Significance
Pseudomonas aeruginosa is a major cause of hospital-acquired infections due to its formation of biofilms that are highly tolerant to antibiotics. Conventional drugs often fail to kill slowly growing biofilms because they do not target the mechanisms that sustain cells in this state; alternative biofilm control strategies are thus urgently needed. One way in which P. aeruginosa builds robust biofilms is through the production of redox-active phenazines such as pyocyanin. We identified an enzyme that degrades pyocyanin but were stymied in studying its potential to combat biofilms due to its poor expression yield. Here we show how protein design can stabilize the enzyme to improve purification yields, enabling physiological studies to reveal a novel enzyme’s therapeutic potential.
Keywords: Pseudomonas aeruginosa, biofilms, antibiotic tolerance, pyocyanin, AffiLib
Abstract
Pseudomonas aeruginosa is an opportunistic human pathogen that develops difficult-to-treat biofilms in immunocompromised individuals, cystic fibrosis patients, and in chronic wounds. P. aeruginosa has an arsenal of physiological attributes that enable it to evade standard antibiotic treatments, particularly in the context of biofilms where it grows slowly and becomes tolerant to many drugs. One of its survival strategies involves the production of the redox-active phenazine, pyocyanin, which promotes biofilm development. We previously identified an enzyme, PodA, that demethylated pyocyanin and disrupted P. aeruginosa biofilm development in vitro. Here, we asked if this protein could be used as a potential therapeutic for P. aeruginosa infections together with tobramycin, an antibiotic typically used in the clinic. A major roadblock to answering this question was the poor yield and stability of wild-type PodA purified from standard Escherichia coli overexpression systems. We hypothesized that the insufficient yields were due to poor packing within PodA’s obligatory homotrimeric interfaces. We therefore applied the protein design algorithm, AffiLib, to optimize the symmetric core of this interface, resulting in a design that incorporated five mutations leading to a 20-fold increase in protein yield from heterologous expression and purification and a substantial increase in stability to environmental conditions. The addition of the designed PodA with tobramycin led to increased killing of P. aeruginosa cultures under oxic and hypoxic conditions in both the planktonic and biofilm states. This study highlights the potential for targeting extracellular metabolites to assist the control of P. aeruginosa biofilms that tolerate conventional antibiotic treatment.
The opportunistic human pathogen, Pseudomonas aeruginosa, is able to form biofilms that are notoriously problematic and difficult to treat. Biofilms are multicellular structures that are characterized by adherence of microorganisms to each other and to surfaces via a secreted polymeric matrix. P. aeruginosa species are known to form biofilms within host tissues (e.g., diabetic foot ulcers and the lungs of cystic fibrosis patients) and on medical devices (e.g., implants and catheters) (1). While the cells within a biofilm are metabolically active, they can tolerate high concentrations of antibiotics (2), making infections difficult to clear. For this reason, studying how P. aeruginosa cells sustain their metabolism in biofilms and how biofilms can be disrupted is of particular interest.
During P. aeruginosa biofilm development, cells produce redox-active metabolites known as phenazines. One such phenazine, pyocyanin (PYO), is blue in color and can be found in pus from wound infections (3, 4) and in lung infection sputum samples (5). PYO confers a competitive advantage for P. aeruginosa by contributing to iron acquisition (6), anaerobic energy conservation (7), signaling (8), and biofilm development (9). It has also been shown that P. aeruginosa cells lacking the ability to synthesize phenazines (10), and PYO in particular (11), are more sensitive to conventional antibiotics when grown both planktonically and in the biofilm state; the mechanisms underpinning these phenomena are beginning to be elucidated (12). Accordingly, we reasoned that enzymatically degrading PYO within P. aeruginosa biofilms might improve conventional drug treatment, representing a strategy for biofilm control.
Previous work in our laboratory identified and isolated a Mycobacterium fortuitum enzyme (PodA) that demethylated pyocyanin to another phenazine derivative, 1-hydroxy-phenazine (SI Appendix, Fig. S1A) (13):
Consistent with the requirement of PYO for P. aeruginosa biofilm formation and metabolic maintenance (14, 15), addition of this enzyme inhibited biofilm development (13). Motivated by these preliminary observations, we sought to further explore PodA’s therapeutic potential under in vitro conditions that were more relevant to chronic infections, such as those that characterize the mucus-filled lungs of individuals living with cystic fibrosis (16). While our preliminary studies indicated that PodA is an effective P. aeruginosa biofilm disruptor (13), its purification through standard heterologous overexpression provided insufficient yield to enable physiological studies, posing a major barrier to exploring its potential synergy with clinical antibiotics.
There are many factors that contribute to protein stability, ranging from exposure of hydrophobic surfaces (17), entropy of folding (18), cellular turnover by proteases (19, 20), potential production of misfolded or aggregated protein (21), and the atomic contacts that stabilize the native state (22). Overexpressing a protein in a heterologous system may also expose incompatibilities between the host’s protein-synthesis machinery and the target protein (21). Attempts to produce stable protein variants through phylogenetic analyses or the application of in vitro evolution have proven very successful. Nevertheless, these conventional strategies are especially difficult to apply to enzymes like PodA, which require time-consuming protein-production protocols that are impossible to implement in medium or high-throughput screening. By contrast, in recent years, computational design methods that use a combination of phylogenetic analysis and atomistic design calculations have exhibited much higher accuracy than conventional optimization strategies (23), enabling the improvement of protein stability, expressibility (24), and activity (25, 26) through the experimental screening of only a few dozen designs.
The goals of this study were three-fold: first, to increase the yield and stability of PodA through the AffiLib protein design method (25); second, with sufficient protein in hand, to characterize the enzymatic activity of designed PodA under a variety of clinically relevant conditions; third, to determine whether the addition of PodA (i.e., PYO depletion) could increase efficacy of tobramycin, one of the most commonly used antibiotics to treat chronic P. aeruginosa infections, in different growth media. Our results demonstrate the potential of PodA to assist the treatment of chronic infections. Due to increasing rates of multidrug resistance and P. aeruginosa biofilms’ extreme tolerance to antibiotics, it is crucial that we find alternative mechanisms for treating such infections.
Results
PodA Activity Is Stable Across Conditions Relevant to Cystic Fibrosis Sputum.
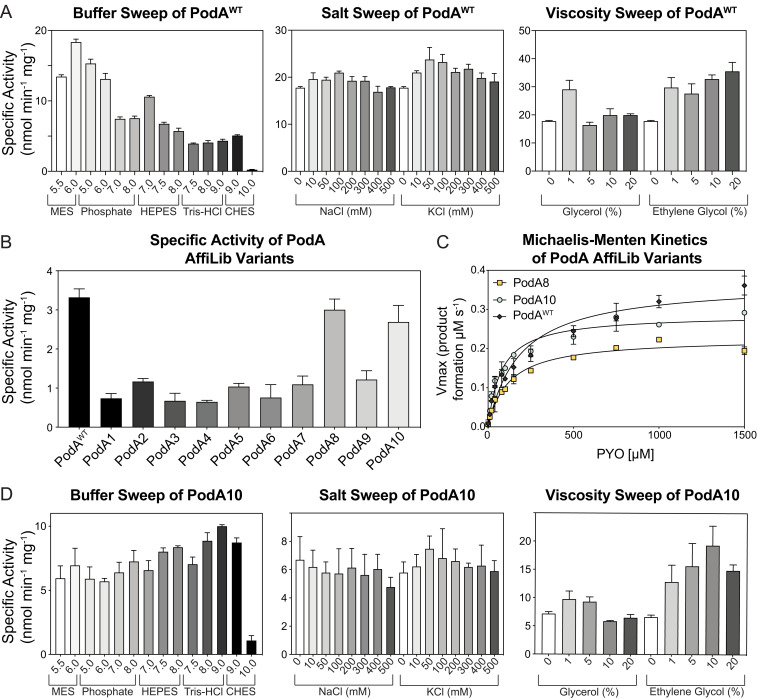
To assess PodA’s potential for therapeutic relevance in the context of human chronic infections, such as those in the cystic fibrosis (CF) airways, we began by quantifying PodA activity and determining its kinetic parameters under a spectrum of physiologically relevant variables (i.e., pH, viscosity, and sodium and potassium concentrations). As done previously to facilitate PodA purification (13), we excluded a predicted N-terminal transmembrane domain, resulting in a wild-type (WT) variant called WT PodA30–162. Moving forward, all purified PodA proteins maintained this deletion. WT PodA30–162 demethylation activity was measured using a spectrophotometric assay that monitors the absorbance of PYO over time. Using a variety of buffers to analyze activity from pH 5.5 to 10.0, we found that WT PodA30–162 had optimal activity at pH 6 (Fig. 1A). Activity of WT PodA30–162 below pH 5.5 was not established due to the pKa of PYO (pKa = 4.9), which changes the chemical and therefore absorptive properties of the substrate. The pH of CF airway surface liquid is acidic and is thought to increase viscosity of CF mucus by influencing mucin electrostatic interactions (27). Using a reaction condition with a pH of 6, a slight increase in viscosity with ethylene glycol or glycerol (1% vol/vol) enhanced WT PodA30–162 activity (Fig. 1A). In the case of glycerol, higher concentrations (5 to 20%) reversed this activity increase. Average ion concentrations of potassium and sodium in cystic fibrosis sputum are 66 mM (Na+) and 15 mM (K+) (16). Addition of these ions in salt form (NaCl and KCl) in this concentration range did not inhibit or enhance WT PodA30–162 activity (Fig. 1A). For these reasons, we established conditions for kinetic analyses to be carried out at a pH of 6 with 1% ethylene glycol.
Fig. 1.
Characterization of WT PodA30–162 and PodA10. (A) WT PodA30–162 specific activity with pH, salt, and viscosity sweeps. WT PodA30–162 (3 µM) was incubated with PYO (100 µM), and activity was monitored by measuring absorbance of PYO (690 nm) over time. (B) PYO (50 µM) demethylation activity of PodA variants (3 µM) was determined using a spectrophotometric assay described in Materials and Methods. (C) Michaelis–Menten kinetics of PodA8 and PodA10 (3 µM of protein per reaction) were found by plotting Vmax values against [PYO], as indicated on the x axis. (D) PodA10 specific activity under varying conditions. PodA10 (3 µM) was incubated with PYO (100 µM), and activity was monitored by measuring absorbance of PYO over time. Activity was analyzed by changing pH, viscosity, or salt concentrations. Error bars in each panel represent SD of triplicates.
The same continuous spectrophotometric assay was used to determine enzyme kinetics of PodA for PYO. Using Michaelis–Menten parameters, WT PodA30–162 exhibited a KM of 0.372 mM ± 0.018, a kcat of 0.176 ± 0.003 s−1, and a catalytic efficiency of 4.73 ± 0.24 × 102 M−1 s−1 (SI Appendix, Fig. S1B). The WT PodA30–162 active-site variants PodAD72A, PodAH121A, PodAH121K, PodAH121R, PodAE154A, PodAY156A, and PodAY156F did not have measurable activity compared to controls, as previously seen (13). A variant inhibiting product release (13), PodAD68A, maintained similar Michaelis–Menten trends for concentrations of PYO under 100 μM, after which Vmax leveled off (SI Appendix, Fig. S1B).
Stabilizing the PodA Trimer Interface Increases Yield by 20-Fold.
Although our characterization of WT PodA30–162 was promising with respect to the enzyme’s activity under a broad range of conditions, our characterization efforts were hampered by poor protein yields (typically ∼0.3 mg⋅L−1 of bacterial culture). Furthermore, we found that PodA purification demands laborious refolding protocols (see below), arguing against using protein-optimization methods that require high- or even medium-throughput experimental screening such as in vitro evolution or deep mutational scanning. Accordingly, we turned to protein design to attempt to increase the stability of PodA in the hopes of efficiently overcoming this experimental bottleneck.
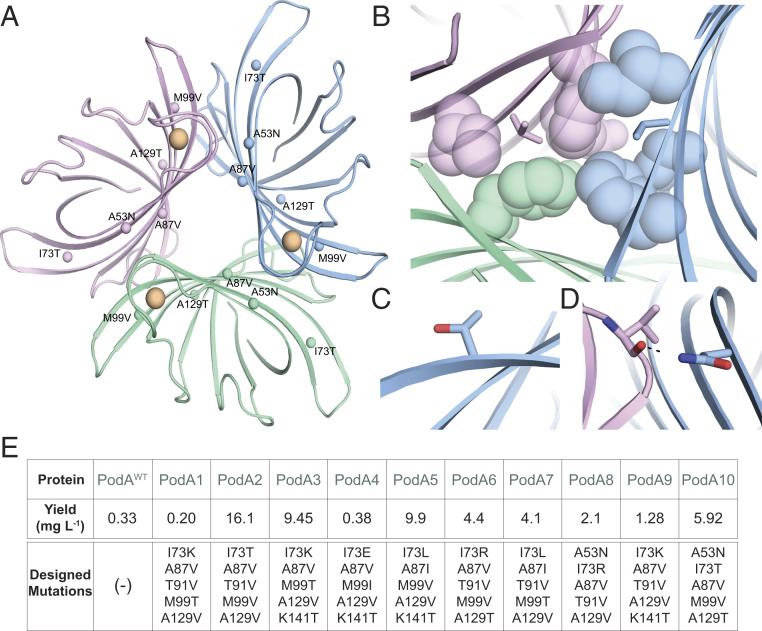
Recently, the AffiLib protein-design method was shown to improve protein binding affinity in designed protein–protein interactions by almost two orders of magnitude (25) and a related method, called AbLIFT, led to dramatic improvement in antibody stability, expressibility, and binding affinity through the design of mutations in the antibody variable domain’s light-heavy chain obligatory interface (28). Because PodA forms symmetric homotrimers, we hypothesized that improving the interactions at these obligatory interfaces may similarly improve complex stability and expressibility. Furthermore, structural analysis of these obligatory trimer interfaces indicated that they may be under-packed due to the presence of three alanine amino acids. The AffiLib design approach is especially relevant for the design of small-to-large mutations, since it uses Rosetta atomistic design calculations to enumerate all of the combinations of mutations that are observed in a phylogenetic analysis of sequence homologs and are individually tolerated according to computational mutation scanning. Each of the enumerated mutants is allowed to relax, including through backbone and rigid-body minimization, to enable the entire trimeric structure to accommodate the introduction of small-to-large mutations. Because PodA is a homotrimer, all modeling and design simulations used symmetric sequence, backbone, and rigid-body sampling. Furthermore, because the available crystallographic structure of PodA shows that each monomer binds a Ca2+ ion (SI Appendix, Fig. S2), we disallowed any change in amino acid identity or conformation at the ion-ligation sites. The designs were ranked according to their computed energies, and the top 35 designs were visually inspected. From those, we chose to test a diverse set of 10 mutants that exhibited improved core packing or hydrogen-bonding interactions across the obligatory interfaces (Fig. 2 A–D).
Fig. 2.
Structure-based design of stabilized PodA variants. (A) PodA forms a symmetric trimer. Each monomer is colored differently, and positions that were subjected to design are labeled with their mutation in PodA10 and marked by spheres. Ca2+ ions are colored orange. (B–D) Selected mutations implemented by AffiLib and their structural context. Colors for each monomer (green, pink, blue) are as in A. (B) Mutation Ala87Val (sticks) increases core packing in a hydrophobic region. (C) Mutation Ile73Thr improves surface polarity. (D) Mutation Ala53Asn form an interfacial hydrogen bond with a backbone carbonyl. (E) Yield of trimer interface designs purified from inclusion bodies per liter of overexpression culture.
Induction studies showed that a majority of the PodA variants were shuttled into inclusion bodies (SI Appendix, Fig. S3). A protocol was developed (Materials and Methods) to denature and purify WT PodA30–162 from inclusion bodies, followed by a refolding procedure. Before moving forward using the inclusion body purification exclusively for the designed variants, we verified that WT PodA30–162 activity and kinetics were not altered after denaturing and refolding the enzyme. Soluble fraction WT PodA30–162 had a catalytic efficiency of 1.05 ± 0.093 × 102 M−1⋅s−1 and WT PodA30–162 purified from inclusion bodies of the same prep had a catalytic efficiency of 1.14 ± 0.25 × 102 M−1⋅s−1.
Remarkably, eight PodA designs exhibited increased protein yields relative to the wild-type protein when purified from inclusion bodies (Fig. 2E). This result is consistent with our working hypothesis that the trimeric interfaces in the wild-type PodA were unstable and that improving packing and interchain contacts at these interfaces might improve protein production yields. Of these eight, wild-type levels of specific activity were maintained for PodA8 and PodA10 (Fig. 1B). We compared the kinetics of these proteins and found no change in catalytic efficiency between PodAWT and PodA8 and a slight increase in efficiency for PodA10 (Fig. 1C). Due to the slightly increased efficiency combined with a higher yield, we decided to use PodA10 to study its effects on P. aeruginosa. As in Fig. 1A with WT PodA30–162, we analyzed PodA10 activity across a broad range of buffers, salts, and viscosity agents. Unlike WT PodA30–162, PodA10 maintained activity over a large range of pH, with no observable preference between a pH of 5.5 and 9 (Fig. 1D), indicating that the design was also more stable to environmental conditions than the wild-type enzyme. PodA10 activity was not altered with addition of salts, and a slight advantage was seen with addition of ethylene glycol (Fig. 2D), similar to WT PodA30–162. Due to the high thermal stability of PodA, we were unable to complete stability assays that rely on monitoring protein unfolding over a temperature gradient. We therefore analyzed the enzyme’s kinetic stability by boiling WT PodA30–162 and PodA10 and measuring activity of each protein after specified boiling times. After 30 and 60 min of boiling, PodA10 had significantly higher activity compared to WT PodA30–162 (SI Appendix, Fig. S4). Thus, the optimized intersubunit interactions in the designed PodA10 resulted in significantly higher Escherichia coli expression levels and kinetic stability compared to the parental enzyme. We also noted that several designed mutations in PodA8 and PodA10 were not prevalent in a multiple-sequence alignment of PodA homologs (SI Appendix, Fig. S5), indicating the importance of the atomistic calculations to design success.
Addition of PodA10 to P. aeruginosa Cultures Enhances Planktonic Killing and Blocks Biofilm Development.
Prior to testing the impact of PodA10 on biofilms, we sought to determine its effects on P. aeruginosa grown planktonically. To ensure that its activity and kinetics would not be compromised in physiologically relevant growth medium, we tested PodA10 PYO demethylation activity in lysogeny broth (LB) and synthetic cystic fibrosis medium (SCFM) (16). We saw no change in catalytic efficiency in either medium compared to experiments performed in buffer (SI Appendix, Fig. S6), allowing us to move forward with studying PodA10 effects on P. aeruginosa. We proceeded to ask whether PodA is synergistic with tobramycin (an antibiotic commonly used to treat P. aeruginosa infections in the clinic). Tobramycin is an aminoglycoside that targets the ribosome, and its efficacy requires a sufficient proton motive force for entry (29, 30). The minimum inhibitory concentration (50%) for tobramycin on Pseudomonas ranges from 1 to 3 µg⋅mL−1 (31), with the target tobramycin concentration in sputum of cystic fibrosis infections being ∼130 µg⋅mL−1 (32).
To assess whether PodA10 is synergistic with tobramycin, we grew planktonic cultures of P. aeruginosa strain PA14 to high densities in LB or SCFM over a range of concentrations of PodA10 (0 to 20 µM) and tobramycin (0 to 100 µg⋅mL−1). Cells were incubated statically. After incubation, cells were resuspended and reinoculated into fresh medium. Lag times were calculated for each well, and we hypothesize that increased lag times reflect increased cell death from the treatment. Based on the results of these screens, we identified synergistic concentrations of PodA and tobramycin to be analyzed in triplicate. A minimum of 10 mg of PodA was needed to complete preliminary experiments in triplicate, followed by targeted experiments in biological triplicate with technical triplicate in LB and SCFM. To perform these studies, we would have had to purify protein from over 30 L of culture for WT PodA30–162 but, due to the improvements in yield observed for PodA10, we were able to achieve our goals using only 1.6 L of culture—a substantial improvement in experimental efficiency.
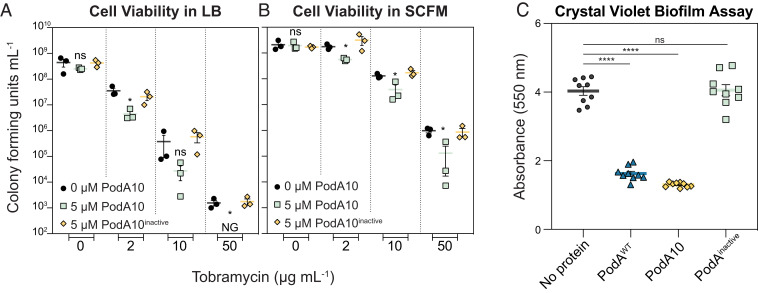
In LB, addition of PodA10 (>1 µM) together with tobramycin increased lag times by 1.5 to 2.0 h compared to PodA (–) controls (SI Appendix, Fig. S7). To verify that lag times were due to increased cell death, cells were plated to count colony forming units (CFUs). PodA10 (5 µM) alone did not increase cell death, but when PodA10 was added with tobramycin, cell viability greatly decreased compared to no protein or inactive PodA10 controls (Fig. 3A). The same experiments were repeated in SCFM, a minimal medium whose composition is based upon that of sputum from cystic fibrosis patients (16) and which elicits a similar response from P. aeruginosa grown in vitro compared to the in vivo CF lung environment (33). While slight differences were seen in lag times when PodA10 was added with tobramycin, the effects were not as dramatic as seen in our experiments with LB (SI Appendix, Fig. S7). However, when plating the same SCFM cultures to calculate viability via CFUs, there was a significant decrease in survival when PodA10 (5 µM) was added with tobramycin compared to no protein or inactive PodA10 controls (Fig. 3B). We speculate that the inconsistent lag times observed in our experiments (SI Appendix, Fig. S7) were due to cells adjusting from being treated in minimal defined medium (SCFM) and then grown in rich medium (LB), an observation that is common in microbiological growth experiments. This indicates that direct cell counts are a better test for analyzing PodA10 and tobramycin synergistic killing.
Fig. 3.
Effects of PodA10 on planktonic viability and crude biofilm development. Cell viability counts of P. aeruginosa liquid media synergy experiments with PodA10 (0 or 5 µM) and tobramycin (concentrations indicated on the x axis) for LB (A) and SCFM (B). Data represent the mean of biological triplicates with error bars representing the SEM. NG, No Growth. (C) Crystal violet biofilm assay. P. aeruginosa cells were inoculated in a 96-well polystyrene plate containing minimal medium with arginine (40 mM) and no PodA, WT PodA30–162, PodA10, or inactive PodA10, followed by incubation at 37 °C. After 24 h of growth, planktonic cells were washed away, and attached cells were stained with crystal violet. Cells were destained with an acetic acid and methanol mixture, after which biomass was quantified based on crystal violet absorbance at 550 nm. Error bars represent SEM. For all graphs, an unpaired t test was used to calculate P values between treatments, ns, not significant; *P < 0.05; ****P < 0.0001. For both panels, PodA10inactive represents the variant, PodA10H121A,E154A,Y156A.
Having established that PodA10 enhances killing of planktonic P. aeruginosa, we wanted to verify that this variant could also inhibit biofilm formation as shown previously for WT PodA30–162 (13). For our first biofilm test, we used the biofilm crystal violet microtiter plate assay to measure the amount of biomass attached to microtiter plate wells after a defined period of time (34). Incubation for 24 h with addition of PodA10 from the beginning of the assay led to a biofilm attachment defect that was comparable to that achieved by WT PodA30–162; addition of inactive PodA10 had no effect on attachment (Fig. 3C).
As a final test of PodA effects being attributable to PYO removal, Pseudomonas cells that could not synthesize PYO (phzM::aacC) were subjected to the same experimental setup as in Fig. 3. For liquid cultures, there were no significant differences when PodA10 was added in combination with tobramycin, compared to the tobramycin-only controls (SI Appendix, Fig. S8). Addition of WT PodA30–162, PodA10, or inactive PodA did not impact attachment of a PYO-deficient Pseudomonas strain compared to no protein controls (SI Appendix, Fig. S8). These results establish that the decreased viability observed for the addition of PodA10 (Fig. 3) is specifically due to the elimination of PYO from the cultures.
Addition of PodA10 to Aggregate Biofilms Together with Conventional Antibiotics Leads to Synergistic Killing.
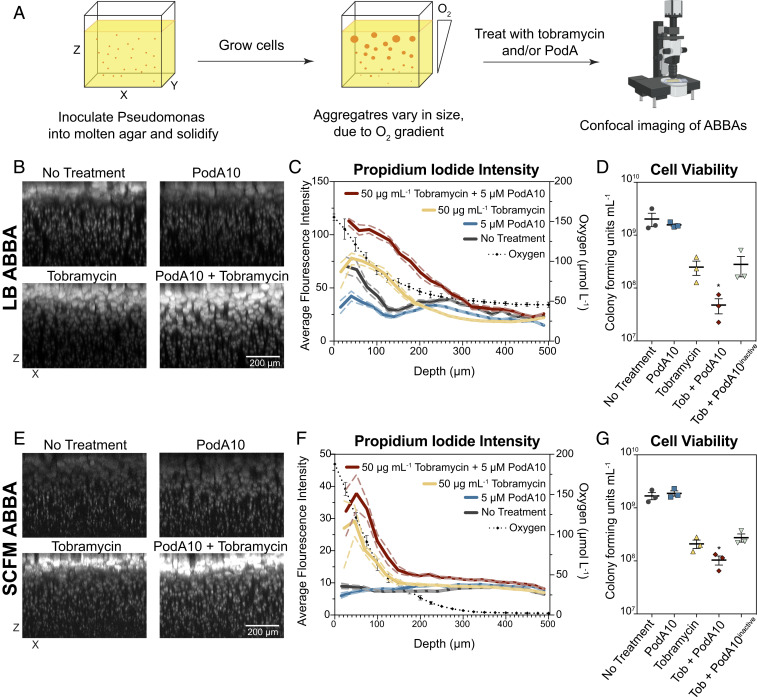
While the crystal violet assay permits biofilm phenotypes to be rapidly screened, it only provides a crude measurement of a biofilm defect related to biofilm development and attachment. To better understand how PodA10 impacts mature biofilms and their viability, we turned to the agar block biofilm assay (ABBA), which was designed to enable the experimental study (13, 35) of the type of aggregate biofilms that characterize human chronic infections (36). Hundreds of distinct biofilm aggregates grow in a single ABBA, allowing for simultaneous observation of biofilms of different sizes. Due to the ABBA being low throughput, we chose to test a single tobramycin concentration (50 µg⋅mL−1) with PodA10 (5 µM) in LB and SCFM, as these concentrations showed the most dramatic synergistic effects in liquid culture (Fig. 3 and SI Appendix, Fig. S7). Cells were inoculated into the agar and allowed to grow into aggregates for 24 h (LB) or 12 h (SCFM), after which PodA10 and tobramycin were added simultaneously and allowed to incubate for 12 h (LB) or 8 h (SCFM) (Fig. 4A). Addition of PodA10 and tobramycin for LB and SCFM was administered at the latest time point for which an oxygen gradient was present (24 h for LB; 12 h for SCFM) (SI Appendix, Fig. S9). This ensured that biofilms were at their most mature state, while still exhibiting metabolic stratification. After treatment, samples were stained with propidium iodide (PI), a fluorescent DNA-binding dye that is incapable of passing through the membrane of highly energized cells and imaged. Generally, higher PI staining indicates lower cell viability due to the compromised membranes of these cells. Aggregates were analyzed via confocal microscopy throughout the depth of the ABBA, with each aggregate representing a distinctly growing Pseudomonas biofilm.
Fig. 4.
Microscopy imaging of propidium-iodide-stained ABBAs treated with PodA10 and tobramycin. (A) Cells were incubated in an ABBA as described in Materials and Methods, followed by treatment with PodA10 (5 µM), tobramycin (50 µg mL−1), or PodA10 and tobramycin with subsequent PI staining. For no treatment conditions, buffer was added in lieu of protein. Cells were grown in either (B) LB or (E) SCFM. (B and E) Maximum-intensity XZ projections where the brightness/contrast in each dataset were normalized to the tobramycin-only sample. (C and F) Average pixel intensity (left y axis) of PI calculated from images in B and E. The solid line represents the mean intensity of aggregates, and the dashed line represents the 95% confidence interval. Both mean and confidence interval were calculated as described in Materials and Methods with a bin size of 20. Each aggregate intensity can be seen in SI Appendix, Fig. S10. The dotted black line represents average oxygen concentration measured at time of treatment as plotted on the right y axis; parallel O2 measurement time points are found in SI Appendix, Fig. S9. Data points (diamonds) are the mean of technical triplicates of biological triplicates, and error bars represent SD. (D and G) PodA and tobramycin ABBA treatments in Eppendorf tubes were homogenized and diluted in series, and dilutions were plated onto LB agar. Cells were counted if a dilution contained 10 to 100 colonies. (D) Aggregates were grown in LB (D) and in SCFM (G). Data represent the mean of technical triplicates from three biological replicates that were treated on different days, with error bars representing the SEM. For D and E, an unpaired t test was used to calculate P values between tobramycin and tobramycin + PodA10 treatments. *P < 0.05.
Incubation of LB-grown aggregates with tobramycin and PodA10 led to PI staining intensity that was significantly greater than PodA10 or tobramycin alone (Fig. 4B). To quantify this visual effect, a three-dimensional mask was generated around each aggregate, and the average PI fluorescence of each aggregate was measured as a function of depth from the top of the agar block (Fig. 4C). In LB, the average PI intensity with PodA10 and tobramycin was higher than the PI intensity of tobramycin alone. To ensure higher intensities were not due to differing aggregate sizes, we plotted the average volume of each aggregate over depth and saw no significant differences in volumes between treatments (SI Appendix, Fig. S10). Plotting the aggregate volume vs. PI intensity of each aggregate further showed that PI staining was highest in the PodA10 + tobramycin treated samples (SI Appendix, Fig. S10). These staining results correlated with cell viability, which was determined by homogenizing ABBAs and plating for CFUs. ABBAs that were incubated with PodA10 and tobramycin had the lowest viable cell counts compared to ABBAs incubated with tobramycin alone or tobramycin and inactive PodA10 (Fig. 4D).
For the LB ABBA, the largest differences in PI staining between tobramycin and tobramycin + PodA10 (Fig. 4 C and F) were between 0 and 300 µm, a region previously characterized as the oxic/hypoxic zone within the agar (13), with oxygen respiration contributing to increased tobramycin efficacy (29). To confirm this, parallel experiments were set up identically, and oxygen profiles were measured using microelectrodes. We observed an oxygen gradient through the depth of the LB ABBA that correlated with the viability patterns observed (Fig. 4C and SI Appendix, Fig. S9). Importantly, the oxygen gradient changed over time: at 12 h, no oxygen was present at a depth of ∼300 μm (SI Appendix, Fig. S9); however, by 24 h, oxygen was again detected at the lower depths. We speculate that these dynamics reflect a change in the limiting nutrient at these different time points: first oxygen, then carbon. Once carbon is depleted, oxygen concentrations rise again due to diffusion and a lack of cellular consumption.
At smaller scales, an oxygen gradient also exists within each single aggregate, due to cellular oxygen consumption at the periphery outpacing O2 diffusion toward the center of the aggregate (37). Additionally, phenazine gradients have been shown to exist within biofilms, with PYO localized to the periphery (38), reflecting the requirement for molecular oxygen in PYO biosynthesis. Analyzing data from Fig. 4B in an XY plane rather than an XZ plane, we find a staining pattern consistent with such gradients: untreated aggregates grown in LB show an outer ring of PI staining that is abolished by addition of PodA10 (SI Appendix, Fig. S11).
Similar synergistic effects between PodA10 and tobramycin treatment were observed for biofilm aggregates grown in SCFM. Qualitative differences in PI staining trends were also seen in this medium (Fig. 4E), with the PI intensity of the PodA10 + tobramycin treatment being higher than tobramycin alone (Fig. 4F). This difference was similar to that seen in LB and was also manifested in the viability counts (Fig. 4G). Microelectrode measurements of 12-h SCFM ABBAs revealed an oxygen gradient decline over depth, mirroring the higher PI staining intensity throughout the upper depths of the ABBA in the presence of tobramycin (Fig. 4F).
Discussion
As a first step toward exploring PodA’s therapeutic potential, we sought to increase its yield during heterologous overexpression. The AffiLib design approach that we utilized was recently applied to the obligatory interfaces in the variable domains of several antibodies, leading to substantial increases in production yields and thermal resistance (28). Applying AffiLib to PodA successfully increased the production yields 20-fold by optimizing contacts across the obligatory homotrimeric interfaces. This enabled us to study the physiological effects of PodA on planktonic and biofilm-grown P. aeruginosa cells under conditions relevant to human infections, revealing significant synergistic killing with the commonly used clinical antibiotic tobramycin.
The phenazine PYO is beneficial to P. aeruginosa biofilm development and metabolic activity (14, 15), and cells that cannot synthesize phenazines have reduced rates of survival in anoxic conditions in the absence of alternative terminal electron acceptors (9, 39). Phenazines must be in an oxidized form to act as electron acceptors, so we wondered whether the conversion of PYOox to 1-OH-PHZred via PodA would be detrimental to P. aeruginosa metabolic vitality within the anoxic zone. Because an oxygen gradient was present throughout the ABBA aggregates, we could examine this effect in deeper regions (>300 µm) of the ABBA. Interestingly, PodA addition alone did not significantly change PI staining at depth (>300 µm, where oxygen is consumed after 12 h). Three possible explanations for this phenotype are: 1) Conversion of PYOox to 1-OH-PHZred still permits sufficient metabolic activity to persist over the time period of our assay (8 to 12 h between PodA addition and PI staining) because 1-OH-PHZred can be abiotically oxidized and used as an electron acceptor; 2) Given the low energetic state previously measured for P. aeruginosa surviving anaerobically by cycling phenazines (7), the membrane potential of cells at this depth of the ABBA system is sufficiently low that PI can be taken up as efficiently by metabolically active cells as by dead cells, thus obscuring a viability readout; and/or 3) PYO is not the primary phenazine sustaining metabolic activity in the anoxic zones; rather, another phenazine, such as phenazine-1-carboxamide [which can also sustain anaerobic survival (40) and localizes to the interior of large anoxic colony biofilms grown on 1% tryptone (38)], may maintain viability under these conditions.
In contrast to the anoxic zone in the ABBAs, PodA addition in combination with tobramycin produced striking effects in the oxic/hypoxic zones of the ABBA assay in LB and SCFM (Fig. 4). Additionally, PodA-only treatments in 24-h LB ABBAs showed a lower PI staining pattern in the oxic region. Importantly, while PYO is beneficial to cells when they are electron-donor replete but oxidant-limited, PYO is toxic to P. aeruginosa under regimes where electron donors are limited but oxidants are replete, such as in the oxic/hypoxic zone of older cultures, where PYO can react with oxygen and cause oxidative stress (41). This fact, combined with the knowledge that PYO is maximally concentrated in the outer regions of large colony biofilms grown on 1% tryptone (38), suggests the following explanation of the effects of PodA on biofilm viability in our 24-h LB ABBA experiments: For untreated cells that are electron donor-limited, PYO causes localized toxicity at the oxic interface of biofilm aggregates. Addition of PodA alone removes PYO from oxic regions, leading to greater metabolic activity of aggregates in these zones, as well as in the outer rings of large aggregates (reflected by lower PI staining). In contrast, when PodA is added together with tobramycin, this enhancement of metabolic activity is a double-edged sword. Greater metabolic activity—specifically, having an inner membrane that is sufficiently energized to be able to take up tobramycin—has been shown to be correlated with greater tobramycin susceptibility (29). Accordingly, in LB and SCFM ABBA regions that have both oxygen and PYO and sufficient carbon, the depletion of PYO by PodA sensitizes these cells to tobramycin treatment. In the LB ABBA, oxygen was consumed by 300 µm, leading to metabolic restriction after 12 h (SI Appendix, Fig. S9), which correlated with no significant differences in PI staining between nontreated and tobramycin-treated samples at these lower depths (Fig. 4C).
It is likely that the differences in effects observed for PodA treatment (in the absence of tobramycin) in LB and SCFM simply reflect differences in timing of treatment and media composition, with varying electron donor:electron acceptor abundance. Notably, SCFM has 3 mM glucose and 9 mM lactate as a carbon source, in contrast to LB that has only amino acids and purines/pyrimidines as carbon sources (42). It is therefore possible that, in the oxic zone at 12 h, cells in SCFM are more protected from PYO toxicity than in LB at 24 h due to a higher electron donor:electron acceptor ratio (41). Finally, the ratios and production of different phenazines (e.g., PYO, PCA, PCN) in P. aeruginosa are known to vary depending on the carbon source in the growth medium (43); such variation could also contribute to the differences that we observed in PodA effects between our media.
Overall, this study demonstrates the utility of protein design methods such as AffiLib to optimize the yield of proteins of interest such as PodA. The significant improvement of protein production yields by optimizing the oligomeric interfaces of PodA and by experimentally testing only 10 designs suggests that other obligatory complexes could similarly be stabilized; this may be quite significant given that a majority of natural proteins form homo-oligomers (44). The availability of a functionally expressible design enabled physiological studies that would have been practically inaccessible otherwise. Because PYO and other phenazines permit P. aeruginosa to adopt metabolic strategies that lead to evasion of antibiotic treatments (10–12), it stands to reason that removal of these metabolites might offer an attractive therapeutic approach. Our results showing enhancement of synergistic killing by a designed PodA and tobramycin provide further motivation and the means to explore PodA’s potential to be used as a biologic therapeutic for treating chronic P. aeruginosa infections. Future experiments will include studying the effects of PodA10 and tobramycin in animal models to determine treatment outcomes.
Materials and Methods
Bacterial Strains, Culture Media, and Chemicals.
Strains used in this study are listed in SI Appendix, Table S1. E. coli BL21 (λDE3) (45) was used for protein overexpression and E. coli DH5α (New England Biolabs) was used for plasmid construction. All E. coli strains were grown at 37 °C in LB (Difco) or Terrific Broth (TB, Difco). Ampicillin for culturing E. coli was used at 100 µg⋅mL−1. P. aeruginosa strain UCBPP-PA14 (hereafter P. aeruginosa) was used for growth analyses and cultivated on LB. The following chemicals were purchased from Sigma-Aldrich unless otherwise noted: glycerol (VWR), Hepes (Gold BioTechnology), and sodium chloride (Fisher Scientific).
Plasmid Construction.
All plasmids used in this work are listed in SI Appendix, Table S2. Primers were synthesized by Integrated DNA Technologies (IDT) and are listed in SI Appendix, Table S3. For heterologous protein expression, the gene coding for PodA was amplified from M. fortuitum strain ATCC 6841 using Phusion High Fidelity DNA Polymerase (ThermoFisher Scientific) per manufacturer’s instructions and cloned into an isopropyl β-d-1-thiogalactopyranoside (IPTG) inducible overexpression vector, pTEV16 (46), using Type II-Restriction BspQI cloning (47). The resulting plasmid was referred to as pPodA1. Catalytic PodA variants were utilized from a previous study (13). For designed proteins, gBlocks were synthesized from IDT to code for relevant amino acid substitutions, with 5′ and 3′ base pairs corresponding to the multiple cloning site (MCS) of pTEV16. One set of primers was used to amplify each gBlock and another set to amplify pTEV16. PCR constructs were isolated using the Monarch PCR & DNA Cleanup Kit (NEB). Genes were then cloned into pTEV16 using Gibson Assembly Master Mix (NEB) per manufacturer’s protocol. The resulting plasmids (pPodA2 to pPodA11) are listed in SI Appendix, Table S2.
Protein Overproduction and Purification of Soluble and Inclusion Body Fractions.
Plasmids coding for proteins of interest were transformed into E. coli BL21 (λDE3). Overnight cultures (10 mL) of transformants were subcultured (1% [vol/vol]) into TB (10 L for WT PodA and 1 L for PodA variants) containing ampicillin. Cultures were grown at 37 °C with shaking to an optical density of 0.4 (OD600nm), and plasmid expression was induced with IPTG (50 µM) and shaken overnight at 16 °C. Cells were harvested by centrifugation at 5,000 x g for 10 min in a Beckman Coulter Avanti J-20 XOI refrigerated centrifuge using a JLA-8.1000 rotor at 4 °C. Cell pellets were stored at −80 °C until used.
For purification, cell pellets were thawed and resuspended in 30 mL buffer A (Hepes [50 mM, pH 7.5], NaCl [500 mM], and imidazole [20 mM]) containing lysozyme (1 mg⋅mL−1), DNase (5 µg⋅mL−1), and protease inhibitor (phenylmethylsulfonyl fluoride, 0.5 mM). Cells were lysed by four passages through an Avestin Emulsiflex C3 (ATA Scientific) at 15,000 psi. Due to difficulties filtering lysate in subsequent steps, additional DNase (5 µg⋅mL−1) was added before centrifugation. Clarified lysates were obtained via centrifugation at 40,000 × g for 30 min at 4 °C in a Beckman Coulter Avanti centrifuge with the JA-25.50 rotor and filtered through a 0.45-µm filter (Millipore). Using an AKTA FPLC, lysate was applied to a pre-equilibrated 5-mL HisTrap FF (Sigma-Aldrich) after which the column was washed with 10 column volumes (CV) of buffer A, 7 CV of 8% buffer B (Hepes [50 mM, pH 7.5], NaCl [500 mM], and imidazole [500 mM]), and a gradient to 100% buffer B over 10 CV. Fractions were run on a sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) gel, and fractions containing PodA were combined and cleaved for 3 h at 25 °C with tobacco etch virus (TEV) protease (1:50 mg:mg ratio of TEV to PodA). TEV was purified and stored as described previously (48). Cleaved protein was dialyzed at 4 °C for 3 h in Hepes (50 mM, pH 7.5), NaCl (500 mM), and EDTA (1 mM), followed by dialysis against Hepes (50 mM, pH 7.5), NaCl (500 mM), and imidazole (20 mM). Cleaved PodA was separated from TEV by passage over a HisTrap FF that was equilibrated with buffer A. For the final dialysis, PodA was dialyzed against 50 mM Hepes (pH 7.5), 100 mM NaCl, and 20% glycerol (vol/vol).
For purification of PodA from inclusion bodies, the pellets remaining following lysis and centrifugation at 40,000 x g were resuspended in 20 mL of detergent buffer (50 mM Hepes [pH 7.5] and 1% Triton-X [vol/vol]) using a Bio-gen PRO200 cell homogenizer (PRO Scientific). Sample was spun for 15 min at 40,000 × g at 4 °C in a Beckman Coulter Avanti centrifuge with a JA-25.50 rotor. Supernatant was discarded, and the remaining cell pellet was resuspended in 5 mL of buffer (Hepes [50 mM, pH 7.5], NaCl (500 mM)] using a cell homogenizer, supplemented with lysozyme (200 µg⋅mL−1) and incubated for 10 min. Volume was increased to 20 mL with Hepes (50 mM, pH 7), and sample was spun for 40,000 × g at 4 °C for 15 min. Supernatant was discarded, and pellet was homogenized in 10 mL of denaturing buffer (Hepes [50 mM, pH 7.5], NaCl [500 mM], and urea [8 M]) and incubating rocking overnight. The sample was spun at 40,000 × g at 25 °C for 30 min, and the supernatant was incubated with 1 mL HisPur Ni-NTA resin (Thermo Fisher Scientific) on a rocking platform for 10 min. Beads were allowed to settle without shaking, supernatant was discarded, and resin was pipetted into glass gravity columns. Resin was washed with 15 mL of denaturing buffer, followed by an elution with denaturing buffer supplemented with imidazole (500 mM). Fractions were analyzed by SDS/PAGE, and those containing protein were combined into SnakeSkin 3.5 kDa molecular weight-cutoff dialysis tubing (Thermo Fisher Scientific). Urea was slowly dialyzed out in Hepes (50 mM, pH 7.5) and NaCl (500 mM) by lowering urea concentrations by 1 M increments and dialyzed at room tempature for 30 min after urea dilution. Each step was dialyzed at room temperature for 30 min. Protein was dialyzed in Hepes (50 mM, pH 7.5), NaCl (500 mM), and imidazole (20 mM) overnight at 4 °C. The following day, protein was cleaved for 3 h at 25 °C with TEV protease (1:50 mg:mg ratio of TEV to PodA). The sample was applied to a HisPur gravity column, and cleaved protein in the flow through was confirmed by SDS/PAGE. Cleaved PodA protein was then dialyzed for storage for 3 h at 4 °C in Hepes (50 mM, pH 7.5), NaCl (250 mM), and finally in Hepes (50 mM, pH 7.5), NaCl (100 mM), and glycerol (20%, vol/vol). Protein concentration was established on a NanoDrop using the extinction coefficient and molecular weight of each protein. Protein was drop-frozen in liquid N2 and stored at −80 °C until use.
Protein Design Calculations.
Using the PodA structure as a starting point (Protein Data Bank ID 5k21), we selected 13 positions in the homotrimeric interfaces for design: 53Ala, 64Val, 67Met, 73Ile, 87Ala, 91Thr, 92Asn, 99Met, 112Glu, 118Leu, 129Ala, 134Thr, and 141Lys. As previously described (25, 26), we generated a Position Specific Scoring Matrix (PSSM) using the default parameters and calculated the tolerated sequence identities at each of the 13 positions with PSSM cutoff ≥ −2 and Rosetta ΔΔG < 6 Rosetta energy units. Next, we enumerated, modeled, and refined in Rosetta all the possible combinations of mutations allowed by the tolerated sequence space that differed from the WT protein by 3 to 5 mutations (a total of 118,424 combinations of mutations). During all Rosetta modeling and design calculations, the structure was modeled with the Ca2+ ions, and their ligating residues were unchanged and subject to C3 symmetry constraints. We then ranked the designs according to Rosetta energy and clustered them, retaining the low-energy designs that exhibited at least two mutations relative to one another. We chose for experimental screening 10 mutants of the top 35 by visual inspection. The command lines and RosettaScripts (49) used to run the symmetric refinement, mutational scanning, and design trajectories are provided in supplemental files refine_auto.xml and filterscan_auto.xml and mutate_auto.xml. The PSSM file used in the design calculations is available as file 5k21_pssm.txt, and the results of the mutational scanning calculations are provided as resfile_ddg6.txt.
A web-accessible version of AffiLib (for nonsymmetric design) is available (http://affilib.weizmann.ac.il/) for academic users.
Synthesis of PYO.
PYO was synthesized from phenazine methosulfate using a protocol described previously (50). Deviations included eliminating purification of PYO via thin-layer chromatography plates and utilizing dichloromethane in the place of chloroform. PYO was analyzed for purity via high-performance liquid chromatography analysis and was found to be >95% pure. Lyophilized PYO was stored at 4 °C until use, after which it was resuspended in 20 mM HCl to make a 5- mM PYO stock and filtered to eliminate insoluble particulate, and PYO stock concentration was calculated by measuring the absorbance at 690 nm and using the extinction coefficient of PYO (4130 M−1⋅cm−1) in combination with Beer’s Law (A = εcl).
Specific Activity and Kinetics of PodA.
To determine specific activity and kinetic parameters of PodA for PYO, a continuous spectrophotometric assay monitoring the absorbance of PYO (690 nm) was utilized. Briefly, assays were performed at 25 °C in 100 µL reaction volumes in 96-well plates, and reactions were monitored at 690 nm over 10 min. Reaction mixtures contained phosphate buffer (50 mM, pH 6.0), ethylene glycol (1%, vol/vol), protein (3 µM), and substrate (for specific activity, 100 µM; for kinetics, varying). For boiled PodA assays, protein was diluted (30 µM) into 50 µL of phosphate buffer and incubated in a PCR thermocycler at 100 °C for time periods listed in SI Appendix, Fig. S4. Reactions were cooled to room temperature for 1 h and were diluted (3 µM) into 100 µL reaction volumes. Data were acquired using the SpectraMax M3 Microplate Reader (Molecular Devices) using the Soft Max Pro software every 10 s over 10 min. Path lengths for each well were calculated using Soft Max Pro endpoint readings, and slopes were corrected for path lengths of 1 cm.
Specific activity was calculated from the slope of the linear range (∆OD690 min−1) using Beer’s Law (A = εcl) with a path length (l) of 1 cm and the molar extinction coefficient (ε) of PYO (4,130 M−1⋅cm−1). Knowing the absorption (A), this equation was solved for c, giving specific activity in nmol⋅min−1⋅mg−1 of PodA. For kinetic parameters, graphs of initial velocity (µM⋅s−1) versus substrate concentration (µM) were plotted using Prism v8 (GraphPad). The Michaelis–Menten kinetics model was used to determine Km and Vmax. The turnover number (kcat) was determined using the following equation: Vmax = kcat[E], where [E] was the concentration of PodA added. All spectrophotometric assays mentioned above were completed three times, each in technical triplicate with a representative dataset shown. Error bars represent SD as calculated by Prism v8 (GraphPad). SD for kcat and kcat KM−1 was calculated using standard formulas for propagation of error.
P. aeruginosa PodA and Tobramycin Liquid Synergy Experiments.
For planktonic synergy experiments, starter cultures were grown 24 h at 37 °C shaking in LB or SCFM supplemented with 30 µM FeSO4. Stationary phase cultures (usually between OD500 4 and 6) were aliquoted (100 µL) into 96-well clear bottom flat well plates (VWR). Tobramycin and PodA were diluted into stock concentrations to pipette 5 µL antibiotic and 2 µL PodA per well. Tobramycin was made fresh daily and was diluted in water, and PodA was diluted in Hepes (50 mM, pH 7.5). Plates were incubated in a humid Tupperware chamber lined with wet paper towel at 37 °C for 8 h. After incubation, wells were pipetted to resuspend, and each well was diluted 1:10 into fresh LB. From that mixture, 10 µL of diluted cells were pipetted into 90 µL fresh LB in 96-well flat bottom plates. Light mineral oil (65 µL undiluted, Sigma-Aldrich) was pipetted onto the top of cultures to prevent dehydration and allow for oxygen diffusion. Microtiter plates were incubated at 37 °C inside a temperature-controlled chamber of a BioTek Synergy 4 plate reader. Plates were continuously shaken (medium setting) and time points (OD500) were taken for 24 h every 30 min. Data were plotted using Prism v8. Growth studies were completed three times with a representation from one experiment shown. For concentrations that led to an increase in lag times, the same experiment was set up but, rather than an outgrowth step, cells were diluted over a dilution series, and seven dilutions (10 µL each) were plated on LB agar to determine CFU mL−1. Cells were plated in biological triplicate to obtain an average number of CFU mL−1 to calculate a SD. Cells from each 10 µL drip dilution were counted if colonies were between 10 and 100.
Crystal Violet Assay.
P. aeruginosa grown overnight in LB were inoculated (1.5%, vol/vol of OD500 = 5 culture) and grown for 24 h at 37 °C in 96-well round bottom plates in minimal medium (100 µL) with arginine (40 mM) as the sole carbon source. PodA (1 µM) was added with cells simultaneously and was present during growth and biofilm development. Planktonic cells were removed, and attached cells were quantified with crystal violet staining (125 µL, 0.1% wt/vol), followed by washes with water and destaining with an acetic acid (30%, vol/vol) mixture in water. Crystal violet was measured using a SpectraMax M3 Microplate Reader (Molecular Devices) at an absorbance of 550 nm. Details of this procedure were published previously (34).
P. aeruginosa PodA and Tobramycin ABBA Synergy Experiments.
Starter P. aeruginosa cultures were grown 24 h at 37 °C shaking in LB and the OD500 was determined by diluting cells 1:10. For inoculation into agar, cells were first diluted to an OD of 1.0, followed by a 1:10 dilution for a final OD of 0.1. Molten LB agar (1% wt/vol) or SCFM agar (1% wt/vol, made by mixing premade 2× SCFM + 30 µM FeSO4 and 2% agar) was incubated at 44 °C, after which 10 µL of OD 0.1 culture was added and mixed into 1 mL of agar for a final starting OD of 0.001. A portion of the mixture (200 µL) was pipetted into a well of an eight-well glass chamber slide for microscopy (Thermo Fisher Scientific #155409) or into a 2-mL Eppendorf tube for cell viability experiments. LB ABBAs were incubated at 37 °C for 24 h in a Tupperware dish lined with wet paper towels, and SCFM ABBAs were incubated for 12 h. Following growth, ABBAs were washed with 200 µL of Hepes (20 mM, pH 7) to rid planktonic cells growing on the surface of the agar. Treatment stocks (100 µL each) were made by diluting PodA10 and/or fresh tobramycin into Hepes (final concentration of 20 mM, pH 7) with the no-treatment control consisting solely of Hepes. Final concentrations of PodA10 (5 µM) and tobramycin (50 µg⋅mL−1) were calculated accounting for the total volume of the agar + liquid. LB ABBAs were treated overnight (12 h), and SCFM ABBAs were treated for 8 h, treatment mixture was decanted, and agar blocks were washed with Hepes (400 µL).
For microscopy experiments, cells were incubated for 30 min with 125 µL of diluted propidium iodide (final concentration of 490 µM, when added to ABBA, taking into account the volume of the agar) and SYTO-9 (final concentration of 16 µM), after which 50 µL of a 1:50 dilution of 5-µm fluorescent beads (Spherotech; CFP-5045-2) were added to each agar block. ABBAs were imaged with a Leica TCS SPE confocal microscope as described elsewhere using a 512- × 512-pixel format (35), with the exception of imaging with a 6-µm step size in the Z-direction.
For cell viability experiments, 300 µL of Hepes (20 mM, pH 7) was pipetted into each Eppendorf ABBA tube, and samples were homogenized on speed 3 of a Bio-Gen PRO200 Homogenizer (PRO Scientific). Samples were briefly spun to rid of air bubbles (<1,000 × g) and diluted in series and plated as described above in liquid synergy experiments. Experiments were completed with biological triplicates and were repeated three times.
ABBA Microscopy Image Analysis.
Images were analyzed in FIJI by reslicing the dataset in the XZ dimension followed by a maximum intensity Z-projection. The brightness/contrast was auto-normalized in FIJI to the sample treated with only tobramycin. These look up table values were then applied to all images in the corresponding datasets as shown in Fig. 4 B and E. Experiments were completed in a minimum of biological triplicate with a representative replicate shown in Fig. 4.
To determine mean pixel intensity and aggregate volume of each ABBA, channels 1 (SYTO-9) and 2 (PI) were combined into a single channel using FIJI. Images were analyzed in Imaris, and aggregate masks were created using the “surface” module of the software. Using the surface Imaris wizard, all options under algorithm settings were unchecked. For surface channel settings, channel 3 (representing combined channels) was chosen, and default settings were maintained. For threshold settings, the no-treatment ABBA sample was analyzed for proper data inclusion, and a value of 60 was used for all LB samples with a threshold value of 40 for all SCFM samples. Finally, data were excluded 0.1 µm from the XY edges to exclude aggregates that were cut off due to the imaging field. Data were not excluded from the top edge of the ABBA, as images were collected from the surface of the agar using fluorescent beads as described above. Exported statistics were used to plot aggregate volume or mean PI intensity against the position of aggregates in the Z-dimension.
To determine mean and 95% confidence intervals of PI staining of ABBAs, aggregates were binned by 25 µm through the depth of the agar. Ten thousand bootstrap replicates were produced per bin, the mean was determined of each of those replicates, and then the 2.5 and 97.5 percentiles were calculated of those collections to calculate the 95% confidence interval.
Oxygen Probe Measurements.
ABBAs were inoculated and set up identically as described in P. aeruginosa PodA and Tobramycin ABBA Synergy Experiments. ABBAs were incubated at 37 °C in a humid chamber and were incubated in a 37 °C sand bath during oxygen probe measurements. Oxygen concentrations were measured using a Clark-type amperometric electrode with a 10-µm tip diameter, which was connected to an amplifier of a multimeter (Unisense). A two-step calibration was performed using an oxygen-free solution (0.1 M NaOH, 0.1 M sodium ascorbate) and an oxygen saturated LB or SCFM solution (with 1% wt/vol salinity). The agar surface was found by decreasing the tip depth by 25 µm until oxygen measurements declined by 2 µmol⋅L−1. The oxygen sensor was positioned 100 µm above the agar interface, and data were collected in 25-µm steps for a total of 700 µm. Technical triplicate measurements were made at each depth, with each measurement taking 3 s with 2 s between measurements. Oxygen profile data are averages from one experiment of technical triplicates of biological triplicates with each experiment done in triplicate on different days. Data were collected using SensorTrace Pro-3.1.3 software and were plotted using Prism v8 (GraphPad).
Supplementary Material
Acknowledgments
This work was supported by the Schwartz/Reisman Collaborative Science Program; NIH Grants 1R01AI127850-01A1 and 1R01HL152190-01; and the Doren Family Foundation. Research in the S.J.F. laboratory was further supported by the Dr. Barry Sherman Institute for Medicinal Chemistry; a Consolidator Grant from the European Research Council (815379); and the Israel Science Foundation (1844). R.L.-S. was supported by a fellowship from the Arianne de Rothschild Women Doctoral Program. We thank Steven Wilbert and Melanie Spero for assistance with ABBA experiments; Steven Wilbert for image analysis; John Ciemniecki for image and statistical analyses; Louise Siskel for experimental assistance; and Scott Saunders for providing growth curve fitting software.
Footnotes
Competing interest statement: C.M.V., R.L.-S., O.K., S.J.F., and D.K.N. are named inventors on patents filed by Caltech and the Weizmann Institute on the design methods.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022012118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Driscoll J. A., Brody S. L., Kollef M. H., The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67, 351–368 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Nickel J. C., Ruseska I., Wright J. B., Costerton J. W., Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob. Agents Chemother. 27, 619–624 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruickshank C. N., Lowbury E. J., The effect of pyocyanin on human skin cells and leucocytes. Br. J. Exp. Pathol. 34, 583–587 (1953). [PMC free article] [PubMed] [Google Scholar]
- 4.Friedheim E. A., Pyocyanine, an accessory respiratory enzyme. J. Exp. Med. 54, 207–221 (1931). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson R., et al., Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56, 2515–2517 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L., et al., Triglyceride-rich lipoprotein lipolysis releases neutral and oxidized FFAs that induce endothelial cell inflammation. J. Lipid Res. 50, 204–213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glasser N. R., Kern S. E., Newman D. K., Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol. Microbiol. 92, 399–412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietrich L. E. P., Price-Whelan A., Petersen A., Whiteley M., Newman D. K., The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61, 1308–1321 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Dietrich L. E. P., et al., Bacterial community morphogenesis is intimately linked to the intracellular redox state. J. Bacteriol. 195, 1371–1380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiessl K. T., et al., Phenazine production promotes antibiotic tolerance and metabolic heterogeneity in Pseudomonas aeruginosa biofilms. Nat. Commun. 10, 762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu K., Chen S., Sysoeva T. A., You L., Universal antibiotic tolerance arising from antibiotic-triggered accumulation of pyocyanin in Pseudomonas aeruginosa. PLoS Biol. 17, e3000573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meirelles L. A., Perry E. K., Bergkessel M., Newman D. K., Bacterial defenses against a natural antibiotic promote collateral resilience to clinical antibiotics. PLoS Biol., in press. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa K. C., Glasser N. R., Conway S. J., Newman D. K., Pyocyanin degradation by a tautomerizing demethylase inhibits Pseudomonas aeruginosa biofilms. Science 355, 170–173 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos I., Dietrich L. E., Price-Whelan A., Newman D. K., Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res. Microbiol. 161, 187–191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saunders S. H., et al., Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms. Cell 182, 919–932.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer K. L., Aye L. M., Whiteley M., Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189, 8079–8087 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halperin I., Wolfson H., Nussinov R., Protein-protein interactions; coupling of structurally conserved residues and of hot spots across interfaces. Implications for docking. Structure 12, 1027–1038 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Brady G. P., Sharp K. A., Entropy in protein folding and in protein-protein interactions. Curr. Opin. Struct. Biol. 7, 215–221 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Parsell D. A., Sauer R. T., The structural stability of a protein is an important determinant of its proteolytic susceptibility in Escherichia coli. J. Biol. Chem. 264, 7590–7595 (1989). [PubMed] [Google Scholar]
- 20.McLendon G., Radany E., Is protein turnover thermodynamically controlled? J. Biol. Chem. 253, 6335–6337 (1978). [PubMed] [Google Scholar]
- 21.Goldenzweig A., Fleishman S. J., Principles of protein stability and their application in computational design. Annu. Rev. Biochem. 87, 105–129 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Honig B., Yang A. S., Free energy balance in protein folding. Adv. Protein Chem. 46, 27–58 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Weinstein J., Khersonsky O., Fleishman S. J., Practically useful protein-design methods combining phylogenetic and atomistic calculations. Curr. Opin. Struct. Biol. 63, 58–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldenzweig A., et al., Automated structure- and sequence-based design of proteins for high bacterial expression and stability. Mol. Cell 63, 337–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Netzer R., et al., Ultrahigh specificity in a network of computationally designed protein-interaction pairs. Nat. Commun. 9, 5286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khersonsky O., et al., Automated design of efficient and functionally diverse enzyme repertoires. Mol. Cell 72, 178–186.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang X. X., et al., Acidic pH increases airway surface liquid viscosity in cystic fibrosis. J. Clin. Invest. 126, 879–891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warszawski S., et al., Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces. PLOS Comput. Biol. 15, e1007207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walters M. C. III, Roe F., Bugnicourt A., Franklin M. J., Stewart P. S., Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47, 317–323 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meylan S., et al., Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem. Biol. 24, 195–206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shawar R. M., et al., Activities of tobramycin and six other antibiotics against Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 43, 2877–2880 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruddy J., et al., Sputum tobramycin concentrations in cystic fibrosis patients with repeated administration of inhaled tobramycin. J. Aerosol Med. Pulm. Drug Deliv. 26, 69–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornforth D. M., Diggle F. L., Melvin J. A., Bomberger J. M., Whiteley M., Quantitative framework for model evaluation in microbiology research using Pseudomonas aeruginosa and cystic fibrosis infection as a test case. MBio 11, e03042-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Toole G. A., Microtiter dish biofilm formation assay. J. Vis. Exp., 47 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spero M. A., Newman D. K., Chlorate specifically targets oxidant-starved, antibiotic-tolerant populations of Pseudomonas aeruginosa biofilms. MBio 9, e01400-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kragh K. N., et al., Role of multicellular aggregates in biofilm formation. MBio 7, e00237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart P. S., Diffusion in biofilms. J. Bacteriol. 185, 1485–1491 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bellin D. L., et al., Electrochemical camera chip for simultaneous imaging of multiple metabolites in biofilms. Nat. Commun. 7, 10535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., Kern S. E., Newman D. K., Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J. Bacteriol. 192, 365–369 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Newman D. K., Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen. Environ. Sci. Technol. 42, 2380–2386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meirelles L. A., Newman D. K., Both toxic and beneficial effects of pyocyanin contribute to the lifecycle of Pseudomonas aeruginosa. Mol. Microbiol. 110, 995–1010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sezonov G., Joseleau-Petit D., D’Ari R., Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189, 8746–8749 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jo J., Price-Whelan A., Cornell W. C., Dietrich L. E. P., Interdependency of respiratory metabolism and phenazine-associated physiology in Pseudomonas aeruginosa PA14. J. Bacteriol. 202, e00700-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodsell D. S., Olson A. J., Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 29, 105–153 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Miroux B., Walker J. E., Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 (1996). [DOI] [PubMed] [Google Scholar]
- 46.VanDrisse C. M., Escalante-Semerena J. C., New high-cloning-efficiency vectors for complementation studies and recombinant protein overproduction in Escherichia coli and Salmonella enterica. Plasmid 86, 1–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galloway N. R., Toutkoushian H., Nune M., Bose N., Momany C., Rapid cloning for protein crystallography using type IIS restriction enzymes. Cryst. Growth Des. 13, 2833–2839 (2013). [Google Scholar]
- 48.Blommel P. G., Becker K. J., Duvnjak P., Fox B. G., Enhanced bacterial protein expression during auto-induction obtained by alteration of lac repressor dosage and medium composition. Biotechnol. Prog. 23, 585–598 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fleishman S. J.et al., RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS one 6, e20161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheluvappa R., Standardized chemical synthesis of Pseudomonas aeruginosa pyocyanin. MethodsX 1, 67–73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.