Abstract
Identifying early indicators of toxicant-induced organ damage is critical to provide effective treatment. To discover such indicators and the underlying mechanisms of toxicity, we used gentamicin as an exemplar kidney toxicant and performed systematic perturbation studies in Sprague Dawley rats. We obtained high-throughput data 7 and 13 h after administration of a single dose of gentamicin (0.5 g/kg) and identified global changes in genes in the liver and kidneys, metabolites in the plasma and urine, and absolute fluxes in central carbon metabolism. We used these measured changes in genes in the liver and kidney as constraints to a rat multitissue genome-scale metabolic network model to investigate the mechanism of gentamicin-induced kidney toxicity and identify metabolites associated with changes in tissue gene expression. Our experimental analysis revealed that gentamicin-induced metabolic perturbations could be detected as early as 7 h postexposure. Our integrated systems-level analyses suggest that changes in kidney gene expression drive most of the significant metabolite alterations in the urine. The analyses thus allowed us to identify several significantly enriched injury-specific pathways in the kidney underlying gentamicin-induced toxicity, as well as metabolites in these pathways that could serve as potential early indicators of kidney damage.
Keywords: aminoaciduria, biomarkers, gentamicin, genome-scale models, metabolic pathways, urine
Exposure to toxic agents, such as drugs, industrial chemicals, and environmental pollutants, in both natural and occupational environments, can result in life-threatening and potentially long-term adverse health effects. Given the active role of the liver and kidneys in drug metabolism and toxicant clearance, tissues from these organs often suffer the most damage after an exposure event (Remmer, 1970; Vree et al., 1992). If the degree of exposure to a toxicant exceeds the capacity of the existing detoxification mechanisms to safely respond to and handle the chemical stress, injuries to these organs may cause irreversible damage, eventual functional failure, and death if left untreated (Lee, 2013; Soderland et al., 2010). In fact, hepatotoxicity and nephrotoxicity are the 2 most common reasons why commercialized drugs are later withdrawn from the market (Kaplowitz, 2005; Onakpoya et al., 2016). Hence, a major concern in both drug development efforts and for establishing safety limits in regulatory guidance is to evaluate chemicals for their potential to cause liver and kidney damage (Lin and Will, 2012). Extensive in vitro and in vivo animal screening studies have established chemical toxicity for a broad range of chemicals, yet current clinical standards for assessing toxicity rely on detecting the presence of already-damaged tissues or organ function. Thus, elevated aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels indicate liver damage and dysfunction (Karmen et al., 1955), whereas elevated creatinine and blood urea nitrogen levels indicate kidney damage (Ferguson and Waikar, 2012). Improving these standards would allow for earlier detection and treatment well before critical damage occurs. In particular, an improved clinical marker should flag potential toxicity at time points that precede the appearance of, or at doses below those that cause, overt tissue damage, toxicity, or disease initiation.
Recent advances in genomic, proteomic, and metabolomic techniques provide promising opportunities to re-examine the mechanisms of liver and kidney toxicity, as well as identify and propose proteins and metabolites as improved indicators of toxicity (Bandara et al., 2003; Collings and Vaidya, 2008; Igarashi et al., 2015; Ramirez et al., 2013; Robertson et al., 2011; Tugwood et al., 2003). These large-scale high-throughput studies can detect subtle disturbances of biochemical pathways (ie, changes in genes, proteins, and metabolites) caused by even mild to low levels of a toxicant. These changes reflect early perturbations in the vital cellular pathways that ultimately undergo toxicant-specific damage in the absence of any treatment and, hence, they may be predictive of emerging toxicity. Global mRNA levels (transcriptomes) are commonly measured to investigate toxicant-induced disturbances at the tissue level, as they are considered the best surrogates of protein levels given the limited throughput of current protein quantification techniques (Chandramouli and Qian, 2009). More recently, global changes in metabolites (metabolomes) are being measured for their potential to serve as organ-injury indicators (Araujo et al., 2017; Boudonck et al., 2009b; Buness et al., 2014; Iruzubieta et al., 2015; Zhang et al., 2012). In the canonical DNA-mRNA-protein function schema, metabolites, which are substrates and products of protein (enzyme) function, reflect the phenotypic changes driven by changes at the genotype level. Yet, few studies to date have concomitantly measured the global toxicant-induced changes of mRNA in tissues and of metabolites in accessible biofluids, such as blood and urine (Fannin et al., 2010; Sun et al., 2013). Moreover, it remains to be seen whether we can link the changes in mRNA to tissue-specific metabolite alterations that are detectable in these biofluids.
Toxicants cause injuries to excretory organs through a wide variety of mechanisms, which complicates the process of identifying indicators common to the detection and progression of injury. Moreover, given that these mechanisms remain unknown for most toxicants, relying on metabolites specific to each toxicant may not be a suitable approach to detect common indicators. Alternatively, we could potentially gain insights into toxicity mechanisms by identifying toxicant-induced perturbations in normal cellular metabolism (as measured by alterations in the transcriptome and metabolome) that predict the impending organ injury. However, this presents us with the daunting challenge of how to integrate such data into a comprehensive, mechanistic framework that accurately characterizes the state of a cell, tissue, or organism in terms of the underlying biological processes. A promising approach is to use genome-scale metabolic models (GSMs), which (1) represent collections of biochemical reactions known to occur in particular cells/tissues, (2) account for gene-protein-reaction-metabolite relationships, and (3) offer a way to mechanistically link and interpret changes in transcriptomic data to changes in metabolism. Indeed, recent studies indicate that GSMs can integrate large-scale high-throughput data, elucidate genotype-phenotype relationships, and identify various biological processes associated with disease states (Agren et al., 2012; Blais et al., 2017; Duarte et al., 2007; Jerby and Ruppin, 2012; Mardinoglu et al., 2014; Pannala et al., 2018, 2019; Shlomi et al., 2009). Thus, GSMs enable the prediction of changes in endogenous metabolites and their subsequent secretion into plasma and urine, where they can be detected. In addition, they can be used to glean further insights on the molecular mechanisms involved, providing an opportunity to distinguish the changes in biofluid metabolites strongly associated with tissue-specific gene-expression changes from all other metabolite alterations (Pannala et al., 2018, 2019).
Here, we used gentamicin, an aminoglycoside antibiotic widely used against infections caused by Gram-negative microorganisms and which selectively affects the kidneys, as an exemplar toxicant to generate global changes in mRNA, metabolites, and absolute fluxes (of central carbon metabolism) in Sprague Dawley rats. Gentamicin-induced nephrotoxicity occurs in 10%–20% of therapeutic regimens; the cellular damage that accumulates in the renal cortex is typically characterized by tubular damage arising from epithelial cell cytotoxicity (Edwards et al., 2007; Li et al., 2009). Using classical indicators of liver and kidney injuries, we determined the minimum dose (0.50 g/kg) that causes kidney injury in Sprague Dawley rats. We then used this dose to evaluate the gene perturbations in the liver and kidney using RNA sequencing (RNA-seq), changes in plasma and urine metabolites (metabolomics), and absolute changes in central carbon metabolism using 2H/13C metabolic flux analysis (MFA).
Building on a previously developed rat GSM (Blais et al., 2017; Pannala et al., 2019), we added new transport and exchange reactions to improve coverage of exchange metabolites and compartmentalized the modified GSM into a multitissue metabolic model in which the liver and kidneys could exchange metabolites via blood and urine compartments. Using this model, we investigated the metabolic differences between control and gentamicin-treated groups and evaluated changes in blood and urine metabolites directly linked to and driven by changes in gene expression in the liver and kidney. Of all the significantly altered metabolites that mapped onto the model, 77% and 70% from the plasma and urine, respectively, were associated with gene-expression changes in the kidneys alone. Conversely, changes originating in the liver were negligible and did not contribute to the alterations in the metabolite profile. Furthermore, our analysis revealed that, compared with blood metabolites, urine metabolites have greater potential to serve as kidney-injury indicators, as several metabolites in the amino acid (AA), carbohydrate, and lipid metabolism pathways increased significantly as early as 7 h after gentamicin exposure. Using the multitissue model, we elucidated potential mechanisms underlying gentamicin toxicity and identified a list of plausible metabolites to be further targeted and clinically assessed for their potential to serve as early indicators of kidney damage. The results show how our platform can be broadly applied to analyze and mechanistically interpret large-scale high-throughput data.
MATERIALS AND METHODS
Animals and study design
We carried out the experiments in accordance with the Guide for the Care and Use of Laboratory Animals of the U.S. Department of Agriculture and the National Institutes of Health, after obtaining protocol approval from the Vanderbilt University Institutional Animal Care and Use Committee and the U.S. Army Medical Research and Development Command Animal Care and Use Review Office. We purchased male Sprague Dawley rats at 10 weeks of age (weighing approximately 250–300 g) from Charles River Laboratories (Wilmington, Massachusetts) and housed them under environmentally controlled conditions (12:12-h light-dark cycle at 23°C). We gave the rats free access to water and a commercially available rodent diet, Formulab Diet 5001 (Purina LabDiet; Purina Miles, Richmond, Indiana) and allowed them to acclimatize to the housing conditions for a week.
Seven days before each experiment, we performed a catheter implantation surgery, as previously described in Shiota (2012). Each surgery involved anesthetizing a rat with isoflurane, followed by 1 of 2 procedures depending on the type of experiment. For experiments to determine the appropriate gentamicin dose and time points of assessment after exposure, as well as those to measure changes in gene expression and plasma metabolite profiles, we cannulated the right external jugular vein with sterile silicone catheters (0.51 mm in inner diameter/0.94 mm in outer diameter). Alternatively, for studies to measure metabolic flux, we cannulated the carotid artery and the right external jugular vein with sterile silicone catheters of the same dimensions. We passed the free end of each catheter subcutaneously to the back of the neck, where it was fixed. Finally, we occluded the catheter with a metal plug following a flush of heparinized saline (200 U heparin/ml). Following the surgery, we housed each rat individually.
Preliminary studies for determining appropriate dose and exposure time of gentamicin
Two days before each study, we moved the rats from their regular housing cages to metabolic cages (Harvard Apparatus, Holliston, Massachusetts). To determine the appropriate dose of gentamicin and the time points of assessment after exposure, we gave rats IP injections of either vehicle (2 ml/kg of saline, n = 6), 0.25 g/kg of gentamicin sulfate (n = 6), or 0.5 g/kg of gentamicin sulfate (n = 6) at 8 am. Subsequently, we collected blood and accumulated urine samples at 8 am and 5 pm daily for 3 days.
Studies for measuring changes in gene expression and plasma metabolite profiles
We selected 0.5 g/kg as the appropriate gentamicin dose and 2 time points after exposure, 1 short (7 h, n = 8) and the other long (13 h, n = 8), based on the results of the dose-response study. Following blood collection, we gave animals IP injections of either vehicle or gentamicin at 7 am, and then moved them to new housing cages where they could access water ad libitum but not food. After collecting blood from each group at 2 pm (7-h group) or 8 pm (13-h group), we anesthetized the animals with an intravenous injection of sodium pentobarbital through the jugular vein catheter, and then immediately performed a laparotomy. We collected urine directly from the bladder. After dissecting the liver and kidneys, we froze them using Wollenberger tongs precooled in liquid nitrogen. Prior to analyses, we kept the collected plasma, urine, and organs frozen in a −80°C freezer.
Measurement of tissue-injury indicators in blood and urine
To evaluate liver injuries, we measured the plasma levels of ALT and AST using ALT and AST activity assay kits (Sigma-Aldrich, St. Louis, Missouri), respectively. Similarly, to evaluate kidney injuries, we measured the kidney-injury molecule-1 (KIM-1) using the KIM-1 Rat ELISA kit (Abcam, Inc, Cambridge, Massachusetts).
Studies for measuring metabolite flux
For flux measurements, we gave rats an IP injection of either gentamicin (0.5 g/kg, n = 8) or vehicle (saline, 2 ml/kg, n = 8) at 7 am on the day of the study, and subsequently removed the food and provided water ad libitum. At 3:50 pm, we anesthetized the animals with isoflurane, and after collecting 200 µl of arterial blood through the carotid artery catheter to determine the natural isotopic abundance of circulating glucose, we injected a bolus of [2H2] water (99.9%) subcutaneously to enrich total body water to 4.5%. At 4 pm (ie, 9 h after dosing), after the rats had recovered from anesthesia, we placed them in bedded containers without food or water and connected them to sampling and infusion lines. Subsequently, we administered a prime-constant infusion of [6,6-2H2] glucose (80 mg/kg prime + 0.8 mg/kg/min infusion) into the systemic circulation through the jugular vein catheter for the duration of the study. Starting 120 min after the [2H2] water bolus, we delivered sodium [13C3] propionate (99%) as a prime-constant infusion (110 mg/kg + 5.5 mg/kg/min infusion). We prepared all infusates in a 4.5% [2H2] water-saline solution unless otherwise specified, and obtained stable isotopes from Cambridge Isotope Laboratories (Tewksbury, Massachusetts).
We monitored blood glucose (AccuCheck, Roche Diagnostics, Indianapolis, Indiana) and infused donor erythrocytes to maintain hematocrit throughout the study. Following a 100-min infusion of [13C3] propionate, we collected 3 blood samples (300 μl each) over a 20-min period. We centrifuged the arterial blood samples in EDTA-coated tubes to yield 3 plasma samples of 100 μl each, which we stored at −80°C prior to glucose derivatization and gas chromatography-mass spectrometry (GC-MS) analysis. Immediately after collecting the final steady-state sample, we quickly euthanized the rats by injecting sodium pentobarbital through the carotid artery catheter.
Preparation of glucose derivatives
We divided the plasma samples into 3 aliquots, and derivatized each separately to obtain di-O-isopropylidene propionate, aldonitrile pentapropionate, and methyloxime pentapropionate derivatives of glucose. To prepare di-O-isopropylidene propionate, we precipitated proteins from 20 µl of plasma using 300 µl of cold acetone, and then evaporated the protein-free supernatant to dryness in screw-cap culture tubes. Derivatization proceeded as previously described in Antoniewicz et al. (2011) to produce glucose 1,2,5,6-di-isopropylidene propionate. For aldonitrile and methyloxime derivatization, we precipitated proteins from 10 µl of plasma using 300 µl of cold acetone and evaporated the protein-free supernatants to dryness in microcentrifuge tubes. Derivatizations then proceeded as previously described in Antoniewicz et al. (2011) to produce glucose aldonitrile pentapropionate and glucose methyloxime pentapropionate. We evaporated all derivatives to dryness, dissolved them in 100 µl of ethyl acetate, and transferred them to GC injection vials with 250-µl glass inserts for GC-MS analysis.
GC-MS analysis
GC-MS analysis involved the use of an Agilent 7890A GC system with an HP-5 MS capillary column (30 m × 0.25 mm × 0.25 μm; Agilent J&W Scientific) interfaced with an Agilent 5975C Mass Spectrometer. We injected the samples into a 270°C injection port in splitless mode, while maintaining helium flow at 0.88 ml/min−1. For analysis of di-O-isopropylidene and aldonitrile derivatives, we held the column temperature at 80°C for 1 min, ramped it up at 20°C/min−1 to 280°C and held it there for 4 min, then ramped it up further at 40°C/min−1 to 325°C. The program for methyloxime derivatives was the same except for the rate of ramp-up to 280°C, which we set to 10°C/min−1. After a 5-min solvent delay, the mass spectrometer collected data in scan mode from m/z 300 to 320 for di-O-isopropylidene derivatives, m/z 100 to 500 for aldonitrile derivatives, and m/z 144 to 260 for methyloxime derivatives. We used a custom MATLAB function (Antoniewicz et al., 2007) to integrate each derivative peak and obtain mass isotopomer distributions (MIDs) for 6 specific ion ranges: aldonitrile, m/z 173–178, 259–265, 284–289, 370–376; methyloxime, m/z 145–149; and di-O-isopropylidene, m/z 301–308. To assess uncertainty, we calculated the root mean squared error by comparing the baseline MID of unlabeled glucose samples with the theoretical MID computed from the known abundances of naturally occurring isotopes.
2H/13C MFA
We employed the in vivo MFA methodology previously described in Hasenour et al. (2015). Briefly, we constructed a reaction network using the INCA software package (Young, 2014) (http://mfa.vueinnovations.com/mfa; Accessed November 19, 2019). This network defined the carbon and hydrogen transitions for biochemical reactions linking hepatic glucose production and associated intermediary metabolic reactions. We estimated the flux through each reaction relative to citrate synthase (fixed at 100) by minimizing the sum of squared residuals (SSRs) between the simulated and experimentally determined MIDs of the 6 fragment ions previously described, and repeating this process 25 times from random initial values. We used the chi-square test to assess goodness-of-fit, and computed 95% confidence intervals (CIs) by evaluating the sensitivity of the SSRs to variations in flux values (Antoniewicz et al., 2006). The average SSR of each experimental group (Control SSR: 22.69 ± 1.83; gentamicin SSR: 27.73 ± 2.17) fell within the 95% CI [13.8–41.9] of the corresponding chi-square distribution with 26 degrees of freedom (ie, the regressions were overdetermined by 26 measurements). We converted the relative fluxes to absolute values using the known [6,6-2H2] glucose infusion rate and rat weights, and then averaged the flux estimates for the steady-state samples to obtain a representative set of values for each rat.
Metabolomics analysis
We sent the samples to Metabolon, Inc (Durham, North Carolina), which performed the metabolomic analyses in a manner similar to a previous study (Hatano et al., 2016). Briefly, individual samples were subjected to methanol extraction and then split into aliquots for analysis by ultrahigh performance liquid chromatography/MS (UHPLC/MS). The global biochemical profiling analysis comprised 4 unique arms: a reverse-phase chromatography positive ionization method optimized for hydrophilic compounds (LC/MS Pos Polar), a corresponding method for hydrophobic compounds (LC/MS Pos Lipid), reverse-phase chromatography with negative ionization conditions (LC/MS Neg), and a hydrophilic interaction liquid chromatography (HILIC) method coupled to negative ionization (LC/MS Polar) (Evans et al., 2014). All methods alternated between full scan MS and data-dependent MSn scans. The scan range varied slightly between methods but generally covered m/z 70–1000.
The identification of metabolites involved automated comparison of the ion features in the experimental samples to a reference library of chemical standard entries, which included retention time, molecular weight (m/z), preferred adducts, in-source fragments, and associated MS spectra, followed by curation via visual inspection for quality control using software developed at Metabolon. Identification of known chemical entities was based on comparison with metabolomic library entries of purified standards (Dehaven et al., 2010).
We performed 2 types of statistical analyses: (1) significance tests and (2) classification analyses. For standard and nonstandard statistical analyses on log-transformed data, we used ArrayStudio and the R program (http://cran.r‐project.org; Accessed November 19, 2019), respectively. Following log transformation and imputation of missing values (if any) with the minimum observed value for each compound, we used Welch’s 2-sample t test to identify biochemicals that differed significantly (p < .05) between experimental groups. We estimated the false discovery rate (FDR) (q value) to account for the multiple comparisons that are typically made in metabolomics-based studies.
RNA isolation and sequencing
Because kidneys are histologically heterogeneous, we powdered frozen whole kidney samples in liquid nitrogen. We isolated total RNA from the liver and powdered kidneys using TRIzol Reagent (Thermo Fisher Scientific, Waltham, Massachusetts) and the direct-zol RNA MiniPrep kit (Zymo Research, Irvine, California). Subsequently, we submitted the isolated RNA samples to the Vanderbilt University Medical Center VANTAGE Core (Nashville, Tennessee) for RNA quality determination and sequencing. Following total RNA quality assessment using a 2100 Bioanalyzer (Agilent, Santa Clara, California), we used at least 200 ng of DNase-treated total RNA of high RNA integrity to generate poly-A-enriched mRNA libraries using KAPA Stranded mRNA sample kits with indexed adaptors (Roche, Indianapolis, Indiana). We then assessed the library quality and quantitated them using 2100 Bioanalyzer (Agilent) and KAPA library Quantification kits (Roche), respectively. We subjected the pooled libraries to 75-bp paired-end sequencing according to the manufacturer’s protocol (Illumina HiSeq3000, San Diego, California). We used Bcl2fastq2 Conversion Software (Illumina) to generate demultiplexed Fastq files.
Analysis of RNA-seq data
We used the RNA-seq data analysis tool Kallisto for read alignment and quantification (Bray et al., 2016). Kallisto pseudo-aligns the reads to a reference, producing a list of transcripts that are compatible with each read while avoiding alignment of individual bases. In this study, we pseudo-aligned the reads to the Rattus norvegicus transcriptome (Rnor_6.0) downloaded from the Ensembl website (Cunningham et al., 2019). Kallisto achieves a level of accuracy similar to that of other competing methods, but is orders of magnitude faster. Its speed allows for the use of a bootstrapping technique to calculate uncertainties of transcript abundance estimates by repeating the analyses after resampling with replacement. Here, we employed this technique to repeat the analysis 100 times. To identify differentially expressed genes (DEGs) from transcript abundance data, we used Kallisto’s companion analysis tool Sleuth, which uses the results of the bootstrap analysis during transcript quantification to directly estimate the technical gene variance for each sample (Pimentel et al., 2017).
Development of a multitissue rat metabolic model
To contextualize the high-throughput data generated from multiple tissues, we converted the rat GSM (iRno model) (Blais et al., 2017) into a multitissue GSM. First, we updated the most recent versions of the rat GSM (Pannala et al., 2018, 2019) to improve the coverage of metabolites exchangeable with the external space based on the evidence of gentamicin-induced global metabolite changes in the plasma and urine in the current study (Supplementary Tables 2 and 3). Specifically, by comparing the predicted and observed changes in metabolites, we identified metabolites that were present in the iRno model but lacked specific transport reactions that would allow their transport into the external space. Furthermore, we corrected the proton imbalance in the molecular formulas for 235 metabolites; these had originally been reported as charged instead of neutral species in the original iRno model (Supplementary Table 9).
We integrated the tissue-specific reconstructions for the liver and kidney into the updated rat GSM to create a multitissue model. First, we renamed the reactions and metabolites in each reconstruction for proper compartmentalization. Then, we constructed new blood and urine compartments that connected the liver to the kidney as well as the kidney to the urine compartment. We removed the exchange reactions in the tissue-specific reconstructions, allowing only gene-associated transporters and free diffusion for intracellular metabolite transport. We then added exchange reactions to allow the blood compartment to exchange metabolites with the external space (ie, to represent the exchange of blood metabolites with other organs of the body). We constructed the kidney exchange reactions with the urine compartment based on the metabolites identified in the global metabolomics data obtained as part of this study (Supplementary Table 3). We allowed metabolite exchange (ie, both uptake and secretion) between the blood compartment and the external system, but only metabolite secretion from the urine compartment to the external system. Overall, the resulting multitissue model, which contained 2324 unique genes and 17 881 reactions connected by 11 543 metabolites, satisfied the biological constraints regarding the connectivity between the liver and kidneys, and successfully reproduced all known functional tasks of these organs. We provide the complete model in SBML format in the Supplementary Information (RMTmodel_SBML).
We tested the consistency of the multitissue model by evaluating 3 functional tasks at the systems level: excretion of urea and creatinine in the urine, and production of glucose in the blood. We evaluated these tasks by keeping the secretion of the respective metabolite under consideration as the objective function in the model for a given input constraint. For example, to simulate the excretion of urea in urine, we set the urea exchange reaction in urine as the objective function and allowed one of the AAs to vary as input to the liver compartment while keeping all other carbon sources zero. A positive flux value for the objective function indicated successful capture of the functionality. We refer the reader to details of this procedure in a recent publication (Pannala et al., 2019).
Algorithm for high-throughput data integration and metabolite predictions
We used the transcriptionally inferred metabolic biomarker response (TIMBR) algorithm (Blais et al., 2017) to integrate the observed gene-expression changes in the liver and kidneys into the multitissue model and make predictions for metabolite alterations in the blood and urine. Briefly, the TIMBR algorithm uses the gene-protein-reaction (GPR) relationships in the model to convert the log2 fold changes of all liver- and kidney-specific alterations in gene expression into reaction weights. It then calculates the global network demand required for producing a metabolite in the blood and urine. The objective function minimizes the weighted sum of fluxes across all reactions for each condition and metabolite, so as to satisfy the associated mass balance and an optimal fraction of the maximum network capability to produce a metabolite. Based on values reported in the literature, we used appropriate uptake and secretion rates for the exchange reactions of the liver and kidney under fed or short-term fasting conditions (Supplementary Table 8). Specifically, we did not find consensus values for uptake and secretion rates of the kidney under short-term fasting conditions and approximated them with the fed-state conditions. Thus, using the gene-expression changes together with the uptake and secretion rates, TIMBR provides a production score (z-score) representing an increase or decrease for each metabolite in the plasma and urine.
We used the experimental log2 fold changes of significantly altered (FDR < 0.10) plasma and urine metabolites from the global metabolic profiling data (Supplementary Tables 2 and 3) and then compared the corresponding TIMBR production scores from the multitissue GSM at 7 or 13 h after gentamicin treatment (Supplementary Table 5). Here, we considered the metabolite levels as having increased or decreased based on TIMBR production score cutoff values of greater than 0.1 and less than −0.1, respectively. We considered metabolites with scores between −0.1 and 0.1 as unchanged. To test the robustness of the multitissue model results, we randomized the original gene-expression data by randomly sorting the gene names and using the resulting data as the input.
KEGG pathway analysis
To understand the biological significance of the alterations in gene-expression levels induced by gentamicin, we used the DEGs derived from Kallisto-Sleuth analyses and identified significantly altered DEGs that were mapped to the rat GSM as input, and used KEGG pathways to identify molecular pathways that were significantly enriched. We used the online tool Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Huang et al., 2009) to perform this task. In addition, we used the aggregated fold change (AFC) method (Yu et al., 2017), which calculates significantly enriched KEGG pathways together with their direction of change, to ascertain that the results were independent of the pathway detection method. Briefly, the AFC method calculates the mean fold change for each gene and defines the KEGG pathway score as the average mean fold change of all genes in the pathway. The sign of the pathway score represents the direction of regulation, with positive values indicating upregulation and negative values indicating downregulation. Similarly, to understand the biological significance of metabolites, we identified those metabolites whose levels were altered by gentamicin exposure and which mapped to the rat GSM as input, and used KEGG pathways to identify molecular pathways that were significantly enriched. We used the pathway analysis functionality in MetaboAnalyst 4.0 (Chong et al., 2018) to perform this task.
RESULTS
Conventional Kidney-Injury Indicators Are Limited in Early Detection of Gentamicin-Induced Kidney Injury
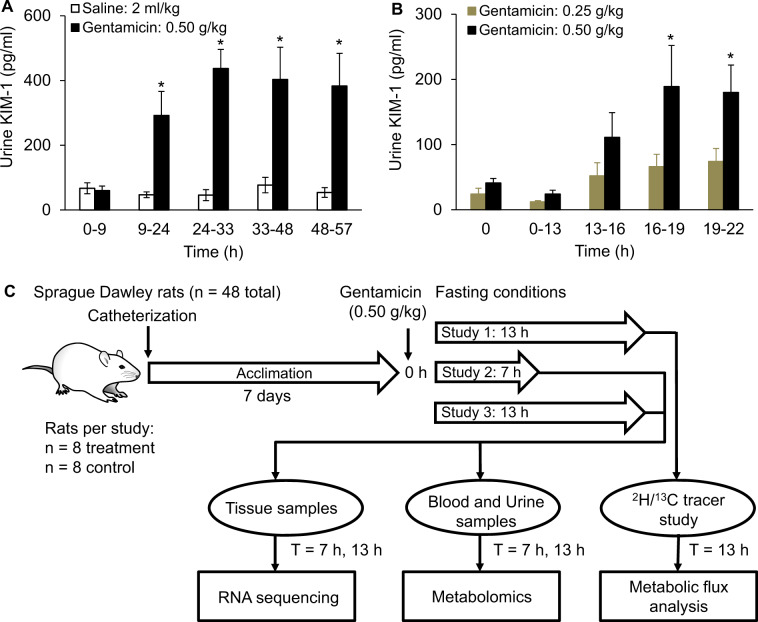
To determine the optimal dose for inducing kidney injury and the time points for injury assessment after initial exposure, we gave a single acute IP injection of either vehicle (2 ml/kg of saline; n = 6) or gentamicin (0.25 or 0.50 g/kg; n = 6 per group) to Sprague Dawley rats and examined the time course of injury in the liver and kidneys for 57 h (Fig. 1 and Supplementary Fig. 1). The classical indicators of liver injury, ALT and AST, did not change significantly in either exposure group relative to the control group (Supplementary Figs. 1A and 1B). Similarly, the classical kidney-injury marker, creatinine, showed no significant change in either the serum or urine, even at 57 h postgentamicin exposure (Supplementary Figs. 1C and 1D). However, levels of urinary kidney-injury molecule 1 (KIM-1), significantly differed between the control and treatment groups as early as 13–16 h after exposure. We observed the maximum levels of KIM-1 between 24 and 33 h postexposure at the high dose (0.50 g/kg; Figs. 1A and 1B). As expected from previous studies (Hoffmann et al., 2010; Luo et al., 2016; Piron et al., 1998), subsequent histopathological observations of liver and kidney tissues 57 h after exposure showed marked kidney injury but no liver injury (results not shown). These results demonstrated that serum creatinine could not detect early-stage gentamicin-induced kidney injury, even though histopathological analysis confirmed injury progression. In contrast, KIM-1, which is currently in the preclinical stage of development (Han et al., 2002; Parikh et al., 2013), differentiated the control group from the treatment groups, indicating the early progression of gentamicin-induced toxic effects on the kidneys.
Figure 1.
Experimental design to measure early perturbations in rat endogenous metabolism, and preliminary studies to determine the dose of gentamicin and time points after exposure to assess injury. Levels of kidney-injury molecule 1 (KIM-1) in urine samples collected at different time windows for rats treated with vehicle (control) or gentamicin (0.50 g/kg) (A) and for rats treated with 0.25 or 0.50 g/kg of gentamicin (B). * Indicates statistically significant differences based on a p value of less than .05. Schematic showing the design of the study, using Sprague Dawley rats exposed to a single dose of 0.50 g/kg of gentamicin under fasting conditions (C). Animals were catheterized 1 week before each experiment and allowed to recover. In Study 1, we administered either gentamicin or vehicle to rats (n = 8 each), and after 13 h, infused them with a combination of 2H/13C tracers to obtain isotope labeling measurements required for metabolic flux analysis. In Studies 2 and 3, we administered gentamicin to rats and observed them for 7 or 13 h (n = 8 in each group with corresponding controls), respectively. We collected samples of liver/kidney tissues and blood/urine after gentamicin exposure (at 7 or 13 h), which we subjected to RNA-sequencing and global metabolic profiling analysis, respectively.
Gentamicin Exposure Leads to Significant Early Changes in Kidney Gene Expression and Metabolites in Urine
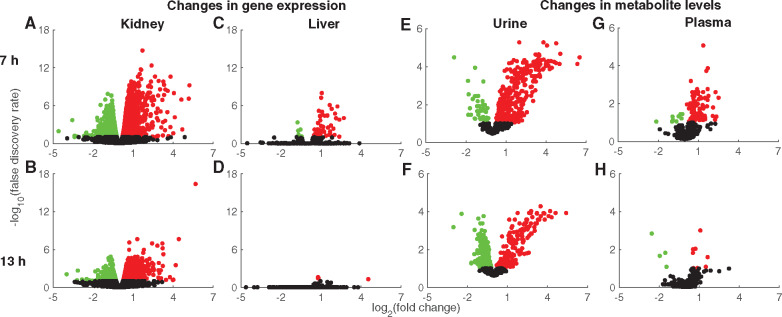
To determine the early perturbations in kidney metabolism that occur before KIM-1 levels change significantly, we selected a gentamicin dose of 0.50 g/kg and 7 and 13 h as the early time points for collecting liver and kidney tissue samples to investigate changes in global mRNA abundance, and blood and urine samples to perform global metabolic profiling (Figure 1C). We also monitored a separate group of rats for 13 h after gentamicin exposure (Study 1) and performed isotope tracer infusions to determine the absolute fluxes in the glucose production and TCA cycle pathways of central carbon metabolism under fasting conditions (Figure 1C; see Materials and Methods section). RNA-sequencing analysis revealed DEGs in the kidney as early as 7 h (Figure 2A), many of which remained differentially expressed 13 h after exposure (Figure 2B). Based on a FDR cutoff value of 0.1, gentamicin significantly changed the expression of 3126 and 1918 kidney genes at 7 and 13 h, respectively (Supplementary Table 1) and consistently altered expression of 1326 genes at both time points. This result suggests that the number of DEGs decreases over time as gentamicin is cleared from the kidneys. In contrast, we observed only 54 and 2 DEGs in the liver at 7 h (Figure 2C) and 13 h (Figure 2D), respectively (see Supplementary Table 1 for a complete list of DEGs). This result is commensurate with the notion that gentamicin acts primarily as a nephrotoxicant and rarely induces liver injury (Faa et al., 2012; Khan et al., 2011).
Figure 2.
Volcano plots for global changes in genes and metabolites induced by gentamicin. Log2 fold changes in gene expression induced by gentamicin compared with those induced by vehicle are plotted against the false discovery rates (FDRs), in kidney tissue after 7 (A) or 13 h (B) or in liver tissue after 7 (C) or 13 h (D). Red and green circles indicate significantly (FDR < 0.1) upregulated and downregulated genes, respectively. Black circles represent genes that did not change significantly. Log2 fold changes in metabolite levels induced by gentamicin relative to those induced by vehicle in urine after 7 (E) or 13 h (F) or in plasma after 7 (G) or 13 h (H). Red and green circles indicate significantly (FDR < 0.1) increased and decreased metabolites, respectively. Black circles represent metabolites that did not change significantly. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Global metabolic profiling analysis revealed significant changes in urine metabolites at both 7 and 13 h after gentamicin exposure (Figs. 2E and 2F). Using a FDR cutoff value of 0.1, approximately 400 urine metabolites changed significantly at both time points. Interestingly, at 7 h, the number of metabolites that significantly increased was higher than the number that decreased (Figure 2E). However, at 13 h, the numbers were comparable for both increased and decreased metabolites (Figure 2F). In contrast, we observed fewer plasma metabolite changes at both time points (Figs. 2G and 2H). Specifically, only 12 metabolites met the significance criteria for the later time point (Figure 2H). These results suggest better detection capability in the urine due to the concentrating effects of metabolites by the kidneys compared with plasma metabolite changes (see Supplementary Tables 2 and 3).
Under fasting conditions, the liver and kidneys maintain plasma glucose levels, with the liver contributing predominantly during short-term fasting while the kidneys contribute increasingly as the fasting duration increases (Gerich et al., 2001; Kaneko et al., 2018). To determine the effect of gentamicin on glucose production in central carbon metabolism, we performed 2H/13C tracer labeling and assessed the major metabolic fluxes in the glucose production pathway 13 h after administering gentamicin (0.50 g/kg) under short-term fasting conditions. These flux estimates represent whole-body contributions to glucose production although the liver is considered as the predominant source. Using the tracer enrichment data and MFA, we calculated the absolute fluxes for the enzymes in the glucose production pathway (Supplementary Table 4). Enzymes in this pathway did not significantly differ between the treatment and control groups (Supplementary Figure 2), consistent with our earlier observation that only 2 DEGs in the liver changed significantly at 13 h (Figure 2D). This result suggests that under short-term fasting, most of the glucose flux is derived from the liver, and that the contribution of the kidneys to glucose production remains either negligible or unaffected by gentamicin, at least for 13 h.
Multitissue Rat GSM Enables High-Throughput Data Integration and Analysis
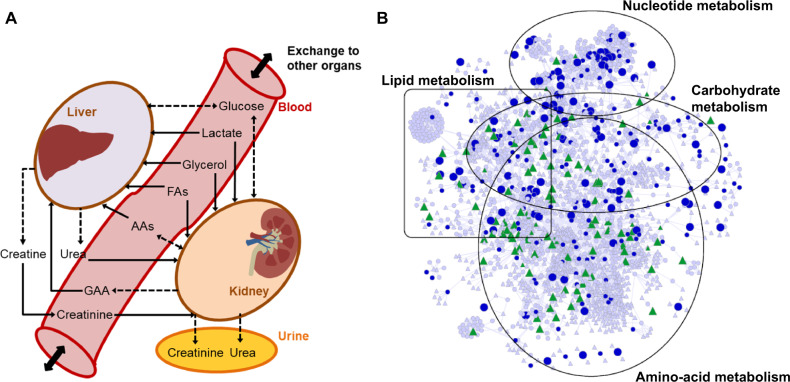
To elucidate the mechanism underlying gentamicin-induced kidney toxicity, we adopted a constraint-based modeling approach to identify major renal metabolic alterations between control and treatment groups. We updated a previously developed rat GSM (Blais et al., 2017; Pannala et al. 2018, 2019) to improve our coverage of exchange metabolites and compartmentalized the modified GSM into a multitissue model in which the liver and kidneys could exchange metabolites via blood and urine compartments (Figure 3A, Materials and Methods section). The uptake rates of AAs, fatty acids, glycerol, lactate, and glucose were approximated for fed conditions, using various sources in the literature (Elhamri et al., 1993; Yamamoto et al., 1974). We assumed that the metabolites in the blood compartment were common to both the liver and kidneys for uptake, whereas the metabolites in the kidney compartment could be exchanged between the blood and urine. We tested whether the multitissue GSM could model 3 physiologically important states of rat metabolism that encompass both the liver and kidney compartments. For the given input conditions, the model accurately captured the excretion of urea in urine, excretion of creatinine in urine, and production of glucose in the blood, suggesting the functional integrity of the developed multitissue model.
Figure 3.
A rat multitissue metabolic model that captures interorgan connectivity. A, A schematic representation of the rat multitissue model comprised of liver and kidney tissues connected via a blood compartment, with the kidneys connected to the urine compartment. Solid, dashed, and dashed bidirectional arrows denote physiological uptake, secretion, and mutual exchange, respectively, of a molecule between compartments. These relationships served as constraints for the kidney and liver to simulate the metabolite alterations. Abbreviations: AAs, amino acids; FAs, fatty acids; GAA, guanidinoacetic acid. B, Network visualization map of differentially expressed genes (DEGs; FDR < 0.1) in the kidneys and significantly altered metabolites in urine 7 h after gentamicin exposure, mapped onto the kidney-specific metabolic network. Circles, triangles, and line denote genes, metabolites, and interconnections, respectively. Blue circles represent DEGs that were significantly upregulated (large) or downregulated (small). Green triangles represent metabolites that significantly increased (large) or decreased (small). Triangles and circles in gray color represent for metabolites and genes that are unchanged. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
RNA-sequencing analysis provides information on DEGs that produce various protein products, including signaling proteins, transcription factors, and enzymes. We used the global gene-expression data as inputs to the multitissue GSM to identify genes that drive the metabolic reactions in the liver and kidneys. For the kidneys, we identified approximately 1800 DEGs that mapped onto the model for both time points. From among these DEGs, we identified 425 and 267 that significantly changed at 7 and 13 h, respectively (FDR cutoff value of 0.1). For the liver, we identified approximately 1700 DEGs that mapped onto the model. However, only 9 and 1 DEGs satisfied the significance criteria at 7 and 13 h, respectively.
Similarly, global metabolic profiling analysis identified 633 and 654 metabolites in the plasma and urine, respectively. However, the coverage of measurements remains sparse for many pathways of interest. Specifically, a comprehensive mapping of all of these metabolites onto the rat multitissue GSM, using both KEGG ID annotations and metabolite names, indicated a coverage of only approximately 40%, suggesting the lack of functional annotation for several identified metabolites in the data set. Figure 3B shows a network visualization map of significant DEGs in the kidneys and significantly altered metabolites in urine at 7 h after gentamicin exposure. The map shows that the majority of the significantly changed genes and metabolites could be grouped into 4 major metabolic super-pathways: those of nucleotides, lipids, carbohydrates, and AAs.
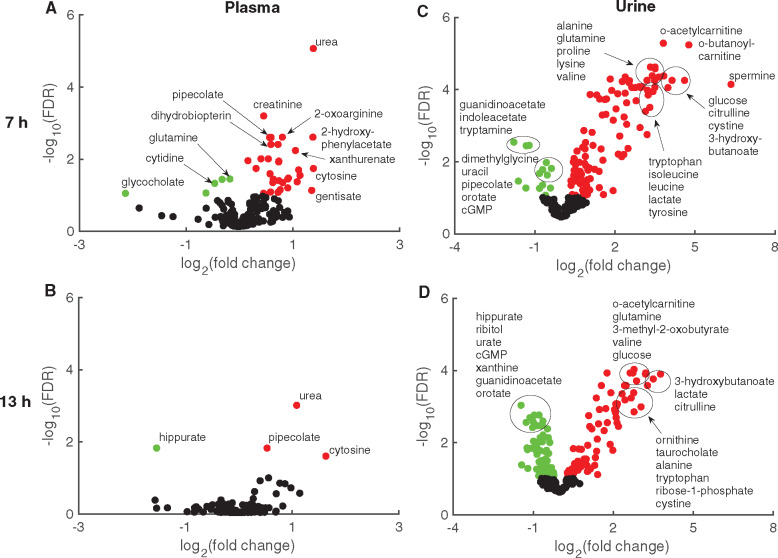
Of all the measured metabolites that mapped onto the model, we identified 37 and 4 in plasma (Figs. 4A and 4B), and 121 and 139 in urine (Figs. 4C and 4D) at 7 and 13 h, respectively, after gentamicin exposure. We consistently detected changes in urea, pipecolate, and cytosine in the plasma samples at both 7 and 13 h (Figs. 4A and 4B, respectively). Although we also detected an increase in the level of plasma creatinine, a classical kidney-injury marker, at 7 h, the magnitude of change was smaller than that in other metabolites and was not significant at 13 h. In contrast, many urine metabolites that mapped onto the model changed significantly (Figs. 4C and 4D). At both time points, many metabolites involved in AA metabolism (eg, citrulline, alanine, glutamine, tryptophan, lysine, valine, and isoleucine) consistently increased whereas many involved in nucleotide metabolism (eg, orotate, guanidinoacetate, and cGMP) consistently decreased, indicating their potential to serve as indicators of kidney injury.
Figure 4.
Volcano plots for measured metabolites mapped onto the multitissue model. Metabolites mapped onto the multitissue model based on KEGG ID annotations from global plasma (A, B) and urine (C, D) samples. Log2 fold changes in metabolite levels induced by gentamicin compared with those induced by vehicle are plotted against false discovery rates (FDRs) after 7 (A, C) or 13 h (B, D). Red and green circles indicate metabolites that significantly (FDR < 0.1) increased and decreased, respectively. Black circles represent metabolites that did not change significantly. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Multitissue Model Predicts Metabolite Alterations Based on Gene-Expression Changes
Metabolites in the plasma and urine change due to several factors, including contributions from all organs involved in their systemic circulation. To determine the tissue of origin for the observed changes in a biofluid metabolite, we used the TIMBR algorithm—a recently developed computational method for integrating high-throughput data—and simulated the changes in plasma and urine metabolites based on concomitantly measured gene-expression changes in liver and kidney tissues. Briefly, TIMBR first summarizes the gene-expression changes as a set of reaction weights that represent the relative cost of carrying flux through each reaction, and then calculates the global network demand required for biomarker production by minimizing the weighted sum of all fluxes across all reactions for each condition (Blais et al., 2017). In this way, for all exchangeable metabolites, it predicts the relative propensity for a metabolite to be elevated or reduced in the plasma or urine, based on the gene-expression changes in the exposure condition relative to those in the control condition. Thus, using the boundary constraints on the uptake and secretion of exchange metabolites for the liver and kidney (see Materials and Methods section) and the TIMBR algorithm, we integrated the gene-expression data to predict the changes in plasma and urine metabolites following exposure to gentamicin. We then compared the log2 fold changes of the metabolites identified from the global metabolic profiling analysis (Supplementary Tables 2 and 3) with the model predictions, and assessed how accurate the model was in evaluating the direction of gentamicin-induced changes in metabolite levels.
The multitissue model correctly evaluated the direction of change (an increase or decrease) for 77% and 70% of the metabolites that significantly changed in the plasma and urine, respectively, at 7 h after gentamicin exposure (Table 1; Supplementary Table 5). In contrast, when we provided the model with random gene-expression changes as the inputs, the corresponding performance levels were 17% and 13%. At 13 h, the model predicted the direction of change for 50% and 48% of the metabolites that significantly changed in the plasma and urine, respectively, showing little difference from the performance of the model (approximately 36%) where we used random gene-expression changes as inputs. The reduced correspondence between metabolite changes and gene-expression changes at 13 h is likely due in part to the reduction in the number of significant DEGs at this time point (Figure 2) and to other factors altering metabolite levels. These results indicate that many of the metabolite changes were correlated with gene-expression changes in the kidney for the early time point (7 h) where the number of genes and their magnitude of change was significant. Furthermore, our model assessments were similar with or without the inclusion of changes in liver gene expression, suggesting that most of the observed metabolite changes were due to gentamicin-induced changes in kidney metabolism.
Table 1.
Summary of the Metabolites Mapped, and Percentage of Mapped Metabolites Predicted Accurately, by the Multitissue Model Using Randomized or Actual Gentamicin-induced Gene-expression Data
| Sample | Time (h) | Metabolites Mapped to the Model | Significantly Changed Metabolites | Model Predictions Based on Gene-Expression Data |
|
|---|---|---|---|---|---|
| Random (%) | Actual (%) | ||||
| Plasma | 7 | 234 | 31 | 17 | 77 |
| 13 | 234 | 2 | 0 | 50 | |
| Urine | 7 | 222 | 113 | 13 | 70 |
| 13 | 222 | 127 | 36 | 48 | |
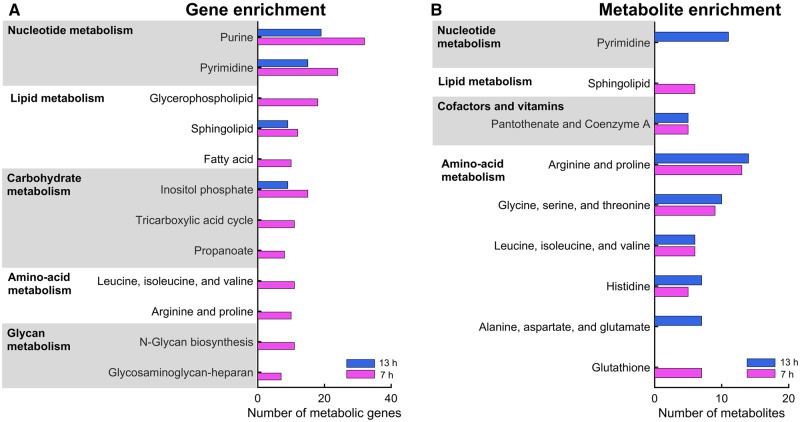
To further ascertain whether the gene-expression changes in the kidney were causally related to the metabolite changes in biofluids, we performed pathway enrichment analysis, using the online tool DAVID and the AFC method, by selecting significant (FDR < 0.1) DEGs from the kidney and metabolites in urine as the inputs (see Materials and Methods section). Our analysis revealed that many genes involved in the metabolism of nucleotides—lipids, carbohydrates, AAs, and glycans—significantly changed as early as 7 h after gentamicin exposure (Figure 5A;Supplementary Table 6). Several metabolites in a subset of these pathways also changed significantly, showing good correlations with the observed gene-expression changes (Figure 5B;Supplementary Table 7). Specifically, genes and metabolites involved in arginine and proline metabolism, as well as in valine, leucine, and isoleucine metabolism, were strongly correlated at the early time point (7 h), whereas those involved in pyrimidine metabolism were strongly correlated at the later time point (13 h).
Figure 5.
Pathway enrichment analysis of significantly altered metabolic genes in the kidney and metabolites in urine. Bar graphs showing the number of metabolic genes (A) and metabolites (B) involved in the significantly enriched pathways, based on genes from the kidney and metabolites from urine, respectively. We used the most significantly altered (FDR < 0.1) differentially expressed genes and metabolites represented in the multitissue model to perform gene and metabolite enrichment analysis.
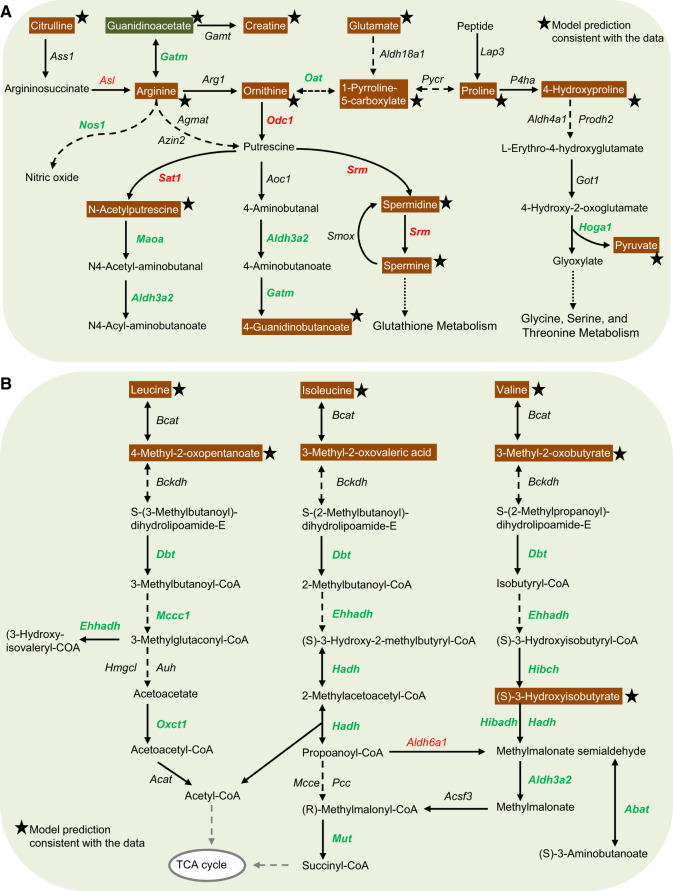
Gentamicin Downregulates Arginine and Proline Metabolism and Leucine, Isoleucine, and Valine Degradation
Gentamicin exposure can lead to aminoaciduria, a condition that involves increased excretion of AAs in urine (Gartland et al., 1989; Macpherson et al., 1991). Proximal tubular transport processes have been implicated in the aminoglycoside toxicity that leads to aminoaciduria and tubular proteinuria (Humes et al., 1982). However, the underlying molecular mechanisms leading to aminoaciduria remain unclear. Using the GPR relationships in the multitissue model, together with pathway enrichment analysis, we further investigated the alterations in genes and metabolites involved in the metabolism of arginine and proline (Figure 6A). None of the transporter genes involved in reabsorption of arginine and proline showed significant changes in expression in the kidneys. We observed increased expression of Asl, the gene responsible for the production of argininosuccinate lyase (the enzyme that converts argininosuccinate to arginine), but downregulation of Nos1, the subsequent gene responsible for the conversion of arginine to nitric oxide. Similarly, we observed increased expression of genes responsible for the production of putrescine, N-acetylputrescine, and spermine from ornithine, but downregulation of the genes that convert them to their respective downstream metabolites (Figure 6A). These changes suggest a major dysregulation in the metabolism of arginine and proline, leading to their accumulation in urine. The multitissue model, which accounts for the entire network demand to calculate the propensity of a metabolite to increase or decrease based on gene-expression changes, accurately predicted the changes for all of these metabolites, most of which were elevated in urine as early as 7 h after gentamicin exposure (Figure 6A, stars).
Figure 6.
Summary of gentamicin-induced alterations in genes in the kidneys and metabolites in the urine in amino-acid metabolism. Mapping of significantly altered genes and metabolites involved in arginine and proline metabolism (A) and leucine, isoleucine, and valine degradation (B). Each arrow indicates the direction of a reaction converting a substrate into a product, with the name of the gene indicated next to the arrow. Upregulated and downregulated genes are shown in red and green, respectively. Increased and decreased metabolites in urine are shown in white text with orange and green backgrounds, respectively. Stars indicate model predictions consistent with the data. Dashed arrows indicate multiple steps involved in a reaction; arrows with dotted lines indicate metabolite precursors involved in other pathways. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Branched-chain amino acids (BCAAs), ie, valine, leucine, and isoleucine, are essential AAs that play a key role in the biosynthesis of peptides, sterols, ketone bodies, and glucose. The first step in BCAA catabolism involves catalysis by the branched-chain aminotransferase (Bcat) isozymes. The distribution of BCAA transamination capacity in rat tissues suggests that the level of activity of Bcat enzymes in the kidneys is second only to that in skeletal muscle (Suryawan et al., 1998). Gentamicin exposure did not significantly alter the expression levels of the first 2 enzymes involved in BCAA catabolism (Figure 6B). However, it significantly downregulated the genes responsible for subsequent BCAA degradation, leading to impaired catabolism resulting in inefficient conversion of BCAAs to precursor molecules necessary for the TCA cycle. Consequently, this significantly elevated BCAAs in urine at both time points after gentamicin exposure (Figure 6B, white text on orange background). Our model simulations further showed that these metabolite changes were correlated with the observed impairments in gene-expression changes in BCAA catabolism (Figure 6B, stars).
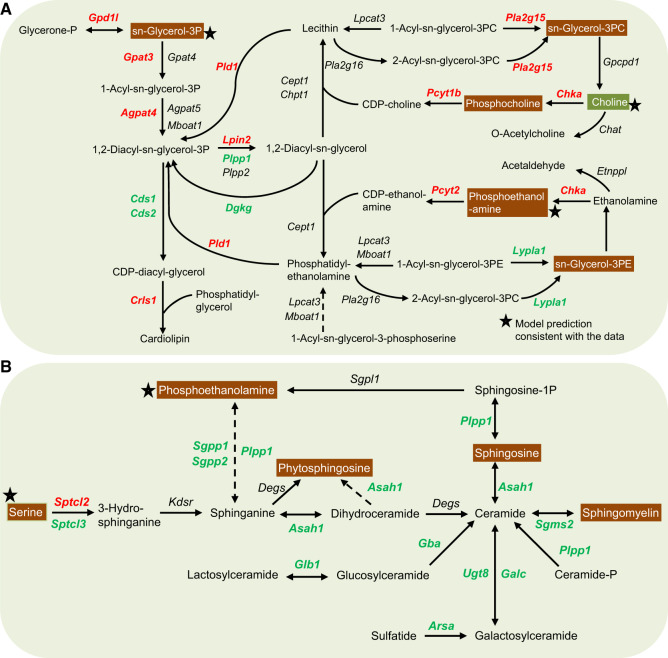
Gentamicin Upregulates Genes Involved in Glycerophospholipid Metabolism and Downregulates Genes Involved in Sphingolipid Metabolism
Gentamicin binds to membrane phospholipids, alters their turnover and metabolism, and thereby causes phospholipidosis (Giuliano et al., 1984; Nonclercq et al., 1992). Diacylglycerols, which are the basic components of tissue membranes and key intermediates in the formation of many glycerolipids, function as second messengers in many cellular processes. Genes responsible for the production of diacylglycerol in glycerophospholipid metabolism significantly increased (Figure 7A, red), indicating alterations in lipid metabolism due to gentamicin-induced toxicity. As a result, several metabolites in this pathway significantly increased in urine (Figure 7A, white text on orange background). Consistent with the model predictions based on gene-expression changes, urine levels of choline significantly increased whereas those of glycerol-3-phosphate and phosphoethanolamine significantly decreased. However, the observed increases in urine levels of phosphocholine, glycerol-3-phosphocholine, and glycerol-3-phosphoethanolamine were inconsistent with the changes in gene expression, suggesting the involvement of other factors that govern these metabolite changes.
Figure 7.
Summary of gentamicin-induced alterations in genes in the kidneys and metabolites in the urine in lipid metabolism. Mapping of significantly altered genes and metabolites involved in the metabolism of glycerophospholipids (A) and sphingolipids (B). Each arrow indicates the direction of a reaction converting a substrate into a product, with the name of the gene indicated next to the arrow. Upregulated and downregulated genes are shown in red and green, respectively. Increased and decreased metabolites in urine are shown in white text with orange and green backgrounds, respectively. Stars indicate model predictions consistent with the data. Dashed arrows indicate multiple steps involved in a reaction. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In contrast to the genes involved in glycerophospholipid metabolism, gentamicin exposure significantly downregulated most genes involved in sphingolipid metabolism (Figure 7B). Specifically, although levels of Sptcl2, the gene responsible for the initial rate-limiting step in sphingolipid biosynthesis, significantly increased, subsequent downstream genes responsible for the synthesis of ceramides significantly decreased at both time points after gentamicin exposure. Correspondingly, the urine levels of many precursor metabolites involved in sphingolipid metabolism significantly increased. However, the global network demand calculated for these metabolites using the multitissue model predicted increased urine levels only for serine and phosphoethanolamine (Figure 7B, stars).
Metabolites in Highly Enriched Pathways Proposed as Indicators of Gentamicin-Induced Kidney Toxicity
Although global metabolic profiling revealed significantly altered metabolites in various biochemical pathways, their relevance to gentamicin toxicity was unclear. By using the multitissue model together with pathway enrichment analysis, however, we were able to determine highly enriched pathways in the kidneys and compile a plausible panel of significantly altered plasma and urine metabolites strongly correlated with changes in gene expression. Tables 2 and 3 show lists of these metabolites in the plasma and urine, respectively, which the model correctly predicted in the highly enriched pathways, arranged according to their main and subordinate pathways. Specifically, our analysis revealed that gentamicin exposure significantly increased urine metabolites involved in AA metabolism as early as 7 h after exposure and that many of them remained elevated at 13 h after exposure (Table 3). These metabolites could potentially serve as early indicators of gentamicin-induced kidney toxicity. Overall, our GSM-based analyses of gene-expression changes in tissues and the corresponding metabolite alterations in biofluids provide a mechanistic interpretation of the observed changes. Thus, they increase our confidence that the identified putative indicators are causally related to the changes induced by exposure to gentamicin.
Table 2.
List of Significantly Altered Metabolites in the Plasma Based on Model Predictions at 7 and 13 h Postgentamicin Exposure
| Main Pathway | Subordinate Pathway | Metabolite | Log2 (Fold Change) |
|
|---|---|---|---|---|
| 7 h | 13 h | |||
| Metabolism of cofactors | Nicotinate and nicotinamide | Quinolinate | 0.92 | |
| N1-methyl-4-pyridone-5-carboxamide | 0.58 | |||
| N1-methyl-2-pyridone-5-carboxamide | 0.58 | |||
| Thiamine | Thiamine monophosphate | 0.63 | ||
| Carbohydrate metabolism | Ascorbate and aldarate | Inositol | 0.57 | |
| Pentose and glucuronate | Glucuronate | 0.74 | ||
| Xylitol | 0.59 | |||
| Ribitol | 0.45 | |||
| Amino sugar and nucleotide sugar | Neu5Ac | 0.72 | ||
| Neu5Gc | 1.08 | |||
| AA metabolism | Arginine and proline | Argininosuccinate | 0.81 | |
| Urea | 1.39 | 1.10 | ||
| 4-Acetamidobutanoate | 0.77 | |||
| Lysine degradation | l-Pipecolate | 0.57 | 0.54 | |
| Cysteine and methionine | 5-Methylthioadenosine | 0.54 | ||
| Phenylalanine | 2-Hydroxyphenylacetate | 1.38 | ||
| Tryptophan | 2,5-Dihydroxybenzoate | 1.36 | ||
| Indoleacetate | 0.91 | |||
| Xanthurenate | 1.05 | |||
| Anthranilate | 0.64 | |||
| Nucleotide metabolism | Pyrimidine | Orotidine-5-phosphate | 0.74 | |
| Cytidine | −0.45 | |||
Table 3.
List of Significantly Altered Metabolites in the Urine Based on Model Predictions at 7 and 13 h Postgentamicin Exposure
| Main Pathway | Subordinate Pathway | Metabolite | Log2 (Fold Change) |
|
|---|---|---|---|---|
| 7 h | 13 h | |||
| AA metabolism | Arginine and proline | Proline | 3.50 | 2.45 |
| Citrulline | 4.15 | 3.30 | ||
| Arginine | 2.80 | 2.10 | ||
| Ornithine | 2.97 | 2.34 | ||
| trans-4-Hydroxyproline | 1.83 | 1.10 | ||
| Creatine | 2.19 | 1.03 | ||
| Guanidinoacetate | −1.31 | −1.25 | ||
| 5-Oxoproline | 0.80 | 0.35 | ||
| Spermidine | 2.34 | |||
| Spermine | 6.36 | |||
| 4-Acetamidobutanoate | 0.58 | |||
| N-Acetylputrescine | 0.41 | |||
| Glycine, serine, and threonine | Glycine | 2.28 | 1.50 | |
| Dimethylglycine | −0.81 | −1.01 | ||
| Serine | 3.03 | 2.13 | ||
| Threonine | 2.54 | 1.46 | ||
| Glycerate | 0.29 | |||
| Pyruvate | 1.90 | 1.13 | ||
| Cysteine and methionine | Methionine | 3.20 | 2.06 | |
| Cystine | 4.60 | 3.05 | ||
| Cysteine | 0.90 | |||
| Valine, leucine, and isoleucine | Leucine | 3.44 | 2.15 | |
| Isoleucine | 3.23 | 2.12 | ||
| Valine | 3.66 | 2.88 | ||
| 4-Methyl-2-oxopentanoate | 1.70 | 1.78 | ||
| 2-Oxo-3-methylvalerate | 1.19 | 1.57 | ||
| 3-Methyl-2-oxobutyrate | 2.75 | 2.77 | ||
| 3-Hydroxyisobutyrate | 1.63 | |||
| Histidine | Histidine | 2.45 | 1.41 | |
| N-Methylhistamine | 0.46 | |||
| Histamine | 1.42 | 1.32 | ||
| Carnosine | 0.91 | |||
| Alanine, aspartate, and glutamate | Alanine | 3.51 | 2.43 | |
| Glutamate | 1.56 | 0.97 | ||
| 1-Pyrroline-5-carboxylate | 1.08 | |||
| Asparagine | 2.40 | 1.21 | ||
| Glutamine | 3.34 | 2.64 | ||
| Phenylalanine and tyrosine | Phenylalanine | 2.92 | 2.11 | |
| Tyrosine | 3.15 | 2.20 | ||
| 4-Hydroxyphenylacetate | 0.92 | |||
| 3-Methoxytyramine | 0.53 | |||
| Vanillylmandelate | 0.90 | |||
| 2-Hydroxyphenylacetate | 0.60 | |||
| 4-Hydroxyphenyllactate | 0.57 | |||
| Tryptophan | Tryptophan | 3.50 | 2.68 | |
| 5-Hydroxyindoleacetate | 1.00 | |||
| 3-Hydroxyanthranilate | 0.86 | |||
| Kynurenine | 2.26 | 1.90 | ||
| Anthranilate | 1.42 | |||
| Lysine | Lysine | 3.41 | 2.17 | |
| l-2-Aminoadipate | 2.35 | |||
| Carbohydrate metabolism | Glycolysis | Glucose | 3.84 | 3.22 |
| Lactate | 3.32 | 3.51 | ||
| Pentose and fructose | Xylitol | 0.45 | ||
| Mannose | 2.56 | |||
| Amino sugar | Glucuronate | 0.70 | ||
| Lipid metabolism | Ketone | (R)-3-hydroxybutanoate | 4.00 | 3.78 |
| Inositol | Inositol | 2.12 | 1.05 | |
| Phospholipid | Choline | −0.48 | ||
| Ethanolamine phosphate | 3.22 | 2.00 | ||
| Glycerolipid | Glycerol-3-phosphate | 1.25 | ||
| Nucleotide metabolism | Pyrimidine | 3-Ureidopropionate | −0.51 | |
| Thymine | 0.71 | |||
| Purine | cGMP | −0.76 | ||
| 2-Deoxyadenosine | −1.19 | |||
DISCUSSION
Unlike inference-based methods of analysis, genome-scale metabolic modeling provides the ability to integrate high-throughput data, enabling increased understanding of genotype-phenotype relationships. Metabolites, the substrates and products of cellular metabolism, are regulated at various levels by genetic makeup, feedback mechanisms, and environmental factors. Moreover, given that most metabolites in the systemic circulation are present in all organs of the body, changes in any particular metabolite will reflect the summed contributions from all organs. Therefore, we need to consider perturbations of metabolites in peripheral compartments (ie, blood and urine) within this context to gain a comprehensive understanding of the causal relationships between genes, metabolites, and toxicity endpoints, and thereby address their potential to serve as indicators of organ injury. Nonetheless, the genes and their protein products that drive the metabolic reactions are one of the major drivers of metabolite perturbations. Hence, using gentamicin as an exemplar toxicant, here we systematically characterized its effects on endogenous cellular metabolism by capturing alterations of genes in rat liver and kidney tissues and of metabolites in the blood and urine.
To address the current lack of early indicators of gentamicin-induced kidney injury, we analyzed global changes in gene expression and metabolite levels and identified significant changes induced by gentamicin as early as 7 h postexposure. Using an updated rat multitissue GSM, we assessed the changes in metabolites in blood and urine that are directly linked to the observed changes in gene expression in liver and kidney metabolism. This systems-level analysis indicated that the gene-expression changes in the kidney primarily drive the metabolite changes in the blood and urine. Finally, using pathway enrichment analysis together with GSMs, we identified pathways in the kidney that significantly changed due to gentamicin exposure and identified metabolites in these pathways as potential early injury indicators.
Several studies using a variety of nephrotoxicants have investigated the potential of various proteins and microRNAs in biofluids to serve as indicators that are more sensitive than established ones, and which are indicative of prerenal damage (Barnett and Cummings, 2018; Fuchs et al., 2014; Ichii and Horino, 2018; Luo et al., 2016; Shin et al., 2014). However, these indicators have yet to be incorporated into clinical practice. Other studies have focused on the identification of metabolites as potential alternatives, which can be sampled readily and detected noninvasively in accessible biofluids earlier than the cellular proteins (Abbiss et al., 2019; Boudonck et al., 2009a; Hanna et al., 2013; Noto et al., 2013; Sun et al., 2012; Uehara et al., 2014). One advantage of global metabolic profiling analysis is its power to observe patterns of metabolites that increase or decrease as a result of exposure to a toxicant. Yet, most studies have examined only a limited number of metabolites, without establishing the causality between their changes in response to toxicant exposure and the ensuing injury in the organ of interest—a necessary step to fully explore the capability of early indicators of organ injury.
Consistent with the known gentamicin nephrotoxicity, our analysis of the global changes at the tissue level, together with the corresponding alterations in biofluids, indicated that gentamicin primarily affected the kidneys without affecting the liver. The lack of impact on the liver is further confirmed by the MFA, where the absolute fluxes for reactions in the glucose production pathway did not change significantly. Furthermore, we observed many significantly changed metabolites that were more readily apparent in urine than in plasma (Figure 2). This result is not surprising, given that gentamicin causes damage to renal proximal tubular epithelial cells, which are primarily involved in the solute reabsorption process, and, hence, any impairment in their function would be apparent in urine but not in plasma. Furthermore, metabolite patterns in blood are more tightly controlled homeostatically compared with urinary changes. Interestingly, most of these changes in urine occurred earlier (at 7 h) than KIM-1, whose levels did not significantly change until 13 h after gentamicin exposure (Figure 1), revealing the potential of global metabolic profiling for early detection of organ injury.
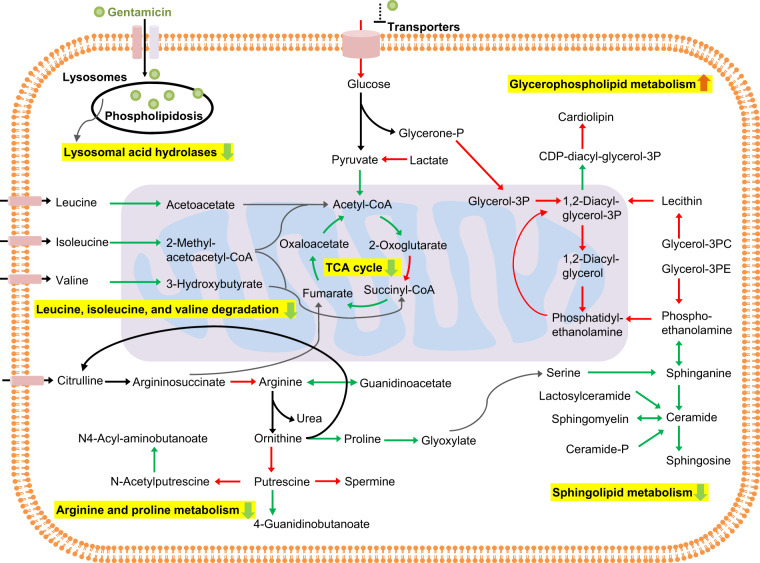
Existing evidence on the mechanism of gentamicin-induced nephrotoxicity suggests that, after being ingested, the drug accumulates in renal tubular cells through a process called receptor-mediated endocytosis (Lopez-Novoa et al., 2011). Excess accumulation of gentamicin within endosomes leads to their disruption and leakage of their contents into the cytosol (Regec et al., 1989). Cytosolic gentamicin then acts on mitochondria directly or indirectly, leading to impairment of ATP synthesis, production of reactive oxygen species, and apoptosis. Our integrated analysis revealed that gentamicin accumulation leads to significant downregulation of most lysosomal genes, indicating the disruption of lysosomal function (Supplementary Table 6). We found significant changes in lipid metabolism reflecting this phenomenon, with upregulation of many genes involved in glycerophospholipid metabolism and downregulation of those involved in sphingolipid metabolism (Figure 8). Similarly, our results indicated mitochondrial damage, with downregulation of most TCA cycle genes, impairment of BCAA degradation, and reduction of arginine and proline metabolism, which provide the precursors (eg, acetyl-CoA and succinyl-CoA) required for energy metabolism. Interestingly, several of the changes in gene expression observed here are in good agreement with toxicogenomic studies in the literature (Amin et al., 2004; Ozaki et al., 2010). For example, using microarray technology, Ozaki et al. evaluated gene-expression changes in the kidneys of rats exposed to a daily dose of 80 mg/kg of gentamicin for 1 week. Their observation that many genes involved in AA metabolism were downregulated is consistent with our study, where we gave rats a single dose of 0.50 g/kg of gentamicin and measured changes in gene expression at 7 h postexposure. These findings reveal the mechanistic basis of the changes in gentamicin-induced kidney damage, and suggest that a targeted measurement of alterations in these pathways could identify novel indicators.
Figure 8.
Summary of gentamicin-induced perturbations in various metabolic pathways in the kidney. Schematic of significantly enriched pathways in the kidney metabolism of rats based on genes significantly altered at 7 h after gentamicin exposure. We propose that gentamicin exposure leads to endosomal membrane disruption and observed downregulation of several lysosomal genes and genes involved in sphingolipid metabolism, as well as upregulation of genes involved in glycerophospholipid metabolism. Subsequent release of gentamicin into the cytosol leads to downregulation of genes involved in the TCA cycle and amino-acid metabolism, which in turn induces mitochondrial damage and impaired energy metabolism. Arrows indicate the reaction directionality, with the color indicating the genes responsible for them either being significantly (FDR < 0.1) upregulated (red), downregulated (green), or unchanged (black). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We identified many metabolites that significantly increased or decreased in the plasma and urine, and whose alterations were driven by gene-expression changes (Tables 2 and 3). Specifically, most urine metabolites associated with arginine and proline metabolism (as well as with leucine, isoleucine, and valine degradation) significantly increased in concordance with the gene-expression changes. Notably, we found evidence indicative of aminoaciduria, in the form of significant (> 5 fold) increases in proline, citrulline, arginine, and ornithine as early as 7 h, whose levels were maintained at 13 h postgentamicin exposure. Importantly, arginine, which is primarily synthesized in the kidneys and is the precursor of nitric oxide, guanidinoacetate, creatine, agmatine, and other polyamines, serves multiple functions in addition to its role in protein synthesis (Barbul, 1986). Downregulation of genes responsible for the synthesis of nitric oxide and guanidinoacetate indicated impaired arginine metabolism, as reflected in the consistent elevation of arginine, creatine, ornithine, and proline, and the reduction of guanidinoacetate in urine (Figure 6A). Altered arginine metabolism has been implicated in end-stage renal disease patients (Lau et al., 2000), with several studies showing a protective effect of arginine on gentamicin-induced nephrotoxicity (Bashan et al., 2014; Miri et al., 2018).
The early changes of AA metabolism observed in our study are consistent with several previous studies under different experimental conditions. For example, newborn rats repeatedly given gentamicin at a dose of 10 or 20 mg/kg for 3 or 7 days showed significant increases in urine levels of tryptophan, kynurenine, glutamine, glycine, and alanine at both 3 and 7 days (Hanna et al., 2013). Similarly, in a urinary metabolite study, Sprague Dawley rats given gentamicin daily at a dose of 40 mg/kg for 1, 5, or 28 days exhibited aminoaciduria, with significant increases in several metabolites (including BCAAs) as observed in our study, suggesting their potential to serve as early indicators of kidney damage (Boudonck et al., 2009b). Furthermore, in a preclinical study (Sun et al., 2012), urinary metabolite changes in rats observed at 11 different time points (1–44 days) revealed an initial increase and a later decrease in metabolites involved in AA metabolism, reflecting the histopathologic changes in the development of, and recovery from, kidney damage, suggesting injury-specific changes in AA metabolism. In addition to metabolites involved in AA metabolism, we identified several others involved in the metabolism of carbohydrates, lipids, and nucleotides (Table 3), which are predicted by the multitissue GSM using the gene-expression changes in the kidney, and which could thereby serve as a potential panel of metabolite indicators. More interestingly, several metabolites identified in our study were also reported to be significantly changed after exposure to cisplatin, a representative of another class of classic nephrotoxicants, indicating a similar mechanism of action in the kidney (Portilla et al., 2006; Xu et al., 2008). Therefore, our analysis suggests that early kidney damage induced by gentamicin may be identified as early as 7 h postexposure by a targeted monitoring of AA-related metabolites in urine among a plausible list of other metabolites.
Using the rat multitissue GSM, we provided a context to interpret the observed metabolite changes by linking them to the changes in gene expression in the liver and kidney. However, our model predicted several metabolites that increase or decrease in biofluids for which experimental data did not show significant changes. Our underlying assumption of a linear relationship between changes in gene expression and metabolite fluxes does not account for metabolite alterations regulated by other biological processes. Therefore, although gene expression is one of the major drivers of changes in metabolites, to reduce false positives, we need further information on the role of other regulatory factors, such as posttranslational modification, gene regulation, and allosteric feedback mechanisms, which take part in the penultimate phenotypic response. Furthermore, currently, our model predictions are restricted only to a subset of metabolites detected in the data (approximately 40%) due to a lack of a more comprehensive reaction network coverage. Despite the aforementioned limitations, our results show notable agreement between model predictions and experimental data and provide opportunities for further curation of GSMs with the addition of more information, on the physiological conditions under which the metabolite modifications occur, to improve the model predictions.
In conclusion, we systematically evaluated toxicant-induced perturbations in the endogenous metabolism of rats and identified global changes in genes in the liver and kidney, metabolites in the plasma and urine, and absolute fluxes in central carbon metabolism. We developed a new multitissue rat metabolic model, used it to elucidate the mechanisms involved in gentamicin-induced kidney damage using transcriptome data, and predicted metabolites in accessible biofluids that were correlated with the gene-expression changes. Our integrated analyses suggest that metabolites involved in the metabolism of AAs, carbohydrates, lipids, and nucleotides are highly correlated with gene-expression changes and could serve as potential early indicators of toxicant-induced kidney damage. Thus, GSMs provide a useful tool to elucidate the underlying mechanisms of chemical-induced toxicity, as well as a potential strategy to identify early indicators of organ injuries.
DECLARATION OF CONFLICTING INTERESTS
The author/authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ACKNOWLEDGMENTS
The Vanderbilt University Medical Center VANTAGE Core provided the genome-wide RNA-seq data; Metabolon, Inc provided the global metabolic profiling data and some technical assistance for this work. The opinions and assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the U.S. Army or of the U.S. Department of Defense, or The Henry M. Jackson Foundation for Advancement of Military Medicine, Inc This paper has been approved for public release with unlimited distribution.
FUNDING
The authors were supported by the U.S. Army Medical Research and Development Command, Ft. Detrick, MD, as part of the U.S. Army’s Network Science Initiative. Dr Jamey Young was supported under Contract No. W81XWH-14-C-0058. VANTAGE is supported in part by a Clinical and Translational Science Awards (CSTA) (5UL1 RR024975-03), the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Vision Center (P30 EY08126), and NIH/NCRR (G20 RR030956).
AUTHOR CONTRIBUTIONS
V.R.P. carried out the RNA-seq analysis, high-throughput data integration, and computational model development and analysis, and wrote the initial draft of the manuscript. K.C.V. contributed to the collection of physiological uptake and secretion rates for the liver and kidney, performed statistical analysis of metabolomics data, and helped edit the manuscript. S.K.E. performed all of the animal studies, including catheterization surgeries. I.T. performed mass spectrometry analysis of plasma samples obtained from stable isotope infusions. T.P.O. performed all of the blood and urine collection and analysis. R.L.P. contributed to RNA extraction from tissue and purifications. J.A.P. supervised and helped to edit the manuscript. J.R. conceived and supervised the study, and helped edit the manuscript. T.O. helped to edit the manuscript. M.S. conceived the study, supervised and carried out the experiments on rats to generate the raw data, and helped write the manuscript. J.D.Y. conceived the study, supervised, performed metabolic flux analysis, and helped write the manuscript. A.W. conceived and supervised the study, analyzed the data, and helped edit and write the final manuscript.
Supplementary Material
REFERENCES
- Abbiss H., Maker G. L., Trengove R. D. (2019). Metabolomics approaches for the diagnosis and understanding of kidney diseases. Metabolites 9, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agren R., Bordel S., Mardinoglu A., Pornputtapong N., Nookaew I., Nielsen J. (2012). Reconstruction of genome-scale active metabolic networks for 69 human cell types and 16 cancer types using INIT. PLoS Comput. Biol. 8, e1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin R. P., Vickers A. E., Sistare F., Thompson K. L., Roman R. J., Lawton M., Kramer J., Hamadeh H. K., Collins J., Grissom S. (2004). Identification of putative gene based markers of renal toxicity. Environ. Health Perspect. 112, 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz M. R., Kelleher J. K., Stephanopoulos G. (2006). Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab. Eng. 8, 324–337. [DOI] [PubMed] [Google Scholar]
- Antoniewicz M. R., Kelleher J. K., Stephanopoulos G. (2007). Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal. Chem. 79, 7554–7559. [DOI] [PubMed] [Google Scholar]
- Antoniewicz M. R., Kelleher J. K., Stephanopoulos G. (2011). Measuring deuterium enrichment of glucose hydrogen atoms by gas chromatography/mass spectrometry. Anal. Chem. 83, 3211–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo A. M., Carvalho M., Carvalho F., Bastos M. L., Guedes de Pinho P. (2017). Metabolomic approaches in the discovery of potential urinary biomarkers of drug-induced liver injury (DILI). Crit. Rev. Toxicol. 47, 633–649. [DOI] [PubMed] [Google Scholar]
- Bandara L. R., Kelly M. D., Lock E. A., Kennedy S. (2003). A correlation between a proteomic evaluation and conventional measurements in the assessment of renal proximal tubular toxicity. Toxicol. Sci. 73, 195–206. [DOI] [PubMed] [Google Scholar]
- Barbul A. (1986). Arginine: Biochemistry, physiology, and therapeutic implications. J. Parenter. Enteral Nutr. 10, 227–238. [DOI] [PubMed] [Google Scholar]
- Barnett L. M. A., Cummings B. S. (2018). Nephrotoxicity and renal pathophysiology: A contemporary perspective. Toxicol. Sci. 164, 379–390. [DOI] [PubMed] [Google Scholar]
- Bashan I., Bashan P., Secilmis M. A., Singirik E. (2014). Protective effect of l-arginine on gentamicin-induced nephrotoxicity in rats. Indian J. Pharmacol. 46, 608–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais E. M., Rawls K. D., Dougherty B. V., Li Z. I., Kolling G. L., Ye P., Wallqvist A., Papin J. A. (2017). Reconciled rat and human metabolic networks for comparative toxicogenomics and biomarker predictions. Nat. Commun. 8, 14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudonck K. J., Mitchell M. W., Német L., Keresztes L., Nyska A., Shinar D., Rosenstock M. (2009a). Discovery of metabolomics biomarkers for early detection of nephrotoxicity. Toxicol. Pathol. 37, 280–292. [DOI] [PubMed] [Google Scholar]
- Boudonck K. J., Rose D. J., Karoly E. D., Lee D. P., Lawton K. A., Lapinskas P. J. (2009b). Metabolomics for early detection of drug-induced kidney injury: Review of the current status. Bioanalysis 1, 1645–1663. [DOI] [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P., Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Buness A., Roth A., Herrmann A., Schmitz O., Kamp H., Busch K., Suter L. (2014). Identification of metabolites, clinical chemistry markers and transcripts associated with hepatotoxicity. PLoS One 9, e97249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli K., Qian P. Y. (2009). Proteomics: Challenges, techniques and possibilities to overcome biological sample complexity. Hum. Genomics Proteomics 1, 239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D. S., Xia J. (2018). MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings F. B., Vaidya V. S. (2008). Novel technologies for the discovery and quantitation of biomarkers of toxicity. Toxicology 245, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F., Achuthan P., Akanni W., Allen J., Amode M. R., Armean I. M., Bennett R., Bhai J., Billis K., Boddu S., et al. (2019). Ensembl 2019. Nucleic Acids Res. 47, D745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehaven C. D., Evans A. M., Dai H., Lawton K. A. (2010). Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J. Cheminform. 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte N. C., Becker S. A., Jamshidi N., Thiele I., Mo M. L., Vo T. D., Srivas R., Palsson B. O. (2007). Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl. Acad. Sci. U.S.A. 104, 1777–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. R., Diamantakos E. A., Peuler J. D., Lamar P. C., Prozialeck W. C. (2007). A novel method for the evaluation of proximal tubule epithelial cellular necrosis in the intact rat kidney using ethidium homodimer. BMC Physiol. 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhamri M., Martin M., Ferrier B., Baverel G. (1993). Substrate uptake and utilization by the kidney of fed and starved rats in vivo. Ren. Physiol. Biochem. 16, 311–324. [DOI] [PubMed] [Google Scholar]
- Evans A. M., Bridgewater B. R., Liu Q., Mitchell M. W., Robinson R. J., Dai H., Stewart S. J., DeHaven C. D., Miller L. A. D. (2014). High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics 4, 1. [Google Scholar]
- Faa G., Ekstrom J., Castagnola M., Gibo Y., Ottonello G., Fanos V. (2012). A developmental approach to drug-induced liver injury in newborns and children. Curr. Med. Chem. 19, 4581–4594. [DOI] [PubMed] [Google Scholar]
- Fannin R. D., Russo M., O'Connell T. M., Gerrish K., Winnike J. H., Macdonald J., Newton J., Malik S., Sieber S. O., Parker J., et al. (2010). Acetaminophen dosing of humans results in blood transcriptome and metabolome changes consistent with impaired oxidative phosphorylation. Hepatology 51, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M. A., Waikar S. S. (2012). Established and emerging markers of kidney function. Clin. Chem. 58, 680–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs T. C., Mally A., Wool A., Beiman M., Hewitt P. (2014). An exploratory evaluation of the utility of transcriptional and urinary kidney injury biomarkers for the prediction of aristolochic acid-induced renal injury in male rats. Vet. Pathol. 51, 680–694. [DOI] [PubMed] [Google Scholar]
- Gartland K. P., Bonner F. W., Nicholson J. K. (1989). Investigations into the biochemical effects of region-specific nephrotoxins. Mol. Pharmacol. 35, 242–250. [PubMed] [Google Scholar]
- Gerich J. E., Meyer C., Woerle H. J., Stumvoll M. (2001). Renal gluconeogenesis: Its importance in human glucose homeostasis. Diabetes Care 24, 382–391. [DOI] [PubMed] [Google Scholar]
- Giuliano R. A., Paulus G. J., Verpooten G. A., Pattyn V. M., Pollet D. E., Nouwen E. J., Laurent G., Carlier M.-B., Maldague P., Tulkens P. M., et al. (1984). Recovery of cortical phospholipidosis and necrosis after acute gentamicin loading in rats. Kidney Int. 26, 838–847. [DOI] [PubMed] [Google Scholar]
- Han W. K., Bailly V., Abichandani R., Thadhani R., Bonventre J. V. (2002). Kidney injury molecule-1 (KIM-1): A novel biomarker for human renal proximal tubule injury. Kidney Int. 62, 237–244. [DOI] [PubMed] [Google Scholar]
- Hanna M. H., Segar J. L., Teesch L. M., Kasper D. C., Schaefer F. S., Brophy P. D. (2013). Urinary metabolomic markers of aminoglycoside nephrotoxicity in newborn rats. Pediatr. Res. 73, 585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenour C. M., Wall M. L., Ridley D. E., Hughey C. C., James F. D., Wasserman D. H., Young J. D. (2015). Mass spectrometry-based microassay of 2H and 13C plasma glucose labeling to quantify liver metabolic fluxes in vivo. Am. J. Physiol. Endocrinol. Metab. 309, E191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano T., Saiki S., Okuzumi A., Mohney R. P., Hattori N. (2016). Identification of novel biomarkers for Parkinson's disease by metabolomic technologies. J. Neurol. Neurosurg. Psychiatry 87, 295–301. [DOI] [PubMed] [Google Scholar]
- Hoffmann D., Adler M., Vaidya V. S., Rached E., Mulrane L., Gallagher W. M., Callanan J. J., Gautier J. C., Matheis K., Staedtler F., et al. (2010). Performance of novel kidney biomarkers in preclinical toxicity studies. Toxicol. Sci. 116, 8–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Humes H. D., Weinberg J. M., Knauss T. C. (1982). Clinical and pathophysiologic aspects of aminoglycoside nephrotoxicity. Am. J. Kidney Dis. 2, 5–29. [DOI] [PubMed] [Google Scholar]
- Ichii O., Horino T. (2018). MicroRNAs associated with the development of kidney diseases in humans and animals. J. Toxicol. Pathol. 31, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y., Nakatsu N., Yamashita T., Ono A., Ohno Y., Urushidani T., Yamada H. (2015). Open TG-GATEs: A large-scale toxicogenomics database. Nucleic Acids Res. 43(Database issue), D921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iruzubieta P., Arias-Loste M. T., Barbier-Torres L., Martinez-Chantar M. L., Crespo J. (2015). The need for biomarkers in diagnosis and prognosis of drug-induced liver disease: Does metabolomics have any role? Biomed. Res. Int. 2015, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerby L., Ruppin E. (2012). Predicting drug targets and biomarkers of cancer via genome-scale metabolic modeling. Clin. Cancer Res. 18, 5572–5584. [DOI] [PubMed] [Google Scholar]
- Kaneko K., Soty M., Zitoun C., Duchampt A., Silva M., Philippe E., Gautier-Stein A., Rajas F., Mithieux G. (2018). The role of kidney in the inter-organ coordination of endogenous glucose production during fasting. Mol. Metab. 16, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplowitz N. (2005). Idiosyncratic drug hepatotoxicity. Nat. Rev. Drug Discov. 4, 489–499. [DOI] [PubMed] [Google Scholar]
- Karmen A., Wroblewski F., Ladue J. S. (1955). Transaminase activity in human blood. J. Clin. Invest. 34, 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. R., Badar I., Siddiquah A. (2011). Prevention of hepatorenal toxicity with Sonchus asper in gentamicin treated rats. BMC Complement. Altern. Med. 11, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau T., Owen W., Yu Y. M., Noviski N., Lyons J., Zurakowski D., Tsay R., Ajami A., Young V. R., Castillo L., et al. (2000). Arginine, citrulline, and nitric oxide metabolism in end-stage renal disease patients. J. Clin. Invest. 105, 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. M. (2013). Drug-induced acute liver failure. Clin. Liver Dis. 17, 575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li Q. X., Xie X. F., Ao Y., Tie C. R., Song R. J. (2009). Differential roles of dihydropyridine calcium antagonist nifedipine, nitrendipine and amlodipine on gentamicin-induced renal tubular toxicity in rats. Eur. J. Pharmacol. 620, 97–104. [DOI] [PubMed] [Google Scholar]
- Lin Z., Will Y. (2012). Evaluation of drugs with specific organ toxicities in organ-specific cell lines. Toxicol. Sci. 126, 114–127. [DOI] [PubMed] [Google Scholar]
- Lopez-Novoa J. M., Quiros Y., Vicente L., Morales A. I., Lopez-Hernandez F. J. (2011). New insights into the mechanism of aminoglycoside nephrotoxicity: An integrative point of view. Kidney Int. 79, 33–45. [DOI] [PubMed] [Google Scholar]
- Luo Q.-H., Chen M.-L., Chen Z.-L., Huang C., Cheng A.-C., Fang J., Tang L., Geng Y. (2016). Evaluation of KIM-1 and NGAL as early indicators for assessment of gentamycin-induced nephrotoxicity in vivo and in vitro. Kidney Blood Press. Res. 41, 911–918. [DOI] [PubMed] [Google Scholar]
- Macpherson N. A., Moscarello M. A., Goldberg D. M. (1991). Aminoaciduria is an earlier index of renal tubular damage than conventional renal disease markers in the gentamicin-rat model of acute renal failure. Clin. Invest. Med. 14, 101–110. [PubMed] [Google Scholar]
- Mardinoglu A., Agren R., Kampf C., Asplund A., Uhlen M., Nielsen J. (2014). Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat. Commun. 5, 3083. [DOI] [PubMed] [Google Scholar]
- Miri S., Safari T., Komeili G. R., Nematbakhsh M., Niazi A. A., Jahantigh M., Bagheri H., Maghool F. (2018). Sex difference in gentamicin-induced nephrotoxicity: Influence of l-arginine in rat model. Int. J. Prev. Med. 9, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonclercq D., Wrona S., Toubeau G., Zanen J., Heuson-Stiennon J.-A., Schaudies R. P., Laurent G. (1992). Tubular injury and regeneration in the rat kidney following acute exposure to gentamicin: A time-course study. Ren. Fail. 14, 507–521. [DOI] [PubMed] [Google Scholar]
- Noto A., Cibecchini F., Fanos V., Mussap M. (2013). NGAL and metabolomics: The single biomarker to reveal the metabolome alterations in kidney injury. Biomed Res. Int. 2013, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onakpoya I. J., Heneghan C. J., Aronson J. K. (2016). Post-marketing withdrawal of 462 medicinal products because of adverse drug reactions: A systematic review of the world literature. BMC Med. 14, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki N., Matheis K. A., Gamber M., Feidl T., Nolte T., Kalkuhl A., Deschl U. (2010). Identification of genes involved in gentamicin-induced nephrotoxicity in rats–a toxicogenomic investigation. Exp. Toxicol. Pathol. 62, 555–566. [DOI] [PubMed] [Google Scholar]
- Pannala V. R., Vinnakota K. C., Rawls K. D., Estes S. K., O'Brien T. P., Printz R. L., Papin J. A., Reifman J., Shiota M., Young J. D., et al. (2019). Mechanistic identification of biofluid metabolite changes as markers of acetaminophen-induced liver toxicity in rats. Toxicol. Appl. Pharmacol. 372, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannala V. R., Wall M. L., Estes S. K., Trenary I., O’Brien T. P., Printz R. L., Vinnakota K. C., Reifman J., Shiota M., Young J. D., et al. (2018). Metabolic network-based predictions of toxicant-induced metabolite changes in the laboratory rat. Sci. Rep. 8, 11678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh C. R., Thiessen-Philbrook H., Garg A. X., Kadiyala D., Shlipak M. G., Koyner J. L., Edelstein C. L., Devarajan P., Patel U. D., Zappitelli M., et al. (2013). Performance of kidney injury molecule-1 and liver fatty acid-binding protein and combined biomarkers of AKI after cardiac surgery. Clin. J. Am. Soc. Nephrol. 8, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H., Bray N. L., Puente S., Melsted P., Pachter L. (2017). Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690. [DOI] [PubMed] [Google Scholar]
- Piron A., Leonard I., Nonclercq D., Toubeau G., Falmagne P., Heuson-Stiennon J. A., Laurent G. (1998). In vitro demonstration of a mitogenic activity in renal tissue extracts during regenerative hyperplasia. Am. J. Physiol. 274, F348–F357. [DOI] [PubMed] [Google Scholar]
- Portilla D., Li S., Nagothu K. K., Megyesi J., Kaissling B., Schnackenberg L., Safirstein R. L., Beger R. D. (2006). Metabolomic study of cisplatin-induced nephrotoxicity. Kidney Int. 69, 2194–2204. [DOI] [PubMed] [Google Scholar]
- Ramirez T., Daneshian M., Kamp H., Bois F. Y., Clench M. R., Coen M., Donley B., Fischer S. M., Ekman D. R., Fabian E., et al. (2013). Metabolomics in toxicology and preclinical research. ALTEX 30, 209–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regec A. L., Trump B. F., Trifilis A. L. (1989). Effect of gentamicin on the lysosomal system of cultured human proximal tubular cells. Endocytotic activity, lysosomal pH and membrane fragility. Biochem. Pharmacol. 38, 2527–2534. [DOI] [PubMed] [Google Scholar]
- Remmer H. (1970). The role of the liver in drug metabolism. Am. J. Med. 49, 617–629. [DOI] [PubMed] [Google Scholar]
- Robertson D. G., Watkins P. B., Reily M. D. (2011). Metabolomics in toxicology: Preclinical and clinical applications. Toxicol. Sci. 120(Suppl. 1), S146–170. [DOI] [PubMed] [Google Scholar]
- Shin Y. J., Kim T. H., Won A. J., Jung J. Y., Kwack S. J., Kacew S., Chung K. H., Lee B. M., Kim H. S. (2014). Age-related differences in kidney injury biomarkers induced by cisplatin. Environ. Toxicol. Pharmacol. 37, 1028–1039. [DOI] [PubMed] [Google Scholar]
- Shiota M. (2012). Measurement of glucose homeostasis in vivo: Combination of tracers and clamp techniques. Methods Mol. Biol. 933, 229–253. [DOI] [PubMed] [Google Scholar]
- Shlomi T., Cabili M. N., Ruppin E. (2009). Predicting metabolic biomarkers of human inborn errors of metabolism. Mol. Syst. Biol. 5, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderland P., Lovekar S., Weiner D. E., Brooks D. R., Kaufman J. S. (2010). Chronic kidney disease associated with environmental toxins and exposures. Adv. Chronic Kidney Dis. 17, 254–264. [DOI] [PubMed] [Google Scholar]
- Sun J., Ando Y., Ahlbory-Dieker D., Schnackenberg L., Xea Y. (2013). Systems biology investigation to discover metabolic biomarkers of acetaminophen-induced hepatic injury using integrated transcriptomics and metabolomics. J. Mol. Biomark. Diagn. S1, 002. [Google Scholar]
- Sun J., Bhattacharyya S., Schnackenberg L. K., Pence L., Ando Y., Zhang J., Stewart S., Rosenzweig B., Rouse R., Portilla D., et al. (2012). Discovery of early urinary biomarkers in preclinical study of gentamicin-induced kidney injury and recovery in rats. Metabolomics 8, 1181–1193. [Google Scholar]
- Suryawan A., Hawes J. W., Harris R. A., Shimomura Y., Jenkins A. E., Hutson S. M. (1998). A molecular model of human branched-chain amino acid metabolism. Am. J. Clin. Nutr. 68, 72–81. [DOI] [PubMed] [Google Scholar]
- Tugwood J. D., Hollins L. E., Cockerill M. J. (2003). Genomics and the search for novel biomarkers in toxicology. Biomarkers 8, 79–92. [DOI] [PubMed] [Google Scholar]
- Uehara T., Horinouchi A., Morikawa Y., Tonomura Y., Minami K., Ono A., Yamate J., Yamada H., Ohno Y., Urushidani T., et al. (2014). Identification of metabolomic biomarkers for drug-induced acute kidney injury in rats. J. Appl. Toxicol. 34, 1087–1095. [DOI] [PubMed] [Google Scholar]
- Vree T. B., Hekster Y. A., Anderson P. G. (1992). Contribution of the human kidney to the metabolic clearance of drugs. Ann. Pharmacother. 26, 1421–1428. [DOI] [PubMed] [Google Scholar]
- Xu E. Y., Perlina A., Vu H., Troth S. P., Brennan R. J., Aslamkhan A. G., Xu Q. (2008). Integrated pathway analysis of rat urine metabolic profiles and kidney transcriptomic profiles to elucidate the systems toxicology of model nephrotoxicants. Chem. Res. Toxicol. 21, 1548–1561. [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Aikawa T., Matsutaka H., Okuda T., Ishikawa E. (1974). Interorganal relationships of amino acid metabolism in fed rats. Am. J. Physiol. 226, 1428–1433. [DOI] [PubMed] [Google Scholar]
- Young J. D. (2014). INCA: A computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics 30, 1333–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Woo H. J., Yu X., Oyama T., Wallqvist A., Reifman J. (2017). A strategy for evaluating pathway analysis methods. BMC Bioinform. 18, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Sun H., Wang P., Han Y., Wang X. (2012). Metabonomics for discovering biomarkers of hepatotoxicity and nephrotoxicity. Pharmazie 67, 99–105. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.