Significance
A timely and appropriate response to genotoxic stress is essential for maintaining genome stability and preventing tumorigenesis. Here, we have established a previously unidentified role of the protein kinase ATM in controlling the extent of DNA end resection during homology-directed repair. Specifically, ATM elicits sequential posttranslational modifications of the key resection factor CtIP and thereby fine-tunes its activity at DSB sites. Our findings highlight the multistep roles of the master kinase ATM in orchestrating DNA damage responses.
Keywords: DNA end resection, homologous recombination, CtIP, ATM, hyperphosphorylation
Abstract
DNA end resection is a critical step in the repair of DNA double-strand breaks (DSBs) via homologous recombination (HR). However, the mechanisms governing the extent of resection at DSB sites undergoing homology-directed repair remain unclear. Here, we show that, upon DSB induction, the key resection factor CtIP is modified by the ubiquitin-like protein SUMO at lysine 578 in a PIAS4-dependent manner. CtIP SUMOylation occurs on damaged chromatin and requires prior hyperphosphorylation by the ATM protein kinase. SUMO-modified hyperphosphorylated CtIP is targeted by the SUMO-dependent E3 ubiquitin ligase RNF4 for polyubiquitination and subsequent degradation. Consequently, disruption of CtIP SUMOylation results in aberrant accumulation of CtIP at DSBs, which, in turn, causes uncontrolled excessive resection, defective HR, and increased cellular sensitivity to DSB-inducing agents. These findings reveal a previously unidentified regulatory mechanism that regulates CtIP activity at DSBs and thus the extent of end resection via ATM-dependent sequential posttranslational modification of CtIP.
DNA double-strand breaks (DSBs) constitute one of the most severe forms of DNA damage and can result in a wide variety of genetic alterations including mutations, deletions, translocations, and chromosome loss (1, 2). Extensive studies have shown that DSBs can be repaired primarily via two pathways, classical nonhomologous end joining and homologous recombination (HR), both of which are highly conserved among all eukaryotes (3–5). Classical nonhomologous end joining, which directly rejoins the two broken ends of a DSB, occurs throughout interphase (3–5). In contrast, DSB repair by HR requires the presence of a sister chromatid and is therefore restricted to the late S and G2 phases of the cell cycle (3–5). HR is initiated by resection of the 5′ strand of the DSB ends, yielding 3′ single-stranded DNA (ssDNA) tails that are initially coated with the replication protein A (RPA) complex (3–5). The resulting RPA-coated ssDNA is an essential intermediate not only in HR repair but also in ATR-CHK1 pathway activation (3–5). Studies conducted in yeast and mammalian cells have established that resection of DSB ends is a two-step process (3–5). First, the conserved MRE11/RAD50/NBS1 complex (MRE11/RAD50/XRS2 in Saccharomyces cerevisiae) cooperates with the key resection factor CtIP (Sae2 in S. cerevisiae, Ctp1 in Schizosaccharomyces pombe) to catalyze limited resection of broken DNA ends (3–6). Second, the resulting short 3′ overhangs are further processed through the action of either the 5′–3′ exonuclease EXO1 or the nuclease–helicase protein complex DNA2–BLM (DNA2–Sgs1 in S. cerevisiae) (3–5). Whereas extensive end resection is required for HR initiation and full checkpoint activation, uncontrolled and excessive processing of DSB ends can have deleterious consequences such as large deletions at DSB sites, persistent checkpoint signaling, and cell death (7–11). However, the mechanisms by which cells precisely control the extent of end resection at DSB sites undergoing homology-directed repair remain obscure.
It has been well established that posttranslational modifications of DNA repair proteins play crucial roles in the cellular response to genotoxic stress (12, 13). For example, phosphorylation of CtIP at threonine 847 (serine 267 in Sae2) by CDK1/2 restricts its activity to the S and G2 phases of the cell cycle (14–17), and promotes its capacity to stimulate the MRE11 endonuclease activity (18) as well as the annealing of broken DNA ends (19). In addition, phosphorylation of CtIP at serine 327 by CDK2 and/or Aurora A is a prerequisite for its interactions with BRCA1 and PLK1 (20–23). Furthermore, CtIP undergoes phosphorylation at threonine 315 by CDK2, and this phosphorylation event regulates CtIP protein stability by facilitating its interaction with the phosphorylation-specific prolyl isomerase PIN1 (24). In addition to acting as a CDK substrate, CtIP can also be hyperphosphorylated by ATM (or ATR in Xenopus) at multiple serine/threonine–glutamine sites in response to DSBs, which is manifested by the appearance of a slow-migrating form of CtIP (25, 26). ATM-dependent hyperphosphorylation of CtIP not only facilitates its association with damaged DNA (27) but also promotes the recruitment of BLM and EXO1 to DSB sites (25). Moreover, a previous study showed that the putative ATM-targeted residues serine 231, serine 664, and serine 745 as well as the CDK-targeted residues serine 276, threonine 315, and serine 347 within CtIP are critical for its endonuclease activity, although the relative contributions of the individual modifications have not been fully characterized (28). In addition to phosphorylation, CtIP is also subject to other posttranslational modifications, such as ubiquitination and acetylation (21, 29–36). Strict regulation of CtIP activity via various posttranslational modifications is crucial for accurate processing and repair of DSBs; however, precisely how these modifications are regulated in a coordinated manner remains unclear.
In this study, we provide evidence that CtIP becomes SUMOylated primarily at lysine 578 upon exposure to DSB-inducing agents, and that this modification controls the activated CtIP level at DSBs and thereby the extent of DSB end resection. CtIP SUMOylation at lysine 578 is dependent on its prior hyperphosphorylation by the protein kinase ATM. SUMO-modified hyperphosphorylated CtIP can be targeted by the SUMO-dependent E3 ubiquitin ligase RNF4 for polyubiquitination and subsequent degradation. As a consequence, cells expressing non-SUMOylatable CtIP mutants exhibit aberrant accumulation of CtIP at DSB sites, uncontrolled excessive end resection, and defective HR. Our results suggest that active CtIP triggers its own SUMOylation and degradation, establishing a negative feedback loop that restricts CtIP activity at DSBs and thereby prevents excessive end resection and genome instability.
Results
CtIP Is SUMOylated on Chromatin in Response to DNA Damage.
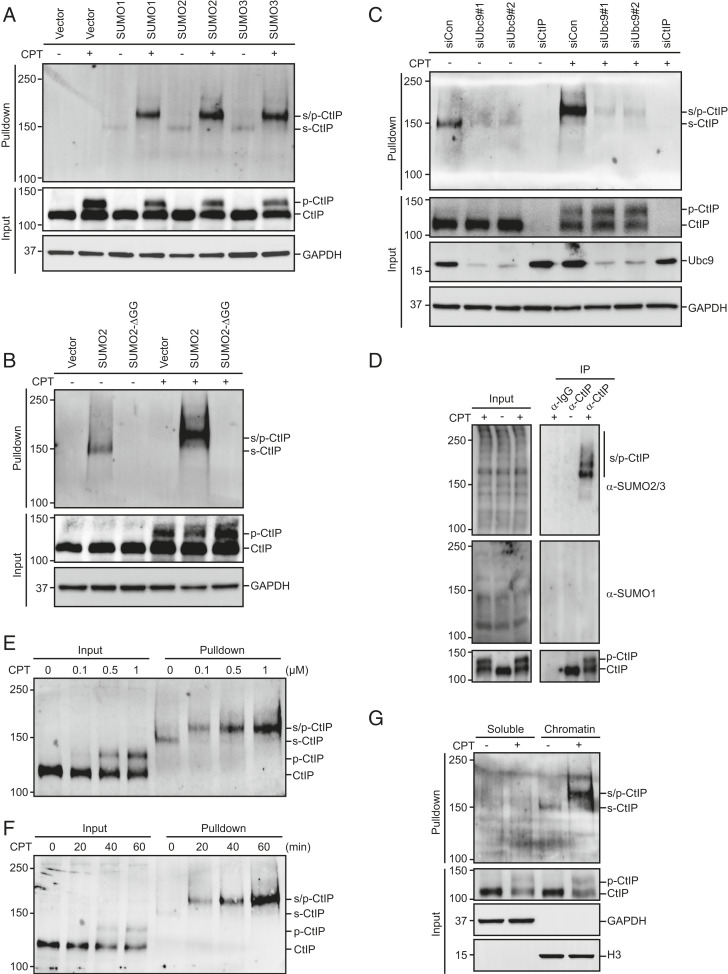
SUMOylation is an important posttranslational modification involved in a variety of cellular processes including transcriptional regulation, cell cycle control, and maintenance of genome integrity (12, 13). To identify proteins involved in the DNA damage response that might be SUMOylated, HEK293T cells stably expressing His-SUMO1, His-SUMO2, or His-SUMO3 were treated with camptothecin (CPT, a topoisomerase I inhibitor) or left untreated. The cells were then lysed under denaturing conditions to preserve SUMO modifications, and SUMOylated proteins were purified using cobalt affinity pulldown and were analyzed using antibodies that specifically recognize endogenous proteins involved in the DNA damage response. To control for specificity, we used a conjugation-defective mutant of SUMO2 (SUMO2-ΔGG) that lacks the essential C-terminal diglycine motif. Interestingly, a high molecular weight form of CtIP was detected in the presence of SUMO1, SUMO2, and SUMO3, but not in the presence of SUMO2-ΔGG (Fig. 1 A and B). This high molecular weight species was almost completely eliminated after knockdown of Ubc9, the sole SUMO-conjugating enzyme in mammalian cells, indicating that it corresponds to SUMOylated CtIP (Fig. 1C). To detect SUMOylation of CtIP at the endogenous level, endogenous CtIP was immunoprecipitated from HEK293T cells in the presence of N-ethylmaleimide (NEM), a cysteine protease inhibitor that allows preservation of SUMOylated cellular proteins, and then blotted with anti-SUMO1 or anti-SUMO2/3 antibodies. As shown in Fig. 1D, several slow-migrating bands immunoreactive to anti-SUMO2/3 but not anti-SUMO1 antibodies were observed, indicating that CtIP is preferentially modified by SUMO2/3 in cells. Notably, CtIP and its S. cerevisiae functional homolog Sae2 have been identified as SUMOylation targets in independent proteomic and biochemical studies (37, 38). Although CtIP SUMOylation was detected in the absence of DNA damage, it was greatly enhanced by CPT treatment in a dose- and time-dependent manner (Fig. 1 E and F). More importantly, CtIP SUMOylation was detected only in the chromatin-enriched fraction (Fig. 1G), indicating that this modification most likely occurs locally at sites of DNA damage and may play an important regulatory role in CtIP-mediated DNA end resection.
Fig. 1.
CtIP is SUMOylated on chromatin in response to DNA damage. (A) CtIP was modified by SUMO. HEK293T cells stably expressing His-SUMO1, His-SUMO2, or His-SUMO3 were treated with 1 μM CPT for 1 h or left untreated. The cells were then lysed under denaturing conditions and subjected to cobalt pulldown. The resulting isolated proteins were assessed by Western blotting for endogenous CtIP; p-CtIP, hyperphosphorylated CtIP; s-CtIP, SUMOylated CtIP; s/p-CtIP, SUMO-modified hyperphosphorylated CtIP. (B) HEK293T cells stably expressing either His-SUMO2 or His-SUMO2-ΔGG were treated with 1 μM CPT for 1 h or left untreated. The cells were then lysed under denaturing conditions and subjected to cobalt pulldown. The resulting isolated proteins were assessed by Western blotting for endogenous CtIP. (C) Ubc9 depletion dramatically reduced CtIP SUMOylation. HEK293T cells stably expressing His-SUMO2 were transfected with the indicated siRNAs and treated with 1 μM CPT for 1 h or left untreated. The cells were then lysed under denaturing conditions and subjected to cobalt pulldown. The resulting isolated proteins were assessed by Western blotting using anti-CtIP antibody. (D) HEK293T cells were treated with 1 μM CPT for 1 h or left untreated. The cells were lysed in SDS buffer and diluted 1:10 in radio-immunoprecipitation assay buffer (RIPA) buffer. Subsequently, cell lysates were incubated with protein A agarose beads conjugated to anti-CtIP antibody. The resulting isolated proteins were assessed by Western blotting using anti-SUMO1 or anti-SUMO2/3 antibody. (E and F) CPT-stimulated CtIP SUMOylation in a dose- and time-dependent manner. HEK293T cells stably expressing His-SUMO2 were treated with the indicated concentrations of CPT for 1 h (E) or with 1 μM CPT for the indicated periods (F). The cells were then lysed and subjected to cobalt pulldown under denaturing conditions. The resulting isolated proteins were assessed by Western blotting for endogenous CtIP. (G) HEK293T cells stably expressing His-SUMO2 were treated with 1 μM CPT for 1 h or left untreated. The cells were then biochemically fractionated, diluted in denaturing buffer, and subjected to cobalt pulldown. The resulting isolated proteins were assessed by Western blotting using anti-CtIP antibody.
ATM-Mediated CtIP Hyperphosphorylation Stimulates Its SUMOylation.
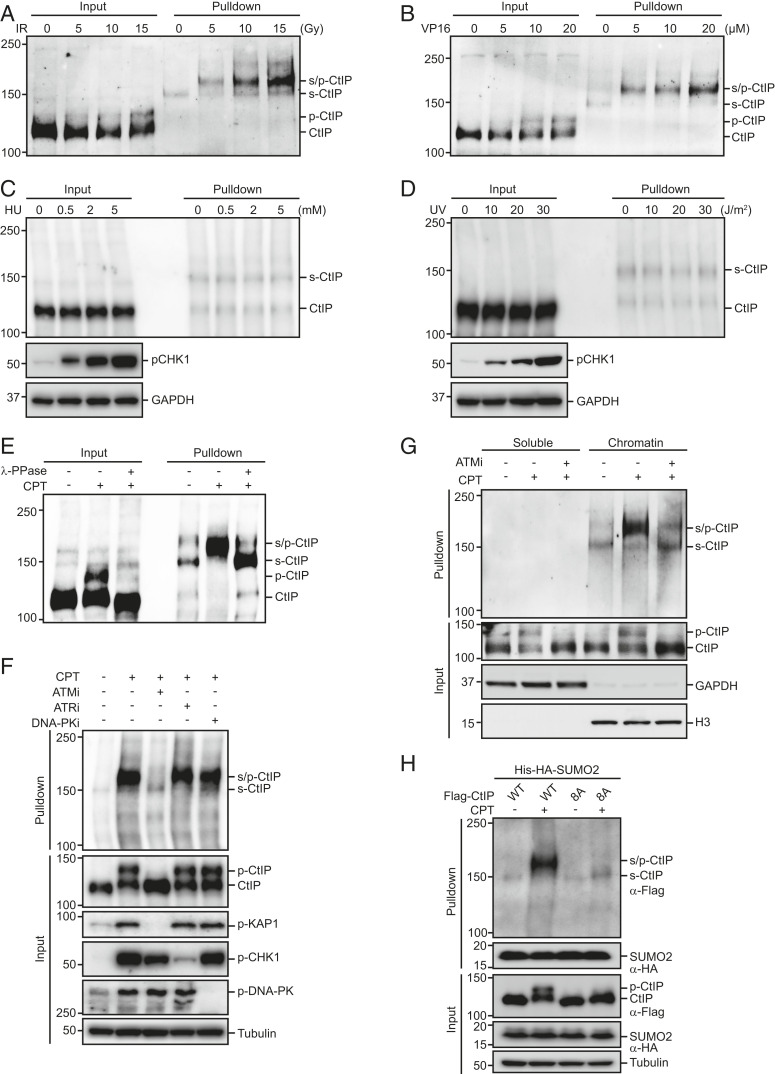
CPT induces replication-associated DSBs. To determine the types of lesions that stimulate CtIP SUMOylation, we treated cells expressing His-SUMO2 with various DNA-damaging agents. As shown in Figs. 2 A and B, both ionizing radiation (IR) and VP16 (a topoisomerase II inhibitor), which exclusively or primarily induce DSBs, also trigger strong dose-dependent induction of CtIP SUMOylation. In contrast, the levels of SUMOylated CtIP were not markedly affected by ultraviolet (UV) irradiation or the DNA replication inhibitor hydroxyurea (HU) (Fig. 2 C and D). Together, these results suggest that DSBs can specifically trigger CtIP SUMOylation. Strikingly, after DSB induction, the major form of SUMOylated CtIP migrated more slowly compared with nontreated control cells (Fig. 2 A and B). Given that DSBs could also trigger ATM-dependent CtIP hyperphosphorylation, and that the extent of CtIP SUMOylation in response to DSBs was positively correlated with its hyperphosphorylation level (Fig. 2 A and B), we speculated that SUMO might preferentially conjugate to hyperphosphorylated CtIP over its nonphosphorylated form. Consistent with this hypothesis, the DSB-induced reduction in the mobility of SUMOylated CtIP was completely reversed by treatment with lambda protein phosphatase (λ-PPase) (Fig. 2E). Furthermore, pharmacological inhibition of ATM using the ATM-specific inhibitor KU-60019 not only fully reversed the DSB-dependent mobility shift of SUMOylated CtIP but also significantly reduced levels of CtIP hyperphosphorylation and SUMOylation (Fig. 2 F and G). In contrast, specific inhibition of ATR or DNA-PK failed to reduce CtIP hyperphosphorylation or SUMOylation (Fig. 2F). More importantly, mutation of all eight putative ATM phosphorylation sites on CtIP (CtIP-8A) (25) dramatically reduced the DSB-induced CtIP mobility shift and SUMOylation (Fig. 2H). Together, these results suggest that DSB-induced ATM-mediated hyperphosphorylation of CtIP stimulates its SUMOylation.
Fig. 2.
DSB-induced ATM-mediated hyperphosphorylation of CtIP stimulates its SUMOylation. (A–D) The SUMOylation of CtIP was enhanced in cells treated with IR (A) or VP16 (B) but not those treated with HU (C) or UV (D). HEK293T cells stably expressing His-SUMO2 were treated with various doses of IR, VP16, HU, or UV. After 1 h, the cells were lysed under denaturing conditions and subjected to cobalt pulldown. The resulting isolated proteins were assessed by Western blotting for endogenous CtIP. (E) SUMO was preferentially conjugated to hyperphosphorylated CtIP. HEK293T cells stably expressing His-SUMO2 were left untreated or treated with 1 μM CPT for 1 h before lysing in NP 40-EDTA-Tris-NaCl (NETN) buffer without EDTA. The cell extracts were then treated with or without λ-phosphatase for 30 min, diluted in denaturing buffer, and subjected to cobalt pulldown. The resulting isolated proteins were assessed by Western blotting using anti-CtIP antibody. (F) Inhibition of ATM but not ATR or DNA-PK greatly reduced CtIP hyperphosphorylation and subsequent SUMOylation. HEK293T cells stably expressing His-SUMO2 were pretreated with the ATM inhibitor KU-60019 (10 μM), the ATR inhibitor VE-821 (10 μM), or the DNA-PK inhibitor NU-7441 (10 μM) for 1 h prior to CPT (1 μM) treatment. After 1 h, the cells were lysed under denaturing conditions and subjected to cobalt pulldown. The resulting isolated proteins were assessed by Western blotting for endogenous CtIP. (G) HEK293T cells stably expressing His-SUMO2 were left untreated or pretreated with the ATM inhibitor KU-60019 (10 μM) for 1 h prior to CPT (1 μM) treatment. After 1 h, cells were biochemically fractionated, diluted in denaturing buffer, and subjected to cobalt pulldown. The resulting isolated proteins were assessed by Western blotting using anti-CtIP antibody. (H) SUMOylation of CtIP required its hyperphosphorylation by ATM. HeLa cells stably expressing Flag-tagged wild-type CtIP or CtIP-8A mutant were infected with a lentiviral vector expressing His-HA-SUMO2. After 24 h, the cells were synchronized in G1/S boundary by 2 mM thymidine for 24 h and released into S phase in fresh medium for 2 h. The cells were then left untreated or treated with 1 μM CPT for 1 h, lysed under denaturing conditions, subjected to cobalt pulldown, and analyzed by Western blotting with the indicated antibodies.
SUMOylation of CtIP at Lysine 578 In Vivo and In Vitro.
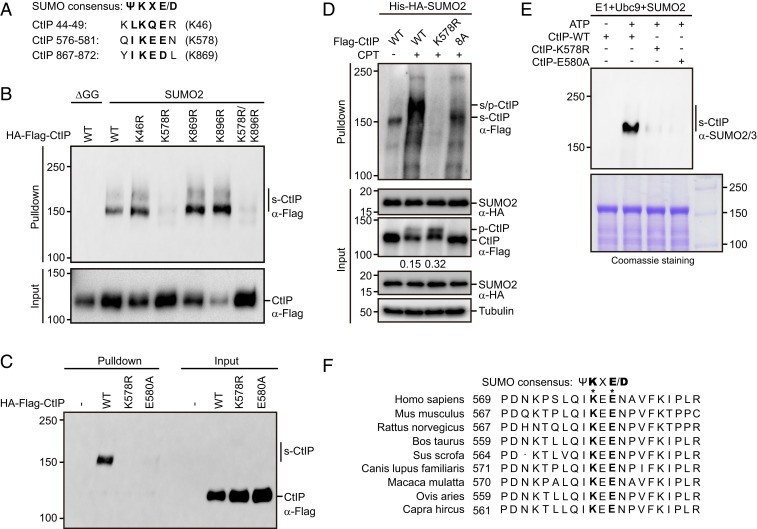
SUMOylation generally occurs on a specific lysine residue within the consensus motif ΨKXE/D, where Ψ represents a large hydrophobic amino acid, and X represents any amino acid. Inspection of the human CtIP protein sequence using the SUMOylation site prediction program SUMOplot revealed three potential SUMO target lysines (lysines 46, 578, and 869) (Fig. 3A). Therefore, we separately mutated each lysine residue to an arginine (K46R, K578R, and K869R) to determine which residue may be SUMOylated. As shown in Fig. 3B, CtIP SUMOylation was not affected by mutation of lysine 46 or lysine 869 to arginine but was significantly suppressed by mutation of lysine 578. In addition, the CtIP mutant E580A (substitution of glutamic acid 580 with alanine), which carries a disrupted IKEE SUMOylation motif in CtIP, also displayed a sharp reduction in SUMOylation (Fig. 3C). More importantly, mutation of lysine 578 to alanine also greatly reduced CPT-induced CtIP SUMOylation (Fig. 3D). These results suggest that CtIP is primarily modified by SUMO at lysine 578. To further confirm this finding, we conducted an in vitro SUMOylation assay using recombinant wild-type and mutant CtIP proteins. As shown in Fig. 3E, in contrast to wild-type CtIP, both the K578R and E580A mutant proteins failed to be efficiently SUMOylated in vitro.
Fig. 3.
CtIP SUMOylation occurs at lysine 578. (A) Candidate SUMO acceptors on CtIP were predicted using SUMO2.0 software (https://www.abcepta.com/sumoplot). (B) Lysine 578 was the major SUMO acceptor site of CtIP. HEK293T cells stably expressing His-SUMO2 or His-SUMO2-ΔGG were transfected with the indicated plasmids for 24 h. The cells were then lysed under denaturing conditions and subjected to cobalt pulldown. The resulting isolated proteins were assessed by Western blotting using anti-Flag antibody. (C) The E580A mutation in CtIP impaired its SUMOylation. HEK293T cells stably expressing His-SUMO2 were transfected with the indicated plasmids for 24 h. The cells were then lysed under denaturing conditions and subjected to cobalt pulldown. The resulting isolated proteins were assessed by Western blotting using anti-Flag antibody. (D) HeLa cells stably expressing Flag-tagged wild-type CtIP, K578R, or CtIP-8A mutant were infected with a lentiviral vector expressing His-HA-SUMO2. After 24 h, the cells were synchronized in G1/S boundary by 2 mM thymidine for 24 h and released into S phase in fresh medium for 2 h. The cells were then left untreated or treated with 1 μM CPT for 1 h, lysed under denaturing conditions, subjected to cobalt pulldown, and analyzed by Western blotting with the indicated antibodies. Ratios of phosphorylated to unphosphorylated CtIP were quantified using ImageJ and are indicated beneath the corresponding lanes. (E) The SUMOylation site was validated in vitro. An in vitro SUMOylation assay was performed using a commercial SUMOylation kit (Abcam, ab139470). MBP-tagged wild-type CtIP, K578R, or E580A mutant proteins purified from E. coli were incubated with SAE1-SAE2 (E1), Ubc9 (E2), ATP, and SUMO2 at 37 °C for 60 min. The reactions were analyzed by Western blotting using anti-SUMO2/3 antibody (Top). Purified CtIP proteins were visualized by Coomassie blue staining (Bottom). (F) The sequence of the region containing the SUMOylation site of CtIP was aligned with those from various species.
A previous study reported that CtIP could be constitutively SUMOylated at lysine 896 by the SUMO E3 ligase CBX4 (35). However, we found that the K896R mutant (substitution of lysine 896 with an arginine) was SUMOylated as efficiently as wild-type CtIP (Fig. 3B). In addition, down-regulation of CBX4 did not markedly affect DSB-stimulated SUMOylation of endogenous CtIP (SI Appendix, Fig. S1A). Taken together, these results suggest that lysine 578 is the major SUMO acceptor site on CtIP. Interestingly, lysine 578 of CtIP and its surrounding residues, but not lysine 896, are highly conserved among mammals (Fig. 3F and SI Appendix, Fig. S1B).
PIAS4 Is the Major SUMO E3 Ligase Required for CtIP SUMOylation.
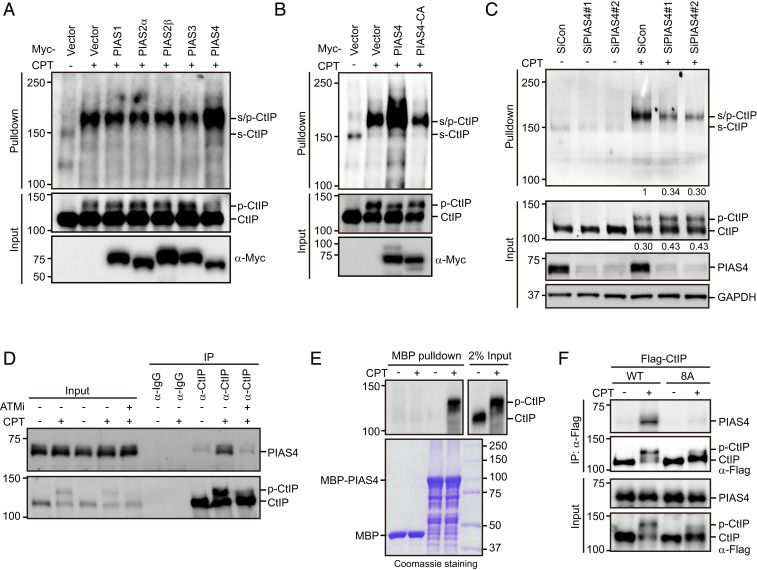
To identify the SUMO E3 ligase responsible for CtIP SUMOylation, we transfected HEK293T cells stably expressing His-SUMO2 with various Myc-tagged SUMO E3 ligases, including PIAS1, PIAS2α, PIAS2β, PIAS3, and PIAS4. As shown in Fig. 4A, PIAS4 greatly enhanced the CPT-induced SUMOylation of CtIP, whereas other SUMO E3 ligases failed to do so. To determine whether this effect requires PIAS4 E3 ligase activity, we mutated the E3 catalytic domain of PIAS4 (substitution of cysteines 342 and 347 with an alanine, PIAS4-CA) and compared its activity toward SUMOylate CtIP with that of wild-type PIAS4. As shown in Fig. 4B, relative to wild-type PIAS4, the catalytic-inactive mutant showed a sharply reduced ability to SUMOylate CtIP. More importantly, ablation of PIAS4 expression significantly inhibited DSB-stimulated SUMOylation of endogenous CtIP (Fig. 4C and SI Appendix, Fig. S1C). These results, together with the observation that codepletion of PIAS4 and CBX4 did not cause any further reduction in CtIP SUMOylation (SI Appendix, Fig. S1D), suggest that PIAS4 encodes the primary SUMO E3 ligase responsible for DSB-induced CtIP SUMOylation. Notably, PIAS4 coimmunoprecipitated with CtIP, and this effect was remarkably stimulated by CPT treatment (Fig. 4D). Furthermore, the stress-induced interaction of PIAS4 and CtIP was efficiently abrogated in the presence of the ATM inhibitor KU-60019 (Fig. 4D). More importantly, the recombinant PIAS4 protein preferentially pulled down hyperphosphorylated species of CtIP (Fig. 4E). These results, along with the observation that SUMOylation of CtIP is stimulated by prior ATM-mediated hyperphosphorylation, suggest that hyperphosphorylation of CtIP by ATM might be essential for its binding to PIAS4. Indeed, mutating ATM phosphorylation sites on CtIP (25) prevented its interaction with PIAS4 (Fig. 4F).
Fig. 4.
PIAS4 interacts with CtIP and stimulates its SUMOylation. (A) PIAS4-stimulated CtIP SUMOylation. HEK293T cells stably expressing His-SUMO2 were transfected with plasmids encoding Myc-tagged PIAS1/2α/2β/3/4 for 24 h. The cells were then left untreated or treated with 1 μM CPT for 1 h, lysed under denaturing conditions, and subjected to cobalt pulldown. The resulting isolated proteins were assessed by Western blotting using anti-CtIP antibody. (B) The SUMO E3 ligase activity of PIAS4 was required for CtIP SUMOylation. HEK293T cells stably expressing His-SUMO2 were transfected with the indicated plasmids for 24 h. The cells were either left untreated or treated with 1 μM CPT for 1 h, lysed under denaturing conditions, and subjected to cobalt pulldown. The resulting isolated proteins were assessed by Western blotting using anti-CtIP antibody. (C) PIAS4 depletion inhibited CPT-induced endogenous CtIP SUMOylation. HEK293T cells stably expressing His-SUMO2 were transfected with the indicated siRNAs. Forty-eight hours after transfection, the cells were either left untreated or treated with 1 μM CPT for 1 h, lysed under denaturing conditions, and subjected to cobalt pulldown. The resulting isolated proteins were assessed by Western blotting using anti-CtIP antibody. SUMO-modified hyperphosphorylated CtIP levels and ratios of phosphorylated to unphosphorylated CtIP were quantified using ImageJ and are indicated beneath the corresponding lanes. (D) CtIP interacted with PIAS4 in a DSB- and ATM-dependent manner. HeLa cells were either mock treated or pretreated with the ATM inhibitor KU-60019 (10 μM) for 1 h prior to CPT (1 μM) treatment. After 1 h, the cells were lysed with NETN buffer in the presence of Benzonase. The cell lysates were incubated with protein A agarose beads conjugated to anti-CtIP antibody, followed by Western blotting. (E) The recombinant PIAS4 protein preferentially pulled down the phosphorylated form of CtIP. HEK293T cells were synchronized in G1/S boundary by 2 mM thymidine for 24 h and released into S phase in fresh medium for 2 h. The cells were then left untreated or treated with 1 μM CPT for 1 h. Subsequently, cells were lysed in NETN buffer containing NaF and protease inhibitors, and the resulting supernatants were incubated with the recombinant MBP-PIAS4 protein at 4 °C for 1 h. The resin was washed three times with NETN buffer, boiled in 2× SDS loading buffer, and resolved on SDS-PAGE. (F) Mutating ATM phosphorylation sites in CtIP impaired its interaction with PIAS4. HeLa cells stably expressing Flag-tagged wild-type CtIP or the 8A mutant were synchronized in G1/S boundary by 2 mM thymidine for 24 h and released into S phase in fresh medium for 2 h. The cells were then left untreated or treated with 1 μM CPT for 1 h. The cell lysates were immunoprecipitated using anti-Flag antibody, and the resulting isolated proteins were subjected to Western blotting.
To map the region of PIAS4 required for mediating its interaction with CtIP, we performed coimmunoprecipitation experiments using a panel of PIAS4 deletion mutants (SI Appendix, Fig. S1 E and F). As shown in SI Appendix, Fig. S1G, the interaction between PIAS4 and CtIP was completely abolished by the deletion of the PINIT motif of PIAS4, indicating that this PINIT motif of PIAS4 is required for its interaction with CtIP. Interestingly, in contrast to wild-type PIAS4, the mutant defective in CtIP binding failed to efficiently stimulate the CPT-induced SUMOylation of CtIP (SI Appendix, Fig. S1H), suggesting that the interaction between PIAS4 and CtIP is critical for PIAS4 function in stimulating CtIP SUMOylation.
The Nuclear Localization Sequence, Rather than SUMOylation of CtIP, Is Responsible for Its Nuclear Localization.
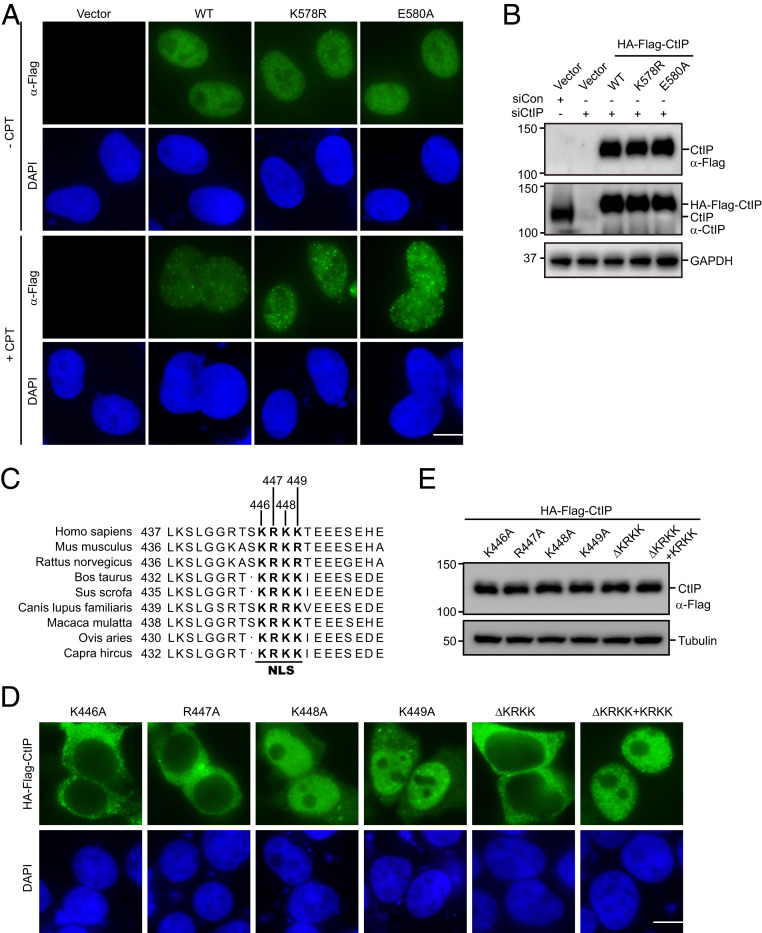
SUMOylation has been shown to orchestrate a variety of cellular processes, in part via control of the nucleocytoplasmic transport of proteins. To examine whether SUMOylation of CtIP at lysine 578 alters its cellular localization, we transfected HA-Flag−tagged small interfering RNA (siRNA)-resistant wild-type CtIP or that of SUMOylation-deficient mutants into HeLa cells following the depletion of endogenous CtIP, and monitored their overall subcellular localization by indirect immunofluorescence staining experimentations. As shown in Fig. 5 A and B, similar to wild-type CtIP, the K578R and E580A mutants were predominantly localized in the nucleus in both mock and CPT-treated cells, indicating that SUMOylation at the identified site had no apparent effect on CtIP nuclear localization.
Fig. 5.
The NLS, not SUMOylation of CtIP, is responsible for its nuclear localization. (A and B) Disruption of CtIP SUMOylation did not alter its cellular localization. HeLa cells transfected with CtIP siRNAs were seeded on a coverslip. Twenty-four hours later, cells were transfected with HA-Flag−tagged siRNA-resistant wild-type CtIP or the SUMOylation-deficient mutants. After 24 h, cells were treated with 1 μM CPT for 1 h or left untreated. Cells were then fixed, permeabilized, and stained with anti-Flag antibody (A). Western blot analysis of the expression of wild-type CtIP or its mutants is shown (B). (Scale bar: 10 μM.) (C) The putative NLS of CtIP was aligned with those from different species. (D and E) The NLS of CtIP was responsible for its nuclear localization. HeLa cells were transfected with HA-Flag−tagged wild-type CtIP or the indicated mutants for 24 h. The cells were then fixed, permeabilized, and stained with anti-Flag antibody (D). Western blot analysis of the expression of wild-type CtIP or its mutants is shown (E). (Scale bar: 10 μM.)
By visual inspection of the CtIP amino acid sequence, we identified a putative nuclear localization sequence (NLS) (amino acid residues 446 to 449, KRKK) in the middle of the CtIP sequence (Fig. 5C). This putative NLS is completely conserved in all known mammalian CtIP homologs (Fig. 5C). To examine whether this putative NLS is necessary for nuclear localization of CtIP, we generated a deletion mutant lacking the entire proposed NLS (CtIP-ΔKRKK) as well as four point mutants with different mutations in the putative NLS motif (substitution of lysine 446, arginine 447, lysine 448, and lysine 449 with an alanine residue K446A, R447A, K448A, and K449A, respectively). As shown in Fig. 5 D and E, whereas the CtIP-ΔKRKK, K446A, and R447A mutants were exclusively present in the cytoplasm, the K448A and K449A mutants were generally localized to the nucleus, with faint cytoplasmic staining. These results indicated that, within the KRKK sequence, the K446 and R447 residues are critical for NLS function, whereas the roles of the other two residues are not apparent. Importantly, addition of the KRKK sequence to the C-terminal end of the CtIP-ΔKRKK construct (ΔKRKK+KRKK) fully restored the nuclear localization of the mutant CtIP-ΔKRKK protein (Fig. 5D). Collectively, these findings indicate that the KRKK sequence within CtIP is necessary and sufficient for its nuclear localization.
RNF4 Targets SUMO-Modified Hyperphosphorylated CtIP for Ubiquitination and Subsequent Degradation.
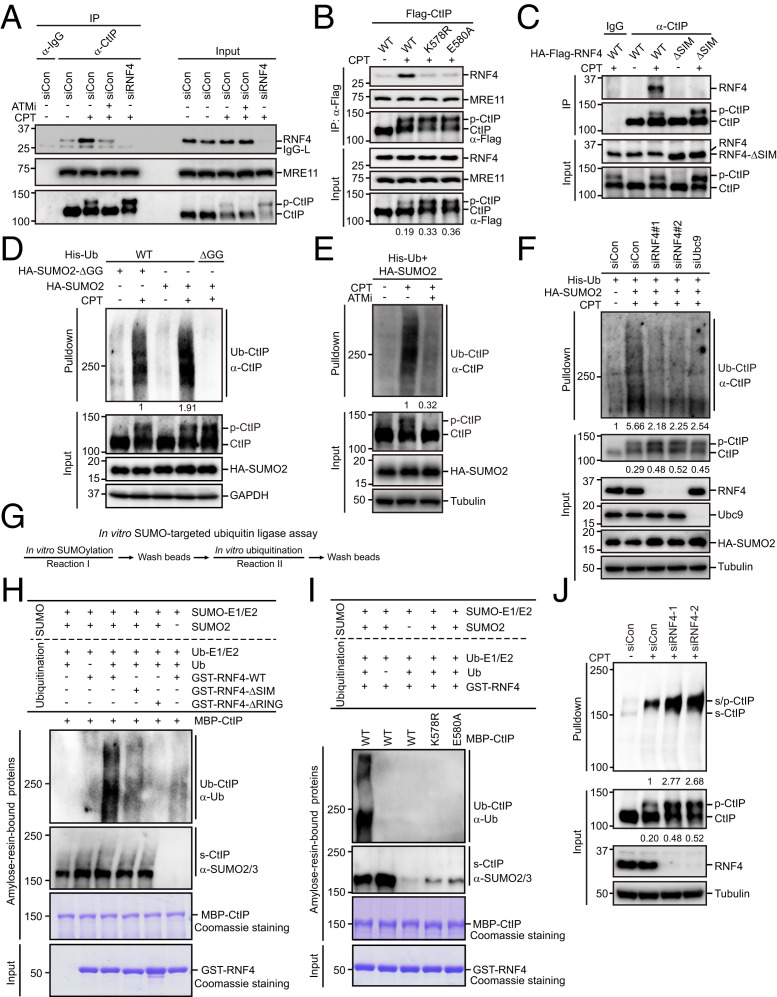
SUMO-targeted ubiquitin ligases (STUbLs), including RNF4 and RNF111 (also known as Arkadia) in vertebrates, are RING ubiquitin ligases that recognize SUMOylated substrates and catalyze their ubiquitination and degradation (39–42) (SI Appendix, Fig. S2 A and B). Therefore, we examined whether RNF4 or RNF111 binds noncovalently to the SUMO moiety of SUMOylated CtIP via their SUMO-interacting motifs (SIMs). As shown in Fig. 6A and SI Appendix, Fig. S2C, CtIP interacted with RNF4 but not RNF111, and the degree of this interaction increased dramatically with CPT treatment. Interestingly, disruption of CtIP SUMOylation or mutation of the SIMs of RNF4 greatly decreased the RNF4–CtIP interaction (Fig. 6 B and C). In accordance with these findings, we found that suppression of ATM kinase activity to block CtIP hyperphosphorylation and subsequent SUMOylation also prevented the interaction of CtIP with RNF4 (Fig. 6A). Together, these data suggest that the RNF4–CtIP interaction is mediated by SIMs and SUMOylation of lysine 578.
Fig. 6.
SUMO-modified hyperphosphorylated CtIP is targeted by RNF4 for polyubiquitination and subsequent degradation. (A) RNF4 interacted with CtIP in a DSB- and ATM-dependent manner. HeLa cells were left untreated or treated with 1 μM CPT for 1 h. The cells were then lysed with NETN buffer in the presence of Benzonase. The cell lysates were incubated with protein A agarose beads conjugated to anti-CtIP antibody and then subjected to Western blotting. (B) SUMOylation of CtIP was required for its interaction with RNF4. HeLa cells stably expressing Flag-tagged wild-type CtIP, K578R, or E580A were synchronized in G1/S boundary by 2 mM thymidine for 24 h and released into S phase in fresh medium for 2 h. The cells were then either left untreated or treated with CPT for 1 h and lysed with NETN buffer in the presence of Benzonase. The resulting lysates were incubated with protein A agarose beads conjugated to anti-Flag antibody and subjected to Western blotting. Ratios of phosphorylated to unphosphorylated CtIP were quantified using ImageJ and are indicated beneath the corresponding lanes. (C) RNF4 interacted with CtIP via its SIMs. HeLa cells stably expressing HA-Flag−tagged wild-type RNF4 or the ΔSIM mutant were either left untreated or treated with 1 μM CPT for 1 h. The cells were then lysed with NETN buffer in the presence of Benzonase. The resulting lysates were incubated with protein A agarose beads conjugated to anti-CtIP antibody and subjected to Western blotting. (D) SUMOylation of CtIP promoted its ubiquitination. HEK293T cells stably expressing His-tagged wild-type ubiquitin or the ΔGG mutant were transfected with HA-tagged wild-type SUMO2 or the ΔGG mutant. After 24 h, the cells were pretreated with 10 μM MG132 for 1 h followed by treatment with 1 μM CPT for 1 h. The cells were then lysed under denaturing conditions and subjected to cobalt pulldown. The resulting isolated proteins were analyzed by Western blotting using the indicated antibodies. Ubiquitinated CtIP levels were quantified by ImageJ and are indicated beneath the corresponding lanes. (E) ATM inhibition greatly reduced CPT-induced SUMO-dependent ubiquitination of CtIP. HEK293T cells stably expressing His-tagged ubiquitin were transfected with HA-tagged wild-type SUMO2. After 24 h, the cells were pretreated with 10 μM MG132 in the presence or absence of the ATM inhibitor KU-60019 (10 μM) for 1 h prior to CPT (1 μM) treatment. After 1 h, the cells were lysed under denaturing conditions and subjected to cobalt pulldown. The resulting isolated proteins were analyzed by Western blotting using the indicated antibodies. Ubiquitinated CtIP levels were quantified by ImageJ and are indicated beneath the corresponding lanes. (F) RNF4 or Ubc9 depletion inhibited CPT-induced SUMO-dependent ubiquitination of CtIP. HEK293T cells stably expressing His-tagged ubiquitin were transfected with the indicated plasmids/siRNAs. Cells were then pretreated with 10 μM MG132 for 1 h prior to CPT (1 μM) treatment. After 1 h, the cells were lysed under denaturing conditions and subjected to cobalt pulldown. The resulting isolated proteins were analyzed by Western blotting using the indicated antibodies. Ubiquitinated CtIP levels and ratios of phosphorylated to unphosphorylated CtIP were quantified using ImageJ and are indicated beneath the corresponding lanes. (G) Schematic diagram of the combined in vitro SUMOylation and ubiquitination assay procedures. (H and I) RNF4-ubiquitinated SUMOylated CtIP in vitro. Bacterially expressed recombinant MBP-CtIP immobilized on amylose resin beads was subjected to in vitro SUMOylation reactions in the presence or absence of SUMO2 and then subjected to in vitro ubiquitination reactions in the presence or absence of bacterially expressed recombinant GST-RNF4-WT, GST-RNF4-ΔSIM, or GST-RNF4-ΔRING, as indicated (H). MBP-CtIP-WT, MBP-CtIP-K578R, or MBP-CtIP-E580A was subjected to in vitro SUMOylation reactions in the presence or absence of SUMO2, followed by in vitro ubiquitination reactions in the presence of bacterially expressed recombinant GST-RNF4-WT, as indicated (I). The ubiquitination and SUMOylation of amylose resin-bound proteins were analyzed by Western blotting using the indicated antibodies. (J) RNF4 depletion increased the levels of both SUMOylated and hyperphosphorylated CtIP. HEK293T cells stably expressing His-SUMO2 were transfected with the indicated siRNAs and treated with 1 μM CPT for 1 h or left untreated. The cells were then lysed under denaturing conditions and subjected to cobalt pulldown. The resulting isolated proteins were analyzed by Western blotting using anti-CtIP antibodies. SUMO-modified hyperphosphorylated CtIP levels and ratios of phosphorylated to unphosphorylated CtIP were quantified using ImageJ and are indicated beneath the corresponding lanes.
We next investigated whether RNF4 promotes CtIP ubiquitination following CPT treatment. As shown in Fig. 6D, overexpression of SUMO2 significantly enhanced CPT-induced polyubiquitination of CtIP. In agreement with this result, the DSB-induced SUMOylation-dependent polyubiquitination of CtIP was greatly reduced upon ATM inhibition (Fig. 6E). Similar results were obtained when the hyperphosphorylation or SUMOylation sites on CtIP were mutated (SI Appendix, Fig. S3A). These results suggest that DSB-induced hyperphosphorylation-dependent SUMOylation of CtIP may stimulate its ubiquitination. The role of SUMOylation in promoting CtIP ubiquitination was further corroborated by our finding where ablation of Ubc9 expression severely limited CPT-induced CtIP ubiquitination (Fig. 6F). Notably, CPT-induced SUMOylation-dependent CtIP ubiquitination was also strongly compromised under the condition of RNF4 depletion (Fig. 6F), suggesting that SUMO-modified hyperphosphorylated CtIP is targeted by RNF4.
To further verify that RNF4 targets SUMOylated CtIP, we conducted an in vitro SUMOylation assay followed by an in vitro ubiquitination assay (Fig. 6G). As shown in Fig. 6H and SI Appendix, Fig. S2A, MBP-tagged recombinant CtIP was efficiently SUMOylated in vitro by SUMO2 and then ubiquitinated by wild-type RNF4, but not by the catalytically inactive mutant RNF4-ΔRING (substitution of cysteines 173 and 176 with serines). In sharp contrast, non-SUMOylated MBP-CtIP was not efficiently ubiquitinated (Fig. 6H, lane 6). Moreover, the SUMOylation-deficient mutants K578R and E580A both failed to be ubiquitinated by RNF4 in vitro (Fig. 6I). More importantly, mutation of the SIM of RNF4 almost completely abrogated its capacity to ubiquitinate CtIP (Fig. 6H). These results indicate that prior SUMOylation of CtIP is required for its ubiquitination by RNF4.
To investigate the biological consequences of RNF4-mediated CtIP ubiquitination, we measured the levels of SUMOylated CtIP and hyperphosphorylated CtIP in cells depleted of RNF4. As shown in Fig. 6J, knockdown of RNF4 led to significantly increased levels of SUMOylated CtIP, accompanied by increased levels of hyperphosphorylated CtIP. Similar results were obtained following treatment with proteasome inhibitor MG132 (SI Appendix, Fig. S3B). These results, combined with the observation that depletion of Ubc9 or PIAS4 impaired CtIP SUMOylation and resulted in higher levels of hyperphosphorylated CtIP after CPT treatment (Figs. 4C and 6F), suggest that SUMO-modified hyperphosphorylated CtIP was targeted by RNF4 for ubiquitination and subsequent degradation. Accordingly, we found that mutating the SUMOylation site of CtIP substantially increased the level of its hyperphosphorylated form after CPT treatment (Fig. 6B).
SUMOylation Limits CtIP Deposition at DSBs and Controls the Extent of End Resection.
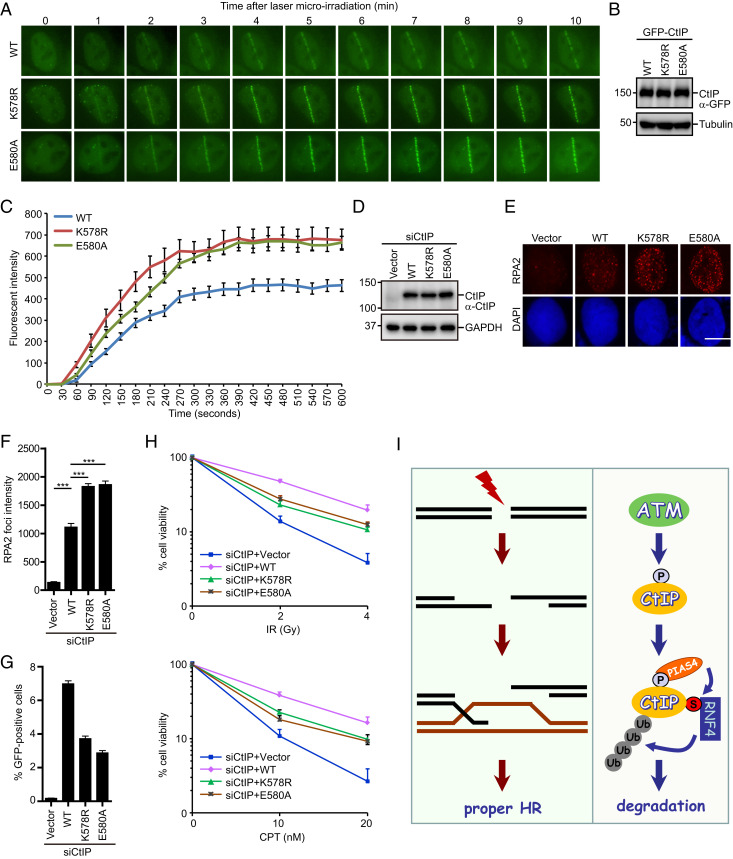
ATM-dependent hyperphosphorylation of CtIP promotes its stable association with sites of DNA damage and subsequent DSB end resection (25–27). Our observation that SUMO-modified hyperphosphorylated CtIP is targeted by RNF4 for degradation raises the possibility that SUMO modification might limit activated CtIP levels at DSBs. To test this possibility, we generated U2OS cell lines stably expressing siRNA-resistant GFP-tagged wild-type CtIP or SUMOylation-defective mutant CtIP and compared their accrual at laser-microirradiated DNA damage sites in the absence of endogenous CtIP. A strikingly high level of SUMOylation-defective CtIP mutant protein localized to laser-induced DNA damage sites when compared with wild-type CtIP (Fig. 7 A–C). These findings, together with the observation that the SUMOylation site is located outside the DNA binding region of CtIP (25–27), indicating that SUMO modification of CtIP regulates its turnover and activity at DSBs.
Fig. 7.
Deregulation of CtIP SUMOylation causes excessive end resection and defective HR. (A–C) SUMOylation-defective mutants of CtIP exhibited enhanced CtIP accumulation at laser-induced DNA damage sites. U2OS cells stably expressing siRNA-resistant GFP-tagged wild-type CtIP, or its SUMOylation-defective mutants were transfected with CtIP siRNAs, laser microirradiated, and then monitored by live cell imaging (A). Magnification: 60×. Western blot analysis of the expression of wild-type CtIP or its mutants is shown (B). The intensity of fluorescence at damaged sites was quantified (C). Data were derived from analysis of at least 20 cells in each experiment and are presented as means ± SEM (SEM). (D–F) Disruption of CtIP SUMOylation resulted in excessive end resection. HeLa cells stably expressing an empty vector (Vector), siRNA-resistant Flag-tagged wild-type CtIP, or its SUMOylation-defective mutants were transfected with CtIP siRNAs and then treated with 1 μM CPT. After 1 h, the cells were processed for RPA2 immunofluorescence. Representative RPA2 foci are shown (E). Quantification results represent the means ± SEM of three independent experiments (F). More than 100 cells were counted in each experiment. (Scale bar: 10 µM.) ***P < 0.001, one-way ANOVA test. Western blot analysis of the expression of wild-type CtIP or its mutants is shown (D). (G) Disruption of CtIP SUMOylation caused defective HR. U2OS DR-GFP cells stably expressing an empty vector (Vector), siRNA-resistant Flag-tagged wild-type CtIP, or its SUMOylation-defective mutants were transfected with CtIP siRNAs and electroporated with pCBASce plasmids for 48 h before assaying for HR efficiency. Data represent means ± SEM of three independent experiments. (H) SUMOylation of CtIP was required for cellular resistance to IR and CPT. HeLa cells stably expressing an empty vector (Vector), siRNA-resistant Flag-tagged wild-type CtIP, or its SUMOylation-defective mutants were transfected with CtIP siRNAs and treated with the indicated doses of IR or CPT. After 24 h, cells were transferred to fresh medium and permitted to grow for 10 d prior to staining. The results are means ± SEM of three independent experiments. (I) Model of the mechanism by which sequential posttranslational modifications regulate CtIP activity at DSBs. P, phosphorylation; S, SUMO; U, ubiquitin.
The findings described above led us to hypothesize that SUMOylation of CtIP may control the extent of end resection. To test this hypothesis, we evaluated the extent of end resection in CtIP-depleted cells reconstituted with either wild-type CtIP or its SUMOylation-defective mutants using RPA2 foci as a surrogate marker. As shown in Fig. 7 D–F, after CPT treatment, the intensity of RPA2 foci was significantly higher in CtIP-depleted cells reconstituted with SUMOylation-defective mutants than in those reconstituted with wild-type CtIP. In line with this, depletion of either RNF4 or UBC9 increases RPA2 foci intensity in CtIP-depleted cells reconstituted with wild-type CtIP but not in cells reconstituted with the SUMOylation-defective mutants (SI Appendix, Fig. S4 A–C). Importantly, disruption of CtIP SUMOylation did not affect cell cycle phase distribution (SI Appendix, Fig. S4D). Taken together, these results suggest that SUMOylation and subsequent ubiquitination of CtIP plays a critical role in determining the extent of end resection.
Disruption of CtIP SUMOylation Results in Defective HR.
As proper regulation of DNA end resection is critical to HR-mediated DSB repair and cell survival after DNA damage, we first employed an established HR reporter system to examine whether disruption of CtIP SUMOylation may cause a defect in the HR repair pathway. To this end, we reconstituted CtIP-depleted U2OS DR-GFP cells with siRNA-resistant wild-type CtIP or SUMOylation-deficient mutant CtIP. As shown in Fig. 7G, CtIP SUMOylation-deficient mutant cells exhibited reduced HR frequency compared with cells expressing wild-type CtIP. Strikingly, wild-type CtIP, but not the SUMOylation-deficient mutants, was able to restore RAD51 foci formation in CtIP-depleted cells (SI Appendix, Fig. S4 E–G), indicating that excessive end resection may prevent RAD51 recruitment to DSBs. It is possible that excessive ssDNAs may not be favorable for BRCA2-dependent RPA-RAD51 exchange. Although one may speculate that extensively resected ssDNA overhangs may preferentially activate the RAD52-mediated single-strand annealing pathway, we found that RAD52 foci also reduced in cells expressing the SUMOylation-defective mutants (SI Appendix, Fig. S4 H–J). The underlying mechanism warrants further investigation. Notably, disruption of CtIP SUMOylation rendered cells more sensitive to the DSB-inducing agents IR and CPT (Fig. 7H). Together, these results suggest that hyperphosphorylation, SUMOylation, and ubiquitination of CtIP act in a sequential manner to ensure the appropriate level of DNA end resection for efficient DSB repair by HR (Fig. 7I).
Discussion
SUMOylation and its crosstalk with other posttranslational modifications have emerged as key mechanisms regulating critical biological processes, including the DNA damage response (12, 13). In this study, we demonstrate that the key resection factor CtIP is SUMOylated by PIAS4 at lysine 578 in response to DSBs, and that this modification is stimulated by its prior ATM-mediated hyperphosphorylation and activation. Failing to SUMOylate CtIP results in excessive end processing, defective HR repair, and hypersensitivity to DSB-inducing agents. Mechanistically, we found that SUMOylation restrains CtIP activity at DSBs via promotion of its polyubiquitination and subsequent degradation. Our results suggest that the interplay among hyperphosphorylation, SUMOylation, and ubiquitination of CtIP determines the activated CtIP level at DSBs and thereby the extent of DNA end resection (Fig. 7I).
A direct link between SUMOylation and protein turnover has recently emerged with the discovery of a family of STUbLs that selectively recognize SUMOylated proteins via SIMs and target them for K48-linked ubiquitination and degradation (39–42). To date, only two STUbLs, RNF4 and RNF111, have been identified in mammalian cells (39–42). Here, we show that RNF4 but not RNF111 is a STUbL for CtIP in the context of genotoxic stress, and that it interacts with the SUMO-modified hyperphosphorylated form of CtIP in an ATM- and SIM-dependent manner. As a consequence, mutation of the SUMOylation site within CtIP or depletion of RNF4 greatly reduced DSB-stimulated SUMO-dependent CtIP ubiquitination, resulting in aberrant accumulation of hyperphosphorylated CtIP. Notably, depletion of RNF4, Ubc9, or PIAS4 caused only a slight increase in the level of hyperphosphorylated CtIP, indicating that only a small fraction of CtIP is SUMOylated and then polyubiquitinated and degraded by RNF4 upon DSB induction. Together, our findings suggest that sequential CtIP posttranslational modifications elicited by ATM are highly coordinated events that are essential to proper CtIP function in DNA end resection and HR repair. In addition to SUMO-modified hyperphosphorylated CtIP, RNF4 can also recognize and ubiquitinate SUMOylated MDC1 and RPA to promote their turnover at DSBs, thereby facilitating HR-mediated DSB repair (43–45). These results strongly suggest that RNF4 can regulate HR repair via multiple processes.
It is noteworthy to mention that knockdown of RNF4 only partially reduced the CPT-induced CtIP ubiquitination (Fig. 6F). We speculate that residual ubiquitination of CtIP observed in RNF4-depleted cells may be attributed to its SUMO-independent ubiquitination by other ubiquitin E3 ligases, including BRCA1, RNF138, Cullin3-KLHL15, APC/C, and SIAH-1 (21, 29, 30, 33, 34). Whereas BRCA1 and RNF138 ubiquitinate CtIP and facilitate its recruitment to DSB sites, RNF4, Cullin3-KLHL15, APC/C, and SIAH-1 target CtIP for ubiquitination and subsequent proteasomal degradation. Interestingly, in contrast to Cullin3-KLHL15, APC/C, and SIAH-1, RNF4 ubiquitinates CtIP for degradation in a manner dependent on ATM-mediated hyperphosphorylation. Thus, it remains formally possible that RNF4 is specifically responsible for fine-tuning CtIP activity at DSBs during the end resection process. Identification of sites on CtIP targeted by different ubiquitin E3 ligases will be necessary to assess the relative contribution of RNF4 to overall CtIP ubiquitination and warrants further investigation.
CtIP and its functional orthologs in various organisms share two evolutionarily conserved sequence motifs in their N and C termini but have highly diverse sequences in other regions (6, 46–48). As proteins in the CtIP family play a conserved role in the early processing of DSB ends, and both human CtIP and its S. cerevisiae homolog Sae2 are SUMOylated in response to DNA damage, it is tempting to speculate that the role of SUMO modification may also be evolutionarily conserved. However, while Sae2 SUMOylation increases the levels of soluble Sae2 to ensure efficient DNA end processing (38), SUMOylation of human CtIP promotes recognition and ubiquitination by RNF4 to avoid uncontrolled excessive resection. In addition, the SUMO attachment site identified in the human CtIP protein is not conserved between mammals and S. cerevisiae, as it is not located within the highly conserved N- and C-terminal regions of CtIP. Moreover, whereas SUMOylation of Sae2 occurs independently of its phosphorylation (38), DSB-induced SUMOylation of human CtIP requires its prior hyperphosphorylation by the ATM protein kinase. In contrast to Sae2, which is constitutively expressed throughout the cell cycle, the expression of human CtIP and its S. pombe ortholog Ctp1 is regulated at the transcriptional level (6, 49, 50). These results suggest that, although the general role of CtIP and its homologs in DNA end resection is conserved from yeast to humans, significant organism-specific differences exist in their regulation.
While this project was in progress, Soria-Bretones et al. (35) reported that CtIP is SUMOylated by the SUMO E3 ligase CBX4 at lysine 896, a residue located within a nonconsensus SUMOylation motif. Surprisingly, although the lysine 896 residue was shown to be related to DNA end resection, Soria-Bretones et al. found that the K896R mutant was SUMOylated to an extent similar to wild-type CtIP (35), which is in accordance with our observations (Fig. 3B). In contrast to lysine 896, we found that mutation of lysine 578 significantly impaired CtIP SUMOylation, irrespective of the presence or absence of DSBs (Fig. 3 B–E). These results demonstrate that lysine 578, but not lysine 896, is the primary SUMOylation site of CtIP. Additionally, Soria-Bretones et al. reported that overexpressed CtIP undergoes SUMOylation in a manner independent of DNA damage (35). In contrast, we found that DSB-inducing reagents caused a significant increase in both endogenous and exogenous CtIP SUMOylation in an ATM- and PIAS4-dependent manner. One possible explanation for this discrepancy is that the SUMOylation experiments in their study were performed in the presence of high levels of ectopically expressed GFP-CtIP, which may bypass the requirement of ATM for DSB-induced SUMOylation of CtIP.
ATM is a master regulator of cellular response to DSBs and belongs to the evolutionary conserved phosphatidylinositol 3-kinase−related protein kinase family. ATM is recruited to DSBs and activated by DNA damage via an interaction with the MRE11/RAD50/NBS1 complex. Growing evidence suggests that ATM is required for efficient DSB end processing, and that phosphorylation of resection factors or resection-related factors might account for at least part of its role in this process. For example, it has been reported that ATM-mediated hyperphosphorylation of CtIP facilitates its stable association with damaged chromatin, thereby stimulating initiation of end resection (27). In addition, hyperphosphorylation of CtIP promotes the recruitment of BLM and EXO1 to sites of DNA damage and is therefore also important for extensive end resection (25). In the present study, we demonstrated that hyperphosphorylation of CtIP by ATM stimulates its SUMOylation and subsequent ubiquitination and degradation, thereby decreasing the level of activated CtIP at DSB sites undergoing homology-directed repair (Fig. 7I). Our findings suggest a dual role of ATM in regulating DSB end processing: 1) ensuring efficient DNA end resection to promote HR-mediated DSB repair and checkpoint activation and 2) suppressing excessive processing of DSB ends to restrain uncontrolled CtIP-mediated end resection (Fig. 7I).
Materials and Methods
In Vitro SUMOylation Assays.
In vitro SUMOylation assays were performed using a SUMOylation kit (Abcam, ab139470) according to the manufacturer’s instructions. Then 200 nM MBP-CtIP-WT−bound, K578R-bound, or E580A-bound beads were incubated at 37 °C for 1 h with the indicated reaction mixture in a total volume of 20 μL. The beads were washed in buffer (20 mM Tris⋅HCl [pH 8.0], 50 mM NaCl, 2 mM DTT, and 1 μg/mL each of leupeptin, aprotinin, and pepstatin) containing 2 mM NEM and boiled in SDS sample buffer or subjected to the in vitro ubiquitination assay.
In Vitro Ubiquitination Assay.
Non-SUMOylated or SUMOylated MBP-CtIP−bound beads were incubated with 50 μM Ub, 200 nM UBA1, 1 μM UBcH5c, and 5 μM RNF4 (wild-type or mutant) in 20 μL of reaction buffer (25 mM Tris⋅HCl [pH 8.0], 50 mM NaCl, 2 mM DTT, 2 mM adenosine 5′-triphosphate [ATP], and 5 mM MgCl2) at 37 °C for 40 min. The beads were then washed in washing buffer (20 mM Tris⋅HCl [pH 8.0], 50 mM NaCl, 2 mM DTT, and 1 μg/mL each of leupeptin, aprotinin, and pepstatin) containing 2 mM NEM and boiled in SDS sample buffer.
Statistics and Reproducibility.
All experiments were repeated at least twice, with similar results. Information about statistical tests and the number of biological repeats is provided in the figure legends.
Supplementary Material
Acknowledgments
We thank the Life Sciences Institute core facilities, Zhejiang University, for technical assistance. We are grateful to M. Jasin for sharing the U2OS DR-GFP cell line and to all members of the J. Huang group for helpful discussions. This work was supported by National Natural Science Foundation of China (Grants 31971220, 31730021, 31700715, and 31961160725), National Key Research and Development Program of China (Grants 2018YFC2000100 and 2018YFC1004900), Fok Ying Tung Education Foundation, and China’s Fundamental Research Funds for the Central Universities.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022600118/-/DCSupplemental.
Data Availability
All other data are available from the corresponding author on request. All study data are included in the article and/or SI Appendix.
References
- 1.Hoeijmakers J. H., DNA damage, aging, and cancer. N. Engl. J. Med. 361, 1475–1485 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A., Elledge S. J., The DNA damage response: Making it safe to play with knives. Mol. Cell 40, 179–204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Symington L. S., Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 51, 195–212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huertas P., DNA resection in eukaryotes: Deciding how to fix the break. Nat. Struct. Mol. Biol. 17, 11–16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu T., Huang J., Quality control of homologous recombination. Cell. Mol. Life Sci. 71, 3779–3797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Limbo O., et al., Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28, 134–146 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H., Lisby M., Symington L. S., RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell 50, 589–600 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossiello F., Herbig U., Longhese M. P., Fumagalli M., d’Adda di Fagagna F., Irreparable telomeric DNA damage and persistent DDR signalling as a shared causative mechanism of cellular senescence and ageing. Curr. Opin. Genet. Dev. 26, 89–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X., et al., 14-3-3 proteins restrain the Exo1 nuclease to prevent overresection. J. Biol. Chem. 290, 12300–12312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paudyal S. C., You Z., Sharpening the ends for repair: Mechanisms and regulation of DNA resection. Acta Biochim. Biophys. Sin. (Shanghai) 48, 647–657 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bordelet H., Dubrana K., Keep moving and stay in a good shape to find your homologous recombination partner. Curr. Genet. 65, 29–39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson S. P., Durocher D., Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Schwertman P., Bekker-Jensen S., Mailand N., Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 17, 379–394 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Huertas P., Cortés-Ledesma F., Sartori A. A., Aguilera A., Jackson S. P., CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455, 689–692 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huertas P., Jackson S. P., Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284, 9558–9565 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferretti L. P., Lafranchi L., Sartori A. A., Controlling DNA-end resection: A new task for CDKs. Front. Genet. 4, 99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manfrini N., Guerini I., Citterio A., Lucchini G., Longhese M. P., Processing of meiotic DNA double strand breaks requires cyclin-dependent kinase and multiple nucleases. J. Biol. Chem. 285, 11628–11637 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anand R., Ranjha L., Cannavo E., Cejka P., Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell 64, 940–950 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Öz R., et al., Phosphorylated CtIP bridges DNA to promote annealing of broken ends. Proc. Natl. Acad. Sci. U.S.A. 117, 21403–21412 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu X., Chen J., DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 24, 9478–9486 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu X., Fu S., Lai M., Baer R., Chen J., BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 20, 1721–1726 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buis J., Stoneham T., Spehalski E., Ferguson D. O., Mre11 regulates CtIP-dependent double-strand break repair by interaction with CDK2. Nat. Struct. Mol. Biol. 19, 246–252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H., et al., PLK1 targets CtIP to promote microhomology-mediated end joining. Nucleic Acids Res. 46, 10724–10739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steger M., et al., Prolyl isomerase PIN1 regulates DNA double-strand break repair by counteracting DNA end resection. Mol. Cell 50, 333–343 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Wang H., et al., The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 9, e1003277 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson S. E., et al., Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol. Cell 49, 657–667 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You Z., et al., CtIP links DNA double-strand break sensing to resection. Mol. Cell 36, 954–969 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makharashvili N., et al., Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol. Cell 54, 1022–1033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Germani A., et al., SIAH-1 interacts with CtIP and promotes its degradation by the proteasome pathway. Oncogene 22, 8845–8851 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Lafranchi L., et al., APC/C(Cdh1) controls CtIP stability during the cell cycle and in response to DNA damage. EMBO J. 33, 2860–2879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H., et al., The deubiquitylating enzyme USP4 cooperates with CtIP in DNA double-strand break end resection. Cell Rep. 13, 93–107 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Wijnhoven P., et al., USP4 auto-deubiquitylation promotes homologous recombination. Mol. Cell 60, 362–373 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt C. K., et al., Systematic E2 screening reveals a UBE2D-RNF138-CtIP axis promoting DNA repair. Nat. Cell Biol. 17, 1458–1470 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferretti L. P., et al., Cullin3-KLHL15 ubiquitin ligase mediates CtIP protein turnover to fine-tune DNA-end resection. Nat. Commun. 7, 12628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soria-Bretones I., et al., DNA end resection requires constitutive sumoylation of CtIP by CBX4. Nat. Commun. 8, 113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajendran P., et al., HDAC turnover, CtIP acetylation and dysregulated DNA damage signaling in colon cancer cells treated with sulforaphane and related dietary isothiocyanates. Epigenetics 8, 612–623 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao Z., et al., System-wide analysis of SUMOylation dynamics in response to replication stress reveals novel small ubiquitin-like modified target proteins and acceptor lysines relevant for genome stability. Mol. Cell. Proteomics 14, 1419–1434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarangi P., et al., Sumoylation influences DNA break repair partly by increasing the solubility of a conserved end resection protein. PLoS Genet. 11, e1004899 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun H., Hunter T., Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. J. Biol. Chem. 287, 42071–42083 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erker Y., et al., Arkadia, a novel SUMO-targeted ubiquitin ligase involved in PML degradation. Mol. Cell. Biol. 33, 2163–2177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poulsen S. L., et al., RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J. Cell Biol. 201, 797–807 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prudden J., et al., SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26, 4089–4101 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo K., Zhang H., Wang L., Yuan J., Lou Z., Sumoylation of MDC1 is important for proper DNA damage response. EMBO J. 31, 3008–3019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galanty Y., Belotserkovskaya R., Coates J., Jackson S. P., RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26, 1179–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin Y., et al., SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26, 1196–1208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sartori A. A., et al., Human CtIP promotes DNA end resection. Nature 450, 509–514 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H., et al., CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded breaks. J. Biol. Chem. 287, 21471–21480 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dubin M. J., et al., Dimerization of CtIP, a BRCA1- and CtBP-interacting protein, is mediated by an N-terminal coiled-coil motif. J. Biol. Chem. 279, 26932–26938 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Liu F., Lee W. H., CtIP activates its own and cyclin D1 promoters via the E2F/RB pathway during G1/S progression. Mol. Cell. Biol. 26, 3124–3134 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang F., Tang H., Jiang Y., Mao Z., The transcription factor GATA3 is required for homologous recombination repair by regulating CtIP expression. Oncogene 36, 5168–5176 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other data are available from the corresponding author on request. All study data are included in the article and/or SI Appendix.