Abstract
Background: Feather pecking is a well-known problem in layer flocks that causes animal welfare restrictions and contributes to economic losses. Birds’ gut microbiota has been linked to feather pecking. This study aims to characterize the microbial communities of two laying hen lines divergently selected for high (HFP) and low (LFP) feather pecking and investigates if the microbiota is associated with feather pecking or agonistic behavior. Methods: Besides phenotyping for the behavioral traits, microbial communities from the digesta and mucosa of the ileum and caeca were investigated using target amplicon sequencing and functional predictions. Microbiability was estimated with a microbial mixed linear model. Results: Ileum digesta showed an increase in the abundance of the genus Lactobacillus in LFP, while Escherichia was abundant in HFP hens. In the caeca digesta and mucosa of the LFP line were more abundant Faecalibacterium and Blautia. Tryptophan metabolism and lysine degradation were higher in both digesta and mucosa of the HFP hens. Linear models revealed that the two lines differ significantly in all behavior traits. Microbiabilities were close to zero and not significant in both lines and for all traits. Conclusions: Trait variation was not affected by the gut microbial composition in both selection lines.
Keywords: gut microbiota, feather pecking, microbiability, laying hen, agonistic behavior
1. Introduction
Feather pecking is a detrimental behavior pattern shown in layer flocks, leading to injured birds and, consequently to the welfare and economic problems. Research over the last few decades revealed the underlying mechanisms of feather pecking (for a review, see Rodenburg et al. [1]). Still, it remains an unsolved problem in the poultry industry worldwide.
It is well known that environmental and genetic factors determine feather pecking. Previous research led to the assumption that the gut microbial composition is also involved in developing the undesired behavior. In laying hen lines divergently selected for feather pecking, supplementation with the essential amino acid L-tryptophan significantly reduced feather pecking by increasing the serotonergic tone [2]. Tryptophan supplementation increases the abundance of non-pathogenic bacteria (Bifidobacteria and Enterococci) known to support gut integrity and health [3]. A higher amount of feather pecking comes with a higher amount of feather eating [4,5,6,7], although raw feathers do not have any nutritional value [8]. Lutz et al. [9] identified a causal effect of feather eating on feather pecking using structural equation models. Meyer et al. [10] found differences in the gut microbiota and their metabolites between laying hen strains fed with different amounts of feathers. Some studies revealed that laying hen lines divergently selected for feather pecking also differed in some aspects of their gut microbial composition [11,12,13]. These findings suggested that the gut microbial composition might be associated with feather pecking and even might be one cause for it.
The ileum represents a major nutrient absorption site of the gastrointestinal (GI) tract in chickens and is dominated by Lactobacillus [14], Streptococcus, and Escherichia coli [15]. The caeca are colonized by a huge diversity of bacterial members, specifically Clostridiaceae, Bacteroidaceae, Lactobacillaceae, Proteobacteria, and butyrate-producing clusters as well as several uncultured bacteria [15,16]. The chicken caeca are an important fermentation site. They are responsible for the digestion of foods rich in cellulose, starch, and resistant polysaccharides, which impact the health and performance of the animals. Therefore, the contributing microbiota has been extensively examined [16,17,18]. Although it is known that the intestinal microbiota differs between mucosa and digesta samples, yet most studies characterized the digesta [19,20]. In all GI sections, mucosa samples showed higher microbial diversity than the digesta samples [15].
The term microbiability [21] describes the part of the phenotypic variance of a trait which is explained by the microbial composition. This parameter can be estimated with microbial mixed linear models. Microbiabilities in a medium-range were estimated for feed-related traits in pigs [22,23]. Verschuren et al. estimated high microbiabilities for the digestibility of several nutrients in fecal samples of pigs [24]. In a study on Japanese quails, medium microbiabilities for feed-related traits were identified [25]. Hence, the usefulness of microbiability to define the gut microbiome’s effect on feed-related traits in pigs and poultry could be revealed successfully.
Research on humans, rodents, and livestock showed that the gut microbiota composition influences behavior, e.g., anxiety-related, social, or feeding behavior [26]. Germ-free quail chicks were selected for high emotional reactivity (measured with tonic immobility test) and received either feces of conventional adults of the same line or a line selected for low emotional reactivity [27]. Germ-free chicks that received gut microbiota of the fearless line showed significantly less emotional reactivity than chicks with the fearful line’s microbiota. After two weeks, the gut microbial composition returned to its equilibrium, which was partially determined by the host genome [27]. Probiotic supplementation reduced fearfulness, improves memory, and reduces agonistic poultry behavior [26,28].
The present study aimed to characterize the gut microbial composition and its predicted functionality from two laying hen lines divergently selected for high (HFP) and low (LFP) feather pecking behavior. A possible influence of the gut microbiota composition toward feather pecking and agonistic behavior was investigated by applying microbial mixed linear models.
2. Materials and Methods
2.1. Birds and Experimental Procedures
The experiment and the experimental population’s establishments are described in Iffland et al. [29]. Briefly, hens of a White Leghorn layer strain were divergently selected for the severe form of feather pecking for 15 generations. Hens were reared together, regardless of the line, and were kept under the same conditions from hatching on. For behavioral observations at around 32 weeks of age, the hens were divided into smaller mixed HFP and LFP groups of about 40 animals and housed in deep litter pens. Observation, by experienced observers, began one week after group formation and took place during four consecutive days [30]. Due to a limited number of pens, two experimental runs were performed phenotyping a total of 492 hens (nHFP = 270, nLFP = 222). Besides others, three behavior traits were recorded, feather pecks delivered (FPD), aggressive pecks delivered (APD), and threats delivered (TD). The ethogram is displayed in Table 1.
Table 1.
Ethograms of the recorded traits feather pecks delivered (FPD), aggressive pecks delivered (APD) and threats delivered (TD).
| Trait | Definition |
|---|---|
| FPD | Non-aggressive severe pecks or pulls are directed to the plumage of conspecifics, sometimes resulting in pulled-out feathers and a recipient, which tolerates or moves away. Therefore, the deliverer does not adopt any special body posture. |
| APD | Pecks delivered in an upright body posture against (mainly) the head and other parts of the recipient’s body. |
| TD | Visual fixation on the recipient in an upright body posture followed by the recipient’s avoidance or withdrawal behavior. |
All recorded traits were BoxCox transformed to reduce their deviation from a normal distribution. After the observation period of each experimental run, the hens were slaughtered at around 35 weeks of age. Both ileum and caeca were longitudinally opened, and digesta was collected with a sterile spoon. The mucosa was washed with a sterile phosphate-buffered saline solution and scraped with a sterile glass slide. Samples were stored in RNAlater at −80 °C until further analysis. The samples were divided into eight groups based on intestinal section (ileum or caecum), type of samples (digesta or mucosa), and line affiliation (HFP or LFP). The number of phenotyped animals with samples is shown separately for the sections and sample types in Table 2.
Table 2.
Number of animals of the high (HFP) and low (LFP) feather pecking line with samples for the respective gut section and type of samples used in the microbial linear mixed model.
| Gut Section and Sample Type | HFP | LFP | ∑ |
|---|---|---|---|
| Ileum mucosa | 96 | 73 | 169 |
| Ileum digesta | 95 | 82 | 177 |
| Caecum mucosa | 48 | 42 | 90 |
| Caecum digesta | 48 | 43 | 91 |
The German Ethical Commission of Animal Welfare of the State Government of Baden-Wuerttemberg, Germany approved the research protocol.
2.2. DNA Extraction Illumina Amplicon Sequencing and Bioinformatic Analysis
DNA was extracted from approximately 250 mg of each digesta and mucosa sample using FastDNATM SPIN Kit for soil from MP Biomedicals (Solon, OH, USA) following the manufacturer instructions. The quality and concentration of DNA were assessed through NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), and DNA was stored until use at −20 °C. The V1-2 region of the 16S rRNA gene was amplified to produce the Illumina sequencing library. The protocol followed the methodology of Kaewtapee et al [31]. Briefly, one microliter of DNA was used as a template in the first PCR, where the forward primer contains a six-nucleotide barcode, and both primers have sequences complementary to the Illumina adapters. Master mixes include the PrimeSTAR® HS DNA Polymerase kit (TaKaRa, Beijing, China). One microliter of the first PCR product was used in a second PCR following the same PCR conditions where both primers were complemented to the sequences of Illumina multiplexing and index primers. Amplicons were verified by agarose gel electrophoresis, purified, and normalized using SequalPrep Normalization Kit (Invitrogen Inc., Carlsbad, CA, USA). Samples and negative controls were sequenced using 250 bp paired-end sequencing chemistry on an Illumina MiSeq platform.
Raw sequence reads obtained from Illumina MiSeq system (Illumina, Inc., San Diego, CA, USA) were analyzed using QIIME v1.9.1 pipeline [32], following a subsampled open-reference operational taxonomic units (OTUs) calling approach [33]. Demultiplexing and trimming of sequencing reads were done using the pipeline’s default parameters with a maximum sequence length of 360 bp [34]. The reads were merged into one FASTA-file and aligned using the SILVA Database (Release 132) [35]. Chimeras were identified and removed using usearch [36]. Reads were clustered at 97% identity into OTUs. Only OTUs present on average abundance higher than 0.0001% and a sequence length of >250 bp were considered for further analysis. The closest representative was manually identified with the seqmatch function of the Ribosomal Database Project. An average of 44,240 reads were obtained per sample. Sequences were submitted to European Nucleotide Archive under the accession number PRJEB40535.
Prediction of functionality was carried out with the R package Tax4Fun2 [37], which relied on the SILVA database [38] and used the Kyoto Encyclopedia of Genes and Genomes (KEGG) hierarchy for the assignations, which comprise gene catalogs from sequenced genomes [39]. The biom table to assign this functionality was obtained from the QIIME pipeline. Genomes from 16S rRNA gene sequences identified in this study were downloaded from the National Center for Biotechnology Information (NCBI) database to produce a case-study-specific database for the ileum and caeca of laying hens.
2.3. Statistical Analysis
Linear discriminant analysis effect size (LEfSe) was applied to observe differences at the OTU level between the HFP and LFP line. The default cutoff was used, including q value < 0.1 and linear discriminant analysis score > 2.0 [40]. Random forest analysis overview was obtained at the OTU level to differentiate the impact of HFP and LFP on the prediction in microbiome data classification. Values by default were 500 trees, and the plots included the out-of-bag error [40].
Datasets were analyzed using PRIMER (version 7.0.9, PRIMER-E, Plymouth Marine Laboratory, Plymouth, UK) [41]. Data was standardized by total, and a similarity matrix was created using the Bray-Curtis coefficient [42]. PERMANOVA analysis, using a permutation method under a reduced model, was used to study the significant differences obtained when the dietary treatments were analyzed and considered significantly different if p < 0.05 [41]. The community similarity structure was depicted through non-metric multidimensional scaling plots. Similarity percentage analysis was used to identify the OTUs responsible for the groups’ differences. Diversity indices (Shannon diversity and Pielou’s evenness) were calculated based on abundance data with PRIMER software.
For estimation of the microbial variance components and the microbiability, the following microbial mixed linear model was applied using ASReml-R (Version 3.0) [43,44]. The model was applied separately for each trait and each gut section.
| (1) |
where y is the vector containing the trait records for the corresponding trait (i.e., FPD, APD, or TD). X is a design matrix for vector b, which contained the line’s fixed effect, and a combination of experimental run and pen, if significant. Vector e denotes the random residual term. The residuals were modeled heterogeneously within the two feather pecking lines. The Vector m contains the random microbiota animal effects with distribution
| (2) |
with M being the microbial relationship matrix and denoting the microbial variance. M was calculated as
| (3) |
with N being the number of OTUs, and X is a matrix, where n is the number of animals. The standardized and log-transformed abundances of the OTUs are contained in X [22]. The microbiabilities for each trait and line l (l = HFP or LFP) were estimated as the fraction of the phenotypic variance in the lines explained by . A likelihood-ratio test on the random microbial animal effect was performed to test the significance of the microbiabilities. The test statistic was calculated as
| (4) |
with being the likelihood of the reduced model, i.e., model (1) without the random microbiota animal effect and the likelihood of the full model. The test statistic D under the null-hypothesis was chi-squared distributed with one degree of freedom. In addition, the two feather pecking lines were analyzed separately with the same model but without a fixed-line effect.
3. Results
3.1. Microbial Community
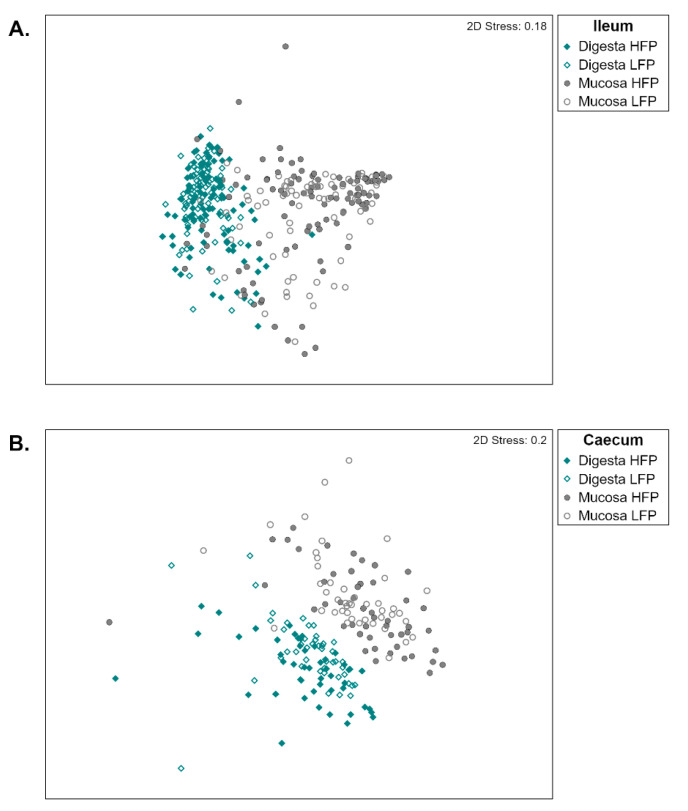
A significant (p = 0.003) difference in section and feather pecking line interaction was demonstrated by PERMANOVA (Table S1A). Samples of both ileum and caeca clustered by mucosa and digesta (Figure 1) and significant differences were obtained for the feather pecking lines and the type of samples (digesta or mucosa) (Table S1B,D,E). The Shannon diversity index showed significant (p ≤ 0.05) differences between ileum and caeca, being higher in the caeca but not between mucosa and digesta samples or the two lines of hens (Figure S1).
Figure 1.
Non-metrical dimensional scaling plot showing the microbial community distribution for ileum (A) and caeca (B) samples of the high (HFP) and low (LFP) feather pecking line.
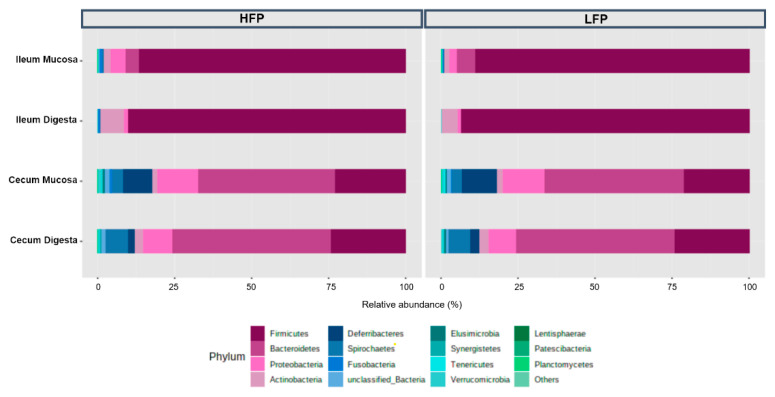
In the ileum digesta, the predominant phylum was Firmicutes with an average relative abundance of 93.5% for the LFP line, in comparison to 89.9% for the HFP line (p ≤ 0.05) (Figure 2). Actinobacteria were detected in the HFP line in higher abundance than in the LFP line (8.0% vs. 5.6%) (p ≤ 0.05). The percentage of Proteobacteria in the HFP line was also slightly higher (1.1%) than in the LFP line (0.8%). Ileum mucosa of LFP birds had more Firmicutes (88.6%) and Bacteroidetes (6.3%) than HFP birds (86.5% and 4.6%, respectively). Proteobacteria was more abundant in the HFP line (4.6%) than in the LFP line (2.2%) (p ≤ 0.05). Fusobacteria (p ≤ 0.05) and Actinobacteria were detected in higher relative abundance in HFP than LFP animals.
Figure 2.
Percentage of relative abundance for phyla distribution in the ileum mucosa, ileum digesta, caeca mucosa, and caeca digesta in the high (HFP) and low (LFP) feather pecking line.
In caeca digesta samples, a significant (p ≤ 0.05) difference was detected for Firmicutes relative abundance (24.2% in HFP compared to 23.9% in LFP). With a percentage lower than 3%, Deferribacteres and Tenericutes increased in HFP hens (p ≤ 0.05) (Figure 2). A significant (p ≤ 0.05) difference was shown in caeca mucosa for Firmicutes (HFP 22.7% compared to LFP 21.1%). In LFP, Elusimicrobia and Fusobacteria were present in less than 2.5% relative abundance, but both increased in HFP hens (p ≤ 0.05) (Figure 2). Only Actinobacteria gave a higher value for the LFP birds (p ≤ 0.05).
Random forest analysis was evaluated based on the global prediction error rate after 500 random forests [45]. After this classification, higher error rates for the microbial communities were obtained in the LFP line for ileum and caeca, mucosa and digesta (Figure S2). This result could imply a more predictable microbial composition in the HFP line since the lowest accuracy was observed in the LFP line.
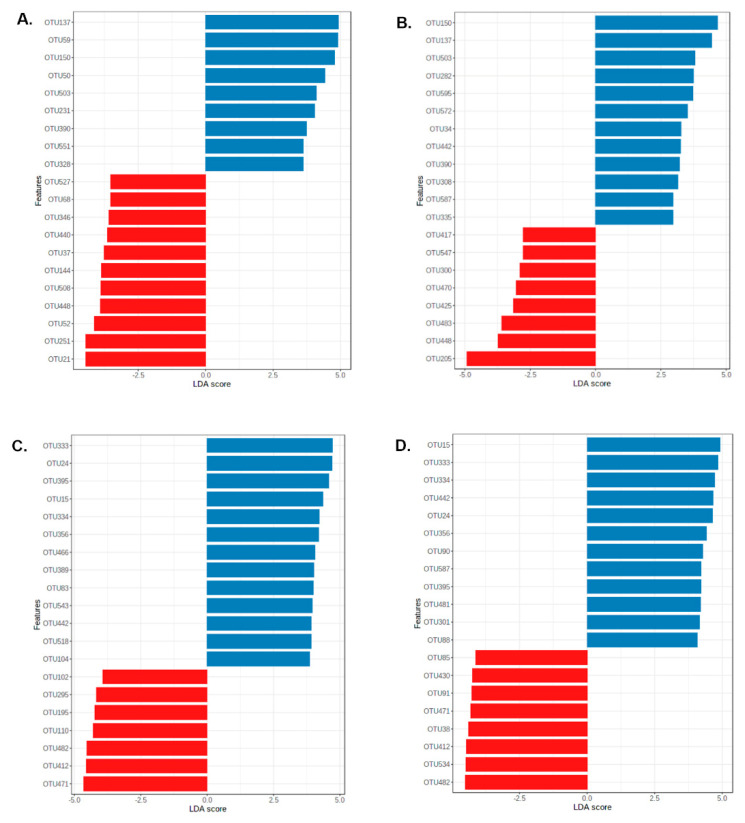
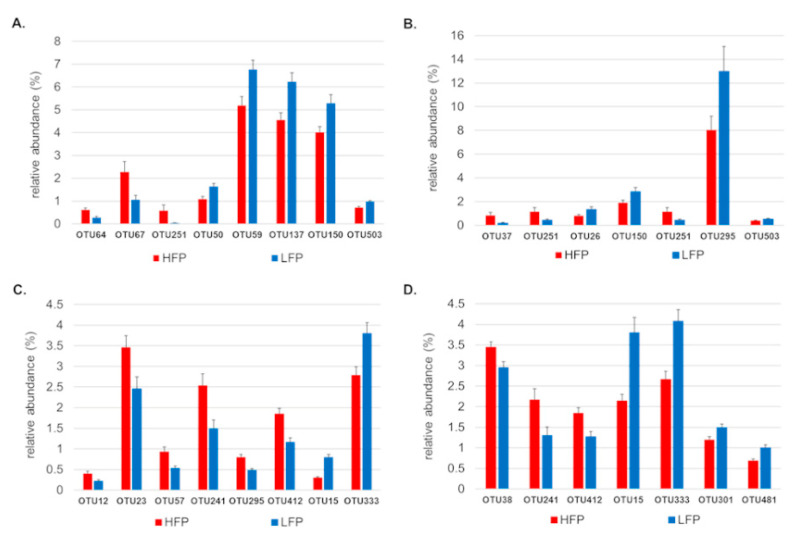
LefSe analysis was consistent, showing differences for the same OTUs in the ileum and caeca of the two feather pecking lines (Figure 3). Lactobacillus species (OTUs: 50, 59, 137, 150, 231, 390, 503, 551) based on LefSe analysis only appeared in the ileum and the occurrence was higher in the LFP line (Figure 3A,B). In the ileum digesta, the relative abundance of the OTUs 64 (Unclassified (Unc.) Olsenella), 67 (Unc. Clostridiaceae 1), and 251 (Clostridium rectum) were higher in the HFP line than in the LFP line (Figure 4). Microbial communities in the HFP hens for the ileum mucosa also included OTU37 (Escherichia coli) and OTU251 (p ≤ 0.05) (Figure 4).
Figure 3.
Linear discriminant analysis effect size (LEfSe) analysis for ileum digesta (A), ileum mucosa (B), caeca digesta (C), and caeca mucosa (D). The linear discriminant analysis (LDA) score is shown. The high feather pecking line is indicated by red and the low feather pecking line by blue.
Figure 4.
Relative abundance for the OTUs showing a significant difference for ileum digesta (A), ileum mucosa (B), caeca digesta (C), and caeca mucosa (D). The high (HFP) feather pecking line is indicated by red and the low (LFP) feather pecking line by blue.
The caeca were colonized by a greater number of bacterial species than the ileum as also represented by a higher diversity index (Figure S1). OTU12 (Lactobacillus kitasatonis), OTU23 (Unc. Paraprevotella), OTU57 (Lactobacillus gallinarum), OTU241 (Unc. Bacteroidales), OTU295 (Unc. Romboutsia), and OTU412 (Unc. Proteobacteria) (p ≤ 0.05) (Figure 4) were detected in higher relative abundance in the HFP line. Less OTUs resulting in significant (p ≤ 0.05) differences were observed for the LFP line; the OTU15 (Unc. Mucispirillum) and OTU333 (Unc. Bacteroidaceae) had higher abundances (Figure 4). In the caecum mucosa of HFP line, OTU38 (Unc. Phascolarctobacterium), OTU241 (Unc. Bacteroidales), and OTU412 (Unc. Proteobacteria) were more abundant (p ≤ 0.05); while for the LFP line again OTU15, OTU 333, and OTU301 (Unc. Suterella) and OTU481 (Unc. Treponema) (p ≤ 0.05) were detected (Figure 4).
Functional prediction showed significant differences for the feather pecking lines in the caeca microbiota, but not in the ileum (Table S2B–E). In the category of amino acid metabolism, tryptophan metabolism, and lysine degradation appeared in both digesta and mucosa, and it was higher in the HFP line. In contrast, cysteine and methionine metabolism and lysine biosynthesis were only predicted in the digesta with increased values in the HFP line. Metabolic pathways of other amino acids were observed in increased abundance in the LFP line in both the digesta and mucosa samples (Figure S3).
In the category of carbohydrate metabolism, 10 out of 15 subcategories resulted in a significant (p ≤ 0.05) difference between the digesta samples of both lines. At the same time, only five were found in the mucosa (Figure S4). In both sections, glycolysis/gluconeogenesis and amino sugar and nucleotide sugar metabolism were higher in the HFP line. LFP hens had more functions related to glyoxylate and dicarboxylate metabolism and C5-branched dibasic acid metabolism. The category of energy metabolism showed enhanced numbers of nitrogen metabolism in LFP digesta and mucosa samples. Oxidative phosphorylation and carbon fixation pathways were only observed in the digesta and enhanced for the LFP line (Figure S5). Membrane transports had higher values for the bacterial secretion system subcategory in LFP birds. ATP-binding cassette (ABC) transporters and phosphotransferase system increased in the HFP birds (Figure S6). LFP birds (digesta and mucosa) showed major significant differences (Figure S7) regarding biosynthesis of secondary metabolites.
Lipid metabolism increased in both digesta and mucosa samples of HFP line for glycerolipid, arachidonic acid, and glycerophospholipid metabolism (p ≤ 0.05) (Figure S8). Cell motility, specifically biofilm formation in E. coli and Pseudomonas aeruginosa, was predicted in the HFP samples (Figure S9).
3.2. Microbial Parameters
The linear models revealed that the lines differ significantly in the three behavior traits in all subsets of animals. The estimations of microbial parameters and microbiabilities for ileum mucosa in the HFP and LFP lines are shown in Table 3. For the agonistic traits APD and TD, low to medium microbial animal effects were estimated, which resulted in low to medium microbiabilities without significance. For FPD in ileum mucosa and all three traits in the other intestinal sections and samples, i.e., ileum digesta, caecum mucosa, and caecum digesta, the microbial animal effect estimators were fixed at the boundary by the algorithm. Hence, the microbial animal effects and thus the microbiabilities were nearly zero and not significant.
Table 3.
Estimated microbial parameters for the ileum mucosa microbial composition of 169 hens of the high (HFP) and low (LFP) feather pecking line for the three behavior traits feather pecks delivered (FPD), aggressive pecks delivered (APD) and threats delivered (TD).
| Ileum Mucosa | ||||||
|---|---|---|---|---|---|---|
| p-Value | ||||||
| FPD | <0.001 (NA) | 0.55 (0.08) | 0.26 (0.05) | <0.001 | <0.001 | 1 |
| APD | 0.08 (0.11) | 1.04 (0.17) | 0.52 (0.12) | 0.07 | 0.13 | 0.54 |
| TD | 0.19 (0.12) | 1.04 (0.17) | 0.35 (0.10) | 0.15 | 0.35 | 0.37 |
The results of the separated analyzes of the two lines (not shown) revealed that none of the microbial animal effects in any of the lines and traits were significant.
4. Discussion
4.1. Microbial Community
The characterization of the intestinal microbiota of both lines used in this study resulted in a similar microbial composition as previously described in laying hens [13] and chickens [15,16], including specific patterns such as the higher diversity in the caeca compared to the ileum. Lactobacillus species are known to be essential inhabitants of the GI tract of animals and are used as probiotic microorganisms due to their health-promoting properties [46,47]. Lactobacillus reduces the GI colonization of pathogens in broiler chickens such as Campylobacter [48], Clostridium [49], and Salmonella [50]. LEfSe analysis showed that in the ileum of LFP laying hens, mainly Lactobacillus species, such as L. johnsonii and L. crispatus, drove the community. La Ragione et al. [49] found that L. johnsonii significantly reduced E. coli colonization in chickens’ small intestine. L. crispatus showed high amylase activity, positively affecting feed conversion and broiler performance [51]. Lactobacillus stimulated serotonin receptors [52] or increased serotonin and dopamine in the brain [53], influencing the locomotor activity or decreased anxiety and depression-related behavior [53,54,55].
The role of Romboutsia species in the small intestines is still unknown due to the limited availability of cultivated representatives [56]. Here, this genus was highly dominating the caeca digesta of HFP birds. The genus Mucispirillum was positively associated with mucus production [57] and therefore related with a healthy intestine [58,59], in the present study it was detected in higher abundance in LFP than in HFP hens.
Random forest analysis is intended to classify and select the microbial data’s main features [40]. It demonstrated that the HFP line comprises less out-of-bag error, which probably indicates a specific microbiota simpler to predict. In contrast, LFP promotes a host-microbiome with more differences leading to higher misclassification rates [45].
In the literature, it was shown that birds fed with feathers differed from control birds in the microbial metabolites and microbial composition. Feather fed birds showed higher numbers of enterobacteria in the ileum and caecum and higher numbers of clostridia in the caecum [10]. Thus, it is expected that feathers’ consumption could change the microbial composition [13] and is assisted by the identified appearance of E. coli in LEfSe analysis in ileum digesta of the HFP hens.
A previous study demonstrated that gut microbes thrive the release of metabolites such as hydrogen sulfide and other sulfur-containing substances or biogenic amines, which are reactive and potentially influence behavior [10]. These findings were also observed in the predicted functions from this study. Another potential influence on behavior was the predicted promotion of biosynthesis of tryptophan in LFP hens. Tryptophan is the precursor of serotonin, and it was assumed that the alteration on the serotonergic system would impact the feather pecking behavior [60]. Indeed, feather pecking was reduced in diets with 2% of tryptophan compared to supplementation of 0.16% [2,60].
4.2. Microbial Parameters
For some of the traits, sample types, and gut sections as for FPD in Table 1, no variance components could be estimated. This was in line with the clustering of the microbial community distribution shown in Figure 1A. Except for ileum mucosa, no cluster separation was observed within and between the lines. For ileum mucosa, a tendency of separation of the two lines was noticeable implying a differentiation of the two lines’ gut microbiota. The limited number of individuals in the present study might be the reason variance components could only be estimated in the ileum mucosa when both lines were analyzed together. No significant effect was determined in the estimated variance components and microbiabilities. Thus, for the behavior traits FPD, APD, and TD, no part of the phenotypic variance could be associated with the gut microbial composition. This means that even though the hens differed significantly in these behavior traits as well as in some fractions of the gut microbial composition, the gut microbiota composition was not associated with the behavior traits. The two feather pecking lines of the 15th generation were genetically distinguishable from each other with huge allele frequency differences between the two lines. This resulted in a mean FST value of 0.16 [30], which was predominately due to drift and only to a minor extent due to selection [30,61]. Hence, these genetic differences might be the cause for the microbial differences as it is known that the microbiota is partially shaped by the host genome [62]. Another explanation might be that HFP hens picked and digested more feathers than LFP hens which altered the gut microbial composition [10].
Besides the idea to repeat the study with larger cohorts, one might apply a similar experimental setup as Kraimi et al. [27], where a microbiota transfer between divergently selected feather pecking lines was conducted, to finally rule out whether the microbiota is responsible for the differences in feather pecking behavior. This setup would also include gut microbiota, which cannot be identified or cultivated with the current techniques. Hence, if there is any influence of the microbiota on feather pecking, it could be revealed by this experiment.
5. Conclusions—Does the Microbial Composition in Ileum or Caecum Influences Feather Pecking Behavior?
No, as far as it is known from the recent results. Although significant differences in the gut microbial composition between the HFP and LFP line were found, it was impossible to show the microbiome’s influence on the behavior traits FPD, APD, and TD.
Acknowledgments
The authors acknowledge support by the High Performance and Cloud Computing Group at the Zentrum für Datenverarbeitung of the University of Tübingen, the state of Baden-Württemberg through bwHPC.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-1729/11/3/235/s1, Figure S1. Shannon diversity index for the ileum and caeca in the mucosa and digesta samples coming from the high (HFP) and low feather pecking (LFP) laying hen lines. Figure S2. Random forest analysis based on the estimation for the out of the error bag (OOB) (y-axis) with a bootstrap of 500 created trees (x-axis), based on abundance information of operational taxonomic units at genus level data in ileum digesta (A), ileum mucosa (B), caeca digesta (C), and caeca mucosa (D). The table explained the classification performance for the high feather pecking line (green), the low feather pecking line (blue), and across both lines (red). Figure S3. Functional predictions for the caeca digesta and mucosa in the subcategory amino acid metabolism in the high and low feather pecking laying hen lines. Figure S4. Functional predictions for the caeca digesta and mucosa in the subcategory carbohydrate metabolism in the high and low feather pecking laying hen lines. Figure S5. Functional predictions for the caeca digesta and mucosa in the subcategory energy metabolism in the high and low feather pecking laying hen lines. Figure S6. Functional predictions for the caeca digesta and mucosa in the subcategory membrane transport in the high and low feather pecking laying hen lines. Figure S7. Functional predictions for the caeca digesta and mucosa in the subcategory biosynthesis of other secondary metabolites in the high and low feather pecking laying hen lines. Figure S8. Functional predictions for the caeca digesta and mucosa in the subcategory lipid metabolism in the high and low feather pecking laying hen lines. Figure S9. Functional predictions for the caeca digesta and mucosa in the subcategory cell motility in the high and low feather pecking laying hen lines. Table S1A–E. Permanova test for the 16S rRNA gene identified bacterial species dataset obtained from the gut microbiome samples of the mucosa and digesta (type) taken either from the ileum or caeca (section) from the high and low feather pecking laying hen lines (line). Table S2A–E. Permanova test for the predicted functions based on 16S rRNA gene identified bacterial species obtained from the gut microbiome samples of the mucosa and digesta (type) taken either from the ileum or caeca (section) from the high and low feather pecking laying hen lines (line).
Author Contributions
Conceptualization: J.S., J.T., W.B., J.B. and A.C.-S., performed the experiment: D.B.-M., H.I., R.M. and S.S., formal analysis: D.B.-M. and H.I., contributed to the statistical analyses: M.S., wrote the paper: D.B.-M., H.I. and A.C.-S., funding acquisition: J.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the German Research Foundation (DFG, Bonn, Germany), Project No. BE3703/8-2.
Institutional Review Board Statement
The study was approved by the German Ethical Commission of Animal Welfare of the State Government of Baden-Wuerttemberg, Germany.
Informed Consent Statement
Not applicable.
Data Availability Statement
Sequences were submitted to European Nucleotide Archive under the accession number PRJEB40535.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rodenburg T.B., van Krimpen M.M., de Jong I.C., de Haas E.N., Kops M.S., Riedstra B.J., Nordquist R.E., Wagenaar J.P., Bestman M., Nicol C.J. The prevention and control of feather pecking in laying hens: Identifying the underlying principles. Worlds Poult. Sci. J. 2013;69:361–374. doi: 10.1017/S0043933913000354. [DOI] [Google Scholar]
- 2.van Hierden Y.M., Koolhaas J.M., Korte S. Chronic increase of dietary l-tryptophan decreases gentle feather pecking behaviour. Appl. Anim. Behav. Sci. 2004;89:71–84. doi: 10.1016/j.applanim.2004.05.004. [DOI] [Google Scholar]
- 3.Bello A.U., Idrus Z., Yong Meng G., Awad E.A., Soleimani Farjam A. Gut microbiota and transportation stress response affected by tryptophan supplementation in broiler chickens. Ital. J. Anim. Sci. 2018;17:107–113. doi: 10.1080/1828051X.2017.1340814. [DOI] [Google Scholar]
- 4.McKeegan D., Savory C.J. Feather eating in layer pullets and its possible role in the aetiology of feather pecking damage. Appl. Anim. Behav. Sci. 1999;65:73–85. doi: 10.1016/S0168-1591(99)00051-9. [DOI] [Google Scholar]
- 5.McKeegan D., Savory C.J. Feather eating in individually caged hens which differ in their prospensity to feather peck. Appl. Anim. Behav. Sci. 2001;73:131–140. doi: 10.1016/S0168-1591(01)00124-1. [DOI] [PubMed] [Google Scholar]
- 6.Harlander-Matauschek A., Bessei W. Feather eating and crop filling in laying hens. Archiv. Geflügelkunde. 2005;69:241–244. [Google Scholar]
- 7.Harlander-Matauschek A., Häusler K. Understanding feather eating behaviour in laying hens. Appl. Anim. Behav. Sci. 2009;117:35–41. doi: 10.1016/j.applanim.2008.11.003. [DOI] [Google Scholar]
- 8.McCasland W., Richardson L.R. Methods for Determining the Nutritive Value of Feather Meals. Poult. Sci. 1966;45:1231–1236. doi: 10.3382/ps.0451231. [DOI] [Google Scholar]
- 9.Lutz V., Kjaer J.B., Iffland H., Rodehutscord M., Bessei W., Bennewitz J. Quantitative genetic analysis of causal relationships among feather pecking, feather eating, and general locomotor activity in laying hens using structural equation models. Poult. Sci. 2016;95:1757–1763. doi: 10.3382/ps/pew146. [DOI] [PubMed] [Google Scholar]
- 10.Meyer B., Bessei W., Vahjen W., Zentek J., Harlander-Matauschek A. Dietary inclusion of feathers affects intestinal microbiota and microbial metabolites in growing Leghorn-type chickens. Poult. Sci. 2012;91:1506–1513. doi: 10.3382/ps.2011-01786. [DOI] [PubMed] [Google Scholar]
- 11.Meyer B., Zentek J., Harlander-Matauschek A. Differences in intestinal microbial metabolites in laying hens with high and low levels of repetitive feather-pecking behavior. Physiol. Behav. 2013;110–111:96–101. doi: 10.1016/j.physbeh.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 12.Birkl P., Bharwani A., Kjaer J.B., Kunze W., McBride P., Forsythe P., Harlander-Matauschek A. Differences in cecal microbiome of selected high and low feather-pecking laying hens. Poult. Sci. 2018;97:3009–3014. doi: 10.3382/ps/pey167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Eijk J.A.J., de Vries H., Kjaer J.B., Naguib M., Kemp B., Smidt H., Rodenburg T.B., Lammers A. Differences in gut microbiota composition of laying hen lines divergently selected on feather pecking. Poult. Sci. 2019;98:7009–7021. doi: 10.3382/ps/pez336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu J., Idris U., Harmon B., Hofacre C., Maurer J.J., Lee M.D. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 2003;69:6816–6824. doi: 10.1128/AEM.69.11.6816-6824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borda-Molina D., Vital M., Sommerfeld V., Rodehutscord M., Camarinha-Silva A. Insights into Broilers’ Gut Microbiota Fed with Phosphorus, Calcium, and Phytase Supplemented Diets. Front. Microbiol. 2016;7:2033. doi: 10.3389/fmicb.2016.02033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanley D., Hughes R.J., Moore R.J. Microbiota of the chicken gastrointestinal tract: Influence on health, productivity and disease. Appl. Microbiol. Biotechnol. 2014;98:4301–4310. doi: 10.1007/s00253-014-5646-2. [DOI] [PubMed] [Google Scholar]
- 17.Pan D., Yu Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes. 2014;5:108–119. doi: 10.4161/gmic.26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oakley B.B., Lillehoj H.S., Kogut M.H., Kim W.K., Maurer J.J., Pedroso A., Lee M.D., Collett S.R., Johnson T.J., Cox N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014;360:100–112. doi: 10.1111/1574-6968.12608. [DOI] [PubMed] [Google Scholar]
- 19.Deusch S., Tilocca B., Camarinha-Silva A., Seifert J. News in livestock research—Use of Omics-technologies to study the microbiota in the gastrointestinal tract of farm animals. Comput. Struct. Biotechnol. J. 2015;13:55–63. doi: 10.1016/j.csbj.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waite D.W., Taylor M.W. Exploring the avian gut microbiota: Current trends and future directions. Front. Microbiol. 2015;6:673. doi: 10.3389/fmicb.2015.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Difford G.F., Plichta D.R., Løvendahl P., Lassen J., Noel S.J., Højberg O., Wright A.-D.G., Zhu Z., Kristensen L., Nielsen H.B., et al. Host genetics and the rumen microbiome jointly associate with methane emissions in dairy cows. PLoS Genet. 2018;14:e1007580. doi: 10.1371/journal.pgen.1007580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camarinha-Silva A., Maushammer M., Wellmann R., Vital M., Preuß S., Bennewitz J. Host Genome Influence on Gut Microbial Composition and Microbial Prediction of Complex Traits in Pigs. Genetics. 2017;206:1637–1644. doi: 10.1534/genetics.117.200782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weishaar R., Wellmann R., Camarinha-Silva A., Rodehutscord M., Bennewitz J. Selecting the hologenome to breed for an improved feed efficiency in pigs-A novel selection index. J. Anim. Breed. Genet. 2020;137:14–22. doi: 10.1111/jbg.12447. [DOI] [PubMed] [Google Scholar]
- 24.Verschuren L.M.G., Schokker D., Bergsma R., Jansman A.J.M., Molist F., Calus M.P.L. Prediction of nutrient digestibility in grower-finisher pigs based on faecal microbiota composition. J. Anim. Breed. Genet. 2020;137:23–35. doi: 10.1111/jbg.12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vollmar S., Wellmann R., Borda-Molina D., Rodehutscord M., Camarinha-Silva A., Bennewitz J. The Gut Microbial Architecture of Efficiency Traits in the Domestic Poultry Model Species Japanese Quail (Coturnix japonica) Assessed by Mixed Linear Models. G3. 2020;10:2553–2562. doi: 10.1534/g3.120.401424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraimi N., Dawkins M., Gebhardt-Henrich S.G., Velge P., Rychlik I., Volf J., Creach P., Smith A., Colles F., Leterrier C. Influence of the microbiota-gut-brain axis on behavior and welfare in farm animals: A review. Physiol. Behav. 2019;210:112658. doi: 10.1016/j.physbeh.2019.112658. [DOI] [PubMed] [Google Scholar]
- 27.Kraimi N., Calandreau L., Zemb O., Germain K., Dupont C., Velge P., Guitton E., Lavillatte S., Parias C., Leterrier C. Effects of gut microbiota transfer on emotional reactivity in Japanese quails (Coturnix japonica) J. Exp. Biol. 2019;222:jeb202879. doi: 10.1242/jeb.202879. [DOI] [PubMed] [Google Scholar]
- 28.Parois S., Calandreau L., Kraimi N., Gabriel I., Leterrier C. The influence of a probiotic supplementation on memory in quail suggests a role of gut microbiota on cognitive abilities in birds. Behav. Brain Res. 2017;331:47–53. doi: 10.1016/j.bbr.2017.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Iffland H., Wellmann R., Schmid M., Preuß S., Tetens J., Bessei W., Bennewitz J. Genomewide Mapping of Selection Signatures and Genes for Extreme Feather Pecking in Two Divergently Selected Laying Hen Lines. Animals. 2020;10:262. doi: 10.3390/ani10020262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iffland H., Schmid M., Preuß S., Bessei W., Tetens J., Bennewitz J. Phenotypic and genomic analyses of agonistic interactions in laying hen lines divergently selected for feather pecking. Appl. Anim. Behav. Sci. 2021;234:105177. doi: 10.1016/j.applanim.2020.105177. [DOI] [Google Scholar]
- 31.Kaewtapee C., Burbach K., Tomforde G., Hartinger T., Camarinha-Silva A., Heinritz S., Seifert J., Wiltafsky M., Mosenthin R., Rosenfelder-Kuon P. Effect of Bacillus subtilis and Bacillus licheniformis supplementation in diets with low- and high-protein content on ileal crude protein and amino acid digestibility and intestinal microbiota composition of growing pigs. J. Anim. Sci. Biotechnol. 2017;8:37. doi: 10.1186/s40104-017-0168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Argüello H., Estellé J., Zaldívar-López S., Jiménez-Marín Á., Carvajal A., López-Bascón M.A., Crispie F., O’Sullivan O., Cotter P.D., Priego-Capote F., et al. Early Salmonella Typhimurium infection in pigs disrupts Microbiome composition and functionality principally at the ileum mucosa. Sci. Rep. 2018;8:7788. doi: 10.1038/s41598-018-26083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borda-Molina D., Roth C., Hérnandez-Arriaga A., Rissi D., Vollmar S., Rodehutscord M., Bennewitz J., Camarinha-Silva A. Effects on the Ileal Microbiota of Phosphorus and Calcium Utilization, Bird Performance, and Gender in Japanese Quail. Animals. 2020;10:885. doi: 10.3390/ani10050885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wemheuer F., Taylor J.A., Daniel R., Johnston E., Meinicke P., Thomas T., Wemheuer B. Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome. 2020;15:1–12. doi: 10.1186/s40793-020-00358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yilmaz P., Parfrey L.W., Yarza P., Gerken J., Pruesse E., Quast C., Schweer T., Peplies J., Ludwig W., Glöckner F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chong J., Liu P., Zhou G., Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020;15:799–821. doi: 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- 41.Clarke K.R., Warwick R.M. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation. 2nd ed. PRIMER-E; Plymouth, UK: 1994. [Google Scholar]
- 42.Bray J.R., Curtis J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957;27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 43.Butler D., Cullis B.R., Gilmour A.R., Gogel B.J. ASReml-R 3 Reference Manual: Mixed Models for S Language Environments. Queensland Government, Department of Primary Industries and Fisheries; Brisbane, Australia: 2009. [Google Scholar]
- 44.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2019. [Google Scholar]
- 45.Roguet A., Eren A.M., Newton R.J., McLellan S.L. Fecal source identification using random forest. Microbiome. 2018;6:185. doi: 10.1186/s40168-018-0568-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson G.R., Probert H.M., van Loo J., Rastall R.A., Roberfroid M.B. Dietary modulation of the human colonic microbiota: Updating the concept of prebiotics. Nutr. Res. Rev. 2004;17:259–275. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- 47.Klaenhammer T.R., Altermann E., Pfeiler E., Buck B.L., Goh Y.-J., O’Flaherty S., Barrangou R., Duong T. Functional genomics of probiotic Lactobacilli. J. Clin. Gastroenterol. 2008;42(Suppl. S3):S160–S162. doi: 10.1097/MCG.0b013e31817da140. [DOI] [PubMed] [Google Scholar]
- 48.Ghareeb K., Awad W.A., Mohnl M., Porta R., Biarnés M., Böhm J., Schatzmayr G. Evaluating the efficacy of an avian-specific probiotic to reduce the colonization of Campylobacter jejuni in broiler chickens. Poult. Sci. 2012;91:1825–1832. doi: 10.3382/ps.2012-02168. [DOI] [PubMed] [Google Scholar]
- 49.La Ragione R.M., Narbad A., Gasson M.J., Woodward M.J. In vivo characterization of Lactobacillus johnsonii FI9785 for use as a defined competitive exclusion agent against bacterial pathogens in poultry. Lett. Appl. Microbiol. 2004;38:197–205. doi: 10.1111/j.1472-765X.2004.01474.x. [DOI] [PubMed] [Google Scholar]
- 50.Pascual M., Hugas M., Badiola J.I., Monfort J.M., Garriga M. Lactobacillus salivarius CTC2197 prevents Salmonella enteritidis colonization in chickens. Appl. Environ. Microbiol. 1999;65:4981–4986. doi: 10.1128/AEM.65.11.4981-4986.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taheri H.R., Moravej H., Tabandeh F., Zaghari M., Shivazad M. Screening of lactic acid bacteria toward their selection as a source of chicken probiotic. Poult. Sci. 2009;88:1586–1593. doi: 10.3382/ps.2009-00041. [DOI] [PubMed] [Google Scholar]
- 52.Horii Y., Nakakita Y., Fujisaki Y., Yamamoto S., Itoh N., Miyazaki K., Kaneda H., Oishi K., Shigyo T., Nagai K. Effects of intraduodenal injection of Lactobacillus brevis SBC8803 on autonomic neurotransmission and appetite in rodents. Neurosci Lett. 2013;539:32–37. doi: 10.1016/j.neulet.2013.01.037. [DOI] [PubMed] [Google Scholar]
- 53.Liu W.-H., Chuang H.-L., Huang Y.-T., Wu C.-C., Chou G.-T., Wang S., Tsai Y.-C. Alteration of behavior and monoamine levels attributable to Lactobacillus plantarum PS128 in germ-free mice. Behav. Brain Res. 2016;298:202–209. doi: 10.1016/j.bbr.2015.10.046. [DOI] [PubMed] [Google Scholar]
- 54.Bravo J.A., Forsythe P., Chew M.V., Escaravage E., Savignac H.M., Dinan T.G., Bienenstock J., Cryan J.F. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. USA. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang S., Wang T., Hu X., Luo J., Li W., Wu X., Duan Y., Jin F. Administration of Lactobacillus helveticus NS8 improves behavioral, cognitive, and biochemical aberrations caused by chronic restraint stress. Neuroscience. 2015;310:561–577. doi: 10.1016/j.neuroscience.2015.09.033. [DOI] [PubMed] [Google Scholar]
- 56.Gerritsen J., Umanets A., Staneva I., Hornung B., Ritari J., Paulin L., Rijkers G.T., de Vos W.M., Smidt H. Romboutsia hominis sp. nov., the first human gut-derived representative of the genus Romboutsia, isolated from ileostoma effluent. Int. J. Syst. Evol. Microbiol. 2018;68:3479–3486. doi: 10.1099/ijsem.0.003012. [DOI] [PubMed] [Google Scholar]
- 57.Belzer C., Gerber G.K., Roeselers G., Delaney M., DuBois A., Liu Q., Belavusava V., Yeliseyev V., Houseman A., Onderdonk A., et al. Dynamics of the microbiota in response to host infection. PLoS ONE. 2014;9:e95534. doi: 10.1371/journal.pone.0095534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Derrien M., Collado M.C., Ben-Amor K., Salminen S., de Vos W.M. The Mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl. Environ. Microbiol. 2008;74:1646–1648. doi: 10.1128/AEM.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodríguez-Piñeiro A.M., Johansson M.E.V. The colonic mucus protection depends on the microbiota. Gut Microbes. 2015;6:326–330. doi: 10.1080/19490976.2015.1086057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Haas E.N., van der Eijk J.A.J. Where in the serotonergic system does it go wrong? Unravelling the route by which the serotonergic system affects feather pecking in chickens. Neurosci. Biobehav. Rev. 2018;95:170–188. doi: 10.1016/j.neubiorev.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 61.Grams V., Wellmann R., Preuß S., Grashorn M.A., Kjaer J.B., Bessei W., Bennewitz J. Genetic parameters and signatures of selection in two divergent laying hen lines selected for feather pecking behaviour. Genet Sel. Evol. 2015;47:77. doi: 10.1186/s12711-015-0154-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spor A., Koren O., Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences were submitted to European Nucleotide Archive under the accession number PRJEB40535.