Abstract
Medically important flaviviruses cause diverse disease pathologies and collectively are responsible for a major global disease burden. A contributing factor to pathogenesis is secreted flavivirus non-structural protein 1 (NS1). Despite demonstrated protection by anti-NS1 antibodies against lethal flavivirus challenge, the structural and mechanistic basis remains unknown. Here we present three crystal structures of full-length dengue virus (DENV) NS1 complexed with a flavivirus cross-reactive NS1-specific monoclonal antibody, 2B7, at resolutions between 2.89 Å and 3.96 Å. They reveal a protective mechanism by which two domains of NS1 are antagonized simultaneously. The NS1 wing domain mediates cell binding while the β-ladder triggers downstream events, both required for dengue, Zika, and West Nile virus NS1-mediated endothelial dysfunction. These observations provide a mechanistic explanation for 2B7 protection against NS1-induced pathology and demonstrate the potential of one antibody to treat infections by multiple flaviviruses.
One Sentence Summary:
A protective and cross-reactive antibody inhibits multiple steps of flavivirus NS1-mediated endothelial dysfunction.
Dengue virus serotypes 1–4 (DENV1–4) are mosquito-borne flaviviruses causing 50–100 million dengue cases and ~500,000 hospitalizations annually, with severe forms of disease manifesting in vascular leak as a result of endothelial dysfunction (1, 2). The triggers of these pathologies are often broadly described as a “cytokine storm” resulting from uncontrolled viral replication and activation of target immune cells, with a direct pathogenic role now characterized for the DENV non-structural protein 1 (NS1) through interactions with endothelial and immune cells (3–5). There are currently no approved therapeutics for dengue, and the only licensed DENV vaccine, Dengvaxia, is now reserved strictly for patients with preexisting DENV immunity due to the risk of predisposing DENV-naïve patients to severe dengue disease, presumably via antibody-dependent enhancement (ADE) (6, 7). This risk has made a successful vaccine targeting the DENV envelope protein (E) challenging; thus, targeting other DENV proteins important for pathogenesis is a critical avenue of investigation.
The multifunctional DENV NS1 protein plays a key role in viral replication as an intracellular dimer (8). NS1 is also secreted from DENV-infected cells as a barrel-shaped hexamer containing lipid cargo (9); it circulates in the blood and serves as a diagnostic antigen and biomarker of severity (10). Extracellular NS1 acts as a virulence factor inhibiting complement, activating platelets and immune cells, and directly interacting with endothelial cells (11–13). This results in disruption of the endothelial glycocalyx layer (EGL) and intercellular junctional complexes, both critical for maintaining endothelial barrier integrity (13–15). NS1-mediated endothelial dysfunction is observed for multiple medically relevant mosquito-borne flaviviruses including Zika (ZIKV), West Nile (WNV), Japanese encephalitis (JEV), and yellow fever (YFV) viruses (16, 17). With a prominent role in flavivirus pathogenesis, NS1 has emerged as a promising vaccine candidate. Indeed, vaccination with NS1 protects against lethal DENV or ZIKV challenge in mice (3, 18–20). As NS1 is conserved and triggers endothelial dysfunction through a comparable mechanism for multiple flaviviruses (16), one broadly reactive flavivirus therapeutic or vaccine targeting NS1 would be an innovative breakthrough.
Flavivirus NS1 has three domains that may possess distinct functions (21). Despite a plethora of structural data (21–25), the mechanistic basis for antibody-mediated protection against NS1-induced endothelial dysfunction and the specific functional domains of NS1 responsible for different pathogenic functions are unknown. We previously identified the anti-DENV NS1 IgG2b mouse monoclonal antibody (mAb) 2B7 as a strong inhibitor of NS1-induced endothelial hyperpermeability (3). We now show that in a DENV2 lethal challenge model, 2B7 was protective in mice in a dose-dependent manner, compared to an IgG isotype control, as was a single chain variable fragment (scFv) of 2B7, suggesting that protection could be achieved in a manner independent of antibody Fc effector functions (Fig. 1A, fig. S1A). In contrast, an anti-E antibody (4G2) given at the same dose was not protective, and in fact led to an accelerated time to death (Fig. 1A). Further, 2B7 blocked DENV NS1-mediated vascular leak in the mouse dermis compared to an IgG isotype control (Fig. 1B–C, fig. S1B–C). We next investigated the protective mechanism of 2B7 in vitro using human pulmonary microvascular endothelial cells (HPMEC) and measuring electrical resistance in a trans-endothelial electrical resistance (TEER) assay. Both 2B7 and its antigen-binding fragment (Fab), but not an IgG isotype control, were sufficient to abrogate NS1-induced endothelial hyperpermeability (Fig. 1D). Further, 2B7 and its Fab were sufficient to abrogate NS1-mediated endothelial dysfunction of HPMEC measured via cathepsin L activation and disruption of the EGL (measured via surface levels of sialic acid) (13, 26) (Fig. 1F–G). We also determined that 2B7 as well as its Fab and scFv were sufficient to block binding of NS1 to HPMEC and 293F cells (Fig. 1H–I, fig. S2A–C).
Fig. 1. Anti-NS1 mAb 2B7 is protective against lethal dengue virus infection and NS1-mediated vascular leak and endothelial dysfunction.
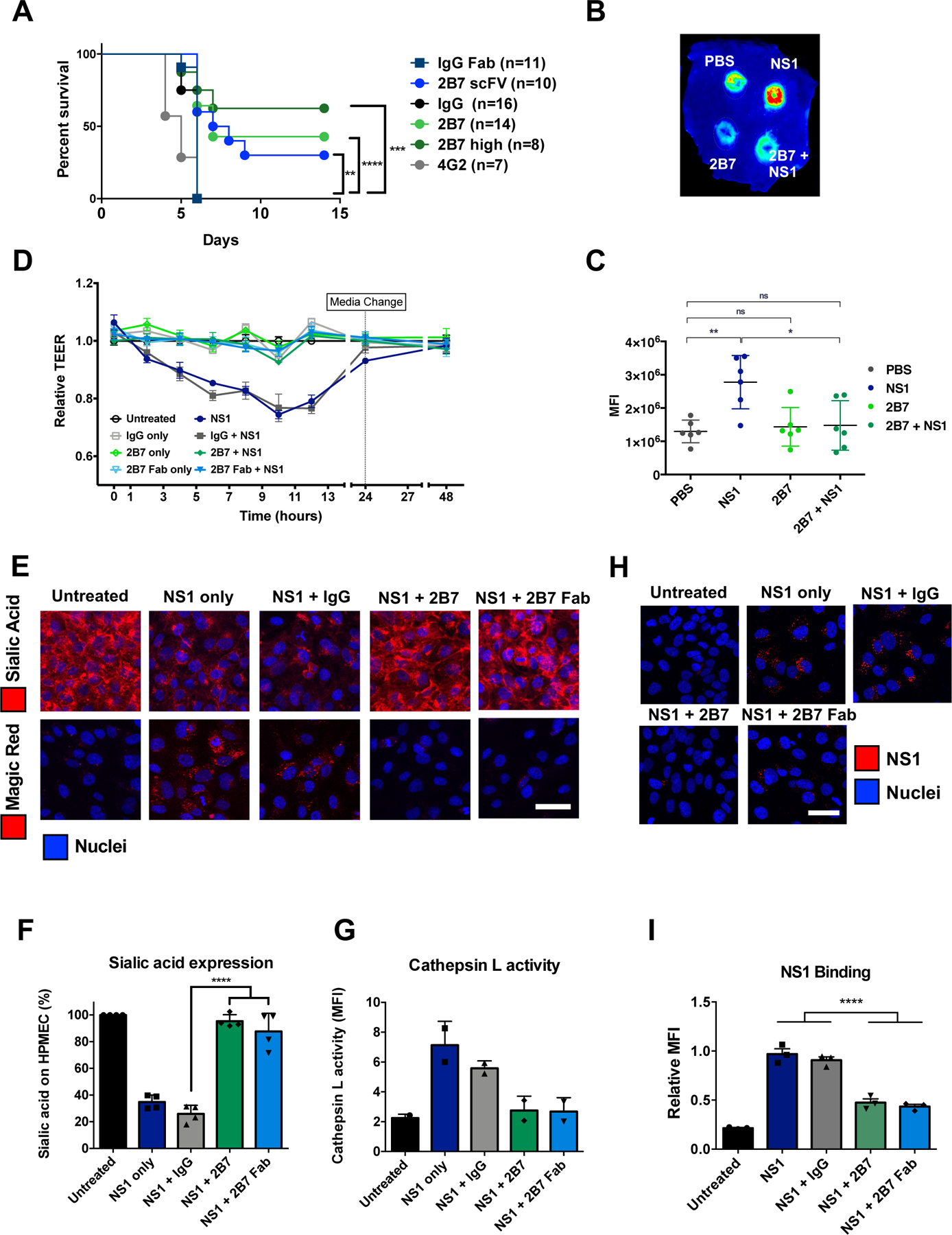
(A) Survival curve of Ifnar−/− mice infected with DENV2-D220. Mice were given two 150-μg doses (300 μg for “2B7 high”) of full-length 2B7, a 2B7 single-chain variable fragment (scFv), an anti-E antibody (4G2), or an isotype control antibody the day before and after infection. Numbers in parentheses indicate the number of mice in each group. (B) Localized leak of the tracer molecule, dextran-647, was measured after dorsal intradermal injection of NS1 with or without 2B7, or the indicated controls, into the shaved backs of mice. One representative experiment of n=6 mice is displayed. (C) Quantification of B as mean fluorescence intensity (MFI). (D) TEER assay of HPMEC hyperpermeability after addition of DENV2 NS1 with or without 2B7, or the indicated controls, at the indicated time-points post-NS1 treatment. n=3 biological replicates. (E) Endothelial dysfunction and EGL disruption was monitored using immunofluorescent microscopy 6 hours post-treatment of DENV2 NS1 with or without 2B7, 2B7 Fab, or the indicated controls. Endothelial cell dysfunction (bottom, n=2 biological replicates) was monitored using the cathepsin L-activity reporter molecule, Magic Red, and EGL disruption (top, n=4 biological replicates) was monitored by staining sialic acid on the cell surface. (F) and (G) Quantification of E. (H) DENV2 NS1 binding to HPMEC in the presence of 2B7, 2B7 Fab, or the indicated controls, was monitored by immunofluorescent microscopy 90 minutes post-NS1treatment. n=3 biological replicates. (I) Quantification of H. For all figures, scale bars are 50 μm. n.s., not significant p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 by one-way ANOVA analysis with multiple comparisons.
Having demonstrated the protective capacity of 2B7 and its Fab against DENV NS1 in vivo and in vitro, we investigated the structural basis of this protection. NS1 has three distinct domains: N-terminal β-roll, wing, and C-terminal β-ladder (21). An ELISA measuring binding of 2B7 to full-length NS1, a recombinant wing domain (residues 38–151), or a recombinant β-ladder domain (residues 176–352) indicated that 2B7 bound strongly to both full-length NS1 and the β-ladder, but not the wing domain (fig. S2D). This observation was confirmed using biolayer interferometry (BLI) (fig. S2E), which also revealed NS1 binding Kd values of 4.8±3.1 nM for 2B7 full-length, 8.3±6.8 nM for the 2B7 Fab, and 5.8±1.1 nM for the 2B7 scFv. We then used the VirScan phage display system with 56-mer overlapping peptides tiled across the DENV2 NS1 polypeptide to identify the epitope target region and found that 2B7 interacted with two overlapping peptides from the C-terminal β-ladder domain (residues 260–316 and 288–344), consistent with our ELISA and BLI results (27) (fig. S3A–B). We visualized the interaction between 2B7 and NS1 in detail by solving crystal structures of the 2B7 Fab in complex with DENV1 NS1 (3.3 Å, Fig. 2A, fig. S3B, fig. S4A, fig. S5B, table S1) and the 2B7 Fab and scFv in complex with DENV2 NS1 (scFv 2.89 Å, Fig. 2B–D, fig. S4B, fig. S5A; Fab 4.2 Å, fig. S5C and D; table S1). Each NS1 dimer binds two copies of the scFv/Fab fragment – one to each distal tip of the β-ladder (Fig. 2A, fig. S4, fig. S5C–E). The NS1:2B7 complex has an overall arch shape, where the antibody fragments form the sides of the arch, with the membrane-facing hydrophobic side of NS1 on the arch inner surface (Fig. 2A, fig. S4). In this configuration, the 2B7 Fab would likely prevent the hydrophobic face of NS1 from interacting with cell surfaces. The 2.89-Å electron density map was of sufficient quality to confidently build the scFv constant and variable regions as well as the side chains of the combining site of the 2B7 scFV and the DENV2 NS1 discontinuous epitope (Fig. 2B–D, fig. S5A). The variable loops of the 2B7 light chain make numerous contacts with NS1 β-ladder residues, consistent with our ELISA, BLI, and phage display results (Fig. 2B–D, fig. S3, fig S4, fig. S5, Table S2). The binding modalities of 2B7 to NS1 were identical in our three structures (fig. S5E), and the NS1 structure was unchanged by 2B7 binding.
Fig. 2. Crystal structure of the 2B7 antigen-binding fragment complexed with DENV NS1 reveals binding to β-ladder.

(A) Perpendicular views of a 3.3-Å crystal structure of 2B7 Fab (heavy chain, dark blue; light chain, light blue) and DENV1 NS1 dimer (β-ladder domains, green; β-roll and wing domains, cyan). The combining site is boxed (yellow) in the lower image, right monomer. (B) Detail of the 2B7 scFv and the DENV2 NS1 combining site highlighting the interacting amino acids. The 2B7 backbone is in blue, and NS1 is in green with key side chains shown as sticks. Pan-flavivirus conserved side chains (orange) are at the center of the discontinuous epitope; DENV-conserved side chains (yellow) are at the epitope periphery; hydrogen bonds are shown as dashed lines. (C) DENV2 NS1 epitope for 2B7 scFv (colored by conservation across flaviviruses according to the key and based on the alignment in Fig. 3A), with surfaces outside the epitope in gray. Sites of mutagenesis in Fig. 3 are labeled. (D) 2B7 scFv complementarity-determining regions (CDRs, in tube rendering) for the heavy chain (dark blue) and light chain (light blue) overlaid on the DENV2 NS1 epitope surface. Surfaces of amino acids conserved among the four DENV serotypes but variable in other flaviviruses are purple, other epitope residues are in green; view as in (C).
The amino acid residues in the 2B7 epitope can be divided into two classes: the epitope core region, composed of residues that are highly conserved across flaviviruses, and the epitope periphery, displaying varying levels of divergence among flaviviruses. (Fig. 2B–D and 3A). Hypothesizing that 2B7 would recognize distinct flavivirus NS1 proteins with affinities that correlated with the extent of conservation with DENV NS1, we performed ELISAs to measure the relative affinities of 2B7 for a panel of flavivirus NS1 proteins including DENV 1–4, ZIKV, Saint Louis encephalitis virus (SLEV), WNV, JEV, tick-borne encephalitis virus (TBEV), Powassan virus (POWV), Usutu virus (USUV), Wesselsbron virus (WSLV), and YFV. 2B7 bound most tightly to NS1 from DENV1–4, followed by ZIKV, SLEV, WNV, JEV, USUV, and WSLV, with minimal binding detected for YFV, POWV, and TBEV. The strength of binding correlated with the degree of conservation with DENV NS1 (Fig. 3B–C). To determine the relative contribution of key flavivirus NS1 amino acid residues on 2B7 binding, we created single, double, triple, or quadruple substitutions of the DENV2 NS1 β-ladder amino acids within the NS1:2B7 epitope (Table S3). We initially screened NS1 mutants produced in 293T cells for candidates that were secreted and displayed diminished binding to 2B7 (fig. S6A–B). We then selected seven DENV NS1 β-ladder single substitutions (NS1-D281P, NS1-T301R, NS1-T301K, NS1-A303W, NS1-G305K, NS1-E326K, and NS1-D327K) for purification and 2B7 binding ELISAs. A direct ELISA revealed that with the exception of NS1-D281P, each of these mutants displayed weaker binding to 2B7 compared to NS1-WT (Fig. 3D–E, fig. S6A, Table S3). To confirm the role of the flavivirus-conserved residues above in 2B7 binding to multiple flavivirus NS1 proteins, we created two single mutants for both WNV and ZIKV NS1 proteins (WNV/ZIKV NS1-T301K and NS1 G305K) (fig. S7A–C). A direct ELISA revealed that these NS1 mutants exhibited a severe binding defect to 2B7 compared to their respective NS1-WT control proteins (Fig. 3F). To test if the NS1 cross-reactivity of 2B7 correlated with function, we used ZIKV and WNV NS1 proteins in NS1-cell binding and TEER assays on human brain microvascular endothelial cells (HBMEC). 2B7, but not an IgG isotype control, blocked binding and abrogated NS1-mediated endothelial hyperpermeability in a dose-dependent manner correlating with conservation to DENV2 NS1, indicating that one cross-reactive NS1 mAb could inhibit pathogenic functions of NS1 from multiple flaviviruses (Fig. 3G–J, fig. S8A–C). As ADE of DENV infection is problematic for anti-E antibodies, we confirmed that anti-NS1 mAb 2B7 was incapable of mediating ADE of DENV, ZIKV, and the WNV strain Kunjin virus (KUNV) infection (fig. S9A–D). Further, as anti-NS1 antibodies have been reported to modulate the clotting cascade (28) as well as bind to endothelial cells, which may directly mediate endothelial dysfunction (29), we tested the capacity of 2B7 to alter clotting time of human plasma as well as to bind to the surface of endothelial cells. These experiments determined that 2B7 does not alter human clotting time and binds to the surface of endothelial cells significantly more than an isotype control only when NS1 is present, suggesting that 2B7 would likely not enhance DENV disease through these specific mechanisms (fig. S10A–C).
Fig. 3. 2B7 cross-reacts with multiple flavivirus NS1 proteins.
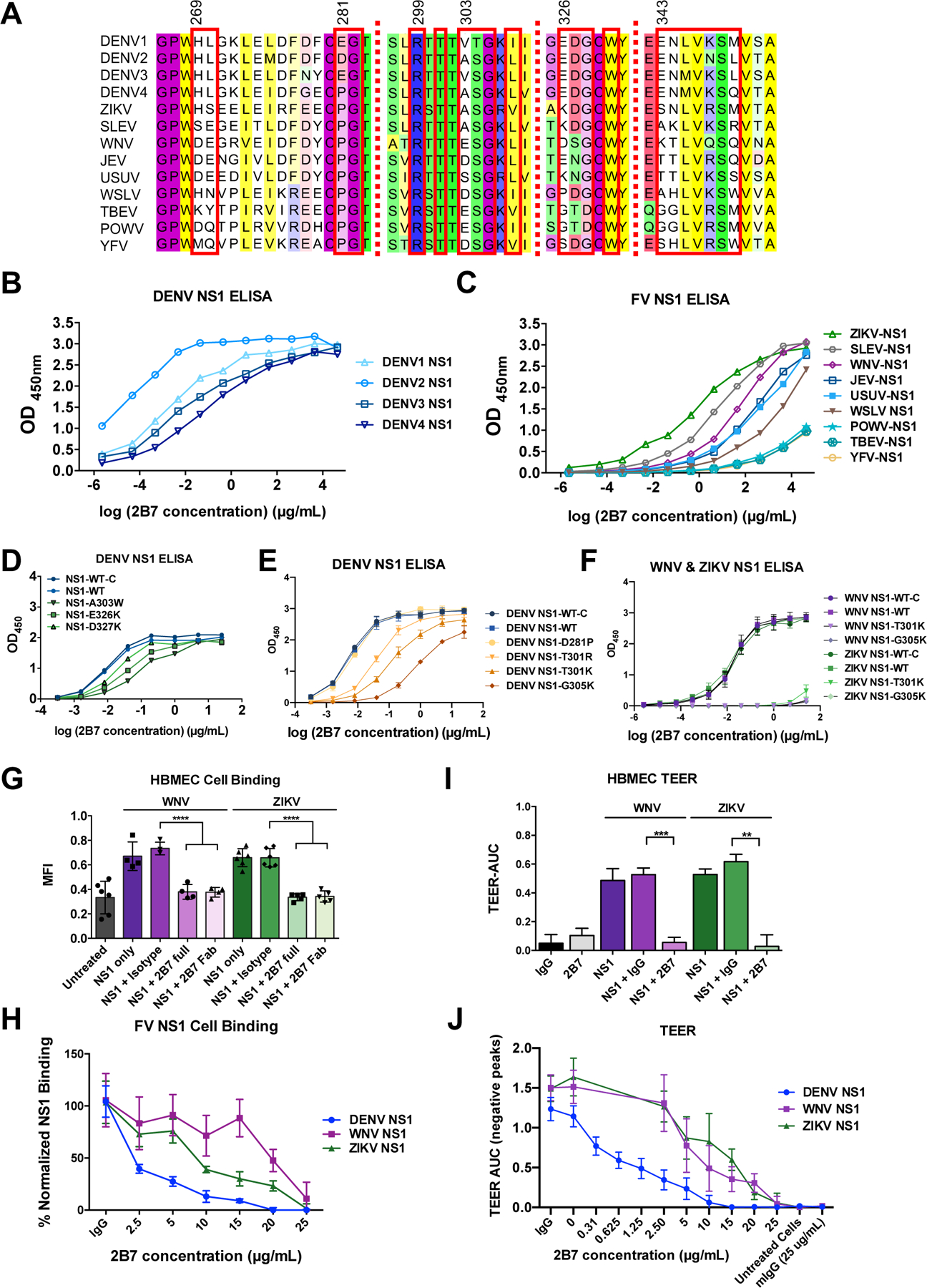
(A) Alignment of amino acids across the discontinuous 2B7 combining site in NS1 from different flaviviruses. Residues in contact with 2B7 are boxed in red. (B) ELISAs measuring the interaction of 2B7 with NS1 from DENV1–4. Data displayed are at least n=3 biological replicates. (C) ELISAs measuring 2B7 interaction with other flavivirus NS1 proteins (at least n=3 biological replicates). (D) and (E) Same as (C) except for the indicated DENV2 NS1 mutagenized proteins compared to the in-house produced (NS1-WT) or commercially purchased (NS1-WT-C) control proteins. Data displayed are n=3 biological replicates. (F) Same as (C) except for the indicated WNV or ZIKV NS1 mutagenized proteins (n=3 biological replicates). (G) WNV or ZIKV NS1 binding to HBMEC in the presence or absence of 25 g/ml 2B7, and the indicated controls, was measured by immunofluorescent microscopy 90 minutes post-NS1 treatment. Data displayed are at least n=4 biological replicates. (H) Same as (G) but with the indicated concentrations of 2B7 (at least n=3 biological replicates). (I) The effect of WNV or ZIKV NS1 on hyperpermeability of HBMEC was measured using TEER, in the presence or absence of 25 g/ml 2B7, or the indicated controls. AUC is the negative area under the curve (fig. S8C) correlating with a drop in endothelial cell monolayer electrical resistance. Data presented are n=2 biological replicates. (J) Same as (I) but with the indicated concentration of 2B7 (at least n=3 biological replicates). **p < 0.01; ***p < 0.001; ****p < 0.0001 by one-way ANOVA analysis with multiple comparisons. FV, flavivirus.
As NS1 is reported to mediate endothelial dysfunction through distinct steps including cell binding, EGL disruption, and endothelial hyperpermeability (13, 14, 16), the mode of 2B7 binding to NS1 provides an opportunity to investigate the molecular basis of NS1-mediated endothelial dysfunction. Although 2B7 binds to the β-ladder, its tilted orientation towards the NS1 hydrophobic surface (Fig. 2A, fig. S4, fig. S5, Fig. 4A–B) predicts that 2B7 would create indirect steric hinderance for the wing domain and would interfere with a predicted interaction between the NS1 wing domain and the cell surface whether NS1 is in the dimeric or hexameric form (22, 30) (Fig. 4A–B, Movie 1). As such, 2B7 is predicted to interfere with both the wing (indirect steric hinderance) and β-ladder (direct binding) of NS1, suggesting that both domains may be critical for NS1 pathology. To test this prediction, we generated mutant expression constructs for NS1 with substitutions predicted to hinder binding to the cell surface or to 2B7. In addition to the DENV NS1 β-ladder mutants described above, we created a DENV NS1 triple substitution within the flavivirus-conserved W115XXW118G119 motif (A115XXA118A119) in an immunodominant region of the wing, which is predicted to interact with the cell surface (22, 30–32) (fig. S11, Fig. 4A–B). We purified six DENV NS1 β-ladder single substitutions (NS1-T301R, NS1-T301K, NS1-A303W, NS1-G305K, NS1-E326K, and NS1-D327K) and the wing domain triple substitution (NS1-WWG>AAA, “NS1-WWG”). All proteins were expressed, secreted, oligomeric, stable, and of purity comparable to wild-type NS1 (NS1-WT) (fig. S12A–F). Further, in contrast to the NS1 β-ladder mutants, the wing domain mutant did not exhibit diminished binding to 2B7 compared to NS1-WT (Fig. 3D–E, fig. S12G). Intriguingly, in a TEER assay on HPMEC with these mutant NS1 proteins, all NS1 mutants except the β-ladder mutants NS1-T301K and NS1-G305K were defective in their capacity to mediate endothelial hyperpermeability, as compared to NS1-WT, with NS1-A303W displaying the greatest functional defect (Fig. 4C, fig. S13A). These data indicate that several residues both in the tip of the β-ladder and, separately, in the wing domain are critical for NS1-mediated endothelial dysfunction. To investigate the mechanism(s) of the functional defects of these NS1 mutants, we conducted an NS1 cell-binding assay using both HPMEC and 293F cells and found that while NS1 with β-ladder substitutions bound to cells comparably to NS1-WT, NS1-WWG possessed a significant cell binding defect (Fig. 4D and fig. S13B–C). In contrast, when we examined steps downstream of NS1 binding, such as activation of cathepsin L, all mutants were defective relative to NS1-WT (focusing only on the DENV conserved β-ladder mutants) (Fig. 4E–F). Taken together, these data suggest that the NS1 wing domain, specifically the WWG motif, is critical for initial attachment of NS1 to cells, whereas the tip of the β-ladder is critical for downstream events required for NS1-mediated endothelial dysfunction.
Fig. 4. Mechanistic insight into 2B7 blockade of NS1-mediated endothelial dysfunction.

(A) 2B7 Fab (blue ribbon) in complex with DENV1 NS1 hexamer above a schematic of the plasma membrane, illustrating 2B7 Fab-mediated steric hinderance of NS1 membrane interaction. The NS1 surface is colored by dimer (light/dark green at right, gray at left and tan in the back), with hydrophobic regions in yellow for all three dimers. The 2B7 Fab is bound to both subunits of the green dimer and occludes cell surface interaction of the yellow wing-domain hydrophobic loop (centered on the WWG motif). (B) Perpendicular views of 2B7 Fab complex with DENV1 NS1 dimer, illustrating Fab interference with membrane interaction of the NS1 hydrophobic face (yellow). The left image shows the dimer hydrophobic face with wing hydrophobic loops at the periphery and central hydrophobic surface of the β-roll domain. This face is inside the hexamer and invisible in (A). (See Movie S1.) The NS1 epitope for 2B7 is in orange. (C) Endothelial hyperpermeability of HPMEC was monitored via TEER for the indicated DENV2 NS1 mutants compared to the commercial NS1-WT (NS1-WT-C) or in-house produced NS1-WT (NS1-WT). Data presented are n=2 biological replicates. (D) Cell binding to HPMEC of NS1 mutants or controls was monitored by immunofluorescent microscopy 90 minutes post-NS1 treatment (n=3 biological replicates). (E) Endothelial dysfunction was monitored using immunofluorescent microscopy and the cathepsin L-activity reporter molecule, Magic Red, 6 hours post-treatment of DENV2 NS1 mutants, or the indicated controls. Data presented are n=2 biological replicates. (F) Quantification of E. n.s., not significant p > 0.05; *p < 0.05; **p < 0.01 by one-way ANOVA analysis with multiple comparisons.
Overall, these findings serve as proof-of-concept that one cross-reactive antibody targeting flavivirus NS1 proteins can provide protection against NS1-mediated pathology from multiple flaviviruses, with no risk of ADE. Further, the structure of the 2B7 scFv/Fab in complex with NS1 revealed that 2B7 obscures the β-ladder through direct binding and the wing domain through indirect steric hinderance of the NS1 dimer and/or hexamer form, suggesting that one anti-NS1 mAb can simultaneously antagonize the cellular interactions of two domains. DENV NS1 mutagenesis implicated the wing domain as critical for initial binding to the endothelial cell surface and the β-ladder as essential for downstream NS1-mediated events, both crucial steps for NS1-triggered pathology. Information gained from the differential binding affinities of 2B7 for multiple flavivirus NS1 proteins provides a molecular roadmap for the rational design of mAbs, or other therapeutics, with even broader flavivirus NS1 cross-reactivity compared to 2B7. In summary, our structural and mechanistic investigations of 2B7-mediated protection reveal the critical and distinct roles of the NS1 wing domain and the β-ladder and highlight the possibility of treating multiple flavivirus infections with one therapeutic targeting flavivirus NS1.
Supplementary Material
Acknowledgments:
We thank Dr. Andrew Sandstrom (UC Berkeley) for assistance with size-exclusion chromatography, and Dr. Sandstrom and Dr. Patrick S. Mitchell (UC Berkeley) for helpful discussion and critical reading of this manuscript. We thank Colin M. Warnes (UC Berkeley) for technical assistance with 2B7 antibody purification. We thank Yumei Leng (Brigham and Women’s Hospital) for assistance processing 2B7 with VirScan. We thank Dr. Michael Diamond (Washington University in St. Louis) for the pmab vector containing the WT DENV2 NS1 protein (strain 16681). Confocal imaging experiments were conducted on a Zeiss LSM 710 at the CRL Molecular Imaging Center, supported by the Gordon and Betty Moore Foundation. This research used the GM/CA@APS facility funded by the NIH NIGMS (AGM-12006) and the NIH NCI (ACB-12002) at the Advanced Photon Source at Argonne National Laboratory (DE-AC02–06CH11357). The Eiger 16M detector was funded by NIH (S10 OD012289).
Funding:
This work was supported by NIAID/NIH grants R01 AI24493 (E.H.), R21 AI146464 (E.H.), U19 AI109761 (E.H., Program Director I. Lipkin) and R56 AI130130 (J.L.S.). S.J.E. was supported by grant 5U24AI116833–02 and the Bill and Melinda Gates Foundation OPP1155863. M.J.M. and S.J.E were supported by a grant from the Value of Vaccine Research Network. S.B.B. was partially supported as an Open Philanthropy Awardee of the Life Sciences Research Foundation.
Footnotes
Competing interests: Authors declare no competing interests.
Data and materials availability: The structures of DENV1 NS1–2B7 Fab, DENV2 NS1–2B7 Fab, and DENV2 NS1–2B7 scFV are deposited in the PDB with accession codes 6WEQ, 6WER, and 7K93, respectively. The sequences of the 2B7 mAb heavy and light chains are submitted to GenBank with accession codes MW246826 and MW246827, respectively. Materials generated in this study are available upon request.
References:
- 1.Bhatt S et al. , Nature 496, 504–507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halstead SB, Lancet 370, 1644–1652 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Beatty PR et al. , Sci Transl Med 7, 304ra141 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Modhiran N et al. , Sci Transl Med 7, 304ra142 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Rothman AL, Nat Rev Immunol 11, 532–543 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Sridhar S et al. , N Engl J Med 379, 327–340 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Katzelnick LC et al. , Science 358, 929–932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scaturro P, Cortese M, Chatel-Chaix L, Fischl W, Bartenschlager R, PLoS Pathog 11, e1005277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutsche I et al. , Proc Natl Acad Sci U S A 108, 8003–8008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paranavitane SA et al. , BMC Infect Dis 14, 570 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avirutnan P et al. , J Exp Med 207, 793–806 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao CH et al. , PLoS Pathog 15, e1007625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puerta-Guardo H, Glasner DR, Harris E, PLoS Pathog 12, e1005738 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glasner DR, Puerta-Guardo H, Beatty PR, Harris E, Annu Rev Virol, (2018). [DOI] [PMC free article] [PubMed]
- 15.Singh S, Anupriya MG, Modak A, Sreekumar E, Journal of General Virology 99, 1658–1670 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Puerta-Guardo H et al. , Cell Rep 26, 1598–1613.e1598 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puerta-Guardo H et al. , J Infect Dis, (2019). [Google Scholar]
- 18.Espinosa DA et al. , J Immunol 202, 1153–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brault AC et al. , Sci Rep 7, 14769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlesinger JJ, Brandriss MW, Walsh EE, J Gen Virol 68 ( Pt 3), 853–857 (1987). [DOI] [PubMed] [Google Scholar]
- 21.Akey DL et al. , Science 343, 881–885 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu X et al. , EMBO J 35, 2170–2178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edeling MA, Diamond MS, Fremont DH, Proc Natl Acad Sci U S A 111, 4285–4290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poonsiri T et al. , J Virol 92, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song H, Qi J, Haywood J, Shi Y, Gao GF, Nat Struct Mol Biol 23, 456–458 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Glasner DR et al. , PLoS Pathog 13, e1006673 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu GJ et al. , Viral immunology. Science 348, aaa0698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuang YC, Lin J, Lin YS, Wang S, Yeh TM, J Immunol 196, 1218–1226 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Lin CF et al. , J Med Virol 69, 82–90 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Brown WC et al. , Nat Struct Mol Biol 23, 865–867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hertz T et al. , J Immunol 198, 4025–4035 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai YC et al. , Sci Rep 7, 6975 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orozco S et al. , J Gen Virol 93, 2152–2157 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montoya M et al. , J Infect Dis 218, 536–545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabata T et al. , 20, 155–166 (2016). [Google Scholar]
- 36.Wang C et al. , PLoS Pathog 15, e1007938 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma M et al. , J Infect Dis, (2019). [Google Scholar]
- 38.Mina MJ et al. , Science 366, 599–606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown WC et al. , Protein Expr Purif 77, 34–45 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DelProposto J, Majmudar CY, Smith JL, Brown WC, Protein Expr Purif 63, 40–49 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabsch W, Acta Crystallogr D Biol Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy AJ et al. , J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K, Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Adams PD et al. , Acta Crystallogr D Biol Crystallogr 58, 1948–1954 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Chen VB et al. , Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashkenazy H et al. , Nucleic Acids Res 44, W344–350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


