Significance
Despite the wide perception that microbes have rapid growth rates, many environments like seawater and soil are often dominated by microorganisms that can only grow very slowly. Our knowledge about growth is necessarily biased toward easily culturable organisms, which tend to be those that grow fast, because microbial growth rates have traditionally been measured using laboratory growth experiments. However, how are potential growth rates distributed in nature? Using genomic data, we predicted the growth rates of over 200,000 organisms, including many as yet uncultivated species. These data reveal how current culture collections are strongly biased toward fast-growing organisms. Finally, we noticed a bimodal distribution of maximal growth rates, suggesting a natural division of microbial growth strategies into two classes.
Keywords: microbial growth, oligotrophy, copiotrophy, codon usage bias
Abstract
Maximal growth rate is a basic parameter of microbial lifestyle that varies over several orders of magnitude, with doubling times ranging from a matter of minutes to multiple days. Growth rates are typically measured using laboratory culture experiments. Yet, we lack sufficient understanding of the physiology of most microbes to design appropriate culture conditions for them, severely limiting our ability to assess the global diversity of microbial growth rates. Genomic estimators of maximal growth rate provide a practical solution to survey the distribution of microbial growth potential, regardless of cultivation status. We developed an improved maximal growth rate estimator and predicted maximal growth rates from over 200,000 genomes, metagenome-assembled genomes, and single-cell amplified genomes to survey growth potential across the range of prokaryotic diversity; extensions allow estimates from 16S rRNA sequences alone as well as weighted community estimates from metagenomes. We compared the growth rates of cultivated and uncultivated organisms to illustrate how culture collections are strongly biased toward organisms capable of rapid growth. Finally, we found that organisms naturally group into two growth classes and observed a bias in growth predictions for extremely slow-growing organisms. These observations ultimately led us to suggest evolutionary definitions of oligotrophy and copiotrophy based on the selective regime an organism occupies. We found that these growth classes are associated with distinct selective regimes and genomic functional potentials.
The growth rates of prokaryotes vary widely, with doubling times ranging from under 10 min for laboratory-reared organisms (1) to several days for oligotrophic marine organisms (2, 3) and even as high as many years for deep subsurface microbes (4–6). Even under optimal nutrient conditions and in the absence of competition, species will vary in their maximal potential growth rates as a function of their ability to rapidly synthesize cellular components and replicate their genomes (7–10). Broad lifestyle differences can be detected across habitats, with many oligotrophic marine systems harboring slow-growing organisms relative to nutrient-rich habitats like the human gut (9, 11). Yet, optimal, or even adequate, culture conditions for the majority of prokaryotic organisms are unknown (12, 13), making it difficult to assess the true diversity of microbial maximal growth rates. Although growth media for some species can be predicted based on their phylogeny (14), cultivation is laborious and impractical in a high-throughput manner for many ecosystems such as deep sea waters. Moreover, as we show here, even comprehensive culturing efforts targeted at a specific ecosystem (e.g., the human gut) tend to be biased toward fast-growing members of the community. By estimating maximal growth rates directly from environmentally derived sequences, it may be possible to build a comprehensive and unbiased snapshot of microbial growth across different habitats.
A beacon of hope, maximal growth rates predicted using genome-wide codon usage statistics (9), appear to capture overall trends in the growth rates of natural communities (15). Because the genetic code is degenerate, genes may vary in their usage of alternative codons for a given amino acid. Highly expressed genes demonstrate a biased usage of alternative codons, optimized to cellular transfer RNA (tRNA) pools (16–21). Vieira-Silva and Rocha (9) showed that among several possible genomic indicators of growth (e.g., ribosomal RNA [rRNA] copy number and proximity to the origin of replication, tRNA copy number, etc.), high codon usage bias (CUB) in genes coding for ribosomal proteins and other highly expressed genes is the best predictor of high maximal growth rates and can be used to make accurate predictions even with partial genomic data. Their growthpred software leverages this bias to predict maximal growth rates from genomic data (9).
We extend the work of Vieira-Silva and Rocha (9) by assessing additional dimensions of codon usage (20, 22). In doing so, we are able to substantially improve our predictive performance. Additionally, we provide a correction based on species abundances to the method when applied to bulk community data from metagenomes, an important but previously neglected correction. Together, we provide an implementation of these methods in an R package (gRodon). Using our method, we assay growth rates in over 200,000 prokaryotic genomes (23–25), including representative reference genomes, environmentally derived metagenome-assembled genomes [MAGs (26–30)], and single-cell amplified genomes [SAGs (31, 32)], in order to survey the natural diversity of prokaryotic growth rates. We provide this comprehensive set of over 200,000 predictions as a compiled database of estimated growth rates (estimated growth rates from gRodon online [EGGO]). This database reveals a strong bias in existing reference genomic databases toward fast-growing organisms. Finally, we observe a bias in growth predictions for slow-growing organisms, ultimately leading us to suggest an evolutionary definition of oligotrophy based on the selective regime an organism occupies.
Results and Discussion
Predicting Maximal Growth Rates.
More than one aspect of codon usage is associated with growth.
We measured three features of codon usage: 1) the CUB of a user-defined set of highly expressed genes relative to an expectation calculated from the genome-wide codon usage pattern (20), 2) the CUB of highly expressed genes relative to an expectation calculated from the codon usage pattern of highly expressed genes only, and 3) the genome-wide codon pair bias (22). Details of these calculations are in Materials and Methods. In practice, we take the set of highly expressed genes to be those coding for ribosomal proteins because these genes are expected to be highly expressed in most organisms (9). The first measure captures CUB in the classical sense, and the measure independent of length and composition (MILC) metric we use (20) controls for overall genome composition as well as gene length. The second measure captures the “consistency” of bias across highly expressed genes, with the intuition that if highly expressed genes are optimized to cellular tRNA pools, then they will share a common bias (low values indicate high consistency). This quantity can be thought of as the “distance” between highly expressed genes in codon usage space, where we expect these genes to be close together when they are highly optimized for growth. The third measure, codon pair bias, captures associations between neighboring codons, which have been suggested to impact translation (22, 33, 34). Specifically, it has been shown that altering the frequency of different codon pairs (but not the overall codon or amino acid usage) can lead to lower rates of translation, and this strategy has been used to produce attenuated polioviruses [potentially to engineer novel vaccines (22)]. Because it is much more difficult to accurately estimate pair bias due to the large number of possible codon pairs, we do so on a genome-wide scale, calculating pair bias over all genes rather than just for highly expressed genes (our R package includes a “partial” mode for when this is not possible due to partial genomic information). Consider that if there are 64 codons, the number of possible ordered pairs is 4,096, and accordingly, far more data will be needed to reliably estimate the frequencies of all of these pairs than the original set of codons.
We fit our model using all available completely assembled genomes in RefSeq (1,415) for the set of 214 species with documented maximal growth rates compiled by Vieira-Silva and Rocha (9). All three of these measures were significantly associated with growth rate in a multiple regression (CUB, ; consistency, ; codon pair bias, ; linear regression). Furthermore, comparing nested models, incorporating first CUB, then consistency, and finally, codon pair bias, we found that each nested model fit the data significantly better than the last (addition of consistency, ; addition of codon pair bias, ; likelihood ratio test).
gRodon accurately predicts maximal growth rates.
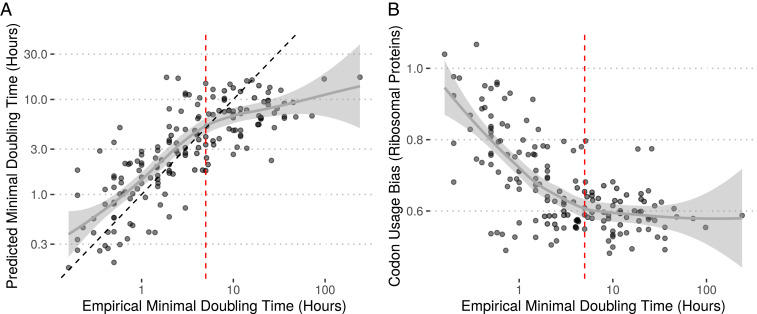
The gRodon model fit the available maximal growth rate data well (adjusted ) (Fig. 1A). Our model demonstrated a significantly better fit to growth data than a linear model fit on the output of growthpred (ANOVA, ) (SI Appendix, Fig. S1). Notably, gRodon provided a better fit to the data than growthpred at both high and low growth rates (SI Appendix, Fig. S2).
Fig. 1.
Predictions from gRodon accurately reflect prokaryotic growth rates, with the caveat that (A) gRodon underestimates doubling times when growth is very slow due to (B) a floor on CUB reached in slow-growth regimes. Vertical dashed red lines at 5 h indicate where the CUB vs. doubling-time relationship appears to flatten. The black dashed line in A is the reference line.
We considered the possibility of overfitting our model to the data, which would inhibit our ability to apply our predictor to new datasets. Overfitting is a particularly relevant concern when dealing with species data since models may end up being fit to underlying phylogenetic structure rather than real associations between variables. In addition to traditional cross-validation (SI Appendix, Fig. S1A), we implemented a blocked cross-validation approach, which effectively controls for phylogenetic structure when estimating model error (35). Under this framework, we take each phylum in our dataset as a fold to hold out for independent error estimation rather than holding out random subsets of our data as in traditional cross-validation. We found that even when predicting growth rates for each phylum in this way (extrapolating from our model fit to all other phyla but excluding the test phylum), we outperformed growthpred’s predictions for the large majority of phyla (SI Appendix, Fig. S1B). Importantly, for this comparison, growthpred’s predictions were based on its fit to the entire dataset (including the test phylum), meaning that gRodon was able to outperform growthpred even when given an unfair disadvantage.
We examined a number of confounding variables that could affect model performance. Observed codon statistics are the result of several interacting evolutionary forces. Selection for rapid growth drives the signal we exploit here, but the effective population size () and the rate of recombination will determine how efficiently selection acts on a given population (36). We found that is correlated with maximal growth rate [as might be expected (37)], as well as our model residuals (SI Appendix, Fig. S3), although the effect is rather weak. For populations with extremely atypical effective population sizes (e.g., intracellular symbionts), we caution that is likely to confound genomic growth rate estimates. Recombination locally increases the efficiency of selection and can lead to weak but significant patterns in guanine–cytosine (GC) content along the genome (38, 39). We found no apparent differences in CUB between genes with or without a signal of recombination, both looking at all genes in a genome (SI Appendix, Fig. S4) and just the ribosomal proteins (SI Appendix, Fig. S5). Finally, especially in oligotrophic marine environments, many microbes experience selection for genome streamlining (high-percentage coding sequence) alongside selection for low genomic GC content (40, 41). While our measures of codon usage should correct for genome nucleotide composition, we wanted to be sure our model’s performance was not affected by these other targets of selection. While percent coding sequence does appear to have some nonlinear association with growth rate, our model residuals were not affected by either percent coding sequence or GC content (SI Appendix, Fig. S6). This is consistent with previous work showing that CUB-based approaches can predict growth rates in low-nutrient marine microcosms (15).
We also assessed the impact of our training set on gRodon’s predictions. The original set of minimal doubling times from Vieira-Silva and Rocha (9) was a carefully hand-curated dataset compiled specifically for this application but includes only a subset of available recorded doubling-time estimates for cultured microbes. Unfortunately, there is no single database describing all known microbial growth rates, but recent work has attempted to compile all available microbial phenotypic data (42), including data on growth rates. We retrained gRodon on the growth rates associated with microbes with completely assembled genomes in the Madin et al. (42) database (464 species with 8,287 genomes). The retrained model yields very similar results to the original gRodon model (SI Appendix, Figs. S7 and S8), despite the two training datasets disagreeing on the maximal growth rates of several species (SI Appendix, Fig. S7). We include this alternative model in the gRodon package alongside the model trained only on the Vieira-Silva and Rocha (9) dataset and include predictions from both models for each entry in the EGGO database.
Prediction from metagenomes.
We implemented a species abundance correction for metagenomes that allows for more accurate prediction of bulk community-wide average maximal growth rates from metagenomes (SI Appendix, Text S1 and Figs. S9–S12).
The problem of slow growers.
For very long doubling times, while gRodon outperforms growthpred it still tends to underestimate the actual doubling time (Fig. 1A and SI Appendix, Fig. S1A). In populations of very slow-growing microbes, selection to optimize transcription of ribosomal proteins is likely quite low, and after the selective coefficient is low enough, drift will dominate the evolutionary process. This expectation is consistent with the pattern seen in Fig. 1B where CUB of the ribosomal proteins reaches a floor at very high doubling times. Importantly, this floor will likely be a problem for many genomic predictors of maximal growth rate where evolutionary optimization is limited by stochastic fluctuations due to drift when selective coefficients are small (43). What can be done in such a scenario? While gRodon cannot accurately differentiate between a doubling time of 10 or 100 h, it can reliably tell us if a doubling time is greater than 5 h long (the threshold at which CUB flattens in Fig. 1B; SI Appendix, Fig. S13). Obviously the degree of CUB will vary to some degree across species and populations for reasons unrelated to growth rate (e.g., as local population size, population structure, selective regimes, recombination rates, etc. vary), but our predictor appears to be largely robust to most confounders (SI Appendix, Figs. S3–S6), and without additional information, 5 h serves well as a pragmatic default. In fact, this threshold suggests a natural definition of an oligotroph as an organism for which selection for rapid maximal growth is low enough so that no signal of growth optimization (e.g., CUB) is observed (discussed in Proposed Evolutionary Definitions of Oligotrophy and Copiotrophy).
The EGGO Database.
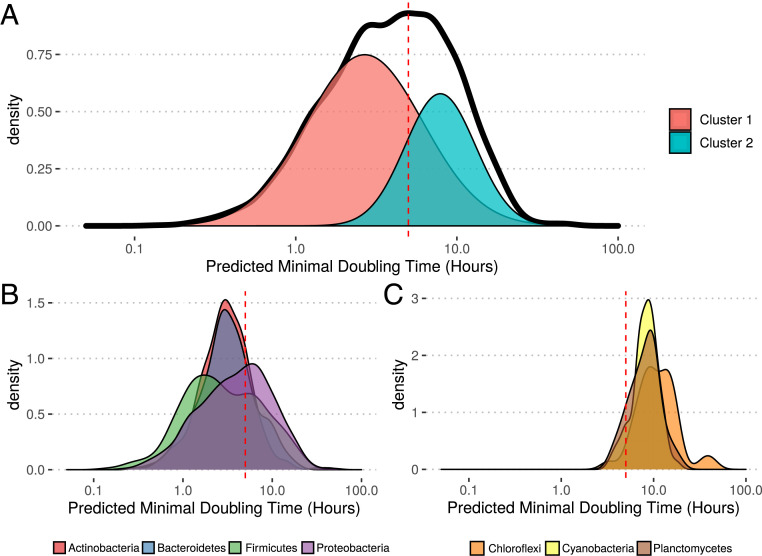
We constructed a database (EGGO) (Table 1) (44–49) of predicted growth rates from 217,074 publicly available genomes, MAGs, and SAGs. Of these, the majority corresponded to RefSeq genome assemblies [184,907 (23, 24)]. The distribution of growth rates across RefSeq was roughly bimodal, with the split between peaks corresponding to the 5-h doubling-time cutoff we proposed above for classifying oligotrophs (Fig. 2A). Additionally, phyla tended to broadly group together in terms of growth rate, and the 5-h divide separated fast- and slow-growing phyla (Fig. 2 B and C). Using a Gaussian mixture model, we obtained two large clusters of microbes, with mean doubling times of 2.7 and 7.9 h, respectively, roughly corresponding to our proposed copiotroph/oligotroph divide (Fig. 2A). We also obtained a third very small and slow-growing cluster, accounting for 0.4% of observations with a mean minimal doubling time of 99 h (too small to plot in Fig. 2A).
Table 1.
Summary of EGGO database
| Source | Type | No. of genomes | Environment |
| RefSeq Assemblies (23) | Isolate | 184,907 | — |
| Parks et al. (26) | MAG | 7,287 | — |
| GORG-tropics (32) | SAG | 7,214 | Marine surface |
| Tully et al. (27) | MAG | 2,266 | Marine |
| Delmont et al. (44) | MAG | 809 | Marine |
| MarRef (45) | Isolate | 725 | Marine |
| Pasolli et al. (46) | MAG | 4,431 | Human microbiome |
| Nayfach et al. (47) | MAG | 4,483 | Human gut |
| Poyet et al. (48) | Isolate | 3,459 | Human gut |
| Zou et al. (49) | Isolate | 1,493 | Human gut |
GORG-tropics, Global Ocean Reference Genomes Tropics.
Fig. 2.
Prokaryotes with sequenced genomes span a broad range of predicted growth rates. (A) Predicted growth rates for assemblies in NCBI’s RefSeq database. Growth rates were averaged over genera to produce this distribution since the sampling of taxa in RefSeq is highly uneven (SI Appendix, Fig. S14 has full distribution) (a small number of genera had inferred doubling times over 100 h, 6 of 2,984). Clusters correspond to the components of a Gaussian mixture model, with area under each curve scaled to the relative likelihood of an observation being drawn from that cluster. (B and C) Growth rate distributions for individual (B) fast- and (C) slow-growing phyla (only showing phyla with genera represented in RefSeq). Vertical dashed red lines in A–C are at 5 h for reference.
We note that this large database of predicted growth rates has many potential applications, including the propagation of growth predictions to microbial taxa on the basis of their 16S rRNA sequence alone (SI Appendix, Text S2 and Figs. S15–S17).
Environmentally derived genomes reveal strong culture biases and ecological insights.
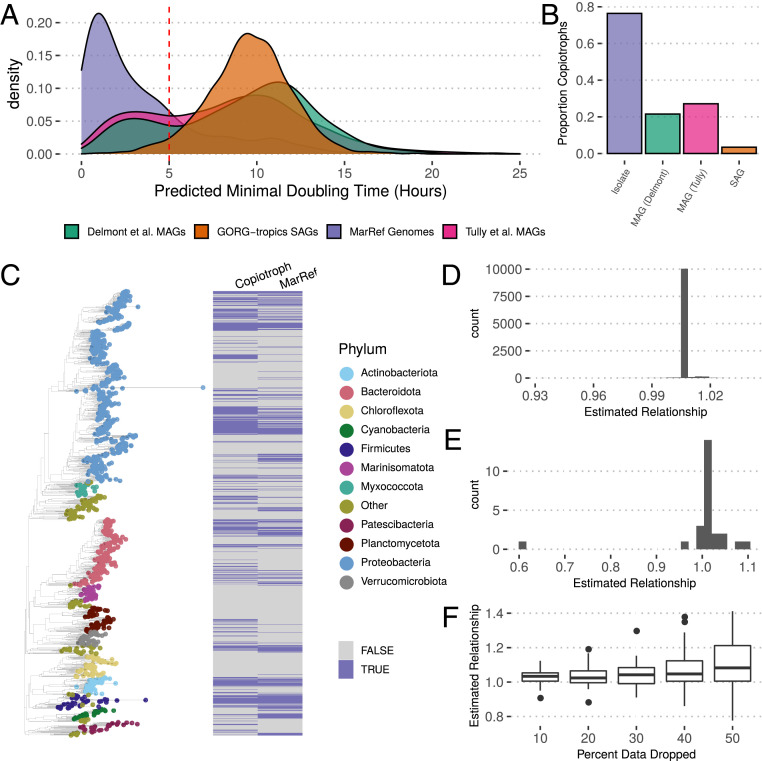
MAGs and SAGs make up a sizable portion of our overall database (26,490) and provide important information about the distribution of growth rates of uncultured organisms. A basic expectation is that cultured microbes from an environment will on average have higher maximal growth rates than the true average across that environment since culturing slow-growing species will in general be more difficult (12, 50). This pattern can be clearly seen in both marine (Fig. 3 A and B and SI Appendix, Fig. S18) and host-associated (SI Appendix, Fig. S19) environments, with isolate collections showing much shorter predicted doubling times than MAGs and SAGs from the same environments. Even in sets of isolates meant to capture the complete taxonomic diversity in an environment (48, 49), we see that they fail to capture the most slowly growing members of the community (SI Appendix, Fig. S19). Illustrating this gap is important, as it shows how existing culture collections are not only incomplete but also biased. These patterns are most apparent when looking within an environment and largely disappear when comparing against MAGs from diverse environments (SI Appendix, Fig. S20) (26).
Fig. 3.
Predicted maximal growth rates in marine environments. Observe that (A and B) genomes from fully sequenced isolates (MarRef) have shorter predicted doubling times on average than MAGs (from the Global Ocean Reference Genomes Tropics [GORG-tropics] database) and SAGs and fail to capture the slow-growing fraction of the community. Additionally, SAGs showed a lower overall growth rate than MAGs, with very few doubling times predicted to be under 5 h, likely due in part to how SAGs were sampled (only at the ocean surface rather than at multiple depths). MAGs generated by distinct research groups showed surprisingly consistent maximal growth rate distributions. (C) Fully sequenced isolates from MarRef are more likely to be copiotrophs (), independent of phylogeny. The tree shown includes one tip sampled per genera, and the corresponding heatmap summarizes whether the mean doubling time was less than 5 h for that genus and whether any representatives of that genus are represented in MarRef (full dataset used for analysis, genus-level summary for visualization only). Phylogenetic logistic regression between isolate status and copiotrophy is robust to (D) the removal of individual species from the analysis, (E) the removal of entire clades from the analysis (the removal of Proteobacteria, the most abundant phylum in the dataset, leads to a weaker but still positive relationship), and (F) the removal of large fractions of the data (up to 50%).
For marine environments in particular, where oligotrophic organisms are prevalent, the existing set of fully sequenced isolates [MarRef (45)] does a poor job of representing the natural distribution of growth rates among taxa (Fig. 3 A and B). We found that these biases are not attributable to simple taxonomic biases in the genomic database. Using phylogenetic logistic regression, we found that whether or not we classify an organism as a copiotroph () (Proposed Evolutionary Definitions of Oligotrophy and Copiotrophy) has a positive impact on whether that strain is represented by a fully sequenced isolate (Fig. 3C) (, ). This relationship is robust to removal of individual species and entire phyla from the dataset (Fig. 3 D and E), as well as to sample size (Fig. 3F) and phylogenetic uncertainty (SI Appendix, Fig. S18). Thus, slow-growing marine organisms are less likely to be represented among completely sequenced genomes than fast-growing organisms, regardless of phylogenetic group. We found the same general pattern in the human gut, although our analysis had less power due to the small number of slow-growing organisms in the gut (, ) (SI Appendix, Fig. S19).
The strong bias shown in Fig. 3 A and B and SI Appendix, Fig. S19 A and B serves to illustrate how important methods for genome-based phenotype prediction are for understanding natural microbial systems. With this in mind, we note that there are many potential use cases for gRodon and the EGGO database, especially when studying subsets of microbes for which additional metadata are available. For example, the very largest cells in marine samples seem to also be those with the highest maximal growth rates (Fisher’s exact test, ) (SI Appendix, Fig. S21). This is consistent with the “nutrient growth law” coined by Schaechter et al. (51), which describes a simple exponential relationship between bacterial cell volumes and their growth rates. In contrast, a similar analysis of growth rate vs. cell size using data from a trait database and cultured isolates (42) was unable to find any association between cell size and growth rate (SI Appendix, Fig. S22). Because maximal growth rate is a basic parameter of microbial lifestyle (10), gRodon and EGGO allow us to build better large-scale comparative studies linking specific traits and habitats to particular microbial life histories.
Proposed Evolutionary Definitions of Oligotrophy and Copiotrophy.
Codon usage may be optimized to promote either translational accuracy, efficiency, or more likely, some combination of the two (21, 52–57). In particular, the strong relationship between maximal growth rate and CUB is thought to be a product of optimization for translational efficiency (9, 52, 56, 58). Our results indicate a clear divide among prokaryotes, where an organism either does or does not experience selection on codon usage to optimize translational efficiency (Fig. 1B). We use this division as the basis for an evolutionary definition of oligotrophy and copiotrophy—defining an oligotroph as an organism for which selection for rapid maximal growth is weak enough so that translation efficiency is not optimized by selection on codon usage (and a copiotroph as an organism that does experience optimization of translation efficiency). This grouping was supported by clustering growth predictions from all organisms in RefSeq (Fig. 2), where we saw two groups naturally emerge with the boundary between them at a doubling time of approximately 5 h, consistent with the CUB optimization cutoff in Fig. 1B.
To illustrate our point that oligotrophy and copiotrophy correspond to two distinct selective regimes, we devised a test for selection to compare the strength of purifying selection on synonymous polymorphisms in highly expressed genes across species. Our test is similar to the statistic, which compares rates of nonsynonymous and synonymous substitution and uses the rate of synonymous substitution () as a null expectation for neutral evolution [although, e.g., ref. 59 has a more developed discussion of the statistic]. For highly expressed genes, where we expect strong purifying selection on codon usage to maintain high translational efficiency, we also expect the rate of synonymous substitution () to be reduced relative to the overall rate of synonymous substitution across the genome. Thus, we define a genome-wide statistic, , which describes the degree of selection at synonymous sites in highly expressed genes relative to the rest of the genome. We predict that this statistic will be significantly lower among copiotrophic organisms as compared with oligotrophs. Using a large database of closely related organisms [Alignable Tight Genomic Clusters (ATGC) database (60)], we made nearly 60,000 pairwise comparisons between organisms to calculate . Using our 5-h doubling-time cutoff, there was a clear difference in selection between copiotrophs and oligotrophs (SI Appendix, Fig. S23) (, Mann–Whitney U test), where copiotrophs had evidence for stronger purifying selection at synonymous sites in highly expressed genes relative to the rest of the genome (), but oligotrophs showed little evidence for optimization of highly expressed genes (). As an important caveat, our test could be confounded by differences in mutation rate across the genome, and if mutation rate is negatively correlated with expression level, we might expect similar results to those shown here even in the absence of selection (assuming that genes encoding highly expressed genes are, in general, even more highly expressed in copiotrophs than oligotrophs). In fact, the opposite is likely true, as it has been shown across diverse organisms that mutation rate increases locally with expression level (61–66). Thus, if anything, differences in mutation rate are probably masking an even stronger difference in the statistic between copiotrophs and oligotrophs than what we see here.
Importantly, our classification redefines copiotrophy and oligotrophy in evolutionary terms, as a specific selective regime that a microbe can occupy. In population genetic terms, we define oligotrophs as existing in a selective regime under which selection on translational efficiency is low enough such that . The boundaries of oligotrophy, in our view, are thus defined both by the selective coefficient () and the effective population size () of a species (as illustrated by the effects of on our model residuals above). Using proxies for and , we found that our statistic was negatively correlated with as expected, particularly in copiotrophs (SI Appendix, Fig. S23). More broadly, it appears that at typical values for microbes [ (37)] (SI Appendix, Fig. S3), codon optimization levels off for maximal doubling times greater than 5 h (Fig. 1B and SI Appendix, Fig. S13). Even for Prochlorococcus marinus, which may have very large effective population sizes [ (67) over a well-mixed marine region, although some estimates of Prochlorococcus are much lower at (37)], growth rates were severely underestimated, although still above our 5-h threshold (predicted doubling time of 6.2 h vs. an actual doubling time of 17 h for strain CCMP1375). This would seem to indicate that for slow-growing organisms like Prochlorococcus, there is essentially no selective advantage to optimizing translational efficiency via codon usage ().
Functional differences between copiotrophs and oligotrophs.
We recognize that our evolutionary definition of copiotrophy/oligotrophy complicates an already murky set of definitions. The terms “oligotroph” and “copiotroph,” as used in the literature, typically conflate two features of microbial lifestyle: resource use and growth rate (41). Giovannoni et al. (41) differentiate the classical definition of oligotrophs and copiotrophs as organisms that grow at low and high nutrient concentrations, respectively, from the more general ecological classes of r and K strategists, which are specialized for either rapid, opportunistic growth or slow and steady growth, respectively (68). Giovannoni et al. (41) emphasize that these nutrient and growth rate definitions need not overlap (not all organisms specialized for growth at high nutrient concentrations must grow quickly), yet theory predicts that, given a rate vs. yield trade-off in adenosine triphosphate (ATP) production, high-resource environments should favor more energetically wasteful but faster growth due to increased competition (69). Thus, we generally expect opportunistic and rapid but wasteful ATP production in nutrient-replete environments vs. slow but relatively energy-efficient ATP production in nutrient-limited environments. Therefore, a natural expectation is that slow- and fast-growing organisms should have distinct resource acquisition strategies aligned to these general niche types (7).
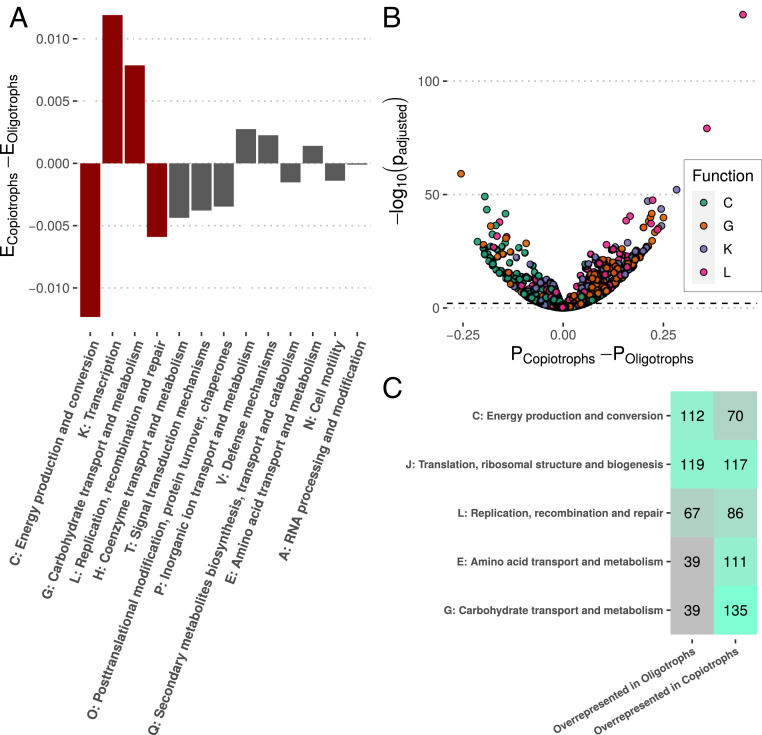
In fact, we found that organisms belonging to our two naturally defined growth rate clusters (Fig. 2), which we refer to here as copiotrophs and oligotrophs, had distinct genomic content reflecting two alternative microbial lifestyles. Gene families involved in transcription and carbohydrate transport and metabolism were strongly overrepresented on copiotroph genomes relative to oligotrophs, corresponding to an overall strategy of rapid acquisition of nutrients and protein production (Fig. 4A). Gene families involved in energy production and conversion and replication, recombination, and repair were overrepresented on oligotroph genomes, corresponding to an overall strategy of energy production and cell maintenance (Fig. 4A). In all, 13 major classes of genes [as defined by the Clusters of Orthologous Genes (COG) database (70)] were significantly differentially enriched on copiotroph or oligotroph genomes. Moreover, many individual gene families were far more prevalent among copiotroph or oligotroph genomes (Fig. 4 B and C and SI Appendix, Fig. S24). Notably, gene families involved in the transport and metabolism of carbohydrates and amino acids were frequently more prevalent among copiotrophs. At the same time, many gene families involved in energy production and conversion were more prevalent among oligotrophs. We also specifically searched for carbohydrate-active enzymes in our two classes of genomes (71) and found that copiotrophs were greatly enriched for these gene families, specifically those involved in breaking down carbohydrates (glycoside hydrolases and polysaccharide lyases) (SI Appendix, Fig. S25).
Fig. 4.
Copiotroph and oligotroph genomes are enriched for different functions. (A) The difference between the average proportion of genes in copiotroph genomes () and the average proportion of genes in oligotroph genomes () assigned to various classes of genes [COG classifications from eggnogmapper (107)]. Positive numbers indicate a functional class is enriched as a percentage of total genes in copiotrophs relative to oligotrophs, and negative values are the opposite. Only significantly differentially enriched classes are shown (with red bars emphasizing classes with larger differences). (B) Volcano plot showing differential prevalence across copiotroph () and oligotroph () genomes of specific gene families belonging to the most differentially enriched gene classes. (C) Table of differentially prevalent gene families from the most commonly differentially prevalent classes (SI Appendix, Fig. S24 has a full table).
Many individual gene families classified as functioning in cellular defense [COG group V (70)] were more prevalent among oligotrophs (SI Appendix, Fig. S24), even as the total percentage of genes in the genome related to defense did not differ a great deal between copiotrophs and oligotrophs (Fig. 4). A closer look at the types of defense genes found in these two groups of organisms revealed stark differences. The majority of defense genes that were copiotroph specific appeared to provide resistance to antimicrobials (e.g., bacteriocins, antibiotics) and oxidants (e.g., hydroperoxides), whereas no such genes were oligotroph specific (55 vs. 0%, , Fisher’s exact test). Many of these genes were transport proteins (e.g., efflux pumps). Many oligotroph-specific genes, on the other hand, were DNA-binding proteins likely involved in antiviral defense, whereas no such genes were copiotroph specific [containing higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domains, pili twitching motility protein N-terminal (PIN) domains, and helix-turn-helix (HTH) domains (72–75); 24 vs. 0%, , Fisher’s exact test]. Similarly, many oligotroph-specific genes were involved in DNA-degrading antiviral defense [e.g., restriction modification systems, CRISPR-Cas systems (76)], whereas few copiotroph-specific genes were (48 vs. 15%, , Fisher’s exact test). Thus, it appears that many genes involved in defending against antimicrobials are copiotroph specific, whereas many forms of antiviral defense are oligotroph specific. Altogether, this suggests that copiotrophs and oligotrophs systematically differ in more than just their growth and resource acquisition strategies.
Conclusions
We produced a community resource in the form of a well-documented R package (gRodon) and comprehensive database (EGGO) for predicting and compiling maximal growth rates across prokaryotes. Using these tools, we show how the set of existing cultured isolates does not fully capture the diversity of prokaryotic lifestyles (although notable culturing efforts have filled in significant gaps) (e.g., ref. 3). We are unlikely to overcome these biases easily, as the slow-growing microbes missing from classically derived culture collections are precisely the ones found to be most difficult (and necessarily very time consuming) to grow in the laboratory (e.g., by dilution to extinction methods). Yet, we have their genomes and may be able to extrapolate their traits from microbes that are more easily cultivable. Growth rate is one example where inference of traits from genomes has clear utility, although we emphasize that genome-wide signals may be confounded by other evolutionary and/or demographic processes and that it is important to assess their robustness and limitations, as we have done here.
It is important to recognize that the relationship of the in situ growth rate and the maximal growth rate of an organism is not clear given the cryptic influence of top-down and bottom-up controls at the sampling time. There are any number of reasons why an organism may not reproduce at its physiological maximal rate (e.g., fluctuating habitat quality, dispersal to suboptimal habitats, etc.). Nevertheless, it is encouraging that recent work using natural communities has shown that CUB-based estimators do a reasonably good job of predicting observed instantaneous growth rates in marine systems (15), even as peak to trough (77–80) methods of estimating growth have been reported to work poorly for marine plankton, with the exception of the most highly abundant copiotrophs (15). Thus, taken together with our benchmarking against nutrient enrichment experiments, the data suggest that CUB-based estimators of maximal growth rate tend to also capture the instantaneous growth rate of a community, likely by approximating the relative proportion of copiotrophs to oligotrophs in a system.
Finally, our analysis of codon usage led us to propose evolutionarily defined growth classes that also align with two distinct functional classes of microbes, with copiotrophs specializing in nutrient acquisition and breakdown and oligotrophs specializing in energy production and cell maintenance (Fig. 4). Environmental resource concentration and growth rate fall along continuous spectra, but microbes appear to fall into two distinct evolutionary regimes in terms of growth optimization (Figs. 1 and 2), corresponding to opposite ends of these spectra. Thus, while in principle oligotrophy and copiotrophy need not correspond to distinct classes and could in fact describe a continuum of life history and resource acquisition strategies, in practice oligotrophs and copiotrophs appear to be discrete groups of organisms.
Materials and Methods
All scripts used to generate figures and analysis, as well as predicted growth rates for various genomic datasets and the full EGGO database, are available at https://github.com/jlw-ecoevo/eggo. The gRodon package, including documentation and a vignette, can be downloaded at https://github.com/jlw-ecoevo/gRodon. All figures were made using R packages ggplot2 and ggpubr (81, 82).
Model Fitting.
For each species with a growth rate listed in the original Vieira-Silva and Rocha dataset [214 (9)], we downloaded all available complete genome assemblies from the National Center for Biotechnology Information (NCBI) RefSeq database [1,415 (23–25)]. For each species, we calculated the mean of each of our three codon usage statistics across all genomes corresponding to that species. Ribosomal protein annotations were taken directly from the annotations generated by NCBI’s default prokaryotic annotation pipeline, and these were the ribosomal proteins passed to both growthpred and gRodon. Importantly, growthpred can also search for ribosomal proteins using a provided database, although we did not use this feature so as to make sure the two prediction methods were compared on identical datasets. For initial model fitting, we excluded thermophiles and psychrophiles from the dataset (31) as these organisms systematically differ in their codon usage patterns (9). Similar to growthpred, we include a temperature option fit using these microbes in the final gRodon package that accounts for optimal growth temperature in the final model, although given the few extremophiles used to fit this model, we caution users against drawing strong conclusions when it is applied to extremophiles (by default, temperature is not used for prediction).
We then fit a linear model to Box–Cox-transformed doubling times [optimal chosen using the MASS package (83)] using our three codon usage measures as predictors. Similarly, we fit models for gRodon’s “partial” (excluding pair bias) and “metagenome” (excluding pair bias and consistency) modes.
For fitting on the Madin et al. (42) training set, we used the same model fitting procedure. We took the minimal recorded doubling time from each species in the “condensed_traits_NCBI.csv” supplementary file (https://doi.org/10.6084/m9.figshare.c.4843290.v1) and where possible, obtained all completely assembled genomes associated with that species from RefSeq. This yielded our training set with 464 species matched to 8,287 genomes. Notably, 130 of these species were either thermophiles or psychrophiles, perhaps making this training set preferable when dealing with extremophiles.
The Gaussian mixture model in Fig. 2 was fit using the Mclust() function in the mclust package with default settings (84). Mclust chooses the optimal mixture of Gaussians based on the Bayesian information criterion (BIC) and finds this optimum (for mean and variance) using an expectation maximization algorithm.
Metagenomic Data.
The raw sequencing data for the metagenomic water samples taken at the end of the Okie et al. (85) experiments were obtained from NCBI under BioProject PRJEB22811. Raw sequencing data for the time series samples taken by Coello-Camba et al. (86) were obtained from NCBI under BioProject PRJNA395437. Adapters and low-quality reads were trimmed using fastp v0.21.0 (87) with default parameters, and only reads longer than 30 base pairs (bp) were kept for further analysis. Okie et al. (85) samples were assembled individually using metaSPAdes v3.10.1 (88). Coello-Camba et al. (86) samples were assembled individually using megahit v1.2.9 (89) with default parameters. We called and annotated open reading frames (ORFs) from assemblies using prokka (90) (with options “–metagenome –compliant –fast”). Reads were mapped to ORFs using bwa 0.7.12 (91), and the number of reads aligned to each ORF was counted using bamcov v0.1.1 (available at https://github.com/fbreitwieser/bamcov). We ran gRodon in weighted and unweighted metagenome modes on each sample, with weights corresponding to mean coverage depth (corrected for gene length). In weighted metagenome mode, the median CUB of the highly expressed genes is taken as a weighed median (weightedMedian in matrixStats R package), with weights corresponding to mean depth of coverage for that gene. One sample from Coello-Camba et al. (86) had a very atypical estimated average minimal doubling time over twice as long as any other estimated doubling time from this dataset (MG078 at 3.1 h, as compared with the second longest doubling time in MG002 at 1.4 h) and strongly disagreeing with a replicate sample from the same experiment and time point (MG073 at 0.35 h). Upon closer inspection, this sample had far fewer bases than the rest (133 mega-bases vs. giga-bases), and only a little over 400 genes were detected in the assembly, far too few for accurate assessment of community-wide growth rate, leading us to omit this sample from further analyses.
EGGO Datasets.
We downloaded all prokaryotic assemblies from RefSeq (23, 24), as well as several collections of isolate genomes (45, 48, 49), MAGs (27, 46, 47), and SAGs (32). Where possible, we used per-existing gene annotations provided by NCBI. For the Pasolli et al. (46) and Nayfach et al. (47) MAGs, gene predictions were not available, and we used prokka to predict ORFs and annotate ribosomal proteins (90). Note that for both of these MAG datasets, we used a subset of all MAGs designated as being representatives of species clusters by the authors. We then ran gRodon on each genome, using partial mode for MAGs and SAGs (which vary in their completeness). Finally, we filtered results from genomes with few ribosomal proteins. Similar to Vieira-Silva and Rocha (9), we found that growth rates were biased when <10 highly expressed genes were used for prediction (SI Appendix, Fig. S26), and we used this cutoff for our MAGs and SAGs. For our isolate genomes, this generally was not an issue, with over 99% of genomes in RefSeq having between 50 and 70 annotated ribosomal proteins. We filtered all genomes outside this range to remove a very small set of obvious problem cases (e.g., one Bacillus genome that had over 1,000 annotated ribosomal proteins). The numbers in Table 1 correspond to postfiltering genome counts.
Measuring Bias.
We use the MILC measure of CUB (20) implemented in the coRdon R package (92). This bias measure behaves slightly better than the Effective Number of Codons Prime (ENC’) measure used by Vieira-Silva and Rocha (9) and automatically accounts for the CUB of genomic background in its calculation (93) [by taking the genome-wide distribution of codons as its expected distribution (20, 92)]. As recommended in the coRdon documentation, genes with fewer than 80 codons were omitted from our calculations. Importantly, we calculate the MILC statistic on a per-gene basis rather than concatenating all of our genes. The contribution () of each amino acid () to the MILC statistic for a gene is calculated as
| [1] |
where is the set of codons coding for , is the observed count of codon , and is the expected count of codon [the original paper has the full calculation of the MILC statistic (20)]. Typically, for a given gene is estimated using the genome-wide frequency of that codon . This is what we mean when we say that for our CUB measurement, the bias of highly expressed genes is calculated “relative to the genomic background.” For calculating the average CUB, we used the median value in order to reduce the influence of any outliers (i.e., misbehaving ribosomal proteins).
For our consistency calculation, MILC was also used but was calculated using the highly expressed proteins as the expected background (using the “subset” option in coRdon). In other words, we estimated the expected count of a codon, , using the frequency of that codon in highly expressed genes only, rather than the genome-wide frequency. For the consistency metric, we took the mean value across ribosomal proteins (rather than the median as with CUB) since we are interested in the distance of all ribosomal proteins from the expected codon usage patterns.
For codon pair bias, we implemented the calculation by Coleman et al. (22) that controls for background amino acid and codon usage when estimating the over-/underrepresentation of codon pairs (figure S1 in ref. 22 has a relevant equation).
Population Parameters.
We obtained estimates of from ref. 37, which are based on dN/dS ratios (the intuition being that selection acts more efficiently in large populations). Gene-specific recombination rates were obtained by applying the PhiPack (94) program for detecting recombination to the ATGC database of closely related genome clusters (60), as described in Weissman et al. (39).
Classification and Phylogeny.
For marine and gut organisms, we classified all genomes, MAGs, and SAGs using GTDB-Tk [v1.3.0 with database release 95 (95, 96)]. For the analyses in Fig. 3C and SI Appendix, Fig. S19C, we used the tree output by GTDB-Tk [built automatically using FastTree (97)]. To assess sensitivity to phylogeny, we built a maximum likelihood tree with 10 bootstrap replicates from the GTDB-Tk alignment using RAxML [v8.2.11, with -k -f a -m PROTGAMMAGTR options (98)]. We performed phylogenetic logistic regression using the R package phylolm (99). We then assessed sensitivity to individual species, entire phyla, sample size, and phylogenic uncertainty using the R package sensiPhy (100). Trees and aligned heatmaps were visualized using R package ggtree (101, 102).
Extrapolating between Closely Related Taxa.
For all genomes used to build EGGO, we extracted all annotated 16S rRNA genes; then, we aligned these sequences and removed poorly aligned columns using ssu-align and ssu-mask [default settings (103)]. We then filtered sequences for which less than 80% of positions were accounted for (i.e., were gaps). We ran fasttree on the resulting alignment [with -fastest, -nt, and -gtr options (97)] to obtain a phylogeny with 192,195 tips representing 60,421 organisms. For phylogenetic prediction of maximal growth rate, we then omitted any tips with EGGO entries where h (13 tips) to minimize the influence of outliers.
To predict growth rate, we first randomly sampled one tip per organism in our tree (to avoid predicting an organisms growth rate from itself). We then iteratively found the five closest tips to each tip in the tree and took the weighted geometric mean of the growth rates associated with these tips. This gave us our predicted maximal growth rate on the basis of 16S rRNA in SI Appendix, Fig. S15A. Weights were calculated as inverse patristic distance, with a small constant added for when organisms had identical 16S sequences (e.g., multiple genomes in EGGO for the same species):
| [2] |
For SI Appendix, Fig. S17, the predicted rate was simply taken as the rate associated with the closest tip on the tree. We identified the closest tips using the castor R package (104).
To produce SI Appendix, Fig. S15B, we sampled 10,000 tips from our tree and calculated all pairwise distances between tips using the cophenetic.phylo() function in the ape R package (105).
Signals of Selection.
We obtained all ATGC clusters of genomes and their core gene alignments (60). We calculated and values for each aligned gene in each cluster of organisms using PAML [v4.9j (106)]. We restricted our final analysis to pairwise comparisons where to ensure that organisms had sufficient substitutions to analyze and that the probability of multiple substitutions at the same site was low (i.e., no saturation). To calculate , we divided the mean of all ribosomal genes by the mean core genome-wide for each pairwise comparison. Previous work has noted that can be used as a proxy measure for since the two values should generally be correlated (37). Additionally, we assume that selection for translation optimization should be proportional to growth rate, such that , in SI Appendix, Fig. S23 C and D.
Functional Analysis.
For each genus in our RefSeq dataset, we randomly sampled a single genome for functional annotation. We assigned a label of “copiotroph” or “oligotroph” on the basis of whether each genome was predicted to have a minimal doubling time of greater or less than 5 h. We ran eggnogmapper [diamond setting, v2.0.0 (107, 108)] on each genome to get COG and archaeal COG (arCOG) annotations and a broad annotation of functional class for each ORF (70, 109). We used hmmer v3.3.1 to search the Carbohydrate-Active Enzyme (CAZy) database against each genome [e-value cutoff ; CAZy database V9 hidden Markov models (HMMs) packaged with the Automated Carbohydrate-Active Enzyme Annotation database (dbCAN2) (71, 110)]. To assess differences in the enrichment of certain functional classes across copiotrophs and oligotrophs, we performed Mann–Whitney tests with a Benjamini–Hochberg correction (). To assess differences in prevalence of individual gene families across copiotrophs and oligotrophs, we performed Fisher’s exact tests with a Benjamini–Hochberg correction ().
For the analysis of defense genes (considering gene families classified broadly under the “V” COG classification that were significantly differentially prevalent among copiotrophs/oligotrophs), we searched for antimicrobial/oxidant resistance among gene family annotations using the key words “resistance,” “transport,” “export,” “efflux,” “beta-lactamase,” “chloramphenicol,” and “hydroperoxide.” We searched for DNA-binding/degrading domains using the key words “HEPN,” “PIN,” and “HTH.” Finally, we searched for antiviral defense function using the key words “nuclease,” “CRISPR,” and “methyl.”
Supplementary Material
Acknowledgments
J.L.W. was supported by a postdoctoral fellowship in marine microbial ecology from Simons Foundation Award 653212. We also acknowledge support from Simons Foundation Collaboration on Computational Biogeochemical Modeling of Marine Ecosystems (CBIOMES) Grant 549943 (to J.A.F.) and US NSF Division of Ocean Sciences (OCE) Grant 1737409 (to J.A.F.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016810118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Eagon R. G., Pseudomonas natriegens, a marine bacterium with a generation time of less than 10 minutes. J. Bacteriol. 83, 736–737 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monod J., The growth of bacterial cultures. Annu. Rev. Microbiol. 3, 371–394 (1949). [Google Scholar]
- 3.Rappé M. S., Connon S. A., Vergin K. L., Giovannoni S. J., Cultivation of the ubiquitous sar11 marine bacterioplankton clade. Nature 418, 630–633 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Colwell F. S., D’Hondt S., Nature and extent of the deep biosphere. Rev. Mineral. Geochem. 75, 547–574 (2013). [Google Scholar]
- 5.Starnawski P., et al. , Microbial community assembly and evolution in subseafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 114, 2940–2945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trembath-Reichert E., et al. , Methyl-compound use and slow growth characterize microbial life in 2-km-deep subseafloor coal and shale beds. Proc. Natl. Acad. Sci. U.S.A. 114, E9206–E9215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauro F. M., et al. , The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 15527–15533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klappenbach J. A., Dunbar J. M., Schmidt T. M., rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66, 1328–1333 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vieira-Silva S., Rocha E. P., The systemic imprint of growth and its uses in ecological (meta) genomics. PLoS Genet. 6, e1000808 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roller B. R., Stoddard S. F., Schmidt T. M., Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat. Microbiol. 1, 16160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sunagawa S., et al. , Structure and function of the global ocean microbiome. Science 348, 1261359 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Rappé M. S., Giovannoni S. J., The uncultured microbial majority. Annu. Rev. Microbiol. 57, 369–394 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Hug L. A., et al. , A new view of the tree of life. Nat. Microbiol. 1, 16048 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Oberhardt M. A., et al. , Harnessing the landscape of microbial culture media to predict new organism–media pairings. Nat. Commun. 6, 8493 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long A. M., Hou S., Ignacio-Espinoza J. C., Fuhrman J. A., Benchmarking microbial growth rate predictions from metagenomes. ISME J. 15, 183–195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikemura T., Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: A proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151, 389–409 (1981). [DOI] [PubMed] [Google Scholar]
- 17.Gouy M., Gautier C., Codon usage in bacteria: Correlation with gene expressivity. Nucleic Acids Res. 10, 7055–7074 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong H., Nilsson L., Kurland C. G., Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260, 649–663 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Hooper S. D., Berg O. G., Gradients in nucleotide and codon usage along Escherichia coli genes. Nucleic Acids Res. 28, 3517–3523 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Supek F., Vlahoviček K., Comparison of codon usage measures and their applicability in prediction of microbial gene expressivity. BMC Bioinf. 6, 182 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frumkin I., et al. , Codon usage of highly expressed genes affects proteome-wide translation efficiency. Proc. Natl. Acad. Sci. U.S.A. 115, E4940–E4949 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coleman J. R., et al. , Virus attenuation by genome-scale changes in codon pair bias. Science 320, 1784–1787 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusova T., Ciufo S., Fedorov B., O’Neill K., Tolstoy I., Refseq microbial genomes database: New representation and annotation strategy. Nucleic Acids Res. 42, D553–D559 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tatusova T., et al. , NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haft D. H., et al. , Refseq: An update on prokaryotic genome annotation and curation. Nucleic Acids Res. 46, D851–D860 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parks D. H., et al. , Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Tully B. J., Graham E. D., Heidelberg J. F., The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci. Data 5, 170203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart R. D., et al. , Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat. Commun. 9, 870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almeida A., et al. , A new genomic blueprint of the human gut microbiota. Nature 568, 499–504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue Y., Jonassen I., Øvreås L., Taş N., Bacterial and archaeal metagenome-assembled genome sequences from Svalbard permafrost. Microbiol. Res. Ann. 8, e00516–e00519 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi J., et al. , Strategies to improve reference databases for soil microbiomes. ISME J. 11, 829–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pachiadaki M. G., et al. , Charting the complexity of the marine microbiome through single-cell genomics. Cell 179, 1623–1635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutman G. A., Hatfield G. W., Nonrandom utilization of codon pairs in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 86, 3699–3703 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchan J. R., Aucott L. S., Stansfield I., tRNA properties help shape codon pair preferences in open reading frames. Nucleic Acids Res. 34, 1015–1027 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts D. R., et al. , Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography 40, 913–929 (2017). [Google Scholar]
- 36.Crow J. F., Kimura M., An Introduction to Population Genetics Theory (Blackburn Press, 1970). [Google Scholar]
- 37.Bobay L. M., Ochman H., Factors driving effective population size and pan-genome evolution in bacteria. BMC Evol. Biol. 18, 153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bobay L. M., Ochman H., Impact of recombination on the base composition of bacteria and archaea. Mol. Biol. Evol. 34, 2627–2636 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weissman J. L., Fagan W. F., Johnson P. L., Linking high GC content to the repair of double strand breaks in prokaryotic genomes. PLoS Genet. 15, e1008493 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swan B. K., et al. , Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc. Natl. Acad. Sci. U.S.A. 110, 11463–11468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giovannoni S. J., Thrash J. C., Temperton B., Implications of streamlining theory for microbial ecology. ISME J. 8, 1553–1565 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madin J. S., et al. , A synthesis of bacterial and archaeal phenotypic trait data. Sci. Data. 7, 170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lynch M., Evolution of the mutation rate. Trends Genet. 26, 345–352 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delmont T. O., et al. , Nitrogen-fixing populations of planctomycetes and proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 3, 804–813 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klemetsen T., et al. , The Mar databases: Development and implementation of databases specific for marine metagenomics. Nucleic Acids Res. 46, D692–D699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pasolli E., et al. , Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nayfach S., Shi Z. J., Seshadri R., Pollard K. S., Kyrpides N. C., New insights from uncultivated genomes of the global human gut microbiome. Nature 568, 505–510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poyet M., et al. , A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat. Med. 25, 1442–1452 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Zou Y., et al. , 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat. Biotechnol. 37, 179–185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vartoukian S. R., Palmer R. M., Wade W. G., Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol. Lett. 309, 1–7 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Schaechter M., Maaløe O., Kjeldgaard N. O., Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. Microbiology 19, 592–606 (1958). [DOI] [PubMed] [Google Scholar]
- 52.Bulmer M., The selection-mutation-drift theory of synonymous codon usage. Genetics 129, 897–907 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akashi H., Synonymous codon usage in Drosophila melanogaster: Natural selection and translational accuracy. Genetics 136, 927–935 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hershberg R., Petrov D. A., Selection on codon bias. Annu. Rev. Genet. 42, 287–299 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Zhou T., Weems M., Wilke C. O., Translationally optimal codons associate with structurally sensitive sites in proteins. Mol. Biol. Evol. 26, 1571–1580 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharp P. M., Emery L. R., Zeng K., Forces that influence the evolution of codon bias. Phil. Trans. Biol. Sci. 365, 1203–1212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yannai A., Katz S., Hershberg R., The codon usage of lowly expressed genes is subject to natural selection. Genome Biol. Evol. 10, 1237–1246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharp P. M., Bailes E., Grocock R. J., Peden J. F., Sockett R. E., Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 33, 1141–1153 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hahn M. W., Molecular Population Genetics (Sinauer Associates, New York, NY, 2019). [Google Scholar]
- 60.Kristensen D. M., Wolf Y. I., Koonin E. V., ATGC database and ATGC-COGS: An updated resource for micro-and macro-evolutionary studies of prokaryotic genomes and protein family annotation. Nucleic Acids Res. 45, D210–D218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polak P., Arndt P. F., Transcription induces strand-specific mutations at the 5′ end of human genes. Genome Res. 18, 1216–1223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lind P. A., Andersson D. I., Whole-genome mutational biases in bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 17878–17883 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park C., Qian W., Zhang J., Genomic evidence for elevated mutation rates in highly expressed genes. EMBO Rep. 13, 1123–1129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen X., Zhang J., No gene-specific optimization of mutation rate in Escherichia coli. Mol. Biol. Evol. 30, 1559–1562 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei W., et al. , Smal: A resource of spontaneous mutation accumulation lines. Mol. Biol. Evol. 31, 1302–1308 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Chen X., Zhang J., Yeast mutation accumulation experiment supports elevated mutation rates at highly transcribed sites. Proc. Natl. Acad. Sci. U.S.A. 111, E4062 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kashtan N., et al. , Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344, 416–420 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Pianka E. R., On r-and k-selection. Am. Nat. 104, 592–597 (1970). [Google Scholar]
- 69.Pfeiffer T., Schuster S., Bonhoeffer S., Cooperation and competition in the evolution of ATP-producing pathways. Science 292, 504–507 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Galperin M. Y., Kristensen D. M., Makarova K. S., Wolf Y. I., Koonin E. V., Microbial genome analysis: The COG approach. Briefings Bioinf. 20, 1063–1070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lombard V., Ramulu H. G., Drula E., Coutinho P. M., Henrissat B., The carbohydrate-active enzymes database (CAZY) in 2013. Nucleic Acids Res. 42, D490–D495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matelska D., Steczkiewicz K., Ginalski K., Comprehensive classification of the pin domain-like superfamily. Nucleic Acids Res. 45, 6995–7020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Makarova K. S., Anantharaman V., Grishin N. V., Koonin E. V., Aravind L., Carf and wyl domains: Ligand-binding regulators of prokaryotic defense systems. Front. Genet. 5, 102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Makarova K. S., et al. , Evolutionary and functional classification of the carf domain superfamily, key sensors in prokaryotic antivirus defense. Nucleic Acids Res. 48, 8828–8847 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anantharaman V., Makarova K. S., Burroughs A. M., Koonin E. V., Aravind L., Comprehensive analysis of the hepn superfamily: Identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct 8, 1–28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bernheim A., Sorek R., The pan-immune system of bacteria: Antiviral defence as a community resource. Nat. Rev. Microbiol. 18, 113–119 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Korem T., et al. , Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349, 1101–1106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brown C. T., Olm M. R., Thomas B. C., Banfield J. F., Measurement of bacterial replication rates in microbial communities. Nat. Biotechnol. 34, 1256 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gao Y., Li H., Quantifying and comparing bacterial growth dynamics in multiple metagenomic samples. Nat. Methods 15, 1041–1044 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Emiola A., Oh J., High throughput in situ metagenomic measurement of bacterial replication at ultra-low sequencing coverage. Nat. Commun. 9, 4956 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wickham H., Ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, New York, NY, 2016). [Google Scholar]
- 82.Kassambara A., ggpubr: ‘ggplot2’ Based Publication Ready Plots (R package Version 0.4.0, 2020). https://rpkgs.datanovia.com/ggpubr/. Accessed 2 March 2021.
- 83.Venables W. N., Ripley B. D., Modern Applied Statistics with S (Springer, New York, NY, ed. 4, 2002). [Google Scholar]
- 84.Scrucca L., Fop M., Murphy T. B., Raftery A. E., Mclust 5: Clustering, classification and density estimation using Gaussian finite mixture models. R J 8, 289–317 (2016). [PMC free article] [PubMed] [Google Scholar]
- 85.Okie J. G., et al. , Genomic adaptations in information processing underpin trophic strategy in a whole-ecosystem nutrient enrichment experiment. Elife 9, e49816 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Coello-Camba A., et al. , Picocyanobacteria community and cyanophage infection responses to nutrient enrichment in a mesocosms experiment in oligotrophic waters. Front. Microbiol. 11, 1153 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen S., Zhou Y., Chen Y., Gu J., fastp: An ultra-fast all-in-one fastq preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nurk S., Meleshko D., Korobeynikov A., Pevzner P. A., Metaspades: A new versatile metagenomic assembler. Genome Res. 27, 824–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li D., Liu C. M., Luo R., Sadakane K., Lam T. W., Megahit: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015). [DOI] [PubMed] [Google Scholar]
- 90.Seemann T., Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 91.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] (2013) https://arxiv.org/abs/1303.3997 (Accessed 3 February 2021).
- 92.Elek A., Kuzman M., Vlahoviček K., Cordon: Codon usage analysis and prediction of gene expressivity. Bioconductor 3, 8 (2019). [Google Scholar]
- 93.Novembre J. A., Accounting for background nucleotide composition when measuring codon usage bias. Mol. Biol. Evol. 19, 1390–1394 (2002). [DOI] [PubMed] [Google Scholar]
- 94.Bruen T. C., Philippe H., Bryant D., A simple and robust statistical test for detecting the presence of recombination. Genetics 172, 2665–2681 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parks D. H., et al. , A complete domain-to-species taxonomy for bacteria and archaea. Nat. Biotechnol. 38, 1079–1086 (2020). [DOI] [PubMed] [Google Scholar]
- 96.Chaumeil P. A., Mussig A. J., Hugenholtz P., Parks D. H., GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Price M. N., Dehal P. S., Arkin A. P., Fasttree 2—approximately maximum-likelihood trees for large alignments. PloS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stamatakis A., Raxml version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ho L. S. T., Ane C., A linear-time algorithm for Gaussian and non-Gaussian trait evolution models. Syst. Biol. 63, 397–408 (2014). [DOI] [PubMed] [Google Scholar]
- 100.Paterno G. B., Penone C., Werner G. D. A., sensiphy: An r- package for sensitivity analysis in phylogenetic comparative methods. Method Ecol. Evol. 9, 1461–1467 (2018). [Google Scholar]
- 101.Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T. Y., ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Method Ecol. Evol. 8, 28–36 (2017). [Google Scholar]
- 102.Yu G., Using ggtree to visualize data on tree-like structures. Curr. Protocol. Bioinform. 69, e96 (2020). [DOI] [PubMed] [Google Scholar]
- 103.Nawrocki E. P., Kolbe D. L., Eddy S. R., Infernal 1.0: Inference of RNA alignments. Bioinformatics 25, 1335–1337 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Louca S., Doebeli M., Efficient comparative phylogenetics on large trees. Bioinformatics 34, 1053–1055 (2018). [DOI] [PubMed] [Google Scholar]
- 105.Paradis E., Schliep K., Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 106.Yang Z., et al. , Paml: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997). [DOI] [PubMed] [Google Scholar]
- 107.Huerta-Cepas J., et al. , Fast genome-wide functional annotation through orthology assignment by eggnog-mapper. Mol. Biol. Evol. 34, 2115–2122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huerta-Cepas J., et al. , Eggnog 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Makarova K. S., Wolf Y. I., Koonin E. V., Archaeal clusters of orthologous genes (arcogs): An update and application for analysis of shared features between thermococcales, methanococcales, and methanobacteriales. Life 5, 818–840 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang H., et al. , dbcan2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.