Abstract
Antimicrobial resistance has been described in all ecosystems, including wildlife. Here we investigated the presence of methicillin-resistant and susceptible staphylococci in both colony-born and wild vervet monkeys (Chlorocebus sabaeus). Through selective isolation, PCR, MALDI-TOF, and whole-genome sequencing, methicillin-resistant and susceptible Staphylococcus spp. isolated from vervet monkeys were characterized. We obtained putatively methicillin-resistant staphylococci from 29 of the 34 nasal samples collected. Strains were identified by MALDI-TOF analysis. Staphylococcus cohnii (n = 15) was the most commonly isolated species, while nine other species were isolated one or two times. PCR analysis indicated that eight [28%] strains were mecA positive. The whole-genome sequencing [WGS] included eight methicillin-resistant strains (S. epidermidis (n = 2), S. cohnii (n = 3), S. arlettae (n = 2) and S. hominis (n = 1)), nine additional S. cohnii strains and two strains that could not be identified by MALDI-TOF, but genetically characterized as one S. cohnii and one S. warneri. Different resistance genes carried by different mobile genetic elements, mainly blaZ (n = 10) and tet(K) (n = 5) were found, while msr(A), cat, fosB, dfrG, erm(C), mph(C) and str were identified in one to three strains. Phylogenetic analysis of the S. cohnii strains based on SNPs indicated four clusters associated with colony born or wild. In addition, one singleton S. cohnii isolated did not form a separate group and clustered within other S. cohnii strains submitted to the NCBI. In this study, we demonstrated the presence of AMR and mobile genetic elements to both colony-born and wild vervet monkeys. We also identified a previously undescribed prevalence of S. cohnii in the nasal flora of these monkeys, which merits further investigation.
Keywords: methicillin resistance, Staphylococcus, whole-genome sequencing, monkey, vervet, Chlorocebus sabaeus
1. Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a major health problem in humans, while in animals, it remains limited to sporadic cases [1]. That being said, the potential for zoonotic transmission of MRSA has been well documented [2,3]. In animals, methicillin-resistant S. pseudintermedius is becoming an imminent health problem in dogs [4]. Specifically, in animals, methicillin-resistance has not been limited to coagulase-positive staphylococci [5,6]. Transfer of the SCCmec element can be mediated by bacteriophages [7], which, based on the zoonotic potential, makes the presence of methicillin-resistance in animal staphylococci a matter of public health concern [2].
Currently, 71 recognized staphylococcal species and 30 subspecies have been described (http://www.dsmz.de/bacterial-diversity/prokaryotic-nomenclature-up-to-date, last accessed on 8 March 2021). However, little is known about the presence of staphylococci in monkeys. Moreover, most studies on staphylococci in monkeys were focused on the isolation of MRSA and S. aureus. We could not find any study dealing with vervet monkeys and staphylococci. In other monkey species, few studies were done on the presence of staphylococci, like a study in gorillas where several staphylococcal species were found in the feces [8] as well as on the conjunctiva [9]. Staphylococci have also been found in the vagina of squirrel monkeys [10], where a new species was discovered [11] and in the nasal, oral, rectal and vaginal flora of tamarins [12,13]. However, the information is fragmentary and does not always include the susceptibility of the strains. Only two studies also reported on antimicrobial resistance, and 13% of the staphylococci from squirrel monkeys proved to be methicillin-resistant [10,13].
In monkeys, as well as other wildlife species, MRSA has been frequently described [2,3,9,14,15,16,17]. However, little is known about methicillin-resistance harbored by other staphylococcal species in monkeys and the zoonotic potential of these methicillin-resistant staphylococci. Therefore, we performed a study on Saint Kitts Island to further investigate the presence of methicillin-resistance in monkey staphylococci and to evaluate their potential to act as a natural reservoir for clinically significant resistance determinants. Saint Kitts is a small island nation in which the monkey population is estimated to exceed the human population leading to frequent direct and indirect contact between humans and monkeys. While the prevalence of MRSA in humans in the hospital is well described, little is known about this pathogen in the wild and captive monkey populations of the island [18]. Such a large population of monkeys on a small island makes this study very well suited for One Health-oriented investigations.
Since methicillin in humans appears to be highly prevalent in humans in Saint Kitts, we hypothesize that this resistance is more widespread in the environment.
In this research, the aim was to determine the presence and characteristics of Staphylococcus spp., isolated from nasal swab samples of vervet monkeys and using cefoxitin supplemented selective media. A second objective was to determine whether there were differences in the frequency of species and diversity of strains in the captive and free-living vervet monkey populations on the island of Saint Kitts
2. Results
Thirty-four samples were taken, of which 21 originated from wild-caught animals and 13 from animals living in captivity. Of the 34 samples, using all methods together, five were negative for staphylococci. In the 29 remaining cultures, the multiplex PCR indicated the isolated strains were staphylococci, and two were S. aureus. Only eight strains showed the presence of the mecA gene (Table 1), five from captive animals (38.5% CI: 13.9–68%) and three from wild animals (14.3% CI: 3–36%). This difference was not statistically significant. MALDI-TOF could not identify two strains, which were subsequently identified by WGS and Kmer analysis. Finally, we detected S. cohnii (n = 15), S. epidermidis (n = 2), S. pettenkoferi (n = 1), S. kloosii (n = 1), S. saprophyticus (n = 1), S. arlettae (n = 2), S. aureus (n = 2), S. xylosus (n = 2) and S. hominis (n = 2) and S. warneri (n = 1) (Table 1).
Table 1.
Isolation results of the samples and DNA analysis of the strains.
| Sample | Origin | mecA | Species | Resistance Genes *** | SCCmec Elements | Plasmids |
|---|---|---|---|---|---|---|
| 1 ** | Captive | + | S. cohnii | blaZ, mph(C) | SCCmecIII(3A) | rep7a rep(pSBK203) U35036 |
| 2 ** | Captive | + | S. epidermidis | fosB, dfrG, erm(C) | SCCmecV(5C2) | rep10 repL(pDLK1) GU562624, rep39 repA(SAP110A) GQ900465 |
| 3 ** | Captive | - | S. cohnii | blaZ, tet(K) | ccrA1:4:LGA251:FR821779 subtype-Vc(5C2&5):10:AB505629 |
rep7a repC(pS0385p1) AM990993 rep21 rep(pSHaeA) AP006717 |
| 4 ** | Captive | - | S. cohnii | blaZ, tet(K) | ccrA1:4:LGA251:FR821779 subtype-Vc(5C2&5):10:AB505629 |
rep7a repC(pS0385p1) AM990993/ repC(Cassette) AB037671 |
| 5 ** | Captive | + | S. cohnii | blaZ, mph(C) | rep7a rep(pSBK203) U35036 | |
| 6 | Captive | - | S. pettenkoferi | none | ||
| 7 ** | Captive | - | S. cohnii | blaZ, tet(K) | ccrA1:4:LGA251:FR821779 subtype-Vc(5C2&5):10:AB505629 |
rep7a repC(pS0385p1) AM990993/repC(Cassette) AB037671/ORF(pKH1) SAU38656 |
| 8 ** | Captive | - | S. cohnii | blaZ | ccrA1:4:LGA251:FR821779 subtype-Vc(5C2&5):10:AB505629 |
rep21 rep(pKH21) EU350088 |
| 9 ** | Captive | - | S. cohnii | blaZ, tet(K) | ccrA1:4:LGA251:FR821779 subtype-Vc(5C2&5):10:AB505629 |
rep7a repC(pS0385p1) AM990993 |
| 10 ** | Captive | + | S. cohnii | blaZ, mph(C) | none | |
| 11 ** | Captive | - | S. warneri * | none | repUS35 A28412072(pvSw2) CP003671 | |
| 12 | Wild | - | S. kloosii | none | ||
| 13 | Wild | - | S. saprophyticus | none | ||
| 14 ** | Wild | - | S. cohnii | none | ccrB3:2:LGA251:FR821779 | rep7a rep(pSBK203) U35036 rep21 rep(pKH21) EU350088 |
| 15 | Wild | - | S. xylosus | none | ||
| 16 ** | Wild | + | S. arlettae | str | rep13 ORF(pC194) V01277 | |
| 17 | Wild | - | S. aureus | none | ||
| 18 ** | Wild | + | S. arlettae | none | none | |
| 19 ** | Wild | + | S. epidermidis | fosB, dfrG | SCCmecV(5C2) | rep10 repL(pDLK1) GU562624 rep39 repA(SAP110A) GQ900465 |
| 20 | Wild | - | S. aureus | |||
| 21 ** | Wild | - | S. cohnii | none | ccrA1:4:LGA251:FR821779 subtype-Vc(5C2&5):10:AB505629 |
rep21 rep(pSHaeA) AP006717 |
| 22 | Wild | - | S. xylosus | |||
| 23 ** | Wild | - | S. cohnii | blaZ, tet(K), cat(pC211) | rep7a rep(pSBK203) U35036/repD(pTZ4) NC010111/ORF(pKH1) SAU38656 | |
| 24 | Wild | - | S. cohnii | |||
| 25 ** | Wild | + | S. hominis | blaZ, msr(A) | rep7a repA(SAP105B) GQ900453 | |
| 26 ** | Captive | - | S. cohnii | none | ccrA1:3:JCSC6943:AB505628 | none |
| 27 ** | Captive | - | S. cohnii * | none | none | |
| 28 | Wild | NA | Negative | |||
| 29 | Wild | NA | Negative | |||
| 30 | Wild | NA | Negative | |||
| 31 | Wild | - | S. hominis | |||
| 32 | Wild | NA | Negative | |||
| 33 | Wild | NA | Negative | |||
| 34 | Wild | - | S. cohnii |
* only identified by WGS. ** WGS performed. *** resistance genes found with ResFinder.
A selection of 19 strains (Table 1) was retained for WGS analysis based on the presence of mecA, no identification by MALDI-TOF and the most prevalent species. Sequence quality control parameters can be found in Table 2. All trimmed raw reads were deposited in NCBI BioProject database and assigned BioProject Temporary SubmissionID PRJNA647767, with accession numbers https://www.ncbi.nlm.nih.gov/biosample/15636413, https://www.ncbi.nlm.nih.gov/biosample/15636414 https://www.ncbi.nlm.nih.gov/biosample/15636415, https://www.ncbi.nlm.nih.gov/biosample/15636416, https://www.ncbi.nlm.nih.gov/biosample/15636417, https://www.ncbi.nlm.nih.gov/biosample/15636418, https://www.ncbi.nlm.nih.gov/biosample/15636419, https://www.ncbi.nlm.nih.gov/biosample/15636420, https://www.ncbi.nlm.nih.gov/biosample/15636421, https://www.ncbi.nlm.nih.gov/biosample/15636422, https://www.ncbi.nlm.nih.gov/biosample/15636423, https://www.ncbi.nlm.nih.gov/biosample/15636424, https://www.ncbi.nlm.nih.gov/biosample/15636425, https://www.ncbi.nlm.nih.gov/biosample/15636426, https://www.ncbi.nlm.nih.gov/biosample/15636427, https://www.ncbi.nlm.nih.gov/biosample/15636428, https://www.ncbi.nlm.nih.gov/biosample/15636429, https://www.ncbi.nlm.nih.gov/biosample/15636430, https://www.ncbi.nlm.nih.gov/biosample/15636431.
Table 2.
Quality parameters of the sequenced strains.
| Sample | Number of Contigs | Mean Contig Length | Median Contig Length | Minimum Contig Length | Maximum Contig Length | Base Count | N50 | Coverage |
|---|---|---|---|---|---|---|---|---|
| 1 | 539 | 4548.83 | 2656 | 137 | 27,277 | 2,451,819 | 9591 | 29 |
| 2 | 469 | 196.37 | 2725 | 137 | 46,151 | 2,210,175 | 9380 | 31 |
| 3 | 514 | 4917.82 | 2570 | 137 | 35,832 | 2,527,758 | 10,396 | 29 |
| 4 | 676 | 152.46 | 2033 | 137 | 30,607 | 2,474,092 | 7349 | 27 |
| 5 | 524 | 187.09 | 2386 | 137 | 54,340 | 2,352,829 | 9277 | 27 |
| 7 | 554 | 4463.89 | 2638 | 137 | 37,050 | 2,472,996 | 8527 | 25 |
| 8 | 674 | 153.83 | 2184 | 137 | 38,142 | 2,488,125 | 7397 | 24 |
| 9 | 614 | 166.17 | 2257 | 137 | 43,634 | 2,448,661 | 8110 | 24 |
| 10 | 433 | 5385.63 | 3060 | 137 | 39,146 | 2,331,979 | 12,088 | 42 |
| 11 | 397 | 248.19 | 2854 | 137 | 44,913 | 2,364,664 | 12,992 | 35 |
| 14 | 543 | 4377.91 | 2462 | 137 | 37,869 | 2,377,204 | 8798 | 38 |
| 16 | 814 | 121.20 | 1738 | 137 | 24,999 | 2,367,457 | 5316 | 48 |
| 18 | 427 | 231.74 | 2963 | 137 | 55,529 | 2,374,768 | 11,863 | 39 |
| 19 | 375 | 6025.86 | 3340 | 137 | 38,343 | 2,259,697 | 12,172 | 40 |
| 21 | 435 | 238.79 | 2540 | 137 | 52,277 | 2,493,071 | 14,825 | 32 |
| 23 | 371 | 279.92 | 3240 | 137 | 71,562 | 2,492,410 | 14,287 | 32 |
| 25 | 386 | 220.87 | 2819 | 137 | 47,447 | 2,046,024 | 10,904 | 34 |
| 26 | 362 | 277.80 | 3243 | 137 | 56,956 | 2,413,478 | 15,510 | 29 |
| 27 | 441 | 228.60 | 3284 | 137 | 55,778 | 2,419,417 | 10,757 | 31 |
By PCR, the mecA gene was identified in eight strains; however, in four cases, ResFinder did not detect the gene, while RAST analysis did not find the gene in two strains. In these two strains, only sequences potentially associated with SCCmec elements could be found. In the two strains where we were able to find the mecA gene through RAST, the gene was located on a very small contig.
We could identify two different SCCmec elements in the collection of strains. The two S. epidermidis strains were SCCmec type V(5C2), and one S. cohnii strain contained type SCCmec type III (3A). Frequently, the SCCmec could not be determined, probably due to fragmentation and gaps in the sequences. On the other hand, in some mecA negative strains, fragments of SCCmec were identified by SCCmecFinder, namely ccrA1:4, part of the LGA251SCCmec in 7 S. cohnii strains, and in six instances also with a part of the subtype-Vc(5C2&5) (Table 1). Further analysis of the contig containing the ccrA1;4 genes indicated that the full contig of 9034 base pairs was also present in an S. nepalensis strain with a coverage of 71% and 95.78% identity. The ccrA gene detected is the ccrA gene present in SCCmec IX. The part containing a portion of the subtype-Vc(5C2&5) was small and related to the presence of the czr gene, encoding zinc resistance, typically associated with LA-MRSA ST398 [1].
In both S. arlettae isolates, mecA was detected by PCR, while ResFinder could only detect the gene in one of the strains. RAST analysis, however, demonstrated the presence of the mecA gene in both strains. No SCCmec could be identified as the contigs on which the mecA gene was located were short; in one strain, it was only the mecA gene on the contig and in the other strain, there were only two additional genes, previously associated with SCCmec, but of insufficient length to determine the type.
In the S. hominis strain, the gene was found with a mecI and mecR gene in one node, while the ccrC1 allele was found on another contig and is associated with SCCmec type II. No definitive SCCmec could be determined.
The resistance genes found by ResFinder were blaZ in ten strains, tet(K) in five, the msr(A) gene in one and the mph(C) gene in three, the cat gene in one, the fosB genes were associated with the two S. epidermidis strains and those two strains carried the dfrG gene. The erm(C) gene was found in one of the two S. epidermidis strains. Finally, the str gene was found in one of the S. arlettae strains (Table 1).
Tetracycline resistance genes tet(K) were located on plasmids, but it was not clear which ones. RAST analysis showed that the tet(K) gene was located on the same plasmid in cluster three S. cohnii strains but was different in cluster two S. cohnii strains. The BLAST analysis showed that the replication initiation protein was most similar to the S. aureus subsp. aureus strain ST20071176 plasmid pT45. The next gene, a type I restriction–modification system, specificity subunit S, had very little homology with other chromosomal genes. The plasmid recombination, mobE mobilization protein gene was very similar to the mobE gene of S. warneri strain WB224 plasmid pWB224_2, and the downstream mobE gene was also 100% identical in that plasmid and several other plasmids of staphylococci. These genes were followed by the tet(K) gene, and the next gene was another replication initiation protein identical to S. aureus strain SR153 plasmid pSR02 replication initiation protein. This was then followed by another type I restriction–modification system (with low similarity) and another mobE gene, similar to the first one. The other location of the tet(K) gene was surrounded by two replication initiation proteins and two hypothetical proteins. tet(K) is frequently associated with IS257; however, this was not the case in our study, as we could not find any insertion sequence in the tet(K) positive strains. None of these sequences were identified as plasmids by the PlasmidFinder. Similarly, chloramphenicol resistance was located on a plasmid as determined by the ResFinder and RAST analysis. The mph(C) gene could not be associated with a mobile genetic element, and the blaZ gene was associated with a Tn552-like element in all but the S. hominis strain. (Table 1).
The erm(C) gene was associated with a gene encoding a replication and maintenance protein. BLAST analysis of this gene showed that it was associated with many staphylococcal plasmids; the contig was, however, too small to find a more exact location.
Several plasmids were identified in the strains using PlasmidFinder (Table 1). Four strains did not contain any plasmids, three S. cohnii strains, of which two had no resistance genes, and one strain carried the blaZ and tet(K), which can be found on the chromosome. The other strain was the mecA positive S. arlettae carrying no other resistance genes. In five strains, two plasmids were detected in each. Eight different replicon types were identified (Table 1). The replicon type 7a was most common (n = 8), though they were not all associated with the same plasmid types. The other seven replicon types were only found one to four times only.
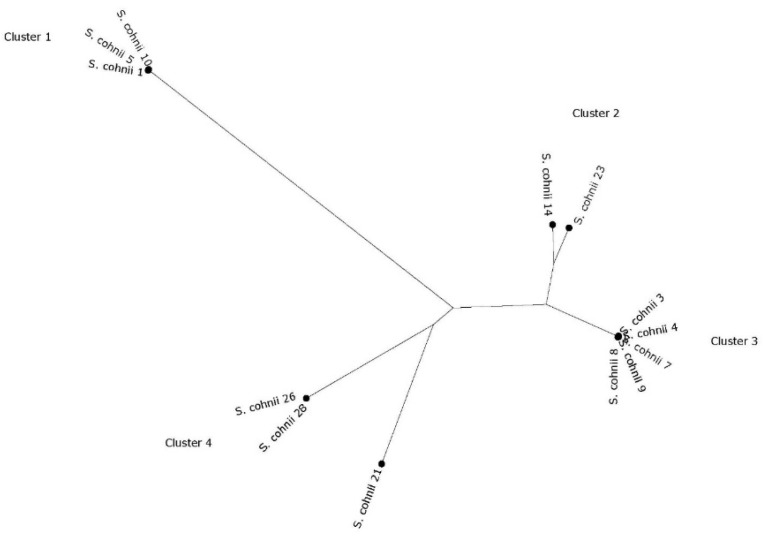
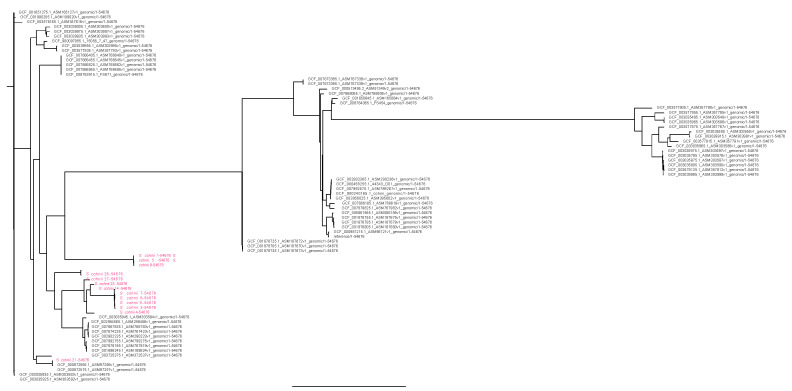
The two S. epidermidis strains were both ST210, and the single S. hominis strain had an unknown MLST profile but with a close match to ST13. No MLST scheme was available for the other species. Further phylogenetic analysis using CSIPhylogeny demonstrated that the two S. epidermidis strains were very similar but not identical with 81 SNP differences (data not shown). Phylogenetic analysis of the S. cohnii strain identified four clusters and a singleton (Figure 1). In cluster one, with three nearly identical strains (between 50 and 68 SNPs different), one strain contained SCCmec type III (3A), while in two others, the mecA gene was found, but the SCCmec type could not be determined. Cluster two contained non-identical strains with a difference of 1368 SNPs. Cluster three is the largest cluster and contained five strains. Differences in SNPs were between 41 and 104. All harbored the same fragments of SCCmec as determined by SCCmecFinder and RAST analysis. However, no mecA gene could be detected in any of the analyses. Cluster four contained two very similar strains with 44 SNP differences, and there was one singleton (strain 21) containing different parts of SCCmec, but it remains undefined. Typically, each cluster was composed of strains only belonging to wild or colony monkeys, indicating a different evolution of the strains with little mixing between wild and colony monkeys. Looking at the phylogenetic tree constructed with all at NCBI available sequenced S. cohnii strains [sequences of 63 strains were available and downloaded on April 29, 2020), the strain clustered among all different S. cohnii strains without any clear pattern (Figure 2). No virulence genes were detected in the S. epidermidis strains.
Figure 1.
Core genome phylogeny of the vervet S. cohnii isolates. Phylogeny was based on SNP differences.
Figure 2.
Rooted phylogenies of all sequenced S. cohnii isolates present in NCBI on 29 April 2020.
3. Discussion
To our knowledge, this is the first study that investigates methicillin-resistant and methicillin-susceptible staphylococcal species isolated from the nasal passage of monkeys. We sampled both wild and captive animals, though we could not detect a significant difference in the prevalence between the two populations. The wild animals were captured at various sites throughout the Island, while the colony-born vervets were living in closer contact. The wild animals had never received antimicrobial treatments, yet their staphylococci harbored a variety of resistance genes, probably picked up from other sources, including human interaction and the environment. However, the presence of the mecA genes in wild animals suggests that the mecA gene is widespread in Saint Kitts and Nevis environments. This may have been through human contact as the prevalence of MRSA in the hospital is very high, with 45% of the S. aureus infections being caused by MRSA or by feeding in human garbage and handouts like in Nepal [2,16]. There are, however, no data on the presence of methicillin-resistant non-aureus staphylococci from humans in Saint Kitts since data on these types of isolates are not generally collected. These are the first data on staphylococci from animals in Saint Kitts. This may suggest that methicillin-resistant non-aureus staphylococci could act as a reservoir for methicillin-resistance determinants.
One of the first surprises was that several of the strains were not methicillin-resistant despite isolation on selective media. This may be due to the presence of the blaZ gene that may have influenced the recovery of such strains; however, that is certainly not a valid hypothesis for all of the strains. It remains unclear what is the reason for the growth of these strains on the selective media and warrants further study. The mecA gene was also identified more frequently by PCR than by WGS, though depending on the analysis method, we could find more mecA genes using RAST analysis rather than ResFinder. This is most probably because RAST has no selection parameters, while in ResFinder, we used the standard parameters. Nevertheless, when the PCR is positive, regardless of the WGS analysis, the strain was regarded as mecA positive as the WGS sequences are generally incomplete.
In this study, a little more than half of the staphylococci isolated were S. cohnii. There is, in general, very limited knowledge about S. cohnii. S. cohnii has been isolated from different animal species without pathology (as dogs, cattle, pet animals), from environmental samples and on rare occasions, S. cohnii has been isolated from infections in humans, indicating that some strains may have some pathogenic potential [19,20,21,22]. It has been shown before that this species can be intrinsically resistant to penicillins and negative for all known β-lactam resistance genes [23]. Furthermore, taking into account the selective isolation method used for isolating these strains, it may have created a bias for the selection of antibiotic-resistant isolates. Nevertheless, there are indications that there is homogeneity in the strains as they are frequently clustered as closely related (Figure 1). It was striking to see that the strains from wild animals had separate clusters from the colony animals, indicating little mixing of the animals and the strains having a different evolution. Nevertheless, looking at all sequenced S. cohnii strains available in the NCBI database, they do not form a specific Saint Kitts vervet cluster in the currently available sequenced strain collection. It would be interesting to see what the prevalence of S. cohnii in other populations is, including humans, in Saint Kitts.
In the pubMLST database (https://pubmlst.org/bigsdb?db=pubmlst_sepidermidis_isolates&page=query, last accessed June 2020), four other S. epidermidis strain have been sequence typed as ST210 strains, strains originating from India, South Korea, and Portugal. All were of human origin, though it should be noted that few animal strains are included in this database, and most of the animal strains are from cases of mastitis in cattle. Former studies have shown that there is a high diversity of strains and no clear delineation between human and animal strains, though this study did not include monkey strains [24]. Moreover, to the best of our knowledge, this is the first study characterizing methicillin-resistant S. epidermidis from monkeys. We could not find any virulence genes associated with either human or animal strains.
Few SCCmec types could be detected. Moreover, we found different fragments pertaining to SCCmec in several of the strains, lacking the mecA gene. Looking for SCCmec, associated genes on the RAST annotated contigs did not solve the problem as exemplified in the S. hominis strain. This could also indicate that there remain uncharacterized SCCmec elements in the strains. The significance of these findings is not clear. Further in-depth sequencing and gap closing could give more insights into this finding. A recent study showed the high prevalence (45%) of the MRSA USA 300 strain among S. aureus infections in Saint Kitts. However, this strain carries SCCmec IVa, which was not found in this study [18].
The tet(K) gene was clearly associated with a plasmid; we could, however not determine, which plasmid it was. Homology searches did not reveal any specific plasmid. tet(K) in staphylococci is frequently associated with an insertion element, though we could not find any linked insertion sequence either.
The msr(A) gene codes for an ABC-F protein, which confers macrolide and streptogramin B resistance. Recently, the action has been shown to be due to ribosomal protection by these proteins [25,26]. The msr(A) gene mediates resistance to macrolides and streptogramin B antibiotics, but not to 16-membered macrolides and lincosamides. This gene has been associated with plasmids, frequently together with erm genes. However, in this S. hominis strain, we could not find any plasmid, nor an erm gene. The chloramphenicol resistance gene was associated with a plasmid that was not identified by the PlasmidFinder software. PC221 is a Rolling-circle plasmid that has been found in several staphylococci [27]. Other elements involved in the mobility of resistance was the Tn552 transposon associated with blaZ.
4. Materials and Methods
This study was approved by the IACUC of RUSVM with file number 17.04.20.
4.1. Sampling
Thirty-four vervet monkeys (Chlorocebus sabaeus) were sampled by nose swabs (BBL CultureSwab plus Amies Medium, Becton Dickinson, New Jersey, US). Of these 34, 13 were animals living in captivity for many generations in the Behavioral Science Foundation, and 21 wild monkeys were caught on the island of Saint Kitts. The captive animals were between 3 and 15 years of age and of both sexes. All were colony-born. Animals did not receive antibiotics prior to this study and were swabbed during routine husbandry and thus randomly selected. The animals are housed in outdoor cages, grouped to the same sex or in breeding groups. Wild animals were trapped in funnel-type traps by professional trappers and according to the animal welfare regulations (and approved by the BSF IACUC). After trapping, they were brought to the facility for quarantine. Estimated ages were between 3 and 15 years. They were trapped at various parts of the island of Saint Kitts, from rural/farming areas. In general, they stay away from humans but share territory at different times; they do not get into contact with sewage nor garbage as this is not their typical behavior. Nasal swabs were taken and transported to the lab for further analysis within 4 h.
4.2. Isolation of Methicillin-Resistant Staphylococci
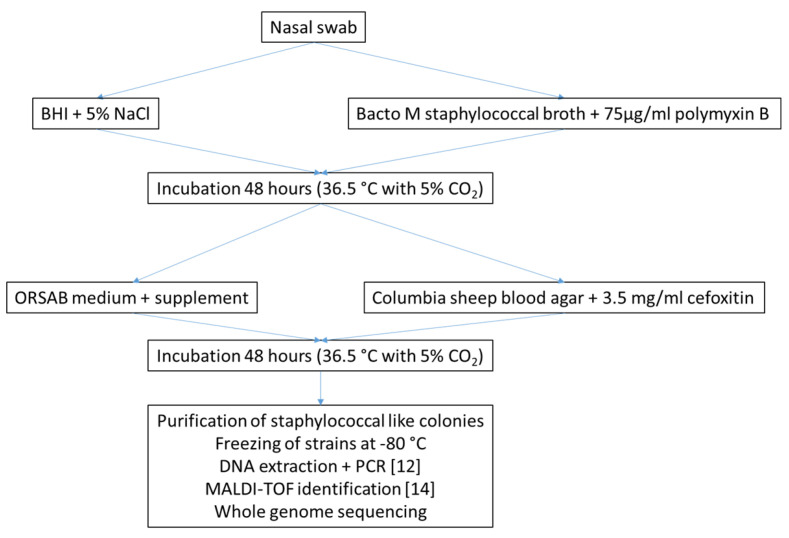
Isolation of methicillin-resistant staphylococci was performed as described before [28], with some adaptations. Briefly, the nasal swabs were inoculated in two different enrichment media, BHI supplemented with 5% NaCl and Bacto M staphylococcal broth (Becton Dickinson, NJ, USA) supplemented with 75 µg/mL of polymyxin B (Sigma-Aldrich, Saint Louis, MO, USA) and incubated for 48 h at 36.5 °C and 5% CO2. Upon growth in these media, a loopful was inoculated on selective plates. One ORSAB plate with SR0195 selective supplement (Oxoid, Basingstoke, UK) for the isolation of MRSA was inoculated, as well as one Columbia sheep blood agar plate supplemented with 3.5 µg/mL of cefoxitin (Sigma, Basingstoke, UK). Colonies resembling staphylococcal morphology were purified on blood agar plates (Figure 3).
Figure 3.
Isolation and identification strategy.
4.3. Bacterial Identification
Purified strains were initially identified with a PCR for the staphylococcal 16S rRNA gene, the nuc gene, specific for S. aureus, as well as the mecA gene [29]. DNA was extracted as previously described [28]. Further identification of the strains was performed using MALDI-TOF Biotyper (Bruker Daltonics, Germany), as previously described [30]. Final identification was based on the whole-genome sequence analysis.
4.4. Whole-Genome Sequencing
Overnight cultures were grown in tryptic soy broth at 37 °C with 200-rpm shaking. Genomic DNA from the staphylococcal strains was isolated using the DNeasy Blood and Tissue kit (Qiagen). DNA purity and concentration were determined using the Nanodrop and Qubit instruments, respectively. Sequencing library preparation was done using the Nextera XT kit and sequenced on a MiSeq using a paired-end 2 × 250 bp sequencing strategy, all following standard Illumina protocols (Illumina, Inc., San Diego, CA, USA).
Adapters were trimmed from the raw reads using the MiSeq program. Initial analyses (de novo assembly) and quality control were performed using Velvet.
The following analysis performed with pipelines from the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/, last accessed 9 March 2021) were used: Kmer analysis to confirm the species identification (KmerFinder), ResFinder v.3.0 for the detection of resistance genes, PlasmidFinder v2.0 for the detection of plasmid replicons, SCCmec was identified using SCCmecFinder, MLST profiles were determined using “MLST”. Genomes of the S. cohnii strains isolated in this study were compared to the 64 S. cohnii strains submitted to the NCBI database using CSIPhylogeny for SNP analysis. Trees were constructed using Figtree V1.4.4 with standard settings.
Strains were annotated using the RAST server using standard settings. Specific contigs were inspected manually and in some cases compared with other sequences using BLAST analysis.
RAST analysis was performed for the detection of virulence genes in S. epidermidis. We specifically looked for the icaRADBC operon, the biofilm-related genes embp, aap, bhp and IS256 [31].
4.5. Statistical Analysis
Groups were compared using Fisher’s exact test.
5. Conclusions
This is the first study into the methicillin-resistant staphylococcal flora of non-human primates. In this study, we isolated several methicillin-resistant staphylococcal species as well as susceptible staphylococci growing on selective methicillin-resistant plates. methicillin-resistance could not always be associated with an SCCmec element, while some strains lacking mecA contained parts of an SCCmec element. Whole-genome analysis showed that there were some well-known resistance genes present, but it could not always be determined with which exact mobile genetic element they were associated. The high number of S. cohnii in these animals requires further investigation on their importance in the nasal flora of vervet monkeys. The resistance genes in association with known mobile genetic elements indicate a potential spillover from other sources as the wild-caught animals do not receive antimicrobials. Our findings confirm a potential public health threat caused by these resistance genes found in pathogens demonstrated to have zoonotic potential.
Acknowledgments
We thank Egle Kudirkiene for the laboratory help in sequencing. We thank the personnel of the Behavioral Science Foundation for the sampling of the vervet monkey.
Author Contributions
Conceptualization, A.H. and P.B.; methodology, A.H., P.B., A.B., M.C.R., A.M. and F.B.; software, P.B.; validation, A.H., F.B., A.B., A.M., M.C.R. and P.B.; formal analysis, P.B.; investigation, A.H. and P.B. resources, P.B.; data curation, P.B.; writing—original draft preparation, A.H. and P.B.; writing—review and editing, A.H., F.B., A.B., A.M., M.C.R. and P.B; visualization, P.B.; supervision, P.B.; project administration, P.B.; funding acquisition, P.B. and F.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by an intramural grant of RUSVM, grant number 41,002–20” The MALDI-TOF MS was financed by the Research Foundation Flanders (FWO-Vlaanderen) as a Hercules project (G0H2516N, AUGE/15/05).
Institutional Review Board Statement
This study was approved by the IACUC of RUSVM with file number 17.04.20.
Informed Consent Statement
Not applicable.
Data Availability Statement
NCBI BioProject database and assigned BioProject Temporary SubmissionID PRJNA647767, with accession numbers https://www.ncbi.nlm.nih.gov/biosample/15636413, https://www.ncbi.nlm.nih.gov/biosample/15636414, https://www.ncbi.nlm.nih.gov/biosample/15636415, https://www.ncbi.nlm.nih.gov/biosample/15636416, https://www.ncbi.nlm.nih.gov/biosample/15636417, https://www.ncbi.nlm.nih.gov/biosample/15636418, https://www.ncbi.nlm.nih.gov/biosample/15636419, https://www.ncbi.nlm.nih.gov/biosample/15636420, https://www.ncbi.nlm.nih.gov/biosample/15636421, https://www.ncbi.nlm.nih.gov/biosample/15636422, https://www.ncbi.nlm.nih.gov/biosample/15636423, https://www.ncbi.nlm.nih.gov/biosample/15636424, https://www.ncbi.nlm.nih.gov/biosample/15636425, https://www.ncbi.nlm.nih.gov/biosample/15636426, https://www.ncbi.nlm.nih.gov/biosample/15636427, https://www.ncbi.nlm.nih.gov/biosample/15636428, https://www.ncbi.nlm.nih.gov/biosample/15636429, https://www.ncbi.nlm.nih.gov/biosample/15636430, https://www.ncbi.nlm.nih.gov/biosample/15636431.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Butaye P., Argudín M.A., Smith T.C. Livestock-Associated MRSA and its current evolution. Curr. Clin. Microbiol. Rep. 2016;3:19–31. doi: 10.1007/s40588-016-0031-9. [DOI] [Google Scholar]
- 2.Roberts M.C., Joshi P.R., Greninger A.L., Melendez D., Paudel S., Acharya M., Bimali N.K., Koju N.P., No D., Chalise M., et al. The human clone ST22 SCCmec IV methicillin-resistant Staphylococcus aureus isolated from swine herds and wild primates in Nepal: Is man the common source? FEMS Microbiol. Ecol. 2018;94:fiy052. doi: 10.1093/femsec/fiy052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts M.C., Joshi P.R., Monecke S., Ehricht R., Müller E., Gawlik D., Diezel C., Braun S.D., Praudel S., Acharya M., et al. Staphylococcus aureus and Methicillin Resistant S. aureus in Nepalese Primates; Resistance to Antimicrobials, Virulence and Genetic Lineages. Antibiotics. 2020;9:689. doi: 10.3390/antibiotics9100689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pires Dos Santos T., Damborg P., Moodley A., Guardabassi L. Systematic review on global epidemiology of methicillin-resistant Staphylococcus pseudintermedius: Inference of population structure from multilocus sequence typing data. Front. Microbiol. 2016;7:1599. doi: 10.3389/fmicb.2016.01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argudín M.A., Vanderhaeghen W., Butaye P. Diversity of antimicrobial resistance and virulence genes in methicillin-resistant non-Staphylococcus aureus staphylococci from veal calves. Res. Vet. Sci. 2015;99:10–16. doi: 10.1016/j.rvsc.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Vanderhaeghen W., Vandendriessche S., Crombé F., Nemeghaire S., Dispas M., Denis O., Hermans K., Haesebrouck F., Butaye P. Characterization of methicillin-resistant non-Staphylococcus aureus staphylococci carriage isolates from different bovine populations. J. Antimicrob. Chemother. 2013;68:300–307. doi: 10.1093/jac/dks403. [DOI] [PubMed] [Google Scholar]
- 7.Miragaia M. Factors contributing to the evolution of mecA-mediated β-lactam resistance in Staphylococci: Update and new Insights from Whole Genome Sequencing (WGS) Front. Microbiol. 2018;9:2723. doi: 10.3389/fmicb.2018.02723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bittar F., Keita M.B., Lagier J.C., Peeters M., Delaporte E., Raoult D. Gorilla gorilla gorilla gut: A potential reservoir of pathogenic bacteria as revealed using culturomics and molecular tools. Sci. Rep. 2014;4:7174. doi: 10.1038/srep07174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang D., Alvarado T.P., Oral D., Vargas J.M., Denena M.M., McCulley J.P. Ophthalmic examination of the captive western lowland gorilla (Gorilla gorilla gorilla) J. Zoo Wildl. Med. 2005;36:430–433. doi: 10.1638/03-121.1. [DOI] [PubMed] [Google Scholar]
- 10.Donato A.C.J., Penna B., Consalter A., Carvalho D.D., Lilenbaum W., Ferreira A.M.R. Antibiotic resistance in Staphylococcus sp. isolated from the vaginal environment of squirrel monkeys (Saimiri spp.) bred ex situ. J. Med. Primatol. 2017;46:75–78. doi: 10.1111/jmp.12258. [DOI] [PubMed] [Google Scholar]
- 11.Pantůček R., Sedláček I., Petráš P., Koukalová D., Švec P., Štětina V., Vancanneyt M., Chrastinová L., Vokurková J., Růžičková V., et al. Staphylococcus simiae sp. nov., isolated from South American squirrel monkeys. Int. J. Syst. Evol. Microbiol. 2005;55:1953–1958. doi: 10.1099/ijs.0.63590-0. [DOI] [PubMed] [Google Scholar]
- 12.Carvalho V.M., Vanstreels R.E., Paula C.D., Kolesnikovas C.K., Ramos M.C., Coutinho S.D., Martins C.S., Pissinatti A., Catão-Dias J.L. Nasal, oral and rectal microbiota of Black lion tamarins (Leontopithecus chrysopygus) Braz. J. Microbiol. 2015;45:1531–1539.D. doi: 10.1590/S1517-83822014000400051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lilenbaum W., Moraes I.A., Cardoso V.S., Varges R.G., Ferreira A.M., Pissinatti A. Antibiotic resistance in Staphylococci isolated from the vaginas of captive female Leontopithecus (Callitrichidae-Primates) Am. J. Primatol. 2006;68:825–831. doi: 10.1002/ajp.20282. [DOI] [PubMed] [Google Scholar]
- 14.Roberts M.C., Feßler A.T., Monecke S., Ehricht R., No D., Schwarz S. Molecular analysis of two different MRSA clones ST188 and ST2368 from primates (Macaca spp.) in a United States primate center. Front. Microbiol. 2018;9:2199. doi: 10.3389/fmicb.2018.02199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soge O.O., No D., Michael K., Dankoff J., Lane J., Vogel K., Smedley J., Roberts M.C. Transmission of MDR MRSA between primates personnel and environment at a United States primate center. J. Antimicrob. Chemother. 2016;71:2798–2803. doi: 10.1093/jac/dkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts M.C., Joshi P.R., Monecke S., Ehricht R., Muller E., Gawlik D., Paudel S., Acharya M., Bhattarai S., Pokharel S., et al. MRSA strains among Nepalese Rhesus macaques (Macaca mulatta) and their environment. Front. Microbiol. 2019;10:2505. doi: 10.3389/fmicb.2019.02505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heaton C.J., Gerbig G.R., Sensius L.D., Patel V., Smith T.C. Staphylococcus aureus epidemiology in wildlife: A systematic review. Antibiotics. 2020;9:89. doi: 10.3390/antibiotics9020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guardabassi L., Moodley A., Williams A., Stegger M., Damborg P., Halliday-Simmonds I., Butaye P. High prevalence of USA300 among clinical isolates of methicillin-resistant Staphylococcus aureus on St. Kitts and Nevis, West Indies. Front. Microbiol. 2019;10:1123. doi: 10.3389/fmicb.2019.01123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz-Ripa L., Feßler A.T., Hanke D., Sanz S., Olarte C., Mama O.M., Eichhorn I., Schwarz S., Torres C. Coagulase-negative staphylococci carrying cfr and PVL genes, and MRSA/MSSA-CC398 in the swine farm environment. Vet. Microbiol. 2020;43:108631. doi: 10.1016/j.vetmic.2020.108631. [DOI] [PubMed] [Google Scholar]
- 20.Heilmann C., Ziebuhr W., Becker K. Are coagulase-negative staphylococci virulent? Clin. Microbiol. Infect. 2019;25:1071–1080. doi: 10.1016/j.cmi.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Wuytack A., De Visscher A., Piepers S., Boyen F., Haesebrouck F., De Vliegher S. Non-aureus staphylococci in fecal samples of dairy cows: First report and phenotypic and genotypic characterization. J. Dairy Sci. 2019;102:9345–9359. doi: 10.3168/jds.2019-16662. [DOI] [PubMed] [Google Scholar]
- 22.Loncaric I., Tichy A., Handler S., Szostak M.P., Tickert M., Diab-Elschahawi M., Spergser J., Künzel F. Prevalence of Methicillin-Resistant Staphylococcus sp. (MRS) in different companion animals and determination of risk factors for colonization with MRS. Antibiotics. 2019;8:36. doi: 10.3390/antibiotics8020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bean D.C., Wigmore S.M., Wareham D.W. Draft Genome Sequence of Staphylococcus cohnii subsp. urealyticus Isolated from a healthy dog. Genome Announc. 2017;5:e01628-16. doi: 10.1128/genomeA.01628-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Argudín M.A., Vanderhaeghen W., Vandendriessche S., Vandecandelaere I., André F.X., Denis O., Coenye T., Butaye P. Antimicrobial resistance and population structure of Staphylococcus epidermidis recovered from animals and humans. Vet. Microbiol. 2015;178:105–113. doi: 10.1016/j.vetmic.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 25.Wilson D.N. The ABC of ribosome-related antibiotic resistance. mBio. 2016;7:e00598-16. doi: 10.1128/mBio.00598-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharkey L.K.R., Edwards T.A., O’Neil A.J. ABC-F Proteins Mediate Antibiotic Resistance through Ribosomal Protection. mBio. 2016;7:e01975. doi: 10.1128/mBio.01975-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018;31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanderhaeghen W., Vandendriessche S., Crombé F., Dispas M., Denis O., Hermans K., Haesebrouck F., Butaye P. Species and staphylococcal cassette chromosome mec (SCCmec) diversity among methicillin-resistant non-Staphylococcus aureus staphylococci isolated from pigs. Vet. Microbiol. 2012;158:123–128. doi: 10.1016/j.vetmic.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 29.Maes N., Magdalena J., Rottiers S., De Gheldre Y., Struelens M.J. Evaluation of a triplex PCR assay to discriminate Staphylococcus aureus from coagulase-negative Staphylococci and determine methicillin resistance from blood cultures. J. Clin. Microbiol. 2002;40:1514–1517. doi: 10.1128/JCM.40.4.1514-1517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cameron M., Perry J., Middleton J.R., Chaffer M., Lewis J., Keefe G.P. Evaluation of MALDI-TOF mass spectrometry and a custom reference spectra expanded database for the identification of bovine-associated coagulase-negative staphylococci. J. Dairy Sci. 2018;101:590–595. doi: 10.3168/jds.2017-13226. [DOI] [PubMed] [Google Scholar]
- 31.Argudín M.A., Vanderhaeghen W., Vandendriessche S., Vandecandelaere I., Denis O., Coenye T., Butaye P. Biofilm formation of ica operon-positive Staphylococcus epidermidis from different sources. APMIS. 2015;123:1081–1089. doi: 10.1111/apm.12472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
NCBI BioProject database and assigned BioProject Temporary SubmissionID PRJNA647767, with accession numbers https://www.ncbi.nlm.nih.gov/biosample/15636413, https://www.ncbi.nlm.nih.gov/biosample/15636414, https://www.ncbi.nlm.nih.gov/biosample/15636415, https://www.ncbi.nlm.nih.gov/biosample/15636416, https://www.ncbi.nlm.nih.gov/biosample/15636417, https://www.ncbi.nlm.nih.gov/biosample/15636418, https://www.ncbi.nlm.nih.gov/biosample/15636419, https://www.ncbi.nlm.nih.gov/biosample/15636420, https://www.ncbi.nlm.nih.gov/biosample/15636421, https://www.ncbi.nlm.nih.gov/biosample/15636422, https://www.ncbi.nlm.nih.gov/biosample/15636423, https://www.ncbi.nlm.nih.gov/biosample/15636424, https://www.ncbi.nlm.nih.gov/biosample/15636425, https://www.ncbi.nlm.nih.gov/biosample/15636426, https://www.ncbi.nlm.nih.gov/biosample/15636427, https://www.ncbi.nlm.nih.gov/biosample/15636428, https://www.ncbi.nlm.nih.gov/biosample/15636429, https://www.ncbi.nlm.nih.gov/biosample/15636430, https://www.ncbi.nlm.nih.gov/biosample/15636431.