Significance
Fasting in humans stimulates a transition from glucose to lipid metabolism via the induction of catabolic programs in response to increases in circulating glucagon and decreases in insulin. The transcription factor CREB and its coactivators, the CRTCs, stimulate the expression of gluconeogenic genes in liver. CREB and CRTC are conserved in Drosophila; they act primarily in neurons to promote energy balance. Mutant flies lacking the Crtc gene are more sensitive to starvation due to reductions in fat and glycogen stores. Using RNA sequencing, we identified CREB/CRTC regulated genes in flies that promote survival during starvation by inhibiting insulin signaling and promoting one-carbon metabolism. Our results provide insight into the mechanisms by which organisms maintain energy balance in response to fasting.
Keywords: CRTC, CREB, fasting metabolism
Abstract
Fasting in mammals promotes increases in circulating glucagon and decreases in circulating insulin that stimulate catabolic programs and facilitate a transition from glucose to lipid burning. The second messenger cAMP mediates effects of glucagon on fasting metabolism, in part by promoting the phosphorylation of CREB and the dephosphorylation of the cAMP-regulated transcriptional coactivators (CRTCs) in hepatocytes. In Drosophila, fasting also triggers activation of the single Crtc homolog in neurons, via the PKA-mediated phosphorylation and inhibition of salt-inducible kinases. Crtc mutant flies are more sensitive to starvation and oxidative stress, although the underlying mechanism remains unclear. Here we use RNA sequencing to identify Crtc target genes that are up-regulated in response to starvation. We found that Crtc stimulates a subset of fasting-inducible genes that have conserved CREB binding sites. In keeping with its role in the starvation response, Crtc was found to induce the expression of genes that inhibit insulin secretion (Lst) and insulin signaling (Impl2). In parallel, Crtc also promoted the expression of genes involved in one-carbon (1-C) metabolism. Within the 1-C pathway, Crtc stimulated the expression of enzymes that encode modulators of S-adenosyl-methionine metabolism (Gnmt and Sardh) and purine synthesis (ade2 and AdSl). Collectively, our results point to an important role for the CREB/CRTC pathway in promoting energy balance in the context of nutrient stress.
In Drosophila, adipokinetic hormone (AKH) and insulin-like peptides (dilps) function as orthologs of glucagon and insulin, respectively (1–6). AKH promotes fasting metabolism by binding to the AKH receptor, which is primarily expressed in the fat body, the fly equivalent of the adipose and liver (7). Ligand binding stimulates cellular cAMP accumulation and PKA activation.
Decreases in insulin signaling during fasting stimulate the dephosphorylation and nuclear translocation of the forkhead protein Foxo (8–12), which promotes the expression of gluconeogenic genes. In parallel, increases in cAMP/PKA signaling stimulate Crtc activity by phosphorylating and inhibiting the salt-inducible kinases (SIKs), which otherwise phosphorylate and sequester cAMP-regulated transcriptional coactivators (CRTCs) in the cytoplasm (13–16). Mammalian studies have revealed an important role for CREB and CRTC2 in promoting gluconeogenic gene expression in liver by binding to relevant promoters (13) in parallel with FOXO.
Crtc is not detectably expressed in Drosophila fat body, where fasting gluconeogenesis and lipolysis occur, however. Rather, Crtc is primarily produced in neurons, where it appears to promote energy balance and resistance to starvation, in part through induction of the neuropeptide Y homolog sNPF (17). In turn, sNPF appears to reduce energy expenditure by promoting gut epithelial integrity. Whether Crtc stimulates other metabolic programs that support starvation resistance is unclear, however.
In Drosophila, fasting up-regulates about 100 genes, many of which are essential for nutrient sensing and for glucose and lipid metabolism (18, 19). Here, we identify Crtc-regulated genes within the core set of fasting-inducible genes. Using flies with a knockout (KO) of Crtc, CrebB, or Sik2, we found a subset of fasting-inducible genes in the fly head that are down-regulated in both CrebB and Crtc KO flies and up-regulated in SIK2 mutants. Most of these genes are likely direct CrebB targets because they have promoter-proximal CrebB binding sites that are conserved among different Drosophila species. These fasting-inducible CrebB/Crtc target genes encode inhibitors of insulin signaling and secretion as well as components of one-carbon (1-C) metabolism that are also up-regulated in response to mitochondrial stress. These results point to a broader involvement of the CREB:CRTC pathway in coordinating the transcriptional response to starvation.
Results
CRTC Is Required for Starvation Resistance.
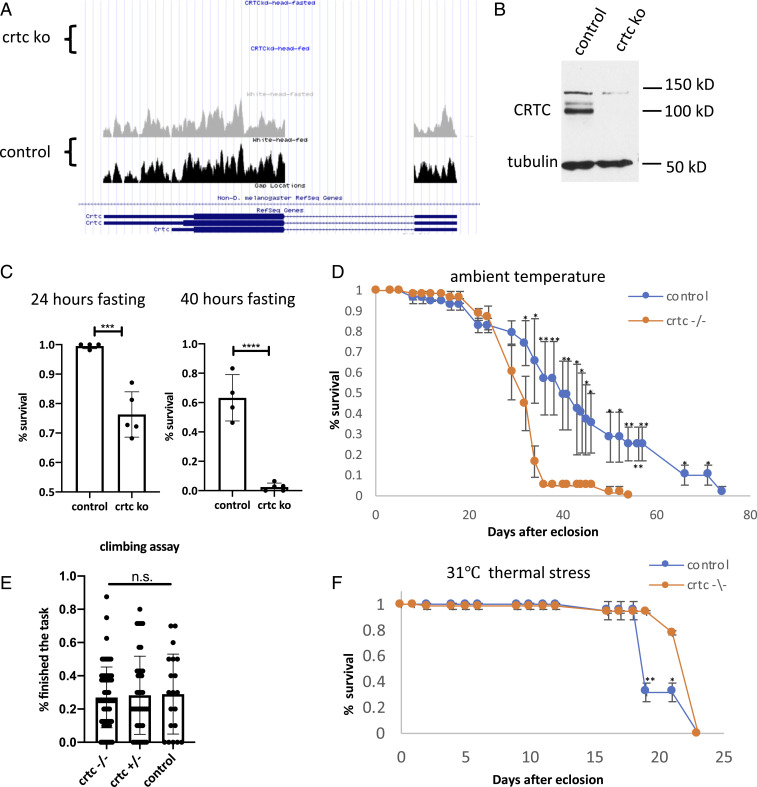
The timely induction of catabolic genes during fasting is critical in maintaining energy balance. Neuronal Crtc is necessary for resistance to starvation and oxidative stress in flies (15). Crtc mutant flies are sensitive to starvation and have shorter lifespans relative to wild type (Fig. 1 A–D). Crtc mutants exhibit normal body size, and they express dilp genes at levels comparable to wild type (SI Appendix, Fig. S1 A and B). Arguing against a sickly phenotype, Crtc mutants display comparable climbing behavior to wild-type flies (Fig. 1E) and they have increased resistance to thermal stress (Fig. 1F and SI Appendix, Fig. S1 C and D).
Fig. 1.
CRTC is required for starvation resistance. (A) UCSC genome browser view showing loss of Crtc transcripts in Crtc KO flies. (B) Western blot showing Crtc protein amounts in wild-type and Crtc KO flies. (C) Survival rates of w1118 control and Crtc KO flies after 24-h and 40-h fasting. (D) Comparison of lifespan at ambient temperature. Blue curve, wild-type flies. Yellow curve, Crtc KO flies. (E) Climbing assay showing mobility of Crtc KO flies relative to Crtc heterozygous flies and wild-type control flies. (F) Effect of 31 °C thermal stress on survival of Crtc KO compared to wild-type flies. ***P < 0.001; ****P < 0.0001; n.s., not statistically significant.
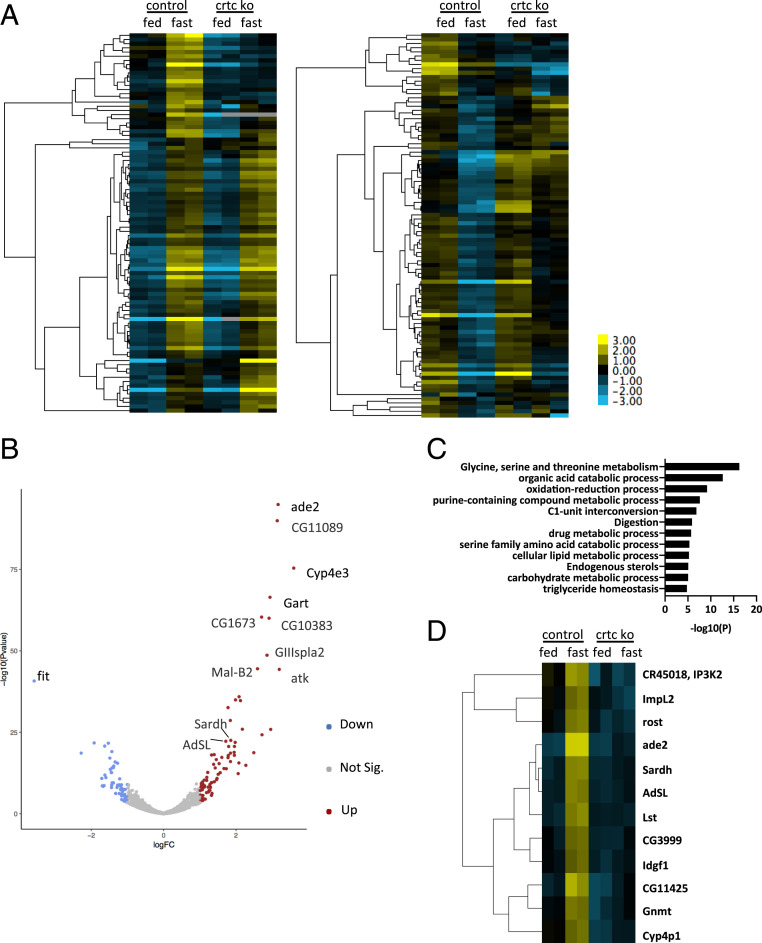
Based on the starvation sensitivity of Crtc mutant flies, we performed RNA sequencing (RNA-seq) studies to identify Crtc-regulated genes that mediate these effects in the head. This analysis revealed 91 genes that were up-regulated twofold or better (fragments per kilobase million [FPKM] ≥10) and 92 that were down-regulated after 16-h fasting relative to ad libitum feeding (Fig. 2A and SI Appendix, Tables S1 and S2).
Fig. 2.
CRTC is required for induction of fasting-inducible genes. (A) Heat map showing fasting-responsive genes in wild-type and Crtc KO flies. Conditions are indicated. The cutoff fold change >2 between feeding and fasting in wild-type flies; FPKM read >10 in at least one (fed/fasted) condition. Ninety-one fasting-inducible genes (Left) and 92 fasting-down-regulated genes (Right) in the control fly head identified under these criteria. Two biological replicates for control and Crtc KO for both feeding and fasting conditions. (B) Volcano plot showing fasting-responsive genes in wild-type fly heads. Blue and red dots in the volcano plot indicate genes with fold change >2 by fasting treatment and adjusted P value <0.05. Blue dots are fasting-down-regulated gene and red dots are fasting-up-regulated genes. (C) GO enrichment analysis with the core list of fasting-inducible genes. Multiple metabolic processes, including amino acid, lipid, and carbohydrate metabolic processes, are enriched by GO analysis. (D) Heat map showing relative expression of 12 Crtc-modulated fasting-inducible genes.
We also identified genes that are modulated during fasting with an adjusted P value of less than 0.05 (fold change ≥2; three replicates for feeding conditions and two replicates for fasting conditions). This analysis yielded 85 fasting-up-regulated genes and 60 fasting-down-regulated genes (Fig. 2B and SI Appendix, Tables S3 and S4). Genes identified by both strategies were assembled into a core list containing 70 fasting-inducible and 51 fasting-inhibited genes (Tables 1 and 2).
Table 1.
Core list of fasting-inducible genes in the fly head
| Gene name | Gene function |
| Aay | Phosphoserine phosphatase |
| ade2 | Phosphoribosylformylglycinamidine synthase |
| Adgf-D | Adenosine deaminase-related growth factor D |
| AdSL | Adenylosuccinate Lyase |
| Amy-d | Alpha-amylase |
| Amy-p | Alpha-amylase |
| Atk | Artichoke (atk) encodes a leucine-rich-repeat extracellular matrix protein |
| Bmm | Triacylglycerol lipase |
| Cyp4ac3 | Cytochrome P450-related |
| Cyp4e2 | Cytochrome P450-related |
| Cyp4e3 | Cytochrome P450-related |
| Cyp4p1 | Cytochrome P450-related |
| Cyp6a2 | Cytochrome P450-related |
| Cyp6a8 | Cytochrome P450-related |
| Cyp6w1 | Cytochrome P450-related |
| Etf-QO | Electron-transferring-flavoprotein dehydrogenase |
| Fbp | Fructose-bisphosphatase |
| GIIIspla2 | Phospholipase A2 |
| GLS | Glutaminase |
| Gnmt | Glycine N-methyltransferase |
| Idgf1 | Imaginal disk growth factor 1 |
| ImpL2 | Negative regulator of insulin signaling |
| Lip4 | Lipase |
| Lst | Negative regulator of insulin signaling |
| mag | Lipase |
| Mal-B2 | Alpha-glucosidase |
| Mcad | Medium-chain acyl-CoA dehydrogenase |
| Nmdmc | NAD-dependent methylenetetrahydrofolate dehydrogenase |
| Pepck2 | Phosphoenolpyruvate carboxykinase |
| pug | Formate–tetrahydrofolate ligase |
| Reg-2 | — |
| Sardh | Sarcosine dehydrogenase |
| Sclp | — |
| Shmt | Serine hydroxymethyl transferase |
| Sirup | Starvation-up-regulated protein |
| Spat | Serine pyruvate aminotransferase |
| Spn88Eb | — |
| Srr | Serine racemase |
| Thor | Translation regulation |
| Tsp42Ed | — |
| yip2 | Acetyl-CoA C-acyltransferase |
| CG10383 | Hydrolase |
| CG11089 | Phosphoribosylaminoimidazolecarboxamide formyltransferase |
| CG11425 | Phosphatidate phosphatase |
| CG11899 | Phosphoserine transaminase |
| CG14022 | Acylphosphatase |
| CG1441 | — |
| CG1673 | Branched-chain amino acid transaminase |
| CG16758 | Purine-nucleoside phosphorylase |
| CG3036 | — |
| CG32687 | — |
| CG34136 | — |
| CG3999 | Glycine dehydrogenase |
| CG42751 | — |
| CG42806 | — |
| CG42876 | — |
| CG5321 | Gamma-butyrobetaine dioxygenase |
| CG5953 | — |
| CG5955 | l-threonine 3-dehydrogenase |
| CG5966 | Carboxylesterase |
| CG6287 | Phosphoglycerate dehydrogenase |
| CG6767 | Phosphoribosyl pyrophosphate synthetase |
| CG7059 | — |
| CG7530 | — |
| CG7763 | — |
| CG8249 | — |
| CG8468 | — |
| CG8654 | — |
| CG9547 | Glutaryl-CoA dehydrogenase |
| CG9757 | — |
Table 2.
Core list of fasting-down-regulated genes in the fly head
| Gene name | Gene function |
| Cbt | Transcription factor |
| CCHa2 | Regulator of Dilps |
| Clect27 | Chitin Binding Protein |
| Cyp313a1 | — |
| ELOVL | Fatty acid elongase |
| fit | Satiety hormone |
| Gbs-70E | Glycogen binding subunit |
| GNBP-like3 | — |
| IM14 | Immune induced molecule |
| Listericin | Antibacterial protein |
| Lsp1beta | Larval serum protein |
| Lsp2 | Larval serum protein |
| MFS9 | Major Facilitator Superfamily Transporter 9 |
| Nplp2 | Neuropeptide-like precursor |
| Nplp3 | Neuropeptide-like precursor |
| Obp99b | Odorant-binding protein 99b |
| Ork1 | Open rectifier potassium channel |
| PGRP-SB1 | N-acetylmuramoyl-l-alanine amidase |
| phu | Alkaline phosphatase |
| Pisd | Phosphatidylserine decarboxylase |
| sug | Transcription factor |
| sxe2 | Carboxylesterase |
| Tig | Extracellular matrix protein |
| tobi | Target of brain insulin |
| TotX | Secreted peptides |
| vkg | A subunit of Collagen IV |
| CG10026 | — |
| CG10516 | — |
| CG11741 | — |
| CG13607 | — |
| CG14298 | — |
| CG14439 | — |
| CG14567 | — |
| CG14688 | — |
| CG15096 | — |
| CG15282 | — |
| CG16772 | — |
| CG18302 | — |
| CG3348 | — |
| CG3630 | — |
| CG4000 | — |
| CG42821 | — |
| CG4462 | — |
| CG4797 | — |
| CG5162 | Carboxylesterase |
| CG5773 | — |
| CG6126 | — |
| CG7203 | — |
| CG9416 | — |
| CG9436 | Alcohol dehydrogenase |
| CG9498 | — |
Gene Ontology (GO) analysis of these 70 fasting-up-regulated genes revealed an enrichment for metabolic pathways, including amino acid and purine metabolism, 1-C metabolism, redox regulation, and lipid and carbohydrate metabolism (Fig. 2C). Indeed, over 40% of fasting-inducible genes (29 of 70) are associated with metabolic pathways by Kyoto Encyclopedia of Genes and Genomes analysis (SI Appendix, Table S5).
CRTC Is Required for Induction of Fasting-Inducible Genes.
To identify metabolic programs that are modulated by Crtc, we selected for genes that are up-regulated by fasting in wild-type and that are down-regulated in Crtc mutant heads relative to control. This analysis yielded 12 putative CREB/CRTC target genes (Fig. 2D and SI Appendix, Table S6). In keeping with the proposed role of CREB, 11 of the 12 target genes have multiple conserved CREB binding sites within 2 kb upstream of the transcription start site (TSS) (SI Appendix, Table S7). A number of these genes, including Gnmt, Sardh, Ade2, and Adsl, encode enzymatic components of the 1-C metabolic pathway. Other genes within this pathway are also induced during fasting, but they appear to be CRTC-independent; these include ade3, pug, Nmdmc, CG11089, and Shmt (CG3011).
We sought to distinguish between genes that are dependent on Crtc uniquely during fasting from genes that are dependent on Crtc more generally under both feeding and fasting conditions. In the fed state, 123 genes are down-regulated in Crtc mutant relative to wild type (SI Appendix, Fig. S1E); in the fasted state, 117 genes are down-regulated in Crtc mutant heads (SI Appendix, Fig. S2A). Of these 117 genes, 70 are expressed at lower levels in Crtc mutants under both feeding and fasting conditions relative to wild type (SI Appendix, Table S8), while 46 are down-regulated in Crtc KO only during fasting (SI Appendix, Table S9). Although some changes in target gene expression are head-specific, others are also present in the body (SI Appendix, Fig. S2B). Nevertheless, fasting-inducible genes in the head appear to be functionally similar to those in the body by GO analysis (SI Appendix, Fig. S2C).
Based on the importance of CREB for recruitment of CRTCs to relevant targets, we evaluated whether the induction of Crtc-inducible genes during fasting is dependent on the Drosophila CREB homolog CrebB. For these experiments we used flies mutant for CrebBΔ400, a deletion in the transactivation and bZIP domains that disrupts the activity of this transcription factor (17). RNA-seq studies of fly head samples revealed six genes that are down-regulated in both Crtc and CrebB mutant flies during fasting (SI Appendix, Table S10). They are Adsl, Sardh, Lst, CR45018, ade2, and ImpL2.
Modulation of Fasting-Inducible Genes by Sik2.
The genes identified above using Crtc and CrebB mutants are likely to include direct targets of CrebB/Crtc as well as genes involved in potential downstream secondary effects. To help distinguish between these, we carried out a genomic analysis of the regulatory regions of the genes. Since CREB/CRTC promote target gene expression by binding to cAMP response elements (CREs), we performed a whole-genome analysis of palindromic CRE sites (TGACGTCA) as well as half-CRE sites (TGACG or CGTCA).
The Drosophila genome contains 2,062 palindromic sites and 141,688 half-CRE sites. To identify binding sites that are more likely to be functional, we examined their conservation across Drosophila species. For each CRE site, a conservation score was assigned according to the mean PhasCons score from 142 insects (20) (SI Appendix, Fig. S3A).
Using all RefSeq genes in the Drosophila DM6 database, we calculated CRE scores for promoters containing CRE sites within 2 kb upstream or 500 bp downstream of the TSS. The CRE scores were determined based on the presence of a full-site or half-site, the distance of the CRE site from the TSS, the conservation of the CRE site among different Drosophila species (20), and the number of sites on the promoter. CRE scores for the fly genes ranged from 0 to 4.43 (SI Appendix, Fig. S3B and Table S11); 85% of fly genes have either no consensus CRE or a poorly conserved CRE site (less than 1.20); 15% have multiple conserved CRE sites (CRE scores greater than or equal to 1.20) (SI Appendix, Fig. S3C).
Within the set of 70 core fasting-inducible genes, 16 have CRE scores greater than or equal to 1.20 (Table 3) and are therefore more likely to function as targets for CREB/CRTC. We found an enrichment of genes with higher CRE scores among fasting-inducible genes compared to genes in the whole genome (hypergeometric distribution P value 0.0241078; SI Appendix, Fig. S3D); 50% of Crtc-dependent fasting-inducible genes have CRE scores greater than 1.20 (SI Appendix, Table S7), suggesting that they are likely direct targets of CREB/Crtc.
Table 3.
Fasting-inducible genes with highly conserved CRE sites
| Gene | CRE score | Encoding |
| GLS | 2.73 | Glutaminase |
| Lst | 2.49 | Limostatin |
| ade2 | 1.99 | Phosphoribosylformylglycinamidine synthase |
| Pepck2 | 1.96 | Phosphoenolpyruvate carboxykinase 2 |
| Nmdmc | 1.85 | NAD-dependent methylenetetrahydrofolate dehydrogenase |
| Srr | 1.66 | Serine racemase |
| pug | 1.5 | Methylenetetrahydrofolate dehydrogenase, methenyltetrahydrofolate cyclohydrolase, formate-tetrahydrofolate ligase |
| CG5966 | 1.45 | Carboxylesterase |
| CG6767 | 1.42 | Ribose-phosphate diphosphokinase |
| ImpL2 | 1.4 | Imaginal morphogenesis protein-Late 2 |
| Adgf-D | 1.34 | Adenosine deaminase |
| CG11425 | 1.31 | Phosphatidate phosphatase |
| Gnmt | 1.26 | Glycine N-methyltransferase |
| Thor | 1.25 | A eukaryotic translation initiation factor 4E binding protein |
| CG1673 | 1.24 | Branched-chain amino acid transaminase |
| CG9547 | 1.2 | N.A. |
N.A., not assessed.
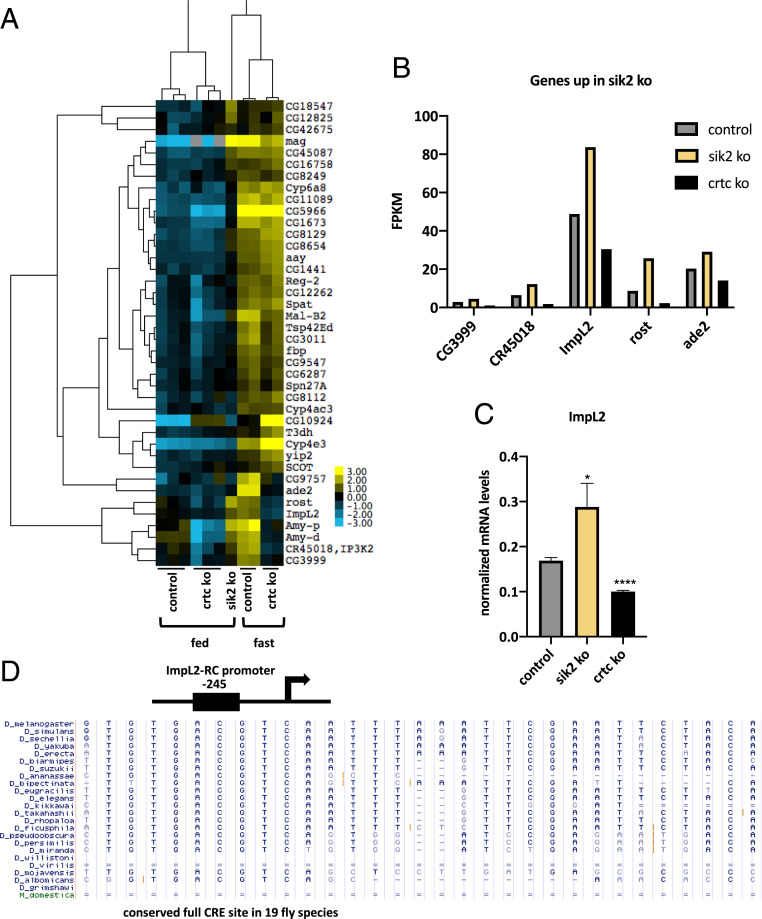
We wondered whether activation of Crtc itself would be sufficient to stimulate expression of fasting-inducible genes. CRTC activity is primarily modulated by SIKs, which phosphorylate and sequester CRTC in the cytoplasm under basal conditions through interaction with 14-3-3 proteins. In mammals, deletion of SIK genes leads to CRTC dephosphorylation, nuclear entry, and constitutive activation of CREB target genes. The fly has two SIK genes (Sik2 and Sik3), and Sik2 has been shown to promote the fasting response by modulating Crtc activity (16). We examined Crtc target gene expression in Sik2 mutant flies, in which the genomic region encoding the catalytic domain is deleted (SI Appendix, Fig. S3E). Sik2 mutants show increased starvation resistance, while Sik2 and Crtc double-mutant flies show starvation sensitivity similar to Crtc mutant flies (16), indicating that Sik2 regulates the fasting response at least in part via a Crtc-dependent mechanism.
Within the list of 91 fasting-inducible genes, 44% are up-regulated in Sik2 mutants under ad libitum feeding conditions (Fig. 3 A–C). Indeed, five of the Crtc-dependent genes are also more highly expressed in Sik2 mutants relative to wild type. Notably, ImpL2 and ade2 genes were identified on this list, both of which contain highly conserved CREs (Fig. 3D). These results indicate that loss of Sik2 in neurons is sufficient to induce expression of a subset of fasting-inducible genes that are targets of Crtc.
Fig. 3.
Modulation of fasting-inducible genes by Sik2. (A) Heat map of fasting-inducible genes that are up-regulated in heads from Sik2 KO flies under feeding conditions. Three biological replicates of head mRNA from control and Crtc KO in the fed state, head sample. Two biological replicates of control and Crtc KO in the fasted state. (B) Examples of CRTC-dependent fasting-inducible genes up-regulated in Sik2 mutant flies in the fed state. Bar graph shows data from RNA-seq studies. (C) RT-qPCR result showing up-regulation of CRTC-dependent fasting-inducible gene, ImpL2, in Sik2 mutant under feeding conditions. *P < 0.05; ****P < 0.0001. (D) Conserved CRE site in the ImpL2-RC promoter of 19 Drosophila species.
Crtc Stimulates Expression of Genes Involved in 1-C Metabolism and Inhibition of Insulin Signaling.
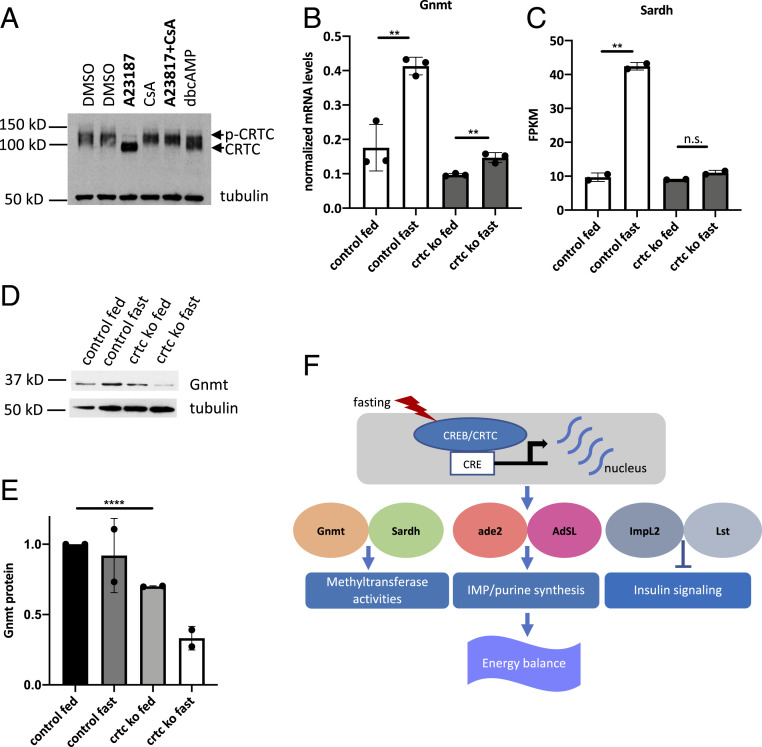
We explored the mechanism of Crtc-dependent induction of target genes. Drosophila Crtc is most similar to mammalian CRTC1 in that both are predominantly expressed in neurons. In mammalian cell lines, increases in intracellular cAMP or cytosolic calcium promote CRTC1 dephosphorylation and nuclear translocation (21). Crtc1 is activated in part by the Ser/Thr phosphatase calcineurin in response to calcium signaling. Exposure to the calcium ionophore, A23187, triggers CRTC1 activation (22).
We compared effects of cAMP and calcium agonists on Crtc activity. Although treatment with dibutyryl cAMP promoted dephosphorylation of CRTC, exposure to calcium ionophore (A23187) appeared more effective in Drosophila Kc167 cells (Fig. 4A). Moreover, Crtc dephosphorylation in response to A23187 was blocked by cotreatment with the calcineurin inhibitor cyclosporin A (CsA), indicating that, in addition to cAMP, increases in Ca2+ signaling also trigger the activation of Drosophila Crtc.
Fig. 4.
Crtc stimulates expression of genes involved in 1-C metabolism and inhibition of insulin signaling. (A) Western blot of Crtc following 0.5-h treatment with db/cAMP, A23187, CsA, or vehicle in Kc167 cells. (B) RT-qPCR assay showing Gnmt mRNA levels in adult fly head. (C) Sardh mRNA levels in adult fly head; genotypes and conditions shown. (D) Immunoblot showing GNMT protein levels in adult fly head. (E) Quantification of GNMT protein levels in adult fly head, normalized to tubulin. (F) Model of neuronal CRTC function in Drosophila. CRTC is activated in response to fasting signals, where it promotes energy balance by mediating induction of CREB target genes that function in 1-C metabolism and inhibition of insulin signaling. **P < 0.01; ****P < 0.0001; n.s., not statistically significant.
The subset of fasting-inducible genes that are CRTC-dependent includes Gnmt and Sardh, which encode glycine N-methyltransferase (Gnmt) and sarcosine dehydrogenase (Sardh), two key enzymes in the 1-C pathway that regulate S-adenosyl-methionine (SAM) metabolism. SAM serves as a methyl donor for a variety of methyltransferases, including enzymes involved in histone methylation. SAM levels are associated with lifespan and aging (23); overexpression of Gnmt, which catalyzes the conversion of SAM to sarcosine, promotes longevity, while decreases in Gnmt, as we observed in Crtc mutants, lead to shorter lifespans. Messenger RNA (mRNA) amounts for both Gnmt and Sardh are significantly down-regulated in Crtc KO flies during fasting relative to wild type (Fig. 4 B and C). Gnmt protein levels are also reduced in Crtc KO flies (Fig. 4 D and E). The dysregulation of Gnmt and Sardh expression in Crtc mutant flies may contribute to their shorter lifespan (Fig. 1D).
Also prominent within the subset of fasting-inducible CRTC targets are ade2 and AdSL, which encode 1-C pathway enzymes for de novo purine synthesis (24, 25). Remarkably, five of the seven genes encoding enzymes for IMP synthesis in Drosophila (Prat, Prat2, ade2, ade3, ade5, AdSL, and CG11089) are induced more than threefold by fasting in the fly head, and at least two of these, ade2 and AdSL, are putative CRTC targets (SI Appendix, Fig. S4A). Taken together, these studies suggest that CRTC modulates enzymes in the 1-C pathway that are required for protein methylation and purine synthesis (Fig. 4F).
In addition to the 1-C enzymes, CRTC targets also include Lst and Impl2 (SI Appendix, Fig. S4 A and B), which function as negative regulators of insulin secretion and signaling (26–32). Both genes have conserved CREB binding sites; they are induced in heads but not bodies of fasted flies (SI Appendix, Fig. S4B). The Crtc-dependent induction of these modulators of insulin action in the head during fasting is consistent with the inhibition of insulin signaling under starvation conditions.
Discussion
Starvation triggers the expression of catabolic programs that maintain energy balance in part by increasing lipid burning. The CREB coactivator Crtc has been shown to function importantly in starvation resistance (15); Crtc mutant flies have decreases in glycogen and triglyceride stores that are partially corrected upon reexpression of Crtc in neurons. To identify metabolic programs that are regulated by neuronal Crtc, we compared the transcriptomes of wild-type and Crtc mutant flies under feeding and fasting conditions. Nearly 20% of fasting-inducible genes in the head are modulated by Crtc. These are likely to include direct targets of CrebB/Crtc as well as genes involved in potential downstream secondary effects. Fifty percent of Crtc-dependent fasting-inducible genes have high CRE scores, suggesting that they are likely direct targets of CREB/Crtc. Future studies in which genes are acutely disrupted could additionally help resolve issues of primary vs. secondary effects.
Superimposed on its effects on sNPF expression (17), neuronal Crtc also appears to stimulate the expression of genes involved in 1-C metabolism (Gnmt, Sardh, ade2, and AdSL) and in inhibition of insulin secretion and signaling (ImpL2 and Lst). Within the set of 1-C enzymes, glycine N-methyltransferase (GNMT) has been found to regulate cellular concentrations of SAM, a methyl donor for many methyltransferase enzymes (33). GNMT reduces cellular SAM levels by converting it to sarcosine. In addition to its induction by Crtc during starvation, Gnmt is also up-regulated by FOXO in the fly fat body (34). Notably, Gnmt overexpression in fat body does not appear to increase starvation resistance, pointing to a potential role for Gnmt expression in other tissues. Reducing SAM levels extends lifespan in worms (35). SAM levels are increased with aging in flies (23). The down-regulation of Gnmt in Crtc mutant flies may contribute to their shorter lifespan. Future studies should reveal the extent to which changes in SAM alter neuronal function in response to fasting.
In addition to its effects on Gnmt and Sardh, Crtc was also found to regulate the expression of 1-C enzymes involved in purine biosynthesis. Mitochondrial impairment due to mutation of the kinase PINK1 promotes mitochondrial biogenesis via the induction of genes in the 1-C pathway (36). Indeed, a number of the same 1-C genes that are induced by fasting are also up-regulated in PINK1 flies. Similarly, knockdown of mitofusin expression causes an impairment in mitochondrial function that triggers up-regulation of the 1-C pathway in Drosophila (37). Changes in mitochondrial activity are thought to trigger a retrograde calcium signaling pathway that promotes expression of nuclear encoded genes for mitochondrial repair. We found that Crtc is efficiently dephosphorylated in response to calcium signals. Future studies should reveal whether induction of the 1-C pathway during fasting is important for mitochondrial biogenesis.
Crtc was also found to promote the expression of two inhibitors of insulin secretion and signaling. ImpL2 was identified as a Drosophila homolog of the mammalian insulin-like growth factor 7 (IGFBP7) that binds to dilp2 and antagonizes insulin action (27). In mammals, circulating IGF-binding proteins (IGFBPs) bind to IGFs, and this binding not only prolongs the half-lives of IGFs (38) but also modulates their activity. Seven IGFBPs (IGFBP1–7) have been identified in mammals; they compete with IGF receptors for binding to IGFs (39). The up-regulation of ImpL2 protein during fasting is detectable in the fly larval fat body; its up-regulation contributes to inhibition of IIS signaling. Our results indicate that the induction of ImpL2 transcription in the head during fasting is dependent on Crtc. Future studies should reveal the extent to which loss of Impl2 in neurons impairs energy balance via an effect on IIS signaling in the brain versus other peripheral tissues.
In addition to its effect on Impl2, Crtc was also found to inhibit insulin signaling via induction of limostatin (Lst), a decretin hormone that appears to block insulin secretion during fasting (32). Although Lst is produced in endocrine cells of the gut, our data indicate that Lst expression in the brain also contributes to effects of this hormone on energy balance.
Collectively, our results demonstrate that, in addition to its effects on sNPF expression and gut barrier integrity, the CREB/CRTC pathway promotes starvation resistance by modulating genes involved in insulin signaling and 1-C function. The identification of relevant Crtc positive neurons in the brain that express these target genes during fasting should provide further insight into this process.
Materials and Methods
Fly Stocks, Starvation Assay, and Climbing Assay.
Fly lines were maintained at 25 °C on standard food medium, except for starvation and thermal stress assays. Crtc and CrebB mutant flies were generated as described previously (15, 17). The control flies are w1118. Fasting treatment was conducted as described previously (15). Male flies 3 to 5 d old were transferred to vials containing 1% agar in phosphate-buffered saline, with filter papers soaked with water. For starvation assay, dead flies were scored every 4 to 8 h.
Male flies 3 to 5 d old were used in the climbing assay. The percentage of flies that finished the task, climbing 5 cm within 10 s, was recorded and compared among different genotypes.
Longevity Assay and Thermal Stress Condition.
Longevity assay was conducted as described previously (40), with some modifications. Newly eclosed adults, 20 to 25 flies per vial, were collected and maintained on standard food medium and transferred to new food twice weekly. For mortality analysis, deaths were scored five or six times every 7 d. For the thermal stress condition flies were maintained at 31 °C.
RNA Extraction and Purification.
Flies used for RNA extraction were collected and frozen in liquid nitrogen. Typically, each sample contains 100 flies. Separation of fly head and body was achieved by forcing the frozen sample through a mesh. One milliliter of TRIzol reagent was then added to each sample and samples were pestled quickly. The mixture was then incubated at room temperature for 5 min, followed by 12,000 rcf centrifugation for 10 min at 4 °C. The supernatant was then transferred to a new tube and 200 μL of chloroform was added. The sample was shaken vigorously by hand, followed by incubation at room temperature for 3 min. The mixture was then centrifuged at 10,000 rcf for 15 min at 4 °C. The upper aqueous phase was transferred to a new RNase-free tube and 0.5 mL of isopropanol was added. The mixture was incubated at room temperature for 10 min, followed by 12,000 rcf centrifugation for 10 min at 4 °C. The supernatant was then discarded and the pellet was washed with 1 mL of 75% ethanol. The mixture was then centrifuged at 7,500 rcf for 5 min at 4 °C. The supernatant was discarded and the sample was centrifuged again briefly to remove the last of liquid. The sample was air-dried for 10 min and the pellet was resuspended in 50 μL RNase-free water.
The extracted RNA was further purified using RNease kit (Qiagen); 350 μL buffer RLT and 250 μL 70% ethanol were mixed with the 50-μL RNA sample. The mixture was loaded on to an RNeasy column in a 2-mL collection tube and centrifuged for 15 s at 8,000 rcf. The column was transferred to a new collection tube and 500 μL buffer RPE was added. The mixture was centrifuged at 8,000 rcf for 15 s and 2 min, and flow-through was discarded after each centrifugation. The column was transferred to a new tube and centrifuged at full speed for 1 min to remove all remaining liquid. Fifty microliters of RNase-free water was added to each sample and the sample was incubated at room temperature for 2 min, followed by 1-min full-speed centrifugation. The RNA concentration was measured using Nanodrop.
RNA-Seq.
RNA-seq libraries were constructed using NEBNext-Ultra kits (New England Biolab) according to the manufacturer’s instructions. Quantitation of libraries was done by Qubit (Invitrogen) and run on a MiSeq instrument with paired-end 75-bp reads using v3 chemistry (Illumina). Data were analyzed by tophat2 and cuffdiff against the Drosophila DM3 genome build for heat maps and STAR and DESeq2 against the Drosophila DM6 genome build for volcano plots. The GEO accession number is GSE156577. GO enrichment analysis was performed on differentially expressed genes using Metascape or WebGestalt.
RT-PCR Analysis.
Total RNA was isolated using TRIzol reagent (Invitrogen), and 1 μg of RNA was used to make complementary DNA (cDNA) with a Transcriptor first-strand cDNA synthesis kit according to the manufacturer’s instructions (Roche). Relative mRNA expression was determined with SYBR green master mix (Roche) by Light-Cycler 480 qPCR machine (Roche). Relative mRNA expression was calculated by 2−ΔΔCT methods. Rp49 mRNA was used as housekeeping gene for mRNA.
The following qPCR primers were used in this study:
rp49-forward (F)-GCTAAGCTGTCGCACAAATG;
rp49-reverse (R)-GTTCGATCCGTAACCGATGT;
Gnmt-(F)-GCCTTCGATAAGTGGGTCAT, Gnmt-(R)-TGCTCCTGAATGTCGTCGTA;
ImpL2-(F)-GGCTCCAAGACCATCTATGC, ImpL2-(R)-GGCGTCAGACGAGAGGAG;
Lst-(F)-AACGGACAGCGTACTCTGATTT, Lst-(R)- TTGATTCGATAATGCGTTCG.
Cell Culture and Treatment.
Kc167 cells were cultured in CCM3 medium (HyClone). Cells were treated with A23187 (5 μM), db-cAMP (100 μM), and CsA (5 μM) as described in the figure legends. For Western blot analysis, cells were treated for 30 min; for RT-qPCR analysis, cells were treated for 2 h.
Western Blot.
Sodium dodecyl sulfate (SDS) protein loading buffer (2×) was added to cells or fly samples. The mixture was then sonicated, followed by 5-min denaturation at 95 °C. After 5-min centrifugation at 13,000 rpm, the supernatant was kept for SDS polyacrylamide gel electrophoresis analysis. The antibodies used were against CRTC, Gnmt, and tubulin (05-829; Millipore Sigma). CRTC antibody was generated as described previously (15). Gnmt antibody was kindly provided by Masayuki Miura, The University of Tokyo, Tokyo, Japan.
Statistical Analysis.
Data are presented as means ± SD. Statistical differences between two groups were determined by unpaired Student’s t test. P values less than 0.05 are considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01 DK083834, the Leona M. and Harry B. Helmsley Charitable Trust, the Clayton Foundation for Medical Research, and the Kieckhefer Foundation.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024865118/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in the NCBI Gene Expression Omnibus (accession no. GSE156577) (41).
References
- 1.Brogiolo W., et al., An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11, 213–221 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Rulifson E. J., Kim S. K., Nusse R., Ablation of insulin-producing neurons in files: Growth and diabetic phenotypes. Science 296, 1118–1120 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Kim S. K., Rulifson E. J., Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature 431, 316–320 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Grönke S., et al., Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 5, e137 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grönke S., Clarke D. F., Broughton S., Andrews T. D., Partridge L., Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 6, e1000857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh Y., et al., A glucose-sensing neuron pair regulates insulin and glucagon in Drosophila. Nature 574, 559–564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bharucha K. N., Tarr P., Zipursky S. L., A glucagon-like endocrine pathway in Drosophila modulates both lipid and carbohydrate homeostasis. J. Exp. Biol. 211, 3103–3110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greer E. L., Brunet A., FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 24, 7410–7425 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Gomis R. R., et al., A FoxO-Smad synexpression group in human keratinocytes. Proc. Natl. Acad. Sci. U.S.A. 103, 12747–12752 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang H., Regan K. M., Lou Z., Chen J., Tindall D. J., CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science 314, 294–297 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Greer E. L., et al., An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 17, 1646–1656 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bakker W. J., Harris I. S., Mak T. W., FOXO3a is activated in response to hypoxic stress and inhibits HIF1-induced apoptosis via regulation of CITED2. Mol. Cell 28, 941–953 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Koo S. H., et al., The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437, 1109–1111 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Dentin R., et al., Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449, 366–369 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Wang B., et al., The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab. 7, 434–444 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi S., Kim W., Chung J., Drosophila salt-inducible kinase (SIK) regulates starvation resistance through cAMP-response element-binding protein (CREB)-regulated transcription coactivator (CRTC). J. Biol. Chem. 286, 2658–2664 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen R., et al., Neuronal energy-sensing pathway promotes energy balance by modulating disease tolerance. Proc. Natl. Acad. Sci. U.S.A. 113, E3307–E3314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zinke I., Schütz C. S., Katzenberger J. D., Bauer M., Pankratz M. J., Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO J. 21, 6162–6173 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grönke S., et al., Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 1, 323–330 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Margulies E. H., Blanchette M., Haussler D., Green E. D.; NISC Comparative Sequencing Program , Identification and characterization of multi-species conserved sequences. Genome Res. 13, 2507–2518 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bittinger M. A., et al., Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr. Biol. 14, 2156–2161 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Altarejos J. Y., et al., The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nat. Med. 14, 1112–1117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obata F., Miura M., Enhancing S-adenosyl-methionine catabolism extends Drosophila lifespan. Nat. Commun. 6, 8332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henikoff S., Multifunctional polypeptides for purine de novo synthesis. BioEssays 6, 8–13 (1987). [DOI] [PubMed] [Google Scholar]
- 25.Holland C., Lipsett D. B., Clark D. V., A link between impaired purine nucleotide synthesis and apoptosis in Drosophila melanogaster. Genetics 188, 359–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sloth Andersen A., Hertz Hansen P., Schäffer L., Kristensen C., A new secreted insect protein belonging to the immunoglobulin superfamily binds insulin and related peptides and inhibits their activities. J. Biol. Chem. 275, 16948–16953 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Honegger B., et al., Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J. Biol. 7, 10 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arquier N., et al., Drosophila ALS regulates growth and metabolism through functional interaction with insulin-like peptides. Cell Metab. 7, 333–338 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Sarraf-Zadeh L., et al., Local requirement of the Drosophila insulin binding protein imp-L2 in coordinating developmental progression with nutritional conditions. Dev. Biol. 381, 97–106 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Bader R., et al., The IGFBP7 homolog Imp-L2 promotes insulin signaling in distinct neurons of the Drosophila brain. J. Cell Sci. 126, 2571–2576 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Roed N. K., et al., Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones. Nat. Commun. 9, 3860 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alfa R. W., et al., Suppression of insulin production and secretion by a decretin hormone. Cell Metab. 21, 323–334 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrossian T. C., Clarke S. G., Uncovering the human methyltransferasome. Mol. Cell. Proteomics 10, 000976 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obata F., et al., Necrosis-driven systemic immune response alters SAM metabolism through the FOXO-GNMT axis. Cell Rep. 7, 821–833 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Cabreiro F., et al., Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153, 228–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tufi R., et al., Enhancing nucleotide metabolism protects against mitochondrial dysfunction and neurodegeneration in a PINK1 model of Parkinson’s disease. Nat. Cell Biol. 16, 157–166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrido-Maraver J., et al., Enhancing folic acid metabolism suppresses defects associated with loss of Drosophila mitofusin. Cell Death Dis. 10, 288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guler H. P., Zapf J., Schmid C., Froesch E. R., Insulin-like growth factors I and II in healthy man. Estimations of half-lives and production rates. Acta Endocrinol. (Copenh.) 121, 753–758 (1989). [DOI] [PubMed] [Google Scholar]
- 39.Allard J. B., Duan C., IGF-binding proteins: Why do they exist and why are there so many? Front. Endocrinol. (Lausanne) 9, 117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Broughton S. J., et al., Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. U.S.A. 102, 3105–3110 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T., Wiater E., Zhang X., Thomas J. B., Montminy M., Drosophila CRTC promotes starvation resistance by modulating one-carbon metabolism and inhibition of insulin signaling. NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156577. Deposited 20 August 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the NCBI Gene Expression Omnibus (accession no. GSE156577) (41).