Neanderthals. Nat. Commun. 8, 16046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prüfer K., et al., The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rougier H., et al., Neandertal cannibalism and Neandertal bones used as tools in Northern Europe. Sci. Rep. 6, 29005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skoglund P., et al., Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc. Natl. Acad. Sci. U.S.A. 111, 2229–2234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peyrégne S., et al., Nuclear DNA from two early Neandertals reveals 80,000 years of genetic continuity in Europe. Sci. Adv. 5, eaaw5873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brown S., et al., Identification of a new hominin bone from Denisova Cave, Siberia using collagen fingerprinting and mitochondrial DNA analysis. Sci. Rep. 6, 23559 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Douka K., et al., Age estimates for hominin fossils and the onset of the Upper Palaeolithic at Denisova cave. Nature 565, 640–644 (2019). [DOI] [PubMed] [Google Scholar]
- 80.Krause J., et al., The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature 464, 894–897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sawyer S., et al., Nuclear and mitochondrial DNA sequences from two Denisovan individuals. Proc. Natl. Acad. Sci. U.S.A. 112, 15696–15700 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Horai S., et al., Man’s place in Hominoidea revealed by mitochondrial DNA genealogy. J. Mol. Evol. 37, 89 (1993). [PubMed] [Google Scholar]
- 83.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Darriba D., Taboada G. L., Doallo R., Posada D., jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guindon S., et al., New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 86.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for Bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bronk Ramsey C., Bayesian analysis of radiocarbon dates. Radiocarbon 51, 337–360 (2009). [Google Scholar]
- 88.Reimer P. J., et al., The IntCal20 Northern Hemisphere radiocarbon age calibration curve (0–55 cal kBP). Radiocarbon 62, 725–757 (2020). [Google Scholar]
- 89.Rasmussen S. O., et al., A new Greenland ice core chronology for the last glacial termination. J. Geophys. Res. 111, D06102 (2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the radiocarbon data generated at the ORAU are archived internally and are also available on the laboratory’s website, along with a link to the paper. The mitochondrial genome from Fonds-de-Forêt is deposited in GenBank with the accession number PRJEB39136. All other study data are included in the article and/or SI Appendix.