Significance
Cell migration is a central process in development, homeostasis, and disease. Among the extracellular signaling molecules that control this process are members of the Wnt family of secreted glycoproteins, which trigger distinct canonical and noncanonical pathways in responding cells. Studies in vertebrates have shown that these pathways can antagonistically regulate migration, but how they functionally interact is not known. Using the migration of the C. elegans QR neuroblast descendants as a tractable model system, we found that the two pathways converge on activators and inhibitors of the Rho family of small GTPases that mediate cell migration. Our results show that through this cross-talk mechanism, Wnt pathways can interact to tightly regulate the migration of cells to their final destinations.
Keywords: C. elegans, Wnt, cell migration, Slt-Robo, pathway interactions
Abstract
Members of the Wnt family of secreted glycoproteins regulate cell migration through distinct canonical and noncanonical signaling pathways. Studies of vertebrate development and disease have shown that these pathways can have opposing effects on cell migration, but the mechanism of this functional interplay is not known. In the nematode Caenorhabditis elegans, a switch from noncanonical to canonical Wnt signaling terminates the long-range migration of the QR neuroblast descendants, providing a tractable system to study this mechanism in vivo. Here, we show that noncanonical Wnt signaling acts through PIX-1/RhoGEF, while canonical signaling directly activates the Slt–Robo pathway component EVA-1/EVA1C and the Rho GTPase–activating protein RGA-9b/ARHGAP, which are required for migration inhibition. Our results support a model in which cross-talk between noncanonical and canonical Wnt signaling occurs through antagonistic regulation of the Rho GTPases that drive cell migration.
Cell migration is a fundamental process in development and adult tissue homeostasis. During embryogenesis, for example, neural crest cells migrate throughout the embryo to form cell types like neurons, bone cells, and the melanocytes of the skin (1). Once development is completed, cell migration remains important, especially in tissue repair and immune functions, while defects in cell migration are closely linked to disease.
Migration is regulated by specific membrane-bound or secreted guidance cues, most of which have been highly conserved during evolution. Among these is the Wnt family of secreted glycoproteins that can trigger different signaling pathways in responding cells. In canonical Wnt signaling, binding of Wnt to the receptors Frizzled and LRP6 leads to stabilization of β-catenin, which in turn interacts with members of the TCF family of transcription factors to coactivate the expression of specific target genes (2). Wnt can also signal independently of β-catenin through different noncanonical pathways. These are activated by binding of Wnt to specific Frizzled members or alternative receptors such as the receptor tyrosine kinases Ror and Ryk (3).
In neural crest cells, Wnt controls migration through a noncanonical pathway that regulates the activity of Rho family GTPases. This polarizes the cell and ensures that protrusive activity is restricted to the lamellipodium at the leading edge of the cell (4). Interestingly, activation of canonical Wnt signaling has been shown to inhibit neural crest cell migration (5), indicating that canonical and noncanonical Wnt signaling have opposing effects on migration. A similar antagonistic relationship has been observed in melanoma progression, where canonical Wnt signaling is associated with a proliferative but noninvasive phenotype, while noncanonical Wnt signaling is correlated with metastasis (6, 7). The mechanism of this functional interplay between canonical and noncanonical Wnt signaling in migration is, however, still poorly understood.
The QR neuroblast lineage of the nematode Caenorhabditis elegans provides a tractable model system to study this cross-talk mechanism at the single-cell level in vivo. During the first stage of larval development, the QR neuroblast divides into an anterior (QR.a) and a posterior (QR.p) daughter cell. Both cells migrate toward the anterior and divide at stereotypic positions to generate three functionally distinct neurons (SI Appendix, Fig. S1A) (8). The migration of the QR descendants depends on Wnt signaling (9–11), and detailed studies of the long-range migration of the QR descendant QR.p revealed that this process is mediated through two parallel acting noncanonical Wnt pathways: an EGL-20/Wnt and CAM-1/Ror–dependent pathway that is required for persistent anterior polarization, and a CWN-1/Wnt and MOM-5/Frizzled–dependent pathway that regulates migration independent of polarity (12). Importantly, once the QR.p daughter cell QR.pa reaches its final position, migration is stopped through a cell intrinsically regulated switch to BAR-1/β-catenin–dependent canonical Wnt signaling.
Here, we have investigated how activation of canonical Wnt signaling stops QR descendant migration. We found that activation of canonical Wnt signaling inhibits migration without affecting the ability of the cell to polarize toward the anterior. Consistently, genetic analysis demonstrated that it counteracts the CWN-1/Wnt–MOM-5/Frizzled–dependent migration pathway and its downstream effector, the Rho activating protein PIX-1. Messenger RNA (mRNA) sequencing of isolated QR descendants showed that canonical Wnt signaling induces a specific transcriptional program. We found that two direct target genes—eva-1, which encodes an essential component of the Slt–Robo pathway, and rga-9 (isoform b), a conserved Rho GTPase activating domain containing protein—are required for migration inhibition. These results show that termination of QR descendant migration is mediated through activation of Slt–Robo signaling and a shift in the balance between the migration-promoting activity of PIX-1/RhoGEF and the migration-inhibiting activity of EVA-1/EVA1C and RGA-9b/ARHGAP. Given the evolutionary conservation of these different signaling components, we speculate that cross-talk at the level of Rho GTPases may also be important in the interplay between canonical and noncanonical Wnt signaling in vertebrate cell migration.
Results
Canonical Wnt/β-Catenin Signaling Inhibits QR.p Migration Cell Autonomously.
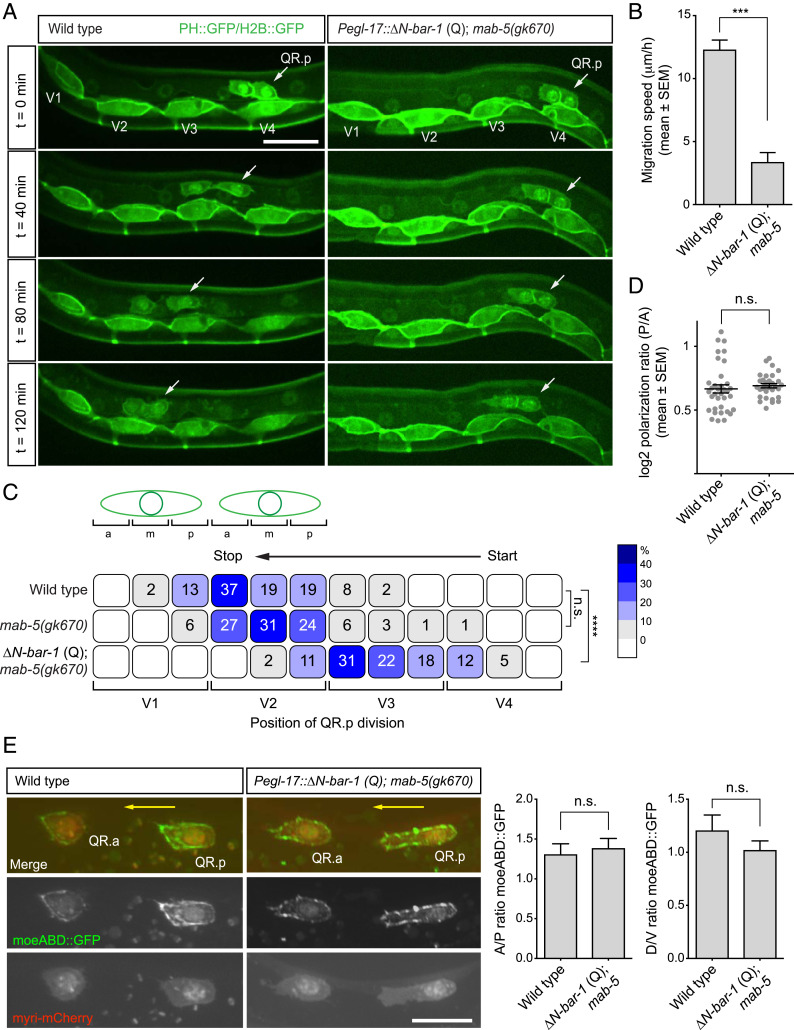
We have previously shown that a switch from noncanonical to canonical Wnt signaling is necessary for terminating the anterior migration of QR.pa (12). QR.pa migrates only a short distance and therefore offers limited possibilities for studying migration dynamics and the mechanism of migration inhibition. We therefore focused on the long-range anterior migration of its precursor, QR.p, which was found to be similarly inhibited by constitutive activation of canonical Wnt signaling (12). In these experiments, canonical Wnt signaling was activated using a mutation in pry-1, an ortholog of the β-catenin destruction complex component Axin (13). During early larval development, pry-1 is expressed in the QR lineage but also in other tissues, including the hypodermal and dorsal body wall muscle cells along which QR.p migrates. We therefore investigated whether the effect of canonical Wnt signaling on QR.p migration is fully cell autonomous. To specifically activate canonical Wnt signaling in the Q cell lineage, we used the egl-17 promoter (14) to express a constitutively active, N terminally truncated form of BAR-1/β-catenin (ΔN-BAR-1 Q) (12, 15). Moreover, a loss-of-function mutation in the Hox gene mab-5 was used to prevent activation in the QR lineage of the posterior migration pathway that is normally induced by canonical Wnt signaling in the related QL lineage (16, 17). As previously observed (12, 16, 18), this did not significantly influence QR descendant migration (Fig. 1C). We found that Q lineage specific expression of ΔN-BAR-1 strongly reduced the speed and distance of QR.p migration (Fig. 1 A–C). Furthermore, the decrease in QR.p migration distance was similar to what has been reported in pry-1/Axin mutants (12). We conclude that canonical Wnt signaling acts cell autonomously in migration termination and that Q lineage specific activation of this pathway inhibits QR.p migration.
Fig. 1.
Activation of canonical Wnt signaling inhibits QR.p migration. (A) Time-lapse imaging of QR.p migration in wild type and animals specifically expressing ΔN-BAR-1 in the Q lineage (ΔN-BAR-1 Q). The seam (V) cells and QR.a and QR.p are marked with nuclear (H2B) and membrane-localized (PH) GFP (huIs63) (38). Anterior is left and dorsal is up (scale bar, 15 μm). (B) The average speed of QR.p during the first hour of migration. The bars represent mean ± SEM (n > 50 for all genotypes). The statistical significance was calculated using an unpaired t test (***P < 0.001). (C) The position of QR.p division with respect to the seam cells. Position at the anterior (a), middle (m), or posterior (p) side of the seam cell are indicated as percentiles of the total number of cells analyzed (n > 50 for all genotypes). The statistical significance was calculated using Fisher’s exact test (n.s., P ≥ 0.05, ****P < 0.0001). (D) Quantification of QR.p polarity as calculated by the ratio of the distance from the nucleus to the posterior and the anterior side of the cell. The black lines indicate mean ± SEM (n > 30 for all genotypes). The statistical significance was calculated using an unpaired t test (n.s., P ≥ 0.05). (E) Representative images of filamentous actin (F-actin) localization in QR.p in wild type and ΔN-BAR-1 (Q)–expressing animals. A moesin actin binding domain fused to GFP (moeABD::GFP) was used to visualize F-actin (44) and myristoylated mCherry as a marker for the Q cells. The ratio of moeABD::GFP along the anteroposterior and dorsoventral axes was quantified. The bars represent mean ± SEM (n > 20 for all genotypes) (scale bar, 10 μm). The statistical significance was calculated using an unpaired t test (n.s., P ≥ 0.05). The ΔN-BAR-1 (Q)–expressing strains contain mab-5(gk670).
Canonical Wnt/β-Catenin Signaling Inhibits the Dynamic Formation and Collapse of Filopodia at the QR.p Membrane.
At the onset of migration, QR.p and its sister cell QR.a polarize and form a lamellipodium-like protrusive front at the anterior side that is maintained until migration stops and the cell enters mitosis (Fig. 1A). Time-lapse and static imaging showed that constitutive activation of canonical Wnt signaling does not influence the polarization of QR.p. Thus, we found that in ΔN-BAR-1–expressing animals, QR.p still forms a major lamellipodium-like protrusion at the anterior side of the cell (Fig. 1 A and E). Moreover, the overall polarity—as measured by the ratio between the posterior and anterior sides of the cell—was similar in wild type and ΔN-BAR-1–expressing animals (Fig. 1D). Consistently, we found that the distribution of filamentous actin and its enrichment at the protrusive front of QR.p was not affected by activation of canonical Wnt signaling (Fig. 1E). Taken together, we conclude that the inhibitory effect of canonical Wnt signaling on QR.p migration is independent of polarization and protrusive front formation.
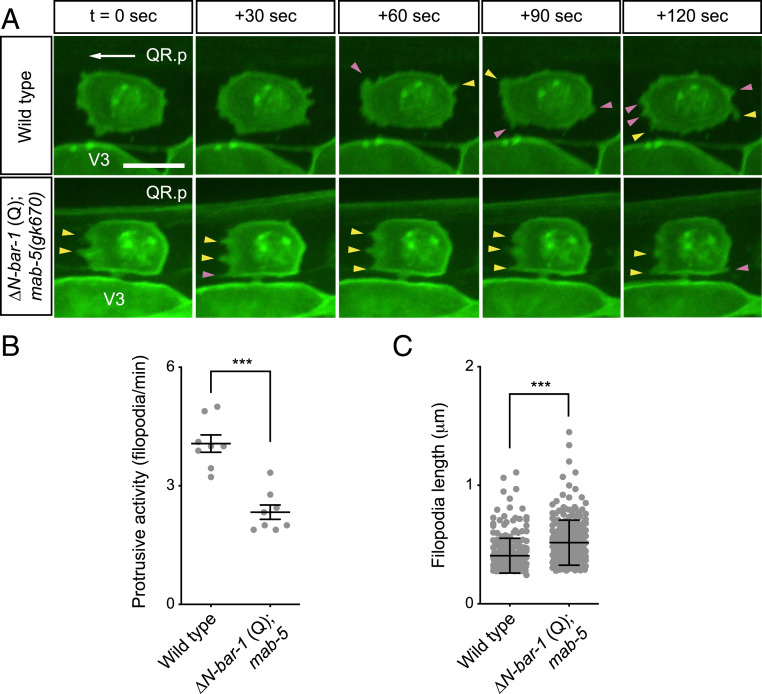
Time-lapse imaging of protrusive activity at the QR.p membrane showed that fewer filopodia-like protrusions were formed when canonical Wnt signaling was activated (Fig. 2 A and B). Furthermore, we found that these filopodia were more stable and significantly longer than in wild-type animals (Fig. 2 A and C). We hypothesize that this reduction in filopodial dynamics contributes to the decrease in QR.p migration speed.
Fig. 2.
Activation of canonical Wnt signaling influences filopodial dynamics. (A) Time-lapse imaging of QR.p in wild type and ΔN-BAR-1 (Q)–expressing animals. Newly formed filopodia-like protrusions are indicated by pink arrowheads, and stable protrusions are indicated by yellow arrowheads. The seam (V) cells and QR.p are marked with nuclear (H2B) and membrane-localized (PH) GFP (huIs63) (38). The ΔN-BAR-1 (Q)–expressing strain contains mab-5(gk670). Anterior is left and dorsal is up (scale bar, 5 μm). (B) Quantification of protrusive activity, as measured by the number of filopodia-like protrusions formed per minute. Data are represented as mean ± SEM (n = 8 for all genotypes). The statistical significance was calculated using an unpaired t test. (C) Length of filopodia. Data are represented as mean ± SD (n > 245 for all genotypes). The statistical significance was calculated using an unpaired t test. ***P < 0.001.
Canonical Wnt/β-Catenin Signaling Genetically Interacts with the CWN-1/Wnt–MOM-5/Frizzled–Dependent Migration Pathway.
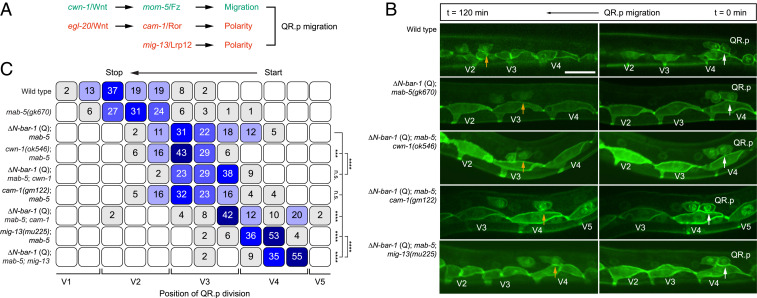
Previous work has shown that QR.p migration is dependent on noncanonical Wnt signaling and a pathway defined by the Lrp12 ortholog MIG-13 (Fig. 3A) (10, 12, 19). However, these pathways control distinct dynamic aspects of the migration process. On the one hand, the anterior polarization of QR.p is dependent on MIG-13 (19) as well as the Wnt ligand EGL-20 and the noncanonical Wnt receptor CAM-1/Ror (12). We found that MIG-13 and CAM-1/Ror are part of distinct pathways, as double mutants of mig-13 and cam-1 show an additive phenotype (Fig. 3C and SI Appendix, Fig. S1B). On the other hand, migration itself is dependent on CWN-1/Wnt and the receptor MOM-5/Frizzled, which are part of a noncanonical Wnt pathway that controls the speed of QR.p migration without affecting its polarity (12). To investigate whether canonical Wnt signaling genetically interacts with these polarity and migration pathways or inhibits QR.p migration through a different mechanism, we combined expression of ΔN-BAR-1 with mutations in cam-1/Ror, mig-13/Lrp12, or cwn-1/Wnt. We found that the ΔN-BAR-1–induced defect in QR.p migration was strongly enhanced by cam-1 (Fig. 3 B and C) and that there was also a clear synergistic interaction between ΔN-BAR-1 and mig-13. Loss of cwn-1 in ΔN-BAR-1–expressing animals, on the other hand, had only a mild additional effect. This is consistent with the phenotypic similarity between ΔN-BAR-1 and cwn-1/Wnt (and mom-5/Frizzled) mutants, both of which inhibit QR.p migration without affecting the ability of the cell to persistently polarize toward the anterior. We did observe a mild but significant effect of ΔN-BAR-1 in the cwn-1 background, which may result from partial redundancy between CWN-1 and other Wnt ligands in the MOM-5/Frizzled pathway (12). We conclude that canonical Wnt signaling functions in the same genetic pathway as cwn-1 but in parallel to the cam-1/Ror and mig-13/Lrp12 polarity pathways.
Fig. 3.
Canonical Wnt signaling counteracts the CWN-1/Wnt–MOM-5/Frizzled–dependent migration pathway. (A) A schematic representation of the parallel Wnt pathways and the mig-13/Lrp12 pathway that control QR.p polarity and migration. (B) Time-lapse imaging of QR.p migration in wild type, ΔN-BAR-1 (Q), and ΔN-BAR-1 (Q) combined with mutations in cwn-1, cam-1, or mig-13. The seam (V) cells and QR.a and QR.p are marked with nuclear (H2B) and membrane-localized (PH) GFP (huIs63) (38). The position of QR.p at time points 0 min and 120 min is indicated by white and yellow arrows, respectively. Anterior is left and dorsal is up (scale bar, 15 μm). (C) Position of QR.p division with respect to the seam cells, indicated as percentiles of the total number of cells analyzed (n ≥ 50 for all genotypes). The ΔN-BAR-1 (Q), cwn-1, cam-1, and mig-13 strains contain mab-5(gk670). The statistical significance was calculated using Fisher’s exact test (n.s., P ≥ 0.05, ***P < 0.001, and ****P < 0.0001).
Activation of Canonical Wnt Signaling Induces a Specific Transcriptional Program in the Q Cell Lineage.
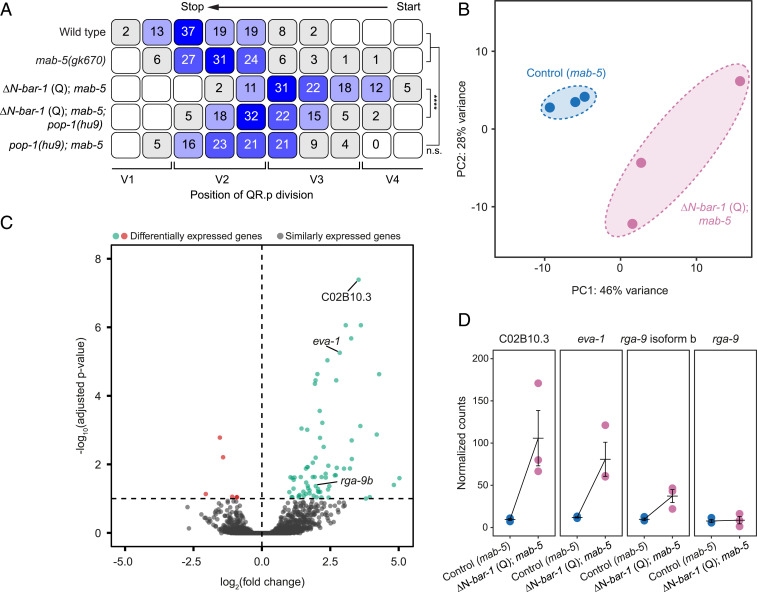
In canonical Wnt signaling, interaction of β-catenin with members of the TCF/Lef1 family of transcription factors induces the expression of specific target genes (2). To investigate whether the ΔN-BAR-1–induced inhibition of QR.p migration is dependent on POP-1—the single C. elegans TCF/Lef1 ortholog—we combined ΔN-BAR-1 with pop-1(hu9). This allele contains a mutation in the BAR-1 binding domain that specifically inhibits canonical Wnt signaling (20). As shown in Fig. 4A, pop-1(hu9) strongly rescued QR.p migration, demonstrating that the effect of ΔN-BAR-1 on QR.p migration is mediated through POP-1/TCF–dependent transcription.
Fig. 4.
Canonical Wnt signaling activates a specific transcriptional program in the Q neuroblast lineage. (A) The position of QR.p division with respect to the seam cells, indicated as percentiles of the total number of cells analyzed (n > 50 for all genotypes). The ΔN-BAR-1 (Q) strains contain mab-5(gk670). The statistical significance was calculated using Fisher’s exact test (n.s., P ≥ 0.05, ****P < 0.0001). (B) Principal component analysis plot of transcriptome similarities of mab-5(gk670) control (Control) and mab-5(gk670); ΔN-BAR-1 Q (∆N-bar-1 Q). (C) Differentially expressed genes between mab-5(gk670) control and mab-5(gk670); ΔN-BAR-1 (Q), using an adjusted P value threshold of 0.1 (Padj > 0.1). (D) The normalized expression levels detected by RNA-seq. The bars represent mean ± SEM.
To gain insight into the transcriptional changes that are induced by canonical Wnt signaling, we performed mRNA sequencing on Q cell descendants isolated from control and ΔN-BAR-1–expressing animals. As detailed in Materials and Methods, similar populations of early Q cell descendants (predominantly Q.a and Q.p) were obtained by tightly synchronizing larvae before generating cell suspensions for fluorescence-activated cell sorting (FACS) (SI Appendix, Fig. S2 A–C). Moreover, since both strains contain a loss-of-function mutation in mab-5, the related QL lineage adopts the same anterior migration program as the QR lineage (16, 17). The isolated cells are therefore a mixture of QR descendants and functionally transformed QL descendants. Three biological replicates were obtained for both control and ΔN-BAR-1–expressing cells, and a global analysis of transcriptional differences showed that these datasets form distinct, nonoverlapping clusters in principal component space (Fig. 4B). Differential gene expression analysis of the two genotypes revealed 78 differentially expressed genes, 72 of which were significantly up-regulated in the ΔN-BAR-1–expressing cells (false discovery rate [FDR] < 0.1) (Fig. 4C, SI Appendix, Fig. S3A, and Dataset S1). Most of these genes (59/78) contain at least one putative TCF binding motif (21) within a 1.5-kb region upstream of the start codon, supporting the notion that these represent direct Wnt target genes (Dataset S2). Gene Ontology term analysis revealed an enrichment of genes involved in neural development and neuronal function (SI Appendix, Fig. S3B), including genes associated with the terminally differentiated state of the final Q cell descendants.
The Canonical Wnt Target Genes eva-1 and rga-9b Are Required for Migration Inhibition.
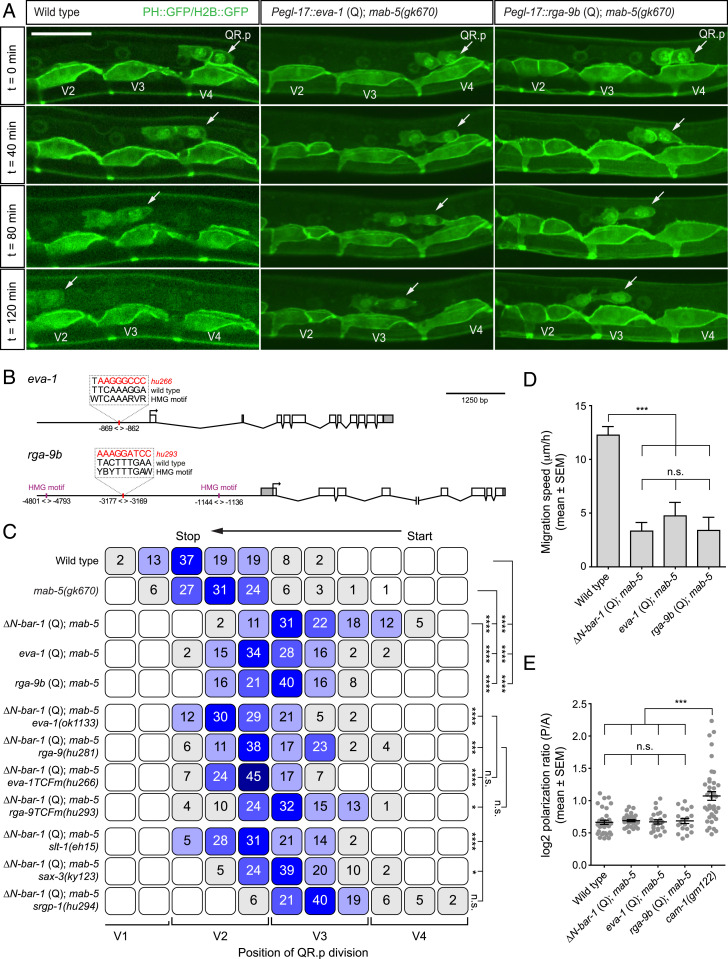
To identify potential mediators of ΔN-BAR-1–induced migration inhibition, we manually selected genes based on domain structure or previously reported roles in cell migration. Among these genes were the following: 1) C02B10.3, an uncharacterized locus predicted to encode a secreted EGF-like repeat containing protein; 2) eva-1, which encodes a conserved transmembrane protein that functionally interacts with the SAX-3/Robo and UNC-40/DCC migration pathways (22, 23); and 3) the isoform b of the uncharacterized gene 2RSSE.1, which encodes a Rho GTPase-activating domain (GAP) containing protein that is related to Drosophila and mammalian ARHGAP family members. This gene was named rga-9. Since C02B10.3, eva-1, and rga-9 isoform b (rga-9b) were significantly up-regulated in ΔN-BAR-1–expressing cells (Fig. 4 C and D), we first investigated whether expression of these genes inhibits QR.p migration. Q lineage–specific expression of C02B10.3 had no effect on QR.p migration (SI Appendix, Fig. S1C). However, we found that migration distance was strongly reduced when eva-1 or rga-9b were expressed (Fig. 5 A and C). Furthermore, and similar to ΔN-BAR-1, expression of eva-1 or rga-9b strongly decreased the speed of QR.p migration (Fig. 5D) without affecting QR.p polarization (Fig. 5E).
Fig. 5.
eva-1/EVA1C and rga-9b/ARHGAP are required for the canonical Wnt pathway–induced inhibition of QR.p migration. (A) Time-lapse imaging of QR.p migration in wild type and animals specifically expressing eva-1 or rga-9b in the Q neuroblast lineage using the egl-17 promoter. The seam (V) cells and QR.p are marked with nuclear (H2B) and membrane-localized (PH) GFP (huIs63) (38) (scale bar, 15 μm). (B) A schematic representation of the eva-1 and rga-9 loci and the predicted TCF binding sites (HMG motifs) that were mutated. Coordinates are bp from the translational start site of the gene. (C) The position of QR.p division with respect to seam cells, indicated as percentiles of the total number of cells analyzed (n > 50 for all genotypes). The statistical significance was calculated using Fisher’s exact test (n.s., P ≥ 0.05, *P < 0.05, ***P < 0.001, and ****P < 0.0001). (D) The average speed of QR.p during the first hour of migration. The bars represent mean ± SEM (n ≥ 50 for all genotypes). The statistical significance was calculated using an unpaired t test (n.s., P ≥ 0.05, ***P < 0.001). (E) Quantification of QR.p polarity as calculated by the ratio of the distance from the nucleus to the posterior and the anterior side of the cell. The black lines indicate mean ± SEM (n ≥ 19 for all genotypes). The statistical significance was calculated using an unpaired t test (n.s., P ≥ 0.05, ***P < 0.001). ΔN-BAR-1 (Q), eva-1 (Q), and rga-9b (Q)–containing strains have mab-5(gk670).
Next, we tested whether eva-1 and rga-9b are required for the ΔN-BAR-1–induced inhibition of QR.p migration. Because no mutant allele of rga-9 was available, we used CRISPR/Cas9-mediated genome editing and homologous recombination to generate a predicted null allele (deletion of the complete coding sequence, see Materials and Methods). Loss of eva-1, rga-9b, or the combined loss of eva-1 and rga-9b did not affect the normal migration of QR.p (SI Appendix, Fig. S1D). However, as shown in Fig. 5C, loss of eva-1 or rga-9b strongly rescued QR.p migration in ΔN-BAR-1–expressing animals.
Both the eva-1 and rga-9 loci contain consensus TCF binding motifs with predicted adjacent helper sites, a hallmark of functional POP-1 binding sites (21). To investigate if eva-1 and rga-9b are direct target genes of canonical Wnt signaling, we used CRISPR-Cas9–mediated genome editing and oligonucleotide mediated repair to specifically mutate these sites (positioned 869 base pairs [bp] upstream of eva-1 and 3,177 bp upstream of rga-9) (Fig. 5B). In the case of eva-1, loss of the predicted POP-1 binding site (hu266) strongly suppressed the ΔN-BAR-1–induced inhibition of QR.p migration (Fig. 5C). Indeed, the phenotype of the TCF binding site mutation was similar to the eva-1 null mutant. Mutation of the predicted TCF site of rga-9 (hu293) weakly suppressed the ΔN-BAR-1–induced inhibition of QR.p migration. Since the rga-9 locus contains two additional predicted TCF binding sites with consensus helper site sequences (one upstream and one downstream with a lower prediction value, Fig. 5B), we hypothesize that POP-1 may act through multiple sites in the rga-9 promoter. Taken together, these results show that eva-1 and rga-9b are canonical Wnt target genes required for inhibiting QR.p migration.
EVA-1/EVA1C and the SLT-1/Slit-SAX-3/Robo Pathway Are Necessary for Canonical Wnt Pathway–Dependent Inhibition of QR.p Migration.
eva-1 encodes an evolutionarily conserved transmembrane protein with predicted galactose-binding lectin domains that is orthologous to human EVA1C (also known as C21ORF63) (22). Previous studies have shown that EVA-1 functionally interacts with both the UNC-40/DCC and SAX-3/Robo guidance pathways (22, 23). In combination with UNC-40, EVA-1 mediates the response of growing axons to the ligand MADD-4 (23). Since neither MADD-4 nor the canonical UNC-40 ligand UNC-6/netrin is required for QR descendant migration (24, 25), we focused on the role of EVA-1 in SAX-3/Robo signaling. EVA-1 physically interacts with SAX-3 and functions as a coreceptor that confers specificity for the ligand SLT-1/Slit (22). Consistent with a shared site of action, we found that sax-3 is expressed in the QR descendants (see Fig. 7B) and that Q-lineage-specific expression of sax-3 (Pegl-17::sax-3, abbreviated as sax-3 Q) rescues the sax-3(ky123) phenotype (see Fig. 7D). Moreover, the ligand slt-1 is expressed in the dorsal body wall muscle cells along which QR.p and QR.pa migrate (26). While a minor role for slt-1 and sax-3 has been reported for QR.ap (AQR) migration (27), a function of these genes in the QR.p lineage has not been described. We found that loss of slt-1 strongly rescued QR.p migration in ΔN-BAR-1–expressing animals (Fig. 5C), a phenotype similar to what we observed in eva-1 null mutants. A slight (but statistically significant) suppression was also found with sax-3(ky123). We hypothesize that this weaker phenotype may result from residual sax-3 activity, as the ky123 allele shows an overall milder phenotype than the deletion allele gk5367 that removes most of the gene. Given the function of EVA-1 as a SLT-1 coreceptor and the similarity in phenotype of the eva-1 and slt-1 null mutants, we conclude that EVA-1–dependent activation of SLT-1-SAX-3/Robo signaling is required for the ΔN-BAR-1–induced inhibition of QR.p migration.
Fig. 7.
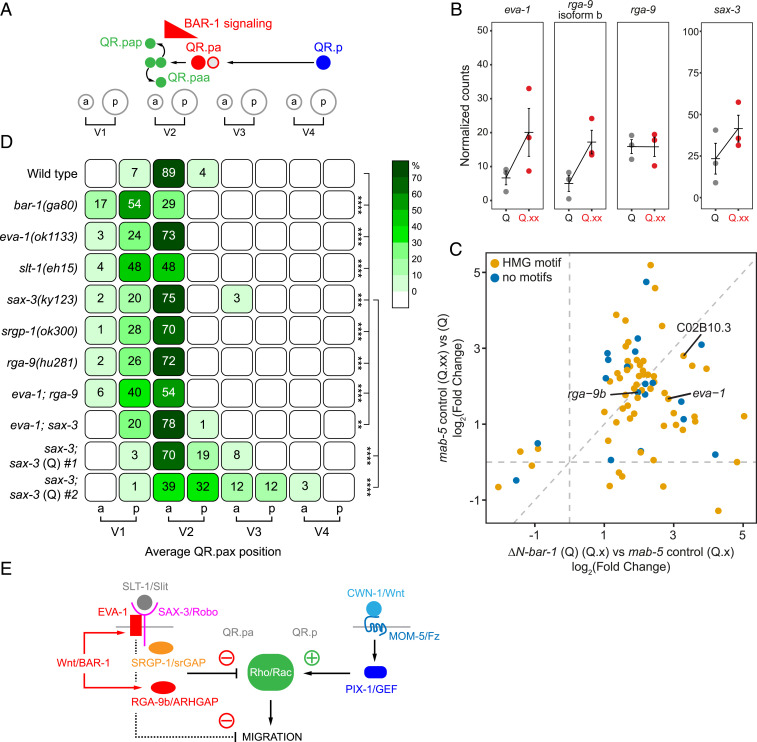
The Wnt target genes eva-1/EVA1C and rga-9b/ARHGAP are up-regulated during QR lineage progression and required for terminating the anterior migration of the final QR descendants. (A) A schematic representation of the QR lineage and the role of canonical Wnt signaling (BAR-1) in stopping the migration of QR.pa. (B) The normalized RNA-seq expression levels in populations enriched for early (Q) and late (Q.pa and Q.ap, [Q.xx]) Q descendants in mab-5(gk670). The bars represent mean ± SEM. (C) Fold change in expression of genes differentially expressed in ΔN-BAR-1 (FDR < 0.1) compared to fold change in expression between populations enriched for Q and Q.xx. (D) The position of the final QR.p descendants QR.paa and QR.pap (QR.pax) with respect to seam cell nuclei, indicated as percentiles of the total number of cells analyzed (n > 50 for all genotypes). The statistical significance (compared to wild type) was calculated using Fisher’s exact test (**P < 0.01, ***P < 0.001, and ****P < 0.0001). (E) A model of cross-talk between noncanonical and canonical Wnt/BAR-1 signaling in QR descendant migration.
RGA-9b/ARHGAP and the CWN-1/Wnt–MOM-5/Frizzled Pathway Component PIX-1/RhoGEF Have Opposing Activities in QR.p Migration.
rga-9b encodes a Rho GAP domain-containing protein that is related to Drosophila and mammalian ARHGAPs. Rho family members—which include different forms of Rho, Rac, and Cdc42—are small GTPases that play a central role in cell polarity and migration (28). They are regulated by specific binding partners that either promote (guanine nucleotide exchange factors [GEFs]) or inhibit (GAPs) their activity. Previous studies have shown that the initial polarization and migration of the Q neuroblasts, and the long-range migration of their descendants, are dependent on the redundantly acting Rho family members MIG-2 and CED-10 (29). Polarization and migration also require UNC-73, a GEF related to Drosophila Trio (25, 29). In unc-73 mutants, the Q neuroblasts fail to form a major lamellipodium-like protrusion, leading to a strong defect in QR polarization (25, 29) and the subsequent migration of QR.p (Fig. 6E). This function in lamellipodium formation and polarization is distinct from ΔN-BAR-1 and the CWN-1/Wnt–MOM-5/Frizzled pathway, which do not influence QR.p polarity (Figs. 1D and 6D).
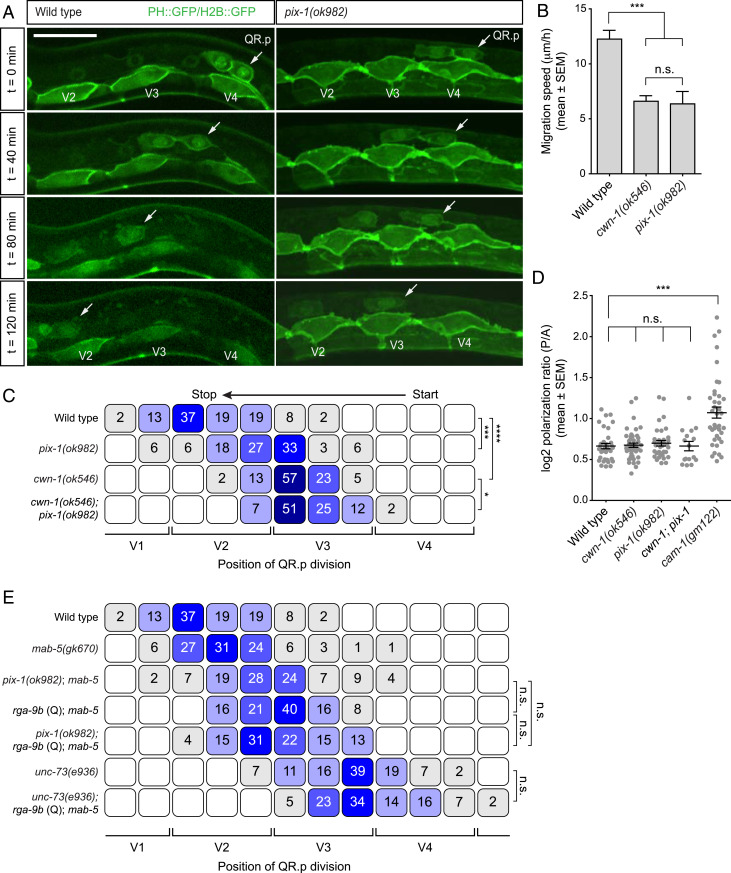
Fig. 6.
The Rho activating protein PIX-1 is part of the CWN-1/Wnt–MOM-5/Frizzled pathway. (A) Time-lapse imaging of QR.p migration in wild type and pix-1(ok982) animals. The seam (V) cells and QR.a and QR.p are marked with nuclear (H2B) and membrane-localized (PH) GFP (huIs63) (38). Anterior is left and dorsal is up (scale bar, 15 μm). (B) The average speed of QR.p during the first hour of migration. The bars represent mean ± SEM (n > 50 for all genotypes). The statistical significance was calculated using an unpaired t test (n.s., P ≥ 0.05, ***P < 0.001). cwn-1(ok546) data are from ref. 12. (C) The position of QR.p division with respect to seam cells, indicated as percentiles of the total number of cells analyzed (n > 50 except pix-1, n = 33). The statistical significance was calculated using Fisher’s exact test (*P < 0.05, ***P < 0.001, and ****P < 0.0001). cwn-1(ok546) data are from ref. 12. (D) Quantification of QR.p polarity as calculated by the ratio of the distance from the nucleus to the posterior and the anterior side of the cell. The black lines indicate mean ± SEM (n > 25 except cwn-1; pix-1, n = 16). The statistical significance was calculated using an unpaired t test (n.s., P ≥ 0.05, ***P < 0.001). cwn-1(ok546) data are from ref. 12. (E) The position of QR.p division with respect to the seam cells, indicated as percentiles of the total number of cells analyzed (n > 50 except unc-73(e936); rga-9b(Q); mab-5(gk670), n = 44). The pix-1 and rga-9b (Q)–containing strains have mab-5(gk670). The statistical significance was calculated using Fisher’s exact test (n.s., P ≥ 0.05).
UNC-73 acts synergistically with another GEF, the p21-activated kinase interacting exchange factor (PIX) ortholog PIX-1 in QR and QR.ap/AQR migration (29). We found that pix-1 null mutants also show a significant decrease in QR.p migration distance (Fig. 6 A and C). Furthermore, the speed of QR.p migration was reduced, but there was no effect on lamellipodium formation and persistent anterior polarization (Fig. 6 A, B, and D). Since this phenotype is comparable to that of cwn-1 and mom-5 mutants (12), we tested if pix-1 is part of the same pathway. As shown in Fig. 6 C and E, QR.p migration and polarity was similar in cwn-1 single and cwn-1; pix-1 double mutants, which is in agreement with a function of pix-1 in the CWN-1/Wnt–MOM-5/Frizzled–dependent migration pathway. Next, we investigated if RGA-9b/ARHGAP counteracts the migration promoting activity of PIX-1. We found that the defect caused by overexpression of rga-9b is similar to that of pix-1 (Fig. 6E). Moreover, overexpression of rga-9b did not further decrease QR.p migration in a pix-1 mutant background, indicating that the inhibitory effect of RGA-9b depends on the Rho activating function of PIX-1.
eva-1 and rga-9b Are Also Necessary for the Canonical Wnt Pathway–Dependent Termination of QR.pa Migration.
Our previous results show that canonical Wnt signaling is activated during the transition from QR.p to QR.pa (Fig. 7A) (12). To investigate whether the transcriptional program that is induced by ΔN-BAR-1 reflects the physiological response to canonical Wnt signaling in QR.pa, we examined normalized mRNA sequencing data from different stages of Q lineage progression (30). As before, we FACS isolated Q descendants from mab-5(gk670) mutants in which the QL lineage adopts a QR-like fate (16, 17) and refer to this mixture of QR and functionally transformed QL cells as QR descendants. Focusing on samples enriched in early Q cells (QR) and late Q cell descendants (QR.ap and QR.pa), we found that 57 of the 78 genes (73%) that were significantly up-regulated (fold change ≥2) by ΔN-BAR-1 were also up-regulated (fold change ≥2) in the QR.pa enriched sample (Fig. 7C), indicating that these genes are part of the endogenous response to canonical Wnt signaling in the QR lineage. A total of 43 of the up-regulated genes have predicted TCF binding sites within 1.5 kb of upstream sequence, but there were also 14 genes in which such sites could not be identified (Dataset S2). Moreover, there were 21 genes (including 16 genes with TCF binding sites) that did not show increased expression. These may be high-threshold target genes that are only induced when canonical Wnt signaling is constitutively activated.
eva-1 and isoform b of rga-9—but not isoform a—were up-regulated in the QR.pa-enriched population (Fig. 7 B and C). Given their essential role in the ΔN-BAR-1–induced inhibition of QR.p migration, we investigated whether eva-1 and rga-9b are also necessary for the canonical Wnt pathway–dependent termination of QR.pa migration. Using the position of QR.paa and QR.pap (abbreviated as QR.pax) as a proxy for the final position of QR.pa, we found that—similar to bar-1/β-catenin null mutants—the QR.pax localized significantly more anterior in eva-1 and rga-9b single mutants. This effect was enhanced in the double mutant (Fig. 7D), consistent with a parallel function of eva-1 and rga-9b in migration regulation. We conclude that eva-1 and rga-9b are key mediators of the canonical Wnt pathway–dependent inhibition of QR.pa migration.
In agreement with the role of EVA-1 in SLT-1/Slit–SAX-3/Robo signaling, loss of slt-1 and sax-3 also resulted in overmigration of the QR.pax, while Q-lineage-specific overexpression of sax-3 resulted in undermigration of these cells (Fig. 7D). Furthermore, the QR.pax overmigration phenotype of eva-1 and sax-3 was not enhanced in the double mutant, confirming their shared function in the Slit–Robo pathway.
A downstream effector of Slit–Robo signaling in mammalian axon guidance is the Rho GTPase activating protein srGAP (31). Given the role of rga-9b/ARHGAP in terminating QR descendant migration, we investigated whether the C. elegans srGAP ortholog srgp-1 is also involved. We found that loss of srgp-1 induced significant QR.pax overmigration (Fig. 7D), which is consistent with a role of srgp-1 in the Slit–Robo pathway–dependent inhibition of QR.pa migration. However, unlike eva-1, slt-1, or sax-3, loss of srgp-1 did not suppress the ΔN-BAR-1–induced inhibition of QR.p migration (Fig. 5C). Therefore, it is likely that other downstream effectors of SLT-1/Slit–SAX-3/Robo signaling are required as well.
Discussion
The migration of the QR neuroblast descendants is regulated through a complex interplay between canonical and noncanonical Wnt signaling (12). During the first phase of migration, in which the QR descendant QR.p moves a relatively long distance toward the anterior, migration is dependent on two noncanonical Wnt pathways that separately control polarity and motility (12). In the second phase, when the QR.p daughter cell QR.pa reaches its final anterior position, migration is stopped through a cell intrinsically regulated switch to canonical Wnt/β-catenin signaling. Here, we have investigated how activation of canonical Wnt signaling inhibits migration. We focused on the long-range anterior migration of QR.p, which offers a more tractable system for studying migration dynamics than QR.pa. Our results show that activation of canonical Wnt signaling, through expression of a constitutively active form of BAR-1/β-catenin (ΔN-BAR-1), inhibits migration without affecting the ability of the cell to polarize or maintain an anteriorly directed lamellipodium-like protrusion. Interestingly, ΔN-BAR-1 did induce a reduction in filopodial dynamics, which may be mechanistically linked to the decrease in migration speed.
To examine the transcriptional response to canonical Wnt signaling, we isolated QR descendants from control and ΔN-BAR-1–expressing animals and performed mRNA sequencing. This revealed a specific set of differentially expressed genes, the majority of which were up-regulated by canonical Wnt signaling. The bias toward up-regulated genes suggests that most of the differentially expressed genes are direct targets of canonical Wnt signaling. This is consistent with the presence of predicted TCF binding motifs in the upstream promoter regions of the majority of the genes and the relatively short time period between pathway activation and cell isolation, which likely precludes the detection of secondary transcriptional effects. Importantly, mRNA sequencing of temporally synchronized Q descendants showed that the majority of these genes were also up-regulated in QR.pa, indicating that the transcriptional program induced by ΔN-BAR-1 closely resembles the endogenous response to canonical Wnt signaling.
Gene Ontology term analysis (SI Appendix, Fig. S3B) revealed an enrichment of genes involved in neuronal development and neuronal function, including genes associated with the terminally differentiated fate of the final Q cell descendants (such as mec-1, mec-3, mec-7, mec-12, and mec-18 for QR.paa/AVM, lad-2 for QR.pap/SDQR, and gcy-32 for QR.ap/AQR). During their migration, the QR descendants transition from large motile neuroblasts into functionally specialized neurons. The up-regulation of these neuronal markers indicates that besides a role in regulating migration, canonical Wnt signaling may also promote neuronal differentiation. However, expression of ΔN-BAR-1 in the early QR descendants did not induce morphological changes that are indicative of premature neuronal differentiation (Fig. 1A). Moreover, in bar-1/β-catenin null mutants, a terminal differentiation marker of the QR.paa/AVM neuron (mec-7) was normally expressed (100% in wild type, 100% in bar-1(ga80), n = 30), and the AVM axon was correctly directed toward the ventral midline (100% in wild type, 100% in bar-1(ga80), n = 30). These results show that canonical Wnt signaling is not necessary for neuronal differentiation but do not rule out the possibility that activation of the pathway at the end of the migration phase may enhance robustness of the neurogenesis process.
To identify potential mediators of the canonical Wnt pathway–dependent inhibition of QR.p migration, candidate genes were examined based on domain structure or previously reported roles in cell migration. Among these genes was eva-1, which encodes an evolutionarily conserved transmembrane protein with extracellular galactose-binding lectin domains that is similar to human EVA1C (also known as C21ORF63) (22). We found that eva-1 is a direct target of canonical Wnt signaling that is necessary for the ΔN-BAR-1–induced inhibition of QR.p migration. Furthermore, eva-1 is also required for the canonical Wnt pathway–dependent termination of QR.pa migration. Thus, consistent with regulation by endogenous canonical Wnt signaling, mRNA sequencing showed that eva-1 expression was increased in samples enriched for QR.pa. Moreover, similar to loss of bar-1/β-catenin, the final position of the QR descendants was significantly more anterior in eva-1 mutants, while the position of QR.p was unaffected. These results show that eva-1 is a mediator of canonical Wnt pathway–dependent inhibition of cell migration and validate our approach of using QR.p as a proxy for studying the endogenous response to canonical Wnt signaling in QR.pa. Previous studies have shown that eva-1 interacts with two of the major neuronal guidance pathways. It functions as a coreceptor of SAX-3/Robo that confers specificity to the ligand SLT-1/Slt (22) and can also associate with the netrin receptor UNC-40/DCC to promote MADD-4–dependent signaling (23). Neither MADD-4 nor the canonical UNC-40 ligand UNC-6/netrin are required for QR descendant migration (24, 25). However, we found that slt-1/Slt and sax-3/Robo are necessary for the ΔN-BAR-1–induced inhibition of QR.p migration. Furthermore, loss of slt-1 and sax-3/Robo induced significant overmigration of the final QR descendants. slt-1 is expressed in dorsal body wall muscle cells (26), while sax-3 is expressed in the QR descendants. However, given the essential role of EVA-1 as a SLT-1 coreceptor (22), SLT-1–dependent SAX-3 signaling will not be activated in the absence of EVA-1. We propose that the Wnt-dependent induction of eva-1 expression functions as a switch that specifically turns on SLT-1–SAX-3/Robo signaling in QR.pa.
In addition to eva-1, the isoform b of the target gene rga-9 (abbreviated as rga-9b) is also required for ΔN-BAR-1 and endogenous canonical Wnt pathway–dependent inhibition of QR descendant migration. rga-9b encodes a Rho GAP domain–containing protein that is related to Drosophila and mammalian ARHGAPs. Rho family members—small GTPases that include Rho, Rac, and Cdc42—are key regulators of cell polarity and migration (28). Their activity is stimulated by specific GEFs (guanine nucleotide exchange factors that promote binding of GTP) and inhibited by GAPs (GTPase-activating proteins that induce hydrolysis of the bound GTP). In case of QR and its descendants, polarization and migration are dependent on the partially redundantly acting Rho family members MIG-2 and CED-10 (29, 32), which are in turn regulated by the GEFs UNC-73/Trio and PIX-1/Pix (25, 29). In the absence of unc-73, QR and its descendants fail to form a major lamellipodium-like protrusion. Loss of pix-1, on the other hand, induces a defect in QR.p migration that is similar to cwn-1 and mom-5 mutants, and double-mutant analysis showed that cwn-1 and pix-1 are part of the same genetic pathway. We found that pix-1 plays a central role in the interaction with canonical Wnt signaling. Loss of pix-1 induces a similar decrease in QR.p migration as expression of ΔN-BAR-1 or rga-9b. Furthermore, we found that pix-1 is required for the ability of rga-9b to inhibit QR.p migration. These results suggest that cross-talk between the CWN-1/Wnt–MOM-5/Frizzled pathway and canonical Wnt signaling occurs at the level of Rho activation by PIX-1 and Rho inhibition by RGA-9b.
We found that the Rho GAP SRGP-1, which has been shown to negatively regulate CED-10 (33), also plays a role in terminating QR.pa migration. In mammalian neurons, the SRGP-1 ortholog srGAP functions as a downstream effector of Slt–Robo signaling. Although SRGP-1 lacks the SH3 domain that is required for direct physical interaction with Robo (34), the similar phenotype of slt-1/Slt, sax-3/Robo, and srgp-1 in endogenous canonical Wnt pathway induced migration inhibition indicates that they are part of the same pathway. This suggests that migration is inhibited through the combined GAP activity of RGA-9b and SRGP-1. However, it should be noted that srgp-1 has a weaker phenotype than slt-1 and sax-3 (loss of srgp-1 is insufficient to suppress the ΔN-BAR-1–induced inhibition of QR.p migration), indicating that SLT-1/Slit–SAX-3/Robo signaling may also inhibit migration through other downstream mechanisms, including Rho-independent effects on migration or adhesion.
Taken together, we propose that QR descendant migration is regulated by a balance between noncanonical Wnt pathway–induced Rho GEF activity, and canonical Wnt pathway induced up-regulation of Rho GAP activity (Fig. 7E). During the long-range anterior migration phase, signaling of CWN-1/Wnt through MOM-5/Frizzled promotes migration through PIX-1–dependent activation of Rho family GTPases, most likely including CED-10 and MIG-2. Once QR.p nears its final position and during the early stage of QR.pa migration, canonical Wnt signaling is activated, leading to the expression of rga-9b and eva-1. RGA-9b may directly inhibit the activity of MIG-2 and CED-10. In addition, expression of eva-1 will trigger SLT-1/Slit–SAX-3/Robo signaling, which may contribute to the inhibition of CED-10 through SRGP-1 and other as yet to be determined downstream mechanisms involved in migration inhibition.
Noncanonical and canonical Wnt signaling also have opposing roles in vertebrate cell migration. For example, activation of canonical Wnt signaling inhibits the Wnt-dependent migration of neural crest cells during Xenopus development (5). Interestingly, and similar to the C. elegans QR neuroblast descendants, neural crest cell migration is inhibited by Slit–Robo signaling (35), indicating that an analogous link between canonical Wnt signaling and Slit–Robo pathway activity may be involved here as well. Another example of the antagonism between canonical and noncanonical Wnt signaling is observed in melanoma progression, where β-catenin signaling is associated with a proliferative but noninvasive phenotype, whereas noncanonical Wnt signaling is correlated with invasion, metastasis, and poor patient survival (6, 7). Future studies will determine whether a similar convergence on Rho family GTPases underlies the antagonistic role of canonical and noncanonical Wnt signaling in vertebrate development and cancer cell migration.
Materials and Methods
C. elegans Strains and Culture.
All C. elegans strains were grown at 20 °C using standard culture conditions. The Bristol N2 strain was used as wild type. The alleles and transgenes used in this work are LGI: mig-1(e1787), pop-1(hu9), pry-1(mu38), eva-1(ok1133), eva-1(hu266), and unc-73(e936); LGII: cam-1(gm122), cwn-1(ok546), rga-9(hu281), rga-9(hu293), and muIs32[Pmec-7::gfp]; LGIII: mab-5(gk670); LGIV: huIs179[Pegl-17::∆N-bar-1; Pmyo-2::mCherry]; srgp-1(ok300), and srgp-1(hu294) LGV: heIs63[Pwrt-2::PH::gfp; Pwrt-2::H2B::gfp; Plin-48::tdTomato] and ayIs9[Pegl-17::gfp; dpy-20(+)]; LGX: bar-1(ga80), mig-13(mu225), huIs166[Pwrt-2::PH::mCherry; Pwrt-2::H2B::mCherry; dpy-20(+)], sax-3(ky123), slt-1(eh15), and pix-1(ok982); and unassigned: casIs328[Pegl-17::myri-mCherry; Pegl-17::mCherry-TEV-his-24, Pegl-17::gfp::moeABD], huEx731 (subsequently integrated and named huIs214) [Pegl-17::eva-1; Plin-32::tdTomato; rol-6(su1006)], huEx737 (subsequently integrated and named huIs221) [Pegl-17::rga-9b; Plin-32::tdTomato; rol-6(su1006)], huEx740[Pegl-17::C02B10.3; Plin-32::tdTomato; rol-6(su1006)], huEx743[Plin-32::tdTomato; rol-6(su1006)], huEx829[Pegl-17::sax-3; Ptoe-2::gfp; Plin-48::tdTomato], and huEx830[Pegl-17::sax-3; Ptoe-2::gfp; Plin-48::tdTomato].
C. elegans Expression Constructs and Transgenesis.
Expression constructs pKN576 [Pegl-17::C02B10.3 cds::unc-54 3′ untranslated region (UTR) in pDESTR4-R3], pKN585 [Pegl-17::rga-9b cds::unc-54 3′ UTR in pDESTR4-R3], and pKN597 [Pegl-17::eva-1 cds::unc-54 3′ UTR in pDESTR4-R3] were injected into KN2591 [mab-5(gk670); heIs63] animals at 10 ng/μL. Coinjection markers pRF4 and pKN611 [Plin-32::tdTomato::unc-54 3′ UTR in pDESTR4-R3] were both injected at 5 ng/μL, and pBluescriptII was added for a total concentration of 150 ng/μL. Expression construct pKN525 [Pegl-17::∆N-bar-1::unc-54 3′ UTR in pDESTR4-R3] was injected into N2 animals at 20 ng/μL with the coinjection marker Pmyo-2::mCherry at 20 ng/μL, and pBluescriptII was added for a total concentration of 150 ng/μL. Expression construct pKN708 [Pegl-17::sax-3 cds::unc-54 3′ UTR in pDESTR4-R3] was injected into sax-3(ky123) animals at 20 ng/μL with the coinjection markers pBC1565 [Ptoe-2::grp::unc-54 3′UTR] at 20 ng/μL and pKN641 [Plin-48::tdTomato::unc-54 3′UTR] at 10 ng/μL, and pBluescriptII was added for a total concentration of 150 ng/μL. The rga-9b sequence cloned for overexpression is the longest transcript among the three rga-9 isoforms and corresponds to 2RSSE.1b.1 on Wormbase. To clone the coding sequences of the genes chosen for overexpression experiments, Gateway compatible primers listed in SI Appendix, Table S1 were used. SI Appendix, Table S2 lists the transgenic reporter strains used in this study.
CRISPR/Cas9-Mediated Genome Editing.
The mutant alleles eva-1(hu266), rga-9(hu281), rga-9(hu293), and srgp-1(hu294) were generated using dpy-10 coconversion and single-stranded DNA repair templates as previously described (36). PCR fragments containing the T7 promoter and the single guide RNA (sgRNA) sequence of interest were used to produced sgRNAs via in vitro transcription. Repair templates and sgRNAs were coinjected with recombinant SpCas9 (37). sgRNAs and repair template sequences are in SI Appendix, Table S1. The sequence of the eva-1(hu266) and rga-9(hu293) mutations are depicted in Fig. 7B. rga-9(hu281) is a deletion of the entire rga-9 coding sequence (∼10-kb deletion). srgp-1(hu294) contains the same 1,406 bp deletion found in srgp-1(ok300).
Analysis of QR Descendant Migration.
The position of QR.p was determined relative to the seam cells V1 to V4 using the seam and Q neuroblast marker heIs63 (38) as described (12). The speed of QR.p migration was measured in synchronized larvae by determining the average distance of migration during the first hour after QR division (12). The final position of the QR.pax was determined with respect to the seam cell daughters V1.a to V6.p in late stage L1 larvae using differential interference contrast microscopy (39).
Imaging.
For static imaging and time-lapse live imaging of QR.p migration, larvae synchronized at 4 to 6 h after hatching were mounted in 1 μL of 0.1-μm diameter polystyrene microspheres in aqueous suspension (Polysciences 00876 2.5% wt/vol aqueous suspension) on a 10% agarose pad (40). Animals were imaged using a PerkinElmer Ultraview Vox spinning disk confocal microscope. Imaging was performed with a 63× objective at 1× zoom, 1× binning, and 4% 488 nm laser power. Z-stacks (0.33 μm) were made every 2 min for a duration of 3 h. Image acquisition was performed using Volocity software, and movies were created using ImageJ software.
FACS Isolation of Q Neuroblast Descendants.
Adult animals were lysed with 1 M NaOH and 5% NaClO for 5 to 7 min. The released eggs were washed two times with sterile M9 and hatched overnight (12 to 15 h) in the same buffer at room temperature. Starved L1 larvae were then fed for 7 h with NA22 bacteria, collected with sterile M9, and washed three times before further processing. Cells were isolated based on a protocol optimized for Q lineage cell isolation (41). Whole animal cell suspensions were collected in cold L-15/fetal bovine serum (FBS) and passed through a 5 μm syringe filter prior to sorting. Using a FACS Aria, green fluorescent protein (GFP), and mCherry, double-positive cells were sorted directly into tubes containing TRIzol and stored at −80 °C before subsequent processing.
Preparation of Sequencing Libraries and Data Analysis.
Each Illumina sequencing library was prepared from a pool of 2,000 cells, according to the cell expression by linear amplification and sequencing (CEL-seq) protocol (42). These libraries were sequenced paired end at 50 bp read length on Illumina HiSEq 2500. The Illumina sequencing reads were aligned to the C. elegans reference genome WS249. Count data were analyzed using R (version 3.3.2). Cosmid IDs were converted to gene names. For genes whose gene names were not available, cosmid ID was used. Only data sets with more than 7,000 genes and 100,000 total transcripts detected were used for differential gene expression analysis. Differential gene expression analysis was done with DESeq2 R-package using an FDR based on adjusted P < 0.1. ggplot2 and pheatmap R-packages were used for data visualization.
Statistical Analysis.
Prior to statistical analysis, QR.p division position measurements were binned into 28 equisized and nonoverlapping bins. Statistical analysis of QR.p division position and final QR.pax position was performed using Fisher’s exact test. A Monte Carlo approximation, iterated 10,000 times using R statistical computing software, was used to estimate significance. Note that for representation purposes, QR.p division position and final QR.pax position were further binned into 14 and 12 bins, respectively. Analysis of differences in QR.p polarization variability was performed using Levene’s test for equal variance. In all other cases, statistical analysis was examined using the unpaired, two-tailed Student’s t test. Results were deemed significant at P < 0.05.
Promoter Motif Analysis.
For each of the differentially expressed genes, sequences of 1,500 bp upstream of the first annotated translation starting site were retrieved from Wormbase. Identification of putative TCF binding sites was done with FIMO (version 5.0.5) (43) by using the TBYTTTGAW consensus motif (21) applying a threshold of P < 0.0001.
Supplementary Material
Acknowledgments
We thank Stefan van der Elst from the Flow Cytometry Core facility and the Hubrecht Imaging Center (Hubrecht Institute) for technical support, and Guangshuo Ou (Tsinghua University, Beijing, China) and Barbara Conradt (University College London, United Kingdom) for reagents. Part of this work was funded by the research program (14NOISE01) of the Foundation for Fundamental Research on Matter, which is financially supported by the Netherlands Organization for Scientific Research. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). We are grateful to members of the Korswagen and Galli groups for helpful discussions and critical reading of the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. O.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013239118/-/DCSupplemental.
Data Availability
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession no. GSE151216 (45).
References
- 1.Szabó A., Mayor R., Mechanisms of neural crest migration. Annu. Rev. Genet. 52, 43–63 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Clevers H., Nusse R., Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Angers S., Moon R. T., Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 10, 468–477 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Carmona-Fontaine C., et al., Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature 456, 957–961 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maj E., et al., Controlled levels of canonical Wnt signaling are required for neural crest migration. Dev. Biol. 417, 77–90 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Hoek K. S., et al., Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 19, 290–302 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Webster M. R., Weeraratna A. T., A Wnt-er migration: The confusing role of β-catenin in melanoma metastasis. Sci. Signal. 6, pe11 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Sulston J. E., Horvitz H. R., Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156 (1977). [DOI] [PubMed] [Google Scholar]
- 9.Harris J., Honigberg L., Robinson N., Kenyon C., Neuronal cell migration in C. elegans: Regulation of Hox gene expression and cell position. Development 122, 3117–3131 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Zinovyeva A. Y., Yamamoto Y., Sawa H., Forrester W. C., Complex network of Wnt signaling regulates neuronal migrations during Caenorhabditis elegans development. Genetics 179, 1357–1371 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Middelkoop T. C., Korswagen H. C., Development and migration of the C. elegans Q neuroblasts and their descendants. WormBook, 1–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mentink R. A., et al., Cell intrinsic modulation of Wnt signaling controls neuroblast migration in C. elegans. Dev. Cell 31, 188–201 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Korswagen H. C., et al., The Axin-like protein PRY-1 is a negative regulator of a canonical Wnt pathway in C. elegans. Genes Dev. 16, 1291–1302 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Branda C. S., Stern M. J., Mechanisms controlling sex myoblast migration in Caenorhabditis elegans hermaphrodites. Dev. Biol. 226, 137–151 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Gleason J. E., Korswagen H. C., Eisenmann D. M., Activation of Wnt signaling bypasses the requirement for RTK/Ras signaling during C. elegans vulval induction. Genes Dev. 16, 1281–1290 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salser S. J., Kenyon C., Activation of a C. elegans Antennapedia homologue in migrating cells controls their direction of migration. Nature 355, 255–258 (1992). [DOI] [PubMed] [Google Scholar]
- 17.Whangbo J., Kenyon C., A Wnt signaling system that specifies two patterns of cell migration in C. elegans. Mol. Cell 4, 851–858 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Josephson M. P., Miltner A. M., Lundquist E. A., Nonautonomous roles of MAB-5/Hox and the secreted basement membrane molecule SPON-1/F-Spondin in Caenorhabditis elegans neuronal migration. Genetics 203, 1747–1762 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., et al., Transmembrane protein MIG-13 links the Wnt signaling and Hox genes to the cell polarity in neuronal migration. Proc. Natl. Acad. Sci. U.S.A. 110, 11175–11180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korswagen H. C., Herman M. A., Clevers H. C., Distinct β-catenins mediate adhesion and signalling functions in C. elegans. Nature 406, 527–532 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Bhambhani C., et al., Distinct DNA binding sites contribute to the TCF transcriptional switch in C. elegans and Drosophila. PLoS Genet. 10, e1004133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujisawa K., Wrana J. L., Culotti J. G., The slit receptor EVA-1 coactivates a SAX-3/Robo mediated guidance signal in C. elegans. Science 317, 1934–1938 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Chan K. K., et al., EVA-1 functions as an UNC-40 Co-receptor to enhance attraction to the MADD-4 guidance cue in Caenorhabditis elegans. PLoS Genet. 10, e1004521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebbing A., Middelkoop T. C., Betist M. C., Bodewes E., Korswagen H. C., Partially overlapping guidance pathways focus the activity of UNC-40/DCC along the anteroposterior axis of polarizing neuroblasts. Development 146, dev180059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honigberg L., Kenyon C., Establishment of left/right asymmetry in neuroblast migration by UNC-40/DCC, UNC-73/Trio and DPY-19 proteins in C. elegans. Development 127, 4655–4668 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Hao J. C., et al., C. elegans slit acts in midline, dorsal-ventral, and anterior-posterior guidance via the SAX-3/Robo receptor. Neuron 32, 25–38 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Josephson M. P., et al., The Caenorhabditis elegans NF2/Merlin molecule NFM-1 nonautonomously regulates neuroblast migration and interacts genetically with the guidance cue SLT-1/Slit. Genetics 205, 737–748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawson C. D., Ridley A. J., Rho GTPase signaling complexes in cell migration and invasion. J. Cell Biol. 217, 447–457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dyer J. O., Demarco R. S., Lundquist E. A., Distinct roles of Rac GTPases and the UNC-73/Trio and PIX-1 Rac GTP exchange factors in neuroblast protrusion and migration in C. elegans. Small GTPases 1, 44–61 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandes Povoa E. E., “Finding the way: transcriptional regulation of migratory neuroblast lineages in C. elegans,” PhD thesis, Utrecht University, Utrecht, The Netherlands: (2020). [Google Scholar]
- 31.Wong K., et al., Signal transduction in neuronal migration: Roles of GTPase activating proteins and the small GTPase Cdc42 in the slit-Robo pathway. Cell 107, 209–221 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Ou G., Vale R. D., Molecular signatures of cell migration in C. elegans Q neuroblasts. J. Cell Biol. 185, 77–85 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neukomm L. J., et al., Loss of the RhoGAP SRGP-1 promotes the clearance of dead and injured cells in Caenorhabditis elegans. Nat. Cell Biol. 13, 79–86 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaidel-Bar R., et al., The F-BAR domain of SRGP-1 facilitates cell-cell adhesion during C. elegans morphogenesis. J. Cell Biol. 191, 761–769 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giovannone D., et al., Slits affect the timely migration of neural crest cells via Robo receptor. Dev. Dyn. 241, 1274–1288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paix A., Folkmann A., Rasoloson D., Seydoux G., High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201, 47–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Astolfo D. S., et al., Efficient intracellular delivery of native proteins. Cell 161, 674–690 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Wildwater M., Sander N., de Vreede G., van den Heuvel S., Cell shape and Wnt signaling redundantly control the division axis of C. elegans epithelial stem cells. Development 138, 4375–4385 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Coudreuse D. Y., Roël G., Betist M. C., Destrée O., Korswagen H. C., Wnt gradient formation requires retromer function in Wnt-producing cells. Science 312, 921–924 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Kim E., Sun L., Gabel C. V., Fang-Yen C., Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS One 8, e53419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernandes Póvoa E. E., Ebbing A. L. P., Betist M. C., van der Veen C., Korswagen H. C., An optimized dissociation protocol for FACS-based isolation of rare cell types from Caenorhabditis elegans L1 larvae. MethodsX 7, 100922 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hashimshony T., Wagner F., Sher N., Yanai I., CEL-seq: Single-cell RNA-seq by multiplexed linear amplification. Cell Rep. 2, 666–673 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Z., et al., Functional coordination of WAVE and WASP in C. elegans neuroblast migration. Dev. Cell 39, 224–238 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–10 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession no. GSE151216 (45).