Abstract
A comprehensive understanding of foodborne pathogen diversity in pre-harvest environments is necessary to effectively track pathogens on farms and identify sources of produce contamination. As such, this study aimed to characterize Listeria diversity in wildlife feces and agricultural water collected from a New York State produce farm over a growing season. Water samples were collected from a pond (N=80) and stream (N=52). Fecal samples (N=77) were opportunistically collected from areas <5m from the water sources; all samples were collected from a <0.5km2 area. Overall, 41% (86/209) and 24% (50/209) of samples were positive for Listeria monocytogenes and Listeria spp. (excluding L. monocytogenes), respectively. For each positive sample, one L. monocytogenes or Listeria spp. isolate was speciated by sequencing the sigB gene, which allowed for additional characterization based on the sigB allelic type (AT). The 86 L. monocytogenes and 50 Listeria spp. isolates represented 8 and 23 different ATs, respectively. A subset of L. monocytogenes isolates (N=44) from pond water and pond-adjacent feces (representing a ~5,000m2 area) were further characterized by PFGE; these 44 isolates represented 22 PFGE types, which is indicative of considerable diversity at a small spatial scale. Ten PFGE types were isolated more than once, suggesting persistence or re-introduction of PFGE types in this area. Given the small spatial scale, the prevalence of L. monocytogenes and Listeria spp., as well as the considerable diversity amongst isolates, suggests traceback investigations may be challenging. For example, traceback of finished product or processing facility contamination with specific subtypes to pre-harvest sources may require collection of large sample sets, and characterization of a considerable number of isolates. Our data also support the adage, “absence of evidence does not equal evidence of absence” applies to L. monocytogenes traceback efforts at the pre-harvest level.
Keywords: Listeria, sigB, PFGE, feces, agricultural water
It is estimated that Listeria monocytogenes is responsible for approximately 1,600 illnesses, 1,455 hospitalizations, and 255 deaths annually in the United States (31), and several high-profile outbreaks and recalls have been traced to L. monocytogenes contamination of fresh produce (8, 24, 44). For example, a 2011 outbreak was traced back to L. monocytogenes contamination of cantaloupes that resulted in 147 illnesses and 33 deaths (2). Additionally, there have been >30 recalls since 2016 due to L. monocytogenes contamination of produce and ready-to-eat produce products (36). While the majority of produce outbreaks and recalls due to L. monocytogenes contamination are traced back to the processing environment, L. monocytogenes can be introduced, or re-introduced, in the processing environment from the pre-harvest environment [e.g., on produce, in soil, on harvesting equipment, or on workers shoes; (32)] or could be introduced onto product in the field and remain on product until consumption.
Following detection of a Listeria-positive sample during routine monitoring or an outbreak investigation, processors or investigators often conduct extensive sampling to identify potential contamination sources (i.e., traceback analysis). Identifying how a Listeria isolate was introduced onto product or into the processing environment is a critical step in performing a root cause analysis and establishing effective corrective actions (to control potential contamination pathways). To determine if an isolate originated in the pre-harvest environment, investigators and operators must collect environmental samples from farms that supply the product. However, the ubiquity of Listeria in the environment (1, 7, 12, 11) may make it challenging to prove a causal relationship between Listeria isolated from the pre-harvest environment and Listeria isolated from post-harvest environments and/or product. Previous studies of pre-harvest environments isolated Listeria from soil (3, 25, 35, 34, 38, 39), wildlife feces (5, 19, 37, 39), agricultural water (1, 6, 17, 20, 33), and biological soil amendments (25). As such, a more comprehensive understanding of the prevalence, diversity, and distribution of Listeria on produce farms is needed to better understand on-farm sources of Listeria, understand transfer of Listeria between sample types on produce farms, and facilitate traceback analysis. While some studies have investigated the diversity of Listeria in pre-harvest produce environments (3, 19, 20, 33, 34, 35), these studies have examined diversity over large geographic areas. For instance, Chapin et al. (3) and Strawn et al. (34) investigated Listeria diversity on 5 New York State (US) farms located up to 205 km from each other, while Stea et al. (33) examined Listeria diversity within an agricultural watershed in Nova Scotia, Canada with a drainage area of 135 km2. As there is limited data on Listeria diversity at small spatial scales (e.g., within individual farms), we aimed, in this study, to address this knowledge gap by examining the diversity of Listeria on a single produce farm.
Agricultural water and wildlife intrusion in produce fields have been identified as key sources and pathways for dispersal of Listeria contamination in pre-harvest environments (14, 26, 37, 38). Multiple studies have isolated L. monocytogenes from agricultural water and wildlife feces collected in produce growing environments (6, 7, 19, 25, 34, 38). For instance, Weller et al. (38) isolated L. monocytogenes from 45% (N=11) of wildlife feces collected on 10 produce farms in New York State. Falardeau et al. (7) isolated L. monocytogenes from 10% (N=223) of agricultural water samples collected from 2 watersheds in British Colombia, Canada. Furthermore, Cooley et al. (6) isolated L. monocytogenes from 43% (N=1405) of water samples collected from the California Central Coast agricultural region. As such, the primary objective of the study reported here was to characterize and compare the L. monocytogenes and Listeria spp., excluding L. monocytogenes, diversity in agricultural water and wildlife feces collected from a single produce farm in New York State.
MATERIALS AND METHODS
Study design and sample collection.
The study reported here was conducted between May and July 2014 on a produce farm in the Finger Lakes region of New York State. Two separate irrigation water sources are located on the farm: (i) a pond and (ii) a stream. The water sources are within a rectangular 0.5km2 (124 acres) area; the sampling sites in the pond and stream are approx. 775 m (2,546 ft) apart.
Agricultural water and fecal samples were collected on 26 different days. Fifty-two and 80 water samples were collected from the stream and pond, respectively; the Listeria prevalence and number of sigB allelic types for the 52 stream samples and the 13 fecal samples collected near the stream were previously reported by Weller et al. (39). However, in order to better understand the Listeria diversity on the studied farm, this data was also included here. All water samples were collected as described in Weller et al. (39). Briefly, 250 mL of water were collected into a sampling cup (Nalgene, Rochester, NY) using a sampling pole and stored on ice until processing. Fecal samples (N=77) located within 5m of the pond (N=64) or stream (N=13) were collected opportunistically, as previously described (39); data for the fecal samples collected near the pond were not previously reported. A 5m buffer around the pond and stream was used for practical reasons. All samples were stored at 4°C and processed within 3h of collection.
Listeria enrichment and isolation.
All samples were processed as previously described (39). Briefly, 10 g of the fecal samples were aliquoted and transferred to separate Whirl-Pak bags (Nasco, Fort Atkinson, WI). The water samples were each passed through separate 0.45 μm filters, and each filter was transferred to a separate Whirl-Pak bag. Each sample was enriched in 90 mL of Buffered Listeria Enrichment Broth (BLEB; Becton Dickinson, Franklin Lakes, NJ) at 30°C, and after 4h, Listeria Selective Enrichment Supplement (Oxoid, Cambridge, UK) was added to each sample. Each sample was incubated for a total of 48h at 30°C. After 24h and 48h of incubation, the enrichments were sub-streaked onto Modified Oxford Agar (MOX; Benton Dickinson) and Listeria monocytogenes Plating Medium (LMPM; Biosynth International, Itasca, IL). MOX plates were incubated at 30°C and LMPM plates were incubated at 35°C for 48h. After incubation, characteristic Listeria colonies on 24 and 48h MOX plates (i.e., grey, donut-shaped colonies) were streaked onto LMPM for isolation and incubated at 35°C for 48h. One colony per presumptive Listeria-positive LMPM plate (up to 4 colonies total; 1 from the 24h LMPM plate, 1 from the 48h LMPM plate, 1 from the LMPM plate sub-streaked from the 24h MOX plate, and 1 from the LMPM plate sub-streaked from the 48h MOX plate) was selected, sub-streaked onto brain heart infusion agar (BHI, Becton Dickinson), and incubated at 37°C for 24h. If blue colonies were present (characteristic of L. monocytogenes, L. ivanovii, hemolytic L. innocua due to phospholipase C action, 29) on a given plate, they were preferentially selected to be sub-streaked on BHI. If a blue colony was not present, a round white colony was selected to be sub-streaked on BHI; this colony phenotype is characteristic of Listeria species other than L. monocytogenes, L. ivanovii, and hemolytic L. innocua. If no round white or blue colonies were present on any of the LMPM plates, the sample was considered negative for Listeria. If a round white or blue colony was present, sigB PCR was performed as a confirmation step and speciation of Listeria, as described below.
sigB sequencing and allelic typing.
All isolates per presumptive-positive Listeria sample (i.e., up to 4 isolates per sample) were confirmed as Listeria via PCR amplification of sigB as described by Nightingale et al. (22); all samples with presumptive-positive Listeria isolates in this study were confirmed as positive via sigB PCR. Sequencing of the sigB gene was then performed on 1 isolate per positive sample for further characterization; if the colony characterized was identified as L. monocytogenes the sample was considered L. monocytogenes positive, if the colony characterized was identified as Listeria spp. other than L. monocytogenes, then sample was considered Listeria spp. positive. While only subtyping 1 isolate per sample may lead to an underestimation of the true diversity present in the sample types investigated here, this approach allowed us to get a preliminary estimate of the diversity present. There were 22 samples with both blue and white colonies present on LMPM plates (out of 209 total samples); for 21 of these samples the blue colony was selected for characterization (2/3 fecal samples, 8/8 stream samples, and 11/11 pond samples). Since only L. monocytogenes was selected for PFGE, preferentially selecting these blue colonies provided us with the larger sample size for further subtyping. A BLAST search was performed against an internal database (http://www.foodmicrobetracker.com/login/intro.aspx) to determine its sigB allelic type (AT) of each sequence. All isolates were archived as 15% glycerol stocks and stored at −80°C. A sigB AT phylogeny was constructed based on the 660 bp nucleotide sequence of the sigB gene using the maximum likelihood method in MEGA7 version 7.0.26; the nucleotide substitution Tamura 3-parameter model was used, and 1,000 bootstraps were performed.
Pulsed field gel electrophoresis.
All confirmed L. monocytogenes isolates from the pond and the feces collected near the pond (N=44) were characterized by pulsed field gel electrophoresis (PFGE). Only isolates from pond water and feces were characterized by PFGE as a subset analysis to investigate diversity over a small spatial scale (i.e., the approx. 5,000m2 around the pond). PFGE was performed using AscI and ApaI restriction enzymes, according to the CDC PulseNet protocol (10).
Statistical analysis.
All statistical analyses were performed using R version 3.5.1 (27). Chi-square tests were used to determine if L. monocytogenes and Listeria spp. (excluding L. monocytogenes) prevalence differed between sample types. The Bonferroni correction was used to account for the 3 comparisons made for Listeria spp. (excluding L. monocytogenes) and the 3 comparisons made for L. monocytogenes (pond vs. fecal, pond vs. stream, fecal vs. stream; α=0.05/3=0.0167). While separate data for the feces collected near the pond and feces collected near the stream are provided in this study, these sample types were combined into a single fecal sample category for analysis due to the small number of fecal samples collected near the stream (N=13 of 77 total fecal samples). To determine the number of unique ATs and PFGE types, AT richness and PFGE type richness were estimated using the breakaway package version 4.6.7 (43), which calculates the transformed species richness estimation with 95% confidence intervals as outlined in Rocchetti et al. (28) and Willis and Bunge (42). To estimate AT and PFGE type diversity for each sample type, Simpsons Index of Diversity was calculated using the vegan package version 2.5–3 (23); 95% confidence intervals (95% CI) were calculated for the Simpson’s Index of Diversity, according to the methods described by Hunter and Gaston (13). The effective number of subtypes was also calculated to communicate Simpson’s Index of Diversity in a more intuitive form (i.e., the estimated number of subtypes present in a sample type given a certain value of Simpson’s Index of Diversity; 15).
RESULTS AND DISCUSSION
Seek and you shall find.
If a reasonably large number of samples are collected from the pre-harvest environment in New York State and tested for Listeria (“seek”), it is likely Listeria will be “found.” The overall prevalence of Listeria in the study reported here was 65% (136/209); the prevalence of Listeria spp. (excluding L. monocytogenes) was 24% (50/209) and the prevalence of L. monocytogenes was 41% (86/209; Table 1). While the Listeria spp. (excluding L. monocytogenes) prevalence in the pond (31%; 25/80) and stream samples (27%; 14/52) were not significantly different (P=0.735), the L. monocytogenes prevalence in the pond (39%; 31/80) and stream samples (63%; 33/52) were significantly different even after the Bonferroni correction (P=0.009; Table 1). The frequency of Listeria isolation from agricultural water has been reported in several previous studies conducted in New York State (3, 34, 35, 38, 40) and other produce-growing regions (1, 6, 7, 18, 33). In previous New York State studies, the L. monocytogenes prevalence ranged between 9% (38) and 59% (35) in pond samples, and between 10% (40) and 50% (38) in stream samples. As such, the frequency of L. monocytogenes isolation from pond samples in the current study falls within the range reported in the literature, while the frequency of L. monocytogenes isolation from the stream samples in the current study is higher than in previous studies. The higher L. monocytogenes prevalence in the stream sampled here, compared to those sampled in other studies, may be due to presence of two dairy operations immediately upstream of the sampling sites along the stream. Additionally, run-off from the dairy operations could explain the significantly higher L. monocytogenes prevalence in the stream compared to the pond. Previous studies have indicated upstream livestock operations are associated with an increased prevalence of L. monocytogenes in stream water (7, 19, 41). Furthermore, another study suggested cattle may increase the prevalence and may distribute L. monocytogenes in the farm environment (22). Of note, the current study only tested 250 mL of water per sample, whereas other studies collected upwards of 1 L per sample. As such, the prevalence reported for the pond and stream investigated in the current study may be an underestimation of the true prevalence. Regardless, while the current study supports agricultural water as a source of L. monocytogenes in the New York State preharvest produce environment, our findings also suggest future studies on agricultural water need to consider the impact of spatiotemporal factors (e.g., upstream land use) on downstream microbial water quality.
Table 1.
Frequency and prevalence of Listeria spp. and L. monocytogenes in fecal and agricultural water samples.
| Sample Typeb | Location | No. of Samples | No. of samples positive for (Prevalence in %) |
||||
|---|---|---|---|---|---|---|---|
| Listeria spp.a | L. innocua | L. seeligeri | L. welshimeri | L. monocytogenes | |||
| Fecal | - | 77 | 11 (14) | 6 (8) | 4 (5) | 1 (1) | 22 (29) |
| Near Pond | 64 | 9 (14) | 4 (6) | 4 (6) | 1 (2) | 13 (20) | |
| Near Stream | 13 | 2 (15) | 2 (15) | 0 (0) | 0 (0) | 9 (69) | |
| Water | - | 132 | 39 (30) | 14 (11) | 20 (15) | 5 (4) | 64 (48) |
| Pond | 80 | 25 (31) | 8 (10) | 15 (19) | 2 (3) | 31 (39) | |
| Stream | 52 | 14 (27) | 6 (12) | 5 (10) | 3 (6) | 33 (63) | |
Listeria spp. includes L. innocua, L. seeligeri, and L. welshimeri.
In addition to the water samples collected from the pond water, one sediment sample was also collected and tested positive for L. innocua.
The prevalence of Listeria spp. (excluding L. monocytogenes) and L. monocytogenes in the fecal samples collected here were 14% (11/77) and 29% (22/77), respectively (Table 1). Based on fecal shape and consistency, the majority of the fecal samples appeared to be produced by Canadian geese (Branta canadenesis; N=71). This is logical given the large flock of Canadian geese that nested in the wetlands adjacent to the pond throughout the study. The remainder of the feces were produced by canids (N=5) and an unknown animal (N=1). The Listeria spp. (excluding L. monocytogenes) prevalence was 13% (9/71) in the goose and 40% (2/5) in the canid fecal samples. The L. monocytogenes prevalence was 28% (20/71) in the goose and 40% (2/5) in the canid fecal samples. Neither Listeria spp. (excluding L. monocytogenes), nor L. monocytogenes, were isolated from the one unidentified fecal sample. While multiple studies have isolated Listeria from wildlife feces (5, 19, 25, 34, 37, 38, 39), relatively few studies have examined the prevalence of L. monocytogenes in Canadian goose feces, specifically (5, 19). The L. monocytogenes prevalence in the goose fecal samples collected as part of the current study was greater compared to previous studies (5, 19). For instance, a study conducted in Ontario, Canada, did not detect L. monocytogenes in any of the 18 goose fecal samples collected (19), while Converse et al. (5) isolated Listeria spp. from 10% (47/495) of goose fecal samples. Although, the prevalence of L. monocytogenes in wildlife fecal samples varied between studies, our findings support previous studies’ findings [e.g., 33, 36, 37] that wildlife can act as source of L. monocytogenes in pre-harvest environments.
Absence of evidence does not equal evidence of absence.
The composition of the Listeria population in the agricultural water and fecal samples was characterized using sigB allelic typing. Allelic typing was used to speciate the isolates and provide a preliminary estimate of diversity. A total of 136 isolates were selected for sigB allelic typing (i.e., one isolate per Listeria-positive sample); L. monocytogenes isolates were preferentially selected for subtyping based on colony morphology (blue colonies on LMPM). Of the 136 isolates, 20, 24, 6, and 86 isolates were identified as L. innocua, L. seeligeri, L. welshimeri, and L. monocytogenes, respectively. We identified 15 sigB ATs among the 50 Listeria spp. (excluding L. monocytogenes) isolates (approx. 1 unique AT per 3 isolates) and 8 ATs among the 86 L. monocytogenes isolates (approx. 1 unique AT per 11 isolates; Tables 2 & 3; Table S1). In addition, we estimated two diversity parameters, richness and the Simpson’s Index for Listeria spp. (excluding L. monocytogenes; Table 2) and L. monocytogenes (Table 3) in the pond, stream, and fecal samples. For example, the estimated L. monocytogenes AT richness for the pond water, stream water and fecal samples were 8 (95% CI= 7, 15), 9 (95% CI= 6, 73) and 5 (95% CI= 5, 8) ATs, respectively, indicating the presence of a considerable diversity on the farm investigated. However, it should be noted that this may be an underestimation of the true diversity present in these sample types on this produce farm, as only one isolate per sample type was characterized by allelic typing. Even with this limitation our data indicate considerable Listeria diversity in the small area of the produce farm investigated here. The proportion of ATs identified relative to the total number of isolates subtyped in the current study was similar to the proportion reported by previous studies conducted on New York State produce farms (3, 34). For example, applying the same analysis as in the current study, Chapin et al. identified 50 sigB ATs among 186 Listeria spp. (excluding L. monocytogenes) isolates [(approx. 1 AT per 4 isolates), (3)] and Strawn et al. identified 12 sigB ATs among 107 L. monocytogenes isolates [(approx. 1 AT per 8 isolates), (34)]; both studies isolated Listeria from soil, drag swabs, water, and fecal samples collected from the five produce farms across New York State. Since all samples collected in the study reported here were from a <0.5 km2 area, compared to past studies which sampled larger areas (between 33 and 205 km apart), the similarity in the ratio of unique ATs relative to the total number of isolates subtyped in this and these previous studies (3, 34) provides Listeria richness may be similar across spatial scales. However, additional research is required to confirm this finding, especially in other geographical areas.
Table 2.
Diversity of Listeria spp. sigB allelic types (AT) in agricultural water and fecal samples.
| Sample Type | No. of Listeria spp.a | No. of ATs | Estimated Subtype Richness (95% CI)b,c | Simpson’s Index (95% CI)c | Effective No. of ATd | |
|---|---|---|---|---|---|---|
| Fecal | - | 11 | 8 | 35 (8, 2871) | 0.84 (0.75, 0.94) | 6 |
| Water | - | 39 | 14 | 23 (14, 259) | 0.84 (0.76, 0.92) | 6 |
| Pond | 25 | 11 | 32 (11, 1445) | 0.79 (0.67, 0.92) | 5 | |
| Stream | 14 | 8 | 11 (8, 39) | 0.84 (0.76, 0.91) | 6 | |
Excluding L. monocytogenes
Estimated using weighted linear regression
95% CI: 95% confidence interval
Effective number of AT is calculated based on Simpson’s Index of Diversity
Table 3.
Diversity of Listeria monocytogenes sigB allelic types and pulsotypes in agricultural water and fecal samples.
| Sample Type | Location | No. of L. monocytogenes | Subtyping Methoda | No. of Subtypes | Estimated Subtype Richness (95% CI)b,c | Simpson’s Index (95% CI)c | Effective No. of Subtyped |
|---|---|---|---|---|---|---|---|
| Fecal | - | 22 | AT | 5 | 5 (5, 8) | 0.63 (0.46, 0.80) | 3 |
| Near Pond | 13 | PT | 7 | 9 (7, 33) | 0.78 (0.63, 0.93) | 5 | |
| Water | - | 64 | AT | 7 | 7 (7, 8) | 0.54 (0.41, 0.68) | 2 |
| Stream | 33 | AT | 6 | 9 (6, 73) | 0.41 (0.20, 0.62) | 2 | |
| Pond | 31 | AT | 7 | 8 (7, 15) | 0.66 (0.49, 0.82) | 3 | |
| Pond | 31 | PT | 19 | 29 (19, 207) | 0.93 (0.91, 0.95) | 15 |
AT = allelic type; PT = PFGE type. PFGE was only performed on Listeria monocytogenes isolates and not on non-pathogenic Listeria species isolates.
Estimated using weighted linear regression
95% CI: 95% confidence interval
The effective number of subtype is calculated based on Simpson’s Index of Diversity.
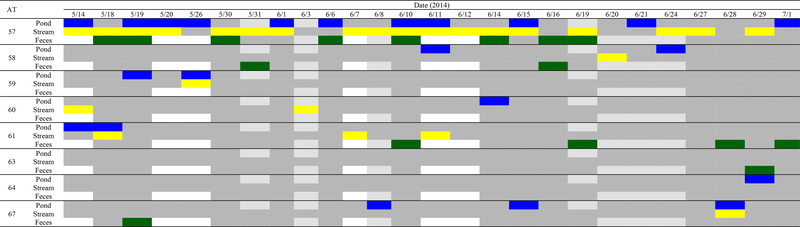
Furthermore, multiple Listeria (including L. monocytogenes) sigB ATs were isolated from a single sample type on a given day on multiple occasions. In the pond, stream, and fecal samples there were an average of 1.9, 1.5, and 1.7 Listeria (including L. monocytogenes) sigB ATs isolated per sampling day, respectively. There was also a maximum of 3, 2, and 4 Listeria (including L. monocytogenes) sigB ATs isolated per sampling day from the pond, stream, and fecal samples, respectively, indicating considerable Listeria diversity in the area sampled even on a given day. Six of the 8 L. monocytogenes sigB ATs were isolated from at least one sample type on multiple dates (see Figure 1, which shows the isolation of L. monocytogenes sigB ATs over time; equivalent data for Listeria spp. [excluding L. monocytogenes] are shown in Figure S1). Most notably, AT 57 was isolated from all three sample types (i.e., pond water, stream water, and feces) on multiple occasions, and was isolated from at least one sample type on 23 of the 26 sampling dates. AT 57 represents 13 PFGE types (Figure 2), consistent with previous reports that PFGE is considerably more discriminatory then sigB allelic typing (4). The high prevalence of a single AT (i.e., AT 57) makes it likely that less prevalent ATs are not isolated, as for example, AT 57 may outcompete other ATs. In addition, the considerable AT diversity observed also make it likely that samplings that do not include large sample numbers will not detect and isolate some Listeria sigB ATs. Hence, traceback investigations could mistakenly conclude that a given Listeria subtype is not present in a given pre-harvest environment, particularly in environments similar to the one investigated here.
Figure 1.
Date of L. monocytogenes sigB AT isolation from water and fecal samples. In total, 31, 33, and 22 isolates from pond, stream, and fecal samples were allelic typed, respectively. The colored squares indicate at least one isolate of the given AT was isolated on the given date from pond  , stream
, stream  , or fecal
, or fecal  samples; dates when a sample was collected, but the specified AT was not isolated are represented by
samples; dates when a sample was collected, but the specified AT was not isolated are represented by  , and dates when a sample was collected but was negative for Listeria are represented by
, and dates when a sample was collected but was negative for Listeria are represented by  . At each sampling event, 3 pond and 3 stream samples were collected. Fecal samples were collected opportunistically (i.e., when feces were present). If no fecal samples were collected, the square is white. The dates of Listeria spp. (excluding L. monocytogenes) sigB AT isolation is shown in Figure S1.
. At each sampling event, 3 pond and 3 stream samples were collected. Fecal samples were collected opportunistically (i.e., when feces were present). If no fecal samples were collected, the square is white. The dates of Listeria spp. (excluding L. monocytogenes) sigB AT isolation is shown in Figure S1.
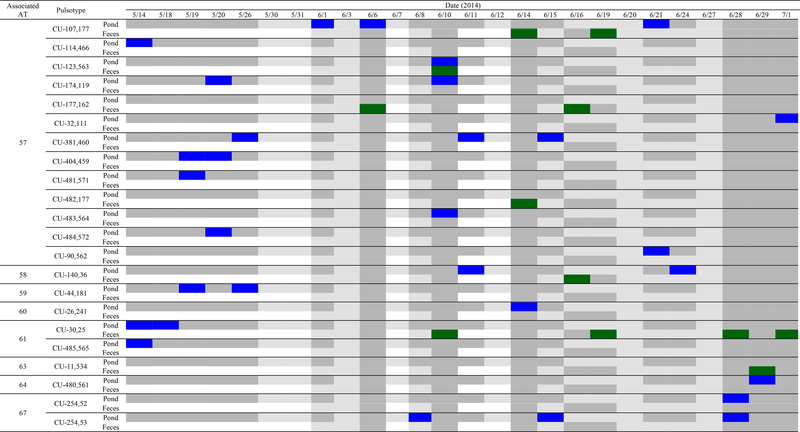
Figure 2.
Date of L. monocytogenes PFGE type isolation from pond and fecal samples collected near the pond. In total, PFGE was performed on 31 and 13 isolates from pond and fecal samples, respectively. The colored squares indicate at least one isolate of the given PFGE type was isolated on the given date from pond  or fecal
or fecal  samples; dates when a sample was collected, but the specified PFGE type was not isolated are represented by
samples; dates when a sample was collected, but the specified PFGE type was not isolated are represented by  , and dates when a sample was collected but was negative for L. monocytogenes are represented by
, and dates when a sample was collected but was negative for L. monocytogenes are represented by  . At each sampling event, 3 pond samples were collected. Fecal samples were collected opportunistically (i.e., when feces were present). If no fecal samples were collected, the square is white.
. At each sampling event, 3 pond samples were collected. Fecal samples were collected opportunistically (i.e., when feces were present). If no fecal samples were collected, the square is white.
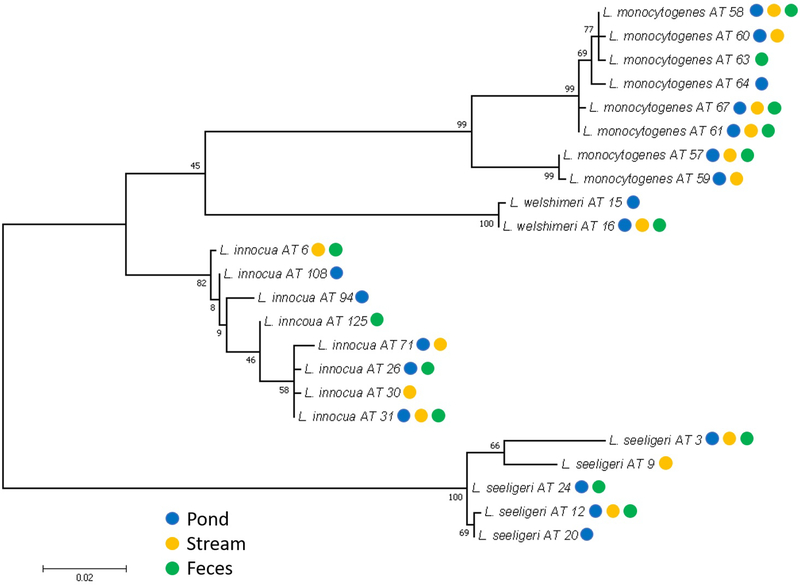
Additionally, a sigB AT phylogeny was constructed to investigate if there was any relationship between sigB AT isolation and sample type (Figure 3). The phylogeny suggests that neither specific sigB ATs nor specific sigB clades are associated with a given sample type. A Fisher’s exact test also found no significant relationship between sigB AT and sample type (P=0.30). While our data could also be interpreted as suggesting that none of the ATs isolated in this study have an enhanced fitness in the different habitats sampled here, future studies with larger data sets are needed to further probe niche adaptation of different Listeria species and clades.
Figure 3.
Maximum likelihood phylogeny of Listeria isolated from pond, stream, and fecal samples from a single produce farm in New York State based on a 660 bp fragment of the sigB gene. Values on the branches are bootstrap values from 1,000 replicate trees. The scale displayed below the tree represents the genetic distance shown in the branch length.
In addition to characterization by sigB AT, we performed PFGE on the L. monocytogenes isolates from the pond (N=31) and feces collected near the pond (N=13) to better understand the diversity of the isolates at a very small spatial scale (approx. 5,000 m2). We identified 19 PFGE types among the 31 L. monocytogenes isolates obtained from pond water samples (approx. 1 PFGE type per 2 isolates) and 7 PFGE types among the 13 L. monocytogenes isolates obtained from fecal samples (approx. 1 PFGE type per 2 isolates). The estimated PFGE type richness for L. monocytogenes isolated from the pond water and fecal samples were 29 (95% CI= 19, 207) and 9 (95% CI= 7, 33) PFGE types, respectively (Table 3). Two concurrent studies conducted in a 200 km2 agricultural watershed in Ontario, Canada also performed PFGE on L. monocytogenes isolates from agricultural water and wildlife feces (19, 20); these studies used the same PFGE protocol as the current study (10). In the two Canadian studies, Lyautey et al., (19, 20) identified 21 PFGE types from 75 L. monocytogenes isolates from water samples (approx. 1 PFGE type per 3 isolates) and 18 PFGE types from 84 L. monocytogenes isolates from fecal samples (approx. 1 PFGE type per 5 isolates). As such, the ratio of PFGE types identified relative to the number of L. monocytogenes isolates subtyped in the current study, which was conducted in a 40,000-fold smaller area, was larger than the ratio found in the Canadian studies (19, 20) for both the fecal samples and water samples. While this may suggest the L. monocytogenes population in the Finger Lakes region of New York is more diverse than Ontario, Canada, the greater diversity on the farm sampled in the study reported here may also be due to factors unique to the sampled farm. For example, since two dairy farms are located uphill of the sampled farm, continuous introduction of L. monocytogenes from the dairies may have resulted in a more diverse population of L. monocytogenes, compared to the other areas. To identify potential drivers of the greater PFGE type diversity observed in the study reported here, compared to Lyautey et al., a systematic survey of Listeria populations across and within regions is needed. Regardless, the diversity of Listeria identified in this study, and other studies, suggests it often may be difficult to determine if an isolate found in a processing environment originated in a particular pre-harvest environment, as multiple subtypes may be present in a single environment. In fact, in order to either rule in or out any particular source in an environment with a considerable diversity, such as the one investigated in the current study, a large number of samples would have to be collected and a large number of isolates would have to be subtyped, including multiple isolates per sample, as previously suggested (9, 30). Consequently, the “absence of evidence” for presence of a given Listeria subtype (i.e., the Listeria subtype was not detected in any of the samples collected) in the pre-harvest environment in New York State and other pre-harvest environments with prevalent Listeria does not necessarily provide evidence that the given subtype is truly absent in this environment, particularly if sample size is small.
Should I stay or should I go.
The L. monocytogenes PFGE data collected here indicate the survival (“stay”) or continuous re-introduction (“go”) of L. monocytogenes into the farm environment. Specifically, 8 of the 19 PFGE types isolated from the pond and 3 of the 7 PFGE types isolated from the feces collected near the pond were isolated on more than one date (Figure 2). The repeated isolation of multiple PFGE types suggests these PFGE types were surviving or were continuously re-introduced in these sample types. For example, PFGE type CU-30,25 was isolated a total of 8 times in the study reported here, making it the most commonly isolated PFGE type (Figure 2). This could indicate PFGE type CU-30,25 has an increased fitness in this environment or there is a source of this PFGE type continuously contaminating the environment. Additionally, 4 out of the 22 PFGE types identified here were isolated from both the pond and feces collected near the pond (Figure 2). Lyautey et al. (19, 20) found similar results in two of their studies, as 5 of the 18 PFGE types from wildlife feces in the South Nation watershed in Ontario, Canada, were also isolated in water samples collected from the same watershed. Since the same PFGE types can be found in agricultural water and feces collected in proximity of the sampled water in both studies, this supports transfer of L. monocytogenes between feces and agricultural water, and/or a common source leading to the contamination of both sample types in the investigated environment.
Work smarter not harder.
Allelic typing of the sigB gene has been used to speciate Listeria isolates and can provide initial characterization of Listeria strains present (16), but is not of high enough discriminatory power to elucidate the actual diversity of a Listeria population in an environment. For example, in the current study AT 57 represents 13 different PFGE types, AT 61 represents 2 different PFGE types and AT 67 represents 2 different PFGE types (Figure 2). Strawn et al. (34) also performed PFGE on L. monocytogenes isolates with matching sigB ATs from a single sample type in a survey of soil, feces, drag swabs, and water samples from 5 farms in New York State (i.e., repeatedly isolated sigB ATs). For 3 out of the 4 repeatedly isolated sigB ATs, they found multiple PFGE types corresponding to each individual AT. PFGE hence clearly provides improved discriminatory power (over sigB allelic typing), facilitating investigations of persistence, transfer between sample types, and continuous re-introduction (4). Interestingly, in the study reported here, all isolates of the same PFGE type also had the same AT (Figure 2). For instance, all isolates of PFGE type CU-371,460 were AT 57, all isolates of PFGE type CU-107,177 were AT 57, all isolates of PFGE type CU-254,53 were AT 67, etc. This finding has not been directly reported in other studies, however, Liao et al. (16) have determined the sigB gene is genetically stable, as it is not a frequent site of homologous recombination or positive selection. As such, we expect this relationship between sigB ATs and PFGE types to hold true across different environments. While sigB allelic typing is not as discriminatory as PFGE, it can still be useful in performing traceback analysis, as it can be used as an initial screen to determine which isolates should be selected for PFGE, or other fine resolution typing methods (e.g., whole genome sequencing, WGS) during a traceback investigation. As a result, to determine which isolates from an environment match an isolate from another environment (e.g., processing environment), allelic typing should first be performed (“work smarter”). Then, isolates from the pre-harvest environment that have the same allelic type as the isolate from the processing environment should be characterized by PFGE or WGS (“work harder”) to determine which are truly the same. This will save time and money, as PFGE and WGS are considerably more expensive and time intensive to perform.
Overall, the data presented here and in previous studies (39) indicate Listeria is prevalent in the New York State pre-harvest produce environment, and as a result, it is likely for Listeria from the pre-harvest environment to contaminate produce, and/or be introduced to the processing environment (from the farm). The considerable Listeria diversity observed in the small area within the produce farm studied, and the evidence of transfer between sample types or continuous re-introduction of Listeria, indicate that it often may be difficult to successfully trace isolates from finished products or the processing environment (e.g., environmental swab) back to farm sources (e.g., feces, water), unless a large number of samples are collected and a large number of isolates are subtyped. However, it should be noted that these conclusions may be limited to similar preharvest produce environments that are continuously contaminated with livestock run-off. As such, additional research is needed to determine if the findings reported as part of this pilot study hold true for other produce farms and produce growing regions.
Supplementary Material
HIGHLIGHTS.
There is considerable Listeria diversity in the farm environment investigated.
Listeria subtypes were re-introduced or persisted over the growing season.
Four L. monocytogenes PFGE types were shared between feces and pond samples.
ACKNOWLEDGMENTS
This work is supported by United States Department of Agriculture (USDA) Specialty Crop Research Initiative grant number 2019-51181-30016 from the National Institute of Food and Agriculture. Manuscript preparation and revision were also partially supported by the National Institute of Environmental Health Sciences, National Institutes of Health (NIH) under award number T32ES007271 and the Virginia Agricultural Experiment Station and the Hatch Program of the National Institute of Food and Agriculture, USDA. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the USDA or NIH.
Footnotes
SUPPLEMENTAL MATERIAL
Supplemental material associated with this article can be found online at: [URL to be completed by the publisher].
REFERENCES
- 1.Bradshaw JK, Snyder BJ, Oladeinde A, Spidle D, Berrang ME, Meinersmann RJ, Oakley B, Sidle RC, Sullivan K, and Molina M. 2016. Characterizing relationships among fecal indicator bacteria, microbial source tracking markers, and associated waterborne pathogen occurrence in water and sediments in a mixed land use watershed. Water Res. 101:498–509. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 27 August 2012. Multistate outbreak of listeriosis linked to whole cantaloupes from Jensen Farms, Colorado (final update). Available at: https://www.cdc.gov/listeria/outbreaks/cantaloupes-jensen-farms/index.html
- 3.Chapin TK, Nightingale KK, Worobo RW, Wiedmann M, and Strawn LK. 2014. Geographical and meteorological factors associated with isolation of Listeria species in New York State produce production and natural environments. J. Food Prot. 77:1919–1928. doi: 10.4315/0362-028X.JFP-14-132 [DOI] [PubMed] [Google Scholar]
- 4.Chen JQ, Regan P, Laksanalamai P, Healey S, and Hu Z. 2017. Prevalence and methodologies for detection characterization and subtyping for Listeria monocytogenes and L. ivanovii in foods and environmental sources. Food Sci Hum Wellness. 6(3):97–120. [Google Scholar]
- 5.Converse K, Wolcott M, Docherty D, and Cole R. 1999. Screening for potential human pathogens in fecal material deposited by resident Canada Geese on areas of public utility. USGS Natl. Wildl. Health Cent. Rep. [Google Scholar]
- 6.Cooley MB, Quinones B, Oryang D, Mandrell RE, and Gorski L. 2014. Prevalence of shiga toxin producing Escherichia coli, Salmonella enterica, and Listeria monocytogenes at public access watershed sites in a California Central Coast agricultural region. Front. Cell. Infect. Microbiol. 4(article 30). doi: 10.3389/fcimb.2014.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falardeau J, Johnson RP, Pagotto F, and Wang S. 2017. Occurrence, characterization, and potential predictors of verotoxigenic Escherichia coli, Listeria monocytogenes, and Salmonella in surface water used for produce irrigation in the Lower Mainland of British Columbia, Canada. PLoS One. 12(9):e0185437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garner D and Kathariou S. 2016. Fresh produce–associated listeriosis outbreaks, sources of concern, teachable moments, and insights. J. Food Prot. 79:337–344. doi: 10.4315/0362-028X.JFP-15-387. [DOI] [PubMed] [Google Scholar]
- 9.Gorski L, Walker S, Liang AS, Nguyen KM, Govoni J, Carychao D, Cooley MB, and Mandrell RE. 2014. Comparison of Subtypes of Listeria monocytogenes isolates from naturally contaminated watershed samples with and without a selective secondary enrichment. PLOS ONE. 9(3): e92467. 10.1371/journal.pone.0092467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graves LM and Swaminathan B. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55–62. [DOI] [PubMed] [Google Scholar]
- 11.Haase JK, Didelot X, Lecuit M, Korkeala H, L. monocytogenes MLST Study Group, and Achtman M. 2014. The ubiquitous nature of Listeria monocytogenes clones: A large-scale multilocus sequence typing study. Environ. Microbiol. 16(2):405–416. doi: 10.1111/1462-2920.12342 [DOI] [PubMed] [Google Scholar]
- 12.Heaton JC and Jones K. 2008. Microbial contamination of fruit and vegetables and the behaviour of enteropathogens in the phyllosphere: A review. J. Appl. Microbiol. 104:613–626. doi: 10.1111/j.1365-2672.2007.03587.x [DOI] [PubMed] [Google Scholar]
- 13.Hunter PR and Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s Index of Diversity. J. Clin. Microbiol. 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jay-Russell MT 2013. What is the risk from wild animals in food-borne pathogen contamination of plants? CAB Rev. 8:1–16. doi: 10.1079/PAVSNNR20138040. [DOI] [Google Scholar]
- 15.Jost L 2006. Entropy and diversity. OIKOS. 113:363–375. [Google Scholar]
- 16.Liao J, Wiedmann M, and Kovac J. 2017. Genetic stability and evolution of the sigB allele, used for Listeria sensu stricto subtyping and phylogenetic inference. Appl. Environ. Microbiol. 83(12). 10.1128/AEM.00306-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linke K, Ruckerl I, Brugger K, Karpiskova R, Walland J, Muri-Klinger S, Tichy A, Wagner M, and Stessl B. 2014. Reservoirs of Listeria species in three environmental ecosystems. Appl. Environ. Microbiol. 80:5583–5592. doi: 10.1128/AEM.01018-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Galvez F, Allende A, Pedrero-Salcedo F, Alarcon JJ, and Gil MI. 2014. Safety assessment of greenhouse hydroponic tomatoes irrigated with reclaimed and surface water. Int. J. Food Microbiol. 191:97–102. 10.1016/j.ijfoodmicro.2014.09.004 [DOI] [PubMed] [Google Scholar]
- 19.Lyautey E, Hartmann A, Pagotto F, Tyler K, Lapen DR, Wilkes G, Piveteau P, Rieu A, Robertson WJ, Medeiros DT, Edge TA, Gannon V, and Topp E. 2007. Characteristics and frequency of detection of fecal Listeria monocytogenes shed by livestock, wildlife, and humans. Can. J. Microbiol. 53: 1158–1167. doi: 10.1139/W07-084. [DOI] [PubMed] [Google Scholar]
- 20.Lyautey E, Lapen DR, Wilkes G, McCleary K, Pagotto F, Tyler K, Hartmann A, Piveteau P, Rieu A, Robertsonm D WJ. Medeiros T, Edge TA, Gannon V, and Topp E. 2007. Distribution and characteristics of Listeria monocytogenes isolates from surface waters of the South Nation River Watershed, Ontario, Canada. Appl. Environ. Microbiol. 73:5401–5410. doi: 10.1128/AEM.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nightingale KK, Schukken YH, Nightingale CR, Fortes ED, Ho AJ, Her Z, Grohn YH, McDonough PL, and Wiedmann M. 2004. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70(8):4458–4467. DOI: 10.1128/AEM.70.8.4458-4467.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nightingale KK, Windham K, and Wiedmann M. 2005. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 187:5537–5551. doi: 10.1128/JB.187.16.5537-5551.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okasanen J, Guillaume Blanchet F, Friendle M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Szoecs E, and Wagner H. 2018. Vegan: Community ecology package. R package version 2.5–3. Available at: https://CRAN.R-project.org/package=vegan. Accessed 4 May 2018. [Google Scholar]
- 24.Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, and Griffin PM. 2013. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg. Infect. Dis. 19:407–415. doi: 10.3201/eid1903.111866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pang H, McEgan R, Mishra A, Micallef SA, and Pradhan AK. 2017. Identifying and modeling meteorological risk factors associated with preharvest contamination of Listeria species in a mixed produce and dairy farm. Food Res. Int. 102:355–363. doi: 10.1016/j.foodres.2017.09.029. [DOI] [PubMed] [Google Scholar]
- 26.Park S, Szonyi B, Gautam R, Nightingale K, Anciso J, and Ivanek R. 2012. Risk factors for microbial contamination in fruits and vegetables at the preharvest level: A systematic review. J. Food Prot. 75:2055–2081. doi: 10.4315/0362-028X.JFP-12-160. [DOI] [PubMed] [Google Scholar]
- 27.R Core Team. 2020. R: A language and environment for statistical computing. Foundation for Statistical Computing, Vienna, Austria. Available at: https://www.R-project.org/. [Google Scholar]
- 28.Rocchetti I, Bunge J, and Bohning D. 2011. Population size estimation based upon ratios of recapture probabilities. Ann. Appl. Stat. 5(2B):1512–1533. doi: 10.1214/10-AOAS436 [DOI] [Google Scholar]
- 29.Restaino L, Frampton EW, Irbe RM, Schabert G, and Spitz H. 1999. Isolation and detection of Listeria monocytogenes using fluorogenic and chromogenic substrates for phosphatidylinositol-specific phospholipase C. J Food Prot. 62(3):244–251. [DOI] [PubMed] [Google Scholar]
- 30.Ryser ET, Arimi SM, Bunduki MMC, and Connelly CW. 1996. Recovery of different Listeria ribotypes from naturally contaminated, raw refrigerated meat and poultry products with two primary enrichment media. Appl. Environ. Microbiol. 62(5):1781–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, and Griffin PM. 2011. Foodborne illness acquired in the United States- major pathogens. Emerg. Infect. Dis. 17:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith A, Moorhouse E, Monaghan J, Taylor C, and Singleton I. 2018. Sources and survival of Listeria monocytogenes on fresh, leafy produce. J. Appl. Microbiol. 125:930–942. doi: 10.1111/jam.14025 [DOI] [PubMed] [Google Scholar]
- 33.Stea EC, Purdue LM, Jamieson RC, Yost CK, and Hansen LT. 2015. Comparison of the prevalences and diversities of Listeria species and Listeria monocytogenes in an urban and a rural agricultural watershed. Appl. Environ. Microbiol. 81:3812–3822. doi: 10.1128/AEM.00416-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strawn LK, Fortes ED, Bihn EA, Nightingale KK, Grohn YT, Worobo RW, Wiedmann M, and Bergholz PW. 2013. Landscape and meteorological factors affecting prevalence of three food-borne pathogens in fruit and vegetable farms. Appl. Environ. Microbiol. 79:588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strawn LK, Grohn YT, Warchocki S, Worobo RW, Bihn EA, and Wiedmann M. 2013. Risk factors associated with Salmonella and Listeria monocytogenes contamination of produce fields. Appl. Environ. Microbiol. 79:7618–7627. doi: 10.1128/AEM.02831-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.U. S. Food and Drug Administration. 14 November 2018. Recalls, market withdrawals, and safety alerts. Available at: https://www.fda.gov/safety/recalls/
- 37.Weis J and Seeliger HPR. 1975. Incidence of Listeria monocytogenes in nature. Appl. Microbiol. 30(1): 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weller D, Wiedmann M, and Strawn LK. 2015. Irrigation is significantly associated with an increased prevalence of Listeria monocytogenes in produce production environments in New York State. J. Food Prot. 78:1132–1141. doi: 10.4315/0362-028X.JFP-14-584. [DOI] [PubMed] [Google Scholar]
- 39.Weller D, Wiedmann M, and Strawn LK. 2015. Spatial and temporal factors associated with an increased prevalence of Listeria monocytogenes in spinach fields in New York State. Appl. Environ. Microbiol. 81:6059–6069. doi: 10.1128/AEM.01286-15. doi:10.1128/AEM.02491–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weller D, Belias A, Green H, Roof S, and Wiedmann M. 2019. Landscape, water quality, and weather factors associated with an increased likelihood of foodborne pathogen contamination of New York streams used to source water for produce production. Conflicts and Compromises between Food Safety Policies and Environmental Sustainability, a special issue of Frontiers in Sustainable Food Systems 3(124). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkes G, Edge TA, Gannon VPJ, Jokinen C, Lyautey E, Neumann NF, Ruecker N, Scott A, Sunohara M, Topp E, and Lapen DR. 2011. Associations among pathogenic bacteria, parasites, and environmental and land use factors in multiple mixed-use watersheds. Water Res. 45:5807–5825. doi: 10.1016/j.watres.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 42.Willis A and Bunge J. 2015. Estimating diversity via frequency ratios. Biometrics. 71(4):1042–1049. [DOI] [PubMed] [Google Scholar]
- 43.Willis A, Martin BD, Trinh D, Barger K, and Bunge J. 2018. Breakaway: Species richness estimation and modeling. R package version 4.6.7. https://adw96.github.io/breakaway/ [Google Scholar]
- 44.Zhu Q, Gooneratne R, and Hussain MA. 2017. Listeria monocytogenes in fresh produce: Outbreaks, prevalence and contamination levels. Foods. 6(21):1–11. doi: 10.3390/foods603002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.