Significance
Virtually all above-ground plant surfaces, such as leaf and stem exteriors, are covered in a cuticle: a wax-infused polyester. This waxy biocomposite is the largest interface between Earth’s biosphere and atmosphere. Its chemical composition is not only highly tuned to mediate nonstomatal water loss, but it also self-assembles to produce superhydrophobic surfaces, protects against UV radiation, and contains bioactive compounds that help resist microbial attack. Developing fundamental knowledge of waxy biocomposites, particularly those on crop species, is a prerequisite for an understanding of their structure–function relationships. Here, we uncover a likely genetic basis for the presence and absence, respectively, of triterpenoids in the leaf waxes of sorghum and maize—compounds previously associated with creating heat-tolerant cuticular water barriers.
Keywords: cuticular wax, drought tolerance, triterpenoids, juvenile-to-adult transition, sorghum bicolor
Abstract
Virtually all land plants are coated in a cuticle, a waxy polyester that prevents nonstomatal water loss and is important for heat and drought tolerance. Here, we describe a likely genetic basis for a divergence in cuticular wax chemistry between Sorghum bicolor, a drought tolerant crop widely cultivated in hot climates, and its close relative Zea mays (maize). Combining chemical analyses, heterologous expression, and comparative genomics, we reveal that: 1) sorghum and maize leaf waxes are similar at the juvenile stage but, after the juvenile-to-adult transition, sorghum leaf waxes are rich in triterpenoids that are absent from maize; 2) biosynthesis of the majority of sorghum leaf triterpenoids is mediated by a gene that maize and sorghum both inherited from a common ancestor but that is only functionally maintained in sorghum; and 3) sorghum leaf triterpenoids accumulate in a spatial pattern that was previously shown to strengthen the cuticle and decrease water loss at high temperatures. These findings uncover the possibility for resurrection of a cuticular triterpenoid-synthesizing gene in maize that could create a more heat-tolerant water barrier on the plant’s leaf surfaces. They also provide a fundamental understanding of sorghum leaf waxes that will inform efforts to divert surface carbon to intracellular storage for bioenergy and bioproduct innovations.
Demand for both food and energy are projected to increase substantially over the coming decades (1). Meeting these needs while minimizing negative impacts on our environment, health, and fresh water supply is one of the largest challenges currently faced by humanity. A component of meeting this challenge is identifying, improving, and deploying high-yield, low-input, multiuse crops (2). One candidate is sorghum (Sorghum bicolor), a multiuse C4 grass crop. Sorghum is exceptionally tolerant of hot and dry climates, being native to arid regions (3, 4), and the genus contains considerable natural diversity that offers great potential for the improvement of existing biomass and grain varieties, as well as sequenced landraces that are adapted to specific, relatively extreme environments (5, 6). Sorghum has a relatively small, sequenced genome (7), its grain is naturally gluten-free (8), and it can be genetically transformed (9, 10). Thus, sorghum is an excellent study system for advancing our understanding of the physiological basis for crop drought tolerance and heat resistance. This is particularly true when comparisons are made between sorghum and its close relative maize (Zea mays), one of the most widely grown crops in the world, but also a crop that suffers major yield losses due to drought. Indeed, investigators in this area have identified a variety of physiological mechanisms underlying sorghum’s positive qualities (11–14). However, there is still a critical need for mechanistic details underlying these processes and how such differ between sorghum and maize. Until these gaps are filled, it will be difficult to further enhance sorghum’s positive qualities, and it will not be possible to transfer these qualities to other crop species such as maize.
Though plants deploy diverse mechanisms to protect themselves against drought and heat, one of the most widespread (found in essentially all land plants), is a hydrophobic, aerial surface coating called the cuticle. This structure is composed of a fatty acid-derived polyester scaffold called cutin, inside and on top of which accumulates wax, a mixture of hydrophobic compounds that seal the surface against the movement of water (15). While cutin makes major contributions to a cuticle’s biomechanical properties and, at least in some species, contributes to pathogen resistance (16, 17), waxes are primarily responsible for preventing water from crossing the leaf–atmosphere interface (18, 19). It is well established that wax mixtures vary between plant species, organs, tissues, cell types, and across leaf developmental stages (20–23). This suggests wax mixtures are tuned to meet the challenges faced by specific plant surfaces in their immediate environment. This notion is supported by experiments on transgenic plants with altered wax amounts and/or wax mixture compositions: They exhibit altered water barrier properties (18, 24–27). These studies also demonstrate that plant drought tolerance may be improved through cuticle engineering, thus further underscoring the importance of understanding the waxes of a drought tolerant plant like sorghum.
Based on the importance of sorghum waxes, the primary objective of this study was to develop our fundamental knowledge in this area, particularly in a comparative context with maize. Previous work has demonstrated that the juvenile-to-adult transition, a developmental event in a plant’s life cycle, can be associated with major changes in surface wax characteristics (28, 29). Accordingly, this investigation began by analyzing sorghum leaf waxes over the course of the plant’s development, then aimed to understand underlying biosynthetic processes as well as the evolutionary history of leaf wax biosynthesis in sorghum, all relative to analogous processes in maize. To accomplish this, the present study combined detailed chemical analyses of the sorghum leaf surface, bioinformatics-guided characterization of sorghum wax synthesis genes by heterologous expression, as well as ancestral state reconstruction and a comparative genomics analysis of critical sorghum wax synthesis genes across six grass species, including sorghum and maize. Considered together, the results indicate that sorghum has maintained a form of wax synthesis likely preset in the common ancestor of maize and sorghum that relies on a neofunctionalized steroid biosynthesis gene capable of generating uncommon triterpenoid wax chemicals. Our analyses show that this gene is present in the maize genome but is truncated and not expressed. This, together with reports of triterpenoids being absent from the maize leaf surface (30), suggest a likely genetic basis for the observed divergence in sorghum and maize leaf wax chemistry.
Results and Discussion
Wax Mixtures from Leaves of Adult Sorghum Plants Contain Uncommon Hopane Triterpenoids.
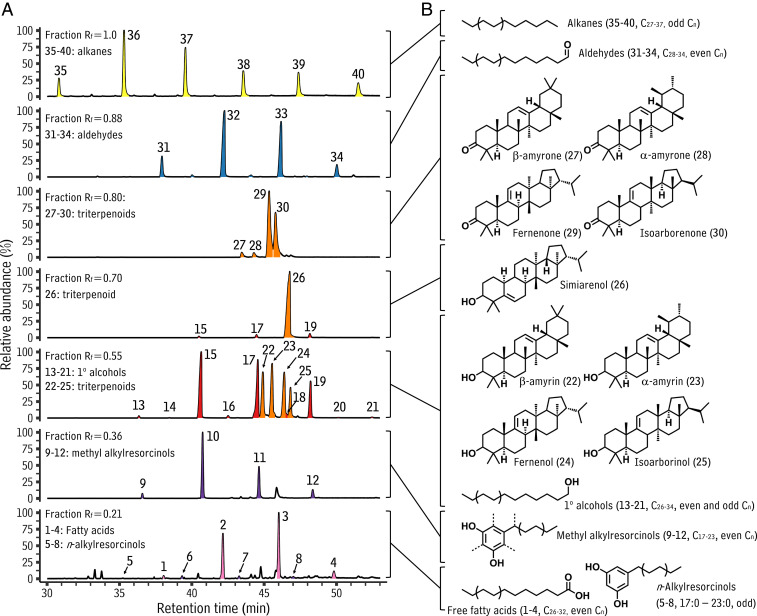
The first objective of this study was to identify the chemical constituents on S. bicolor leaf surfaces. For this, the highest leaf with an exposed ligule on each of 13 sorghum plants 14 to 98 d of age was collected, pooled, and waxes were extracted. An aliquot of this extract was separated using thin layer chromatography (TLC), resulting in seven prominent bands that were each subsequently removed from the plate. The contents of each band were then analyzed using gas chromatography-mass spectrometry (GC-MS), resulting in the detection of 40 different wax compounds (compounds 1 through 40, Fig. 1A). The identities of these 40 compounds were determined by comparing their mass spectra against published mass spectra of authentic standards and using multiple derivatization reactions, as described briefly below.
Fig. 1.
Leaf wax constituents from S. bicolor. (A) Line plot depicting total ion GC-MS chromatograms of TLC-separated waxes of pooled S. bicolor leaves from plants at a range of ages (14 to 98 d after planting). Plotted is the relative abundance (y axis) of the signal as a function of retention time (x axis). Peak fill corresponds to compound class, yellow: alkanes, blue: aldehydes, orange: triterpenoids, red: primary alcohols, purple: methyl and n-alkylresorcinols, and pink: fatty acids. (B) Diagram showing the structure of each compound or homologous series identified, each of which is linked, using brackets, to the TLC fraction in which it was most abundant.
The first TLC fraction (Rf = 0.21) contained a homologous series of free fatty acids (compounds 1 to 4, even total carbon numbers C26 to C32; Fig. 1A, pink peaks), as well as a second homologous series with prominent mass spectral peaks at m/z 268 (SI Appendix, Fig. S1) that identified them as n-alkylresorcinols (compounds 5 to 8, 17:0 to 23:0; Fig. 1A, purple peaks). The second fraction (Rf = 0.36) contained a homologous series with prominent mass spectral peaks at m/z 282 (SI Appendix, Fig. S2) that identified them as methyl alkylresorcinols (compounds 9 through 12, 17:0 to 23:0; Fig. 1A, purple peaks). The exact position of the methyl group in these structures could not be determined by our analyses.
The third TLC fraction (Rf = 0.55) contained a combination of homologous primary alcohols (compounds 13 to 21, even and odd total carbon numbers C26 to C34; Fig. 1A, red peaks) and four triterpenoids (compounds 22 to 25; Fig. 1A, orange peaks). Two of these triterpenoids had mass spectra that identified them as the common triterpenoid alcohols β-amyrin and α-amyrin (compounds 22 and 23, Fig. 1A, orange peaks). Compounds 24 and 25 had the same Rf values as the amyrins (Fig. 1A) and mass spectral features suggesting triterpenoid structures, including prominent peaks at m/z 241 (SI Appendix, Fig. S3). These suggested triterpenoid alcohol structures with double bonds between C-9 and C-11, thus matching the molecular features of fernenol and isoarborinol, compounds previously reported as constituents of sorghum leaf wax mixtures via the analysis of their acetate derivates (31, 32). To verify these structures, compounds 24 and 25 were acetylated and analyzed again. The mass spectra of their acetate derivatives matched those of fernenol and isoarborinol (SI Appendix, Fig. S3), confirming these structural assignments.
The fourth TLC fraction (Rf = 0.70) contained a compound with a mass spectrum matching that of simiarenol (SI Appendix, Fig. S4), a previously reported sorghum leaf wax compound (31) and was thus identified as such (compound 26; Fig. 1A, orange peaks). The fifth fraction (Rf = 0.80) contained four compounds, two of which, compounds 27 and 28, had mass spectra identifying them as β-amyrone and α-amyrone (SI Appendix, Fig. S5), and thus oxidized forms of the co-occurring (but not TLC comigrating) pair of triterpenoid alcohols β-amyrin and α-amyrin. Compounds 29 and 30 had the same Rf values as the amyrones and mass spectral features suggesting triterpenoid structures, including prominent peaks at m/z 241, suggesting these also as ketonic forms of another pair of sorghum triterpenoid alcohols, likely fernenol and isoarborinol. To test this hypothesis, compounds 29 and 30 were reduced with NaBH4. This resulted in compounds with retention times and mass spectra identical to fernenol and isoarborinol (SI Appendix, Fig. S6), and thus identifying compounds 29 and 30 as fernenone and isoarbornone. The sixth fraction (Rf = 0.88) contained a homologous series of four very-long-chain aldehydes (compounds 31 to 34, even total carbon numbers C28 to C34; Fig. 1A, blue peaks). The final fraction (Rf = 1.0) contained a homologous series of six n-alkanes (compounds 35 to 40, odd total carbon numbers C27 to C37; Fig. 1A, yellow peaks).
Thus, 40 different sorghum leaf wax compounds were identified, including 1) very-long-chain linear aliphatics that varied in head group oxidation state and chain length; 2) alicyclic alkylresorcinols that varied in chain length and methylation status; and 3) cyclic triterpenoid compounds that varied in oxidation state and backbone configuration, specifically as amyrin and hopane triterpenoid alcohols and ketones. Very-long-chain linear aliphatics are ubiquitous in plant cuticular wax mixtures. In contrast, cuticular alkylresorcinols and triterpenoids occur in species-specific patterns. Alkylresorcinols similar to those reported here have been detected in wax mixtures of, for example, Triticum aestivum (33) and Secale cereale (34, 35); and amyrin alcohols and ketones are found in the wax mixtures of, for example, Artemisia annua (36), Citrus paradisi (37), and Ilex aquifolium (38). Wax mixtures with compounds similar to the sorghum surface hopane triterpenoids are uncommon and seem to only have been reported from Euphorbia peplus so far (39). Thus, sorghum wax mixtures contain compounds that are ubiquitous as well as quite rare. This prompted a more detailed investigation of how such may relate to potential changes in cuticular wax mixtures associated with sorghum’s juvenile-to-adult transition.
The Juvenile-To-Adult Transition in Sorghum, Unlike in Maize, Is Characterized by a Shift to Triterpenoid-Dominated Surface Chemistry.
With the major wax compounds that sorghum leaves can generate identified, the next goal was to examine specific wax mixtures on leaves produced by sorghum plants at different stages of development. For this, leaf tips were removed from the newest leaf with an exposed ligule on multiple S. bicolor plants each week over the course of plant development from 14 to 98 d after planting. In total, the cuticular waxes from 74 leaf tissue samples were extracted and analyzed using GC-MS. In these samples, it was possible to identify and quantify all of the compounds found in the TLC fractions except C32 aldehyde (which coeluted with high abundance triterpenoids) as well as 1) C34 aldehyde; 2) the four primary alcohols with odd total carbon numbers; 3) the two amyrones; and 4) the four n-alkylresorcinols, all of which had very low abundances in the total wax mixtures. In addition, C24 fatty acid was also detected and quantified, bringing the total number of compounds quantified in each sample to 29 and the total number of measurements in the dataset to more than 2,100. A principal components analysis revealed that the major drivers of variance in this dataset were the wax chemical characteristics of samples from juvenile versus adult plants (SI Appendix, Fig. S7).
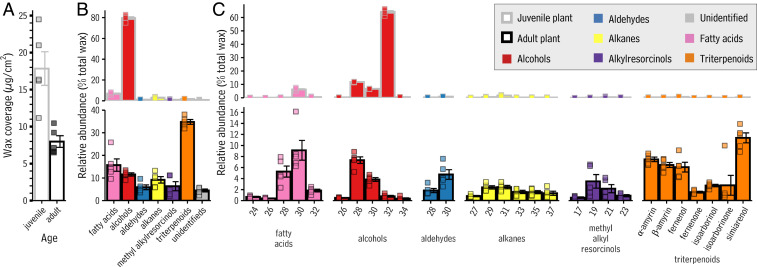
To define the chemical characteristics of the wax mixtures on leaves produced by juvenile versus adult sorghum plants, wax compounds on the newest leaf with an exposed ligule of the five youngest plants (between 14 and 21 d after planting) and the newest leaf with an exposed ligule of the five oldest plants (between 88 and 96 d after planting) were investigated. Leaves produced by juvenile plants bore 17.8 ± 2.3 μg/cm2 wax (Fig. 2A and Dataset S1), which was composed almost entirely of primary alcohol compounds (84.4% of total wax on leaves of juvenile plants; Fig. 2 B, Top), together with small amounts of fatty acids (7.3%). Within the alcohol compound class, the C32 homolog dominated (Fig. 2C, Top), and within the fatty acid compound class the C30 homolog was the most abundant.
Fig. 2.
Coverage and composition of wax on juvenile and adult leaves of S. bicolor. (A) Bar chart showing total wax coverage (in μg/cm2; y axis) on leaves produced by juvenile (Left bar) and adult (Right bar) S. bicolor plants. Each bar is given a border to reflect plant maturity, where those colored gray correlate to data for juvenile plants and those bordered black are adult plants. (B) Bar chart showing the relative abundance (percent total wax from that leaf; y axis) of each compound class (x axis) present on leaves produced by juvenile (Top, gray bordered bars) and adult (Bottom, black bordered bars) S. bicolor plants. Bars are color filled according to compound class, and as defined in Fig. 1A: yellow: alkanes, blue: aldehydes, orange: triterpenoids, red: primary alcohols, purple: methyl alkylresorcinols, pink: fatty acids, and gray: unidentified. (C) Bar chart showing the relative abundance (percent total wax from that leaf; y axis) of each compound identified on juvenile (Top, gray bordered bars) and adult (Bottom, black bordered bars) leaves of S. bicolor. Numbers below each bar indicate the total carbon number of that compound, and the text below each group of bars indicates the compound class represented by those bars. The chemical structures of these compounds are presented in Fig. 1B. Throughout, bar heights and error bars represent the mean and SE of five independent samples, respectively.
Leaves produced by adult plants yielded 8.0 ± 0.8 μg/cm2 wax (Fig. 2A), which was made up primarily of triterpenoids (39.6%; Fig. 2 B, Bottom), together with fatty acids (17.8%), alcohols (13.2%), alkanes (10.4%), methyl alkylresorcinols (7.3%), aldehydes (6.7%), and a small amount of wax compounds that could not be identified (5.1%). The triterpenoid compound class was dominated by simiarenol (11.7% of the total wax; Fig. 2 C, Bottom), followed by α-amyrin (7.7%), β-amyrin (6.7%), and fernenol (6.2%) as well as fernenone, isoarborinol, and isoarborninone (2 to 3% each). Within the fatty acid, aldehyde, and alcohol classes the C28 and C30 homologs were most abundant, the C29 and C31 homologs had a slight predominance in the alkane class, and the C19:0 and C21:0 homologs were most abundant in the methyl alkylresorcinol class. Waxes from leaves of mature sorghum plants grown in a field and a greenhouse under similar conditions were nearly identical (SI Appendix, Fig. S14).
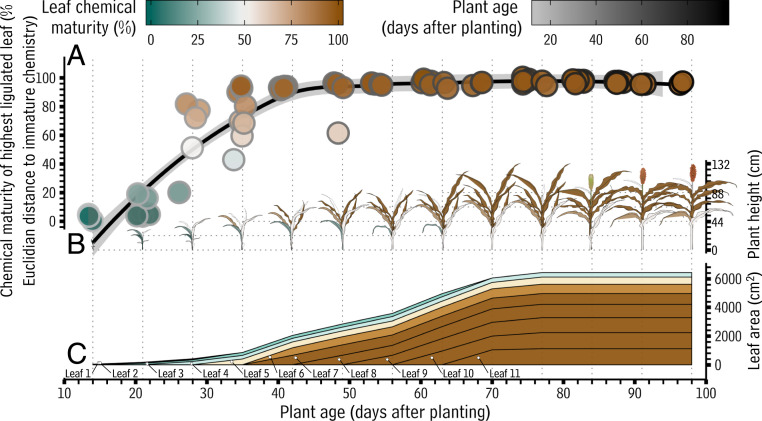
Typically, a sorghum plant produces five to six juvenile phase leaves before transitioning to adulthood and producing adult phase leaves. So far, the data presented here indicated there was a considerable difference in the chemical composition of leaves produced by juvenile versus adult S. bicolor plants, but it was still unclear when during development this shift took place. To determine when these changes occurred in the context of plant development, each of the 74 leaf wax samples was placed on a scale defined by the Euclidian distance from juvenile to adult chemistry: end points that were defined by the average chemical composition of the samples from the five youngest and five oldest plants, respectively (i.e., the compositions shown in Fig. 2). On this scale, a sample with wax chemistry very similar to that of the leaves from the youngest plants in the dataset will have a low Euclidian distance from juvenile cuticular chemistry (∼0%). Conversely, a sample with wax chemistry similar to the waxes from leaves produced by adult plants will have a high Euclidian distance from juvenile cuticular chemistry (∼100%). The position of each sample on this scale was then examined as a function of plant age. Until 21 d after planting (the five-leaf stage, three leaves with exposed ligules), leaves produced by S. bicolor were covered with wax mixtures very similar to the leaves from the youngest plants tested (less than 20% distant on the Euclidian scale; Fig. 3A). At 28 d after planting (the six-leaf stage, four leaves with exposed ligules), some plants had produced leaves with intermediate wax compositions (ranging from 20 to 80% distant from juvenile chemistry on the Euclidian scale). Following 42 d after planting (the seven-leaf stage, five leaves with exposed ligules), virtually all leaves produced by S. bicolor had chemistry largely indistinguishable from that of the chemistry covering the highest leaves with exposed ligules of the oldest plants sampled (∼100% distant from juvenile chemistry on the Euclidian scale). This revealed that the shift from the production of leaves with juvenile chemistry to leaves with adult chemistry occurs largely between 25 and 42 d after planting and is monotonic. We also examined the abundances of each chemical compound before, during, and after this shift. This revealed that free fatty acids, in addition to primary alcohols and triterpenoids, underwent a considerable shift in abundance during the juvenile-to-adult transition (SI Appendix, Fig. S15). This may be attributable to the layer of visible epicuticular wax crystals that can, on mature leaves of some sorghum varieties, form on portions of the abaxial surface. Though the triterpenoid class as a whole is more than twice as abundant on mature sorghum leaves than the fatty acid class, it cannot be ruled out that these fatty acids contribute to leaf wax function.
Fig. 3.
Surface area covered by juvenile versus adult wax chemistry on developing S. bicolor plants. In this figure, a previously defined color scheme (gray to black: juvenile-to-adult plant age) is used along with a new scheme (green to brown). The new scheme defines the progression of leaf chemical maturity, rather than plant age, as indicated in the green to brown legend. This legend indicates the meaning of the colors of 1) the points in A, 2) the leaves in B, and 3) the layers in C. (A) Scatterplot showing the surface wax chemical maturity of the highest leaf with an exposed ligule on developing S. bicolor plants. Each point represents one gas chromatography-mass spectrometry sample prepared from one sorghum leaf, with each being plotted according to its Euclidian distance from juvenile to adult chemistry (“leaf chemical maturity,” Left y axis, juvenile and adult chemistry defined as the profiles shown in Fig. 2C). These points are also plotted as a function of the age of the plant from which the sample was taken (“plant age,” x axis). Thus, each point represents with both color and y position the chemical maturity of the leaves with the same color at the same position on the x axis in the plant diagrams below. (B) Diagram showing plant height (in cm, Top Right y axis) at each week during development. Leaf color corresponds to the chemical maturity of the leaf’s surface, as indicated in the legend. Plant images in B were reprinted with permission from Kansas State University Agricultural Experiment Station and Cooperative Extension Service. (C) Area chart depicting the cumulative leaf surface area (cm2; Lower Right y axis) of a sorghum plant over the course of development. Each leaf is represented with a track, as indicated with the “leaf 1”–“leaf 11” labels. The height of each track corresponds to the area of that leaf as a function of time, with the total height of all tracks corresponding to the total leaf area of the plant. Each track is shaded according to the chemical maturity of that leaf’s surface, with green corresponding to juvenile leaf chemistry and brown corresponding to adult leaf chemistry, as indicated in the legend. Leaf surface areas shown are the average of three independent plants.
There are drastic differences in the size of the sorghum leaves bearing juvenile chemistry versus adult surface chemistry (Fig. 3B). Since leaf chemistry is a surface phenomenon and thus distributed over a particular surface area, this raised the question of what proportion of a sorghum plant’s surface area, over the course the plant’s life, is protected by juvenile versus adult leaf chemistry. To answer this, the average surface area of each leaf on a sorghum plant over the course of development was measured. This information was used to determine the surface area covered by juvenile versus adult chemistry. Leaves 1 through 4 (counting from the first leaf with an exposed ligule that appears on the plant) had essentially juvenile surface chemistry (less than 15% triterpenoids, SI Appendix, Fig. S16) and reached an average maximum surface area of 149 cm2 (Fig. 3C and Dataset S2). Typically, many of these leaves died and fell off the plants around 40 to 70 d after planting. Generally speaking, leaf 5 had an intermediate chemical phenotype (around 20 to 25% triterpenoids, SI Appendix, Fig. S16; a roughly 1:1 mixture of the chemical phenotypes of juvenile and mature leaves, SI Appendix, Fig. S17) and reached an average maximum size of 504 cm2. Leaves 6 through 11 had essentially adult surface chemistry (triterpenoids as the majority compound class at >30% of the total wax, SI Appendix, Fig. S16) and reached an average maximum size of 935 cm2. Thus, the vast majority (>80%) of sorghum leaf surface area over time is protected by a triterpenoid-rich wax mixture.
Overall, our chemical analyses revealed that leaves made by juvenile plants had around twice the total amount of wax as leaves made by adult plants and were covered mainly with C32 primary alcohols, while leaves made by adult plants were covered with a mixture dominated by pentacyclic triterpenoids, chiefly simiarenol. A shift in wax coverage and composition has also been observed as part of the maize juvenile-to-adult transition. Like sorghum, leaves of juvenile maize plants also bear more wax than their adult counterparts and are covered mainly with C32 primary alcohols (40). However, after the transition to adulthood, maize plants produce leaves covered with alkyl esters and alkanes (30). On leaves made by adult maize plants, triterpenoids are trace compounds. Thus, sorghum is markedly different from maize in that the majority of its surface area over time is protected by triterpenoid-rich surface chemistry, rather than a chemistry dominated by very-long-chain aliphatic compounds. This difference prompted an investigation of the genetic basis for sorghum triterpenoid biosynthesis, and how it may differ from that of maize.
The Majority of Sorghum Cuticular Triterpenoids Are Synthesized by a Neofunctionalized Steroid Biosynthesis Gene that Is Present, but Disrupted, in the Maize Genome.
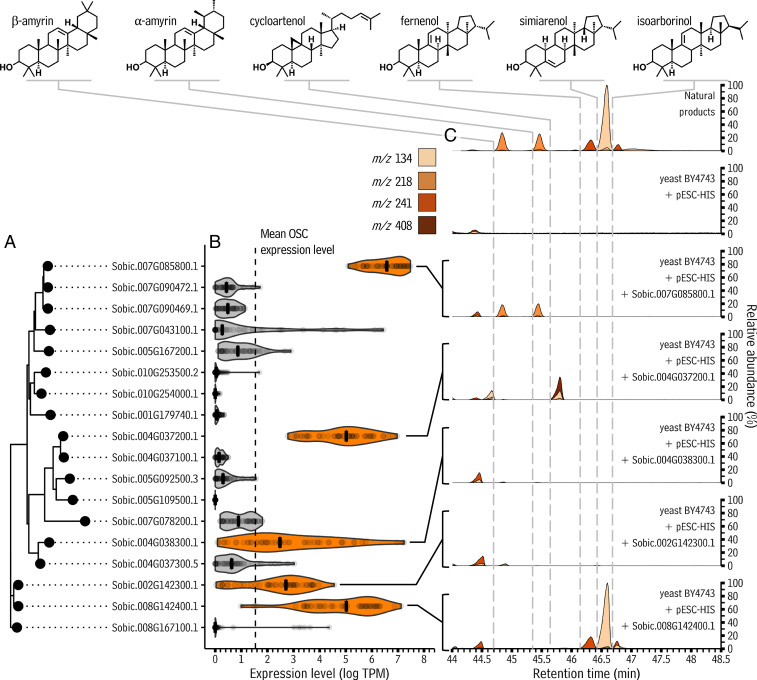
In other species, the accumulation of cuticular triterpenoids is controlled by oxidosqualene cyclase (OSC) genes. Enzymes encoded by OSC genes accept 2,3-oxidosqualene substrates and can cyclize them into one or more diverse triterpenoids (41). To identify candidate sorghum leaf triterpenoid biosynthesis genes, a BLAST search was performed on the S. bicolor genome using an Arabidopsis OSC as query. This revealed 18 OSC-like sequences annotated in the sorghum genome (Fig. 4A). To determine which of these might be active in leaves, and thus potentially in leaf wax synthesis, public sorghum expression data were mined from the National Center for Biotechnology Information (NCBI)’s Sequence Read Archive. An initial set of 150 sorghum leaf RNA-sequencing samples obtained from the Sequence Read Archive were filtered to retain only those samples with a mapping rate to the sorghum reference genome of ≥90%, producing a set of 50 samples for quantifying sorghum OSC expression levels (Dataset S4). The expression level of each candidate sorghum OSC was determined using Salmon (42). Across these samples, the expression of each sorghum OSC gene was relatively consistent, generally speaking, and five OSCs (shown in orange, Fig. 4B) had median expression levels greater than the mean expression level of all OSCs across all samples (dotted line, Fig. 4B). In a previous study of sorghum leaf gene expression over the course of development (43), these same five genes were the most highly expressed OSCs until 45 d after planting, a period in which leaves are undergoing rapid growth and actively synthesizing wax chemicals for cuticle construction, and four of the five remain the most highly expressed OSCs throughout the time course (SI Appendix, Fig. S18). These five genes (Sobic.007G085800.1, Sobic.004G037200.1, Sobic.004G038300.1, Sobic.002G142300.1, and Sobic.008G142400.1) were thus considered as high-probability leaf triterpenoid biosynthesis candidates.
Fig. 4.
Bioinformatic analysis and heterologous expression of OSCs from S. bicolor. (A) An unrooted phylogenetic tree showing relationships between OSCs annotated in the S. bicolor genome. (B) Violin plot showing the distribution of expression levels of each S. bicolor OSC as determined from 50 different publicly available RNA-sequencing samples. The mean OSC expression level across all samples is shown with a vertical dotted line, and the median expression level of each OSC is shown with a vertical cross bar inside each violin. OSCs with a median expression level greater than the mean expression level of all OSCs across all samples are highlighted in orange. (C) Line plot showing single ion chromatograms from gas chromatography-mass spectrometry analyses of transgenic S. cerevisiae cultures expressing the S. bicolor OSC genes highlighted in B as indicated by lines and brackets. Peak fill corresponds to the single ion trace from which they arise, as indicated in the legend. Vertical, dotted gray lines indicate the retention times of each peak of interest: the time point at which the signal leaves the baseline to form the beginning of each peak. To preclude batch effects, heterologous expression experiments in yeast were performed in two independent batches of three replicates each, yielding similar results both times.
The next goal was to determine if the protein products encoded by any of the identified candidate OSC genes were capable of producing sorghum leaf cuticular triterpenoids, particularly the hopane triterpenoids, the most abundant type of triterpenoids in the wax mixture of leaves made by adult sorghum plants. OSC cyclization of 2,3-oxidosqualene into triterpenoid products is a process that occurs in organisms from many kingdoms of life, and it is well established that functional characterization of OSCs from one organism can be performed by transferring them into another organism (44, 45). For example, plant OSCs are frequently functionally characterized by expressing them in a heterologous yeast system (46, 47), which is convenient as yeast produce endogenous 2,3-oxidosqualene that can be used by foreign OSC enzymes. To invoke this approach in the present study, each candidate gene was placed into an inducible, binary expression vector and transformed into Saccharomyces cerevisiae. Yeast harboring the empty vector produced none of the triterpenoids found on the sorghum leaf surface. However, GC-MS traces of extracts of yeast transformed with Sobic.007G085800.1 contained two peaks not present in the negative control that had retention times (Fig. 4C) and mass spectra (SI Appendix, Figs. S8 and S9) identical to β-amyrin and α-amyrin, respectively. The average ratio of these two products in three induced yeast cultures independently transformed with Sobic.007G085800.1 was 1:1. Thus, these data indicated that the product of Sobic.007G085800.1 was a mixed β- and α-amyrin synthase. Extracts analyzed with GC-MS from yeast transformed with Sobic.004G037200.1 resulted in a peak not present in the negative control, nor on the sorghum leaf surface. This peak had a mass spectrum identifying it as cycloartenol (SI Appendix, Fig. S10), a precursor to plant steroid-type hormones. The GC-MS traces from yeast transformed with Sobic.004G038300.1 or Sobic.002G142300.1 contained no peaks not found in the negative control. The analysis of yeast transformed with Sobic.008G142400.1 revealed three peaks not in the negative control and with retention times (Fig. 4C) and mass spectra (SI Appendix, Figs. S11–S13) identical to those of the fernenol, simiarenol, and isoarborinol triterpenoid products on the leaf surface. The average ratio of these three in three induced yeast cultures independently transformed with Sobic.008G142400.1 was 2:6:1. Thus, the data indicated that together, Sobic.007G085800.1 (a mixed amyrin synthase) and Sobic.008G142400.1 (a mixed hopane triterpenoid synthase with primarily simiarenol product specificity) were able to generate the main triterpenoids (i.e., the triterpenols) found on the sorghum leaf surface. Previous work demonstrated that plant triterpenones can be derived from their corresponding triterpenols via cytochrome P450 oxidation (36), making it highly likely that, in sorghum, fernenone and isoarborinone are derived from fernenol and isoarborinol, respectively. This means the mixed hopane triterpenoid synthase Sobic.008G142400.1 almost certainly participates in the formation of sorghum leaves’ fernenone and isoarborinone, and thus that, in total, Sobic.008G142400.1 is likely involved in generating the majority (64%) of sorghum leaf surface triterpenoids.
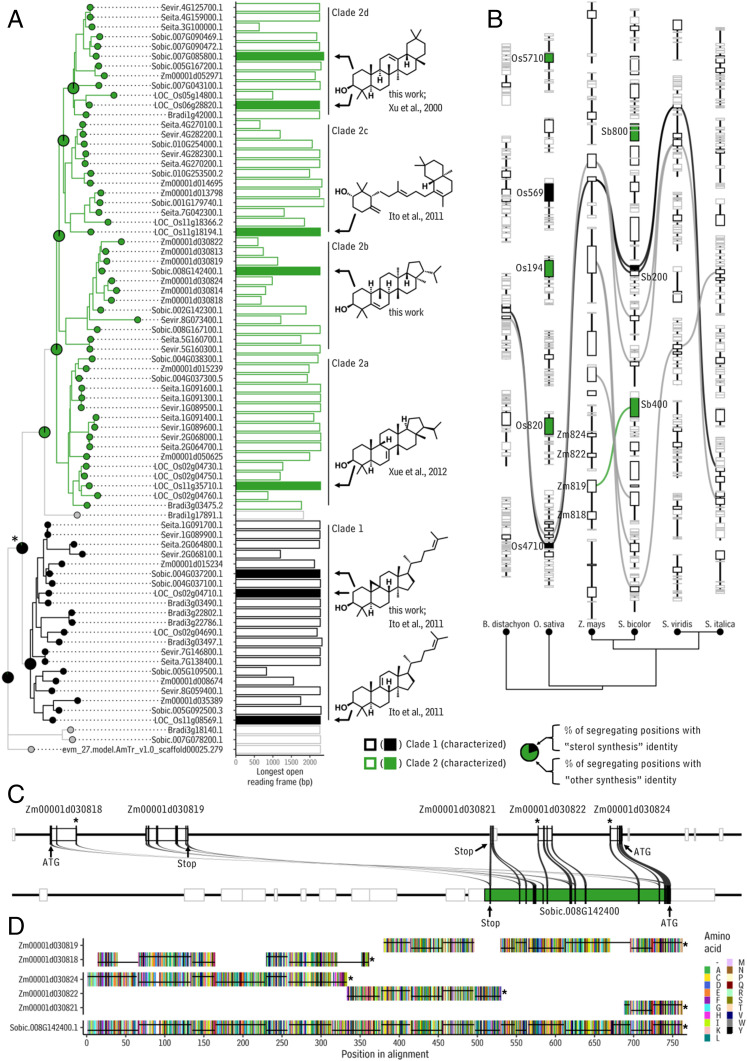
To explore relationships between OSC genes in sorghum and other members of the grass family (particularly maize), genome assemblies for S. bicolor, Z. mays, Brachypodium distachyon, Oryza sativa, Setaria viridis, and Setaria italica were searched for OSC sequences using a BLAST, then the resulting sequences were aligned and visualized with a phylogenetic tree rooted using the sequence of an OSC gene from the distantly related angiosperm Amborella trichopoda. This revealed two major clades of cyclases. The first clade was a smaller clade (“clade 1,” black branches and tips, Fig. 5A). It contained 21 genes of which 3, after including the data presented here, have been characterized: the cycloartenol synthases Sobic.004G037200.1 and LOC_Os02g04710.1, as well as a parkeol synthase LOC_Os11g08569.1 (48). Reexamining published information about syntenic relationships among in these species (49) revealed that Sobic.004G037200.1 and LOC_Os02g04710.1 (the two characterized cycloartenol synthases) both belong to a syntenic set of OSC orthologs spanning all six grass species (Fig. 5B, black synteny lines). This indicates that these cycloartenol synthases predate the divergence of these species, which is consistent with cycloartenol as an intermediate in the biosynthesis of plant sterols, compounds with diverse roles in plant physiology across a massive range of plant species (50).
Fig. 5.
Evolution of monocot OSCs. (A) Rooted phylogenetic tree (Left) showing relationships between annotated OSC-like gene models in six grass genomes (B. distaychon, accessions “Bradi[…]”; O. sativa, accessions “Loc_Os[…]”; Z. mays, accessions “Zm[…]”; S. bicolor, accessions “Sobic.[…]”; S. viridis, accessions “Sevir.[…]”; and S. italica, accessions “Seita.[…]”). The bar chart shows the longest open reading frame within each gene. Color-filled bars correspond to functionally characterized genes, with the structures of their major products shown to the Right of the bar chart. Colored tip labels, branches, and bars denote particular clades within the tree as indicated by clade markers (vertical lines to the Right of the bar chart) and as denoted in the legend. (B) Diagram showing syntenic relationships between OSCs in the six grass genomes (Top). Bezier curves connect syntenic orthologs and are colored if one of the orthologs has been functionally characterized, according to the product profile of the enzyme encoded by that gene. Phylogenetic tree showing the relationships between the species in the synteny diagram (Bottom). Abbreviations: Sb200: Sobic.004G037200.1, Sb400: Sobic.008G142400.1, Sb800: Sobic.007G085800.1, Zm819: Zm00001d030819, Os710: LOC_Os11g35710.1, Os569: LOC_Os11g08569.1. In both B and C, black lines represent chromosomal segments, while boxes on those lines indicate gene boundaries. Boxes with black outlines are OSC genes, while boxes with gray outlines are not OSC genes. Color-filled boxes indicate functionally characterized genes as denoted in the legend. (C) Diagram showing portions of the maize genome with high homology to Sobic.008G142400.1. Black bars superimposed on top gene boxes indicate coding sequence segments. Gray curves connect regions with high homology. Asterisks indicate premature stop codons in the coding sequences of the maize homologs of Sobic.008G142400.1. (D) Multiple sequence alignment of amino acid sequences encoded by Sobic.008G142400.1, Zm00001d030818, Zm00001d030819, Zm00001d030821, Zm00001d030822, and Zm00001d030824. Colored vertical bars correspond to amino acids as indicated in the legend. Black lines in each lane connect amino acids encoded by the same exon. Asterisks indicate stop codons.
The second, larger clade in the phylogenetic tree of grass OSCs (“clade 2,” green branches and tips; Fig. 5A) contained 51 genes of which 5, after including the data presented here, have been characterized: the hopane triterpenoid synthases Sobic.008G142400.1 and LOC_Os11g35710.1 (51), the amyrin synthases Sobic.007G085800.1 and LOC_Os06g28820.1 (52), and an achilleol B synthase, LOC_Os11g18194 (48). Considering functionally characterized genes, members of this second clade 1) are not conserved at syntenic locations across the six grasses analyzed here (Fig. 5B), with the exception of the simiarenol synthase Sobic.008G142400.1 (the primary gene likely generating sorghum leaf wax), which is syntenic with the maize gene Zm00001d030819 (Fig. 5B, green synteny line); and 2) catalyze reactions leading to diverse pentacyclic and tricyclic products in species-specific patterns. Overall, these features suggest that the second clade exhibits both high rates of birth (gene duplication) and death (gene deletion). This is consistent with gene family members catalyzing diverse and species-specific reactions as a result of repeated neofunctionalization, which is a common mode of functional divergence in grasses (53). Finally, we conducted an expanded analysis of OSC sequences using two additional monocot genome assemblies, that of Ananas comosus (pineapple, a nongrass member of the Poales) and Musa acuminata (banana, a monocot not in the Poales). Each of these species contained one full-length OSC sequence, both of which diverged before the radiation of clade 1 and clade 2 OSCs in the grass family (SI Appendix, Fig. S19). This suggests that the expansion of the OSC gene family into clades 1 and 2 may have occurred largely within the grasses after they diverged from other monocot species.
To further explore the evolutionary history of OSCs in the grass family, we used ancestral state reconstruction to determine the most likely sequences of genes at ancestral nodes in the grass family OSC phylogenetic tree. For this, we first examined a multiple sequence alignment of the functionally characterized members of the two clades (functionally characterized genes in clade 1 versus clades 2a–d, Fig. 5A) and identified 98 positions in the alignment that segregated completely for the activities encoded by the two clades (i.e., “sterol synthesis” versus “other triterpenoid synthesis”). We then used ancestral state reconstruction to determine the most likely identity of ancestral sequences at those same positions. Filtering out positions for which the ancestral state could not be determined with greater than 95% confidence (for example, two of the 98 positions for the starred node in Fig. 5A), we found that the ancestor of the two clades had the bases associated with sterol synthesis activity in more than 90% of the segregating positions (pie chart at the starred node, Fig. 5A). All the data considered together indicate that the ancestral cyclase functioned in sterol (likely cycloartenol) biosynthesis from which genes encoding other product specificities (isotriterpenol, amyrin, achilleol) have arisen as divergent copies. We note that the product profiles of terpene synthases can change in response to single amino acid substitutions—a property that can complicate evolutionary studies. Here however, we are predicting generalized product classes (sterol-like specificity versus nonsterol specificity), an aim less subject to complications from enzyme plasticity. It is informative to simultaneously consider the results from: 1) our chemical analyses, which indicated that simiarenol is the most abundant sorghum leaf wax compound over the plant’s life span (Figs. 2 and 3); 2) our biochemical analyses, which revealed that Sobic.008G142400.1 is a simiarenol synthase (Fig. 4); and 3) the ancestral state reconstruction, which suggests that Sobic.008G142400.1 is a diverged copy of a sterol biosynthesis gene. Together, these results suggest that the simiarenol synthase Sobic.008G142400.1 generates the major wax compound present on the sorghum leaf surface over time, which in turn could mean that the wax component of the sorghum leaf wax biocomposite is primarily produced by the activity of a divergent sterol biosynthesis gene.
The sorghum triterpenoid synthesizing gene Sobic.008G142400.1 resides in a clade (clade 2b, Fig. 5A) together with genes from Z. mays, S. viridis, and S. italica, including the maize gene Zm00001d030819, which was detected as a syntenic ortholog of Sobic.008G142400.1 (Fig. 5B). This strongly suggests the duplication and neofunctionalization of this gene happened sometime after the divergence of the common ancestor of these species from O. sativa (Fig. 5B). However, the longest coding sequences within the maize genes in this clade are only around half the length of the functionally characterized OSCs from the grass family. A closer inspection of homologous regions of the sorghum and maize genomic regions containing Sobic.008G142400.1 and Zm00001d030819, respectively, revealed that Zm00001d030819, together with the adjacent gene Zm00001d030818, contain most of the major exons present in Sobic.008G142400.1 (Fig. 5C). However, Zm00001d030819 contains an in-frame stop codon (asterisk, Fig. 5D) on the same exon and immediately downstream from nucleotides encoding many amino acids that are homologous with a region of the functional sorghum gene Sobic.008G142400.1. This suggests that the stop codon is due to a mutation and is not an artifact of genome annotation. This is consistent with Zm00001d030818/Zm00001d030819 being remnants of the gene inherited by maize that in sorghum became Sobic.008G142400.1. A similar situation was observed for Zm00001d030824, Zm00001d030822, and Zm00001d030821, which, together, form a stop-codon-interrupted, inverted copy of Zm00001d030818/Zm00001d030819. PCR amplification and subsequent sequencing of the regions containing these stop codons verified their presence in the maize genome (SI Appendix, Fig. S21). Overall, this means that maize has not maintained its counterpart(s) of the ancestral form of Sobic.008G142400.1 that both maize and sorghum inherited from their common ancestor. This is further supported by: 1) an analysis of 50 RNA sequencing samples of maize leaves downloaded from NCBI’s Sequence Read Archive (Dataset S6) in which nearly all these truncated genes were not expressed (SI Appendix, Fig. S20), and 2) the absence of triterpenoids in the wax mixture present on leaves made by adult maize plants (30). Overall, the evidence supports the hypothesis that sorghum, but not maize, creates its waxy surfaces using a cuticular triterpenoid-synthesizing gene it inherited from a maize–sorghum common ancestor after the neofunctionalization of a steroid synthesis gene created the capability to produce cuticular triterpenoids in that lineage. This also suggests that the ancestor of maize and sorghum probably had the ability to create a triterpenoid-rich wax mixture.
Sorghum Triterpenoids Are Present in the Water Barrier–Forming Intracuticular Wax Layer.
It is interesting to consider what factors may have caused sorghum, but not maize, to maintain the simiarenol synthase Sobic.008G142400.1 that is capable of generating compounds for a triterpenoid-rich cuticle. Plant waxes are present in two layers, an “epicuticular” layer that is exposed to the plant–environment interface, as well as an underlying “intracuticular” layer that is embedded inside the polyester cutin scaffold (54). There is mounting evidence that the intracuticular waxes establish the water barrier (55, 56), and that elevated temperatures cause the efficacy of the water barrier to decrease (57, 58). It has been postulated that this may be due to mechanical stresses placed on the cuticle by: 1) the swelling of the polysaccharide cell walls underneath the cuticle (58) or 2) differential thermal expansion of the waxes and the cutin (59), either or both of which could cause cracks in the water barrier–forming intracuticular wax layer. In either case, it seems likely that desert plants may deploy unique strategies to mitigate this problem. Indeed, it has been shown that intracuticular triterpenoids, which contribute mechanical strength to the cuticle (60), are particularly abundant in the cuticle of the nonsucculent desert tree Rhazya strica, which, at elevated temperatures, is more than twice as resistant to the movement of water than the triterpenoid-devoid cuticle of the temperate tree Juglans regia (61). Thus, the evidence suggests that intracuticular triterpenoids strengthen the cuticle in a way that protects the plant against water loss in hot climates. Relative to maize, sorghum was domesticated in a particularly hot and dry environment (3, 4, 62), meaning that if sorghum leaf triterpenoids are present in the intracuticular layer where they could contribute mechanical strength to the cuticle and thus aid in maintaining low cuticular permeability in hot climates, such a trait would likely be of more benefit to sorghum than maize. Based on previous analyses of plant cuticles, triterpenoids may be present in both the epicuticular and intracuticular layer or just one of the two (63–65). Accordingly, the final objective of this study was to determine if the triterpenoids on sorghum leaves were present in the intracuticular layer in substantial amounts.
To determine the layer-specific wax composition of sorghum leaves, the epicuticular wax compounds (compounds at the very interface of the plant surface and atmosphere) were isolated via four sequential applications and removals of gum arabic paste to the leaf surface, the adaxial (upper) and abaxial (lower) leaf sides of the leaf being treated independently. The resulting peels were spiked with internal standard and extracted with chloroform to obtain epicuticular wax isolates, then the exposed, underlying surface wax was extracted with chloroform to obtain intracuticular wax isolates. Wax isolates were analyzed with GC-MS in the same way as the whole wax samples, and all the compounds that could be quantified in the whole wax samples were also identified and quantified in the epicuticular and intracuticular wax samples. For both the adaxial and abaxial surfaces, the amount of wax recovered from the fourth gum arabic peel was less than or equal to only 10% of the wax that had been recovered in the first three peels (SI Appendix, Fig. S22), indicating that the epicuticular wax was nearly entirely removed by the four peels, and overall, that this approach generated data enabling a high-quality assessment of wax amounts and composition in the two wax layers on either side of the sorghum leaf.
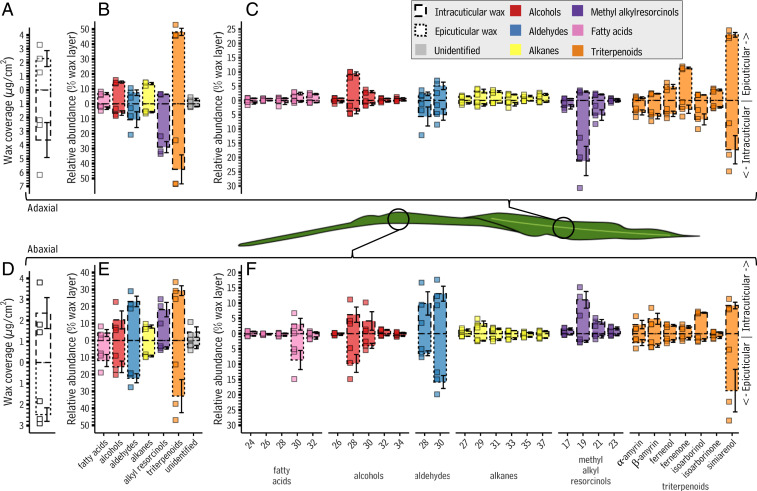
The epicuticular and intracuticular wax data from the adaxial surface were examined first. On this surface, epicuticular and intracuticular wax accumulated to 2.30 ± 0.6 μg/cm2 and 3.63 ± 1.3 μg/cm2, respectively (Fig. 6A and Dataset S3). Triterpenoids comprised the majority of both these layers (48.2% and 43.9% of the two wax layers, epicuticular and intracuticular, respectively, Fig. 6B), with methyl alkylresorcinols (6.4% and 29%), alcohols (14.6% and 6.2%), aldehydes (7.4% and 10.6%), alkanes (13.6% and 5.6%), and fatty acids (6.9% and 2.9%) present in much smaller amounts. All these compounds were roughly evenly distributed between the two layers except methyl alkylresorcinols, which accumulated primarily in the intracuticular layer. In both layers, each of these compound classes was composed of multiple homologs, or in the case of triterpenoids, isomers. Simiarenol (compound 26, Fig. 1) predominated in the triterpenoid compound class (23.0% and 17.3% of terpenoids in the epicuticular and intracuticular layers, respectively, Fig. 6C). On the abaxial surface, epicuticular and intracuticular wax accumulated to 2.5 ± 0.3 μg/cm2 and 2.4 ± 0.7 μg/cm2, respectively (Fig. 6D and Dataset S3). Triterpenoids were the most abundant compounds in both layers (32.8% and 29.3% of epicuticular and intracuticular layer, respectively; Fig. 6E), followed by aldehydes (22.3% and 23.0%), fatty acids (11.9% and 4.4%), alcohols (15.5% and 12.1%), alkanes (9.2% and 8.2%), and methyl alkylresorcinols (4.5% and 18.2%). Two compound classes exhibited layer-specific enrichment: fatty acids (11.9% of the epicuticular but only 4.4% of the intracuticular), and methyl alkylresorcinols (4.5% of the epicuticular, 18.3% of the intracuticular). Simiarenol was the most abundant triterpenoid in both layers (18.7% and 9.2%, Fig. 6F). Thus, triterpenoids, simiarenol in particular, were major components of the intracuticular wax layer on both sides of the leaf, a location where they could act as strengthening nanofillers (60) and improve the performance of the leaf cuticle at elevated temperatures (61).
Fig. 6.
Epicuticular and intracuticular wax composition on leaves produced by adult S. bicolor plants. Bar charts showing the abundance and composition of epicuticular and intracuticular waxes from both the adaxial and abaxial sides of leaves made by adult sorghum leaves. Throughout, bar correspondence with intracuticular versus epicuticular wax components is coded using both the direction the bar projects from the x axis (toward center: intracuticular wax, away from center: epicuticular wax) and the bar border (dashed border: intracuticular wax, dotted border: epicuticular wax). The direction of bar projection in each panel series (A–C and D–F) reflects the location of each epicuticular and intracuticular layer when viewing a S. bicolor leaf through a sagittal plane. The borders for both epicuticular and intracuticular observations are dots and dashes, respectively; this formatting is used to show both epicuticular (dots) and intracuticular (dashes) components. Together, these make up the total wax coverage observed (solid border, shown in Fig. 4) of S. bicolor leaves. Bars are colored according to compound class, as set precedent by Fig. 1A: yellow: alkanes, blue: aldehydes, orange: triterpenoids, red: primary alcohols, purple: methyl alkylresorcinols, and pink: fatty acids, gray: unidentified. Numbers below each bar indicate the total carbon number of that compound, and the text below each group of bars indicates the compound class represented by those bars. Shown is the total amount of epicuticular and intracuticular wax on (A) the adaxial surface and (D) the abaxial surface (y axis; shown in mg/cm2), the relative abundance of each wax compound class in the epicuticular and intracuticular wax layers on (B) the adaxial surface and (E) the abaxial surface, and the relative abundance of each wax compound in the epicuticular and intracuticular layer on (C) the adaxial surface and (F) the abaxial surface. Bar heights and error bars indicate the mean and SE of three independent analyses, respectively. To preclude batch effects, the entire experiment was performed in two batches of three replicates each, yielding similar results both times.
Conclusions.
The biochemical and chemical data as well as bioinformatic analyses presented here led to multiple main findings: 1) cuticular triterpenoid biosynthesis in sorghum is likely carried out by a neofunctionalized steroid biosynthesis gene inherited by both sorghum and maize; 2) in sorghum, but not maize, this gene’s activity has been maintained as part of the juvenile-to-adult transition, likely enabling a triterpenoid-rich cuticle on adult leaves; and 3) these triterpenoids are a major component of the sorghum intracuticular wax layer, where literature suggests they could contribute to the mechanical strength of the sorghum cuticle and thus help maintain the efficacy of the water barrier at high temperatures. This is consistent with sorghum’s hot and dry native climate as well as the drought tolerance for which the crop is known. Previous studies have also indicated that, at nonelevated temperatures, triterpenoids may increase the permeability of the cuticle to water (66) or have a negligible role in forming the water barrier (55), thus indicating that future studies on the functional role(s) of cuticular triterpenoids should consider that such may be temperature dependent. Additionally, the present data suggest that the ancestor of maize and sorghum had the capacity to create a triterpenoid-rich cuticle that may have been advantageous in hot climates. Considered together, these data also outline a potential future scenario in which cuticle triterpenoid-synthesizing genes could be resurrected in maize to create a more heat-tolerant water barrier on the plants’ leaf surfaces. We are currently endeavoring to generate transgenic lines to test this hypothesis. Finally, the information presented on the temporal content and composition of sorghum leaf wax will inform efforts to tap surface carbon allocations for vegetative storage and conversion to high-value biofuels and bioproducts for feedstock enhancement.
Materials and Methods
Plant Material and Tissue Harvesting.
S. bicolor BTx623 plants were grown under a standard protocol in the University of Nebraska–Lincoln’s Beadle Greenhouse Center. Seeds were planted individually in 4-inch clay pots, then adolescents were transferred to 10-inch plastic pots (3 plants per 10″ pot) containing ProMix BX soil mixture, supplemented with Biofungicide and MicroMax micronutrients. The plants also received fertilizer (500 ppm Ca, 350 ppm N, 60 ppm Fe) once per week. Plants were watered once per week and spot watered as needed in between scheduled waterings. The greenhouse’s temperature ranged from 27 to 29 °C during the day and from 19 to 21 °C, during the night. The humidity was ∼65%. Light was supplemented with high pressure sodium (400 W and 1,000 W) as well as metal halide (400 W and 1,000 W) lamps to maintain a 16-h day length. S. bicolor seeds were planted every week so that plants at each developmental stage were available for analysis throughout the study. Field-grown plants were cultivated in a field on the East Campus of the University of Nebraska–Lincoln from late May until late August, when leaves were sampled. During this period, the average temperature was roughly 26 °C and the average humidity was about 70%. Thus, while these conditions of course varied over the course of the 3 mo, they were not markedly different from the conditions in the greenhouse. When leaf tissue from a plant of particular age was needed, the highest leaf with a fully exposed ligule on that plant was selected. Flag leaves were not examined as part of this study.
Bulk Extraction of Sorghum Leaf Waxes and Thin Layer Chromatography Separation.
Sorghum leaf waxes were extracted in bulk by harvesting one leaf each from plants separated by 1 wk in age at 14 to 98 d after planting. The leaves were combined in a beaker (500 mL), chloroform was added (200 mL), and a glass rod was used to make sure the leaves were immersed for at least 30 s each. The leaves were removed from the beaker, the solvent was evaporated, leaving the wax behind as a solid residue. Thin layer chromatography separation and gas chromatography-mass spectrometry analysis of resulting bands was performed as described previously (67). Full details also appear in SI Appendix, Supplementary Methods.
Preparation of Wax Samples.
Each total leaf wax sample was prepared from a square of tissue (1.25 cm2) that had been excised from a separate leaf. Leaf squares were placed in scintillation vials, spiked with tetracosane internal standard (25 μL; 0.1 mg/mL), extracted twice with CHCl3, then the CHCl3 was evaporated to leave the wax as a residue. Full details appear in SI Appendix, Supplementary Methods. To prepare epicuticular and intracuticular wax samples, four successive applications of gum arabic paste (1g gum arabic/1 mL of ddH2O) were used to remove the epicuticular wax layer and expose the intracuticular waxes, which were then extracted using CHCl3. Epicuticular waxes were obtained by dissolving the gum arabic in water and extracting with diethyl ether. Full details appear in SI Appendix, Supplementary Methods.
Gas Chromatography-Mass Spectrometry Analyses.
TLC band samples, whole leaf wax samples, epicuticular wax samples, and intracuticular wax samples were all analyzed using the same GC-MS system (Agilent 7890A GC). The method used was as previously described (68). Full details also appear in SI Appendix, Supplementary Methods.
Analysis of Public RNA Sequencing Data.
Raw RNA sequencing data were downloaded from NCBI’s Sequence Read Archive. A list of samples downloaded and used in this project is provided in Dataset S4. A set of reference transcripts for S. bicolor (Sbicolor_454_v3.1.1.transcript_primaryTranscriptOnly.fa) was downloaded from Phytozome. A local version of BLAST+ (69) was used to extract S. bicolor OSC sequences using the Arabidopsis thalaniol cyclase (At5g48010) as a query. Salmon (42) was used to determine the number of transcripts per million for each gene in each sample. Samples with mapping rates less than 90% were excluded from further analysis. The remaining 50 samples were used to determine which S. bicolor OSC genes had median expression levels above mean OSC expression across all samples using R (70).
Expression of Sorghum Oxidosqualene Cyclases in S. cerevisiae.
Candidate OSC genes in pYES2-HIS expression vectors were obtained through the gene synthesis and subcloning services of Twist Biosciences. These vectors were then transformed into competent BY7473 yeast cells using the Zymogen Yeast Transformation Kit according to manufacturer instructions. An aliquot (50 µL) of the transformed cells was then grown on plates with yeast growth media that lacked histidine (28 °C; 5 d). Three independent colonies from each plate were then isolated, transferred into a Falcon tube (50 mL) containing selection media (1 mL; 2% raffinose carbon source), and allowed to incubate to metabolize residual glucose (28 °C, 48 h; 100 rpm). Next, each yeast culture was centrifuged (700 × g, 1 min), the liquid raffinose media was removed from the tube with a pipette, then residual media was removed by washing each pellet three times with sterile water (2 mL). Finally, transgene expression was induced in each cell culture by resuspending each in liquid induction media (8 mL, 2% galactose solution carbon source), then incubating at a cool temperature (18 °C, 96 h, 100 rpm) to minimize production of endogenous yeast triterpenoids.
To analyze the triterpenoid composition of the yeast cultures, each sample was transferred to a 7.5-mL glass screw-cap tube. The glass tubes were centrifuged (4,000 × g, 4 min), the induction media was discarded, and the cells were lysed by adding glass beads (150 µL) and methanol (200 µL) and vortexing vigorously. Once homogenized, the cell cultures were extracted by adding water (1 mL) and hexane (2 mL), vortexing, allowing the layers to separate, then transferring the upper, organic phase into a new 8-mL glass tube. The extraction was performed twice, then the hexane extracts from each culture were pooled and evaporated to dryness at 70 °C under N2. To remove saponifiable lipids, ethanol (2 mL) was added to the dry extracts, the tubes were heated (90 °C, 5 min), the tubes were cooled, then potassium hydroxide (166 µL, 80% wt/vol) was added, the tubes were vortexed, then incubated (90 °C, 10 min), with another vortex step halfway through the incubation period. Immediately after the incubation, the tubes were placed on ice, ice-cold water (1 mL) was added to each, then each was vortexed and returned to ice. These contents were then extracted twice with the addition of hexane (1 mL) and separated by pooling the hexane phases in a GC vial. The extracts then underwent the derivatization portion of the GC sample preparation. The initial hexane extracts were partially evaporated under N2 until there was ≤200 µL remaining, then the extracts were transferred into a GC vial insert, and the remaining hexane was evaporated completely. Then, 30 µL of derivatization agents were added to the insert (15 µL of N,O-Bis(trimethylsilyl)trifluoroacetamide [BSTFA] and pyridine), after which the GC vials were capped, vortexed, and incubated at 70 °C for 60 min. After incubation, the derivatization reagents were evaporated under N2. The contents remaining in the GC vial insert were resuspended in 10 µL of CHCl3 and vortexed. From the solution, 2 µL were injected into the GC column for chemical analysis.
Evolutionary Analysis of Oxidosqualene Cyclases in the Grass Family.
Genome assemblies for S. bicolor, Z. mays, B. distachyon, O. sativa, S. viridis, S. italica, A. comosus, and M. acuminata were downloaded from the sources listed in Dataset S5. BLAST searches were performed using BLAST+ (69) using Sobic.008G167100.1 and the Arabidopsis thalaniol cyclase as queries, resulting in 92 hits. The sequence of the hit Os06g28820 from O. sativa contained a frame shift mutation causing a premature stop codon, but a previous report found that this stop codon is not present when the gene is cloned, at least for some rice varieties (52); accordingly, we replaced the sequence of the hit Os06g28820 with the sequence reported for that cloned gene. The 92 sequences were then filtered by length. Hits shorter than 75% of the mean hit length (573 bp) were removed from the analysis, as were hits that did not contain an open reading frame with a stop codon (Zm00001d053889) and hits with insertions larger than 300 bp in their coding sequences (Bradi3g17507.6), leaving 78 hits. Hit sequences were aligned using the R package “msa” (71), trimmed using gBlocks (72), a phylogeny was constructed using the R package “phangorn” (73) which was then visualized using the R package “ggtree” (74). Ancestral state reconstruction was performed using the ancestral.pml function from the R package phangorn.
Confirmation of Stop Codons in Maize Oxidosqualene Cyclase Genes.
Approximately 1-kb regions of the maize B73 genome containing sequence flanking each of the three reported stop codons in the ZEAMMB73_Zm00001d030818, ZEAMMB73_Zm00001d030822, and ZEAMMB73_Zm00001d030824 genes were amplified by PCR using maize B73 genomic DNA as template. Primer sequences are provided in SI Appendix, Supplementary Methods. PCR reactions were conducted with Phusion DNA polymerase (Thermo Fisher Scientific) and consisted of 35 cycles of 15 s at 98 °C, 15 s at 59 °C, and 60 s at 72 °C and followed by 10 min at 72 °C. Amplification of ∼1-kb products for each reaction was confirmed by agarose gel analysis. The product of each reaction were ligated directly into the pCR-BluntII plasmid using the Zero Blunt TOPO PCR cloning kit (Invitrogen) and transformed into One-Shot Escherichia coli cells according to the manufacturer’s protocol. Inserts from miniprepped plasmid from two independent colonies from each PCR were sequenced in the forward and reverse directions and compared with the reported genomic sequence of the ZEAMMB73_Zm00001d030818, ZEAMMB73_Zm00001d030822, and ZEAMMB73_Zm00001d030824 genes to confirm the stop codon occurrences.
Supplementary Material
Acknowledgments
We thank Samantha Link for the highest quality assistance with growing sorghum plants, Dr. Ignacio A. Ciampitti at Kansas State University for permission to reproduce with alteration his image “Sorghum Growth and Development,” and the Holland Computing Center at the University of Nebraska–Lincoln for computing resources. We are also grateful to Rebecca E. Cahoon for providing comments on a draft of this manuscript. L.B. would like to thank E.B.C. as well as David R. Holding and Chi Zhang for their mentorship. L.B. acknowledges support in the form of a fellowship from the NSF Plant Genome Research Program (NSF PRFB IOS-1812037). E.S. was supported by funding from the NSF (Grant OIA-1557417). E.B.C. acknowledges support from the U.S. Department of Energy Center for Advanced Bioenergy and Bioproducts Innovation (Award DE-SC0018420) for sorghum feedstock improvement.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022982118/-/DCSupplemental.
Data Availability
Sequence data used in this work can be found in the Phytozome/GenBank databases under the accession numbers listed in Dataset S4). All accession codes are provided in Supplemental Datasets. Figs. 1–4 and 6 have associated raw data that are available at Dryad (https://doi.org/10.5061/dryad.m0cfxpp3f). The program R (v. 3.5.3) was used for all data analysis in this study. Specific R packages used are listed in Materials and Methods. The only custom script was the one used to determine which sites in the multiple sequence alignment should be used in the ancestral state reconstruction is an R script that is available on GitHub as part of the R package phylochemistry (https://thebustalab.github.io/phylochemistry/phylochemistry.html).
References
- 1.Alexandratos N., Bruinsma J., “World agriculture towards 2030/2050, the 2012 revision” (2012).
- 2.Foley J. A., et al., Solutions for a cultivated planet. Nature 478, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Winchell F., Stevens C. J., Murphy C., Champion L., Fuller D., Evidence for sorghum domestication in fourth millennium BC Eastern Sudan: Spikelet morphology from Ceramic impressions of the Butana group. Curr. Anthropol. 58, 673–683 (2017). [Google Scholar]
- 4.Wet J. M. J. D., Harlan J. R., The origin and domestication of Sorghum bicolor. Econ. Bot. 25, 128–135 (1971). [Google Scholar]
- 5.Morris G. P., et al., Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. U.S.A. 110, 453–458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mace E. S., et al., Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat. Commun. 4, 2320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paterson A. H., et al., The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Schober T. J., Messerschmidt M., Bean S. R., Park S., Arendt E. K., Gluten‐free Bread from sorghum: Quality differences among hybrids. Cereal Chem. 82, 394–404 (2005). [Google Scholar]
- 9.Gao Z., Xie X., Ling Y., Muthukrishnan S., Liang G. H., Agrobacterium tumefaciens-mediated sorghum transformation using a mannose selection system. Plant Biotechnol. J. 3, 591–599 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Zhao Z. Y., et al., Agrobacterium-mediated sorghum transformation. Plant Mol. Biol. 44, 789–798 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Borrell A., et al., “Drought adaptation in sorghum” in Drought Adaptation in Cereals (The Hawath Press Inc, 2006), pp. 335–399. [Google Scholar]
- 12.Borrell A. K., et al., Drought adaptation of stay-green sorghum is associated with canopy development, leaf anatomy, root growth, and water uptake. J. Exp. Bot. 65, 6251–6263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris K., et al., Sorghum stay-green QTL individually reduce post-flowering drought-induced leaf senescence. J. Exp. Bot. 58, 327–338 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Zegada-Lizarazu W., Zatta A., Monti A., Water uptake efficiency and above- and belowground biomass development of sweet sorghum and maize under different water regimes. Plant Soil 351, 47–60 (2012). [Google Scholar]
- 15.Yeats T. H., Rose J. K. C., The formation and function of plant cuticles. Plant Physiol. 163, 5–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isaacson T., et al., Cutin deficiency in the tomato fruit cuticle consistently affects resistance to microbial infection and biomechanical properties, but not transpirational water loss. Plant J. 60, 363–377 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Fich E. A., Segerson N. A., Rose J. K. C., The plant polyester cutin: Biosynthesis, structure, and Biological roles. Annu. Rev. Plant Biol. 67, 207–233 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Vogg G., et al., Tomato fruit cuticular waxes and their effects on transpiration barrier properties: Functional characterization of a mutant deficient in a very-long-chain fatty acid β-ketoacyl-CoA synthase. J. Exp. Bot. 55, 1401–1410 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Schönherr J., Water permeability of isolated cuticular membranes: The effect of cuticular waxes on diffusion of water. Planta 131, 159–164 (1976). [DOI] [PubMed] [Google Scholar]
- 20.Busta L., Jetter R., Moving beyond the ubiquitous: The diversity and biosynthesis of specialty compounds in plant cuticular waxes. Phytochem. Rev. 17, 1275–1304 (2018). [Google Scholar]
- 21.Busta L., Budke J. M., Jetter R., The moss Funaria hygrometrica has cuticular wax similar to vascular plants, with distinct composition on leafy gametophyte, calyptra and sporophyte capsule surfaces. Ann. Bot. 118, 511–522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busta L., Hegebarth D., Kroc E., Jetter R., Changes in cuticular wax coverage and composition on developing Arabidopsis leaves are influenced by wax biosynthesis gene expression levels and trichome density. Planta 245, 297–311 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Guo Y., Busta L., Jetter R., Cuticular wax coverage and composition differ among organs of Taraxacum officinale. Plant Physiol. Biochem. 115, 372–379 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Lee S. B., Kim H., Kim R. J., Suh M. C., Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Rep. 33, 1535–1546 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Bourdenx B., et al., Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156, 29–45 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J.-J., et al., Mutation in Wilted Dwarf and Lethal 1 (WDL1) causes abnormal cuticle formation and rapid water loss in rice. Plant Mol. Biol. 74, 91–103 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Aharoni A., et al., The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16, 2463–2480 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bianchi G., Avato P., Salamini F., Surface waxes from grain, leaves, and husks of maize (Zea mays L.). Cereal Chem., 45–47 (1984). [Google Scholar]
- 29.Moose S. P., Sisco P. H., Glossy15 controls the Epidermal Juvenile-to-adult phase transition in maize. Plant Cell 6, 1343–1355 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourgault R., et al., Constructing Functional Cuticles: Analysis of Relationships between Cuticle Lipid Composition, Ultrastructure and Water Barrier Function in Developing Adult Maize Leaves (Ann Bot-london, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heupel R. C., Varietal similarities and differences in the polycyclic isopentenoid composition of sorghum. Phytochemistry 24, 2929–2937 (1985). [Google Scholar]
- 32.Heupel R. C., Sauvaire Y., Le P. H., Parish E. J., Nes W. D., Sterol composition and biosynthesis in sorghum: Importance to developmental regulation. Lipids 21, 69–75 (1986). [DOI] [PubMed] [Google Scholar]
- 33.Adamski N. M., et al., The inhibitor of wax 1 locus (Iw1) prevents formation of β- and OH-β-diketones in wheat cuticular waxes and maps to a sub-cM interval on chromosome arm 2BS. Plant J. 74, 989–1002 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Sun Y., et al., Characterization of an alkylresorcinol synthase that forms phenolics accumulating in the cuticular wax on various organs of rye (Secale cereale). Plant J. 102, 1294–1312 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Ji X., Jetter R., Very long chain alkylresorcinols accumulate in the intracuticular wax of rye (Secale cereale L.) leaves near the tissue surface. Phytochemistry 69, 1197–1207 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Moses T., et al., OSC2 and CYP716A14v2 catalyze the biosynthesis of triterpenoids for the cuticle of aerial organs of Artemisia annua. Plant Cell 27, 286–301 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szakiel A., Pączkowski C., Pensec F., Bertsch C., Fruit cuticular waxes as a source of biologically active triterpenoids. Phytochem. Rev. 11, 263–284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niemann G. J., Baas W. J., The composition of the lipid constituents of Ilex aquifolium L. Aquifoliaceae) in relation to the age of the leaf. I. The leaf wax J. Plant Physiol. 118, 219–226 (1985). [DOI] [PubMed] [Google Scholar]
- 39.Hemmers H., Gülz P.-G., Marner F.-J., Triterpenoids in epicuticular waxes of three European Euphorbia species. Zeitschrift Für Naturforschung C 43, 799–805 (1988). [Google Scholar]
- 40.Li L., et al., The maize glossy6 gene is involved in cuticular wax deposition and drought tolerance. J. Exp. Bot. 70, erz131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thimmappa R., Geisler K., Louveau T., O’Maille P., Osbourn A., Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 65, 225–257 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varoquaux N., et al., Transcriptomic analysis of field-droughted sorghum from seedling to maturity reveals biotic and metabolic responses. Proc. Natl. Acad. Sci. U.S.A. 116, 27124–27132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abe I., Prestwich G. D., Molecular cloning, characterization, and functional expression of rat oxidosqualene cyclase cDNA. Proc. Natl. Acad. Sci. U.S.A. 92, 9274–9278 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haralampidis K., et al., A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc. Natl. Acad. Sci. U.S.A. 98, 13431–13436 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z., et al., Two oxidosqualene cyclases responsible for biosynthesis of tomato fruit cuticular triterpenoids. Plant Physiol. 155, 540–552 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Busta L., et al., Oxidosqualene cyclases involved in the biosynthesis of triterpenoids in Quercus suber cork. Sci. Rep. 10, 8011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ito R., et al., Triterpene cyclases from Oryza sativa L.: Cycloartenol, parkeol and achilleol B synthases. Org. Lett. 13, 2678–2681 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y., et al., Differentially regulated orthologs in sorghum and the subgenomes of maize. Plant Cell 29, 1938–1951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaller H., The role of sterols in plant growth and development. Prog. Lipid Res. 42, 163–175 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Xue Z., et al., Divergent evolution of oxidosqualene cyclases in plants. New Phytol. 193, 1022–1038 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Sun J., Xu X., Xue Z., Snyder J. H., Qi X., Functional analysis of a rice oxidosqualene cyclase through total gene synthesis. Mol. Plant 6, 1726–1729 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Jiang X., Assis R., Rapid functional divergence after small-scale gene duplication in grasses. BMC Evol. Biol. 19, 97 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jetter R., Schäffer S., Riederer M., Leaf cuticular waxes are arranged in chemically and mechanically distinct layers: Evidence from Prunus laurocerasus L. Plant Cell Environ. 23, 619–628 (2000). [Google Scholar]
- 55.Jetter R., Riederer M., Localization of the transpiration barrier in the Epi- and intracuticular waxes of eight plant species: Water transport resistances are associated with fatty acyl rather than alicyclic components. Plant Physiol. 170, 921–934 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeisler-Diehl V., Müller Y., Schreiber L., Epicuticular wax on leaf cuticles does not establish the transpiration barrier, which is essentially formed by intracuticular wax. J. Plant Physiol. 227, 66–74 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Schreiber L., Effect of temperature on cuticular transpiration of isolated cuticular membranes and leaf discs. J. Exp. Bot. 52, 1893–1900 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Riederer M., Thermodynamics of the water permeability of plant cuticles: Characterization of the polar pathway. J. Exp. Bot. 57, 2937–2942 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Schreiber L., Schönherr J., Phase transitions and thermal expansion coefficients of plant cuticles: The effects of temperature on structure and function. Planta 182, 186–193 (1990). [DOI] [PubMed] [Google Scholar]
- 60.Tsubaki S., Sugimura K., Teramoto Y., Yonemori K., Azuma J., Cuticular membrane of Fuyu persimmon fruit is strengthened by triterpenoid nano-fillers. PLoS One 8, e75275 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schuster A.-C., et al., Effectiveness of cuticular transpiration barriers in a desert plant at controlling water loss at high temperatures. AoB Plants 8, plw027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wendorf F., et al., Saharan exploitation of plants 8,000 years BP. Nature 359, 721–724 (1992). [Google Scholar]
- 63.Racovita R. C., Jetter R., Composition of the epicuticular waxes coating the adaxial side of Phyllostachys aurea leaves: Identification of very-long-chain primary amides. Phytochemistry 130, 252–261 (2016). [DOI] [PubMed] [Google Scholar]
- 64.van Maarseveen C., Jetter R., Composition of the epicuticular and intracuticular wax layers on Kalanchoe daigremontiana (Hamet et Perr. de la Bathie) leaves. Phytochemistry 70, 899–906 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Markstädter C., Federle W., Jetter R., Riederer M., Hölldobler B., Chemical composition of the slippery epicuticular wax blooms on Macaranga (Euphorbiaceae) ant-plants. Chemoecology 10, 33–40 (2000). [Google Scholar]
- 66.Buschhaus C., Jetter R., Composition and physiological function of the wax layers coating Arabidopsis leaves: β-amyrin negatively affects the intracuticular water barrier. Plant Physiol. 160, 1120–1129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Busta L., et al., Identification of genes encoding enzymes catalyzing the early steps of carrot polyacetylene biosynthesis. Plant Physiol. 178, 01195.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Busta L., Jetter R., Structure and biosynthesis of branched wax compounds on wild type and wax biosynthesis mutants of Arabidopsis thaliana. Plant Cell Physiol. 58, 1059–1074 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Camacho C., et al., BLAST+: Architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R Core Team , R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria). 5 October 2020.
- 71.Bodenhofer U., Bonatesta E., Horejš-Kainrath C., Hochreiter S., msa: An R package for multiple sequence alignment. Bioinformatics 31, 3997–3999 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Talavera G., Castresana J., Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Schliep K. P., phangorn: Phylogenetic analysis in R. Bioinformatics 27, 592–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T., ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data used in this work can be found in the Phytozome/GenBank databases under the accession numbers listed in Dataset S4). All accession codes are provided in Supplemental Datasets. Figs. 1–4 and 6 have associated raw data that are available at Dryad (https://doi.org/10.5061/dryad.m0cfxpp3f). The program R (v. 3.5.3) was used for all data analysis in this study. Specific R packages used are listed in Materials and Methods. The only custom script was the one used to determine which sites in the multiple sequence alignment should be used in the ancestral state reconstruction is an R script that is available on GitHub as part of the R package phylochemistry (https://thebustalab.github.io/phylochemistry/phylochemistry.html).