Significance
The mammalian inner ear uses sensory hair cells and neurons for both auditory and vestibular function. Precursor cells for hair cells and neurons are derived from Sox2+ proneurosensory progenitors specified in the otic ectoderm. Here, we demonstrate that the inner ear neurosensory-specific transcription factor Six1 and its coactivator Eya1 collaborate with chromatin remodelers to promote the initial specification of a proneurosensory-restricted progenitor cell from an ectodermal cell within the otic ectoderm by inducing Sox2 activation. This process is mediated through three distal 3′ Sox2 enhancers whose otic-restricted activity depends on Six1. Our findings have broad implications for understanding the architecture of genetic regulatory circuits that induce the initial events of otic neurosensory lineage establishment and maintenance.
Keywords: otic neurosensory lineage, chromatin remodelers, transcription factors, Sox2 enhancers, regulation of otic Sox2 expression
Abstract
Specification of Sox2+ proneurosensory progenitors within otic ectoderm is a prerequisite for the production of sensory cells and neurons for hearing. However, the underlying molecular mechanisms driving this lineage specification remain unknown. Here, we show that the Brg1-based SWI/SNF chromatin-remodeling complex interacts with the neurosensory-specific transcriptional regulators Eya1/Six1 to induce Sox2 expression and promote proneurosensory-lineage specification. Ablation of the ATPase-subunit Brg1 or both Eya1/Six1 results in loss of Sox2 expression and lack of neurosensory identity, leading to abnormal apoptosis within the otic ectoderm. Brg1 binds to two of three distal 3′ Sox2 enhancers occupied by Six1, and Brg1-binding to these regions depends on Eya1-Six1 activity. We demonstrate that the activity of these Sox2 enhancers in otic neurosensory cells specifically depends on binding to Six1. Furthermore, genome-wide and transcriptome profiling indicate that Brg1 may suppress apoptotic factor Map3k5 to inhibit apoptosis. Together, our findings reveal an essential role for Brg1, its downstream pathways, and their interactions with Six1/Eya1 in promoting proneurosensory fate induction in the otic ectoderm and subsequent neuronal lineage commitment and survival of otic cells.
The mammalian inner ear uses sensory hair cells for mechanotransduction of both vestibular and auditory stimuli and transmits this information to the brain via sensory neurons. Precursor cells for hair cells and neurons are specified within the otocyst, which develops from the otic placode, ectoderm thickening that forms around embryonic day 8.5 (E8.5) and differentiates to form all inner ear structures. The neurosensory domain within the ventral region of the otocyst is defined by transcription factors (TFs) Eya1, Six1, and Sox2 (1). A subpopulation of their daughter cells is induced to become Neurog1+ neuroblasts at ∼E9.0 (2, 3), which differentiate to become Neurod1+ progenitors and then delaminate into the mesenchyme to form the vestibular and spiral ganglion neurons (4–6). In contrast, the prosensory progenitors continue to divide to reach a defined number for each sensory organ before differentiating into either hair cells or underlying supporting cells within each sensory epithelium around E12 in vestibula and E14 in cochlea in mice (7, 8). Sox2 is a crucial TF necessary for prosensory cell specification (9) and inner ear neurogenesis (10). The transcription-coactivator Eya1 and the homeodomain protein Six1 form a key transcriptional complex and forced expression of both in cochlear explant culture induces nonsensory epithelial cells to differentiate into hair cells or spiral neurons through interaction with Sox2 or with chromatin remodelers, respectively (11, 12). Mutations in these TFs cause congenital sensorineural hearing loss in humans and loss of neurosensory structures in mice (13–18). However, whether Eya1/Six1 interact to induce Sox2 activation to initiate Sox2+ proneurosensory fate specification within the otic ectoderm and how these TFs operate to establish subsequent neuronal or sensory cell identity remain elusive.
During lineage progression, the activity of TFs is mediated by lineage-specific promoters and cis-regulatory elements (CREs)/enhancers through interaction with developmental cues and epigenetic factors. The SWI/SNF-like BAF chromatin-remodeling complex plays a vital role in promoting depletion of nucleosomes, which requires the ATPase activity of the catalytic subunit Brm or Brg1 encoded by Smarca2/Brm or Smarca4/Brg1, respectively (19). Our early study showed that overexpression of Eya1-Six1 with Brg1-BAF170/155 in cochlear explant culture induces neuronal differentiation (11), but there is no direct evidence of the need for the BAF complex in inner ear neurosensory cell development in any species.
Through conditional deletion of the ATPase subunit Brg1 or inactivation of both Eya1/Six1, our studies demonstrate a key role of Brg1 or both Eya1/Six1 in proneurosensory fate induction within the otic ectoderm. We identified a cooperative interaction between Eya1/Six1 and Brg1 in regulating Sox2 expression in neurosensory-lineage through cobinding to distal 3′ Sox2 CREs/enhancers whose activity in otic neurosensory cells specifically depends on Six1. In addition, we identified molecular pathways downstream of Brg1 that control cell apoptosis, proliferation, and otic neurosensory cell development. Our results provide mechanistic insight into how disruption of a delicate balance between TFs Eya1/Six1 and Brg1-based SWI/SNF complex can lead to aberrant activation of genes, thus resulting in defective neurosensory cell development in the inner ear.
Results
Brg1 Specifies Otic Proneurosensory Fate and Subsequent Neuronal Lineage Commitment.
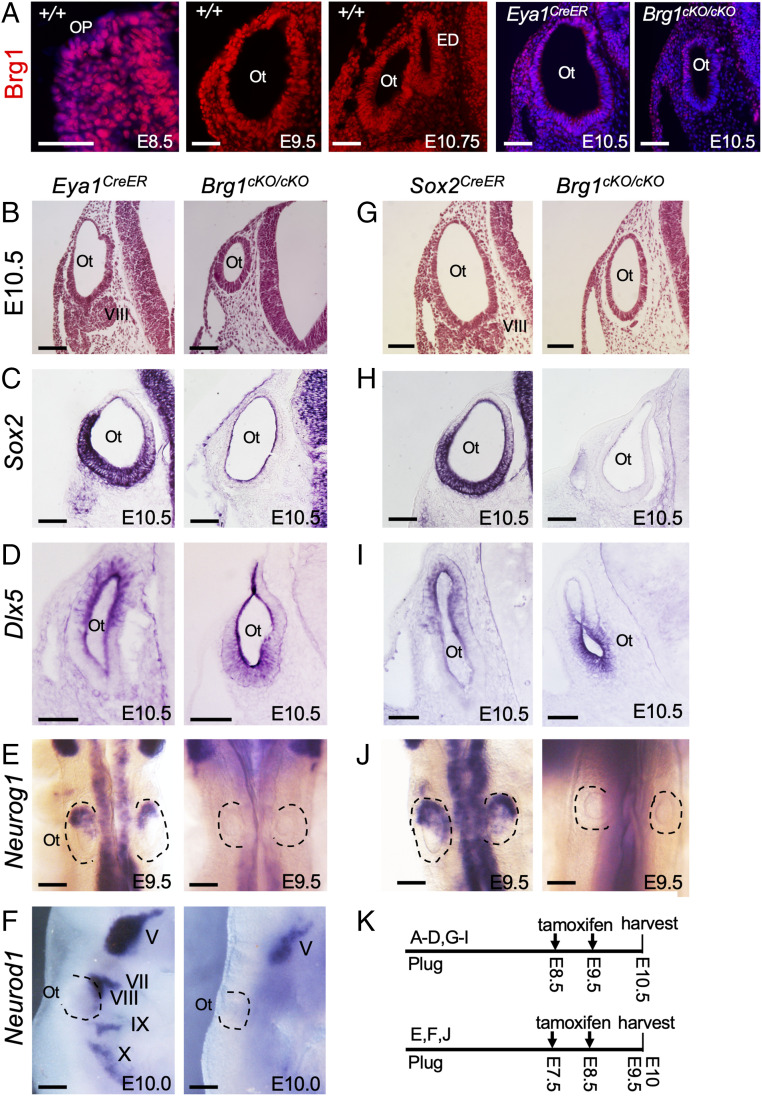
To test Brg1 function in establishing proneurosensory lineage within the otocyst, we first examined Brg1 expression from otic placode stage at E8.5 and found that Brg1 is ubiquitously expressed in the otic placode and otocyst (Fig. 1A), overlapping with Eya1 and Six1 expression in the otic placode and the ventral otocyst from which all neurosensory structures develop (14, 18, 20). At later stages, Brg1 expression overlaps with Eya1 and Six1 in all inner ear sensory epithelia and ganglion neurons (SI Appendix, Fig. S1A). To achieve proneurosensory lineage-specific ablation of Brg1, we crossed Eya1CreER (21–23) with Brg1fl/fl mice (24) and administered tamoxifen at E8.5 to E9.5 to delete Brg1 in the otic placode/otocyst. Immunostaining confirmed selective depletion of Brg1 in Eya1-expressing domain of the ventral otocyst (Fig. 1A). To alleviate potential defects due to Eya1 heterozygosity, we directly compared Brg1cKO/cKO (Eya1CreER;Brg1fl/fl) with Brg1cKO/+ (Eya1CreER;Brg1fl/+) and Eya1CreER/+ littermates. Newborn Brg1cKO/+ mice did not display obvious enhancement of inner ear abnormalities, whereas Brg1cKO/cKO displayed rudimentary inner ears without recognizable neurosensory structures (SI Appendix, Fig. S1B). Histological analysis of inner ears at E15.5 confirmed that Brg1cKO/cKO only had a degenerated cavity-like structure compared with their wild-type, Eya1CreER or Brg1cKO/+ littermates (SI Appendix, Fig. S1C). Analysis at E10.5 revealed an absence of the VIIIth ganglion and a noticeably smaller otocyst in Brg1cKO/cKO (Fig. 1B). Thus, Brg1 appears to be essential for inner ear neurosensory structure formation.
Fig. 1.
Deletion of Brg1 using Eya1CreER or Sox2CreER in otic placode results in lack of neurosensory cell fate specification. Sections are transverse and dorsal is up. For whole-mount images, anterior is up. (A) Anti-Brg1 immunostaining on sections showing Brg1 expression in otic placode (Op) and otocyst (Ot). The Left is a merge of red and Hoechst nuclear staining. (Scale bars, 50 μm.) (B and G) H&E-stained sections of otocyst of Eya1CreER and Brg1cKO/cKO (Eya1CreER;Brg1fl/fl) or Sox2CreER and Brg1cKO/cKO (Sox2CreER;Brg1fl/fl). (Scale bars, 50 μm.) (C, D, H, and I) ISH on sections for Sox2 or Dlx5. (Scale bars, 50 μm.) (E, F, and J) Whole-mount ISH for Neurog1 (dorsal view, E and J) or Neurod1 (lateral view, F) in otic and cranial ganglia. (Scale bars, 120 μm.) (K) Schedules for tamoxifen administration. n = 6 embryos for each genotype. ED, endolymphatic duct; V to X, Vth to Xth cranial ganglion.
To define the basis for the lack of neurosensory structure formation associated with Brg1-deficiency, we performed marker gene analysis. Sox2 is strongly expressed in the ventral region of the otocyst, from which all inner ear neurosensory structures form (Fig. 1C). However, in Brg1cKO/cKO otocyst, Sox2 expression was reduced to background level (Fig. 1C). The expression of Dlx5, a marker for nonsensory cells in the dorsal otocyst (25), was shifted to the ventral region in Brg1cKO/cKO (Fig. 1D). Thus, in the absence of Sox2-active cell fate, the otic epithelium is filled with Dlx5+ cells, suggesting acquisition of a nonsensory fate.
To determine if lack of VIIIth ganglion formation is due to lack of neuroblast specification or differentiation, we examined the expression of Neurog1 and Neurod1. Neurog1 marks neuroblasts, which are specified within the neurogenic domain located in anteroventral region of the otocyst (Fig. 1E). However, Neurog1 expression was undetectable in Brg1cKO/cKO (Fig. 1E). Neurod1 acts downstream of Neurog1 and is expressed in differentiating neuronal precursors in all cranial ganglia (5) (Fig. 1F), whereas its expression in the VIIIth—along with VIIth, IXth, and Xth ganglia—was absent in Brg1cKO/cKO (Fig. 1F). Thus, Neurog1+ neuroblasts are not specified within Brg1cKO/cKO otocyst.
To confirm these observations, we used Sox2CreER (26) to ablate Brg1 and Brg1cKO/cKO also lacked the VIIIth ganglion and had a smaller otocyst (Fig. 1G) with similar alterations in the expression of Sox2 (Fig. 1H), Dlx5 (Fig. 1I), and Neurog1 (Fig. 1J). Together, these results indicate that Brg1 is essential for proneurosensory fate induction and subsequent neuronal lineage commitment in vivo.
Ablation of Brg1 Leads to Loss of Both Eya1 and Six1 Expression, which Are Necessary for the Initial Activation of Sox2 in the Otic Ectoderm.
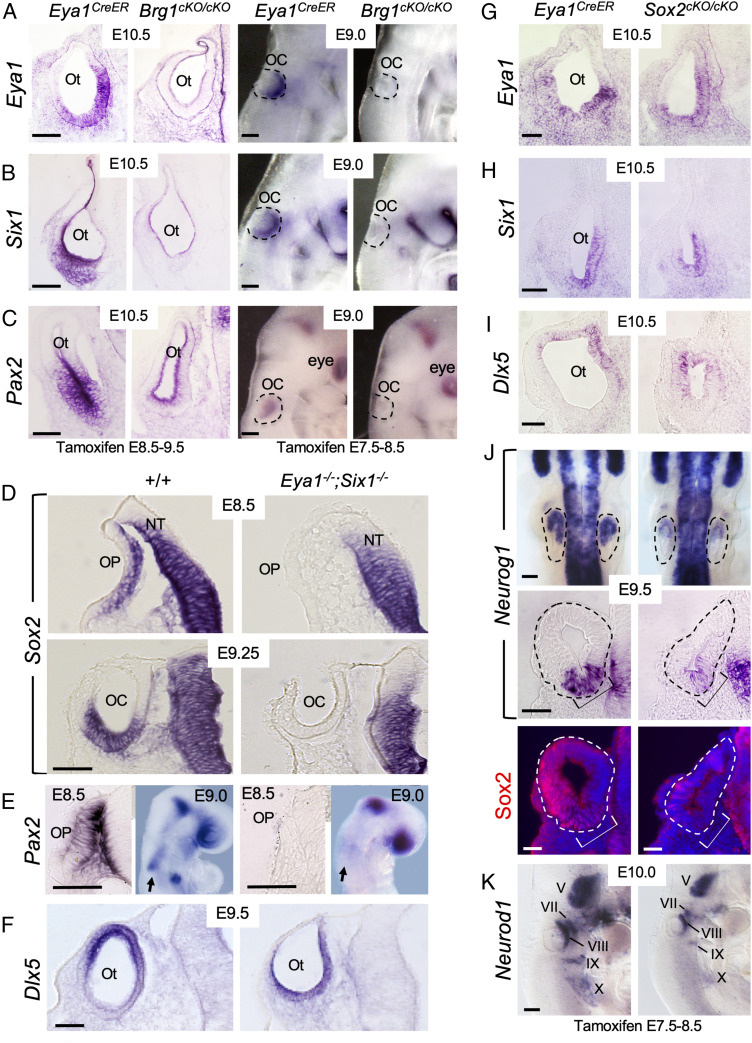
Previously, we have shown that like in Brg1cKO/cKO, Neurog1-Neurod1 are not expressed in Eya1−/−;Six1−/− double mutant (27). This led us to test if loss of Neurog1-Neurod1 expression in Brg1-cKO could be due to loss of both Eya1/Six1 expression. Indeed, marked reduction of Eya1/Six1 expression was observed in the E10.5 otocyst of Brg1cKO/cKO (Fig. 2 A and B). When examined at earlier stages, both Eya1 and Six1 expression were similarly reduced to background level in the invaginating otic placode/cup (Fig. 2 A and B). Expression of two additional neurosensory markers, Pax2 (28) and Hmx3 (29), was also displayed undetectable expression in the ventral otocyst of Brg1cKO/cKO (Fig. 2C and SI Appendix, Fig. S2A), but Pax2 expression in the brain and eye (Fig. 2C) or Hmx3 expression in the cranial and pharyngeal or caudal regions of the embryo (SI Appendix, Fig. S2A) were unaffected. Thus, in the absence of Brg1, Eya1+Six1+ ventral neurosensory fate is not specified in the otocyst.
Fig. 2.
Brg1 is necessary for the expression of Eya1/Six1, both of which are necessary for the activation of Sox2 expression in otic ectoderm. Sections are transverse and dorsal is up. For whole-mount images, anterior is up. (A–C) Left panels are section views of otic vesicle (Ot) and Right panels are whole-mount lateral views of invaginating otic placode (OP)/cup (OC) (outlined by dashed lines) showing ISH for Eya1, Six1, and Pax2. Note that sections were cut from ISH embryos. (Scale bars, 50 μm.) (D) Section ISH for Sox2. Note that Sox2 expression in the neural tube (NT) was not affected. (Scale bar, 50 μm.) (E) Section (E8.5) and whole-mount (E9.0, lateral view) ISH for Pax2. Arrows indicate Pax2 mRNA in otic ectoderm in wild-type and no detectable expression in Eya1−/−;Six1−/−. (Scale bars, 50 μm.) (F) Section ISH for Dlx5. (Scale bar, 50 μm.) (G–J) Deletion of Sox2 does not alter Eya1+Six1+ ventromedial (G and H) or Dlx5+ dorsal (I) region in the otocyst but leads to reduction in Neurog1 expression within the neurogenic domain (brackets) of the otocyt (dashed lines). Upper panels in J are dorsal views of whole-mount embryos, and Middle panels are section view. Bottom panels are dark-field images showing sections of the Middle panels coimmunostained with anti-Sox2. A few Sox2+ cells were observed in the dorsal region of the otocyst but not in the neurogenic Neurog1+ domain (brackets). (Scale bars, 50 μm for Bottom and 120 μm for Upper panels.) (K) Lateral view of whole-mount embryos stained for Neurod1 riboprobe. Note the reduction of Neurod1 in V-X ganglia is due to loss of Sox2. (Scale bars, 120 μm.).
We previously detected reduced Sox2 expression in the otocyst of Eya1−/− or Six1−/− single mutant (14, 30). As Neurog1 or Neurod1 expression is also present in Eya1−/− or Six1−/− but lost in Eya1−/−;Six1−/− otocyst (11), we thus hypothesized that Eya1/Six1 may interact to induce Sox2 activation to specify Sox2+ otic proneurosensory progenitors. Remarkably, Sox2 expression was not activated in Eya1−/−;Six1−/− otic placode and cup ∼E8.5 to E9.25 (Fig. 2D), but its expression in adjacent neural tube was unaffected. Pax2 expression in the otic placode and cup ∼E8.5 to E9.25 was also lost in Eya1−/−;Six1−/− (Fig. 2E), but its expression in the brain and eye regions was preserved. In contrast, Dlx5 displayed expression in otic placode (SI Appendix, Fig. S2D) and the ventral otocyst (Fig. 2F). Brg1 expression in the otocyst of either Eya1−/− single or Eya1−/−;Six1−/− double mutant was also present (SI Appendix, Fig. S2E). These data suggest that Eya1/Six1 may collaborate to induce Sox2 expression in otic ectodermal progenitors to promote the Brg1-Eya1/Six1-mediated proneurosensory fate specification.
Next, to identify the cellular basis of these phenotypes, we performed TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) and EdU (5-ethynyl-2′-deoxyuridine) incorporation assays and found increased apoptosis (SI Appendix, Fig. S2B) and decreased proliferation rate (SI Appendix, Fig. S2C) in Brg1cKO/cKO otocyst. This suggests that Brg1 may have a role for both cell proliferation and survival in the otocyst.
Deletion of Sox2 Does Not Alter Eya1+Six1+ Proneurosensory Domain but Results in Reduced Neurog1 Expression and Smaller VIIIth Ganglion.
We next asked if shift of Dlx5 expression to the ventral otocyst in Brg1cKO/cKO or Eya1−/−;Six1−/− otocyst is due to lack of Sox2 expression. To test this, we conditionally deleted Sox2 from E7.5 to E8.5 using Eya1CreER and observed neither ventral shift of Dlx5 expression (Fig. 2I) nor loss of Eya1/Six1 expression in the ventral region of Sox2cKO/cKO otocyst (Fig. 2 G and H). This confirms that Eya1/Six1 function upstream of Sox2 and that in the presence of Eya1/Six1, loss of Sox2 does not cause ventral progenitors to acquire a nonsensory fate.
Previous studies implicated a role of Sox2 in otic neurogenesis. However, its role in neuronal lineage establishment has been controversial. One study using Sox2CreER showed that Sox2 was required in the initial events in otic neuronal specification (31), but in a more recent study using Foxg1Cre its depletion had no effect on early ear neurogenesis (32). To clarify this discrepancy, we examined Neurog1 expression in Sox2cKO/cKO and found that while reduced, Neurog1 expression was detectable throughout the prospective anteroventral neurogenic domain compared with control littermates (Fig. 2J). Anti-Sox2 immunostaining on sections hybridized with Neurog1 riboprobe detected very few Sox2+ cells outside of the Neurog1+ domain (Fig. 2J and SI Appendix, Fig. S2F), confirming that the low level of Neurog1 mRNA detected in Sox2cKO/cKO is independent of Sox2 function. Interestingly, the Neurog1+ progenitors were capable of differentiating into Neurod1+ progenitors, despite reduced size of the Neurod1+ Vth to Xth ganglia compared with controls (Fig. 2K). While other members of the Sox gene family may have a redundant function with Sox2, our observation is consistent with a previously published study, which showed that early inner ear neurogenesis can proceed after deletion of Sox2 (32). Nonetheless, our results demonstrate that Eya1/Six1 is necessary for Sox2 activation, which may in turn synergize with Eya1/Six1 to mediate neuronal commitment.
Transcriptome Profiling Reveals the Requirement of Brg1 for Specifying Otic Neurosensory Lineage and Suppressing Expression of Genes for Muscle/Heart Contraction and Apoptotic Signaling.
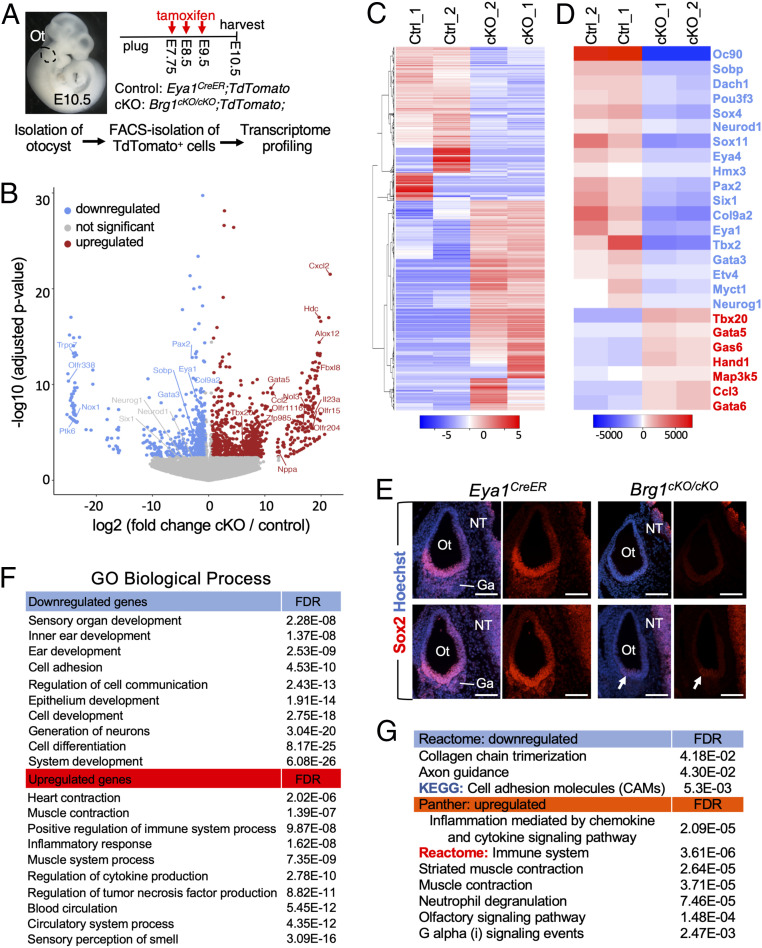
To gain mechanistic insights into how loss of Brg1 function compromises otic cell lineages, we sequenced RNA isolated from TdTomato+ otic cells FACS-purified from control and Brg1cKO/cKO otocysts, respectively (Fig. 3A) (n = 2 biological replicates for each genotype). The transcriptome analysis identified 3,092 differentially expressed genes (1,622 with P adjusted < 0.05 and 1,470 with log2FoldChange > 1.2 and raw P < 0.05) with 1,308 down-regulated and 1,784 up-regulated genes in Brg1cKO/cKO compared with control (Fig. 3 B and C, with 1,470 genes presented in gray color, and SI Appendix, Table S1). Genes characteristic of the neurosensory fate were conspicuously down-regulated including Oc90 (-log10 P > 80) (SI Appendix, Fig. S3A), Sobp (downstream and interacting factor of Six/So family), Col9a2, Eya1, Pax2, Sox9, Sox11, Eya4, Otx1, Chd7, and Pou3f3 (Fig. 3 B and D and SI Appendix, Fig. S3 A and B). However, while reduced, changes in the expression of Sox2, Six1, Hmx3, Neurog1, Neurod1, and Sox4 were not found to be statistically significant (P adjusted > 0.05) (Fig. 3 B and D). This may reflect the sensitivity of deep sequencing as Brg1 activity may not be completely depleted by tamoxifen administration. Statistical power analysis using powsimR (33) indicated that the true positive rate (TPR) for differential genes (P adjusted < 0.05) is close to 40% for a sample size of two per group, which increases to a value of ∼40% or 62% for three or five replicates per group, while the false-discovery rate (FDR) is close to 50% for a sample size of two per group and decreases to ∼40% or ∼25% for three or five replicates per group (SI Appendix, Table S2).
Fig. 3.
Transcriptome analysis reveals the requirement of Brg1 for specifying otic neurosensory lineage and a repressive function of nonotic gene expression. (A) Schematic drawing of TdTomato+ cell isolation. Otocysts (n = 8) were isolated from control and Brg1cKO/cKO littermates (n = 4 for each genotype harvested from 3 pregnant females) and dissociated into single cells. Five thousand FACS-sorted TdTomato+ cells were used for each biological replicate (samples “1” and “2”). Magnification, 6×. (B) Volcano plot showing transcripts differentially responding to depletion of Brg1. (C and D) Heatmap showing expression of all 3,092 (C) or 25 selected (D) differentially expressed genes. Blue or red indicates down- or up-regulated genes in Brg1cKO/cKO. Twenty-five selected genes include otic genes (in blue) known to be important for neurosensory progenitor development and TFs critical for heart as well as apoptotic and inflammatory proteins (in red). (E) Anti-Sox2 immunostaining on sections at E10.5. Arrows point to weak Sox2 signal in a few cells in the ventral otocyst (Ot). No Sox2+ progenitors in VII–VIIIth ganglionic region (Ga) were detectable in Brg1cKO/cKO, but Sox2 expression in the neural tube (NT) is unaffected. Scale bars, 100 μm. (F and G) GO (F) and pathway (G) enrichment analysis for differentially expressed genes. Blue or red indicates down- or up-regulated genes. GO, Panther and reactome pathway annotation tools (geneontology.org), heatmap and KEGG pathway tools (bioinformatics.sdstate.edu).
We next performed anti-Sox2 immunostaining to further validate that otic neurosensory cell lineage specification depends on Brg1 activity. Although Sox2 expression level in the neural tube adjacent to the otocyst was comparable between control and Brg1cKO/cKO (Fig. 3E), weak Sox2 expression was detectable only in a few cells in the ventral region of Brg1cKO/cKO otocyst and no Sox2+ cells were observed in the VII–VIIIth ganglionic region (Fig. 3E), thus confirming that Sox2 expression is Brg1-dependent.
Other otic genes such as Gata3, Tbx2, and Dach1 and numerous cell-adhesion molecules, including cadherins, Sfrp1, Wnt7b, Fat3, Nrxn1, and Epcam were also down-regulated in Brg1cKO/cKO (Fig. 3 B and D and SI Appendix, Fig. S3 B and C). While changes in Myc and Ccnd1/2 expression were not statistically significant, Myc downstream genes Myct1/Ndrg1 and interacting gene Mycbp2 displayed decreased expression (Fig. 3D and SI Appendix, Table S1). In contrast, Cdkn1c and numerous key TFs and signaling molecules for neural crest cell-derived muscle system/heart contraction including Tbx5 (>200×)/Tbx20 (>70×), Gata4 (>25×)/Gata5 (>200×)/Gata6 (>7.5×), Hand1 (>7×), Pax3 (>200×), Pdlim3 (>50×), Heyl (>3.5×), Osr1 (>300×), Nkx2-5 (>13×), Tcf21 (>13×), Bmp2 (>21×), and Wnt3a (>500×) exhibited increased expression in Brg1cKO/cKO (Fig. 3 B and D and SI Appendix, Fig. S3 B and C). We noted that expression levels of the apoptotic factor Map3k5 (∼6×, also known as Ask1, apoptosis signal-regulating kinase 1), tumor necrosis factors such as Tnf (∼18×), Gas6 (growth arrest factor), and the oncogenic TF Runx3 (>80×) (34), as well as other factors involved in tumor necrosis and tumorigenesis were increased (Fig. 3D and SI Appendix, Fig. S3B). Additionally, genes involved in sensory perception of smell, such as Olfr15/204/1116, Mkks, and Gnat1/2 appeared to be up-regulated (Fig. 3B and SI Appendix, Fig. S3 A and C). We next selected Map3k5, Gata5, Tbx20, and the top otic neurosensory gene Oc90 and performed reverse-transcription and real-time PCR (RT-qPCR) to confirm up-regulation of Map3k5, Tbx20, and Gata5 and down-regulation of Oc90 in the cKO cells (SI Appendix, Fig. S3D).
Gene ontology (GO) analysis displayed an enrichment of down-regulated genes related to generation of neurons, epithelium/ear/inner ear/sensory organ development, cell adhesion, and cell differentiation (Fig. 3F), while sensory perception of smell, blood circulation, tumor necrosis factor production, cytokine production, muscle system, muscle/heart contraction, and inflammatory response were enriched in up-regulated genes (Fig. 3F and SI Appendix, Fig. S3B). Pathway enrichment analysis identified collagen chain trimerization, axon guidance, and cell-adhesion molecules associated with down-regulated genes and inflammation mediated by chemokine and cytokine signaling pathways, muscle contraction, and olfactory signaling pathways in up-regulated genes (Fig. 3G and SI Appendix, Fig. S3B). Collectively, these analyses suggest that depletion of Brg1 in Eya1+ progenitors may provide a permissive environment through dysregulation of several pathways that may promote aberrant activation of nonotic ectodermal genes and trigger otic cellular growth arrest and apoptosis.
Genome-Scale Chromatin Immunoprecipitation-Sequencing Reveals Brg1 Binding to Active and Repressed Regulatory Regions in the Otocyst.
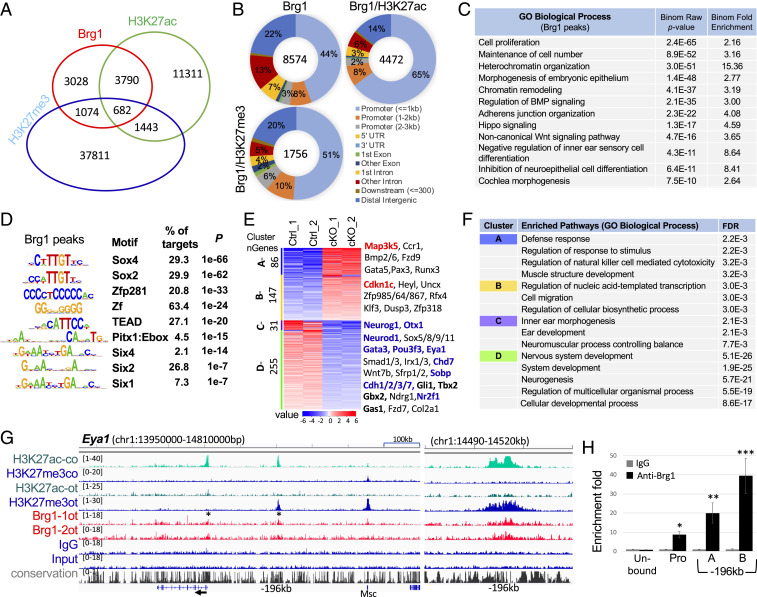
Chromatin remodelers are critical coregulators of transcription through direct interaction with either histone modification enzymes or TFs at gene promoters or distal CREs (35–37). To examine if Brg1 cooperates with Eya1 and Six1 to regulate Sox2 expression to promote otic proneurosensory fate, we prepared chromatin from otocysts isolated from E10.5 embryos and performed chromatin immunoprecipitation sequencing (ChIP-seq) to characterize Brg1 genome-wide occupancy. We also performed ChIP-seq for the histone marks H3K27ac associated with active promoters and enhancers (38) and H3K27me3 associated with chromatin condensation and transcriptional repression (39). In peak calling against both genomic input DNA and IgG ChIP-seq controls with the default setting, we generated a bed file for 8,574 regions from three datasets for further analysis (Fig. 4A and SI Appendix, Fig. S4A). GREAT analysis identified 4,064 genes by assigning 8,574 peaks to their single nearest genes within 500-kb of the nearest gene’s transcription start site (TSS) (Dataset S1). While 1,756 regions showed association with H3K27me3 deposition, 4,472 regions were active promoters/putative enhancers that displayed the deposition of H3K27ac (Fig. 4 A and B and Dataset S2). Although Brg1 near TSSs displayed a higher binding density, ∼76.6% of these regions were associated with H3K27ac (Fig. 4B). Notably, ∼43% of Brg1 peaks were detected in intronic and intergenic regions and ∼29% of these peaks were accompanied by H3K27ac, representing an active class of regulatory regions/enhancers (Fig. 4B). GO analysis indicated that genes related to cell proliferation, cell number maintenance, heterochromatin organization, morphogenesis of embryonic epithelium, chromatin remodeling, BMP/adherens junction organization/hippo/Wnt signaling pathways, negative regulation of inner ear sensory and neuroepithelial cell differentiation, and cochlea/ear morphogenesis were overrepresented in Brg1 targets (Fig. 4C and SI Appendix, Fig. S4B).
Fig. 4.
Genome-wide occupancy by Brg1 in the otocyst. (A) Venn diagram indicating overlap of Brg1-binding sites and of H3K27ac or H3K27me3 deposition. (B) Pie charts (Galaxy toolkits) showing genomic distribution. UTR, untranslated region. (C) GREAT analysis showing association of Brg1-enriched regions with terms in GO database. (D) Sequence logos of the significantly enriched top motifs from Homer Known motif analysis, letter size indicates nucleotide frequency. Percent of target sites in Brg1 peaks with significance of motif occurrence (P value) are indicated. (E) Heatmap (Kmeans clustering) showing expression of 4 clusters of the 519 Brg1 target genes (identified in ChIP-seq datasets). Selected genes in each cluster were also listed on the right. Red or blue indicates up- or down-regulated genes. Several important genes related to apoptosis, cell-cycle or otic neurosensory cell development are shown in bold. (F) Kmeans GO enrichment analysis for each cluster of the 519 genes. (G) Genomic browser visualization of Brg1-bound sites at proximal-promoter and an upstream intergenic region at −196 kb of Eya1 (asterisks). Higher magnification of the −196-kb region on (Right) showing Brg1-enrichment with H3K27me3 deposition in the otocyst (ot) but with H3K27ac deposition in E13.5 cochlea (co). (H) ChIP-qPCR for the proximal-promoter and the −196-kb regions. “A” and “B” represent two different primer sets (SI Appendix, Table S3) within the peak region. Brg1 unbound region was used as a negative control. qPCR was performed in triplicates and repeated three times for each group. Data were normalized with mock IgG control, which was considered to be onefold. *P < 0.05, **P < 0.01, and ***P < 0.001.
De novo motif analysis revealed that consensus binding sites for SOX family proteins, Zinc finger proteins, TEAD, homeobox and bHLH proteins are among the most enriched motifs at the sites occupied by Brg1 (Fig. 4D and SI Appendix, Fig. S4C). TFs with these motifs are known to interact with Brg1-containing SWI/SNF and act as Brg1’s transcriptional coregulators (36, 37, 40). Motifs for Six1/2/4, all of which are known to bind to Six1/2/4 proteins with different affinities (41), were overrepresented motifs at a relatively lower rank. Thus, this genome-wide analysis suggests a direct involvement of the Brg1-based SWI/SNF complex in regulating gene expression and provides insight into potential DNA-binding TFs that Brg1 interacts with to target CREs.
Comparison with RNA-sequencing (RNA-seq) data found 519 differentially expressed genes targeted by Brg1 (Dataset S3). We clustered RNA-seq data for the 519 genes to four groups using Kmeans clustering analysis (Fig. 4E) and found that Eya1, Sobp, Gata3, Pou4f3, and Neurod1 were among the 255 genes in cluster D. Other factors known to be important for inner ear neurosensory structure formation, such as Otx1, Chd7, Nr2f1, Gbx1/2, Gli1, and Gas1, cadherins (Cdh1/2/3/7), the cell cycle regulators Cdk15 (∼3.5× decrease) and Cdkn1c (∼1.4× increase), and the apoptotic factor Map3k5 were also among the 519 genes. GO enrichment analysis identified defense response, muscle development (cluster A), and regulation of transcription (cluster B) for up-regulated genes and inner ear and neurogenesis for down-regulated genes (cluster C, D) (Fig. 4F). Analysis for enrichment of biological pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) identified MAPK, focal adhesion, Wnt, and Hippo signaling pathways and signaling pathways regulating pluripotency of stem cells for cluster D and no significant pathways were identified for other clusters (SI Appendix, Fig. S4D). We noted that Map3k5 and all genes involved in muscle/heart contraction with increased expression in Brg1cKO/cKO had strong H3K27me3 deposition at their promoters (SI Appendix, Figs. S3E and S4E and Dataset S4), suggesting that these genes are not fully active.
At the Eya1, peak calling identified Brg1 binding to the proximal-promoter and a distal intergenic region 196-kb upstream from the promoter (Fig. 4G). Interestingly, comparison of ChIP-seq data between E10.5 otocyst and E13.5 cochlea found that the intergenic region at −196 kb from the Eya1 promoter was associated with strong H3K27me3 deposition in the otocyst but with strong H3K27ac deposition in the cochlea, suggesting that the activity of this element is temporally controlled as development proceeds. Since Brg1 does not directly bind to DNA, we found that Brg1 peaks were not as high (SI Appendix, Fig. S4A) as that of DNA-binding TFs or histone marks. We then performed ChIP-qPCR to confirm Brg1-enrichment at the Eya1 promoter (−453 to +701 bp) and the distal region −196 kb (Fig. 4H). Clusters of Brg1 peaks were also identified at the loci of Six1 and Sox2 (Dataset S1). Peak calling also identified Brg1 enrichment at cell cycle regulators, such as Mycn, Ccnd1/2, and Cdkn1b, several genes that are known to cause deafness or inner ear abnormalities, including Spry2/Ptk7/Vangl2 (Wnt signaling), Emx2, Fgfr1/3, Jag1/Dll1/Notch1/Hes1/5 (Notch signaling), Map3K1/4/7/13 (MAPK signaling), Gli3/Smo (Shh signaling), Rnd3 (Rho GTPase), and Nog (SI Appendix, Fig. S4E and Dataset S1). Thus, this genome-wide analysis revealed Brg1-binding not only at the loci of key proneurosensory genes, but also genes involved in cell cycle regulators, deafness, and multiple signaling pathways including the apoptosis signal-regulating kinase 1 (Map3k5). These data indicate a direct involvement of Brg1 in the regulation of these gene expression. Thus, Brg1 may suppress Map3k5 expression to inhibit apoptosis.
Brg1, Six1, and Eya1 Cooperatively Regulate Sox2 Expression through Three Distal 3′ Sox2 Enhancers.
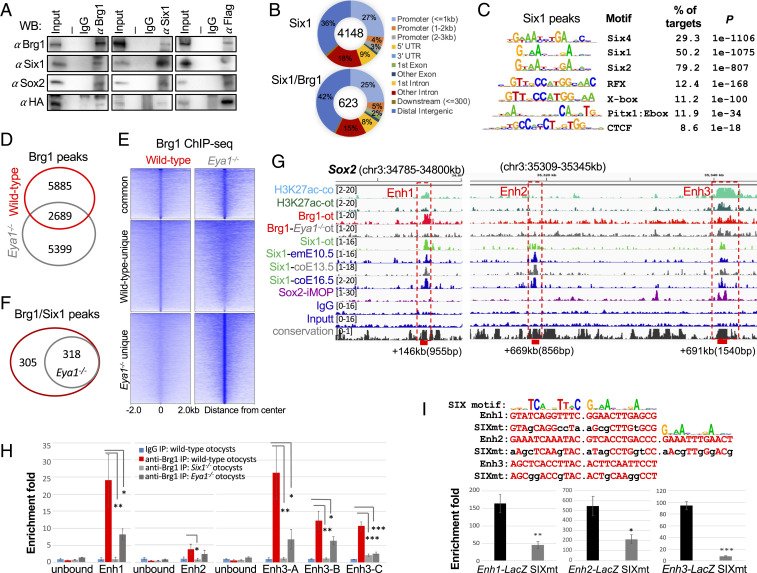
We previously found that both Eya1 and Six1 interact with Brg1/BAF170 and Sox2 in vitro pull-down assays and in E13.5 cochlea independent of DNA as addition of ethidium bromide did not affect protein complex formation (11). Given the critical role of Six1-Eya1 in otic proneurosensory fate specification, we hypothesized that Brg1 cooperates with Six1-Eya1 to target Sox2 and other proneurosensory promoting genes. To test this, we performed coimmunoprecipitation (coIP) using nuclear extracts from otocysts isolated from E10.5 embryos and demonstrated that endogenous Brg1 interacts with Eya1 and Six1 in the otocyst (Fig. 5A and SI Appendix, Fig. S5A), thus supporting the idea that these proteins may form a functional complex at enhancers. Consistent with the view that Sox2 may synergize with Eya1/Six1 to coregulate downstream events during neurosensory lineage specification and the identification of Sox2 motif enriched in Brg1 peaks, coIP revealed physical association of Sox2 with Brg1, Eya1, and Six1 (Fig. 5A and SI Appendix, Fig. S5A). In E13.5 cochlea, consistent with our previous observation (11), Eya1-Six1 or Sox2 form a protein complex with Brg1 (SI Appendix, Fig. S5B). Interestingly, coIP revealed physical association of Eya1 and Six1 with the subunits BAF60a/c but not with BAF60b, while all three subunits coimmunoprecipitated by anti-Brg1 in E13.5 cochlea (SI Appendix, Fig. S5B).
Fig. 5.
Brg1 physically forms protein complex with Eya1/Six1 and Brg1 enrichment at distal 3′ Sox2 enhancer regions is disrupted in the absence of Eya1 or Six1. (A) CoIP analysis using nuclear extracts prepared from E10.5 otocysts of Eya1HA-Flag embryos. Anti-Brg1, -Flag, or -Six1 was used for IP and antibodies used for Western blot are indicated. Input was 5% of the amount used for IP. Row Western blots are shown in SI Appendix, Fig. S5A. (B) Pie charts showing genomic distribution of Six1-binding sites and its overlapping peaks with Brg1. (C) Sequence logos of the significantly enriched top motifs from Homer Known motif analysis, letter size indicates nucleotide frequency. Percent of target sites in Brg1 peaks with significance of motif occurrence (P value) are indicated. (D) Venn diagram indicating overlap of Brg1 sites in wild-type and Eya1−/− otocyst. (E) Heatmaps of Brg1 peaks shown in D within a −2-kb/+2-kb window centered on Brg1 peaks. (F) Venn diagram indicating overlap of sites cooccupied by Brg1/Six1. (G) Genome browser visualization of overlapping occupancy of H3K27ac, Brg1, Six1, and Sox2 to the three distal 3′ Sox2 enhancer elements (boxes). Samples used for comparison: H3K27ac-co, E13.5 cochleae; H3K27ac-ot, E10.5 otocysts (ot); Brg1-ot, E10.5 otocysts; Brg1-Eya1−/−-ot, Eya1−/− otocysts; Six1-ot, E10.5 otocysts; Six1-emE10.5, E10.5 embryos; Six1-coE13.5 or -coE16.5, E13.5 or E16.5 cochleae; Sox2-iMOP, immortalized iMOP. (H) ChIP-qPCR confirms Brg1-binding to the Enh1 to -3 regions in E10.5 otocyst and a large decrease in Brg1-enrichment in Eya1−/− or Six1−/− otocyst. Primers for two Brg1 unbound regions as negative controls and three different primer sets (SI Appendix, Table S3) within the Enh3 region (”A,” “B,” and “C”) were used for qPCR. qPCR was performed in triplicates and the enrichment of mock IP (IgG) was considered onefold. *P < 0.05, **P < 0.01, and ***P < 0.001 determined by Student’s t test. (I) Sox2 Enh1 and Enh3 contain two SIX motifs, while Enh2 has three SIX motifs. Mutations introduced into the predicted SIX-motifs for each enhancer is indicated. Six1-binding to these reporters were assessed by ChIP-qPCR using chromatin prepared from 293 cells cotransfected with His-Six1 expression plasmid and each reporter, respectively. The enrichment of mock IP was considered onefold. *P < 0.05, **P < 0.01, and ***P < 0.001 determined by Student’s t test.
Since Eya1 does not bind DNA directly and acts as a transcriptional activator through interaction with DNA-binding protein Six1 (1), we thus carried out Six1 ChIP-seq to identify Six1-binding sites in the otocyst and its cooccupancy with Brg1. Peak calling identified 4,148 regions (2,912 genes) (Fig. 5B and Dataset S5). GO analysis revealed overrepresentation of genes related to BMP signaling, cranial ganglion/nose/ectoderm/otic vesicle/ectodermal placode development, regulation of cell fate commitment/catenin import into nucleus, and maintenance of cell number (SI Appendix, Fig. S5C). As expected from a direct association of Six1-DNA, the most enriched known motifs matched to the Six1/2/4-binding motifs (Fig. 5C). A proportion of Six1 peaks lacked SIX-binding consensus sequences, suggesting an indirect association of Six1 to DNA through interactions with other DNA-binding proteins. Similar to that observed in E13.5 to E16.5 cochlea (42), RFX and CTCF were among the top five most enriched motifs in Six1-binding sites in the otocyst. Comparison of the ChIP-seq data identified 623 regions of Six1 overlapped with Brg1-occupancy (Fig. 5B and Dataset S6), while comparison of 2,912 Six1-bound and 4,064 Brg1-bound genes identified 1,321 common targets for both Brg1/Six1 (Dataset S7). Among the 623 overlapping peaks, overlapping binding of Brg1/Six1 was identified at a proximal-promoter region of Mycn (−403 bp) or Sobp (+3,510 bp) (SI Appendix, Fig. S5D); both genes are necessary for normal neurosensory cell production (43, 44). At the Sox2 locus, Six1 peaks were identified at several distal intergenic regions in the otocyst and embryo at E10.5 and cochlea at E13.5 and E16.5 (SI Appendix, Fig. S6). While Six1 showed no binding to regions at +31 kb and +755 kb occupied by Brg1 with weak H3K27ac deposition, Brg1 showed no enrichment at a region bound by Six1 (+669 kb, Enh2). Notably, however, two Six1-bound intergenic regions displayed overlapping patterns of occupancy by Brg1 (Enh1 +146 kb with weak H3K27ac deposition and Enh3 +691 kb with strong H3K27ac-enrichment). Six1 binding to these regions was also identified in E10.5 embryo, and E13.5 and E16.5 cochlea (SI Appendix, Fig. S6). We compared with published Brg1 ChIP-seq from E11.5 forebrain and face (45) and found Brg1-binding at +31 kb, +691 kb (Enh3), and +755 kb in the forebrain and face (SI Appendix, Fig. S6), but no Brg1-occupancy to the Enh1 and Enh2 regions in both samples. Consistent with the physical interaction between Sox2, Six1, Eya1, and Brg1, Sox2 ChIP-seq from immortalized multipotent otic progenitor (iMOP) cells (46) also identified Sox2-binding to the Enh3 region (SI Appendix, Fig. S6).
To determine if Eya1 activity is needed for Brg1 to access to these distal 3′ Sox2 regions, we examined Brg1-occupancy in Eya1−/− otocysts. Peak calling identified 8,088 Brg1 peaks in Eya1−/− otocysts (Dataset S8). Normalized coverage comparison analysis via deepTools (47) showed that Brg1 signals appeared to be stronger in Eya1−/− than in wild-type otocysts (SI Appendix, Fig. S4A), which is likely due to experimental variation between different samples. Among the Brg1 peaks, 2,689 peaks were found in both wild-type and Eya1−/− otocysts (Fig. 5 D and E). Of 623 peaks cooccupied by Brg1/Six1 in wild-type otocyst, peak calling failed to detect Brg1-enrichment at 305 regions in Eya1−/− otocyst (Fig. 5F), suggesting that Brg1-binding to these sites may depend on Eya1 activity. At the Sox2 locus, Brg1 barely bound to Enh1 and its enrichment at Enh3 appeared to be reduced (Fig. 5G). To confirm this and test if Six1 may regulate Brg1 to target Sox2 enhancers, we prepared chromatin from Six1−/− otocysts in which Eya1 expression is preserved (14) and otocysts of wild-type or Eya1−/− embryos. We then used an equal amount of chromatin for ChIP with anti-Brg1 or IgG. qPCR confirmed that Brg1 enrichment to the Enh2 region was very weak compared with its binding to Enh1 and Enh3 in wild-type and that its binding to Enh1 and Enh3 regions was largely reduced in Six1−/− or Eya1−/− (Fig. 5H). Thus, Brg1-binding to the 3′ Sox2 enhancer regions corresponds to Eya1 or Six1 activity. To ensure the functional association of Six1 in Sox2 gene regulation, we constructed reporters by inserting 955 bp of Sox2 +146 kb/Enh1, 856 bp of Sox2 +669 kb/Enh2, and 1,540 bp of Sox2 +691 kb/Enh3 fragment into a LacZ reporter driven by Hsp68 minimal promoter flanked by H19 insulators (42), respectively, and mutant reporters in which all predicted Six1-motifs were mutated (Fig. 5I). ChIP assays using chromatin prepared from HEK293 cells transfected each wild-type or SIXmt reporter with Six1 expression plasmid, respectively, (Methods) confirmed Six1-binding to these regions and disruption of Six1-binding to all three mutated reporters (Fig. 5I).
Six1-Binding Is Necessary for Activation of the Three Brg1/Six1-Bound 3′ Sox2 Enhancers in Otic Neurosensory Cells In Vivo.
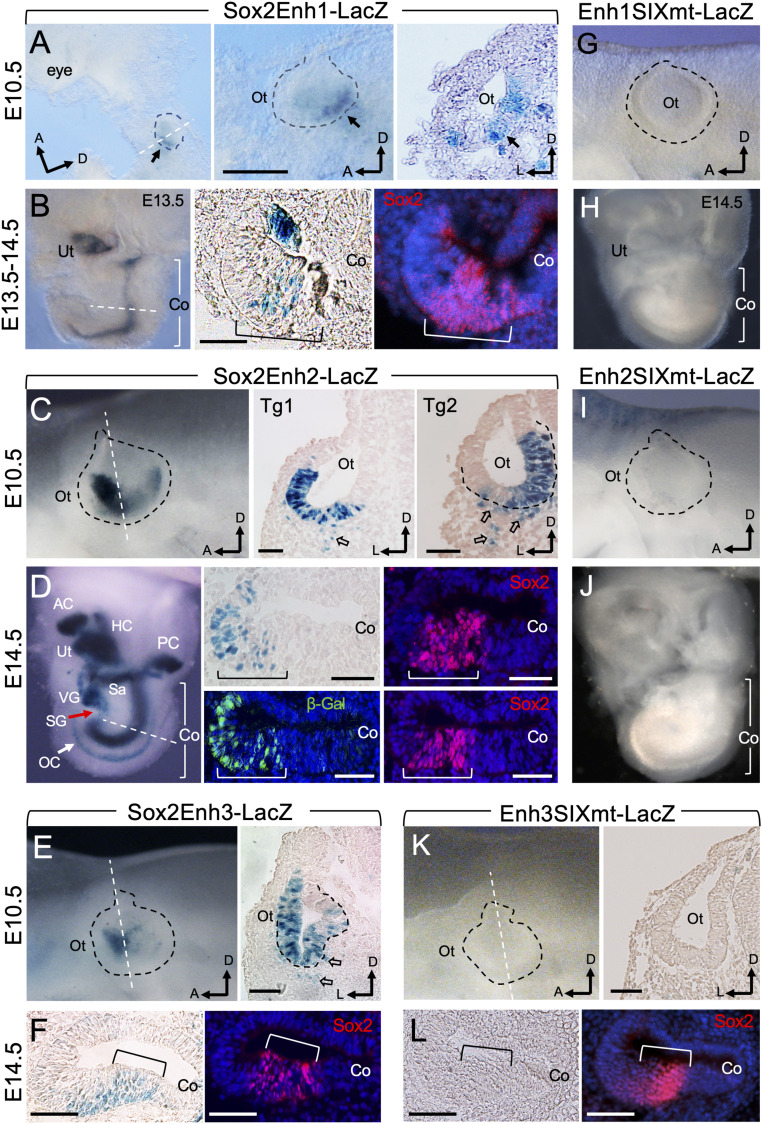
To examine the functional roles of the three distal 3′ Sox2 enhancer regions in mediating expression in otic neurosensory cells, we assayed their activity in vivo using an established mouse transient (G0) transgenic (Tg) assay. The 955 bp of Enh1 +146 kb specifically drove β-Gal expression restricted to the posteroventral domain of the otocyst but not in the anteroventral domain, including the neurogenic domain in all three Tg lines (Fig. 6A). We further clarified whether β-Gal+ otic progenitors only give rise to a subset of sensory structures at later stages by analyzing transgene expression in the E14.5 inner ear. β-Gal activity was restricted to the prosensory epithelia of the cochlea marked by Sox2 and utricle in all three Tg lines (Fig. 6B). Fewer β-Gal+ cells were also present in the anterior crista, but not in the other vestibular organs or ganglion neurons (Fig. 6B and SI Appendix, Fig. S7A), thus confirming that Enh1 only mediates expression in inner ear sensory subsets.
Fig. 6.
Brg1/Six1-bound 3′ Sox2 distal enhancers direct overlapping or complimentary patterns of expression in otic neurosensory cells in response to binding of Six1. (A–F) X-gal–stained G0 transgenic embryo at E10.5 (A, C, and E) or inner ear at E14.5 (B, D, and F) of each transgene driven by Sox2 Enh1-3. E14.5 cochlear sections were stained with anti-Sox2 (red) to label the prosensory domain (brackets). Otocysts are outlined by black dashed line and orientation of anterior (a), dorsal (d), or lateral (l) is indicated. Dashed white lines indicate regions where sections were generated. (A) Left and Center (higher-magnification panel) are whole-mount and Right is section view showing β-Gal+ cells (arrows) in the otocyst (Ot). Scale bar, 200 μm. (B) Whole inner ear (Left) showing β-Gal+ cells in the utricle (Ut) and cochlea (Co) sensory epithelium, which is marked by Sox2 on cochlear sections (Center and Right). Note that diffuse Sox2 staining was due to longer duration of the samples in the X-gal staining solution, which contains detergents that may cause leakage of nuclear proteins. (C) Whole-mount (Left) and section (Center and Right) view showing β-Gal+ cells in the ventral region of the otocyst (Ot) with strong activity in the anterior neurogenic domain. Open arrows point to migratory neuroblast progenitors. (D) Whole-mount inner ear (Left) and section (Center and Right) view showing β-Gal+ cells in all neurosensory structures. Arrow points to the cochlear organ of Corti. (E) Whole-mount (Left) and section (Right) view showing β-Gal activity in the otocyst (Ot) with stronger activity in the anterior neurogenic domain. Open arrows point to migratory neuroblasts. (F) Sections of Enh3-LacZ cochlea (Co) costained with anti-Sox2. (G–K) X-gal–stained G0 transgenic embryo at E10.5 (G, I, and K) or inner ear at E14.5 (H, J, and L) of each transgene driven by the different SIXmt enhancer. Transgenic lines: Enh1SIXmt, E10.5 (n = 4) and E14.5 (n = 5); Enh2SIXmt, E10.5 (n = 5) and E14.5 (n = 4); Enh3SIXmt, E10.5 (n = 3) and E14.5 (n = 3). (G–J) Whole-mount views of E10.5 otocyst or E14.5 inner ear. (K) Whole-mount (Left) or section (Right) view of otocyst (K) or cochlear sections (L). All transgenic lines showed consistent expression in the sensory progenitors. (Scale bars, 50 μm.)
The 856 bp of Enh2 element (+669 kb) was active in the entire ventral region of the otocyst, including the anterior neurogenic domain where the neuroblasts are specified in all three Tg lines Fig. 6C). On sections, β-Gal+ cells were detected in the otocyst and delaminating neuronal progenitors all three Tg lines (Fig. 6C). Additionally, Enh2 was active in the olfactory epithelium (SI Appendix, Fig. S7B), where Six1 has a crucial role as olfactory epithelium development arrests at E12.5 in the Six1−/− embryo (48). In the E14.5 inner ear, β-Gal activity was detected in the Sox2+ progenitors in all inner ear sensory organs and both vestibular and spiral ganglion (Fig. 6D and SI Appendix, Fig. S7B). The activity of Enh2 was also observed in the greater epithelial ridge region where Sox2 becomes down-regulated, which is likely due to the absence of cis-acting repressive elements in the 856-bp fragment but present in the intact Sox2 locus.
The 1,540 bp of Enh3 (+691 Kb) with strong H3K27ac deposition displayed activity in the ventral domain of the otocyst with more β-Gal+ cells in the anteroventral neurogenic domain in all three Tg lines (Fig. 6E and SI Appendix, Fig. S7C). On sections, β-Gal+ cells were observed in both the otocyst and neuroblasts (Fig. 6E). This enhancer was also active in the brain and mesenchymal cell surrounding the olfactory epithelium (SI Appendix, Fig. S7C), which is consistent with the published report showing Brg1-binding to this region with H3K4me1 deposition in E11.5 forebrain and face (45). In the E14.5 inner ear, β-Gal+ cells were overlapping with Sox2+ prosensory progenitors of all inner ear sensory organs and of both vestibular and spiral ganglion in all four Tg lines (Fig. 6F and SI Appendix, Fig. S7C).
Remarkably, however, SIXmt in each enhancer region abolished activity in the otocyst and neurosensory structures of E14.5 inner ear in all Tg lines (Fig. 6 G–L and SI Appendix, Fig. S7 D–F). In contrast, some β-Gal+ cells were observed in the brain for Enh1SIXmt-LacZ in three of four Tg lines (SI Appendix, Fig. S7D), while β-Gal activity of Enh2SIXmt-LacZ transgene was detected in some mesenchymal cells dorsal to the otocyst in all five Tg lines (SI Appendix, Fig. S7E). For the Enh3, SIXmt did not disrupt its activity in the brain and mesenchyme surrounding the olfactory epithelium in all three Tg lines (SI Appendix, Fig. S7F). In the E14.5 inner ear, no β-Gal+ cells were observed in two of three Tg embryos, but some ectopic β-Gal+ cells were detected in mesenchymal cells surrounding vestibular or cochlear epithelium in one of three Tg embryo (SI Appendix, Fig. S7F). Altogether, these results demonstrate that the three distal 3′ Sox2 enhancers cooperatively mediate the transcriptional activities of Six1-, Eya1-, and Brg1-based SWI/SNF complex to regulate Sox2 expression in “time and space” during inner ear neurosensory lineage commitment.
Discussion
Chromatin remodelers act to establish nucleosome-depleted regions at promoters (49) and promote nucleosome depletion to permit TFs access to their cognate binding sites (50, 51). Their tissue-specific functions during development depends on interaction with tissue-specific TFs. However, how chromatin remodelers interact with lineage-specific TFs to establish de novo enhancer repertoires to activate lineage-specific programs of gene expression during inner ear development remains unexplored. Six1 and its coactivator Eya1 are transcriptional regulators long known to be involved in inner ear neurosensory structure formation as mutations in either gene in human cause branchio-oto or branchio-oto renal syndrome, and no inner ear neurosensory structure forms in Eya1−/− or Six1−/− mice (1). However, whether Eya1 and Six1 cooperate to initiate the earliest event of neurosensory fate specification within the otic ectoderm and how this process is linked to chromatin-remodeling have not been studied. Here, we provide evidence that Eya1/Six1 collaborate with the Brg1-based SWI/SNF chromatin-remodeling complex to promote the initial specification of a proneurosensory-restricted progenitor cell from an ectodermal cell within the otic ectoderm by inducing Sox2 activation.
We identified Brg1 binding at two (Enh1 and Enh3) of three Six1-bound distal 3′ Sox2 CREs/enhancers that direct overlapping pattern of expression in otic neurosensory progenitors. The activity of these enhancers is associated with Six1-binding, as mutations of putative SIX-motifs in each enhancer specifically compromised their activity in the otic ectoderm or inner ear and abolished their binding to Six1 in HEK293 cells. Consistent with Brg1-binding to the Enh3 but not to Enh1 and Enh2 in E11.5 brain or face tissue (45), we found Enh3-LacZ expression in the brain and mesenchyme surrounding the olfactory epithelium (SI Appendix, Fig. S7C). Importantly, however, the activity of Enh3 in these two tissues is not associated with Six1 as Enh3SIXmt did not affect the reporter expression either in the brain or the mesenchyme (SI Appendix, Fig. S7F). As Sox2 also binds to this region in iMOP cells (SI Appendix, Fig. S6) and this region contains multiple SOX sites (SI Appendix, Table S4), Sox2 alone without Six1 may be incapable of directing expression in the otic cells. While our results show that Eya1 or Six1 depletion compromised Brg1-enrichment at the 3′ Sox2 enhancer regions, it is unclear whether Eya1 cooperates with Six1 to recruit Brg1, as Eya1 depletion results in reduced Six1 expression (14). These low levels of Six1 present in Eya1−/− otocyst, in which Brg1 is expressed, may be capable of assisting weak Brg1–BAF binding to the enhancer elements to interact with the promoter to weakly activate Sox2 expression. Consistent with this view, Brg1-binding to these enhancer regions is more reduced in Six1−/− than in Eya1−/− otocyst (Fig. 5G). This also suggests that Eya1 in the absence of Six1 is incapable of recruiting Brg1 to these CREs as it is expressed in Six1−/− otocyst (14).
Based on our genetic and molecular analyses, we speculate that these proteins directly interact or Eya1 as a coactivator may bridge Six1 and the Brg1–BAF complex to enable DNA bending to form a compact active complex capable of inducing transcriptional activation of Sox2 via binding to multiple distal CREs. In the absence of Eya1, Six1 may not form a stable complex with Brg1–BAFs, which then abrogates Brg1 recruitment, thus alone is incapable of inducing robust transcriptional activation. This view is in agreement with our previous observations of Eya1 dosage-dependent Sox2 expression in the otocyst (30) and very few Neurog1+ cells present in Eya1−/− otocyst (11, 30). Although Brg1 occupies other regions near the Sox2 gene body and at +755 kb cooccupied by Sox2, these regions may not mediate otic-restricted Sox2 expression as Brg1 binding at these regions was also identified in the E11.5 brain or face (SI Appendix, Fig. S6). Even if Brg1 interacts with other TFs to up-regulate otic Sox2 expression, the expression of such factors may be lost in Eya1−/−;Six1−/−. This explains why Brg1–BAFs without both Eya1 and Six1 fail to promote Sox2 activation in the otic ectoderm. After Sox2 is turned on, it may synergize with Eya1-Six1 to establish the precise gene-expression state that defines otic prosensory or neuronal identity. As Sox2 also physically interacts with Brg1 and Sox4/2 motifs are enriched in Brg1-peaks, Sox2 may not only cooperate with Eya1/Six1 but also with a Brg1-based SWI/SNF complex to jointly regulate subsequent neuronal or sensory lineage commitment.
Previous studies identified two conserved elements NOP1 (−10,256 to −10,510 bp) and NOP2 (+5.3 to +5.65 kb) that drive expression in the otocyst of chicken embryos (49, 52, 53). However, neither Six1 nor Brg1 occupancy was identified at these sites in our ChIP-seq datasets (Dataset S1). It is worth noting that previous studies identified two allelic mouse Sox2 mutants (9), Lcc induced by X-ray irradiation (54) and Ysb caused by a transgene insertion (55). Lcc/Lcc mice completely lack inner ear sensory organs (9) and spiral ganglion neurons (10), while Ysb/Ysb mice are severely hearing impaired (9). Notably, all three Sox2 enhancers (Enh1 to -3) are located within the regions between either the insertion points in Ysb or the breakpoints in Lcc (56). Thus, future studies to test whether each of the enhancers alone or all three together can complement Ysb and/or Lcc or whether deletion of these enhancers through CRISPR/Cas9 disrupts inner ear neurosensory structure formation will provide further evidence for their function in regulating Sox2 expression in vivo.
Interestingly, at the Eya1 locus, we identified Brg1-binding at −196 kb associated with H3K27me3 in the E10.5 otocyst but with H3K27ac in E13.5 cochlea, suggesting that Brg1 binding to this region is involved in transcriptional repression and activation of Eya1 during development. Motif search of this region identified numerus motifs, including GATA, C/EBP, Oct1, YY1, TBP, Sp1, Id1/3, AP1/2, IRF, c-Jun, c-Fos, and Myc. Future studies of assaying the function of this CRE and other Brg1-binding sites in vivo will shed light on its role in the control of otic neurosensory cell development.
Our RNA-seq failed to detect quantitative significance (P adjusted > 0.05) in differential expression of many Brg1 targets identified by ChIP-seq due to the variation between replicates. It is possible that Brg1 activity was not completely depleted in some cells by tamoxifen administration, which may lead to gene expression in those cells. Such expression may not be detected by in situ hybridization (ISH) but can be detected by deep-sequencing. Our RNA-seq highlighted aberrant activation of numerous TFs and signaling molecules necessary for muscle/heart contraction, all of which had strong H3K27me3 deposition at their promoter region without H3K27ac deposition. This may result in up-regulation of other muscle/heart related genes, such as Six1, which is also a critical regulator for smooth muscle cell development (57). Nonetheless, down-regulation of a subset of top otic neurosensory genes, including Oc90, Col9a2, Eya1, Eya4, Pax2, Sobp, Otx1, Gli1, Gas1, and Nr2f1 was evident in RNA-seq datasets. Our observation of aberrant up-regulation of oncogenic factors, such as Runx3 and tumor necrosis factors, is consistent with published studies showing that Brg1 acts as a cell cycle regulator and promotes both tumor-suppressive and oncogenic activities at distinct stages in some tumors (58, 59). Of particular interest, our RNA-seq identified increased expression of Map3k5, a direct target of Brg1, in Brg1cKO/cKO cells. As Brg1 binding to the proximal-promoter region of Map3k5 is accompanied by H3K27me3, the aberrant activation of Map3k5 is likely to trigger abnormal apoptosis, which was observed in Brg1cKO/cKO otocyst (SI Appendix, Fig. S2B). Thus, Brg1 may play a direct role in repressing Map3k5 expression to inhibit apoptosis. As abnormal apoptosis was also observed in Eya1−/− (13) or Six1−/− (14) otocyst, it is possible that Brg1 may interact with Eya1-Six1 to regulate otic cell survival.
In conclusion, our studies have provided crucial insights into gene-expression programs necessary for neurosensory fate induction within the otic ectoderm and repression of nonotic genes. These insights have broad implications for understanding the architecture of genetic regulatory circuits that induce the initial events of otic neurosensory lineage commitment. The present work provides a framework for understanding how the functional specificity of Brg1-containing BAF complexes is regulated by tissue-specific TFs Eya1 and Six1 to regulate the initial activation of Sox2 expression specifically in the otic ectoderm to specify proneurosensory fate. Future studies on how the chromatin remodelers regulate landscape of otic neurosensory-restricted enhancers will provide a deeper understanding of how the epigenome of otic cells is dynamically programmed to ensure neurosensory structure formation.
The importance of the Brg1-based BAFs in proneurosensory cell fate specification and cell survival highlights their potential as a target for therapeutic intervention in generation or conversion of general otic epithelial cells into neurosensory cells.
Methods
Animals, Tamoxifen Administration, Histology, Immunostaining, Antibodies, ISH, EdU, and TUNEL Assays.
All animal protocols were approved by Animal Care and Use Committee of the Icahn School of Medicine at Mount Sinai (protocol #06-0807). Brg1fl (24), Eya1CreER (21, 22), Sox2CreER (26), Sox2fl (60), Eya1+/− (13), Six1+/− (14), and TdTomato (Jackson laboratory, Stock No. 007914) mice were maintained on a 129/Sv and C57BL/6J mixed background. The 2×HA-3×Flag-Eya1 knockin allele was generated similarly to Eya1-LacZ knockin allele (30). Mice were bred using timed mating, and noon on the day of vaginal plug detection was considered as E0.5. Detailed protocols are described in SI Appendix, Methods.
Otocyst Single Cell Isolation, FACS, RNA-Seq, and RT-qPCR.
Otocysts were isolated from E10.5 Eya1CreER;TdTomato or Eya1CreER;Brg1fl/fl;TdTomato embryos and dissociated to single-cell suspension for FACS to isolate TdTomato+ cells. For each sample, we used ∼5,000 cells for total RNA preparation in replicates for RNA-seq analysis. Next, 100 to 500 ng of total RNAs were used for RT-qPCR. Detailed protocols are described in SI Appendix, Methods.
ChIP-Seq, ChIP, Peak Calling, Bioinformatic, and CoIP Analyses.
The ChIP assay, bioinformatic and coIP analyses were performed as previously described (42). Briefly, otocysts were dissected from E10.5 embryos, fixed, homogenized, and lysed for chromatin preparation as previously described (42). Detailed protocols are described in SI Appendix, Methods.
Transient (G0) Transgenic Analysis of Enhancer Activity.
The GFP or LacZ reporter driven by Hsp68 minimal promoter flanked by the H19 insulators (42) was used to generate reporter construct by inserting each individual enhancer element upstream of the Hsp68 minimal promoter. Detailed protocols are described in SI Appendix, Methods.
Statistical Analysis.
To determine the P values for all the experiments performed, Student’s t test was performed. A P value of <0.05 was assumed as statistically significant. P < 0.05, P < 0.01, and P < 0.001 are represented with single, double, and triple asterisks, respectively. All statistical analyses were performed with Microsoft Excel.
Supplementary Material
Acknowledgments
We thank the mouse genetics and gene targeting facility at Icahn School of Medicine for pronuclear injection and the flow cytometry facility for assistance with cell sorting. This work was supported by NIH Grant R01DC014718 (to P.-X.X.). B.F. was supported by NIH Grant RO1AG060504.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025196118/-/DCSupplemental.
Data Availability
All data needed to evaluate the conclusion are present in the paper, SI Appendix, and Datasets S1–S8. Brg1, Six1, H3K27ac, and H3K27me3 ChIP-seq and RNA-seq data reported in this paper have been deposited to the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE166588 and GSE158167). Public Sox2 ChIP-seq in iMOP (GSE62514), Flag-Brg1 ChIP-seq in E11.5 brain and face (GSE37151), and Six1 ChIP-seq in E10.5 embryo and cochlea (GSE108130) were used for comparison. Protocols and materials are available upon request.
References
- 1.Wong E. Y., Ahmed M., Xu P. X., EYA1-SIX1 complex in neurosensory cell fate induction in the mammalian inner ear. Hear. Res. 297, 13–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma Q., Chen Z., del Barco Barrantes I., de la Pompa J. L., Anderson D. J., neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron 20, 469–482 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Ma Q., Anderson D. J., Fritzsch B., Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J. Assoc. Res. Otolaryngol. 1, 129–143 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macova I., et al., Neurod1 is essential for the primary tonotopic organization and related auditory information processing in the midbrain. J. Neurosci. 39, 984–1004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim W. Y., et al., NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development 128, 417–426 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu M., et al., Loss of BETA2/NeuroD leads to malformation of the dentate gyrus and epilepsy. Proc. Natl. Acad. Sci. U.S.A. 97, 865–870 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruben R. J., Development of the inner ear of the mouse: A radioautographic study of terminal mitoses. Acta Otolaryngol. 220, 1–44 (1967). [PubMed] [Google Scholar]
- 8.Chen P., Segil N., p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development 126, 1581–1590 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Kiernan A. E., et al., Sox2 is required for sensory organ development in the mammalian inner ear. Nature 434, 1031–1035 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Puligilla C., Dabdoub A., Brenowitz S. D., Kelley M. W., Sox2 induces neuronal formation in the developing mammalian cochlea. J. Neurosci. 30, 714–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmed M., Xu J., Xu P. X., EYA1 and SIX1 drive the neuronal developmental program in cooperation with the SWI/SNF chromatin-remodeling complex and SOX2 in the mammalian inner ear. Development 139, 1965–1977 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed M., et al., Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev. Cell 22, 377–390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu P. X., et al., Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet. 23, 113–117 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Zheng W., et al., The role of Six1 in mammalian auditory system development. Development 130, 3989–4000 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruf R. G., et al., SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc. Natl. Acad. Sci. U.S.A. 101, 8090–8095 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdelhak S., et al., A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat. Genet. 15, 157–164 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Hagstrom S. A., et al., SOX2 mutation causes anophthalmia, hearing loss, and brain anomalies. Am. J. Med. Genet. A. 138A, 95–98 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Ozaki H., et al., Six1 controls patterning of the mouse otic vesicle. Development 131, 551–562 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Ho L., Crabtree G. R., Chromatin remodelling during development. Nature 463, 474–484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou D., Silvius D., Rodrigo-Blomqvist S., Enerbäck S., Xu P. X., Eya1 regulates the growth of otic epithelium and interacts with Pax2 during the development of all sensory areas in the inner ear. Dev. Biol. 298, 430–441 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J., et al., Identification of mouse cochlear progenitors that develop hair and supporting cells in the organ of Corti. Nat. Commun. 8, 15046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J., et al., Eya1 interacts with Six2 and Myc to regulate expansion of the nephron progenitor pool during nephrogenesis. Dev. Cell 31, 434–447 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang T., Xu J., Maire P., Xu P. X., Six1 is essential for differentiation and patterning of the mammalian auditory sensory epithelium. PLoS Genet. 13, e1006967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sumi-Ichinose C., Ichinose H., Metzger D., Chambon P., SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol. Cell. Biol. 17, 5976–5986 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merlo G. R., et al., The Dlx5 homeobox gene is essential for vestibular morphogenesis in the mouse embryo through a BMP4-mediated pathway. Dev. Biol. 248, 157–169 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Arnold K., et al., Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9, 317–329 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou D., Silvius D., Fritzsch B., Xu P. X., Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development 131, 5561–5572 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burton Q., Cole L. K., Mulheisen M., Chang W., Wu D. K., The role of Pax2 in mouse inner ear development. Dev. Biol. 272, 161–175 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Wang W., Grimmer J. F., Van De Water T. R., Lufkin T., Hmx2 and Hmx3 homeobox genes direct development of the murine inner ear and hypothalamus and can be functionally replaced by Drosophila Hmx. Dev. Cell 7, 439–453 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Zou D., et al., Eya1 gene dosage critically affects the development of sensory epithelia in the mammalian inner ear. Hum. Mol. Genet. 17, 3340–3356 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steevens A. R., Sookiasian D. L., Glatzer J. C., Kiernan A. E., SOX2 is required for inner ear neurogenesis. Sci. Rep. 7, 4086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dvorakova M., et al., Early ear neuronal development, but not olfactory or lens development, can proceed without SOX2. Dev. Biol. 457, 43–56 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vieth B., Ziegenhain C., Parekh S., Enard W., Hellmann I., powsimR: Power analysis for bulk and single cell RNA-seq experiments. Bioinformatics 33, 3486–3488 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Chen F., Liu X., Bai J., Pei D., Zheng J., The emerging role of RUNX3 in cancer metastasis (Review). Oncol. Rep. 35, 1227–1236 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Hu G., et al., Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 21, 1650–1658 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trotter K. W., Archer T. K., The BRG1 transcriptional coregulator. Nucl. Recept. Signal. 6, e004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Y., et al., Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 152, 248–261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heintzman N. D., et al., Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal T., Salmon-Divon M., Gerlitz G., The heterochromatin landscape in migrating cells and the importance of H3K27me3 for associated transcriptome alterations. Cells 7, 205 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marathe H. G., et al., BRG1 interacts with SOX10 to establish the melanocyte lineage and to promote differentiation. Nucleic Acids Res. 45, 6442–6458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spitz F., et al., Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc. Natl. Acad. Sci. U.S.A. 95, 14220–14225 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J., et al., Dynamic changes in cis-regulatory occupancy by Six1 and its cooperative interactions with distinct cofactors drive lineage-specific gene expression programs during progressive differentiation of the auditory sensory epithelium. Nucleic Acids Res. 48, 2880–2896 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kopecky B., Santi P., Johnson S., Schmitz H., Fritzsch B., Conditional deletion of N-Myc disrupts neurosensory and non-sensory development of the ear. Dev. Dyn. 240, 1373–1390 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Z., et al., Jxc1/Sobp, encoding a nuclear zinc finger protein, is critical for cochlear growth, cell fate, and patterning of the organ of corti. J. Neurosci. 28, 6633–6641 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Attanasio C., et al., Tissue-specific SMARCA4 binding at active and repressed regulatory elements during embryogenesis. Genome Res. 24, 920–929 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwan K. Y., Shen J., Corey D. P., C-MYC transcriptionally amplifies SOX2 target genes to regulate self-renewal in multipotent otic progenitor cells. Stem Cell Reports 4, 47–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afgan E., et al., The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen B., Kim E. H., Xu P. X., Initiation of olfactory placode development and neurogenesis is blocked in mice lacking both Six1 and Six4. Dev. Biol. 326, 75–85 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tolkunov D., et al., Chromatin remodelers clear nucleosomes from intrinsically unfavorable sites to establish nucleosome-depleted regions at promoters. Mol. Biol. Cell 22, 2106–2118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan H. Y., He X., Kingston R. E., Narlikar G. J., Distinct strategies to make nucleosomal DNA accessible. Mol. Cell 11, 1311–1322 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Ramirez-Carrozzi V. R., et al., Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 20, 282–296 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uchikawa M., Ishida Y., Takemoto T., Kamachi Y., Kondoh H., Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell 4, 509–519 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Okamoto R., Uchikawa M., Kondoh H., Sixteen additional enhancers associated with the chicken Sox2 locus outside the central 50-kb region. Dev. Growth Differ. 57, 24–39 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Lyon M. F., Phillips R. J., Fisher G., Dose-response curves for radiation-induced gene mutations in mouse oocytes and their interpretation. Mutat. Res. 63, 161–173 (1979). [DOI] [PubMed] [Google Scholar]
- 55.Dong S., et al., Circling, deafness, and yellow coat displayed by yellow submarine (ysb) and light coat and circling (lcc) mice with mutations on chromosome 3. Genomics 79, 777–784 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Xu P.-X., Cheah K. S. E., “SOX2 in neurosensory fate determination and differentiation in the inner ear” in Sox2, Lovell-Badge R., Ed. (ScienceDirect, 2016), pp. 263–280. [Google Scholar]
- 57.Delgado-Olguín P., et al., Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat. Genet. 44, 343–347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muthuswami R., et al., BRG1 is a prognostic indicator and a potential therapeutic target for prostate cancer. J. Cell. Physiol. 234, 15194–15205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roy N., et al., Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. 29, 658–671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shaham O., et al., Pax6 is essential for lens fiber cell differentiation. Development 136, 2567–2578 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusion are present in the paper, SI Appendix, and Datasets S1–S8. Brg1, Six1, H3K27ac, and H3K27me3 ChIP-seq and RNA-seq data reported in this paper have been deposited to the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE166588 and GSE158167). Public Sox2 ChIP-seq in iMOP (GSE62514), Flag-Brg1 ChIP-seq in E11.5 brain and face (GSE37151), and Six1 ChIP-seq in E10.5 embryo and cochlea (GSE108130) were used for comparison. Protocols and materials are available upon request.