Significance
The retina is derived from the neuroepithelium in the vertebrate. During retinal development, three spatially patterned domains are formed, namely the ciliary margin zone, the optic disc, and a region that contains retinal progenitor cells. Little is known about the mechanisms that regulate this patterning and specification. In this study, we show that retinal regionalization is impaired in the Meis1 and Meis2 compound-null retina in mice. By analyzing transcriptomic and epigenomic profiles, we map and characterize the Meis-dependent gene regulatory network within the embryonic retina. We propose that Meis1 and Meis2 function redundantly to promote expression of retinal progenitor cell-specific genes, while simultaneously restricting ciliary margin zone- and optic disc-specific genes.
Keywords: retina, Meis, development
Abstract
The vertebrate eye is derived from the neuroepithelium, surface ectoderm, and extracellular mesenchyme. The neuroepithelium forms an optic cup in which the spatial separation of three domains is established, namely, the region of multipotent retinal progenitor cells (RPCs), the ciliary margin zone (CMZ)—which possesses both a neurogenic and nonneurogenic potential—and the optic disk (OD), the interface between the optic stalk and the neuroretina. Here, we show by genetic ablation in the developing optic cup that Meis1 and Meis2 homeobox genes function redundantly to maintain the retinal progenitor pool while they simultaneously suppress the expression of genes characteristic of CMZ and OD fates. Furthermore, we demonstrate that Meis transcription factors bind regulatory regions of RPC-, CMZ-, and OD-specific genes, thus providing a mechanistic insight into the Meis-dependent gene regulatory network. Our work uncovers the essential role of Meis1 and Meis2 as regulators of cell fate competence, which organize spatial territories in the vertebrate eye.
The retinal neuroepithelium is derived from the anterior region of the embryonic neural tube that coexpresses several eye field transcription factors (1, 2). The first morphological indication of eye development is the formation of the optic vesicle, which is derived from this eye-specified neuroepithelium. The optic vesicle subsequently invaginates in coordination with the lens placode to form a double-layered optic cup, while the inner layer of the neuroepithelium gives rise to the neural retina (3). The peripheral part of the optic cup, called the ciliary margin zone (CMZ), contains both neural retinal and nonneuronal ciliary epithelial progenitor cells. The latter cell population gives rise to the ciliary body and iris, while the former population of CMZ progenitors, being distinct from classic retinal progenitor cells (RPCs), contribute to mammalian retinogenesis (4). RPCs that reside in the inner layer of the optic cup are multipotent and generate six types of neurons, as well as the Müller glia in an evolutionarily conserved time order. RPCs are heterogeneous and change their competence in order to generate specific cell types at the appropriate developmental stages. The temporal identity of RPCs is primarily controlled by intrinsic factors (5–7). Relatively little is known regarding how the transition of RPC competence is controlled; however, previous studies have identified several transcription factors that are involved in this process (8–12). The centrally located part of the optic cup, the optic stalk, is initially formed as a hollow structure that maintains the connection of the optic cup to the diencephalon. Once the axons of retinal ganglion cells (RGCs) reach the optic stalk, they grow into it, forming the optic nerve. The interface between the optic stalk and the retina constitutes the optic disk (OD), which develops from the edges of the optic fissure. OD precursors represent a unique population of Bmp7-dependent Pax2+ cells (13, 14). Progenitor cells located at the OD, the interface between the optic stalk and the neural retina, express multiple axon guidance molecules (15). One such molecule, Netrin-1 (Ntn1), acts via its receptor Dcc on the RGC axons to promote their growth out of the eye (16). Numerous genetic and molecular studies of retina development have focused on various aspects of RPC, CMZ, or OD biology. However, the molecular mechanisms underlying the regionalization of the developing optic cup into CMZ, RPC, and OD zones remain poorly understood.
Meis genes encode the TALE superclass of atypical homeodomain-containing transcription factors. Meis proteins can bind to DNA on their own, or in cooperation with a wide range of transcription factors. For example, Meis proteins heterodimerize with the PBC subclass of TALE proteins and the complex cooperatively associates with DNA along with additional tissue-specific transcription factors, such as Hox transcription factors. It has been proposed that the main function of the TALE complex is to poise regulatory regions for transcription, which is then triggered by the binding of HOX and possibly other transcription factors (17–20). Three Meis homologs—Meis1, Meis2, and Meis3—have been identified in mice (18, 21). Both Meis1- and Meis2-null mutations are embryonically lethal at midgestation due to hematopoetic and cardiovascular defects (22–24). Recently, Meis1 and Meis2 have been shown to act redundantly or functionally compensate for each other in the lens, as expected from the high sequence homology between them (25). During retinal development in mice, both Meis1 and Meis2 are expressed in the optic vesicle (26). Previous studies have shown that Meis1 plays a critical role in early retinal development (18, 22, 27). A Meis1-null mutation results in microphthalmia, exhibiting lens vesicle reduction and partial duplication of the retina in mice (22, 27). Furthermore, Meis1 controls proliferation, dorsoventral and ventrodistal patterning of the optic cup in a dose-dependent manner, and also regulates components of the Notch signaling pathway, as well as genes associated with human microphthalmia (26, 27). Meis1-null mutants die between embryonic day (E)11.5 and E14.5 due to hematopoietic and vascular system defects (22), making it impossible to examine the role of Meis1 at later stages of retinal development.
In this study, we examined the role of Meis1 and Meis2 homeobox genes during retinal development by conditional genetic ablation in the developing optic cup. Our results demonstrate that Meis1 and Meis2 are redundantly required for optic cup regionalization by promoting RPC fate, while simultaneously restricting the CMZ and OD ones. By combining chromatin immunoprecipitation-sequencing (ChIP-seq) and bulk RNA-sequencing (RNA-seq) experiments, we identified Meis-dependent target genes that are expressed during optic cup regionalization and early stages of retinal neurogenesis. Furthermore, we identified the transcription factor Lhx2 and members of the basic helix–loop–helix (bHLH) family as the key coregulators involved in the Meis-dependent gene regulatory network that underlie cell fate decisions in the embryonic retina.
Results
Meis1/2 Deletion Results in Hypocellular Retina due to Prolonged Cell Cycle Length and Increased Cell Death.
To investigate the role of Meis1 and Meis2 during the retinal development, we first examined the spatiotemporal Meis1 and Meis2 protein expression. Meis1 was detected throughout the optic cup and maintained both in the neuroblastic layer and in the ganglion cell layer at later stages (SI Appendix, Fig. S1). At the optic cup stages, Meis2 was detected in the neuroepithelium. At later stages, the Meis2 protein was strongly expressed in the CMZ, OD, and a subset of cells in the outer neuroblastic layer and ganglion cell layer (SI Appendix, Fig. S1). In order to overcome the embryonic lethality of Meis1−/− and Meis2−/− embryos, we generated mice containing a Meis1 floxed allele (Meis1f/f) (Materials and Methods) and crossed with Meis2f/f (23) and mRx-Cre mice. Cre recombinase is active in the retina-committed progenitor cells from E9.0 onwards (28); however, residual Meis1 and Meis2 proteins were detected by E13.5, probably due to protein stability, which dictates target-inactivation time. These proteins were specifically deleted in the mRx-Cre;Meis1f/f;Meis2f/f (Meis1/2 conditional knockout [cKO]) retina at E14.5 (SI Appendix, Fig. S1). mRx-Cre is active in the presumptive retinal pigment epithelium as well; however, Meis proteins did not appear to be efficiently deleted in the tissue and the retinal pigment epithelium was virtually normal (SI Appendix, Figs. S1 and S2). To investigate the consequences of Meis1 and Meis2 inactivation in RPCs, frontal sections of Meis1/2 cKO embryos were stained with hematoxylin and eosin. The Meis1/2 cKO retina was virtually normal at E11.5; however, the retina subsequently became hypocellular and exhibited coloboma (SI Appendix, Fig. S1). At postnatal day 1, Meis1/2 cKO embryos developed a thin retinal layer and displayed dysplasia of the iris and ciliary body, accompanied with aberrant expression of ciliary body and iris-specific genes (SI Appendix, Figs. S1 and S2). The mRx-Cre;Meis1f/f (Meis1 cKO) retina became progressively hypocellular at postnatal stages, while the mRx-Cre;Meis2f/f (Meis2 cKO) retina was virtually normal (SI Appendix, Fig. S3).
Meis1 regulates proliferation in the retina, at least partially through the down-regulation of Ccnd1, which promotes progression through the G1 phase of the cell cycle (26, 27). Consistent with this, we found that Ccnd1 was down-regulated in the Meis1/2 cKO retina (SI Appendix, Fig. S4). RPCs of the Ccnd1−/− retina progress through the cell cycle at a slower rate (29). To determine whether the cell cycle length is altered in the Meis1/2 cKO retina, we utilized window-labeling based on two distinct thymidine analogs. We labeled S-phase RPCs by administering pregnant dams with sequential BrdU and EdU pulses separated by a 1.5-h interval. The lengths of the whole cell cycle (Tc) and S-phase (Ts) were calculated as previously described (29, 30). The length of the whole cell cycle in the Meis1/2 cKO retina at E12.5 was 26.5 ± 4.5 h, which was significantly longer than that of the retina in the control (Tc = 17.0 ± 0.9 h), while the length of S-phase was 10.4 ± 1.7 h, which was comparable to that of the control (Ts = 9.4 ± 0.6 h). Similar results were obtained at E14.5 (SI Appendix, Fig. S4). At the same time, we observed significantly increased cell death, as assayed by the antibody against cleaved caspase 3 (SI Appendix, Fig. S4). Taking these data together, we conclude that the slower rate of proliferation and increased cell death contribute to the hypocellularity of the Meis1/2 cKO retina.
Meis1 and Meis2 Are Required for Maintenance of the RPC Expression Program.
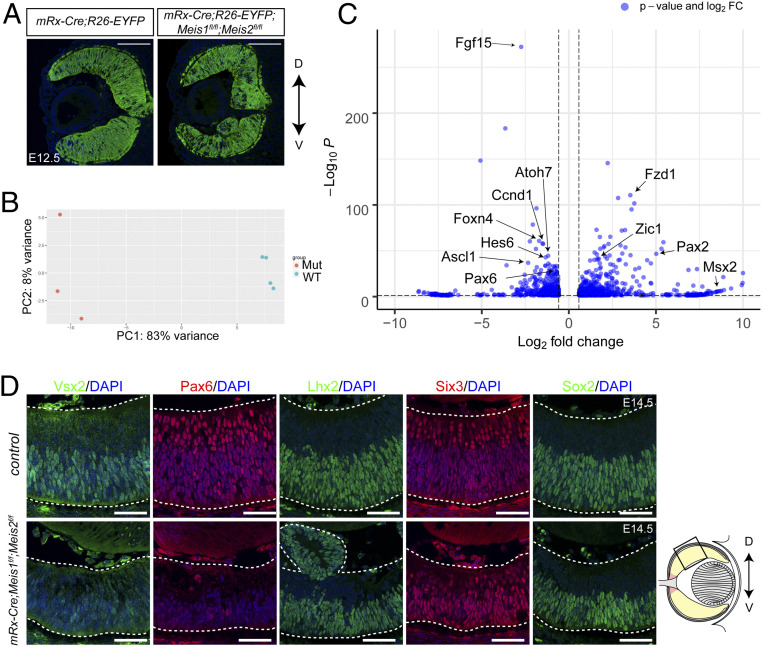
To study the role of Meis1 and Meis2 in RPC gene expression, we performed bulk RNA-seq of the wild-type and Meis1/2 cKO embryonic neural retina. We crossed R26-EYFP mice with mRx-Cre or mRx-Cre;Meis1f/f;Meis2f/f (Fig. 1A) and isolated EYFP+ RPCs and their progeny from E14.5 retina by fluorescence-activated cell sorting. Principal component analysis of gene expression of Meis1/2-deficient and control retinae showed that individual samples were clustered according to genotype (Fig. 1B). Next, we examined differentially expressed genes between Meis1/2 cKO and control retina and identified 722 down-regulated genes (log2FC [fold-change] < −0.55, q-value < 0.05) and 839 up-regulated genes (log2FC > 0.55, q-value < 0.05) (Fig. 1C and Dataset S1).
Fig. 1.
The effects of Meis1/2 deletion on the transcriptomes of the RPCs. (A) Frontal sections showing EYFP expression of mRx-Cre;R26-EYFP and mRx-Cre;Meis1f/f;Meis2f/f;R26-EYFP at E12.5. (Scale bars in A, 100 μm.) (B) Principal component (PC) analysis of bulk RNA-seq data. Four samples from control embryos and three samples of Meis1/2 cKO embryos were used for RNA-seq. (C) Volcano plot of control versus Meis1/2 cKO retina at E14.5. Significantly misregulated genes (Padj < 0.05, |log2FC| > 0.58) are indicated in blue. Selected differentially expressed genes are labeled. (D) Immunostaining with Vsx2, Pax6, Lhx2, Six3, and Sox2 antibodies in frontal sections of control and Meis1/2 cKO embryos at E14.5. Schematic representation of frontal sections of the eyes. The boxed area indicates the area where representative confocal microscopic images were taken. The dorsal-ventral axis is indicated by an arrow. D, dorsal; V, ventral. (Scale bars in D, 50 μm.)
The inactivation of Meis1 and Meis2 resulted in hypocellularity due to the prolonged length of the cell cycle and increased apoptosis. Therefore, we investigated whether the expression of cell cycle regulators was altered in the Meis1/2 cKO retina. Consistent with our immunohistochemistry data, we found that Ccnd1 mRNA was significantly down-regulated in the Meis1/2 cKO retina. The Meis1/2 cKO retina displays lengthening of the cell cycle time, as observed in the Ccnd1−/− retina (29). However, in contrast to the Meis1/2 cKO retina, the laminar structure is maintained and proliferating cells persist beyond the normal period of RPC proliferation in the Ccnd1−/− retina (29, 31). Furthermore, transfection with a Ccnd1 construct only partially rescued the RPC proliferation defect caused by forced expression of a dominant-negative Meis1 construct in the chick retina (26). These studies indicate that Meis1/2 likely regulate additional cell cycle regulatory genes. Indeed, we found that cell cycle regulators Myb and Plag1 were significantly down-regulated in the Meis1/2 cKO retina (Dataset S1). Taken together, these results indicate that Meis1/2 maintain RPC pools by controlling cell cycle regulators.
Clark et al. (12) showed that neuroretinal progenitors go through three distinct stages: neuroepithelial state, in which Notch signaling is essentially absent and the rates of promotion are very low; early-stage primary retinal progenitors, in which Notch signaling is relatively low; and late-stage primary progenitors, where Notch signaling is very high. At E14.5, RPCs are primarily composed of early RPCs and neurogenic RPCs. Therefore, we examined the expression of “canonical” RPC genes, early RPC genes, neurogenic RPC genes, and Notch signaling components. The expression levels of some of the canonical RPC genes, such as Pax6 and Vsx2, which are excluded from the OD, were down-regulated, while the expression of Rax and Six3, which are expressed in both RPCs and the OD (32), was apparently unchanged. Genes characteristic of early RPCs, such as Fgf15, and neurogenic RPCs, such as Atoh7, were also significantly down-regulated (Dataset S1). Furthermore, Notch signaling components, such as Dll1, Dll3, Dll4, and Rbpj, in addition to Notch targets Hes and Hey, were down-regulated in the Meis1/2 cKO retina (Dataset S1). Our results indicate that transition of RPCs’ developmental competence was impaired in the Meis1/2 cKO retina.
To confirm our bulk RNA-seq data, we examined the expression of Pax6, Vsx2, Lhx2, Six3, and Sox2 by immunohistochemistry. Consistent with our bulk RNA-seq data, Pax6 and Vsx2 protein levels were reduced, whereas other canonical RPC-specific transcription factors Six3, Sox2, and Lhx2 were maintained at an apparently normal level in the Meis1/2 cKO central retina (Fig. 1D). Moreover, we found that RGC-, amacrine cell- (AC), and cone photoreceptor cell (CP)-specific genes, such as Ptf1a, were also down-regulated in the Meis1/2 cKO retina (SI Appendix, Fig. S5). It is important to note that we did not observe down-regulated expression of Pax6 and Vsx2 in the mRx-Cre;Meis1f/f (Meis1 cKO) and mRx-Cre;Meis2f/f (Meis2 cKO) retinae (SI Appendix, Fig. S3), indicating that Meis1 and Meis2 function redundantly to control RPC identity. Together, these data show that Meis1/2 initiates early stages of neurogenesis and developmental competence by promoting Notch signaling, neurogenic bHLH factors, and RPC gene expression.
Meis1/2 Directly Regulate the RPC-Specific Genes.
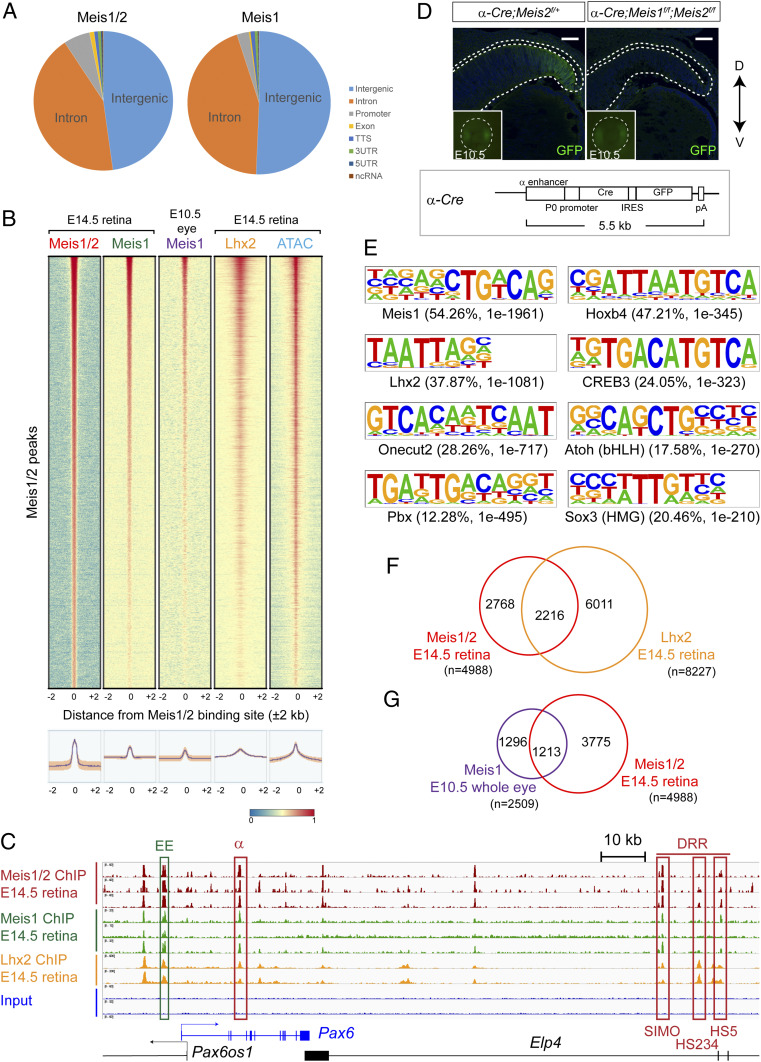
To provide the mechanistic understanding of the genetic program governed by Meis1 and Meis2 during retinal neurogenesis, we performed ChIP-seq using chromatin prepared from the neural retina at E14.5. We made use of antibodies that recognize either Meis1 alone, or both Meis1 and Meis2. In accordance with previous reports (19, 27, 33), the majority of Meis binding sites identified in retina samples were located in intergenic and intronic regions (Fig. 2A). To study the chromatin status at Meis1/2 and Meis1 target loci, we analyzed the correlation of Meis1/2 ChIP-seq peaks with ATAC-seq peak (34). We found that the presence of Meis-bound regions was frequently associated with open chromatin (Fig. 2B).
Fig. 2.
Meis1/2 directly regulate RPC-specific genes in concert with Lhx2. (A) Distribution of E14.5 retina Meis1/2 and Meis1 ChIP-seq peaks in the genome. (B) Density plots showing the distribution of Meis1 binding within the E10.5 whole eye [GSE62786 (27)], Meis1/2, Meis1 (E-MTAB-10112, present study), Lhx2 binding [GSE99818 (49)], and open chromatin region [GSE87064 (34)] in E14.5 neural retina at Meis1/2-bound regions (±2 kb from binding sites). The color scale shows the intensity of the distribution signal. The plotted lines were aligned according to E14.5 Meis1/2 ChIP-seq peaks. (C) ChIP-seq showing Meis1/2, Meis1, or Lhx2 binding in the E14.5 retina at Pax6. ChIP-seq peaks located within previously identified enhancers and promoters of Pax6 (downstream regulatory region [DRR] and α) are indicated by red boxes. (D) Frontal sections of E14.5 α-Cre;Meis2f/+ (control) and α-Cre;Meis1f/f;Meis2f/f embryos showing GFP signal. Insets show GFP signals from the heads of E10.5 embryos. The areas around the eye are shown in the Insets (Microscopic field: 400 × 400 μm). Schematic representation of α-Cre transgene. (Scale bar: 50 μm.) (E) DNA-binding motifs found in the Meis1/2-bound region by Homer. Percentage of target sequences with the motif and P value are indicated. (F) Venn diagram showing overlapping binding regions shared between Meis1/2 (E-MTAB-10112, present study) and Lhx2 [GSE99818 (48)] in E14.5 retina. (G) Venn diagram showing overlapping binding regions shared between Meis1/2 in E14.5 retina (E-MTAB-10112, present study) and Meis1 in E10.5 whole eye (27).
As described above, the expression of multiple cell cycle regulators was down-regulated in the Meis1/2 cKO retina. We, therefore, investigated whether Meis1/2 directly interacts with the regulatory regions of the corresponding genes. We found that Meis transcription factors bind to the regulatory regions of several cell proliferation-related genes, most notably Ccnd1, Myb, and Plag1, which were down-regulated in the Meis1/2 cKO retina (SI Appendix, Fig. S6), indicating that Meis1 and Meis2 directly regulate genes that are involved in cell cycle progression.
Our bulk RNA-seq data demonstrated that Meis1 and Meis2 are required for the maintenance of the RPC-specific gene-expression program. Hence, we next investigated whether the regulatory regions of RPC-specific genes down-regulated in the Meis1/2 cKO retina are bound by Meis1/2 in vivo. Indeed, we identified Meis1/2 interaction with several loci encoding RPC-specifying factors, early RPC, or neurogenic RPC markers (Fig. 2C and SI Appendix, Fig. S7). Interestingly, Meis1/2-bound regions were also found at the loci of late RPC-specific genes Nfia and Nfib, which are not expressed in the E14.5 retina (SI Appendix, Fig. S8). In addition, Nfib was 1.95-fold up-regulated in the Meis1/2 cKO retina (Dataset S1). Thus, Meis1/2 might premark retinal-specific enhancers of late-expressing genes or they may repress late RPC-specific genes at early stages.
In order to provide further evidence that Meis1/2 directly regulate the expression of RPC-specific genes, we determined whether the Meis-bound regions are located within the previously identified enhancers of the corresponding genes. Meis1/2 occupancy regions correlated with a well-characterized set of RPC enhancers. For example, at the Pax6 locus, we found that Meis transcription factors were bound to SIMO, HS234, and HS5 enhancers within the downstream regulatory region, as well as the α-enhancer, all of which have been shown to drive PAX6 expression in the developing retina (Fig. 2C) (35–37). To assess whether the α-enhancer is responsive to Meis1 and Meis2, we crossed Meis1f/f;Meis2f/f with α-Cre mice. In this Cre line, cre and gfp expression is under the control of the α-enhancer, which is active in the retina as early as E10.5 (38) (Fig. 2D). GFP signal was detected both in the α-Cre;Meis2f/+ and α-Cre;Meis1f/f;Meis2f/f retinae at E10.5. At E14.5, GFP+ cells were present in the peripheral retina of α-Cre;Meis2f/+ mice but not in the α-Cre;Meis1f/f;Meis2f/f retina (Fig. 2D), indicating that the α-enhancer is under the control of Meis1/2 transcription factors. In addition to Pax6 enhancers, we found that Meis transcription factors were bound to the regions corresponding to the U9 chicken SOX2 enhancer (39), Six3 promoter (40), CR4 enhancer of Foxn4 (41), and CNS enhancer of Ascl1 (42), which are active in the embryonic retina, and Atoh7 remote shadow enhancer (43, 44), which controls Atoh7 expression in the retina (SI Appendix, Fig. S7). In addition to RPC-specific genes, Meis1/2 ChIP-seq peaks were found at the loci of genes characteristic of the RGC, AC, and CP (SI Appendix, Fig. S5). These observations strongly indicate that Meis1/2 directly regulate a defined set of transcription factor genes previously implicated in RPC development and retinal differentiation.
In order to examine if Meis transcription factors bind to the same loci in the neuroepithelium at E10.5 and in the RPC at E14.5, we analyzed whether E14.5 retina Meis1/2 ChIP-seq peaks were colocalized with E10.5 whole-eye Meis1 ChIP-seq peaks (27). Only 24% of E14.5 retina Meis1/2 ChIP-seq peaks colocalized with E10.5 whole-eye Meis1 ChIP-seq peaks (Fig. 2G), which was likely due to the distinct transcriptional profiles observed between the neuroepithelium and RPCs. Both E10.5 whole-eye Meis1 ChIP-seq and E14.5 retina Meis1/2 ChIP-seq peaks were observed at Six3, which is expressed in both the neuroepithelium and RPC. In contrast, E14.5 retina Meis1/2 ChIP-seq peaks were observed at neurogenic RPC genes, such as Atoh7, and late RPC-specific genes, such as Nfia, as well as genes that are required for retinal differentiation, such as Prdm1 (SI Appendix, Figs. S5, S7, and S8).
Meis1 and Meis2 Regulate the RPC-Specific Genes in Cooperation with Lhx2.
Meis proteins can bind DNA directly or in cooperation with a diverse range of transcription factors, most notably with other homeodomain-containing proteins but also members of more distant transcription factor families (17, 18). For example, Hox transcription factors were shown to choose particular Meis-bound enhancers, which stabilize Meis binding to generate a functional output of enhancer activity (45). Partners of Meis1/2 in the embryonic retina are currently unknown. To identify transcription factors that potentially cooperate with Meis1/2, we performed de novo motif analysis by Homer using the whole-genome set of retina-specific Meis1/2-bound sites. As expected, the most enriched motif was the Meis1 motif itself (54.3% of the Meis1/2 ChIP-seq peaks contained the Meis consensus; P = 1E-1961) (Fig. 2E). With the exception of Meis1, the DNA regions bound by Meis1/2 were enriched with motifs of the homeobox transcription factor Lhx2, TALE class homeobox transcription factor Pbx3, Onecut2 (Hnf6b), CREB3, bHLH transcription factor Atoh1, and SRY-related HMG Box transcription factor Sox3 (Fig. 2E). Sox1, Sox2, and Sox3 belong to the SoxB1 subgroup. One member of the group compensates for the loss of another and shares the same binding motif (46). bHLH transcription factors, such as Atoh7 and Ascl1, which are expressed in RPCs, bind to the same E-box motif as Atoh1 (the CAGCTG motif) (47, 48), suggesting that these transcription factors may cooperate with Meis1/2. Only half of Meis1/2-bound regulatory regions in the retina contained the Meis motif, which indicated that Meis1/2 must bind DNA through noncanonical motifs in cooperation with other transcription factors. Interestingly, when analyzed in silico, 37.5% of Meis1/2 ChIP-seq peaks contained the Lhx2 binding motif (P value = 1E-1081) (Fig. 2E). Lhx2 was previously shown to primarily bind to open chromatin regions of early RPC genes (49).
We hypothesized that Meis1/2 cooperate with Lhx2 to regulate RPC-related genes. We therefore analyzed whether Meis1/2 ChIP-seq peaks were colocalized with Lhx2 ChIP-seq peaks (49). We found that Meis1/2 ChIP-seq peaks frequently colocalized with Lhx2 ChIP-seq peaks and that 44.6% of the Meis1/2 binding regions were also occupied by Lhx2 (Fig. 2F). We further tested the statistical significance for the spatial concordance of Meis1/2 ChIP-seq sites with Lhx2 motifs and Lhx2 ChIP-seq sites using a sliding window approach (Materials and Methods). We found significant enrichment of the Lhx2 motif at Meis1/2 ChIP-seq sites (Fisher’s exact test, P < 2.2E-16, odds ratio = 1.89, 95% CI 1.79 to 1.98). Similarly, cooccurrence of Meis1/2 ChIP-seq peaks and Lhx2 ChIP-seq peaks was highly significant (Fisher’s exact test, P < 2.2E-16, odds ratio = 146.1, 95% CI 137.9 to 153.4). Thus, the cooccupancy of Lhx2 and Meis1/2 at this frequency appeared unlikely to occur by chance. Meis1/2 and Lhx2 cooccupy the retinal specific enhancers of canonical RPC genes, such as Pax6, and neurogenic RPC genes, such as Foxn4 and Atoh7 (Fig. 2C and SI Appendix, Fig. S7). Lhx2 expression in RPCs is required for maintaining the optic cup identity and its morphogenesis and for RPC competence transition. The Lhx2 inactivation causes the reduction of the RPC population and alters the RPC competence (50, 51), which in fact resembled the phenotype observed in the Meis1/2 cKO retina. Taken together, our data indicate that Meis1/2 functions in cooperation with Lhx2 to regulate RPC-specific genes.
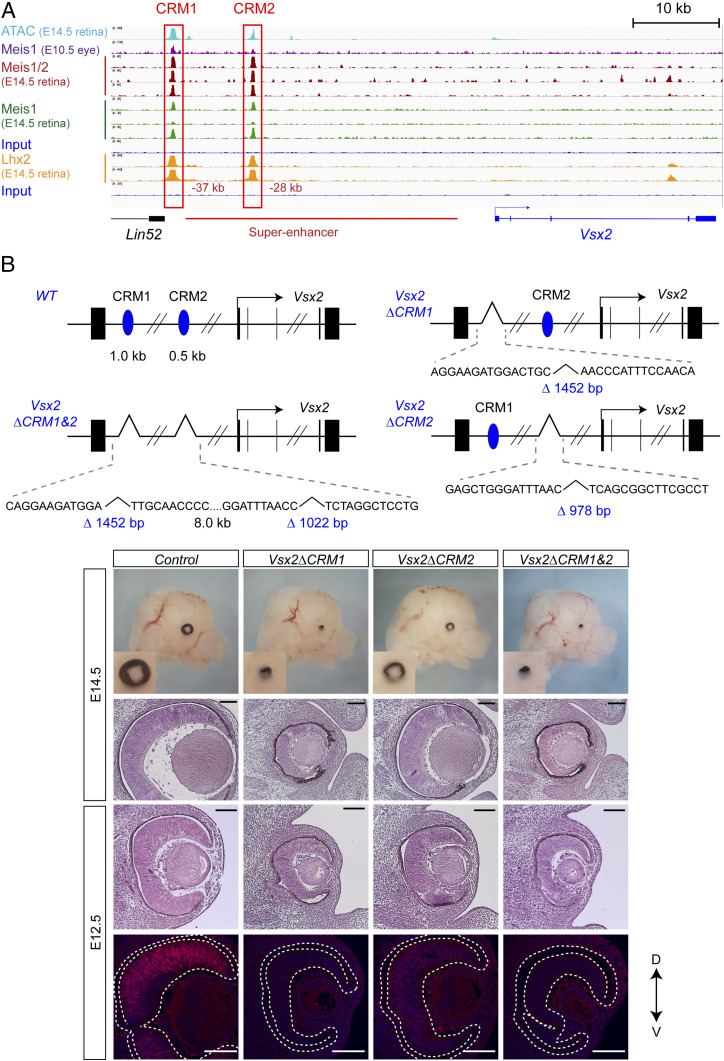
There were two regions that contained Meis1/2 and Lhx2 binding sites at −27 kb and −38 kb upstream of the transcription start site of Vsx2 (Fig. 3A), designated cis-regulatory module 1 and 2 (CRM1 and CRM2). CRM2 was located within the 32-kb Vsx2 core regulatory circuit superenhancer (52). CRM1 and CRM2 were found to be evolutionarily conserved among chicken, mouse, and human and contained Meis1/2 and Lhx2 motifs (SI Appendix, Fig. S9). Furthermore, two open chromatin regions were present at the Vsx2 locus in the retina at E14.5 and were found to be associated with CRM1 and CRM2 (Fig. 3A). Vsx2 was down-regulated in both Meis1/2 cKO and Lhx2 cKO retinae (Fig. 1) (51), suggesting that Meis1/2 and Lhx2 regulates the expression of Vsx2 through these enhancers. In order to examine the functionality of the CRMs, we deleted these CRMs in mice using CRISPR/Cas9 (Materials and Methods and Fig. 3B). Vsx2ΔCRM1, Vsx2ΔCRM2, and Vsx2ΔCRM1&2 embryos showed a microphthalmic phenotype as observed in the Vsx2-null mutant (53), although Vsx2ΔCRM2 displayed a less severe phenotype (Fig. 3B). Vsx2 was down-regulated in Vsx2ΔCRM2, while its expression was abolished in Vsx2ΔCRM1 and Vsx2ΔCRM1&2 retinae (Fig. 3B). Both Meis1/2 and Lhx2 were bound to CRM1 and CRM2 and Vsx2 was down-regulated in the Lhx2 cKO as well as the Meis1/2 cKO retina; thus, Meis and Lhx2 are likely to cooperatively regulate Vsx2 expression through the CRMs.
Fig. 3.
Meis1/2 regulate Vsx2 expression through the CRM1 and 2. (A) ChIP-seq showing Meis1/2, Meis1 (E-MTAB-10112, present study), Lhx2 binding [GSE99818 (49)], and ATAC-seq [GSE87064 (34)] in the E14.5 retina and Meis1 binding in the E10.5 whole eye [GSE62786 (27)] at Vsx2. Previously identified Vsx2 superenhancer (52) is indicated by a red line. (B) Schematic representation of Vsx2ΔCRM1, Vsx2ΔCRM2, and Vsx2ΔCRM1&2. CRM1 and CRM2 are depicted by blue ovals. The size of deletions and sequences across the deletion junctions are indicated in the panel. Haematoxylin and eosin-stained frontal sections, immunostaining with Vsx2 antibody in frontal sections, and whole head of control and Vsx2ΔCRM1, Vsx2ΔCRM2, and Vsx2ΔCRM1&2 at indicated stages. (Scales bars, 100 μm.)
Meis Deficiency Results in Expansion of the CMZ at the Expense of the Neural Retina.
By analyzing the bulk RNA-seq data from wild-type and Meis1/2 cKO retinae, we found that CMZ-specific genes, such as Otx1, were up-regulated in Meis1/2 cKO mice (SI Appendix, Fig. S10). To confirm bulk RNA-seq data, we analyzed the expression of CMZ markers Cdo, Zic1, Msx1, Aqp1, and FoxP2 (4, 54) by immunohistochemistry. CMZ is composed of the proximal and distal parts, which are distinguished by the expression of Msx1 and Aqp1, respectively, whereas Cdo marks the entire CMZ (4).
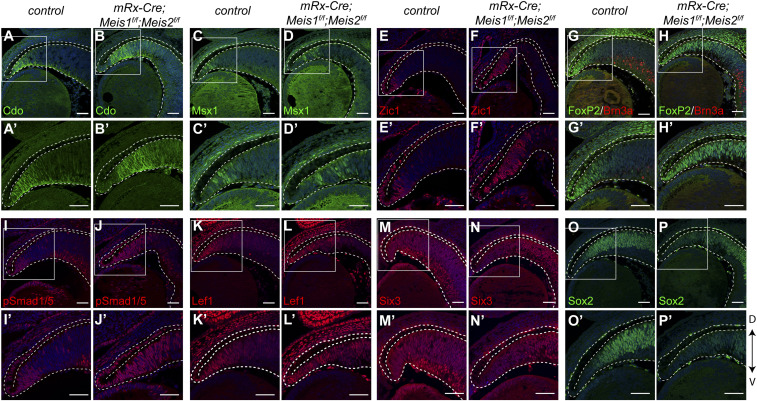
In control mice, Cdo and Zic1 were restricted to the peripheral retina and FoxP2 was present in the CMZ, as well as in the RGC subset. In the Meis1/2 cKO retina, the expression of Cdo, Zic1, and FoxP2 proteins was expanded toward the central part of the retina. We confirmed that FoxP2+ cells in the expanded CMZ area were not colocalized with Brn3a, which excludes the possibility that FoxP2+ cells in the CMZ were RGCs. In addition to Cdo, Zic1, and FoxP2, Msx1+ cells were also located more centrally in the Meis1/2 cKO retina compared to the control, whereas expression of Apq1 was apparently unchanged (Fig. 4 A–H′ and SI Appendix, Fig. S11). These results indicate that the proximal CMZ is expanded while the distal CMZ is unaffected. Although the CMZ markers were centrally expanded in the Meis1/2 cKO retina, they were not detected in the central retina at E14.5. Notably, the expression of Cdo, Msx1, and Aqp1 proteins was not changed in either Meis1 cKO or Meis2 cKO (SI Appendix, Fig. S11). In addition, hematoxylin and eosin staining demonstrated that the structure of the CMZ was not altered in the Meis1- and Meis2-deficient retina (SI Appendix, Fig. S11). These data indicate that Meis1 and Meis2 function redundantly to restrict the CMZ area.
Fig. 4.
Meis1/2 deletion results in expansion of the CMZ. (A−P′) Immunostaining with Cdo, Msx1, Zic1, FoxP2, Brn3a, phosphorylated Smad1/5, Lef1, Six3, and Sox2 antibodies in frontal sections of control and Meis1/2 cKO embryos at E14.5. Magnified views indicated by the white boxed areas of top images are shown below. The dorsal-ventral axis is indicated by an arrow. (Scale bars, 50 μm.)
Meis Proteins Restrict CMZ by Modulating Wnt/β-Catenin and Bmp Signaling in the Retinal Periphery.
The precise regulation of Wnt/β-catenin and Bmp signaling activity is essential for determining the boundary between the neural retina and CMZ (55, 56). We analyzed the expression of Wnt/β-catenin and Bmp signaling components. Multiple Bmp ligands, such as Bmp2, its receptor Bmpr1b, Wnt ligands including Wnt7b, and Wnt receptors, such as Fzd1, were up-regulated in the Meis1/2 cKO retina (SI Appendix, Fig. S10). In order to investigate whether the up-regulation of Bmp ligands causes the activation of Bmp signaling in the Meis1/2 cKO retina, we performed immunohistochemistry with antiphosphorylated Smad1/5. Phosphorylated Smad1/5 was restricted to the peripheral region in the control retina, while it was detected more centrally in the Meis1/2 cKO retina (Fig. 4 I–J′), correlating with the observed expansion of CMZ marker genes. To examine the activity of Wnt/β-catenin signaling, we analyzed the expression of Lef1, which is directly regulated by this signaling in the CMZ (55). We observed that Lef1 expression was expanded centrally in the Meis1/2 cKO (Fig. 4 K–L′), correlating with the observed expansion of CMZ marker genes and Bmp signaling activity.
We next investigated if Meis transcription factors could directly regulate genes encoding some of the components of Wnt/β-catenin and Bmp signaling pathways. Indeed, by analyzing the ChIP-seq data, we identified Meis occupancy within the putative regulatory regions of Wnt7b and Bmp4 (SI Appendix, Fig. S12). In addition, we also found that Meis transcription factors were bound to the loci of CMZ-specific genes, such as Cdo and FoxP2 (SI Appendix, Fig. S12). Since Six3/6 and Sox2 are known to suppress Wnt/β-catenin signaling in the retina in order to maintain the pool of RPCs (57–59), we analyzed the expression of Sox2 and Six3 in the Meis1/2 cKO retina. We found that Sox2 and Six3 were maintained at normal levels in the central retina; however, they were down-regulated in the peripheral retina of Meis1/2 cKO embryos (Fig. 4 M–P′), suggesting that Meis1/2 may be required for Sox2 and Six3 expression. It has been shown that Meis1 interacts with Sox2 (60). Furthermore, the homeodomain core motif, which can be bound with Six3 (61) and Sox motifs, are enriched in the Meis1/2 ChIP-seq peaks (Fig. 2E). We hypothesized that Six3 and Sox2 suppress CMZ-specific genes and Wnt/β-catenin signaling regulators in concert with Meis1/2. We therefore analyzed whether the Meis1/2-bound regions of these genes contained a Sox2 and Six3 consensus. We confirmed that these motifs are present within Meis1/2-bound enhancers of CMZ-specific genes and Wnt/β-catenin signaling regulators (SI Appendix, Fig. S12), suggesting that Meis1/2 cooperatively restrict the CMZ fate by inhibiting Wnt/β-catenin signaling components in concert with Sox2 and Six3.
Taken together, our data suggest that Meis1 and Meis2 in the retina periphery regulate the activities of Bmp and Wnt/β-catenin signaling pathways in order to maintain the population of RPCs.
Meis Deficiency Results in Expansion of the OD at the Expense of the Neural Retina.
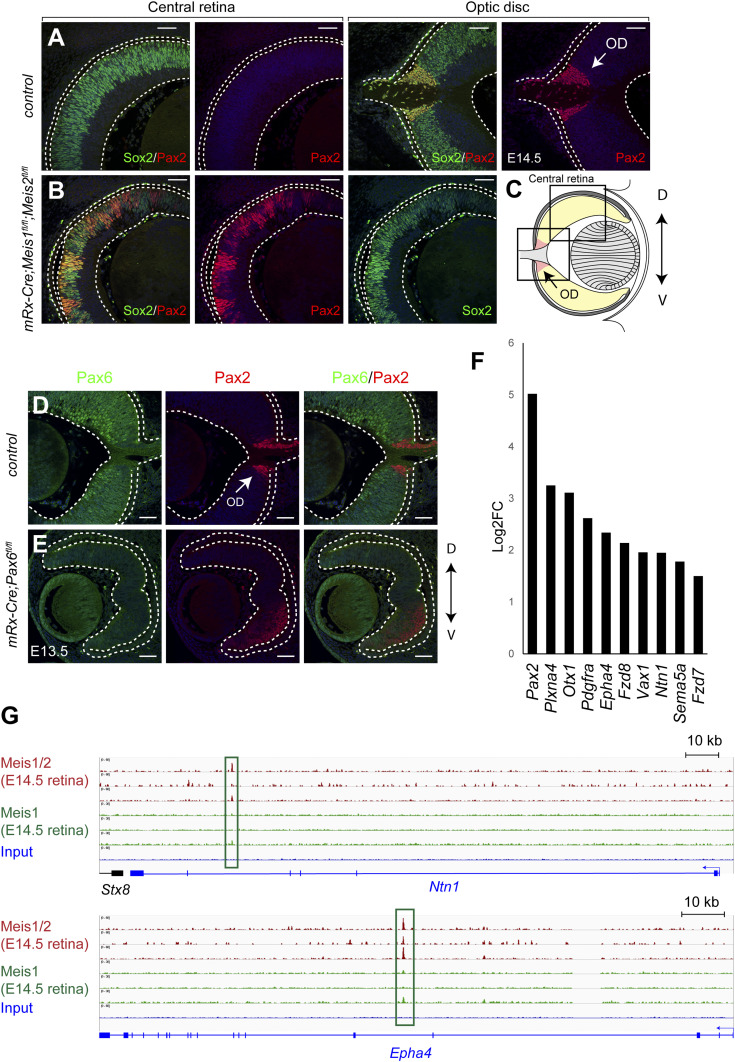
Surprisingly, by searching the RNA-seq data, we found that OD marker Pax2 was up-regulated 32-fold in the E14.5 Meis1/2 cKO retina compared to the control. We first validated the mRNA data by performing immunohistochemistry for the Pax2 protein. We found that in the Meis1/2 cKO retina, the Pax2 protein was ectopically expressed (Fig. 5 A–C), most likely at the expense of typical RPC markers Vsx2. The expression of Pax6 is normally excluded from the OD, and the optic stalk/optic cup boundary is established by reciprocal transcriptional repression of Pax2 and Pax6 (62). To determine whether Pax2 up-regulation in the Meis1/2 cKO retina is mediated by Pax6 down-regulation, we crossed mRx-Cre with Pax6f/f mice to inactivate Pax6 in RPCs and analyzed the expression of Pax2. At E13.5, the Pax2 protein was ectopically expressed in the ventral mRx-Cre;Pax6f/f retina (Fig. 5 D and E), which indicates that Meis1/2 may restrict the expression of Pax2 to the OD, at least in some regions, through the action of Pax6. Since Pax2 is required for OD formation, we next analyzed the expression of additional genes that are characteristic of the OD fate. Interestingly, we found that several OD-specific genes were up-regulated in the Meis1/2 cKO retina (Fig. 5F) (32, 63–68). Furthermore, we found that Meis1/2 directly bound to the putative regulatory regions of the up-regulated OD-specific genes, such as Ntn1 and Epha4 (Fig. 5G). From these results, we conclude that Meis1 and Meis2 are required to suppress the OD fate in the developing retina.
Fig. 5.
Meis1/2 deletion results in expansion of the optic disk. (A and B) Immunostaining with Pax2 and Sox2 antibodies in frontal sections of control and Meis1/2 cKO embryos at E14.5. (C) Schematic representation of a frontal section of the eye. Boxed areas indicate the central retina and the OD that are shown in A and B. (D and E) Immunostaining with Pax6 and Pax2 antibodies in frontal sections of control and Pax6 cKO embryos at E13.5. (F) OD-specific genes that are up-regulated in the Meis1/2 cKO (log2FC > 0.58, q-value < 0.05). (G) ChIP-seq showing Meis1/2 or Meis1 binding the loci of Ntn1 and Epha4. Arrows indicate the OD. The dorsal-ventral axis is indicated by an arrow. (Scale bars, 50 μm.)
Discussion
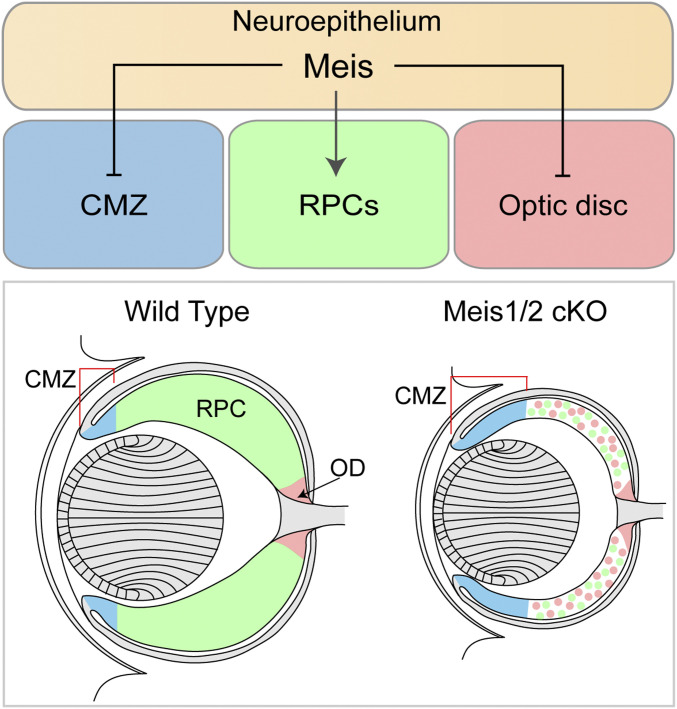
Here we have proposed, based on the genetic manipulation of mice, that Meis1 and Meis2 function redundantly to promote the expression of RPC-specific genes, while they simultaneously restrict CMZ- and OD-specific genes (Fig. 6). The molecular mechanism underlying this phenomenon likely involves direct DNA binding of Meis1/2, as well as temporally dependent and cell-type–specific cooperation with other transcription factors. To this end, we demonstrated that Meis transcription factors bind the regulatory regions of RPC-, CMZ-, and OD-specific genes and act in cooperation with other transcription factors, such as Lhx2, bHLH, Six3, and Sox2, depending on the context. Meis1/2 are expressed throughout the retina and Meis1/2-bound regulatory regions contain motifs of multiple classes of transcription factors. Thus, Meis1/2 likely cooperate with additional RPC-, CMZ-, and OD-specific transcription factors to control gene expression. Indeed, previous studies have indicated that the outputs of Meis1/2-bound regulatory regions are tightly controlled by other tissue-specific transcription factors, such as Hox (19, 45, 69). These tissue-specific transcription factors determine the functional outputs of the enhancers and promoters of Meis target genes. For example, during branchial arch development, Meis1 binds to the regulatory regions that are common to all branchial arches; however, only a subset of them is activated in a tissue-specific manner. In the secondary branchial arch, Hoxa1 is recruited to Meis1-premarked enhancers that contain Hox motifs, which facilitates Meis binding to Hoxa1 target genes. The increased Meis1 binding subsequently leads to gene activation (19). Although Hox genes are not expressed in the retina, it is reasonable to assume that one of the modes of action by which transcription factors cooperate with Meis1/2 in the retinal tissue may also involve increased DNA binding. This view is supported by the fact that only about one half of the retina-specific Meis1/2 ChIP-seq peaks contained the Meis consensus logo, indicating that the other half of ChIP-seq peaks represent indirect Meis1/2 DNA binding, or contain a relaxed consensus predisposed to regulatory mechanisms, such as increased DNA-binding affinity mediated by cooperating factors.
Fig. 6.
Schematic representation of the retinal phenotype in the Meis1/2-deficient retina. Meis1 and Meis2 redundantly promote RPC-specific genes and simultaneously restrict CMZ and OD fate. Inactivation of Meis1 and Meis2 in the retina results in expansion of CMZ and OD at the expense of RPC.
RPCs are heterogeneous and change their developmental competence over time in order to generate different types of neurons and Müller glia (5). Primary immature RPCs harbor competence to give rise to all types of retinal cells. Early RPCs mostly generate early-born cell-type RGCs, ACs, horizontal cells, and CPs, while late-born cell types, bipolar cells and Müller glia, originate from late RPCs. The temporal identity of RPCs is primarily controlled by intrinsic signals, including transcription factors. However, intrinsic factors that govern the RPC competence transition to the neurogenic RPC are poorly understood (5, 7). In this study, we suggest that Meis1/2 transcriptional factors play a pivotal role in this process. The neurogenic RPC-specific genes are down-regulated in Meis1/2 cKO, suggesting that RPC competence is impaired in the Meis1/2 cKO retina. Our data also indicate that Meis1/2 regulate RPC-specific genes directly through the set of previously well-characterized retina-specific enhancers. Global analysis of transcription factor occupancy provided strong evidence for the cooperation of Meis1/2 with the homeodomain-containing transcription factor, Lhx2, to control RPC-specific genes. Lhx2 allows the establishment of open chromatin regions in the vicinity (49), indicating that Lhx2 controls chromatin accessibility at Meis1/Lhx2 binding sites.
Meis1/2-bound regulatory regions are also highly enriched with the motif of bHLH transcription factors, which are known to control the competence of RPCs. Thus, our observations indicate that Meis1/2 cooperate with the bHLH transcription factors and act as intrinsic factors to control the RPCs’ competence by directly regulating the expression of transcription factors that influence the temporal identity of RPCs. Among RPC-specific regulators, Sox2 has been shown to interact with Meis transcription factors (60). Meis1/2-bound enhancers of CMZ-specific genes, such as Cdo and FoxP2, contain Sox2 and Six3 binding motifs. TALE factors are known to access the regulatory regions to poise them for later activation. For example, the TALE complex occupies the hoxb1a promoter already during early blastula stages when the expression of hoxb1a is inhibited. At the gastrula stage, Hoxb1b is recruited to the TALE complex at the hoxb1a promoter, which triggers the expression of hoxb1a (20). We have shown that Meis1/2 bind to late RPC-specific genes, such as Nfia and Nfib, in the retina at E14.5, when these genes are not yet expressed in the RPCs. Thus, Meis1/2 probably premarks or supresses the regulatory regions of late RPC-specific genes in the RPCs to ensure the expression of these genes at the appropriate stage of retinal development. These observations have lead us to propose that RPC-specific transcription factors—including Lhx2, bHLH, Sox2, Six3, and Nfia/Nfib—determine the output of Meis1/2-premarked enhancers within the retina, paralleling a mechanism that has been previously observed in other biological contexts. Further studies are nevertheless necessary for the identification of additional transcription factors that determine the functional outputs of Meis1/2-bound regulatory regions.
Meis1/2 and Pax6 are coexpressed in the vertebrate retina during embryogenesis. In Drosophila, the Pax6 homolog ey, Meis homolog hth, and Tzhz homolog tsh function as a complex in eye development (70). In vertebrates, Meis2 directly interacts with Pax6 and Dlx2 to functionally cooperate during subventricular zone neurogenesis (71). The Pax6/Meis interaction may represent an evolutionarily conserved mechanism.
Marcos et al. (27) have shown that Meis1 regulates components of the Notch signaling pathway and genes associated with human microphthalmia in the lens and neuroepithelium by ChIP-seq and bulk RNA-seq with E10.5 whole eye. In this study, we investigated the Meis gene regulatory network (GRN) in RPCs by ChIP-seq and bulk RNA-seq with E14.5 neural retina. At E10.5 (i.e., prior to the onset of neurogenesis), the presumptive retina is primarily composed of neuroepithelium, while the neural retina at E14.5 is mainly formed of RPCs and the genesis of early retinal cell types has been initiated (5). Single-cell RNA-seq analysis of retinal development shows a clear transcriptional distinction between the neuroepithelium and RPCs (12). Thus, Meis-dependent GRN is expected to reflect transcriptional changes. Indeed, 51% of E10.5 whole-eye Meis1 ChIP-seq peaks and 76% of E14.5 retina Meis1/2 ChIP-seq peaks were unique at each stage. Meis proteins bind to the genes expressed in both the neuroepithelium and RPCs at E10.5 as well as E14.5. However, only E14.5 retinal samples allowed the identification of Meis1/2 ChIP-seq peaks at neurogenic RPC genes, late RPC-specific genes, or genes required for retinal differentiation. Analysis of our current and previous data allowed us to reveal a more comprehensive Meis-dependent GRN in retinal development during embryogenesis.
Our data strongly suggest that Meis1/2 function in cooperation with Lhx2 to regulate RPC-specific genes. Lhx2 binding motifs are highly enriched within Meis1/2 ChIP-seq peaks and about half of Meis1/2 in vivo binding sites are in close vicinity of experimentally identified Lhx2 binding sites. As observed in Meis1/2 cKO, Lhx2 cKO embryos display an RPC proliferation defect and a substantially reduced expression of RPC-specific genes, such as Vsx2 and Foxn4, and a range of genes selectively expressed in the differentiating retinal cells (49, 51). Both Meis1/2 and Lhx2 bind the Vsx2 enhancers CRM1 and CRM2, which are required for the expression of Vsx2 (Fig. 3). Vsx2 is down-regulated in the Meis1/2 cKO retina even though Lhx2 is present (Fig. 1), suggesting that Lhx2-mediated expression of Vsx2 requires Meis1/2. Lhx2 cKO embryos do not exhibit the OD/CMZ expansion phenotype but, rather, the up-regulation of a set of genes that are normally expressed in the thalamic eminence and anterodorsal hypothalamus (51). Chx10-Cre;Lhx2f/f mice show an early arrest in lens fiber development due to the down-regulation of Fgfs in the retina (72). In contrast, the lens is smaller but no defects in lens fiber differentiation are evident in Meis1/2 cKO mice (Fig. 1 and SI Appendix). Fgf3, Fgf9, and Fgf15 are down-regulated in the Chx10-Cre;Lhx2f/f retina (72). Fgf3/15 were down-regulated, however, Fgf9 expression was unchanged in the Meis1/2-deficient retina (Dataset S1). In addition, Fgf8 was up-regulated in the Meis1/2 cKO retina (Dataset S1). Presumably, down-regulation of Fgf3/15 is compensated by Fgf8/9, which maintains development of the lens.
mRx-Cre;Meis1f/f displayed a distinct retinal phenotype that was observed in Meis1-deficient mice, probably due to delayed depletion of Meis1 protein. In addition, a truncated Meis1 protein that has been predicted to act as a dominant-negative form is produced in Meis1−/− mice (22). In another Meis1-deficient mouse model, Meis1aER, a Meis1–ERT2 fusion protein, is present in the cytoplasm (24). The phenotypic discrepancies in eye development among Meis1-deficient mice indicates the potential effects of Meis1–ERT2 or truncated Meis1 protein in vivo (22, 24, 27). No truncated Meis1 protein was detected in mRx-Cre;Meis1f/f;Meis1f/f, suggesting that Meis1f/f would represent a true loss-of-function allele, albeit with a delayed time course.
By E11.5, dorsal-ventral and distal-proximal patterning of the optic cup is disrupted in Meis1-deficient mice Meis1aER, which is accompanied with abnormal expression of Pax6, Pax2, Otx1, Tbx5, and Vax2 (27). Furthermore, proliferation is reduced and the area in which Tuj1, Isl1/2, and Otx2 are expressed is less than that found in the control, suggesting that Meis1 is required for retinal differentiation and proliferation (27). Meis1-deficient mice display phenotypes both in the lens and in the retina (22, 27), which adds a complication to the study of the possible function in the retina using these embryos. For example, the signals from the lens has been shown to influence retinal development by controlling CMZ identity and the initial direction of RGC axon outgrowth (73, 74). In order to overcome early embryonic lethality and provide further insight into retinal differentiation, as well as CMZ development, the conditional deletion of Meis genes in the retina is required. Meis1 and Meis2 proteins were depleted at E14.5 in Meis1/2 cKO mutants, thus the optic cup patterning would be presumably maintained by the presence of these proteins at earlier stages. It is complicated to study dorsal-ventral and distal-proximal patterning at later stages due to the OD/CMZ expansion phenotype in the Meis1/2 cKO. In addition, our bulk RNA-seq data did not provide clear evidence of the patterning defect. We did not observe the OD/CMZ expansion phenotype in mRx-Cre;Meis1f/f and mRx-Cre;Meis2f/f. These data combined show that Meis1 controls optic cup patterning at early stages of retinal development, and at later stages Meis1 and Meis2 jointly control the regionalization of CMZ/OD/RPC in the retina.
CMZ/OD expansion in the Meis1/2 cKO may reflect a direct role of Meis1/2 in suppression of OD/CMZ fate, or the phenotype might arise indirectly from the depletion of RPCs in the proliferative zone. We are in favor of the first model. Multiple transcription factors, such as Lhx2, Vsx2, Hes1, Sox2, and Pax6 are required for RPC maintenance but inactivation of these genes does not result in OD/CMZ expansion (38, 51, 75, 76). A Vsx2-null mutation results in transdifferentiation of the neural retina into pigmented cells (75, 76). Pax6 inactivation in RPCs leads to exclusive generation of ACs (38). The optic cup/stalk boundary is disrupted in the Hes1 cKO (77). Only CMZ is expanded at the expense of the RPC in the Sox2 cKO and Six3/6 cKO embryos (57, 59). Therefore, simple depletion of RPCs probably does not correlate with CMZ/OD expansion. Our Meis ChIP-seq was performed using E14.5 neural retina, which is primarily composed of RPCs. CMZ and OD represent a small proportion of the neural retinal cell population. Therefore, Meis1/2 ChIP-seq peaks found at CMZ- and OD-specific genes probably represent Meis1/2 binding sites in RPCs. The binding of Meis does not induce transcription of its target genes. Meis proteins act as a hub, which assists combinatorial assembly of other transcription factors, which define transcriptional output of selected enhancers (45). Meis1/2 appear to cooperate with other retina-specific transcription factors in order to maintain the RPC pool and simultaneously suppress CMZ/OD fate in RPCs.
The molecular mechanisms underlying regionalization of the developing optic cup into CMZ, RPC, and OD zones remain poorly understood. Nevertheless, the involvement of signaling pathways has been well documented. OD development is controlled by Bmp, Shh, and Fzd signaling. In Bmp7−/− and Smad4−/− retinae, the OD-specific factors Pax2 and Ntn1 are lost (13, 78, 79). In addition, the Bmp receptor, Bmp1b, is required for RGC axon guidance to the OD (79). Shh is absent in the OD, however it is expressed in RGCs. Shh derived from these cells modulates the size of the Pax2+ cell population in the OD (67). We found that Bmp7, its receptor Bmpr1b, Fzd8, and Fzd5 were up-regulated in the Meis1/2 cKO retina, suggesting that Meis1/2 maintain the boundary between the OD and neural retina, at least in part, by controlling the expression of these signaling components. CMZ specification requires Wnt/β-catenin and BMP signaling activities (55, 56). Wnt/β-catenin signaling is antagonized by Sox2 and Six3/Six6 to suppress CMZ fate. Sox2 and Six3/Six6 inactivation results in the expansion of CMZ at the expense of the neural retina, accompanied with ectopic activation of Wnt/β-catenin signaling (57–59). We have shown that Sox2 and Six3 are down-regulated and Wnt/β-catenin signaling is aberrantly activated in the peripheral retina of the Meis1/2 cKO retina. Our ChIP-seq analysis revealed that Meis1/2 is bound to the Six3 promoter and Sox2 enhancers, which were described to drive Six3 and Sox2 expression in the retina. In addition, several Wnt/β-catenin signaling components are expressed in the developing retina and especially Tcf7, Fzd1, and Fzd7 are enriched in the CMZ. Furthermore, Fzd1 and Tcf7 are also significantly up-regulated in the Six3/Six6- and Sox2-deficient retina, respectively (57, 58). In combination, the available data strongly suggest that Meis1/2 regulate Six3 and Sox2 expression and suppresses CMZ fate in cooperation with Six3/6 and Sox2.
Single-cell RNA-seq analysis of the developing mouse retina has revealed that CMZ-enriched genes, such as Msx1, are also expressed in the early neuroepithelium (12). This result raises two major possible explanations regarding why CMZ-specific genes are up-regulated in the Meis1/2 cKO retina. First, the failure of RPC competence transition from the optic cup or optic vesicle neuroepithelium is responsible for the elevated expression of CMZ-like markers. Second, the up-regulation of CMZ markers is due to the expansion of the CMZ territory. At present, we prefer the latter possibility. In our conditional mouse model, Meis1 and Meis2 proteins are depleted only by E14.5, when the transition from the neuroepithelium should already have been completed. Canonical RPC genes Pax6 and Vsx2, which are also expressed in the early neuroepithelium, are down-regulated in the Meis1/2 cKO retina. Furthermore, Wnt/β-catenin signaling is not active in the early neuroepithelium and becomes active subsequently in the CMZ, where this signaling controls the transcription of CMZ-specific genes (80). In the Meis1/2 cKO retina, Wnt/β-catenin signaling is aberrantly activated in the peripheral retina. The retinal explant culture propagated in the presence of Wnt3a and GSK3β inhibitor shows up-regulation of CMZ/specific genes in the peripheral retina but not in the central retina (57). Thus, it appears that only the RPCs located in the peripheral retina are competent to adopt the CMZ fate (57). Despite the deletion of Meis1/2 throughout the entire neural retina, the expansion of CMZ-specific genes was restricted to the peripheral retina in the Meis1/2 cKO. These observations favor the possibility that the state of the Meis1/2 cKO retina is compatible with that of the CMZ rather than the early neuroepithelium.
Nevertheless, we cannot currently exclude the scenario in which Meis1 and Meis2 control the competence transition from the early neuroepithelium. First of all, both Meis1 and Meis2 are expressed in the neuroepithelium. Although the complete depletion of Meis proteins was typically achieved by E14.5, when the transition from the neuroepithelium should have already been completed, mRx-Cre is active in the eye-specified neuroepithelim from E8.5 onwards and we have observed patches of diminished levels of Meis proteins as early as E11.5. We have shown that Meis1/2 directly bind regulatory regions of CMZ-specific genes; however, it remains elusive whether these putative enhancers are active in the early neuroepithelim, CMZ, or both. Further studies are highly warranted to clarify whether Meis1/2 facilitate competence transition from the neuroepithelium.
Materials and Methods
All experiments with mice were performed in compliance with the European Communities Council Directive of 24 November 1986 (86/609/EEC) and national and institutional guidelines. Animal care and experimental procedures were approved by the Animal Care Committee of the Institute of Molecular Genetics. Vsx2 ΔCRM1, Vsx2 ΔCRM2, and Vsx2 ΔCRM1&2 mice were generated using the CRISPR/Cas9 approach. Details are described in SI Appendix. Immunofluorescence, ChIP-seq, and bulk RNA-seq were performed according to published protocols. More detail regarding specific experimental procedures, a list of antibodies, and primers are provided in the SI Appendix.
Supplementary Material
Acknowledgments
We thank S. Blackshaw for critical comments on the manuscript; A. Buchberg for antibodies; S. Takacova and J. Manning for proofreading the manuscript; and Institute of Molecular Genetics core facilities for their outstanding support, in particular the Transgenic and Archiving Module for help in generation of Vsx2 mutant lines. This work was supported by the Czech Science Foundation (GACR) 18-20759S to Z.K. and Ministry of Education, Youth, and Sports of the Czech Republic (LQ1604-BIOCEV: from Fundamental to Applied Research). Mouse costs were partially covered by the Czech Center for Phenogenomics infrastructure supported by LM2018126, OP VaVpI CZ.1.05/2.1.00/19.0395, and CZ.1.05/1.1.00/02.0109.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. T.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013136118/-/DCSupplemental.
Data Availability
RNA-seq and ChIP-seq data have been deposited in the ArrayExpress database (accession nos. E-MTAB-9143 and E-MTAB-10112). Dataset S1 is available as part of the submission. All other study data are included in the article and supporting information.
References
- 1.Zuber M. E., Gestri G., Viczian A. S., Barsacchi G., Harris W. A., Specification of the vertebrate eye by a network of eye field transcription factors. Development 130, 5155–5167 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Esteve P., Bovolenta P., Secreted inducers in vertebrate eye development: More functions for old morphogens. Curr. Opin. Neurobiol. 16, 13–19 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Fuhrmann S., Eye morphogenesis and patterning of the optic vesicle. Curr. Top. Dev. Biol. 93, 61–84 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bélanger M. C., Robert B., Cayouette M., Msx1-positive progenitors in the retinal ciliary margin give rise to both neural and non-neural progenies in mammals. Dev. Cell 40, 137–150 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Cepko C., Intrinsically different retinal progenitor cells produce specific types of progeny. Nat. Rev. Neurosci. 15, 615–627 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Cayouette M., Mattar P., Harris W. A., Progenitor competence: Genes switching places. Cell 152, 13–14 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Javed A., Cayouette M., Temporal progression of retinal progenitor cell identity: Implications in cell replacement therapies. Front. Neural Circuits 11, 105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brzezinski J. A. 4th, Kim E. J., Johnson J. E., Reh T. A., Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development 138, 3519–3531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brzezinski J. A. 4th, Prasov L., Glaser T., Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev. Biol. 365, 395–413 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott J., Jolicoeur C., Ramamurthy V., Cayouette M., Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron 60, 26–39 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Mattar P., Ericson J., Blackshaw S., Cayouette M., A conserved regulatory logic controls temporal identity in mouse neural progenitors. Neuron 85, 497–504 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark B. S., et al., Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron 102, 1111–1126.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morcillo J., et al., Proper patterning of the optic fissure requires the sequential activity of BMP7 and SHH. Development 133, 3179–3190 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Patel A., Sowden J. C., Genes and pathways in optic fissure closure. Semin. Cell Dev. Biol. 91, 55–65 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Erskine L., Herrera E., Connecting the retina to the brain. ASN Neuro 6, 1759091414562107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deiner M. S., et al., Netrin-1 and DCC mediate axon guidance locally at the optic disc: Loss of function leads to optic nerve hypoplasia. Neuron 19, 575–589 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Bobola N., From DNA binding to transcriptional activation: Is the TALE complete? J. Cell Biol. 216, 2603–2605 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulte D., Geerts D., MEIS transcription factors in development and disease. Development 146, dev174706 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Amin S., et al., Hoxa2 selectively enhances Meis binding to change a branchial arch ground state. Dev. Cell 32, 265–277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choe S. K., Ladam F., Sagerström C. G., TALE factors poise promoters for activation by Hox proteins. Dev. Cell 28, 203–211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blasi F., Bruckmann C., Penkov D., Dardaei L., A tale of TALE, PREP1, PBX1, and MEIS1: Interconnections and competition in cancer. BioEssays 39, (2017). [DOI] [PubMed] [Google Scholar]
- 22.Hisa T., et al., Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 23, 450–459 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machon O., Masek J., Machonova O., Krauss S., Kozmik Z., Meis2 is essential for cranial and cardiac neural crest development. BMC Dev. Biol. 15, 40 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azcoitia V., Aracil M., Martínez-A C., Torres M., The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev. Biol. 280, 307–320 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Antosova B., et al., The gene regulatory network of lens induction is wired through Meis-dependent shadow enhancers of Pax6. PLoS Genet. 12, e1006441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heine P., Dohle E., Bumsted-O’Brien K., Engelkamp D., Schulte D., Evidence for an evolutionary conserved role of homothorax/Meis1/2 during vertebrate retina development. Development 135, 805–811 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Marcos S., et al., Meis1 coordinates a network of genes implicated in eye development and microphthalmia. Development 142, 3009–3020 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Klimova L., Lachova J., Machon O., Sedlacek R., Kozmik Z., Generation of mRx-Cre transgenic mouse line for efficient conditional gene deletion in early retinal progenitors. PLoS One 8, e63029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das G., Choi Y., Sicinski P., Levine E. M., Cyclin D1 fine-tunes the neurogenic output of embryonic retinal progenitor cells. Neural Dev. 4, 15 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martynoga B., Morrison H., Price D. J., Mason J. O., Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol. 283, 113–127 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Das G., Clark A. M., Levine E. M., Cyclin D1 inactivation extends proliferation and alters histogenesis in the postnatal mouse retina. Dev. Dyn. 241, 941–952 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai Z., et al., Deficient FGF signaling causes optic nerve dysgenesis and ocular coloboma. Development 140, 2711–2723 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penkov D., et al., Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Rep. 3, 1321–1333 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Aldiri I.et al.; St. Jude Children’s Research Hospital—Washington University Pediatric Cancer Genome Project , The dynamic epigenetic landscape of the retina during development, reprogramming, and tumorigenesis. Neuron 94, 550–568.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleinjan D. A., et al., Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum. Mol. Genet. 10, 2049–2059 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Kleinjan D. A., et al., Long-range downstream enhancers are essential for Pax6 expression. Dev. Biol. 299, 563–581 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kammandel B., et al., Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev. Biol. 205, 79–97 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Marquardt T., et al., Pax6 is required for the multipotent state of retinal progenitor cells. Cell 105, 43–55 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Okamoto R., Uchikawa M., Kondoh H., Sixteen additional enhancers associated with the chicken Sox2 locus outside the central 50-kb region. Dev. Growth Differ. 57, 24–39 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Furuta Y., Lagutin O., Hogan B. L., Oliver G. C., Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis 26, 130–132 (2000). [PubMed] [Google Scholar]
- 41.Islam M. M., Li Y., Luo H., Xiang M., Cai L., Meis1 regulates Foxn4 expression during retinal progenitor cell differentiation. Biol. Open 2, 1125–1136 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verma-Kurvari S., Savage T., Smith D., Johnson J. E., Multiple elements regulate Mash1 expression in the developing CNS. Dev. Biol. 197, 106–116 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Miesfeld J. B., et al., The Atoh7 remote enhancer provides transcriptional robustness during retinal ganglion cell development. Proc. Natl. Acad. Sci. U.S.A. 117, 21690–21700 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghiasvand N. M., et al., Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat. Neurosci. 14, 578–586 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bridoux L., et al., HOX paralogs selectively convert binding of ubiquitous transcription factors into tissue-specific patterns of enhancer activation. PLoS Genet. 16, e1009162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamachi Y., Kondoh H., Sox proteins: Regulators of cell fate specification and differentiation. Development 140, 4129–4144 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Hernandez J., et al., Highly conserved sequences mediate the dynamic interplay of basic helix-loop-helix proteins regulating retinogenesis. J. Biol. Chem. 282, 37894–37905 (2007). [DOI] [PubMed] [Google Scholar]
- 48.VandenBosch L. S., et al., Developmental changes in the accessible chromatin, transcriptome and Ascl1-binding correlate with the loss in Müller glial regenerative potential. Sci. Rep. 10, 13615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zibetti C., Liu S., Wan J., Qian J., Blackshaw S., Epigenomic profiling of retinal progenitors reveals LHX2 is required for developmental regulation of open chromatin. Commun. Biol. 2, 142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gordon P. J., et al., Lhx2 balances progenitor maintenance with neurogenic output and promotes competence state progression in the developing retina. J. Neurosci. 33, 12197–12207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy A., et al., LHX2 is necessary for the maintenance of optic identity and for the progression of optic morphogenesis. J. Neurosci. 33, 6877–6884 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Norrie J. L., et al., Nucleome dynamics during retinal development. Neuron 104, 512–528.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burmeister M., et al., Ocular retardation mouse caused by Chx10 homeobox null allele: Impaired retinal progenitor proliferation and bipolar cell differentiation. Nat. Genet. 12, 376–384 (1996). [DOI] [PubMed] [Google Scholar]
- 54.Trimarchi J. M., Cho S. H., Cepko C. L., Identification of genes expressed preferentially in the developing peripheral margin of the optic cup. Dev. Dyn. 238, 2327–2329 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu H., et al., Ciliary margin transdifferentiation from neural retina is controlled by canonical Wnt signaling. Dev. Biol. 308, 54–67 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Zhao S., Chen Q., Hung F. C., Overbeek P. A., BMP signaling is required for development of the ciliary body. Development 129, 4435–4442 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Diacou R., Zhao Y., Zheng D., Cvekl A., Liu W., Six3 and Six6 are jointly required for the maintenance of multipotent retinal progenitors through both positive and negative regulation. Cell Rep. 25, 2510–2523.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heavner W. E., Andoniadou C. L., Pevny L. H., Establishment of the neurogenic boundary of the mouse retina requires cooperation of SOX2 and WNT signaling. Neural Dev. 9, 27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsushima D., Heavner W., Pevny L. H., Combinatorial regulation of optic cup progenitor cell fate by SOX2 and PAX6. Development 138, 443–454 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jolma A., et al., DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527, 384–388 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Zhu C. C., et al., Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development 129, 2835–2849 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Schwarz M., et al., Spatial specification of mammalian eye territories by reciprocal transcriptional repression of Pax2 and Pax6. Development 127, 4325–4334 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Bertuzzi S., Hindges R., Mui S. H., O’Leary D. D., Lemke G., The homeodomain protein vax1 is required for axon guidance and major tract formation in the developing forebrain. Genes Dev. 13, 3092–3105 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu C., Bakeri H., Li T., Swaroop A., Regulation of retinal progenitor expansion by Frizzled receptors: Implications for microphthalmia and retinal coloboma. Hum. Mol. Genet. 21, 1848–1860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petros T. J., Williams S. E., Mason C. A., Temporal regulation of EphA4 in astroglia during murine retinal and optic nerve development. Mol. Cell. Neurosci. 32, 49–66 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Oster S. F., Bodeker M. O., He F., Sretavan D. W., Invariant Sema5A inhibition serves an ensheathing function during optic nerve development. Development 130, 775–784 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Dakubo G. D., et al., Retinal ganglion cell-derived sonic hedgehog signaling is required for optic disc and stalk neuroepithelial cell development. Development 130, 2967–2980 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Liu H., Mohamed O., Dufort D., Wallace V. A., Characterization of Wnt signaling components and activation of the Wnt canonical pathway in the murine retina. Dev. Dyn. 227, 323–334 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Ladam F., et al., TALE factors use two distinct functional modes to control an essential zebrafish gene expression program. eLife 7, e36144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bessa J., Gebelein B., Pichaud F., Casares F., Mann R. S., Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 16, 2415–2427 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Agoston Z., et al., Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development 141, 28–38 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Thein T., et al., Control of lens development by Lhx2-regulated neuroretinal FGFs. Development 143, 3994–4002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thompson H., Camand O., Barker D., Erskine L., Slit proteins regulate distinct aspects of retinal ganglion cell axon guidance within dorsal and ventral retina. J. Neurosci. 26, 8082–8091 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith J. N., et al., Lens-regulated retinoic acid signalling controls expansion of the developing eye. Development 145, dev167171 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Rowan S., Chen C. M., Young T. L., Fisher D. E., Cepko C. L., Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development 131, 5139–5152 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Horsford D. J., et al., Chx10 repression of Mitf is required for the maintenance of mammalian neuroretinal identity. Development 132, 177–187 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Bosze B., Moon M. S., Kageyama R., Brown N. L., Simultaneous requirements for Hes1 in retinal neurogenesis and optic cup-stalk boundary maintenance. J. Neurosci. 40, 1501–1513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murali D., Kawaguchi-Niida M., Deng C. X., Furuta Y., Smad4 is required predominantly in the developmental processes dependent on the BMP branch of the TGF-β signaling system in the embryonic mouse retina. Invest. Ophthalmol. Vis. Sci. 52, 2930–2937 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu J., Wilson S., Reh T., BMP receptor 1b is required for axon guidance and cell survival in the developing retina. Dev. Biol. 256, 34–48 (2003). [DOI] [PubMed] [Google Scholar]
- 80.Liu H., Thurig S., Mohamed O., Dufort D., Wallace V. A., Mapping canonical Wnt signaling in the developing and adult retina. Invest. Ophthalmol. Vis. Sci. 47, 5088–5097 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ChIP-seq data have been deposited in the ArrayExpress database (accession nos. E-MTAB-9143 and E-MTAB-10112). Dataset S1 is available as part of the submission. All other study data are included in the article and supporting information.