Significance
Genomic imprinting is essential for human development and occurs in germ cells before fertilization. The noncoding RNA, nc886, is the only known example of more than 100 such human genes which shows variable frequencies of maternal imprinting. Here, we show that the DNA methylation imprint is present in oocytes and that the probability of imprinting increases as a function of maternal age. Importantly, we demonstrate that alcohol consumption but not cigarette smoking is associated with a lower frequency of imprinting. While most studies focus on the postconceptional developmental time, our work indicates that maternal age and exposures the year prior to pregnancy may alter the epigenome and therefore the developing child.
Keywords: imprinting, nc886, oocyte
Abstract
Genomic imprinting occurs before fertilization, impacts every cell of the developing child, and may be sensitive to environmental perturbations. The noncoding RNA, nc886 (also called VTRNA2-1) is the only known example of the ∼100 human genes imprinted by DNA methylation, that shows polymorphic imprinting in the population. The nc886 gene is part of an ∼1.6-kb differentially methylated region (DMR) that is methylated in the oocyte and silenced on the maternal allele in about 75% of humans worldwide. Here, we show that the presence or absence of imprinting at the nc886 DMR in an individual is consistent across different tissues, confirming that the imprint is established before cellular differentiation and is maintained into adulthood. We investigated the relationships between the frequency of imprinting in newborns and maternal age, alcohol consumption and cigarette smoking before conception in more than 1,100 mother/child pairs from South Africa. The probability of imprinting in newborns was increased in older mothers and decreased in mothers who drank alcohol before conception. On the other hand, cigarette smoking had no apparent relationship with the frequency of imprinting. These data show an epigenetic change during oocyte maturation which is potentially subject to environmental influence. Much focus has been placed on avoiding alcohol consumption during pregnancy, but our data suggest that drinking before conception may affect the epigenome of the newborn.
Maternal age and environmental exposures during pregnancy are known to affect the developing child and have been extensively studied (1, 2). In utero exposures to teratogens, drugs, alcohol, cigarettes, and maternal diet, particularly folic acid, can severely affect early human embryonic development, showing that this phase is sensitive to perturbation (1, 3–6). However, the preconception time period, specifically the ∼6-mo interval of oocyte maturation, is also a unique window when maternal characteristics and behavior could impact oogenesis and therefore health outcomes in children. Since changes in the oocyte are inherited in every somatic cell of the conceptus, these can have widespread sequelae. For example, increasing maternal age is a well-known risk factor for miscarriage and adverse health outcomes in offspring, particularly due to chromosomal aneuploidies such as Down’s syndrome (7).
Maternal genomic imprinting is established during oocyte maturation, where de novo DNA methylation marks an extended region of parental-specific differentially methylated DNA (DMR) causing allele-specific gene expression in all somatic cells of offspring (8, 9). A marked exception to the rule that genomic imprinting is almost 100% penetrant in humans is the DMR containing the noncoding RNA, nc886, which shows DNA methylation of the maternal allele in about 75% of individuals in all populations studied (10–13). Nc886 is an RNA polymerase 3 transcribed noncoding RNA that is demonstrably silenced by DNA methylation and can be activated by treatment with DNA methylation inhibitors (13). Increased DNA methylation is associated with an unambiguous loss of chromatin accessibility showing that it has functional consequences in terms of transcriptional inhibition (14). A failure to imprint nc886 in the oocyte, is associated with a higher body mass index (BMI) in 5 y olds (15) and increased survival of patients with acute myelogenous leukemia (13) implying that gene dosage, regulated by imprinting, might be causative of important phenotypes. We show here that the probability of nc886 imprinting is significantly increased as a function of maternal age at conception. We also show that alcohol consumption before conception decreases the probability of nc886 imprinting, whereas it is unaffected by cigarette smoking.
Results
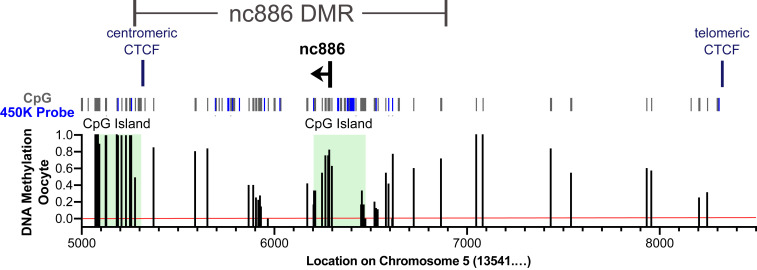
Nc886 is included in an ∼1,600-bp differentially methylated region (DMR) that was previously defined in Carpenter et al. (10), using whole genome bisulfite sequencing. The nc886 DMR includes a CpG island that overlaps a CTCF binding site on the centromeric side and a CpG island that overlaps the nc886 gene and associated promoter. Here, we examined DNA methylation of the nc886 DMR in 202 pooled human oocytes from the dataset from Okae et al. (16) and found roughly 75% methylation levels in the CpG island containing nc886 and its promoter (Fig. 1). Even though coverage of the DMR is low, CpG methylation is clearly present, suggesting that methylation begins in the oocyte (Fig. 1).
Fig. 1.
DNA methylation across the nc886 DMR in pooled human oocytes. Existing whole genome bisulfite sequencing data for pooled human oocytes from Okae et al. (16), was analyzed across the nc886 DMR and flanking regions (Chr5:135415000 to 135418500). Coverage of this region is somewhat low, with an average read depth of 8.16 reads. All CpGs are identified using gray or blue bars, where blue bars represent CpGs present on the 450K array and used for analysis in Fig. 2 and SI Appendix, Fig. S1. Red line at 0% methylation indicates where reads are present that equal 0% methylation vs. no coverage for a particular CpG. CpG islands are depicted by green highlight.
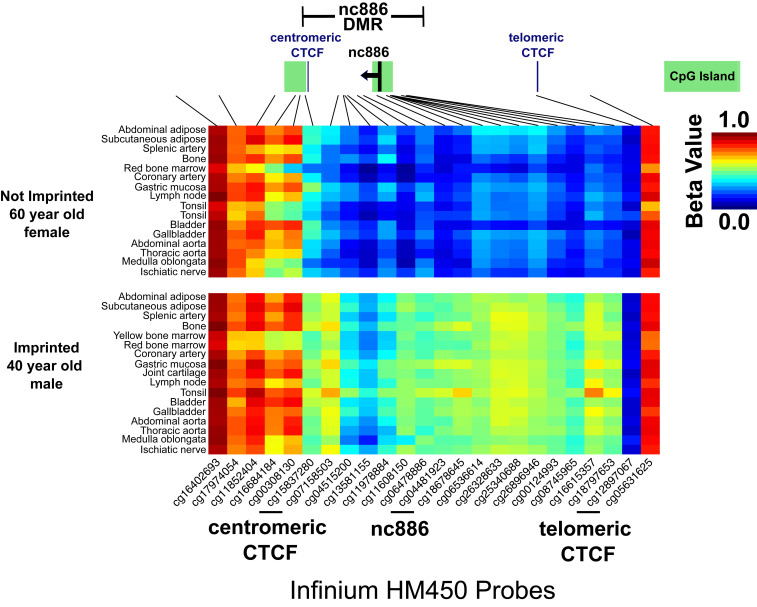
Given that nc886 is methylated in the oocyte, we expected the same pattern of DNA methylation in all somatic tissues in a given individual. Previously, Silver et al. (12) showed intertissue correlations for DNA methylation at three CpG sites in the nc886 DMR in brain, kidney, and liver in 17 cadavers. To confirm that the entire nc886 DMR maintains its imprint in adults, we analyzed genome-wide DNA methylation data from 17 somatic tissues, collected from four individuals at autopsy (17). Using these data (Fig. 2) and data from oocytes (Fig. 1), we redefined the nc886 DMR from our previous work (10) to ∼1,600 bp beginning with the CTCF binding motif, located centromeric to nc886. We found that imprinting status of the nc886 DMR was consistent across all tissue types of an individual whether that individual was imprinted or not imprinted (Fig. 2 and SI Appendix, Fig. S1). Additionally, all four individuals were more than 40 y of age, confirming that DNA methylation at the nc886 DMR, whether or not imprinted, is maintained throughout life in many tissues.
Fig. 2.
Imprinting status across the nc886 DMR is consistent across tissue types within single individuals. Beta values for DNA methylation were reanalyzed from Lokk et al. (17). Data are from Illumina Infinium 450K arrays across chr5:136075450 το 136084330 (hg38) and are plotted for two individuals from somatic tissues collected at autopsy. For the not imprinted 60-y-old, female, yellow bone marrow and joint cartilage tissues were not available. CpG islands are indicated by green bars. The nc886 DMR was defined in Carpenter et al. (10), as ∼1,900 bp using whole genome bisulfite sequencing data from blood samples but here, we redefined the DMR to about 1,600 bp beginning with the centromeric CTCF motif. Data were obtained from GSE50192 (31).
Previous work demonstrated that periconceptual maternal environment is linked to the likelihood that the nc886 DMR is imprinted in children at birth and children born to teenage mothers have a lower probability of imprinting (10). We used a South African cohort to confirm if maternal age is linked to imprinting of the nc886 DMR, and if preconceptual smoking and alcohol use might also have such a relationship. Our study utilized survey data from mothers enrolled in the Safe Passage Study (SPS), as part of the Prenatal Alcohol and Sudden Infant Death Syndrome (SIDS) and Stillbirth (PASS) Network (18) with associated DNA from newborns to avoid potential impacts of the postnatal environment on the epigenome. For the SPS, mothers were asked to recall drug, alcohol, and cigarette usage the year prior to pregnancy (19) and newborn samples were divided into groups based on maternal age or behaviors the year prior to pregnancy.
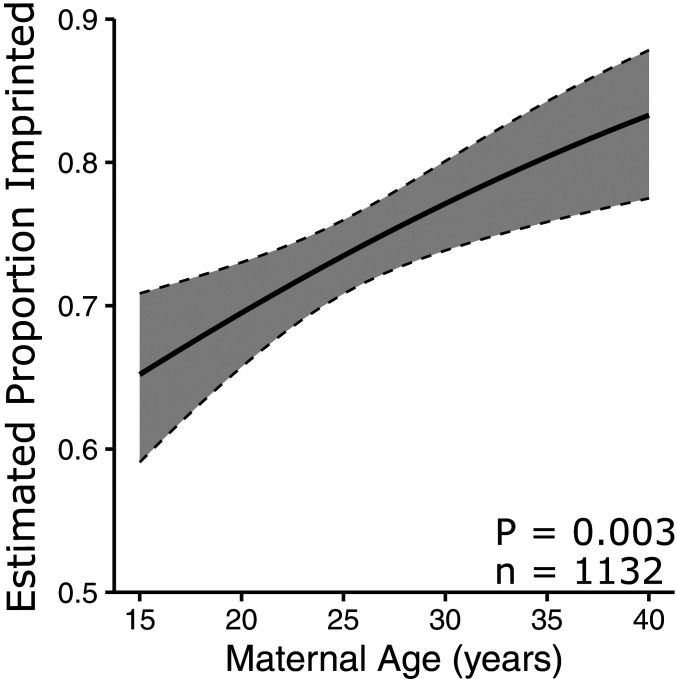
We used high resolution melt curve analysis (13) (SI Appendix, Figs. S2 and S3) on blood spots or cord blood samples from 1,132 South African mother/child pairs and found a positive correlation between increasing maternal age and the probability of nc886 imprinting in offspring (Fig. 3; P = 0.003). A 1-y increase in maternal age increases the odds of imprinting in the child by 3.9% (95% CI = 1.6 to 6.4%). Previously, we analyzed a Norwegian dataset (20) and similarly, found that children born to teenage mothers were less likely to be imprinted at nc886 (10). Upon reanalyzing this Norwegian dataset (20) as a continuous variable (SI Appendix, Fig. S4; P = 0.075; n = 827) we found that a 1-y increase in maternal age increases the odds of imprinting in a child by 2.96% (95% CI = −0.3 to 6.4%). The remarkably similar percent increase in odds between these two different populations indicates a strong positive correlation between increasing maternal age and imprinting of nc886.
Fig. 3.
Maternal age is positively correlated with imprinting of the nc886 DMR in children at birth in South Africa. Plot of proportion of individuals with imprinting at nc886 with increasing maternal age, estimated using standard logistic regression with maternal age as the only covariate. Data represented are from children born to South African mothers, collected in this study and pooled without regard to drinking and smoking status. Estimates were calculated. The shaded gray area is the 95% confidence interval ribbon.
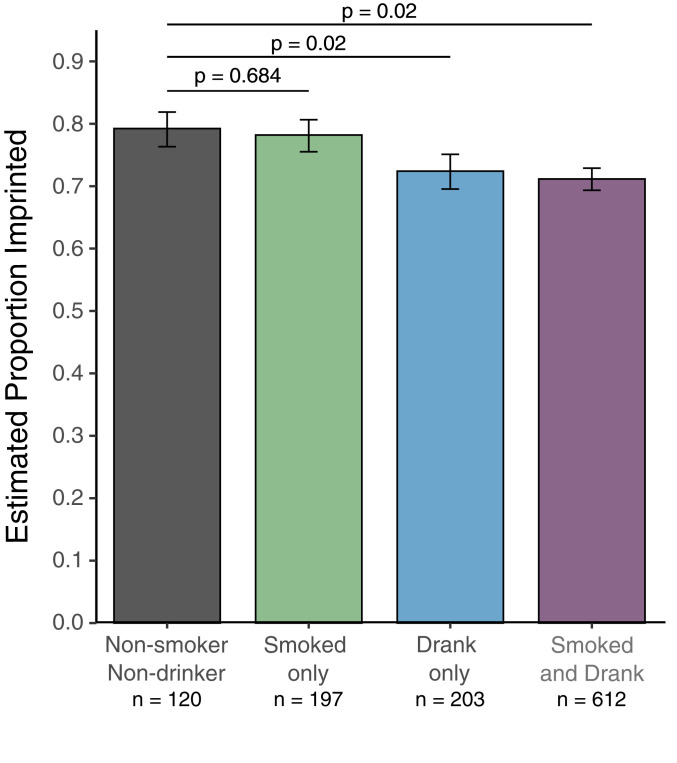
The landmark study by Silver et al. (12) showed that season of conception and by association maternal nutrition, is associated with altered methylation of individual CpG sites at nc886 in children, implying that diet might influence nc886 methylation. Folic acid supplementation, which is recommended for prenatal care, also shifts individual CpG methylation at nc886, as measured in adults (21). We hypothesized that alcohol and cigarette exposure may be associated with an altered likelihood that the nc886 DMR would be imprinted in children. We focused on exposures the year before the last menstrual period (LMP) because mouse studies show that maternally imprinted genes acquire DNA methylation during the nondividing oocyte growth phase, corresponding to up to 6 mo prior to ovulation in humans (22). We categorized mothers and associated newborn DNA into four groups based on maternal behavior the year prior to LMP: nonsmokers and nondrinkers, smoked only, drank only, and smoked and drank. We calculated the percentage of newborn samples in each group that were imprinted and used logistic regression to determine the expected proportion imprinted after correcting for effects of maternal age. When comparing “smoked only” to “nonsmokers and nondrinkers” there was no difference in the expected proportion of children that were imprinted (Fig. 4). However, mothers in the “drank only” group are less likely to have children who are imprinted than nonsmokers and nondrinkers (P = 0.02) independent of the effect of maternal age on imprinting (SI Appendix, Fig. S5A). There was not a further decrease in the expected proportion imprinted in the “smoked and drank” group over the drank only group, indicating that maternal smoking had no impact on the likelihood of imprinting in children.
Fig. 4.
Maternal drinking the year prior to pregnancy is associated with a decreased probability of imprinting at nc886. Plot of the estimated proportion of individuals with imprinting at the nc886 DMR separated by nonsmokers and nondrinkers (gray), maternal smoking (green), maternal drinking (blue), both behaviors (purple). Estimates were calculated using standard logistic regression with drinking status (yes/no in the year prior to pregnancy), smoking status (yes/no in the year prior to pregnancy), and maternal age as covariates. Error bars represent the SE of the estimated proportions.
Allele-specific DNA methylation is associated with genetic polymorphisms (23, 24). The nc886 DMR contains a CTCF binding site with a single nucleotide polymorphism (SNP) that disrupts the CTCF motif on the centromeric side of the DMR. We previously showed that an A nucleotide at SNP rs2346018 was associated with higher local DNA methylation density than a C nucleotide (10). In a random subset of 759 samples from the South African population, SNP rs2346018 had no impact on the probability of imprinting at the nc886 DMR (SI Appendix, Fig. S6). The discrepancy is likely because we previously analyzed local DNA methylation density at the CTCF site which may impact local CTCF binding but, in this study, found that the SNP does not directly alter imprinting status of the DMR. This gives us confidence that the variability in imprinting of the nc886 DMR is derived primarily from maternal age and preconceptual alcohol exposure.
Discussion
Our current data on DNA methylation of the nc886 DMR in oocytes and consistent methylation across tissue types, when combined with earlier studies (10, 11, 25), definitively establish nc886 as a bona fide maternally imprinted gene. This is strongly supported by the fact that monozygotic but not dizygotic twins are concordant for imprinting status at nc886, indicating that DNA methylation must be applied in the gametes or just prior to twinning (10, 26). In this sense, nc886 is not a metastable allele which refers to an epigenetic state established during embryogenesis as opposed to during oogenesis (27). The distinction is important because variations in levels of DNA methylation at individual CpGs within the DMR have often been referred to as metastable alleles in epidemiologic studies (12, 28). The fact that the imprint is applied during oogenesis, allowed us to track the relationships between maternal age, alcohol and cigarette consumption, and an epigenetic change in the oocyte. Therefore, even though most subjects continued their drinking and smoking behaviors after conception, we can be sure that it was the preconceptual behavior which was related to establishment of the imprint rather than a postfertilization condition in the fetus. This is important because the unique nature of variable imprinting at nc886 allows us to track epigenetic influences on the human germline which is distinct from changes induced postconceptually.
We find a strong positive correlation between increasing maternal age and the likelihood that an imprint will be established at nc886 in the ovary—as measured in children at birth. We confirmed this by reanalyzing the nc886 data included in the work of Markunas et al. (20) who first showed that maternal aging was associated with reduced DNA methylation at four contiguous CpGs in the gene KLHL35. However, the timing of this change in KLHL35 could not be determined, whereas the timing of imprinting is known to be during oogenesis. Our analyses do not establish causation. However, given the well-known effects of age on oocyte quality and increased incidence of aneuploidies (7), it is certainly possible that this shift in nc886 imprinting is a reflection of increased maternal age.
We also determined that maternal alcohol usage the year prior to the LMP was associated with a decreased probability that the child would be imprinted at nc886 at birth. Smoking the year prior to LMP had no impact on the probability that nc886 would be imprinted, indicating that a change in imprinting is specific for certain types of exposure. The negative effects of alcohol exposure in utero are well established and include increased risk of still birth, miscarriage, and delayed or impaired intellectual and developmental abilities (5, 19). However, our study clearly demonstrates an alcohol-associated epigenetic change that occurs in the oocyte growth phase, when imprints are established, that could potentially impact the long-term health of the child.
The biological functions of nc886 remain unclear but it does have the ability to bind to and modulate the activity of the double-stranded RNA-dependent protein kinase, PKR (29). PKR has been implicated in a host of functions including cancer, metabolism, the antiviral immune response, and response to alcohol exposure (30). Unfortunately, the lack of representation of nc886 in the mouse genome limits functional studies to discern phenotypic consequences of imprinting of the nc886 DMR. Nevertheless, the fact that nonimprinted newborns are twice as likely to be in the top quartile for BMI at age 5 (15) and nonimprinted adults with acute myelogenous leukemia have a better prognosis (13), strongly suggests that nc886 methylation has measurable phenotypic consequences.
Epidemiologic studies using DNA methylation often use surrogates such as blood for a target tissue and struggle to link a function to DNA methylation changes. The use of an imprinted gene overcomes many of these problems, given that the nc886 imprint is essentially the same in all somatic cells (Fig. 2 and SI Appendix, Fig. S1) and that methylation of the DMR functions to directly silence transcription of nc886 (13, 14). Unfortunately, nc886 is the only imprinted gene identified to date which can be used for these analyses, given that it is polymorphically imprinted. However, there may be more such genes which have not yet been identified and we provide a glimpse of germline-induced changes in the oocyte. Nevertheless, our data show a strong association between maternal age and maternal alcohol consumption before conception and imprinting of the nc886 DMR. The epigenome of the oocyte might therefore be subject to environmental influence with potential phenotypic outcomes.
Materials and Methods
Sample Collection and Ethical Approvals.
This study used newborn blood specimens and data collected from mothers in conjunction with the larger SPS (19), which is a part of the PASS Network. The SPS was conducted in accordance with the principles outlined in the Declaration of Helsinki (Declaration of Helsinki. Bull World Health Organ, 2001) (4). Voluntary written informed consent was obtained from the mother or caregiver of the child. Participants also gave informed consent for specific study components, which included collection of newborn Guthrie spots and cord blood samples. Options were provided so that participants could choose to decline aspects of the study or withdraw from the study at any time.
For this study, we received 1,148 SPS specimens (893 Guthrie blood spot and 255 cord blood) from Fisher Biorepository after obtaining approval from the PASS Network, Stellenbosch University Health Research Ethics Committee 2 (HREC2), and the Van Andel Institute Institutional Review Board (IRB). Specimens were processed and analyzed using HREC2 and IRB approved protocols. Group information was removed, and samples were randomized before further processing.
The modified alcohol timeline follow-back survey was conducted up to four times during pregnancy and once after delivery: 1) gestational age (GA) −15 to 15 d based on LMP; 2) GA 0 to 97 d, trimester 1; 3) GA 98 to 195 d, trimester 2; 4) GA 196 d to delivery, trimester 3 (19). Mothers were also asked to recall drinking and smoking behaviors up to 1 y prior to the LMP. Mothers ranked cigarette and alcohol usage the year prior to LMP by indicating a value of 0 to 5 where 0 = “none,” 1 = “monthly or less,” 2 = “2 to 4 d a month,” 3 = “2 to 3 d a week,” 4 = “4 to 6 d a week,” or 5 = “7 d a week.” When it became clear that no dosing effect could be identified and complications due to an inability to determine the exact number or nature of cigarettes/drinks consumed (e.g., binges vs. occasional drinks), the groups were combined into binary yes/no to limit biases. We classified nonsmokers and nondrinkers as mothers who indicated 0 for both behaviors. Smoked only had a value of 1 or greater for smoking and 0 for drinking. Drank only had a value of 1 or greater for drinking and 0 for smoking. Smoked and drank had a value of 1 or greater for both behaviors. Based on the obtained information, samples were separated into nonsmokers and nondrinkers, smoking only, drinking only, and both smoking and drinking. Any amount of smoking or drinking behavior in the year prior to LMP classified a sample into the smoking or drinking group.
Sodium Bisulfite Sequencing for Control DNA.
Genomic DNA was isolated from cancer cell lines using the DNeasy Blood and Tissue Kit (69504, Qiagen). Bisulfite conversion and clean-up of 200 ng of genomic DNA was performed by using the EZ DNA Methylation Kit (D5002, Zymo Research) according to the manufacturer’s protocol. Locus-specific PCR was performed by using primers specific for bisulfite-converted DNA and PCR products cloned using the pGEM-T Vector System (A3600, Promega) and NEB 5-alpha–competent Escherichia coli (C2987I, New England Biolabs). Colonies were screened for positive inserts by PCR and sequencing performed using the M13 promoter. Primers for nc886: F, tagtaaagttaaaagggataaaaaataata; R, acctcccctccaataaataaatttt.
DNA Extraction from Guthrie Blood Spot and Cord Blood Specimens.
Cord blood specimens.
DNA was extracted from 100 mL of thawed cord blood using the QiaAmp DNA Micro Kit (56304, Qiagen) according to the manufacturer’s protocol for “Isolation of Genomic DNA from Small Volumes of Blood” with minor modifications. Briefly, samples were lysed in Buffer AL with proteinase K while incubating at 56 °C for 15 min at 500 rpm on a Thermomixer (Thermo Fisher). DNA was extracted and purified using QiaAmp spin columns according to the manufacturer’s protocol. Samples were eluted in 60 μL of 5 mM Tris/HCl, pH 8.5 (buffer BE, 740306.100, Macherey-Nagel) using two serial elutions of 30 μL each. The DNA was quantitated using a Nanodrop (Thermo Fisher) and stored at 4 °C.
Blood spot specimens.
Sterile metal ticket punchers were used to collect one dried blood spot (DBS) from the Guthrie card of each specimen. DNA was extracted using the GenSolve-Qiagen protocol described by Ghantous et al. with minor modifications (31). Lysis solution and proteinase K from the GenSolve DNA Recovery Kit (NBGVR1027, Nova Biostorage) were used to lyse and remove blood cells while incubating at 56 °C for 1.5 to 2.0 h at 1,000 rpm on a Thermomixer, followed by extraction and purification of the DNA using the QiaAmp DNA Micro Kit (56304, Qiagen). Lysis solution and DBS punches were transferred to a spin basket (Investigator Lyse & Spin Basket Kit, 19597, Qiagen) and centrifuged. Recovery solution B and 100% ethanol were added to the filtrate and the sample was loaded onto a QiaAmp spin column and processed according to the QiaAmp DNA Micro Kit protocol. Samples were eluted in 80 μL of 5 mM Tris/HCl, pH 8.5 (buffer BE, 740306.100, Macherey-Nagel) using two serial elutions of 40 μL each. The DNA was quantitated using a Nanodrop (Thermo Fisher) and stored at 4 °C.
High-Resolution Melt Curve Analysis.
The EZ DNA Methylation kit (D5002, Zymo Research) was used to convert 200 ng of control and sample DNA according to manufacturer’s protocol and eluted in 40 μL DNase-free water. A total of 10 μL of Precision Melt Supermix for High-Resolution Melt Analysis (1725112, Bio-Rad) was combined with 7 μL of converted DNA, 2 μL of primers specific for bisulfite converted DNA (2 mM for nc886; 5 mM for RPL30), and 1 μL water. DNA was amplified with a melt temp of 60 °C and 39 cycles followed by melting analysis from 65 °C to 95 °C with 0.1 °C increments (SI Appendix, Figs. S2 and S3). To control for bisulfite conversion, we used a region of DNA that is always unmethylated, in the gene RPL30. Melt curves for this region should remain the same across all samples and if complete conversion is achieved, there should be normal PCR amplification of this region as well as nc886 (SI Appendix, Fig. S3). Primers for nc886 were as follows: F, gtcgtttatttaggtttagtagttaggttgttgagg; R, cacaaaacccctacctttaacaaattcataaactac. Primers for RPL30 were as follows: F, cacaaaacccctacctttaacaaattcataaactac; R, accatcttaacgactactattaataaataaactcctac.
Raw relative fluorescence units (RFUs) were plotted for melting temperature. The temperature range to analyze was selected by choosing the minimum temperature where all samples have similar, or ideally the same, RFU values. The maximum temperature is chosen where all samples have completely melted and are at their lowest RFU value. For nc886, samples were analyzed at 69 to 77 °C. For RPL30, samples were analyzed at 69 to 75 °C. The following Excel equation was used to normalize melt curves: (X1 – X$81)/(X$1-X$81), where X is the location of the sample, 1 is the lowest melt temp chosen, and 81 is the highest melt temp chosen, with the number (e.g., 81) of high values changing depending on the temperature range. Samples were normalized to a chosen sample, here RKO, using the following equation: (X85 - $B85), where X is the location of the sample, 85 is the normalized RFU value for corresponding temperature, B is the chosen reference sample.
Normalized RFU values relative to RKO were plotted. For every experiment, known controls were used as reference curves for unknown samples. Based on the normalized melt curves, unknowns were defined as “imprinted” or “not imprinted.” For each smoking and drinking group statistical analysis was performed based on percentage of each group that was not imprinted.
Polymorphism Analysis for rs2346018.
For cord blood DNA, 150 ng of genomic DNA was analyzed by high-resolution melt analysis using 1x Precision Melt Supermix (1725112, Bio-Rad) combined with 250 nM forward and reverse primers. DNA was amplified with a melt temp of 60 °C and 40 cycles followed by melting analysis from 65 °C to 95 °C with 0.1 °C increments. Primers flanking rs2346018 were as follows: F, GGTCAAGGGACAAGAAGACGAT; R, GAGAATGCAAGCCCAGAGGC.
For blood spot DNA, 150 ng of genomic DNA was amplified by standard PCR using 1X MyTaq Red Mix (BIO-25044, Meridian Bioscience) combined with 250 nM forward and reverse primers. DNA was PCR amplified for 35 cycles with a melt temp of 60 °C. 1X NEB buffer, 10 units of BSOB1 restriction enzyme (R0586S, New England Biolabs), and water was added to each sample and incubated at 37 °C for 1 h. Restriction digests were run on a 1% agarose gel and genotype was determined by fragment length. Primers flanking rs2346018 were as follows: F, GGTCAAGGGACAAGAAGACGAT; R, GAGAATGCAAGCCCAGAGGC.
Statistical Analysis.
To determine if the proportion of individuals imprinted differed based on smoking or drinking behavior in the year prior to pregnancy, a logistic regression with a logit link and false discovery rate adjusted linear contrasts was fit via R v 3.6.0 (https://cran.r-project.org/). Logistic regressions with log(maternal age) and sqrt(maternal age) were also compared to the untransformed fit. The Akaike information criterion (AIC) and Bayesian information criterion (BIC) between these transformed models were improved by a negligible amount (e.g., AIC: 1,299 original vs. 1,298 log vs. 1,298 sqrt). This improvement in accuracy was not sufficient to justify the added complexity these transformations would impart on model interpretations.
This model was also adjusted for maternal age. A two-way interaction between smoking and drinking was originally included to determine if individuals who both smoked and drank had odds that differed from an additive effect of the two. No evidence was found of an interaction effect (P = 0.86), thus the final model included only main effects of drinking status (yes drank/did not drink in the year prior to pregnancy), smoking status (yes smoked/did not smoke in the year prior to pregnancy), and maternal age. An analysis adjusting the model for education (years), drugs used (yes/no), and body weight Z-score was run to determine if the “maternal age” and “yes drank” findings were sensitive to possible confounders; this model’s results were not meaningfully different from the reduced model (P values changed by ∼0.002, likelihood ratio test P = 0.74).
Supplementary Material
Acknowledgments
We thank the Prenatal Alcohol and SIDS and Stillbirth Network for providing the blood spot and cord blood samples, as well as Dan Rogers and Ryan Burgos for their assistance in procuring the samples. The Van Andel Institute Bioinformatics and Biostatistics Core was instrumental in providing statistical support. Dr. Wanding Zhou provided and analyzed whole genome bisulfite sequencing data. Funding for this study was provided by the Van Andel Institute to P.A.J. and grant 1F32GM129987 from the National Institute of General Medical Sciences to B.L.C. The Safe Passage Study was funded by grants from the National Institute on Alcohol Abuse and Alcoholism, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and the National Institute on Deafness and Other Communication Disorders (U01 HD055154, U01 HD045935, U01 HD055155, U01 HD045991, and U01 AA016501 to H.J.O.).
Footnotes
Competing interest statement: P.A.J. is a paid consultant for Zymo Research, whose kits were used in this study.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026580118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Frick A. P., Advanced maternal age and adverse pregnancy outcomes. Best Pract. Res. Clin. Obstet. Gynaecol. 70, 92–100 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Tarín J. J., Brines J., Cano A., Long-term effects of delayed parenthood. Hum. Reprod. 13, 2371–2376 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Borge T. C., Aase H., Brantsæter A. L., Biele G., The importance of maternal diet quality during pregnancy on cognitive and behavioural outcomes in children: A systematic review and meta-analysis. BMJ Open 7, e016777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Smidt J. J. A., et al., In utero teratogen exposure and cardiometabolic risk in 5-year-old children: A prospective pediatric study. J. Matern. Fetal Neonatal Med., 10.1080/14767058.2019.1692337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramoney S., Eastman E., Adnams C., Stein D. J., Donald K. A., The early developmental outcomes of prenatal alcohol exposure: A review. Front. Neurol. 9, 1108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osterhues A., Ali N. S., Michels K. B., The role of folic acid fortification in neural tube defects: A review. Crit. Rev. Food Sci. Nutr. 53, 1180–1190 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Hassold T., Hunt P., Maternal age and chromosomally abnormal pregnancies: What we know and what we wish we knew. Curr. Opin. Pediatr. 21, 703–708 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartolomei M. S., Ferguson-Smith A. C., Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 3, a002592 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaneda M., et al., Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Carpenter B. L., et al., Mother-child transmission of epigenetic information by tunable polymorphic imprinting. Proc. Natl. Acad. Sci. U.S.A. 115, E11970–E11977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paliwal A., et al., Comparative anatomy of chromosomal domains with imprinted and non-imprinted allele-specific DNA methylation. PLoS Genet. 9, e1003622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silver M. J., et al., Independent genomewide screens identify the tumor suppressor VTRNA2-1 as a human epiallele responsive to periconceptional environment. Genome Biol. 16, 118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Treppendahl M. B., et al., Allelic methylation levels of the noncoding VTRNA2-1 located on chromosome 5q31.1 predict outcome in AML. Blood 119, 206–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helbo A. S., Lay F. D., Jones P. A., Liang G., Grønbæk K., Nucleosome positioning and NDR structure at RNA polymerase III promoters. Sci. Rep. 7, 41947 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Dijk S. J., et al., DNA methylation in blood from neonatal screening cards and the association with BMI and insulin sensitivity in early childhood. Int. J. Obes. 42, 28–35 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Okae H., et al., Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet. 10, e1004868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lokk K., et al., DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 15, r54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dukes K. A.et al.; PASS Research Network , The safe passage study: Design, methods, recruitment, and follow-up approach. Paediatr. Perinat. Epidemiol. 28, 455–465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dukes K.et al.; PASS Network , A modified timeline followback assessment to capture alcohol exposure in pregnant women: Application in the safe passage study. Alcohol 62, 17–27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markunas C. A., et al., Maternal age at delivery is associated with an epigenetic signature in both newborns and adults. PLoS One 11, e0156361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richmond R. C., et al., The long-term impact of folic acid in pregnancy on offspring DNA methylation: Follow-up of the aberdeen folic acid supplementation trial (AFAST). Int. J. Epidemiol. 47, 928–937 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obata Y., Kono T., Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J. Biol. Chem. 277, 5285–5289 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Chandler L. A., Ghazi H., Jones P. A., Boukamp P., Fusenig N. E., Allele-specific methylation of the human c-Ha-ras-1 gene. Cell 50, 711–717 (1987). [DOI] [PubMed] [Google Scholar]
- 24.Tycko B., Allele-specific DNA methylation: Beyond imprinting. Hum. Mol. Genet. 19, R210–R220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romanelli V., et al., Variable maternal methylation overlapping the nc886/vtRNA2-1 locus is locked between hypermethylated repeats and is frequently altered in cancer. Epigenetics 9, 783–790 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Baak T. E., et al., Epigenetic supersimilarity of monozygotic twin pairs. Genome Biol. 19, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rakyan V. K., Blewitt M. E., Druker R., Preis J. I., Whitelaw E., Metastable epialleles in mammals. Trends Genet. 18, 348–351 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Gonseth S., et al., Epigenomic profiling of newborns with isolated orofacial clefts reveals widespread DNA methylation changes and implicates metastable epiallele regions in disease risk. Epigenetics 14, 198–213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calderon B. M., Conn G. L., Human noncoding RNA 886 (nc886) adopts two structurally distinct conformers that are functionally opposing regulators of PKR. RNA 23, 557–566 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe T., Imamura T., Hiasa Y., Roles of protein kinase R in cancer: Potential as a therapeutic target. Cancer Sci. 109, 919–925 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghantous A., Hernandez-Vargas H., Herceg Z., “DNA methylation analysis from blood spots: Increasing yield and quality for genome-wide and locus-specific methylation analysis” in DNA Methylation Protocols, Tost J., Ed. (Methods in Molecular Biology, Springer, 2018), pp. 605–619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.