Abstract
Gliomas are the most common primary neoplasm of the central nervous system. A promising frontier in the definition of glioma prognosis and treatment is represented by epigenetics. Furthermore, in this study, we developed a machine learning classification model based on epigenetic data (CpG probes) to separate patients according to their state of immunosuppression. We considered 573 cases of low-grade glioma (LGG) and glioblastoma (GBM) from The Cancer Genome Atlas (TCGA). First, from gene expression data, we derived a novel binary indicator to flag patients with a favorable immune state. Then, based on previous studies, we selected the genes related to the immune state of tumor microenvironment. After, we improved the selection with a data-driven procedure, based on Boruta. Finally, we tuned, trained, and evaluated both random forest and neural network classifiers on the resulting dataset. We found that a multi-layer perceptron network fed by the 338 probes selected by applying both expert choice and Boruta results in the best performance, achieving an out-of-sample accuracy of 82.8%, a Matthews correlation coefficient of 0.657, and an area under the ROC curve of 0.9. Based on the proposed model, we provided a method to stratify glioma patients according to their epigenomic state.
Keywords: immunosuppression, tumor microenviroment, neural network, genome-wide methylation model, glioma, extracellular matrix
1. Introduction
Gliomas are brain tumors that arise from glial precursor cells. According to their pathological features, gliomas are subdivided in glioblastomas (GBMs), which have the highest grade (IV), and low-grade gliomas (LGGs), a heterogeneous group composed by various tumor types, such as astrocytic, oligodendroglial and ependymal tumors. Gliomas have a heterogeneous clinical outcome with the worse course happening in the GBM group, whereas LGGs are generally less severe. Several biomarkers have been proposed to predict the clinical outcome and response to treatments of gliomas, including genetic and epigenetic ones such as IDH mutation and methylation of the MGMT promoter. A detailed characterization of glioma-associated molecular signatures has made possible the development of novel therapies, including the use of tyrosine kinase inhibitors. On the other hand, based on the results obtained in the context of other tumors, the use of immune checkpoint inhibitors (ICIs) has been proposed for gliomas, including GBMs. However, despite the recently proposed novel targeted therapy and immunotherapy treatment approaches, treatment strategies for gliomas are in the majority of cases still conventional. In particular, for GBMs, the current standard of care still consists of surgical resection, followed by radiotherapy and chemotherapy [1]. Moreover, so far no immunotherapeutic approach against GBM has demonstrated efficacy in a controlled clinical trial [2,3,4].
The clinical outcome of gliomas is strictly related with the composition and cell cross-talk of tumor microenvironment (TME), in particular with the immune texture in terms of the distinct immune cell types as well as the different immunosuppressive cell populations, such as T regulatory cells (Tregs), myeloid-derived suppressor cells (MDSCs), tumor-associated macrophages (TAMs), dendritic cells and antigen-presenting cells specific to the brain such as microglia [5,6,7]. A significant infiltration of Tregs can be detected in a large fraction of gliomas, in particular in the GBM group. In this context, the activity of IDO can contribute to the immunosuppressive state of the TME by creating a tryptophan shortage, which contributes to the suppression of T cell activation and proliferation [8]. Within glioma tumors, microglia and macrophages can represent up to the 12% of the tumor mass [9,10,11,12].
With respect to the macrophages displaying the M1 phenotype, M2 macrophages are more strongly involved in the maintenance of an immunosuppressive state in the TME. Notably, M2 macrophages are generally characterized by the peculiar expression of several cell surface markers including CD163 [13,14,15,16]. The extracellular matrix (ECM) components such as glycosaminoglycans, glycoproteins, proteoglycans, play a crucial role in the invasion mechanisms of gliomas, mainly through promoting angiogenesis and tumor cell migration. Hypervascularity is a characteristic of gliomas with an increment in angiogenesis compared to healthy brain tissue. This tumor-associated vasculature is not completely formed with leaky vessels and associated with an increase in the interstitial fluid pressure [17].
The degree of immunosuppression of the glioma TME can be associated with a peculiar immunosuppressive signature, with the most accentued immunosuppressive state happening in the case of GBMs [18]. Moreover, specific immunosuppressive features such as depletion of tumor infiltrating lymphocytes (TIL), high PD-L1 expression, and a reduced IIFN signature have been associated with recurrent genomic mutations, such as IDH1, TP53, NF1, PTEN EGFR and MAPK pathway mutations. Epigenetic modifications including alteration of histone patterns, chromatin structure, changes in microRNA expression levels and DNA methylation status at specific promoters are involved in the modulation of the TME by allowing cells to grow and to escape from immune surveillance. Thus, the immunosuppressive state can be recapitulated by epigenetic regulation, in particular by DNA methylation influencing the expression of transcription factors and regulatory genes related to the immune cell transcriptome. Since DNA methylation plays an important role in cancers, many studies have utilized DNA methylated sequences as biomarkers for cancer detection, including CpG markers and promoter markers. In particular, DNA methylation has been demonstrated to resolve cell of origin of peripheral blood cells [19] and cell-free DNA [20,21,22], and was introduced as a complementary approach to classify central nervous system (CNS) tumors [23]. Moreover, irregular methylations in promoters of cancer-related genes could serve as biomarkers for early cancer diagnosis and prognosis. An example of this is MGMT promoter methylation that was demonstrated to be a predictive biomarker for cancer prognosis in GBMs and response to chemotherapy with temozolomide [24,25]. In this context, DNA methylation can be useful to more adequately understand the distribution of the different immune cell subtypes in the context of the TME [26,27,28]. In this study, we fed DNA methylation data into a machine learning model to classify gliomas over their immunosuppression state. We used methylation data as features for our dataset. The target is a novel binary indicator of the immunosuppression state. Due to the limited number of cases available in public datasets, we resorted to both expert and data-driven selection to shrink the number of features and decrease the noise. Given the large number of features and the possibly non-linear nature of the problem, we adopted properly tuned random forest (RF) and deep neural network as classifiers.
We found that the multi-layer perceptron deep outperforms the RF and that a proper feature selection is capable of improving the accuracy of the model. In light of the result of the study, a proper discussion of the biological implications of our study was provided. This classification model could be useful to predict the responsiveness of glioma-affected patients to novel immunotherapeutic approaches, such as the use of ICIs.
2. Materials and Methods
2.1. Data
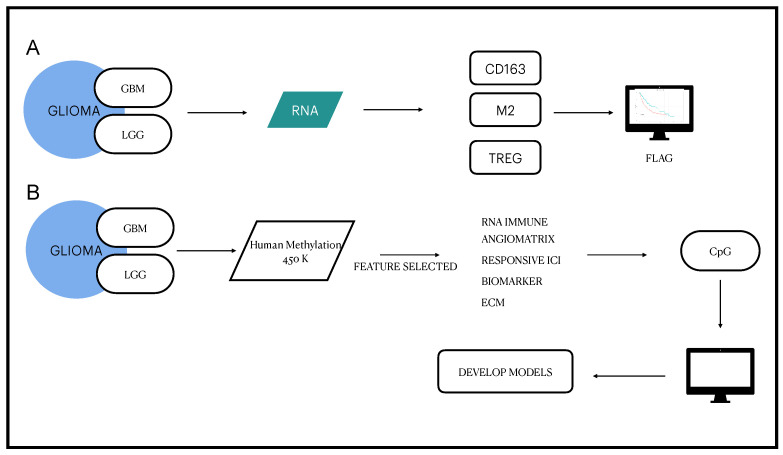
The complete workflow, from raw data to the predictive model, is presented in Figure A2.
Our dataset is derived from The Cancer Genome Atlas (TCGA) data hub, available on Xena https://xenabrowser.net (accessed on 12 March 2020). From this source, we extracted the count (FPKM-UQ) of RNA sequencing (RNA-seq) and DNA methylation data for LGG and GBM. The clinical and pathological information of the patients was also gathered from TCGA and the Consortium publication on glioma [29]. The selection of the cases was based on the following criteria: (i) presence of a diagnosed GBM or LGG, (ii) availability of the DNA methylation and RNA sequencing data. A total of 573 cases of brain tumors were enrolled (Table 1).
Table 1.
Cases included in the study from The Cancer Genome Atlas (TCGA) cohorts for Glioma cancer types.
| Cohort | Cancer Type | Cases | Cases Flagged as EDISON Positive |
|---|---|---|---|
| LGG | Brain lower grade glioma | 506 | 271 |
| GBM | Glioblastoma multiforme | 47 | 10 |
The input to our machine learning model was made only of methylation data, while the RNA-seq data and the information about the patients were only used in the construction of the target or in ancillary analysis. The methylation at each 5′—C—phosphate—G—3′ (CpG) site is described by the value, defined as the ratio between the intensity of the methylated probe and the intensity of the total probe. A total of 482,421 CpG sites throughout the genome were assessed and filtered using the procedure described by Bourgon et al. [30], resulting in an initial dataset containing 355,314 CpG probes. We called this dataset AllCpGs.
Taking into account the relevance of M2 macrophages and TReg populations in the modulation of immunosuppression in the context of the TME, cases were labeled for their putative capability of escaping an immunosuppressive state. To do this, we evaluated the immune cells in the TME using immunedecov [31]. First, the data relative to RNA-seq were log-transformed and standardized to zero mean and unit variance. We then defined three different criteria based on RNA-seq: (i) expression of the CD163 gene, (ii) expression of M2 macrophage signature (Macropage M2), and (iii) expression of Tregs signature (T cell regulatory Tregs). The two latter signatures were evaluated using quantiseq [32] and xCell [33]. The three parameters, (i) to (iii), were used to label cases based on their putative capability of escaping an immunosuppressive state. A case was labelled EvaDe Immune SuppressiON (EDISON) positive if it had more than two out of the three parameters, (i) to (iii), below the first quartile of expression. For the evaluation of the interactions between the immune system and the TME, we leveraged the signatures published on the “Immune-Subtype-Clustering” GitHub repository [34], as proposed in our previous study [35]. The EDISON label was used as a target for our classification models.
2.2. Feature Selection
Due to the high number of variables in the DNA methylation data compared to the number of cases, before applying any classification model, we opted to reduce the dimensionality of the input via feature selection. At first, we applied expert selection.
We included in the dataset the CpGs related to the genes which were shown to play crucial roles in gliomas. Specifically, we chose:
The genes linked to the putative response of immune suppression in the study by Thorston et al. [18];
The genes with the angiomatrix signatures [36];
The genes associated with the putative response for ICIs in GBMs [37];
The genes reported as with prognostic value for gliomas by Mesrati et al. [38];
The genes related to the extracellular matrix (ECM) recently linked to the glioma by Zhao et al. [39].
In order to evaluate the predictive power of different sets of genes, two different datasets were obtained. We called ImmuneAngioICIs the one containing the genes described in points 1, 2, and 3, while we called ImmuneAngioICIsMesECM the dataset containing the genes described in points 1, 2, 3, 4, and 5.
In order to assess the soundness and effectiveness of our expert selection, we also considered a dataset containing all the CpG probes without any filtering. Our results will show that including all the probes does not result in a better modelling: conversely, the additional features bring noise and worsen the predictive power of our models.
The expert selection reduces the number of CpG in the dataset by a factor of 50. Still, many uninformative features might be present. Given the limited number of available cases, the inclusion of uninformative features results in an increase in the noise and may have detrimental effects on the accuracy of the machine learning model. Therefore, we opted to adopt also a data-driven selection procedure. On each dataset, we applied the Boruta algorithm to detect the set of most relevant features [40]. A scheme with a 10-fold cross-validation and 100 repetitions was adopted. We called AllCpGs + BORUTA the dataset resulting after the application of Boruta to AllCpGs, ImmuneAngioICIs + BORUTA the dataset resulting after the application of Boruta to ImmuneAngioICIs, and ImmuneAngioICIsMesECM + BORUTA the dataset resulting after the application of Boruta to ImmuneAngioICIsMesECM. A summary of the datasets is presented in Table 2.
Table 2.
Summary of the datasets, with the number of CpGs included in each one.
| Dataset | CpG Count |
|---|---|
| AllCpGs | 355,314 |
| ImmuneAngioICIs | 6368 |
| ImmuneAngioICIsMesECM | 6754 |
| AllCpGs + BORUTA | 3554 |
| ImmuneAngioICIs + BORUTA | 512 |
| ImmuneAngioICIsMesECM + BORUTA | 338 |
2.3. Modelling
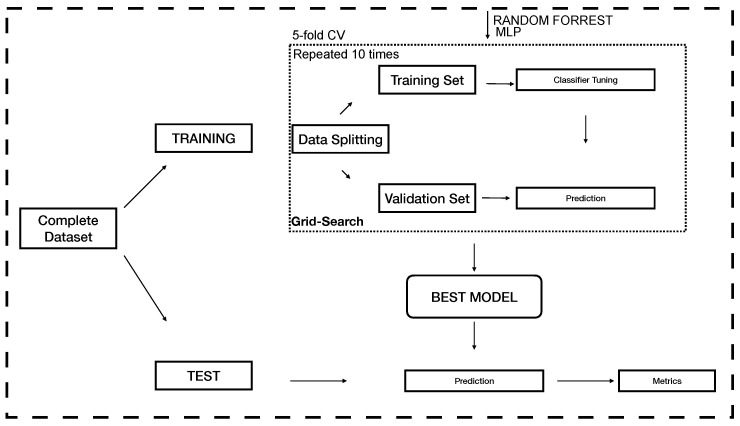
To allow a proper evaluation of the machine learning models, each of the the available datasets, d, {AllCpGs, ImmuneAngioICIs, ImmuneAngioICIsMesECM, AllCpGs + BORUTA, ImmuneAngioICIs + BORUTA, ImmuneAngioICIsMesECM + BORUTA}, was split into a training set , containing 80% of the samples, and a test set , including the remaining 20%. The feature selection and the tuning of model hyperparameters were allowed to only take advantage of the training set , while samples in were left apart for the final evaluation. It is important to note that the training sets only differ in the inputs, while the target variable and the target sample are the same irrespective of d. The same holds for the test sets . This point is critical to allow for a sound comparison among the performance of the models.
On each dataset, the classification models were then tuned and trained. At first, we considered a RF model. We optimized the hyperparameters, such as the number of trees in the forest, the maximum depth of a tree and the minimum number of samples in a leaf, using a grid-search cross-validation. The tuning procedure followed the one described in Vadalas et al. [41].
On the dataset leading to the best performance metrics, namely ImmuneAngioICIsMes ECM + BORUTA, two more models were trained. We selected two architectures of deep neural networks: a multi-layer perceptron (MLP) and a convolutional neural network (CNN). For both models, the hyperparameters such as number of hidden layers, neurons in each layer, and learning rate, were optimized using a grid-search cross-validation.
To further evaluate the complex regulation of methylation effect in different genomic localization, we investigated if the EDISON classification model could be improved by dividing ImmuneAngioICIsMesECM and ImmuneAngioICIs by regional sites and by applying the RF model.
2.4. Evaluation
In addition to the standard accuracy (ACC), we considered the Matthews Correlation Coefficient (MCC), and the area under the receiver operating characteristic (AUC) as performance metrics. First introduced by B.W. Matthews to assess the performance of the prediction of protein secondary structure [42], the MCC has become a widely used measure in biomedical research [43,44]. Due to their large popularity and simple interpretation, MCC and AUC were selected in the US FDA-led initiative MAQC-II, aimed at reaching a consensus on the best practices for the development and validation of predictive models for personalized medicine [43].
The evaluation metrics were computed both in cross-validation, on samples belonging to the train sets , and on the samples of the test set . For the cross-validation metrics, the 95% confidence intervals (CIs) were also computed. In order to substantiate the results, the McNemar test was used to assess the significance in performance difference among classifiers [45].
2.5. Evaluation of the 338 CpG Probes Used for the Model as Survival Prognosticator
We evaluated the prognostic role of the CpG probes used by the best performing model, i.e., the ones included in ImmuneAngioICIsMesECM + BORUTA with survival analysis. In particular, we adopted a random survival forest, an ensemble tree method for the analysis of censored survival data, described by Wang et al. [46]. The hyperparameters of the model were chosen with a randomized search and the feature importance was extracted from the best model using permutation importance.
2.6. Definition of a Possible CpG Signature Useful for Liquid Biopsy
The CpG probes used by the best performing model (ImmuneAngioICIsMesECM + BORUTA) were also analyzed using the Blood–Brain Epigenetic Concordance (BECon) to assess their possible use in liquid biopsy (https://redgar598.shinyapps.io/BECon/ (accessed on 12 March 2020)). We first chose the CpGs that presented a percentile rank of CpG Change Beta over 75. Then, we applied the least absolute shrinkage and selection operator (LASSO) Cox regression to develop an optimal risk signature with the minimum number of CpGs [47,48]. The correlation of the CpGs with gene expression was also evaluated.
2.7. Correlation Analysis between CpGs and Genes
To examine the impact of DNA methylation on the local regulation of gene expression, the Pearson correlation between the values of the CpGs and the normalized expression of the corresponding genes was calculated. Moreover, in order to investigate the distant regulation of gene expression, we computed the correlation between the values of CpGs of differentially methylated and expressed genes and the normalized expression of differentially expressed genes.
2.8. PPI Network Analysis of DNA Methylation-Driven Genes
The 338 CpG probes used by the best performing model (ImmuneAngioICIsMesECM + BORUTA) were mapped by Search Tool for the Retrieval of Interacting Genes (STRING) database (version 10.5 [49] ) by using Cytoscape (3.8.2) [50]. The PPI network was generated based on the medium confidence score of 0.40.
2.9. Computational Details
The classification pipeline was built on top of the Scikit Learn library, version 0.20.3 [51] and Python 3.6. All the experiments were run on a 32-core Intel Core i7 workstation with 128GB of RAM running CentOS 7.5. Cox regression and Kaplan–Meier survival curves were computed using R (version 3.6.1) with the survival and survminer packages. The Wilcoxon rank-sum test was used to compare the difference between the groups, while Kruskal–Wallis (K-W) test was adopted to evaluate the differences in risk scores across three or more groups.
3. Results
3.1. Definition of the EDISON Classification Flag
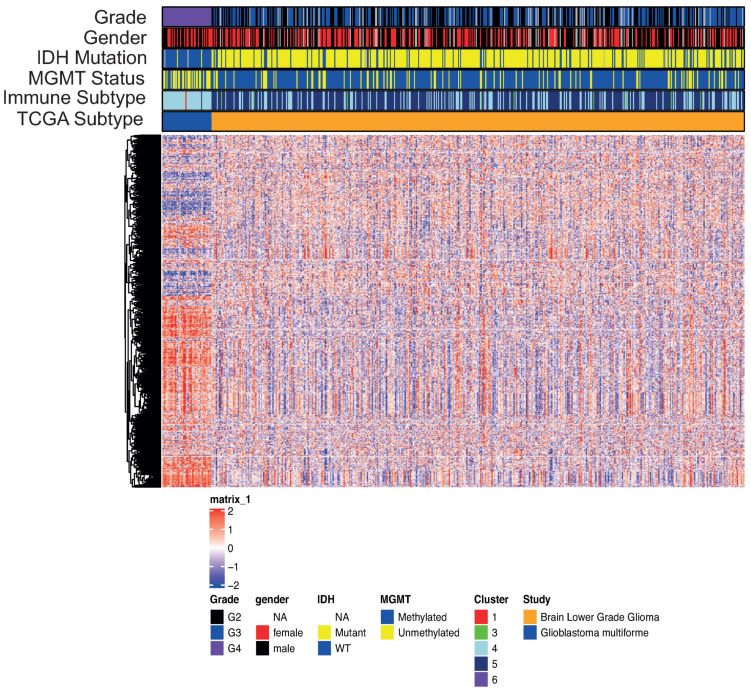
We analyzed publicly available datasets of primary glioma samples for which transcriptomic and epigenomic molecular profiles were available. We collected a total of 573 cases, of which 47 cases were GBMs and 506 cases were LGGs. This series of 573 glioma cases was used to develop the model irrespective of being GBMs or LGGs. Figure A2 represents the adopted workflow. Considering the transcriptomics to explore the immune environments landscape (Figure 1), we observed how the different subpopulations of gliomas based on the grade can be described by the the differential expression of some genes, capable of segregating GBMs from LGGs. The LGG group is enriched in IDH mutated cases. This is in keeping with previous published results showing that IDH mutations are associated with favorable immune composition within the TME and decreased leukocyte chemotaxis, leading to fewer tumor-associated immune cells and better outcome [52]. On the other hand, the GBM group is characterized by a high number of MGMT unmetylated cases [24]. Moreover, we evaluated all the cohort for the immune subtype classification, as described in Thorston et al. [18]. With this approach we found that the set of glioma cases employed in the present study is enriched in cases belonging to the subtype 4 (lymphocyte Depleted) and 5 (Immunologically Quiet). These results were in agreement with what previously described showing that the gliomas included in cluster subtype 4 are characterized by a more prominent macrophage signature, with a high M2 response and suppression of the Th1 T cell population, as well as that the glioma cases included in the cluster subtype 5 exhibit the lowest lymphocyte population and the highest macrophage response dominated by M2 macrophages [18,53,54,55].
Figure 1.
Transcriptomics landscape of patients with either glioblastoma (GBM) or low-grade glioma (LGG). The 2365 genes shown were used to develop the immune cluster subtype by Thorston et al. [18].
Based on these characteristics, peculiar of an immune suppressive TME, we chose to assess the immune-related signatures of the 573 sample RNA-seq data by using immunedecov (xCell tools) to comprehensively evaluate the transcriptome-based cell-type quantification [31].
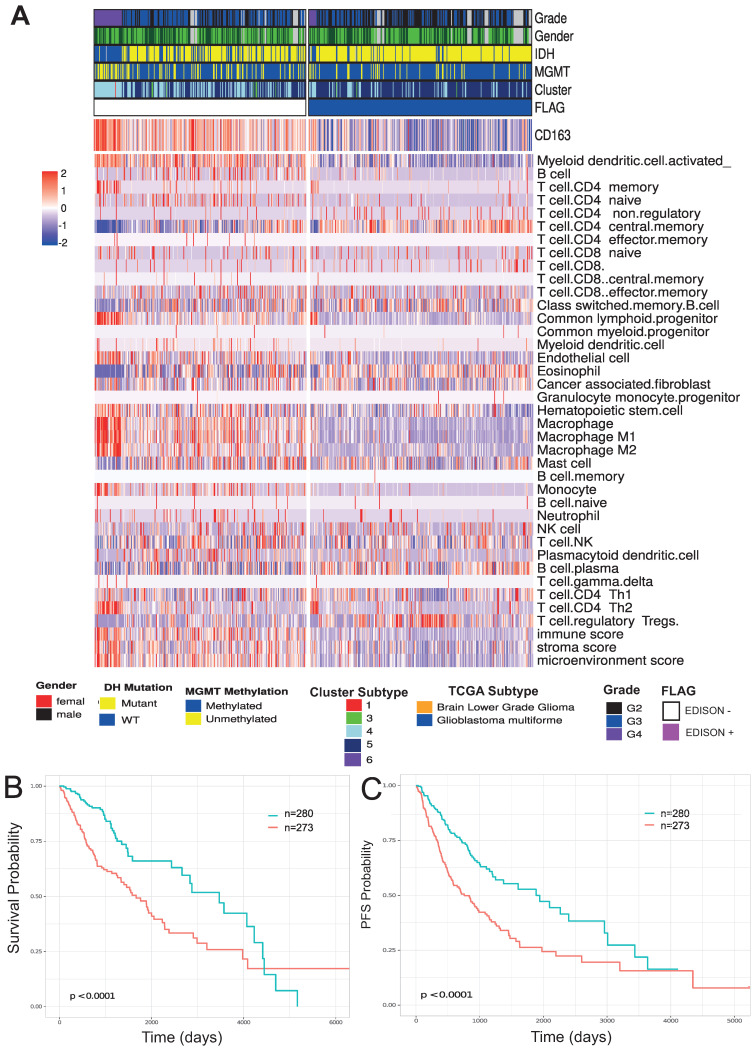
Figure 2 shows the immune-cell-related gene expression signatures for the glioma cases included in the study. In this context, increasing evidence indicates that TME plays a critical role in supporting the progression of gliomas. In fact, the majority of immune-related cells within brain tumors are macrophages, often comprising up to 30% of the tumor mass [10]. Most TAMs are considered to have M2 phenotype. Increased infiltration of TAMs correlated with improved glioma progression and tumor grade, and predicts poor prognosis in GBM patients. This raises the intriguing possibility that targeting TAMs may be a successful therapeutic strategy for intractable gliomas and GBMs [21]. On the other hand, the capacity to evade the anti-tumoral immune response is associated to the subset of T cells termed CD4+ CD25+ regulatory T cells (Treg), that have been shown to inhibit the actions of the effector T lymphocytes [5,56]. Thus, we considered the possible influence of two different cell populations, i.e., Tregs and M2 TAMs by evaluating RNA-seq data for gene expression signatures associated with the immunosuppressive role of these two populations. Moreover, we also evaluated the expression of CD163 itself, being CD163 one of the most important surface markers of M2 TAMs, that has been recently associated to a prognostic role [14]. We labeled cases as Evade Immune SuppressiON (EDISON) positive with a low immunosuppression state if at least two among these three parameters—CD163, M2 TAMs and Tregs—were below the first quartile. The resulting classification describes the possibility that a patient evades the immuno-suppression state and for this reason we called the flag EDISON (EvaDe Immune SuppressiON) positive. Consistently, as reported in Figure 2, EDISON positive cases showed less immunesuppressive phenotypes with both low values of the stromal signature score and the microenviroment signature score, as well as low endothelial signature score [57]. GBM was shown to be characterized by extensive endothelial hyperplasia [58] and the related signatures reported in Figure 2 confirmed this peculiar state.
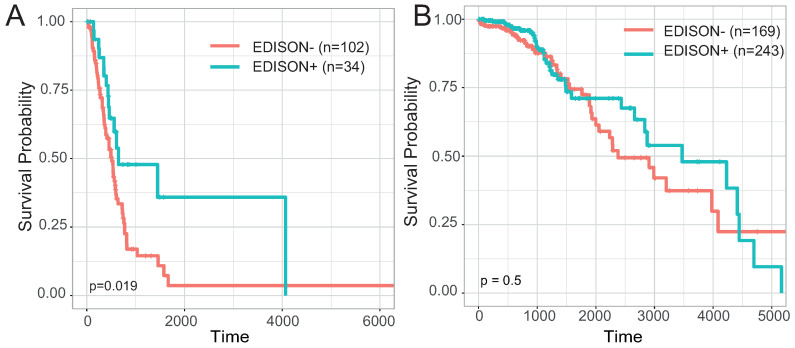
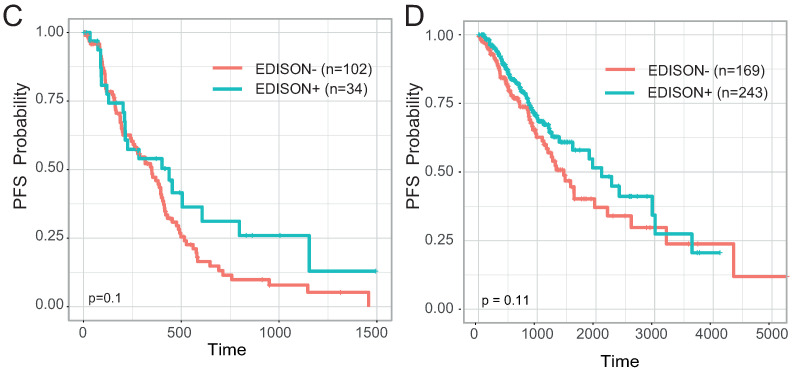
Figure 2.
Immune landscape of glioma patients. (A) Heatmap of immune signature computed on glioma cohorts from the TCGA study. The signature was calculated using immunodeconv (xCell) and the expression of gene CD163. The mutational status and immuno subtype are reported. (B) Kaplan–Meier survival curves showing OS interval based on the previously calculated flag on TCGA glioma patients. Time is reported in days. (C) Kaplan–Meier survival curves showed progression-free survival (PFS) intervals based on the previously calculated flag on TCGA glioma patients. Time is reported in days.
We also evaluated the capability of the EDISON classification by Kaplan–Meyer for assessing a prognostic significance using both overall survival (OS) and progression-free survival (PFS) intervals. We found that the EDISON positive cases showed significantly longer OS and PFS intervals than EDISON negative cases, thus confirming the importance of the immuno-suppressive-related parameters included in the EDISON flag (Figure 2B,C and Table 3). Figure A1 shows the EDISON classification in the context of IDH mutatant or IDH wildtype cases taken separately for both OS and PFI intervals.
Table 3.
Univariate Cox regression analysis of OS and PFS in the entire cohort included in the study using classification derived from RNA-seq data.
| Endpoint | Status | Number of Samples | HR | 95% CI for HR | p Value |
|---|---|---|---|---|---|
| OS | EDISON+ | n = 553 | 0.55 | 0.39–0.77 | <0.01 |
| PFI | EDISON+ | n = 553 | 0.57 | 0.43–0.75 | <0.01 |
Abbreviations: OS, overall survival; PFI, progression-free survival; HR, hazard ratio; CI, confidence interval.
3.2. From RNA Genes to the Classification Model
The procedure adopted to process the epigenetic data, that includes the creation the EDISON label for the immunosuppressive state, the development of the classification models and their evaluation, is summarized in Figure A2, while a focus on the machine learning models is provided in Figure A3. As described in Section 2.1, we considered a dataset where the input features are values from CpG probes and the target is a binary label corresponding to the EDISON flag. Starting from the genes used in Thorston et al. [18], we extracted the more informative genes to classify the immunosuppressive state [54,55,59,60,61]. We included also genes associated with the angiogenic signature, according to the prominent role of macrophages in tumor growth and angiogenesis [62], by including the angiomatrix signature reported by Langlois et al. [36]. Moreover, based on the fact that the response of ICIs has been shown to be relevant in both GBM and LGG [63], we evaluated a series of genes putatively related to responsiveness to ICIs, according to the GBM-associated signature reported in Zhao et al. [37]. More precisely, we compared the gene expression of the six GBM cases reported as Responsive against six GBM cases reported as Not Responsive and we obtained that 490 genes were differentially expressed, with adjusted p-values lower than 0.01.
The CpG beta values from 450 k Human DNA methylation microarray analysis consisted of 485,577 CpG methylation probes that were pre-processed by applying different basic filters to remove the useless probes, resulting in a final series of 355,314 CpG probes. A total of 6387 CpG probes were included in the overall signature we created and we labeled this set ImmuneAngioICIs. On such 6387 CpG probes, a first RF was created (Figure A3), and an out-of-sample MCC of 523 was obtained on the test set (see Table 4).
Table 4.
Metrics obtained for the random forest model on different datasets. The metrics were computed both in cross-validation (CV) on the train set (mean with 95% confidence intervals) and in out-of-sample evaluation on the test set . In bold, the best performer.
| Dataset | ACC CV (CI) | ACC Test | MCC CV (CI) | MCC Test |
|---|---|---|---|---|
| AllCpGs | 0.713 (0.676–0.747) | 0.756 | 0.435 (0.359–0.502) | 0.538 |
| ImmuneAngioICIs | 0.7155 (0.679–0.754) | 0.716 | 0.436 (0.368–0.512) | 0.523 |
| ImmuneAngioICIsMesECM | 0.710 (0.674–0.748) | 0.739 | 0.429 (0.354–0.504) | 0.490 |
| AllCpGs + BORUTA | 0.736 (0.699–0.770) | 0.755 | 0.478 (0.404–0.547) | 0.532 |
| ImmuneAngioICIs + BORUTA | 0.717 (0.681–0.752) | 0.729 | 0.443 (0.373–0.511) | 0.469 |
| ImmuneAngioICIsMesECM + BORUTA | 0.747 (0.713–0.780) | 0.793 | 0.498 (0.432–0.563) | 0.589 |
Based on a recent review evaluating prognostic genes for GBM [38], we evaluated the possibility of including a second model called ImmuneAngioICIsMesECM as described in Section 2.2 [17,39,48]. This procedure created a new set of 6754 CpG probes that were evaluated to classify EDISON positive cases. This second model resulted in an out-of-sample MCC of 0.490.
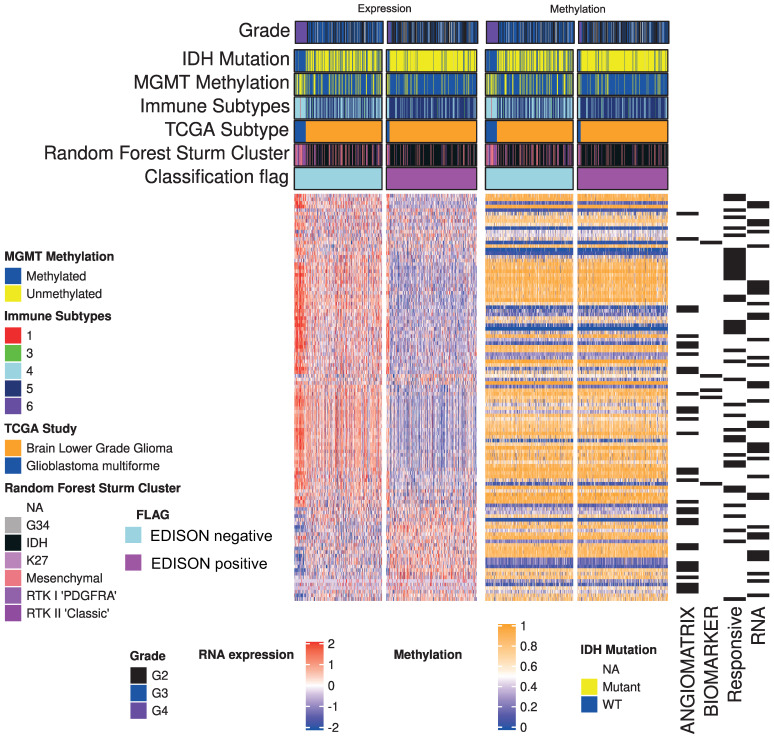
Figure 3 shows the expression of genes included in the model (left panel), and average mean value for each gene (right panel). While a clearly different expression can be explained for the EDISON classification, the average value for methylation seemed not to be sufficient to capture the methylation status. This result is in agreement with the complex modulation operated by the epigenetic regulation on gene expression. The resulting performance metrics are reported in Table 4. The model trained on ImmuneAngioICIsMesECM achieved a better out-of-sample accuracy, but a worse MCC.
Figure 3.
Genome-wide mean methylation status and matched transciptomic landscape from glioma cohort used in this study.
The application of a further step of feature selection, with the adoption of Boruta, resulted in an improvement of the metrics achieved by the RF classifiers, with the best results achieved with the dataset ImmuneAngioICIsMesECM + BORUTA. The 338 CpGs are listed in Table A2. As reported in Table 4, by using these features selected by Boruta in the datasets ImmuneAngioICIs + BORUTA and ImmuneAngioICIsMesECM + BORUTA, we obtained an out-of-sample MCC on and of 0.469 and 0.589, respectively.
Moreover, we evaluated the model fed by all the CpGs, either with or without the adoption of Boruta, and we observed a deterioration in the metrics with respect to our best performing model, trained on ImmuneAngioICIsMesECM + BORUTA (Table 4). This evidence substantiates the validity and the effectiveness of the expert selection.
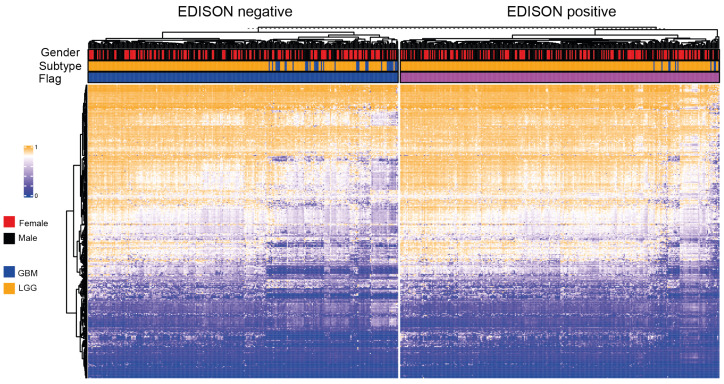
To further improve the model, we also considered the regional studies of the principal genomic localization such as CpG islands, shores, shelves and open sea. However, by this approach, no improvement in performance was obtained (Table A1). However, shore regions showed a better predictive power with respect to the other regions. This is consistent with previous studies which showed that these regions are more correlated with the regulation of gene expression. Figure 4 shows the genome-wide methylation landscape based on the selected 338 CpG probes, divided by the EDISON flag. Several differences in methylation can be appreciated between EDISON negative and EDISON positive cases. Moreover, in both EDISON positive and EDISON negative categories, GBM and LGG show different behaviours.
Figure 4.
Genomic landscape of the 338 CpG probe selected for the classification model according to the EDISON classification flag.
3.3. Deep Learning for the EDISON Classification
We evaluated the adoption of a deep learning model in place of the RF. Fixing the dataset to ImmuneAngioICIsMesECM + BORUTA, we tested both a feed-forward multilayer perceptron (MLP) and a 1D convolutional architecture. We observed better results with an MLP consisting of the input layer (338 neurons), two hidden layers (128 neuron each) and the output layer (1 neuron). Such MLP achieved an out-of-sample MCC of 0.658 and an accuracy of 0.828 on the test set (Table 5), outperforming the RF model.
Table 5.
Metrics obtained for the random forest and the MLP model on dataset ImmuneAngioICIsMesECM + BORUTA. The metrics were computed both in cross-validation (CV) on the train set (mean with 95% confidence intervals) and in out-of-sample evaluation on the test set . In bold, the best performer.
| Model | ACC CV (CI) | ACC Test | MCC CV (CI) | MCC Test |
|---|---|---|---|---|
| RF | 0.747 (0.713–0.780) | 0.793 | 0.498 (0.432–0.563) | 0.589 |
| MLP | 0.807 (0.795–0.819) | 0.828 | 0.625 (0.601–0.647) | 0.657 |
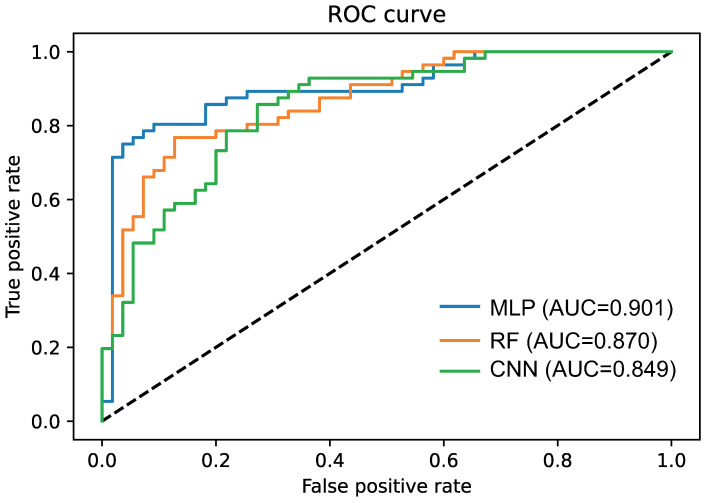
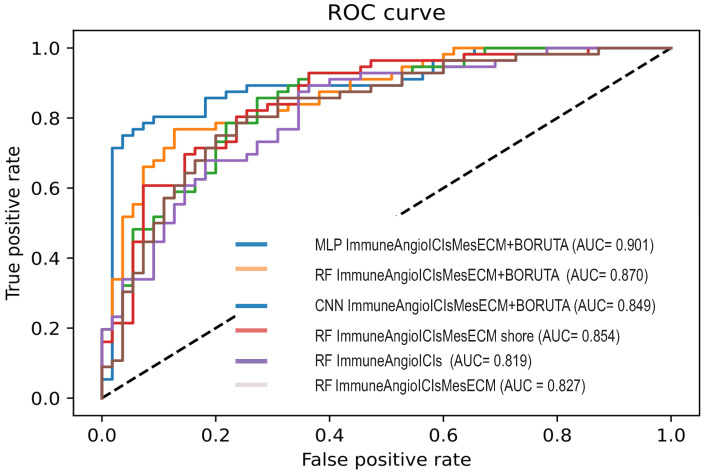
To assess the significance of the difference, we applied the McNemar test. We found that the difference in performance is significant, with a p value of 0.00952. This fact can also be visually appreciated by comparing the ROC curves (Figure 5).
Figure 5.
ROC curves of 3 models for EDISON classification using multilayer perceptron (MLP), convolutional neural network (CNN) and random forest (RF). All the models were trained on the dataset ImmuneAngioICIsMesECM + BORUTA. The out-of-sample AUC calculated on the test is also reported.
3.4. Biological Significance of the Selected CpG Probes
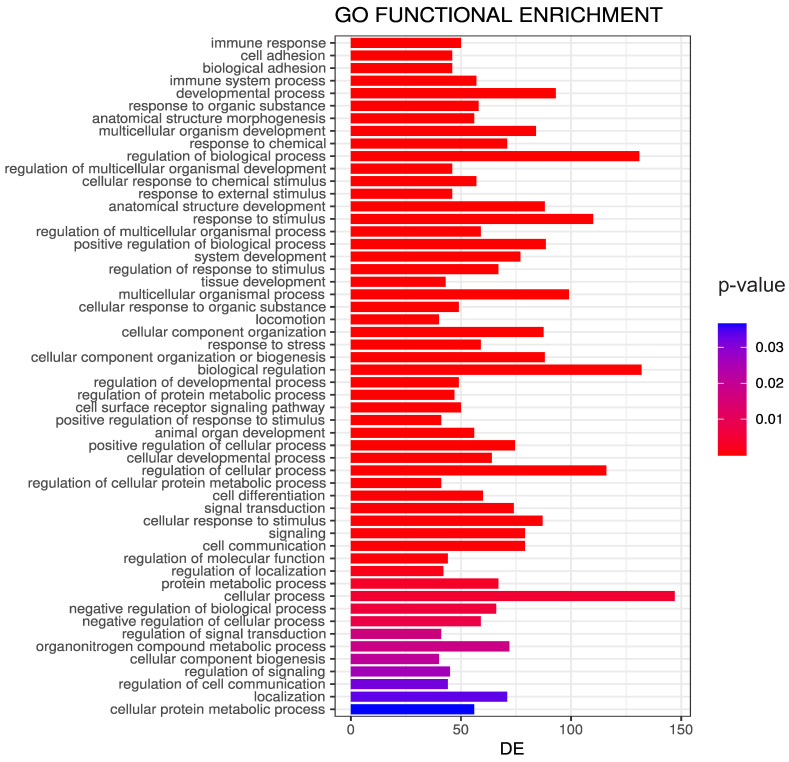
To gain insight into the biological significance of the model, we verified if the selected CpGs in ImmuneAngioICIsMesECM + BORUTA were correlated with the phenotype we tried to predict by our models. To do so, we applied the g-profile tool [64] to search for an enrichment in GO terms associated with the 338 CpG probes translated in genes. As expected, the selected go-terms were mainly associated with ECM organization, immune response, and regulation of cell adhesion (see Table 6 and Figure A5).
Table 6.
Top 30 terms’ signatures from enrichment analysis using gProfile on 338 CpG probe from the best model [64].
| #Term ID | Term Description | Observed Gene Count | Background Gene Count | Strength | False Discovery Rate |
|---|---|---|---|---|---|
| GO:0030198 | extracellular matrix organization | 31 | 296 | 1.14 | |
| GO:0006955 | immune response | 43 | 1560 | 0.56 | |
| GO:0002376 | immune system process | 49 | 2370 | 0.43 | |
| GO:0030155 | regulation of cell adhesion | 23 | 623 | 0.68 | |
| GO:0048514 | blood vessel morphogenesis | 18 | 381 | 0.79 | |
| GO:0001568 | blood vessel development | 19 | 464 | 0.73 | |
| GO:0007155 | cell adhesion | 25 | 843 | 0.59 | |
| GO:0009653 | anatomical structure morphogenesis | 40 | 1992 | 0.42 | |
| GO:0001525 | angiogenesis | 15 | 297 | 0.82 | |
| GO:0035239 | tube morphogenesis | 21 | 615 | 0.65 | |
| GO:0048583 | regulation of response to stimulus | 59 | 3882 | 0.3 | |
| GO:0010033 | response to organic substance | 48 | 2815 | 0.35 | |
| GO:0035295 | tube development | 23 | 793 | 0.58 | |
| GO:0002684 | positive regulation of immune system process | 24 | 882 | 0.55 | |
| GO:0071310 | cellular response to organic substance | 40 | 2219 | 0.37 | |
| GO:2000026 | regulation of multicellular organismal development | 36 | 1876 | 0.4 | |
| GO:0007492 | endoderm development | 8 | 76 | 1.14 | |
| GO:0050896 | response to stimulus | 91 | 7824 | 0.18 | |
| GO:0050776 | regulation of immune response | 23 | 873 | 0.54 | |
| GO:0045765 | regulation of angiogenesis | 13 | 277 | 0.79 | |
| GO:0045321 | leukocyte activation | 23 | 894 | 0.53 | |
| GO:0002443 | leukocyte mediated immunity | 19 | 632 | 0.59 | |
| GO:0070887 | cellular response to chemical stimulus | 44 | 2672 | 0.33 | |
| GO:0002274 | myeloid leukocyte activation | 18 | 574 | 0.61 | |
| GO:0010757 | negative regulation of plasminogen activation | 4 | 6 | 1.94 | |
| GO:0051239 | regulation of multicellular organismal process | 45 | 2788 | 0.32 | |
| GO:0002682 | regulation of immune system process | 29 | 1391 | 0.43 | |
| GO:0006027 | glycosaminoglycan catabolic process | 7 | 62 | 1.17 | |
| GO:0050778 | positive regulation of immune response | 18 | 589 | 0.6 |
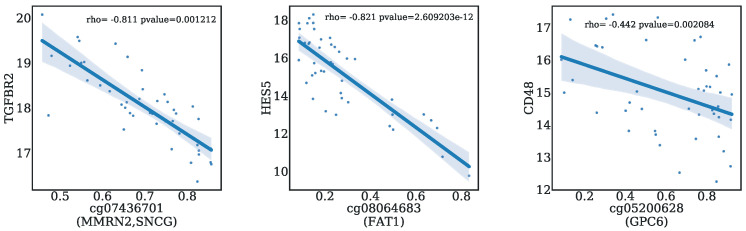
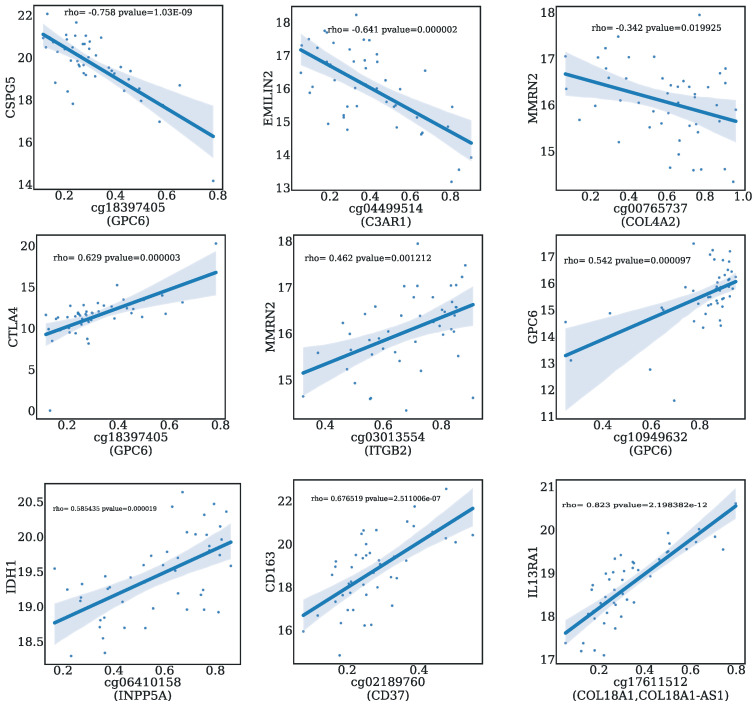
Moreover, we performed an analysis of the genes related to the 338 CpG probes of ImmuneAngioICIsMesECM + BORUTA using STRING in the Cytoscape app (Figure A6). We found that the genes resulted in a linked network of protein–protein interaction (PPI) of 165 nodes and 4058 edges (Figure A5). We also evaluated the involvement of CpG methylation genes in the modulation of the gene expression of gliomas. In Table A8, the CpG probes highly correlated with gene expression are reported. Among these CpGs, we found correlation with genes belonging to angiogenesys pathway, ECM organization, immune response and checkpoint molecules. In Figure A7, several examples of positive and negative correlation are shown.
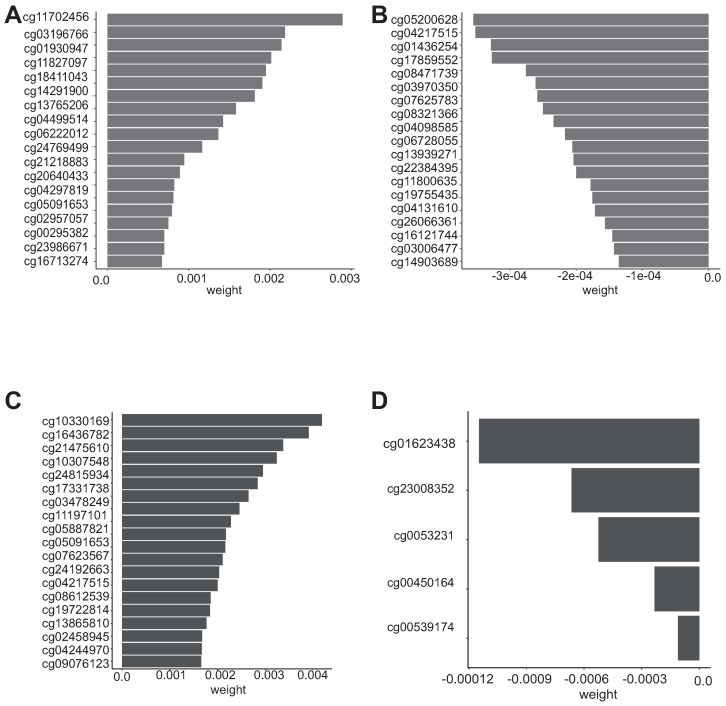
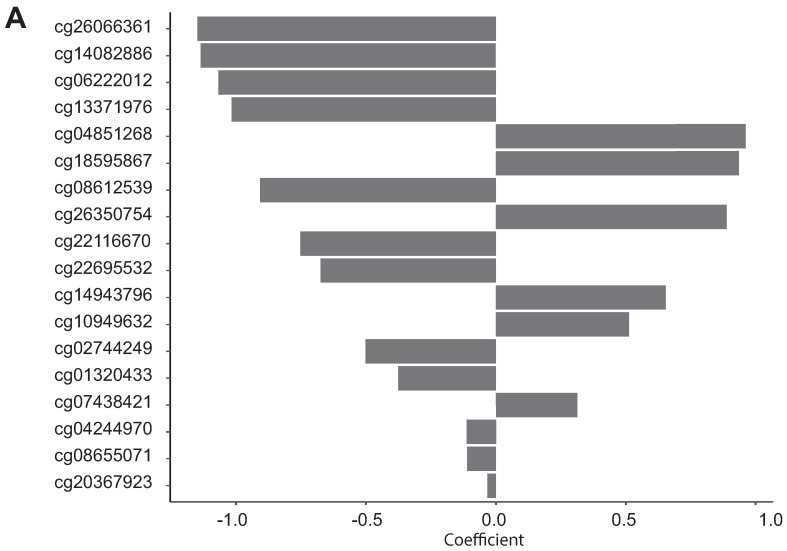
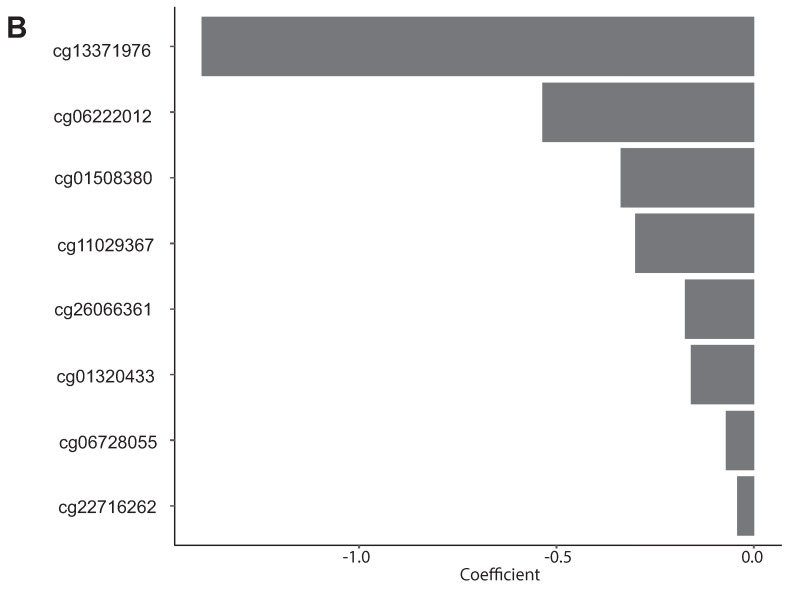
To perform a further selection of the most important CpG among the 338 in ImmuneAngioICIsMesECM + BORUTA, we applied random survival forest. The importance values obtained by the permutation analysis are depicted in Figure 6, while overall survival and progression free intervals are reported in Table A3 and Table A4, respectively.
Figure 6.
Variable importance of random survival forest model. (A) Top 20 CpG probes are reported with positive value influencing the OS interval, (B) Top 20 CpG probes are reported with negative influence OS interval, (C) Top 20 CpG probes are reported with positive value influencing the progression free survival, (D) Top 5 probes are reported with positive value influencing the progression-free survival.
3.5. Evaluation of the Transferability of the CpG Methylation Signature in Liquid Biopsy Samples
The methylation signature discussed in this study was obtained from primary glioma samples. However, although DNA methylation is tissue-specific, surrogate tissues such as blood are necessary due to the inaccessibility of human brain samples. Thus, we evaluated the possibility to obtain the genome-wide methylation using the blood to implement a liquid biopsy approach.
BECon (Blood–Brain Epigenetic Concordance; https://redgar598.shinyapps.io/BECon/ (accessed on 12 March 2020) is a tool that allows one to evaluate the concordance of CpGs between blood and brain, and to estimate how strongly a CpG is affected by the cell composition in both blood and brain. To perform such analyses, we imported the 338 CpGs of ImmuneAngioICIsMesECM + BORUTA on the BECon software tool and we selected the CpGs which varied in the most consistent way in the blood and in the brain. BECon select 113 CpG probes among 338. A LASSO Coxnet feature selection was then performed to detect the CpGs that can best explained both the overall survival (Figure A8, panel A) and the progression-free interval (Figure A8, panel B). Eighteen CpG probes were selected for the OS interval and eight for PFS (Table A6). The coefficients obtained from LASSO Coxnet were reported in Table A6. Positive values of coefficients were considered risk-associated in contrast to negative values which considered protective-associated. The GO terms analysis performed in positive and negative CpG associated probes is reported in Table A5.
4. Discussion
Gliomas are among the most common and aggressive primary tumors in adults [65]. Despite improved insight into the underlying molecular mechanisms, they are still hard to be treated and the prognosis of patients remains poor due to fast progress and scarcity of effective treatment strategies. The highly heterogeneous TME plays a substantial role in tumor malignancy and treatment responses. It is also related to the resistance of glioma cells to chemotherapy [10,59,66,67]. The glioma TME exerts a key role in tumor progression, in particular by providing an immunosuppressive state, with low number of TILs and of other immune effectors cell types as well as a high number of M2 macrophages, that contribute to tumor proliferation and growth [68]. Among the different processes regulating immune escape, TME-associated soluble factors, and/or cell surface-bound molecules are mostly responsible for dysfunctional activity of tumor-specific CD8+T cells. This TME immunosuppression could be involved in the capability of gliomas to respond to ICI treatment. A good understanding of TME and its mutual effects with tumor is important to reveal the treatment resistance mechanisms but also provide new strategies to improve the efficacy of these treatments including immunotherapies [61,69,70,71]. In this study, we systematically evaluated the possibility of creating an epigenetic model to stratify patients according to their capability to evade the immunosuppressive state peculiar of gliomas. We proposed the novel EDISON (EvaDe Immune SuppressiON) flag to summarize the contribution of macrophage M2 and Tregs in the immune suppressive state of gliomas. By comparing a random forest and two different neural network classifiers we showed the superiority of a multi-layer perceptron composed by two hidden layers. Such result is in agreement with that reported by other recent studies [72]. For most of the considered datasets and the models, we recorded higher metrics in the out-of-sample evaluation on the test set with respect to the cross-validation on the train set. This is a symptom of underfitting in the models. The most obvious and effective way to solve the issue, would be to include more samples in the dataset. Unfortunately, we were not able to find larger datasets to integrate our analysis. This could be considered as a limitation even if in an attempt to address the lack of an independent validation set, we followed the recommendations described in Shi et al. [43]. Moreover, further experiments are needed.
The proposed model could be used to predict the capability of the glioma patients to respond to immunotherapy such as ICIs. In this context, the employment of DNA methylation in place of RNA-seq data seems to provide a faster and more cost-effective approach.
Based on the results of the modelling, we defined a set of CpGs to be used as features: we proposed a final series of 338 CpGs related to genes belonging to ECM organization, immune response, angiogenesis and regulation of cell adhesion. Notably, the model trained on the 338 CpGs of ImmuneAngioICIsMesECM + BORUTA achieved better out-of-sample metrics than the ones trained on AllCpGs and AllCpGs + BORUTA. This evidence substantiates the validity and the effectiveness of the expert selection. Finally, we proposed a methylation signature that could be useful in the prediction of the clinical outcome of gliomas when liquid biopsy samples are used. Liquid biopsy represents a minimally invasive procedure that can provide similar information to what is usually obtained from a tissue biopsy samples. We found a small set of CpG (18 CpGs belong OS C and 8 CpGs PFS) that could be easily transferable to the laboratory routine for the classification of glioma patient by using BECon, a tool for interpreting DNA methylation features from blood. This could be useful in the management of glioma patients during the treatments. Moreover, several further suggestions could be highlighted regarding the involvement of the epigenetic modulation of the genes defined by the proposed model in key processes and mechanisms affecting the glioma pathogenesis and progression, such as ECM organization, immune response, angiogenesis and regulation of cell adhesion.
5. Conclusions
Despite the advances of molecular understanding and therapies that can be used for glioma treatment, clinical benefits have remained limited. A revelant role in treatment response is exerted by the TME in which the number of TILs and M2 macrophages is responsible for the degree of immunosuppression. In the present study, we proposed an epigenetic model to stratify patients according to their capability to evade the immune suppressive state called EDISON (EvaDe Immune SuppressiON) peculiar of gliomas. We demonstrated the superiority of the neural network composed by two hidden layers to classify the immunosuppressive state with respect to the random forest and convolutional approach. We also proposed a methylation signature that could be useful in the prediction of the clinical outcome of gliomas when liquid biopsy samples are used.
Acknowledgments
All the results here showed are based on data generated by the TCGA Research Network: https:/cancer.gov/tcga (accessed on 12 March 2020). For the processing of the data, tools provided by the Garr consortium were used as part of the agreement with the Ministry of Health for IRCCS, through the Garr Cloud Platform, a GDPR-compliant private-cloud system certified ISO 27001, ISO 27017 and ISO 27018 for information protection.
Abbreviations
The following abbreviations are used in this manuscript:
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three-letter acronym |
| LD | Linear dichroism |
| TME | Tumor microenvironment |
| EDISON | Evade immunosuppresion |
| TCGA | The Cancer Genome Atlas |
| GBM | Glioblastoma |
| LGG | Low-grade glioma |
| CI | Confidence interval |
| MLP | Multi-layer percerptron |
| RF | Random Forest |
| CNN | Convolutional neural network |
| ECM | Extracellular matrix |
| ICIs | Immune checkpoint inhibitors |
| BECon | Blood–Brain Epigenetic Concordance |
| MCC | Matthew correlation coefficient |
| ACC | Accuracy |
| Tregs | T regulatory cells |
| MDSCs | Myeloid-derived suppressor cells |
| TAMs | Tumor-associated macrophages |
Appendix A
Figure A1.
(A) Kaplan–Meier survival curves showing OS interval based on the previously calculated flag on TCGA glioma patients with IDH wild-type status. Time is reported in days. (B) Kaplan–Meier survival curves showing OS intervals based on the previously calculated flag on TCGA glioma patients with IDH mutated. Time is reported in days. (C) Kaplan–Meier survival curves showing PFI interval based on the previously calculated flag on TCGA glioma patients. with IDH wild-type status. Time is reported in days. (D) Kaplan–Meier survival curves showing PFS intervals based on the previously calculated flag on TCGA glioma patients with IDH mutated. Time is reported in days.
Figure A2.
Workflow for the development of a methylation-based machine learning model to predict the immune suppressive state responsiveness of glioma patients. (A) Development of the EDISON classification flag using transcriptomic data; (B) model construction on methylation dataset (Complete description present in Materials and Methods (Section 2.2).
Figure A3.
Machine learning workflow for developing the classification model starting by glioma dataset composed by Human Methylation data (450 k) composed by brain low-grade glioma (LGG) patients and glioblastoma (GBM).
Figure A4.
ROC curves of some models for EDISON classification with the indication of dataset used and the machine learning model used. The out-of-sample AUC calculated on the test is also reported. Random Forest (RF); multi-layer perceptron (MLP); convolutional neural network (CNN).
Table A1.
Model metrics in cross-validation (mean with confidence intervals) and on the test set using CpG probes derived from RNA. ACC: accuracy; MCC: Matthews Correlation, prec: Precision, recal: Recall Coefficient; CI: 95% studentized bootstrap confidence interval; RF: Random Forest.
| Model | Regions | ACC (CI) | ACC Test | MCC (CI) | MCC Test |
|---|---|---|---|---|---|
| RF | IImmuneAngioICIsMesECM-ISLAND | 0.724 (0.688–0.769) | 0.747 | 0.460 (0.389–0.549) | 0.522 |
| RF | ImmuneAngioICIsMesECM-OPENSEA | 0.725 (0.689–0.762) | 0.691 | 0.456 (0.386–0.533) | 0.469 |
| RF | ImmuneAngioICIsMesECM-SHORE | 0.749 (0.705–0.790) | 0.774 | 0.501 (0.417–0.583) | 0.553 |
| RF | ImmuneAngioICIsMesECM-SHELF | 0.758 (0.722–0.789) | 0.747 | 0.529 (0.459–0.593) | 0.510 |
| RF | ImmuneAngioICIs-ISLAND | 0.756 (0.716–0.792) | 0.758 | 0.514 (0.439–0.587) | 0.518 |
| RF | ImmuneAngioICIs-SHORE | 0.753 (0.710–0.798) | 0.734 | 0.509 (0.422–0.598) | 0.536 |
| RF | ImmuneAngioICIs- OPENSEA | 0.729 (0.757–0.700) | 0.738 | 0.463 (0.406–0.520) | 0.490 |
| RF | ImmuneAngioICIs-SHELF | 0.725 (0.685–0.763) | 0.720 | 0.457 (0.373–0.537) | 0.543 |
Table A2.
338 CpG probes included in best model to classify patient according to the EDISON flag.
| CpG | Gene |
|---|---|
| cg01681098 | SENCR, FLI1 |
| cg24457026 | GRN |
| cg13909178 | RP11-744N12.3 FLI1 |
| cg03531211 | XXbac-BPG181M17.5, HLA-DMA |
| cg04917472 | CTSZ |
| cg21012874 | MMRN2, SNCG |
| cg13662634 | RALGPS1, ANGPTL2 |
| cg17054708 | FBLN2 |
| cg10453850 | AL645941.1, HLA-DMB, XXbac-BPG181M17.5 |
| cg23008352 | COL4A1 |
| cg24421410 | XXbac-BPG181M17.5, HLA-DMA |
| cg07852825 | GHSR |
| cg04499514 | C3AR1 |
| cg16436782 | RP11-212E4.1, COL4A1 |
| cg03677069 | MMRN2, SNCG |
| cg00215182 | C1QB |
| cg13353679 | AFF3, AC092667.2 |
| cg14082886 | CD44 |
| cg09552892 | MMRN2, SNCG |
| cg04275881 | SLAMF8 |
| cg02072495 | ANXA2 |
| cg00338116 | EPSTI1 |
| cg10762214 | INPP5A |
| cg10070185 | SERPINA1 |
| cg13810673 | GPR65 |
| cg07857225 | PLXND1 |
| cg11037750 | TGFB1 |
| cg07450037 | HOTAIRM1, HOXA1, HOTAIRM1_1 |
| cg22568423 | MYO1F |
| cg01436254 | CD86 |
| cg17451419 | CYR61 |
| cg18273417 | S100A4 |
| cg18837947 | CCNG2 |
| cg27565899 | AMPD2 |
| cg07625783 | SLAMF8 |
| cg13371976 | PRELP |
| cg24815934 | ITGB2 |
| cg17599241 | VCAN-AS1, VCAN |
| cg10518264 | HLA-DMB, XXbac-BPG181M17.5 |
| cg11800635 | DOK1, LOXL3 |
| cg26357596 | GZMA |
| cg09456094 | SP100 |
| cg11827097 | SP100 |
| cg04131610 | CCR5, RP11-24F11.2 |
| cg00609834 | SPON1 |
| cg08076018 | RALGPS1, ANGPTL2 |
| cg06746774 | KIAA1522 |
| cg13700051 | TTC33 |
| cg17928895 | CTSZ |
| cg15550100 | ATG4B |
| cg07251141 | ADAM12 |
| cg26969179 | ADAM12 |
| cg18245281 | CTSZ |
| cg00539174 | CTSZ |
| cg17571335 | FLI1 |
| cg25428929 | ATG4B |
| cg01536987 | EPSTI1 |
| cg20694619 | TRAF3IP3 |
| cg03970350 | PES1, TCN2 |
| cg13765206 | EMILIN2 |
| cg04217515 | ITGB2 |
| cg14994258 | PXDN |
| cg11029367 | HEG1 |
| cg00765737 | COL4A2 |
| cg07464217 | CTSZ |
| cg03075156 | PRKCE |
| cg08655071 | TRAF3IP3 |
| cg00295382 | MYCL |
| cg14903689 | COL18A1 |
| cg19408145 | CD48 |
| cg17420036 | HSPG2 |
| cg18274749 | HSPG2 |
| cg07436701 | MMRN2, SNCG |
| cg02744249 | CTSZ |
| cg22116670 | CTB-113P19.1, SPARC |
| cg24192663 | HSPA6, RP11-25K21.6, FCGR2A |
| cg13785221 | ANXA2 |
| cg17801352 | PXDN |
| cg05887821 | INPP5A |
| cg18411043 | LAPTM5 |
| cg03478249 | EPSTI1 |
| cg21936552 | BAHCC1 |
| cg05200628 | CD48 |
| cg01930947 | C1orf111, RP11-565P22.6, C1orf226 |
| cg10330169 | DIS3L2 |
| cg10587741 | LGALS1 |
| cg24539923 | SERPINE1 |
| cg10768321 | CTC-301O7.4, CD37 |
| cg09538921 | IL27RA, CTB-55O6.4 |
| cg18968623 | INPP5A |
| cg08064683 | FAT1 |
| cg06330722 | PCOLCE, PCOLCE-AS1 |
| cg10307548 | SOD3 |
| cg09707038 | CALM2, RP11-761B3.1 |
| cg16024530 | FLI1 |
| cg13790288 | CD28 |
| cg08139855 | CSF1 |
| cg19919590 | LAPTM5 |
| cg20600379 | HLA-DMB, XXbac-BPG181M17.5 |
| cg24375627 | S100A6 |
| cg12339920 | TGFBI |
| cg27617132 | INPP5A |
| cg03682712 | LOXL1, LOXL1-AS1 |
| cg21746573 | PRKCE |
| cg19506628 | CEP72 |
| cg17319576 | CYR61 |
| cg17911539 | C3orf22, CHST13 |
| cg04232128 | TMEM173 |
| cg05360958 | C12orf60, MGP |
| cg04755674 | IL27RA, CTB-55O6.4 |
| cg03013554 | ITGB2 |
| cg04297819 | HSPG2 |
| cg00799121 | ADAMTS2 |
| cg08321366 | MMP14 |
| cg19722814 | SERPINE1 |
| cg14943796 | BAHCC1 |
| cg04771838 | COL4A2 |
| cg11581627 | CD33 |
| cg14991595 | MB21D2 |
| cg15347156 | MMRN2 |
| cg04153551 | FBLN5 |
| cg06222012 | AC078941.1, AC023115.2 |
| cg04244970 | SLAMF7 |
| cg22704788 | PRELP |
| cg21043746 | ADAMTS2 |
| cg26532826 | PES1, TCN2 |
| cg13962321 | HIST2H2BB, RP5-998N21.7, RP5-998N21.10 |
| cg11702456 | SP100 |
| cg09076123 | NCF2, SMG7 |
| cg08825225 | FLI1 |
| cg17713010 | LAIR1 |
| cg15522984 | LAMC1 |
| cg08682341 | INPP5A |
| cg03813885 | CFAP97, SNX25 |
| cg10845380 | SLC7A7 |
| cg12613839 | ADAMTS2 |
| cg02588309 | TTC33 |
| cg02189760 | CTC-301O7.4, CD37 |
| cg16925003 | PXDN |
| cg07947930 | PRELP |
| cg06410158 | INPP5A |
| cg24644113 | TADA1 |
| cg27547543 | POU5F1 |
| cg21860679 | DUSP6, RP11-823E8.3 |
| cg27329371 | ALDH3A1 |
| cg00771084 | ATG4B |
| cg11594010 | INPP5A |
| cg11301254 | TTC33 |
| cg09926389 | TGFB1 |
| cg03982087 | RAB31 |
| cg02286081 | HLA-DPA1, HLA-DPB1 |
| cg26025068 | PPP1R8 |
| cg00078334 | MMP2 |
| cg23638686 | INPP5A |
| cg19755435 | GPR65 |
| cg08530414 | RP4-607I7.1, CD44 |
| cg15999547 | TMEM54, HPCA |
| cg26214645 | SECTM1 |
| cg25206536 | MIR572 |
| cg20502977 | COL6A3 |
| cg23659056 | FOXD2, FOXD2-AS1 |
| cg23986671 | ADAMTS5 |
| cg26138144 | LGALS1 |
| cg07855465 | BAHCC1 |
| cg03196766 | THBS1 |
| cg17859552 | INPP5A |
| cg18900669 | RP11-186B7.4, CD68 |
| cg19915711 | EPSTI1 |
| cg10974980 | LOXL1 |
| cg08612539 | CTA-833B7.2, NCF4 |
| cg18397405 | GPC6 |
| cg00450164 | TRAF3IP3 |
| cg26650846 | ADAMTS2 |
| cg04098585 | CD28 |
| cg16826739 | INPP5A |
| cg24767336 | TGFB1, CTC-435M10.3, TMEM91 |
| cg03006477 | CD109 |
| cg16713274 | COL18A1, LL21NC02-21A1.1 |
| cg25450450 | CTB-118N6.2, SEMA6A |
| cg09277376 | FOXD2-AS1 |
| cg03440588 | FOXD2, FOXD2-AS1 |
| cg24129356 | XXbac-BPG181M17.5, HLA-DMA |
| cg16121744 | COL18A1 |
| cg14139008 | DNM1 |
| cg24226528 | TMEM37 |
| cg11875119 | PES1, TCN2 |
| cg01508380 | MMP14 |
| cg09280946 | CTSC |
| cg02543462 | IL1RN |
| cg00142150 | LGALS1 |
| cg21005525 | ARF1 |
| cg07697770 | TGFBI |
| cg03930369 | COL4A2 |
| cg06671298 | BAHCC1 |
| cg15254671 | MYO1F |
| cg00292662 | LGALS1 |
| cg21236655 | TNC |
| cg07724259 | EMILIN2 |
| cg23865240 | HOTAIRM1, HOXA1 |
| cg09321817 | HLA-DPA1 |
| cg18595867 | FOXD2-AS1 |
| cg22158252 | BMP8A |
| cg27438456 | INPP5A |
| cg07085815 | SERPINE2 |
| cg18644834 | ANKRA2, UTP15 |
| cg24287218 | HLA-DPA1 |
| cg24707889 | ITGB2, ITGB2-AS1 |
| cg09269866 | FOXD2, FOXD2-AS1 |
| cg03753191 | EPSTI1 |
| cg22716262 | MPP7 |
| cg22595235 | SUMF1, LRRN1 |
| cg19575208 | HLA-DRB1 |
| cg06507307 | INPP5A |
| cg13939271 | DNM1 |
| cg23225572 | RP11-565P22.6, NOS1AP, C1orf226 |
| cg11197101 | KIAA1522 |
| cg21869219 | ARHGAP31 |
| cg10954654 | CTSS |
| cg20481110 | SECTM1 |
| cg11804789 | CST7 |
| cg25214684 | AKIRIN1 |
| cg15114672 | VCAN |
| cg00516966 | ALDH3A1 |
| cg14791054 | RP11-66B24.4, ALDH1A3 |
| cg00816609 | FBLN2 |
| cg03055440 | MS4A6A |
| cg21218883 | PRKCE |
| cg02458945 | MMP2 |
| cg22118297 | ADAMTS9, ADAMTS9-AS1 |
| cg20640433 | LAMA2 |
| cg12689670 | LAMC1 |
| cg03573861 | BAHCC1 |
| cg07438421 | SERPINF1 |
| cg05822532 | ELN |
| cg15849060 | ALDH3A1 |
| cg02784696 | C2orf44, MFSD2B |
| cg26399819 | MIER3 |
| cg18832223 | CEP72 |
| cg09777237 | ELN |
| cg15504747 | PLXND1 |
| cg01338658 | LAMC1 |
| cg00894134 | DNM1 |
| cg25306579 | INPP5A |
| cg00532319 | RPN1 |
| cg07906179 | BAHCC1 |
| cg24493834 | LAMA2, MESTP1 |
| cg22136020 | CSPG4 |
| cg01320433 | XXyac-YX65C7_A.2, THBS2 |
| cg10989879 | CFAP97, SNX25 |
| cg15459165 | LAPTM5 |
| cg01623438 | CTSZ |
| cg12253414 | ITGB5 |
| cg00777079 | SERPINF1 |
| cg08638320 | FOXD2, FOXD2-AS1 |
| cg05831823 | CR2 |
| cg12630520 | SPARCL1 |
| cg23446438 | MYO1F |
| cg06728055 | WWTR1 |
| cg05492532 | INPP5A |
| cg09545579 | BAHCC1 |
| cg26204079 | RP11-400N9.1, DGKD |
| cg14291900 | SLC7A7 |
| cg21475610 | CCNG2 |
| cg07575373 | CTC-301O7.4, CD37 |
| cg05658236 | FOXD2-AS1 |
| cg15046675 | CTC-301O7.4, CD37 |
| cg22216491 | CASP6 |
| cg05091653 | SP100 |
| cg11076970 | HLA-DOA |
| cg26262232 | XXbac-BPG181M17.5, HLA-DMA |
| cg25645491 | HLA-DRA |
| cg23173573 | DUSP10 |
| cg14880894 | CNOT6L |
| cg02316283 | MMP14 |
| cg05041061 | BAHCC1 |
| cg12937501 | AC106875.1, LPIN1 |
| cg26034531 | LPPR5, RP5-896L10.1 |
| cg06390079 | ALDH3A1 |
| cg01120369 | PLXND1 |
| cg18764513 | SLC7A7 |
| cg05830842 | COL14A1 |
| cg11728145 | PXDN |
| cg07659054 | HOTAIRM1, HOXA1 |
| cg13802966 | CASP1 |
| cg13865810 | COL15A1, RP11-92C4.6 |
| cg07623567 | HLA-DMB, XXbac-BPG181M17.5 |
| cg11912272 | SPATS2L |
| cg17016011 | INPP5A |
| cg00416645 | AC007563.5, IGFBP5 |
| cg01997629 | TRAF3IP3 |
| cg10928302 | RBM6, RBM5 |
| cg02957057 | NID1 |
| cg17081489 | RP4-798P15.3, SEC16B |
| cg10001720 | LAPTM5 |
| cg20407868 | INPP5A |
| cg24769499 | TMEM37 |
| cg26350754 | HLA-DPA1, HLA-DPB1 |
| cg10949632 | GPC6 |
| cg22905097 | EPSTI1 |
| cg26066361 | CLEC7A |
| cg09099927 | RP11-333E13.4 |
| cg17611512 | COL18A1, COL18A1-AS1 |
| cg13477614 | BAHCC1 |
| cg25913233 | CTB-113P19.1, SPARC |
| cg07616471 | CCR5, RP11-24F11.2 |
| cg04654716 | CTD-2377O17.1, FAM169A |
| cg08471739 | PLXND1 |
| cg27297192 | INPP5A |
| cg04851268 | GHSR |
| cg24931346 | C1QB |
| cg21784272 | FAT1 |
| cg22987448 | MYO1F |
| cg22164238 | AMPD2, GNAT2 |
| cg08288016 | FAT1 |
| cg21398469 | CCNG2 |
| cg22384395 | RP11-66B24.9, ALDH1A3 |
| cg05710142 | KIAA1522 |
| cg21904489 | ARHGAP31 |
| cg01975495 | SERPINE1 |
| cg12917072 | ADAMTS12 |
| cg03393607 | AFF3, AC092667.2 |
| cg01821226 | PXDN |
| cg05955301 | PRELP |
| cg27470554 | FCGR2A |
| cg06238491 | LAIR1 |
| cg22695532 | RP11-475O6.1 |
| cg00742851 | SUMF1, LRRN1 |
| cg27553626 | PPP1R8 |
| cg25394505 | INPP5A |
| cg08735211 | XXbacBPG181M17.5, HLA-DMA |
| cg09983885 | TRIM21 |
| cg26514080 | KIAA1522 |
| cg05886789 | PLXDC2 |
| cg05826823 | CIZ1, DNM1 |
| cg20367923 | XXyac-YX65C7_A.2, THBS2 |
| cg24023498 | NR4A2 |
| cg16239257 | LTBP2 |
| cg17331738 | NES |
Table A3.
Top features detected by permutation analysis from Random Survival Forest using PFS selected by 338 CpG probes.
| Feature | Weight | std | Gene | Direction |
|---|---|---|---|---|
| cg02458945 | 0.00167149 | 0.0004095 | MMP2 | Positive |
| cg03478249 | 0.00245778 | 0.00088256 | EPSTI1 | Positive |
| cg04131610 | 0.00323843 | 0.00146985 | CCR5, RP11-24F11.2 | Positive |
| cg04217515 | 0.00200311 | 0.00073152 | ITGB2 | Positive |
| cg04244970 | 0.00166519 | 0.00067762 | SLAMF7 | Positive |
| cg05091653 | 0.00216262 | 0.00032063 | SP100 | Positive |
| cg05887821 | 0.00216972 | 0.00032553 | INPP5A | Positive |
| cg07623567 | 0.00210498 | 0.00037064 | HLA-DMB, XXbac-BPG181M17.5 | Positive |
| cg08612539 | 0.00185325 | 0.00028535 | CTA-833B7.2, NCF4 | Positive |
| cg09076123 | 0.00165326 | 0.0007419 | NCF2, SMG7 | Positive |
| cg10307548 | 0.00294971 | 0.00098171 | SOD3 | Positive |
| cg10330169 | 0.00418932 | 0.00097532 | DIS3L2 | Positive |
| cg11197101 | 0.00227763 | 0.00036055 | KIAA1522 | Positive |
| cg13865810 | 0.00176505 | 0.00045972 | COL15A1, RP11-92C4.6 | Positive |
| cg16436782 | 0.00390891 | 0.00123599 | RP11-212E4.1, COL4A1 | Positive |
| cg17331738 | 0.00264999 | 0.00031466 | NES | Positive |
| cg19722814 | 0.00184012 | 0.00034522 | SERPINE1 | Positive |
| cg21475610 | 0.00337569 | 0.00100237 | CCNG2 | Positive |
| cg24192663 | 0.00203206 | 0.00052522 | HSPA6, RP11-25K21.6, FCGR2A | Positive |
| cg24815934 | 0.00283711 | 0.00098436 | ITGB2 | Positive |
| cg00450164 | −2.33 | 0.00011302 | TRAF3IP3 | Negative |
| cg00532319 | −5.25 | 9.97 | RPN1 | Negative |
| cg00539174 | −1.10 | 0.0001254 | CTSZ | Negative |
| cg01623438 | −0.0001147 | 0.00016634 | CTSZ | Negative |
| cg23008352 | −6.65 | 0.00011621 | COL4A1 | Negative |
Table A4.
Top features detected by permutation analysis from Random Survival Forest using OS interval selected by 338 CpG probes.
| Feature | Weight | std | Gene | Direction |
|---|---|---|---|---|
| cg01436254 | −0.0003291 | 0.00023658 | CD86 | Negative |
| cg03006477 | −0.000143 | 0.00015906 | CD109 | Negative |
| cg03970350 | −0.0002618 | 0.0001385 | PES1, TCN2 | Negative |
| cg04098585 | −0.0002341 | 8.09 | CD28 | Negative |
| cg04131610 | −0.0001756 | 0.00013193 | CCR5, RP11-24F11.2 | Negative |
| cg04217515 | −0.0003524 | 0.00027475 | ITGB2 | Negative |
| cg05200628 | −0.0003555 | 0.00013671 | CD48 | Negative |
| cg06728055 | −0.0002169 | 0.00021168 | WWTR1 | Negative |
| cg07625783 | −0.0002587 | 8.14 | SLAMF8 | Negative |
| cg08321366 | −0.0002504 | 0.0001037 | MMP14 | Negative |
| cg08471739 | −0.0002763 | 0.00018392 | PLXND1 | Negative |
| cg11800635 | −0.0001998 | 0.00016978 | DOK1, LOXL3 | Negative |
| cg13939271 | −0.0002059 | 0.00018542 | DNM1 | Negative |
| cg14903689 | −0.0001355 | 0.00015751 | COL18A1 | Negative |
| cg16121744 | −0.0001452 | 0.00012391 | COL18A1 | Negative |
| cg17859552 | −0.0003278 | 0.00012804 | INPP5A | Negative |
| cg19755435 | −0.0001786 | 0.00014323 | GPR65 | Negative |
| cg22384395 | −0.0002037 | 0.00018963 | RP11-66B24.9, ALDH1A3 | Negative |
| cg24421410 | −0.0001716 | 6.39 | XXbac-BPG181M17.5, HLA-DMA | Negative |
| cg26066361 | −0.0001566 | 0.00031627 | CLEC7A | Negative |
| cg00295382 | 0.00069785 | 0.00031361 | MYCL | Positive |
| cg00777079 | 0.00194878 | 0.00093506 | SERPINF1 | Positive |
| cg01930947 | 0.00214195 | 0.00092059 | C1orf111, RP11-565P22.6, C1orf226 | Positive |
| cg02957057 | 0.00074669 | 0.00018464 | NID1 | Positive |
| cg03196766 | 0.00218507 | 0.00059328 | THBS1 | Positive |
| cg04297819 | 0.00081885 | 0.00047748 | HSPG2 | Positive |
| cg04499514 | 0.00142254 | 0.00078907 | C3AR1 | Positive |
| cg05091653 | 0.00080697 | 0.00048847 | SP100 | Positive |
| cg06222012 | 0.00135962 | 0.00059195 | AC078941.1, AC023115.2 | Positive |
| cg11702456 | 0.0028937 | 0.00103411 | SP100 | Positive |
| cg11827097 | 0.00201346 | 0.00095397 | SP100 | Positive |
| cg13765206 | 0.00158138 | 0.00079435 | EMILIN2 | Positive |
| cg14082886 | 0.00079245 | 0.00050107 | CD44 | Positive |
| cg14291900 | 0.00181238 | 0.00080766 | SLC7A7 | Positive |
| cg16713274 | 0.00066793 | 0.00025659 | COL18A1, LL21NC02-21A1.1 | Positive |
| cg18411043 | 0.0019083 | 0.00079707 | LAPTM5 | Positive |
| cg20640433 | 0.00088705 | 0.00032218 | LAMA2 | Positive |
| cg21218883 | 0.00094513 | 0.00049267 | PRKCE | Positive |
| cg23986671 | 0.00069697 | 0.00025722 | ADAMTS5 | Positive |
| cg24769499 | 0.00116293 | 0.00021175 | TMEM37 | Positive |
Table A5.
Gene ontology (GO) that define biological function. The GO annotations, accompanied by evidence-based statements describe specific gene product and specific ontology term (biological function). g-profile enrichment terms obtained from positive and negative values of 18 CpGs selected from BECon on overal survival by LASSO procedure. All the data have p value low than 0.05. MF: molecular function, CC: cellular component, BP: biological process.
| Source | Term_NAME | Term_id | Adjusted_p_Value | Negative_log10_of _Adjusted_p_Value | Direction Coefficient |
|---|---|---|---|---|---|
| GO:MF | glycosaminoglycan binding | GO:0005539 | 0.008788287085578 | 2.05609576449095 | Negative |
| GO:CC | collagen-containing extracellular matrix | GO:0062023 | 0.048474322092945 | 1.31448825575832 | Negative |
| KEGG | ECM-receptor interaction | KEGG:04512 | 0.037233188408127 | 1.42906977203396 | Negative |
| CORUM | CD44-LRP1 complex | CORUM:7535 | 0.049698019554143 | 1.30366091738024 | Negative |
| GO:MF | growth hormone secretagogue receptor activity | GO:0001616 | 0.049775611543161 | 1.3029833955258 | Positive |
| GO:BP | regulation of neurotransmitter receptor localization to postsynaptic specialization membrane | GO:0098696 | 0.000167004924934 | 3.77727072142761 | Positive |
| GO:BP | regulation of receptor localization to synapse | GO:1902683 | 0.001168645063196 | 2.93231737121765 | Positive |
| GO:BP | protein localization to postsynaptic specialization membrane | GO:0099633 | 0.001335544908879 | 2.8743415038609 | Positive |
| GO:BP | neurotransmitter receptor localization to postsynaptic specialization membrane | GO:0099645 | 0.001335544908879 | 2.8743415038609 | Positive |
| GO:BP | regulation of protein localization to synapse | GO:1902473 | 0.003070843427737 | 2.51274232624767 | Positive |
| GO:BP | protein localization to postsynaptic membrane | GO:1903539 | 0.007006418620425 | 2.15450391795907 | Positive |
| GO:BP | protein localization to postsynapse | GO:0062237 | 0.009117775996435 | 2.04011108165666 | Positive |
| GO:BP | response to dexamethasone | GO:0071548 | 0.009117775996435 | 2.04011108165666 | Positive |
| GO:BP | regulation of postsynaptic membrane neurotransmitter receptor levels | GO:0099072 | 0.013616524667261 | 1.86593372285098 | Positive |
| GO:BP | receptor localization to synapse | GO:0097120 | 0.013616524667261 | 1.86593372285098 | Positive |
| GO:BP | protein localization to synapse | GO:0035418 | 0.033345288657273 | 1.47696551870962 | Positive |
Figure A5.
Enrichment of GO terms from 338 CpG probes obtained from the best model. GO terms are plotted according to adjusted p-values (BH). Bar sizes represent the number of CpGs translated as genes that fall within a GO category; DE and colour represent the adjusted p-values (BH).
Figure A6.
Protein–protein interaction (PPI) network of the genes from 338 CpG probes of ImmuneAngioICIsMesECM + BORUTA using STRING in the Cytoscape app [50].
Figure A7.
Correlation between CpG probes selected among 338 CpGs with gene expression of some revelant genes. On the x-axis, the methylation status is reported, on the y-axis, RNA expression values are reported. Correlation values by Pearson and p-value are reported for each panel.
Figure A8.
Feature selection using LASSO COXNET of the 113 CpGs selected by BECon. The 18 CpGs selected for OS (A) and 8 for PFS interval (B) are reported.
Table A6.
Feature selection with the coefficient value and gene name of CpG probe selected by LASSO COXNET in the context of the developed signature for liquid biopsy based on BECon.
| CpG | COEFFICIENT | GENE | INTERVAL |
|---|---|---|---|
| cg01320433 | −0.1592041 | XXyac-YX65C7_A.2, THBS2 | PFS |
| cg01508380 | −0.33756425 | MMP14 | PFS |
| cg06222012 | −0.5359208 | AC078941.1, AC023115.2 | PFS |
| cg06728055 | −0.07162224 | WWTR1 | PFS |
| cg11029367 | −0.30043008 | HEG1 | PFS |
| cg13371976 | −1.39825624 | PRELP | PFS |
| cg22716262 | −0.04181589 | MPP7 | PFS |
| cg26066361 | −0.17464321 | CLEC7A | PFS |
| cg01320433 | −0.37578276 | XXyac-YX65C7_A.2, THBS2 | OS |
| cg02744249 | −0.5024627 | CTSZ | OS |
| cg04244970 | −0.11253157 | SLAMF7 | OS |
| cg04851268 | 0.96144542 | GHSR | OS |
| cg06222012 | −1.06780331 | AC078941.1, AC023115.2 | OS |
| cg07438421 | 0.31295409 | SERPINF1 | OS |
| cg08612539 | −0.90719367 | CTA-833B7.2, NCF4 | OS |
| cg08655071 | −0.10973404 | TRAF3IP3 | OS |
| cg10949632 | 0.5123932 | GPC6 | OS |
| cg13371976 | −1.01620416 | PRELP | OS |
| cg14082886 | −1.13578356 | CD44 | OS |
| cg14943796 | 0.65287911 | BAHCC1 | OS |
| cg18595867 | 0.93613126 | FOXD2-AS1 | OS |
| cg20367923 | −0.03248293 | XXyac-YX65C7_A.2, THBS2 | OS |
| cg22116670 | −0.75216037 | CTB-113P19.1, SPARC | OS |
| cg22695532 | −0.67502749 | RP11-475O6.1 | OS |
| cg26066361 | −1.14858784 | CLEC7A | OS |
| cg26350754 | 0.88852671 | HLA-DPA1, HLA-DPB1 | OS |
Table A7.
Correlation of CpG probes with genes that have high correlation values from 338 CpGs used to create the model.
| CpG | geneCpG | Gene | rho | p Value | Correlation Strength |
|---|---|---|---|---|---|
| cg13353679 | AFF3, AC092667.2 | HAVCR2 | 0.67520241 | 2.65 | high |
| cg13353679 | AFF3, AC092667.2 | CCR4 | 0.62698564 | 3.13 | high |
| cg13353679 | AFF3, AC092667.2 | TGFB1 | 0.72533605 | 1.19 | high |
| cg13353679 | AFF3, AC092667.2 | IL10 | 0.75708106 | 1.14 | high |
| cg22568423 | MYO1F | HAVCR2 | 0.6391164 | 1.75 | high |
| cg22568423 | MYO1F | TGFB1 | 0.61936567 | 4.45 | high |
| cg22568423 | MYO1F | IL10 | 0.65815888 | 6.66 | high |
| cg17599241 | VCAN-AS1, VCAN | HAVCR2 | 0.6225838 | 3.84 | high |
| cg17599241 | VCAN-AS1, VCAN | TGFB1 | 0.66056385 | 5.87 | high |
| cg17599241 | VCAN-AS1, VCAN | IL10 | 0.71015235 | 3.25 | high |
| cg08064683 | FAT1 | CCR4 | 0.60946563 | 6.94 | high |
| cg08064683 | FAT1 | TGFB1 | 0.7482646 | 2.26 | high |
| cg08064683 | FAT1 | IL10 | 0.66059345 | 5.86 | high |
| cg00799121 | ADAMTS2 | HAVCR2 | 0.6833261 | 1.67 | high |
| cg00799121 | ADAMTS2 | TGFB1 | 0.69708683 | 7.37 | high |
| cg00799121 | ADAMTS2 | IL10 | 0.71008968 | 3.26 | high |
| cg04153551 | FBLN5 | HAVCR2 | 0.6642975 | 4.81 | high |
| cg04153551 | FBLN5 | CCR4 | 0.63922606 | 1.74 | high |
| cg04153551 | FBLN5 | TGFB1 | 0.64462765 | 1.33 | high |
| cg04153551 | FBLN5 | IL10 | 0.75654189 | 1.19 | high |
| cg22704788 | PRELP | HAVCR2 | 0.66998995 | 3.53 | high |
| cg22704788 | PRELP | TGFB1 | 0.64874971 | 1.08 | high |
| cg22704788 | PRELP | IL10 | 0.71501411 | 2.37 | high |
| cg12613839 | ADAMTS2 | HAVCR2 | 0.70546862 | 4.38 | high |
| cg12613839 | ADAMTS2 | TGFB1 | 0.70966683 | 3.35 | high |
| cg12613839 | ADAMTS2 | PDCD1LG2 | 0.60313469 | 9.15 | high |
| cg12613839 | ADAMTS2 | IL10 | 0.7323874 | 7.25 | high |
| cg02189760 | CTC-301O7.4, CD37 | HAVCR2 | 0.64276143 | 1.46 | high |
| cg02189760 | CTC-301O7.4, CD37 | TGFB1 | 0.65060293 | 9.85 | high |
| cg02189760 | CTC-301O7.4, CD37 | IL10 | 0.68557785 | 1.46 | high |
| cg07947930 | PRELP | HAVCR2 | 0.66424332 | 4.82 | high |
| cg07947930 | PRELP | TGFB1 | 0.68739449 | 1.32 | high |
| cg07947930 | PRELP | PDCD1LG2 | 0.62213951 | 3.92 | high |
| cg07947930 | PRELP | IL10 | 0.6692665 | 3.68 | high |
| cg27329371 | ALDH3A1 | HAVCR2 | 0.63023007 | 2.68 | high |
| cg27329371 | ALDH3A1 | TGFB1 | 0.666298 | 4.32 | high |
| cg27329371 | ALDH3A1 | PDCD1LG2 | 0.6318309 | 2.49 | high |
| cg27329371 | ALDH3A1 | IL10 | 0.69220582 | 9.90 | high |
| cg25206536 | MIR572 | HAVCR2 | 0.65967075 | 6.15 | high |
| cg25206536 | MIR572 | TGFB1 | 0.65401204 | 8.27 | high |
| cg25206536 | MIR572 | IL10 | 0.71059003 | 3.16 | high |
| cg20502977 | COL6A3 | IL10 | 0.66111585 | 5.70 | high |
| cg18397405 | GPC6 | CTLA4 | 0.62938967 | 2.79 | high |
| cg18397405 | GPC6 | HAVCR2 | 0.64510535 | 1.30 | high |
| cg18397405 | GPC6 | CCR4 | 0.65623064 | 7.37 | high |
| cg18397405 | GPC6 | TGFB1 | 0.75578234 | 1.26 | high |
| cg18397405 | GPC6 | IL10 | 0.76613612 | 5.48 | high |
| cg16121744 | COL18A1 | HAVCR2 | 0.76562289 | 5.72 | high |
| cg16121744 | COL18A1 | TGFB1 | 0.72925722 | 9.04 | high |
| cg16121744 | COL18A1 | IL10 | 0.77214535 | 3.31 | high |
| cg15254671 | MYO1F | HAVCR2 | 0.73827055 | 4.76 | high |
| cg15254671 | MYO1F | IL10 | 0.70599224 | 4.24 | high |
| cg22595235 | SUMF1, LRRN1 | CTLA4 | 0.61078204 | 6.55 | high |
| cg09777237 | ELN | HAVCR2 | 0.67250205 | 3.08 | high |
| cg09777237 | ELN | TGFB1 | 0.61588628 | 5.21 | high |
| cg09777237 | ELN | IL10 | 0.68055003 | 1.96 | high |
| cg21475610 | CCNG2 | TGFB1 | 0.68792815 | 1.28 | high |
| cg21475610 | CCNG2 | IL10 | 0.6470373 | 1.18 | high |
| cg11076970 | HLA-DOA | CCL22 | 0.68567625 | 1.46 | high |
| cg17611512 | COL18A1, COL18A1-AS1 | HAVCR2 | 0.67446387 | 2.76 | high |
| cg17611512 | COL18A1, COL18A1-AS1 | TGFB1 | 0.75562488 | 1.28 | high |
| cg17611512 | COL18A1, COL18A1-AS1 | IL10 | 0.76622066 | 5.44 | high |
| cg22987448 | MYO1F | HAVCR2 | 0.70835256 | 3.65 | high |
| cg22987448 | MYO1F | TGFB1 | 0.63248797 | 2.41 | high |
| cg22987448 | MYO1F | IL10 | 0.70835142 | 3.65 | high |
| cg21398469 | CCNG2 | TGFB1 | 0.61126793 | 6.41 | high |
| cg05955301 | PRELP | HAVCR2 | 0.64915158 | 1.06 | high |
| cg05955301 | PRELP | TGFB1 | 0.658497 | 6.55 | high |
| cg05955301 | PRELP | IL10 | 0.69042987 | 1.10 | high |
| cg00742851 | SUMF1, LRRN1 | HAVCR2 | 0.64108721 | 1.59 | high |
| cg00742851 | SUMF1, LRRN1 | CCR4 | 0.65102622 | 9.64 | high |
| cg00742851 | SUMF1, LRRN1 | TGFB1 | 0.73293612 | 6.98 | high |
| cg00742851 | SUMF1, LRRN1 | IL10 | 0.75167928 | 1.74 | high |
Table A8.
Correlation among 338 CpG probes with gene expression of paired sample that present a high correlation value and significant p value.
| CpG | Gene | rho | p Value | CpGgene | Magnitude |
|---|---|---|---|---|---|
| cg02957057 | DEFB126 | 0.99984824 | NID1 | high | |
| cg20640433 | LRRIQ4 | −0.8429582 | LAMA2 | high | |
| cg02957057 | ZDHHC8P1 | −0.8399142 | NID1 | high | |
| cg23986671 | KRTAP6-3 | 0.83840023 | ADAMTS5 | high | |
| cg20640433 | TXK | −0.8184023 | LAMA2 | high | |
| cg20640433 | NLRP14 | −0.8100527 | LAMA2 | high | |
| cg20640433 | DEFB126 | 0.80488085 | LAMA2 | high | |
| cg16713274 | OR56A5 | 0.79052422 | COL18A1 | high | |
| cg02957057 | RFESD | −0.7883283 | NID1 | high | |
| cg02957057 | MMACHC | −0.7871875 | NID1 | high | |
| cg20640433 | GRP | −0.7852917 | LAMA2 | high | |
| cg02957057 | ISPD | −0.7804697 | NID1 | high | |
| cg02957057 | OSBPL9 | −0.7789042 | NID1 | high | |
| cg20640433 | FBXO17 | −0.7746792 | LAMA2 | high | |
| cg16121744 | IL10 | 0.77214535 | COL18A1 | high | |
| cg18397405 | CCR5 | 0.76767048 | GPC6 | high | |
| cg18397405 | CD96 | 0.7668386 | GPC6 | high | |
| cg17611512 | IL10 | 0.76622066 | COL18A1 | high | |
| cg18397405 | IL10 | 0.76613612 | GPC6 | high | |
| cg16121744 | HAVCR2 | 0.76562289 | COL18A1 | high | |
| cg02957057 | PCGEM1 | 0.76170335 | NID1 | high | |
| cg02957057 | ANKRD7 | −0.7606609 | NID1 | high | |
| cg13353679 | IL10 | 0.75708106 | AFF3, AC092667.2 | high | |
| cg04153551 | IL10 | 0.75654189 | FBLN5 | high | |
| cg18397405 | TGFB1 | 0.75578234 | GPC6 | high | |
| cg17611512 | TGFB1 | 0.75562488 | COL18A1, COL18A1-AS1 | high | |
| cg18397405 | ITGB2 | 0.75480536 | GPC6 | high | |
| cg20640433 | FAHD2B | −0.7544591 | LAMA2 | high | |
| cg00742851 | IL10 | 0.75167928 | SUMF1, LRRN1 | high | |
| cg23986671 | TAF1L | −0.7511669 | ADAMTS5 | high | |
| cg08064683 | TGFB1 | 0.7482646 | FAT1 | high | |
| cg20640433 | IL22RA1 | −0.7447273 | LAMA2 | high | |
| cg18411043 | GIMAP5 | 0.74431857 | LAPTM5 | high | |
| cg02957057 | FRMPD2 | −0.7389076 | NID1 | high | |
| cg02957057 | FAHD2B | −0.7387551 | NID1 | high | |
| cg15254671 | HAVCR2 | 0.73827055 | MYO1F | high | |
| cg18397405 | CD163 | 0.73748155 | GPC6 | high | |
| cg16713274 | C6orf132 | −0.7366047 | COL18A1 | high | |
| cg20640433 | ZDHHC8P1 | −0.7353041 | LAMA2 | high | |
| cg02957057 | MAP1LC3A | −0.7341328 | NID1 | high | |
| cg00742851 | TGFB1 | 0.73293612 | SUMF1, LRRN1 | high | |
| cg12613839 | IL10 | 0.7323874 | ADAMTS2 | high | |
| cg18411043 | WDR76 | −0.7298891 | LAPTM5 | high | |
| cg16121744 | TGFB1 | 0.72925722 | COL18A1 | high | |
| cg18411043 | SALL3 | −0.7280974 | LAPTM5 | high | |
| cg20640433 | GUCY2D | −0.7279513 | LAMA2 | high | |
| cg20640433 | ALDH7A1 | −0.7273038 | LAMA2 | high | |
| cg13353679 | TGFB1 | 0.72533605 | AFF3, AC092667.2 | high | |
| cg18397405 | CD74 | 0.72520583 | GPC6 | high | |
| cg14291900 | SFMBT2 | 0.72226357 | SLC7A7 | high | |
| cg22704788 | IL10 | 0.71501411 | PRELP | high | |
| cg02957057 | DPEP3 | −0.7149296 | NID1 | high | |
| cg20640433 | C17orf82 | −0.7134266 | LAMA2 | high | |
| cg18411043 | KCNK6 | 0.71200028 | LAPTM5 | high | |
| cg02957057 | N6AMT2 | −0.7119956 | NID1 | high | |
| cg02957057 | SLC25A20 | −0.7113793 | NID1 | high | |
| cg18397405 | CD14 | 0.71124306 | GPC6 | high | |
| cg25206536 | IL10 | 0.71059003 | MIR572 | high | |
| cg02957057 | ITPRIPL1 | −0.7102111 | NID1 | high | |
| cg17599241 | IL10 | 0.71015235 | VCAN-AS1, VCAN | high | |
| cg00799121 | IL10 | 0.71008968 | ADAMTS2 | high | |
| cg20640433 | SVOPL | −0.7100842 | LAMA2 | high | |
| cg12613839 | TGFB1 | 0.70966683 | ADAMTS2 | high | |
| cg22987448 | HAVCR2 | 0.70835256 | MYO1F | high | |
| cg22987448 | IL10 | 0.70835142 | MYO1F | high | |
| cg18397405 | CD68 | 0.70783649 | GPC6 | high | |
| cg18411043 | TGFBR2 | 0.70621641 | LAPTM5 | high | |
| cg15254671 | IL10 | 0.70599224 | MYO1F | high | |
| cg12613839 | HAVCR2 | 0.70546862 | ADAMTS2 | high | |
| cg04499514 | PDIA6 | −0.7048045 | C3AR1 | high | |
| cg20640433 | C9orf64 | −0.7042363 | LAMA2 | high | |
| cg02957057 | SLC35F3 | −0.7035707 | NID1 | high | |
| cg02957057 | POTEA | 0.70296616 | NID1 | high | |
| cg18411043 | IGFBP6 | 0.70161792 | LAPTM5 | high | |
| cg23986671 | TWIST2 | −0.7011775 | ADAMTS5 | high | |
| cg02957057 | OR10G7 | 0.70091288 | NID1 | high | |
| cg14291900 | FGD3 | 0.70047149 | SLC7A7 | high | |
| cg18397405 | GPR65 | 0.70013203 | GPC6 | high | |
| cg23986671 | MYOZ2 | −0.6993028 | ADAMTS5 | high | |
| cg23986671 | PDE6C | −0.6980551 | ADAMTS5 | high | |
| cg00799121 | TGFB1 | 0.69708683 | ADAMTS2 | high | |
| cg20640433 | AREG | −0.6964744 | LAMA2 | high | |
| cg20640433 | NMNAT3 | −0.6951494 | LAMA2 | high | |
| cg20640433 | XKR8 | −0.6950749 | LAMA2 | high | |
| cg20640433 | SLC25A44 | 0.69436599 | LAMA2 | high | |
| cg02957057 | ANKK1 | −0.6934215 | NID1 | high | |
| cg18397405 | GRN | 0.69317267 | GPC6 | high | |
| cg18411043 | TNFRSF10D | 0.6931024 | LAPTM5 | high | |
| cg18411043 | KIF22 | −0.6926606 | LAPTM5 | high | |
| cg20640433 | HDHD3 | −0.6922066 | LAMA2 | high | |
| cg27329371 | IL10 | 0.69220582 | ALDH3A1 | high | |
| cg18411043 | C19orf57 | −0.6912927 | LAPTM5 | high | |
| cg18411043 | SERPINB9 | 0.69077738 | LAPTM5 | high | |
| cg05955301 | IL10 | 0.69042987 | PRELP | high | |
| cg18411043 | CACNA2D4 | 0.6878496 | LAPTM5 | high | |
| cg21475610 | TGFB1 | 0.68792815 | CCNG2 | high | |
| cg20640433 | SSH3 | −0.6876975 | LAMA2 | high | |
| cg07947930 | TGFB1 | 0.68739449 | PRELP | high | |
| cg18411043 | CLDN23 | 0.68678752 | LAPTM5 | high | |
| cg02957057 | ZNF683 | −0.6867046 | NID1 | high | |
| cg02189760 | IL10 | 0.68557785 | CTC-301O7.4, CD37 | high | |
| cg11076970 | CCL22 | 0.68567625 | HLA-DOA | high | |
| cg13765206 | AMN | −0.6852093 | EMILIN2 | high | |
| cg02957057 | ISG20L2 | 0.68458461 | NID1 | high | |
| cg20640433 | MAP1LC3A | −0.683593 | LAMA2 | high | |
| cg18397405 | CCL5 | 0.68360635 | GPC6 | high | |
| cg00799121 | HAVCR2 | 0.6833261 | ADAMTS2 | high | |
| cg18411043 | MKS1 | −0.6816571 | LAPTM5 | high | |
| cg18411043 | IL4R | 0.68111791 | LAPTM5 | high | |
| cg09777237 | IL10 | 0.68055003 | ELN | high | |
| cg18411043 | GIMAP6 | 0.68005964 | LAPTM5 | high | |
| cg02957057 | STK33 | −0.6799005 | NID1 | high | |
| cg02957057 | PYDC2 | 0.67988313 | NID1 | high | |
| cg20640433 | MYD88 | −0.6798431 | LAMA2 | high | |
| cg14291900 | PIK3IP1 | 0.6795888 | 2.07 | SLC7A7 | high |
| cg18411043 | NUSAP1 | −0.6794355 | 2.08 | LAPTM5 | high |
| cg23986671 | RFPL3S | −0.6794146 | 2.09 | ADAMTS5 | high |
| cg20640433 | HEBP1 | −0.6789805 | 2.14 | LAMA2 | high |
| cg04499514 | RUNX1 | −0.6782038 | 2.23 | C3AR1 | high |
| cg18397405 | GZMA | 0.67785253 | 2.28 | GPC6 | high |
| cg02957057 | FAM19A1 | −0.6772002 | 2.37 | NID1 | high |
| cg02957057 | SPRR1A | 0.67668058 | 2.44 | NID1 | high |
| cg20640433 | MSN | −0.6761721 | 2.51 | LAMA2 | high |
| cg11827097 | PTK6 | 0.67530072 | 2.63 | SP100 | high |
| cg13353679 | HAVCR2 | 0.67520241 | 2.65 | AFF3, AC092667.2 | high |
| cg20640433 | SH3RF2 | −0.6749581 | 2.68 | LAMA2 | high |
| cg17611512 | HAVCR2 | 0.67446387 | 2.76 | COL18A1, COL18A1-AS1 | high |
| cg20640433 | PACSIN3 | −0.6738627 | 2.85 | LAMA2 | high |
| cg02957057 | CMBL | −0.673238 | 2.95 | NID1 | high |
| cg18411043 | MCTP2 | 0.67300302 | 2.99 | LAPTM5 | high |
| cg07436701 | CD74 | −0.6729908 | 2.99 | MMRN2, SNCG | high |
| cg14082886 | PPP1R15A | −0.6727144 | 3.04 | CD44 | high |
| cg18411043 | NCAPD3 | −0.6726225 | 3.06 | LAPTM5 | high |
| cg09777237 | HAVCR2 | 0.67250205 | 3.08 | ELN | high |
| cg02957057 | TYSND1 | −0.6716086 | 3.23 | NID1 | high |
| cg04499514 | TSPO | −0.6709273 | 3.35 | C3AR1 | high |
| cg18411043 | TMEM87B | 0.67084118 | 3.37 | LAPTM5 | high |
| cg23986671 | ZBTB32 | −0.6707326 | 3.39 | ADAMTS5 | high |
| cg14291900 | ZNF71 | −0.669991 | 3.53 | SLC7A7 | high |
| cg22704788 | HAVCR2 | 0.66998995 | 3.53 | PRELP | high |
| cg18411043 | B3GNT2 | 0.66993926 | 3.54 | LAPTM5 | high |
| cg07947930 | IL10 | 0.6692665 | 3.68 | PRELP | high |
| cg18411043 | MAPK13 | 0.66886709 | 3.76 | LAPTM5 | high |
| cg20640433 | SHROOM1 | −0.6687824 | 3.77 | LAMA2 | high |
| cg14291900 | ZNF134 | −0.6665228 | 4.27 | SLC7A7 | high |
| cg27329371 | TGFB1 | 0.666298 | 4.32 | ALDH3A1 | high |
| cg07436701 | CCR5 | −0.6662434 | 4.33 | MMRN2, SNCG | high |
| cg18411043 | CYP1B1 | 0.66507974 | 4.61 | LAPTM5 | high |
| cg18411043 | EMB | 0.66449718 | 4.76 | LAPTM5 | high |
| cg04153551 | HAVCR2 | 0.6642975 | 4.81 | FBLN5 | high |
| cg07947930 | HAVCR2 | 0.66424332 | 4.82 | PRELP | high |
| cg04499514 | MAPT | 0.66400429 | 4.89 | C3AR1 | high |
| cg13765206 | ITCH | 0.66292628 | 5.18 | EMILIN2 | high |
| cg20640433 | RFESD | −0.6627852 | 5.22 | LAMA2 | high |
| cg14082886 | CLVS2 | 0.66221087 | 5.38 | CD44 | high |
| cg18411043 | CHEK1 | −0.6614723 | LAPTM5 | high | |
| cg11702456 | TAGLN2 | −0.6614648 | SP100 | high | |
| cg18397405 | CD244 | 0.66144045 | GPC6 | high | |
| cg20502977 | IL10 | 0.66111585 | COL6A3 | high | |
| cg18411043 | PAPSS2 | 0.66103523 | LAPTM5 | high | |
| cg00295382 | MKRN3 | −0.6607885 | MYCL | high | |
| cg08064683 | IL10 | 0.66059345 | FAT1 | high | |
| cg17599241 | TGFB1 | 0.66056385 | VCAN-AS1, VCAN | high | |
| cg23986671 | GCOM1 | −0.660442 | ADAMTS5 | high | |
| cg18411043 | LYVE1 | 0.6597996 | LAPTM5 | high | |
| cg25206536 | HAVCR2 | 0.65967075 | MIR572 | high | |
| cg14082886 | RGS9 | 0.65942308 | CD44 | high | |
| cg14082886 | NEK6 | −0.6591273 | CD44 | high | |
| cg18411043 | NUMB | 0.65897549 | LAPTM5 | high | |
| cg20640433 | SLC43A3 | −0.6588459 | LAMA2 | high | |
| cg23986671 | VTCN1 | −0.6588003 | ADAMTS5 | high | |
| cg20640433 | RFPL2 | −0.6584724 | LAMA2 | high | |
| cg05955301 | TGFB1 | 0.658497 | PRELP | high | |
| cg22568423 | IL10 | 0.65815888 | MYO1F | high | |
| cg18397405 | CCR4 | 0.65623064 | GPC6 | high | |
| cg18397405 | CCR4 | 0.65623064 | GPC6 | high | |
| cg14291900 | YPEL2 | 0.65585997 | SLC7A7 | high | |
| cg20640433 | ZDHHC1 | −0.6557248 | LAMA2 | high | |
| cg18411043 | MAP3K8 | 0.6551703 | LAPTM5 | high | |
| cg02957057 | HSD17B7 | −0.6541632 | NID1 | high | |
| cg25206536 | TGFB1 | 0.65401204 | MIR572 | high | |
| cg18411043 | GAB1 | −0.6539384 | LAPTM5 | high | |
| cg18411043 | OIP5 | −0.6532665 | LAPTM5 | high | |
| cg04499514 | LGALS1 | −0.6531144 | C3AR1 | high | |
| cg23986671 | HYALP1 | 0.65295592 | ADAMTS5 | high | |
| cg02957057 | SCAMP3 | 0.65296148 | NID1 | high | |
| cg20640433 | DYNLT3 | −0.6527851 | LAMA2 | high | |
| cg04499514 | CD63 | −0.6526897 | C3AR1 | high | |
| cg04499514 | CD63 | −0.6526897 | C3AR1 | high | |
| cg18411043 | HIST1H4A | −0.652456 | LAPTM5 | high | |
| cg18397405 | IGF1 | 0.65236854 | GPC6 | high | |
| cg14291900 | ZNF787 | −0.6523244 | SLC7A7 | high | |
| cg20640433 | SH2D4A | −0.6513244 | LAMA2 | high | |
| cg23986671 | MMP1 | −0.6510743 | ADAMTS5 | high | |
| cg07436701 | ITGB2 | −0.6510564 | MMRN2, SNCG | high | |
| cg00742851 | CCR4 | 0.65102622 | SUMF1, LRRN1 | high | |
| cg04499514 | RPS6KA5 | 0.65061879 | C3AR1 | high | |
| cg02189760 | TGFB1 | 0.65060293 | CTC-301O7.4, CD37 | high | |
| cg18411043 | CD59 | 0.65051987 | LAPTM5 | high | |
| cg18411043 | ST3GAL1 | 0.64986187 | LAPTM5 | high | |
| cg18411043 | ZNF620 | −0.6492829 | LAPTM5 | high | |
| cg04499514 | CRELD2 | −0.6492545 | C3AR1 | high | |
| cg05955301 | HAVCR2 | 0.64915158 | PRELP | high | |
| cg20640433 | ACSF2 | −0.6487996 | LAMA2 | high | |
| cg02957057 | PARVA | −0.6487478 | NID1 | high | |
| cg22704788 | TGFB1 | 0.64874971 | PRELP | high | |
| cg16713274 | BCL2L10 | −0.6485899 | COL18A1 | high | |
| cg20640433 | SLC35F3 | −0.6482366 | LAMA2 | high | |
| cg14291900 | KLHL32 | 0.6481981 | SLC7A7 | high | |
| cg11702456 | RIPK1 | −0.6478933 | SP100 | high | |
| cg11702456 | PTK6 | 0.64795892 | SP100 | high | |
| cg14291900 | ZNF473 | −0.6477937 | SLC7A7 | high | |
| cg13765206 | CRNKL1 | 0.64734872 | EMILIN2 | high | |
| cg14291900 | AKAP8 | −0.6473027 | SLC7A7 | high | |
| cg18411043 | PSTPIP2 | 0.64723289 | LAPTM5 | high | |
| cg21475610 | IL10 | 0.6470373 | CCNG2 | high | |
| cg11702456 | RAB34 | −0.6468892 | SP100 | high | |
| cg02957057 | XKR8 | −0.6468834 | NID1 | high | |
| cg18411043 | LTBP2 | 0.64651112 | LAPTM5 | high | |
| cg18411043 | WHSC1 | −0.6464405 | LAPTM5 | high | |
| cg04499514 | SMAGP | −0.6459406 | C3AR1 | high | |
| cg11827097 | RIPK1 | −0.6456769 | SP100 | high | |
| cg18411043 | B4GALT1 | 0.64554786 | LAPTM5 | high | |
| cg02957057 | UCHL1 | −0.6451568 | NID1 | high | |
| cg18397405 | HAVCR2 | 0.64510535 | GPC6 | high | |
| cg11702456 | EMP3 | −0.6447434 | SP100 | high | |
| cg18411043 | LILRB2 | 0.64470941 | LAPTM5 | high | |
| cg04153551 | TGFB1 | 0.64462765 | FBLN5 | high | |
| cg18411043 | PLK4 | −0.6445754 | LAPTM5 | high | |
| cg18411043 | TNFRSF10A | 0.64435812 | LAPTM5 | high | |
| cg13765206 | HPS1 | −0.6438577 | EMILIN2 | high | |
| cg02957057 | PPP1R3C | −0.6439269 | NID1 | high | |
| cg13765206 | KLHDC7B | −0.643828 | EMILIN2 | high | |
| cg18411043 | GPSM2 | −0.6430751 | LAPTM5 | high | |
| cg18411043 | POLA2 | −0.6428379 | LAPTM5 | high | |
| cg02189760 | HAVCR2 | 0.64276143 | CTC-301O7.4, CD37 | high | |
| cg18411043 | MCM2 | −0.6424966 | LAPTM5 | high | |
| cg04499514 | HSP90B1 | −0.6419106 | C3AR1 | high | |
| cg14291900 | HPN | 0.64190596 | SLC7A7 | high | |
| cg04499514 | EMILIN2 | −0.6416321 | C3AR1 | high | |
| cg04499514 | EMILIN2 | −0.6416321 | C3AR1 | high | |
| cg14082886 | HSPA5 | −0.6412986 | CD44 | high | |
| cg18411043 | ASGR2 | 0.6412585 | LAPTM5 | high | |
| cg18411043 | PRKCD | 0.64119535 | LAPTM5 | high | |
| cg00742851 | HAVCR2 | 0.64108721 | SUMF1, LRRN1 | high | |
| cg18411043 | FAM181B | −0.6409853 | LAPTM5 | high | |
| cg00295382 | ZNF292 | −0.6404944 | MYCL | high | |
| cg11702456 | TSEN34 | −0.6397343 | SP100 | high | |
| cg04153551 | CCR4 | 0.63922606 | FBLN5 | high | |
| cg22568423 | HAVCR2 | 0.6391164 | MYO1F | high | |
| cg04499514 | SPRR2A | 0.63900466 | C3AR1 | high | |
| cg20640433 | CRHR2 | −0.6389645 | LAMA2 | high | |
| cg14291900 | ERMN | 0.63877586 | SLC7A7 | high | |
| cg16713274 | VWDE | −0.6386629 | COL18A1 | high | |
| cg20640433 | SCAMP3 | 0.63863905 | LAMA2 | high | |
| cg02957057 | SMG5 | 0.63832232 | NID1 | high | |
| cg18411043 | CDCA5 | −0.637901 | LAPTM5 | high | |
| cg18411043 | SMC2 | −0.6376489 | LAPTM5 | high | |
| cg23986671 | GPS1 | 0.63753383 | ADAMTS5 | high | |
| cg20640433 | OR10G7 | 0.63739025 | LAMA2 | high | |
| cg20640433 | VNN3 | −0.6368153 | LAMA2 | high | |
| cg18411043 | RNF144B | 0.63671956 | LAPTM5 | high | |
| cg02957057 | NMNAT3 | −0.6360154 | NID1 | high | |
| cg18411043 | FANCC | −0.6359325 | LAPTM5 | high | |
| cg14291900 | SLC46A3 | 0.63593824 | SLC7A7 | high | |
| cg04499514 | TSEN34 | −0.6356583 | C3AR1 | high | |
| cg14082886 | PCYT1A | −0.6351289 | 2.12 | CD44 | high |
| cg18411043 | ARPC1B | 0.63501952 | 2.13 | LAPTM5 | high |
| cg18411043 | GPR132 | 0.63497935 | 2.14 | LAPTM5 | high |
| cg02957057 | ELOVL3 | −0.6344793 | 2.19 | NID1 | high |
| cg13765206 | C2CD4D | −0.6344131 | 2.20 | EMILIN2 | high |
| cg14291900 | SEMA4A | 0.63440849 | 2.20 | SLC7A7 | high |
| cg18411043 | KIF15 | −0.6340036 | 2.24 | LAPTM5 | high |
| cg18411043 | NCF4 | 0.63389721 | 2.25 | LAPTM5 | high |
| cg23986671 | DCST1 | −0.6335087 | 2.30 | ADAMTS5 | high |
| cg00777079 | N4BP2 | −0.6335114 | 2.30 | SERPINF1 | high |
| cg23986671 | CLEC4F | −0.6330094 | 2.35 | ADAMTS5 | high |
| cg04499514 | DUSP4 | −0.632834 | 2.37 | C3AR1 | high |
| cg14291900 | TCF3 | −0.6328327 | 2.37 | SLC7A7 | high |
| cg14291900 | ZNF416 | −0.6328258 | 2.37 | SLC7A7 | high |
| cg18411043 | CD1D | 0.63278019 | 2.38 | LAPTM5 | high |
| cg22987448 | TGFB1 | 0.63248797 | 2.41 | MYO1F | high |
| cg20640433 | GLIS3 | −0.6319628 | 2.47 | LAMA2 | high |
| cg04499514 | SEC24D | −0.631827 | 2.49 | C3AR1 | high |
| cg02957057 | NLRX1 | −0.6317766 | 2.49 | NID1 | high |
| cg27329371 | PDCD1LG2 | 0.6318309 | 2.49 | ALDH3A1 | high |
| cg18411043 | PSMC3IP | −0.631709 | 2.50 | LAPTM5 | high |
| cg23986671 | GOLGA4 | −0.6315958 | 2.52 | ADAMTS5 | high |
| cg14291900 | U2AF2 | −0.6314722 | 2.53 | SLC7A7 | high |
| cg18411043 | CEP72 | −0.6310813 | 2.58 | LAPTM5 | high |
| cg18411043 | NCAPH | −0.6308592 | 2.61 | LAPTM5 | high |
| cg18411043 | TRIM38 | 0.63055942 | 2.64 | LAPTM5 | high |
| cg27329371 | HAVCR2 | 0.63023007 | 2.68 | ALDH3A1 | high |
| cg04499514 | S100A11 | −0.6301523 | 2.69 | C3AR1 | high |
| cg18411043 | GIMAP8 | 0.63012277 | 2.70 | LAPTM5 | high |
| cg18411043 | LMNB1 | −0.6298277 | 2.74 | LAPTM5 | high |
| cg04499514 | SEMA3D | 0.62956973 | 2.77 | C3AR1 | high |
| cg18397405 | CTLA4 | 0.62938967 | 2.79 | GPC6 | high |
| cg14291900 | DOCK5 | 0.62929209 | 2.81 | SLC7A7 | high |
| cg14291900 | ACSM5 | 0.62920997 | 2.82 | SLC7A7 | high |
| cg04499514 | TWF2 | −0.6290565 | 2.84 | C3AR1 | high |
| cg18411043 | MRC1 | 0.62895863 | 2.85 | LAPTM5 | high |
| cg18411043 | TNFRSF1B | 0.62892778 | 2.86 | LAPTM5 | high |
| cg18411043 | MEN1 | −0.6288512 | 2.87 | LAPTM5 | high |
| cg18411043 | RAB11FIP1 | 0.62865829 | 2.89 | LAPTM5 | high |
| cg18411043 | F13A1 | 0.62865476 | 2.89 | LAPTM5 | high |
| cg18411043 | TESC | 0.62858843 | 2.90 | LAPTM5 | high |
| cg14291900 | LGALS9C | 0.62857927 | 2.90 | SLC7A7 | high |
| cg18411043 | GIPC2 | 0.62849757 | 2.91 | LAPTM5 | high |
| cg02957057 | LRRIQ4 | −0.6285485 | 2.91 | NID1 | high |
| cg14291900 | NKAIN2 | 0.6284529 | 2.92 | SLC7A7 | high |
| cg01930947 | TACR1 | 0.62834753 | 2.93 | C1orf111 | high |
| cg14082886 | COL4A3 | 0.62833753 | 2.94 | CD44 | high |
| cg04499514 | RPS6KA3 | −0.6282354 | 2.95 | C3AR1 | high |
| cg14291900 | LHPP | 0.62822146 | 2.95 | SLC7A7 | high |
| cg11702456 | GALNS | −0.6280356 | 2.98 | SP100 | high |
| cg18411043 | AMICA1 | 0.6278709 | 3.00 | LAPTM5 | high |
| cg20640433 | ISG20L2 | 0.62790094 | 3.00 | LAMA2 | high |
| cg13765206 | PLEKHG6 | −0.6278379 | 3.01 | EMILIN2 | high |
| cg04499514 | TTC38 | −0.6274235 | 3.06 | C3AR1 | high |
| cg20640433 | RAB36 | −0.627427 | 3.06 | LAMA2 | high |
| cg20640433 | CST3 | −0.6274226 | 3.06 | LAMA2 | high |
| cg18411043 | MLKL | 0.62716867 | 3.10 | LAPTM5 | high |
| cg02957057 | C9orf64 | −0.627194 | 3.10 | NID1 | high |
| cg11702456 | S100A13 | −0.626989 | 3.13 | SP100 | high |
| cg01930947 | TMEFF2 | 0.62681323 | 3.15 | C1orf111 | high |
| cg18411043 | MAN1A1 | 0.62676193 | 3.16 | LAPTM5 | high |
| cg02957057 | FBXO17 | −0.6267928 | 3.16 | NID1 | high |
| cg02957057 | SH3BP2 | −0.6266051 | 3.18 | NID1 | high |
| cg05091653 | SERPINF2 | −0.6263606 | 3.22 | SP100 | high |
| cg18411043 | TRPM2 | 0.62624593 | 3.24 | LAPTM5 | high |
| cg18411043 | CD33 | 0.62613919 | 3.25 | LAPTM5 | high |
| cg18411043 | CD46 | 0.62591061 | 3.29 | LAPTM5 | high |
| cg14291900 | NR2C2AP | −0.6258545 | 3.30 | SLC7A7 | high |
| cg20640433 | PALM2 | −0.6257442 | 3.32 | LAMA2 | high |
| cg18411043 | P2RY6 | 0.62560017 | 3.34 | LAPTM5 | high |
| cg24769499 | FGF6 | 0.62560159 | 3.34 | TMEM37 | high |
| cg00295382 | UBE4A | −0.6253707 | 3.37 | MYCL | high |
| cg04499514 | FBLIM1 | −0.6251549 | 3.41 | C3AR1 | high |
| cg00777079 | RFWD3 | −0.6250366 | 3.43 | SERPINF1 | high |
| cg18411043 | CTSB | 0.62483558 | 3.46 | LAPTM5 | high |
| cg16713274 | NXPH2 | −0.624851 | 3.46 | COL18A1 | high |
| cg18411043 | INCENP | −0.6247948 | 3.47 | LAPTM5 | high |
| cg14291900 | CDK6 | −0.624404 | 3.53 | SLC7A7 | high |
| cg02957057 | DEFB125 | 0.62421976 | 3.56 | NID1 | high |
| cg18411043 | CSF1R | 0.62400537 | 3.60 | LAPTM5 | high |
| cg18411043 | TIGD3 | −0.6238521 | 3.62 | LAPTM5 | high |
| cg23986671 | ATP6V0D2 | −0.6237079 | 3.65 | ADAMTS5 | high |
| cg20640433 | RIT1 | 0.62363135 | 3.66 | LAMA2 | high |
| cg14082886 | SLCO1A2 | 0.62356807 | 3.67 | CD44 | high |
| cg18411043 | ALOX5 | 0.62349813 | 3.68 | LAPTM5 | high |
| cg18411043 | MSI1 | −0.6234219 | 3.69 | LAPTM5 | high |
| cg23986671 | DBH | −0.6231996 | 3.73 | ADAMTS5 | high |
| cg00295382 | NDUFB2 | 0.62314436 | 3.74 | MYCL | high |
| cg20640433 | EVC2 | −0.6228847 | 3.79 | LAMA2 | high |
| cg17599241 | HAVCR2 | 0.6225838 | 3.84 | VCAN-AS1, VCAN | high |
| cg18411043 | RUNX2 | 0.62236407 | 3.88 | LAPTM5 | high |
| cg13765206 | TCHH | −0.6221834 | 3.91 | EMILIN2 | high |
| cg07947930 | PDCD1LG2 | 0.62213951 | 3.92 | PRELP | high |
| cg18411043 | POLD3 | −0.6218415 | 3.97 | LAPTM5 | high |
| cg04499514 | MFSD5 | −0.621664 | 4.01 | C3AR1 | high |
| cg18411043 | MPP1 | 0.62163839 | 4.01 | LAPTM5 | high |
| cg18411043 | HRH2 | 0.62163838 | 4.01 | LAPTM5 | high |
| cg18411043 | TOP2A | −0.621607 | 4.02 | LAPTM5 | high |
| cg18411043 | IRAK3 | 0.62139951 | 4.06 | LAPTM5 | high |
| cg18397405 | GPC2 | −0.6210758 | 4.12 | GPC6 | high |
| cg18411043 | OAF | 0.62104659 | 4.12 | LAPTM5 | high |
| cg04499514 | SPRY4 | −0.6209967 | 4.13 | C3AR1 | high |
| cg18411043 | C1S | 0.6209673 | 4.14 | LAPTM5 | high |
| cg02957057 | ALDH7A1 | −0.6209514 | 4.14 | NID1 | high |
| cg14291900 | CNOT3 | −0.6209149 | 4.15 | SLC7A7 | high |
| cg02957057 | TXK | −0.6207198 | 4.18 | NID1 | high |
| cg07436701 | IGF1 | −0.6206649 | 4.19 | MMRN2, SNCG | high |
| cg18411043 | KIF18B | −0.6205685 | 4.21 | LAPTM5 | high |
| cg18411043 | ARHGAP30 | 0.6203351 | 4.26 | LAPTM5 | high |
| cg18411043 | AIF1 | 0.62025359 | 4.27 | LAPTM5 | high |
| cg00295382 | TGM2 | 0.62016995 | 4.29 | MYCL | high |
| cg14291900 | SLC26A9 | 0.62000399 | 4.32 | SLC7A7 | high |
| cg23986671 | TMEM52 | −0.6197762 | 4.37 | ADAMTS5 | high |
| cg18411043 | LHFPL2 | 0.61974103 | 4.38 | LAPTM5 | high |
| cg14291900 | SLC1A7 | 0.6196898 | 4.39 | SLC7A7 | high |
| cg04499514 | DUSP6 | −0.619504 | 4.42 | C3AR1 | high |
| cg11702456 | APOBEC3F | −0.6195267 | 4.42 | SP100 | high |
| cg22568423 | TGFB1 | 0.61936567 | 4.45 | MYO1F | high |
| cg14082886 | ADAM22 | 0.61922422 | 4.48 | CD44 | high |
| cg11827097 | TAGLN2 | −0.6185018 | 4.63 | SP100 | high |
| cg02957057 | CSRP1 | −0.6184356 | 4.64 | NID1 | high |
| cg21218883 | HSPBP1 | −0.618324 | 4.67 | PRKCE | high |
| cg00295382 | HGFAC | −0.618239 | 4.69 | MYCL | high |
| cg14291900 | GRWD1 | −0.6179975 | 4.74 | SLC7A7 | high |
| cg18411043 | CMKLR1 | 0.61781132 | 4.78 | LAPTM5 | high |
| cg18411043 | CYTH4 | 0.61755945 | 4.83 | LAPTM5 | high |
| cg02957057 | ACADS | −0.6173054 | 4.89 | NID1 | high |
| cg18411043 | FANCI | −0.6170511 | 4.95 | LAPTM5 | high |
| cg20640433 | LGALS8 | −0.616979 | 4.96 | LAMA2 | high |
| cg04499514 | CD276 | −0.6168248 | 5.00 | C3AR1 | high |
| cg18411043 | TNFSF10 | 0.61660766 | 5.05 | LAPTM5 | high |
| cg18411043 | VENTX | 0.61649043 | 5.07 | LAPTM5 | high |
| cg04499514 | CASC3 | 0.61596213 | 5.20 | C3AR1 | high |
| cg09777237 | TGFB1 | 0.61588628 | 5.21 | ELN | high |
| cg18411043 | SIGLEC10 | 0.61564503 | 5.27 | LAPTM5 | high |
| cg14291900 | FAM124A | 0.61534899 | 5.34 | SLC7A7 | high |
| cg11702456 | APOBEC3C | −0.6153217 | 5.35 | SP100 | high |
| cg20640433 | KHNYN | −0.6151494 | 5.39 | LAMA2 | high |
| cg14291900 | RAB40B | 0.61510576 | 5.40 | SLC7A7 | high |
| cg04499514 | TAGLN2 | −0.6149763 | 5.43 | C3AR1 | high |
| cg18411043 | RAC3 | −0.6149957 | 5.43 | LAPTM5 | high |
| cg14082886 | EMP1 | −0.6149246 | 5.44 | CD44 | high |
| cg14291900 | MRVI1 | 0.61493337 | 5.44 | SLC7A7 | high |
| cg14082886 | TAGLN2 | −0.614913 | 5.45 | CD44 | high |
| cg18411043 | FGD2 | 0.61480269 | 5.47 | LAPTM5 | high |
| cg18411043 | DSE | 0.6147844 | 5.48 | LAPTM5 | high |
| cg23986671 | UBP1 | −0.6147229 | 5.49 | ADAMTS5 | high |
| cg23986671 | XKR5 | −0.6145796 | 5.53 | ADAMTS5 | high |
| cg18411043 | POLD4 | 0.61428878 | 5.60 | LAPTM5 | high |
| cg18411043 | FMNL1 | 0.61431112 | 5.60 | LAPTM5 | high |
| cg04499514 | SPAG9 | 0.61418162 | 5.63 | C3AR1 | high |
| cg18411043 | EZH2 | −0.6141715 | 5.63 | LAPTM5 | high |
| cg14291900 | EFHD1 | 0.61415875 | 5.63 | SLC7A7 | high |
| cg18411043 | TPX2 | −0.6140728 | 5.66 | LAPTM5 | high |
| cg11702456 | EFEMP2 | −0.6138811 | 5.70 | SP100 | high |
| cg04499514 | APBA1 | 0.61375515 | 5.74 | C3AR1 | high |
| cg01930947 | DNM3 | 0.61362978 | 5.77 | C1orf111 | high |
| cg14082886 | DAAM2 | 0.61357396 | 5.78 | CD44 | high |
| cg04499514 | SDF2L1 | −0.6132695 | 5.86 | C3AR1 | high |
| cg02957057 | ACSF2 | −0.6130867 | 5.91 | NID1 | high |
| cg18411043 | TMEM97 | −0.6127385 | 6.00 | LAPTM5 | high |
| cg18411043 | CDC25A | −0.6126424 | 6.03 | LAPTM5 | high |
| cg18411043 | GIMAP7 | 0.61247867 | 6.07 | LAPTM5 | high |
| cg14291900 | ZNF45 | −0.6125104 | 6.07 | SLC7A7 | high |
| cg02957057 | SH2D4A | −0.6123811 | 6.10 | NID1 | high |
| cg04499514 | ATP1A4 | 0.61222663 | 6.14 | C3AR1 | high |
| cg18411043 | KIF2C | −0.6121091 | 6.17 | LAPTM5 | high |
| cg18411043 | SLC20A1 | 0.61194867 | 6.22 | LAPTM5 | high |
| cg20640433 | MMACHC | −0.6119354 | 6.22 | LAMA2 | high |
| cg18411043 | ECM1 | 0.61187776 | 6.24 | LAPTM5 | high |
| cg00295382 | C5orf51 | −0.6118449 | 6.25 | MYCL | high |
| cg18411043 | CMTM7 | 0.61176944 | 6.27 | LAPTM5 | high |
| cg04499514 | EHD4 | −0.611663 | 6.30 | C3AR1 | high |
| cg18411043 | CRISPLD2 | 0.61159373 | 6.32 | LAPTM5 | high |
| cg00295382 | ATF7IP | −0.6113318 | 6.39 | MYCL | high |
| cg07436701 | CD163 | −0.6113281 | 6.39 | MMRN2, SNCG | high |
| cg21398469 | TGFB1 | 0.61126793 | 6.41 | CCNG2 | high |
| cg07436701 | CD244 | −0.6112525 | 6.41 | MMRN2, SNCG | high |
| cg14291900 | ABCG1 | 0.61123795 | 6.42 | SLC7A7 | high |
| cg14291900 | ZNF761 | −0.6110213 | 6.48 | SLC7A7 | high |
| cg18411043 | HHEX | 0.61088519 | 6.52 | LAPTM5 | high |
| cg22595235 | CTLA4 | 0.61078204 | 6.55 | SUMF1, LRRN1 | high |
| cg24769499 | IL22 | 0.61065046 | 6.59 | TMEM37 | high |
| cg18411043 | MNDA | 0.61034153 | 6.68 | LAPTM5 | high |
| cg18411043 | FAH | 0.61025131 | 6.71 | LAPTM5 | high |
| cg11702456 | SP100 | −0.6102275 | 6.71 | SP100 | high |
| cg23986671 | DUOXA1 | −0.6102538 | 6.71 | ADAMTS5 | high |
| cg00295382 | PANK3 | −0.6101989 | 6.72 | MYCL | high |
| cg18411043 | CLEC10A | 0.61013964 | 6.74 | LAPTM5 | high |
| cg18411043 | TRAF3IP3 | 0.61007507 | 6.76 | LAPTM5 | high |
| cg13765206 | CAPN8 | −0.6097062 | 6.87 | EMILIN2 | high |
| cg14291900 | PAQR8 | 0.60959693 | 6.90 | SLC7A7 | high |
| cg02957057 | SDC4 | −0.609594 | 6.90 | NID1 | high |
| cg20640433 | ISPD | −0.6095796 | 6.91 | LAMA2 | high |
| cg08064683 | CCR4 | 0.60946563 | 6.94 | FAT1 | high |
| cg04499514 | AP2S1 | −0.6092851 | 7.00 | C3AR1 | high |
| cg04499514 | ITPRIP | −0.60917 | 7.03 | C3AR1 | high |
| cg04499514 | ADHFE1 | 0.60916978 | 7.03 | C3AR1 | high |
| cg00295382 | ARPC1B | 0.60912481 | 7.05 | MYCL | high |
| cg18411043 | ZNF90 | −0.6090191 | 7.08 | LAPTM5 | high |
| cg00295382 | CREBZF | −0.6089524 | 7.10 | MYCL | high |
| cg14291900 | DPEP2 | 0.60894355 | 7.10 | SLC7A7 | high |
| cg02957057 | CCDC163P | −0.608834 | 7.14 | NID1 | high |
| cg04499514 | AKAP1 | 0.60866034 | 7.19 | C3AR1 | high |
| cg20640433 | SLC2A10 | −0.6085969 | 7.21 | LAMA2 | high |
| cg14291900 | TPD52L1 | 0.60852744 | 7.24 | SLC7A7 | high |
| cg11827097 | PRMT2 | −0.6081478 | 7.36 | SP100 | high |
| cg23986671 | PRPH2 | −0.6081063 | 7.37 | ADAMTS5 | high |
| cg18397405 | ITGB1 | 0.60795654 | 7.42 | GPC6 | high |
| cg02957057 | RRP12 | 0.60790296 | 7.44 | NID1 | high |
| cg18411043 | ADAP2 | 0.60781942 | 7.46 | LAPTM5 | high |
| cg18411043 | CCR1 | 0.60783567 | 7.46 | LAPTM5 | high |
| cg18411043 | IL15RA | 0.60780418 | 7.47 | LAPTM5 | high |
| cg11702456 | CMTM3 | −0.6077214 | 7.50 | SP100 | high |
| cg04499514 | FBXW12 | 0.60765521 | 7.52 | C3AR1 | high |
| cg14291900 | ADRBK2 | 0.60758355 | 7.54 | SLC7A7 | high |
| cg18411043 | WDR34 | −0.6072402 | 7.66 | LAPTM5 | high |
| cg18411043 | LAIR1 | 0.60714771 | 7.69 | LAPTM5 | high |
| cg00295382 | ZBTB44 | −0.6070874 | 7.71 | MYCL | high |
| cg13765206 | NRAP | −0.6067666 | 7.82 | EMILIN2 | high |
| cg14291900 | SLCO1A2 | 0.60635008 | 7.96 | SLC7A7 | high |
| cg16713274 | OLFM3 | −0.6062793 | 7.99 | COL18A1 | high |
| cg00295382 | FAM166A | −0.6061765 | 8.02 | MYCL | high |
| cg02957057 | RAB36 | −0.6061395 | 8.03 | NID1 | high |
| cg14291900 | TMEM86A | 0.60607985 | 8.05 | SLC7A7 | high |
| cg14291900 | EVI2A | 0.60605156 | 8.06 | SLC7A7 | high |
| cg18411043 | CTSZ | 0.60597807 | 8.09 | LAPTM5 | high |
| cg13765206 | NCR3 | −0.6057964 | 8.16 | EMILIN2 | high |
| cg13765206 | KRTAP5-9 | −0.605737 | 8.18 | EMILIN2 | high |
| cg18411043 | HES5 | −0.6056663 | 8.20 | LAPTM5 | high |
| cg11702456 | ARSI | −0.6056573 | 8.20 | SP100 | high |
| cg18411043 | MFSD1 | 0.60562112 | LAPTM5 | high | |
| cg00295382 | ZG16 | −0.6055812 | MYCL | high | |
| cg20640433 | DPEP3 | −0.6054516 | LAMA2 | high | |
| cg18411043 | MAP2 | −0.6053901 | LAPTM5 | high | |
| cg18411043 | ADAMTS14 | 0.60538276 | LAPTM5 | high | |
| cg04499514 | KDELR1 | −0.605255 | C3AR1 | high | |
| cg04499514 | RALGPS1 | 0.60522199 | C3AR1 | high | |
| cg18411043 | BRIP1 | −0.6052277 | LAPTM5 | high | |
| cg14291900 | DLEU7 | 0.60520069 | SLC7A7 | high | |
| cg18411043 | RNF149 | 0.60510698 | LAPTM5 | high | |
| cg18411043 | LEPROT | 0.60482116 | LAPTM5 | high | |
| cg18411043 | GIMAP4 | 0.60467954 | LAPTM5 | high | |
| cg00295382 | RGS19 | 0.60458767 | MYCL | high | |
| cg18411043 | IL10RA | 0.60450736 | LAPTM5 | high | |
| cg18411043 | SLCO2B1 | 0.60445283 | LAPTM5 | high | |
| cg00295382 | TTC38 | 0.60437946 | MYCL | high | |
| cg14291900 | PTBP1 | −0.6043814 | SLC7A7 | high | |
| cg18411043 | IL16 | 0.60434727 | LAPTM5 | high | |
| cg04499514 | PPIB | −0.6040378 | C3AR1 | high | |
| cg18411043 | MAPKAPK2 | 0.60396534 | LAPTM5 | high | |
| cg04499514 | TGFBI | −0.6038334 | C3AR1 | high | |
| cg04499514 | IGFBP2 | −0.6037505 | C3AR1 | high | |
| cg11702456 | SLC2A4 | 0.60343179 | SP100 | high | |
| cg04499514 | IKBIP | −0.603409 | C3AR1 | high | |
| cg04499514 | ETV5 | −0.603302 | C3AR1 | high | |
| cg12613839 | PDCD1LG2 | 0.60313469 | ADAMTS2 | high | |
| cg04499514 | KIAA1324L | 0.60307321 | C3AR1 | high | |
| cg18411043 | FMN1 | 0.60306718 | LAPTM5 | high | |
| cg18411043 | SH3TC1 | 0.6029736 | LAPTM5 | high | |
| cg02957057 | LEKR1 | −0.6029529 | NID1 | high | |
| cg18411043 | GRB2 | 0.60266775 | LAPTM5 | high | |
| cg04499514 | PSD2 | 0.60262221 | C3AR1 | high | |
| cg11702456 | CASP8 | −0.6025684 | SP100 | high | |
| cg04499514 | IFNGR2 | −0.6025283 | C3AR1 | high | |
| cg14082886 | DCAF8 | 0.60237965 | CD44 | high | |
| cg02957057 | C10orf107 | −0.6023697 | NID1 | high | |
| cg04499514 | CKAP4 | -0.6023102 | C3AR1 | high | |
| cg02957057 | RTP2 | 0.60230352 | NID1 | high | |
| cg18411043 | RFC5 | −0.6022106 | LAPTM5 | high | |
| cg11827097 | TSEN34 | −0.6020259 | SP100 | high | |
| cg18411043 | YWHAZ | 0.60194076 | LAPTM5 | high | |
| cg02957057 | TMIE | −0.6019364 | NID1 | high | |
| cg02957057 | GLIS3 | −0.6017402 | NID1 | high | |
| cg00295382 | TRO | −0.60165 | MYCL | high | |
| cg20640433 | FAM19A1 | −0.6014471 | LAMA2 | high | |
| cg18411043 | LCORL | −0.6013862 | LAPTM5 | high | |
| cg20640433 | PAOX | −0.6011955 | LAMA2 | high | |
| cg14291900 | SAE1 | −0.6011065 | SLC7A7 | high | |
| cg18411043 | DENND1C | 0.60107737 | LAPTM5 | high | |
| cg00295382 | ZNF510 | −0.6009576 | 1.01 | MYCL | high |
| cg18411043 | MED24 | −0.6008814 | 1.01 | LAPTM5 | high |
| cg14082886 | ATP8A1 | 0.60058342 | 1.02 | CD44 | high |
| cg18411043 | RAD54L | −0.600678 | 1.02 | LAPTM5 | high |
| cg18411043 | SP4 | −0.6005993 | 1.02 | LAPTM5 | high |
| cg04499514 | TIMP1 | −0.6001084 | 1.04 | C3AR1 | high |
| cg14291900 | RASGEF1B | 0.60012159 | 1.04 | SLC7A7 | high |
| cg02957057 | ZDHHC1 | −0.6000678 | 1.04 | NID1 | high |
| cg02957057 | HRASLS5 | −0.6000243 | 1.05 | NID1 | high |
| cg07436701 | CD96 | −0.5993043 | 1.08 | MMRN2, SNCG | high |
| cg07436701 | CCR4 | −0.5972957 | 1.18 | MMRN2, SNCG | high |
| cg18397405 | ITGA4 | 0.59418147 | 1.34 | GPC6 | high |
| cg03677069 | CD74 | 0.59295197 | MMRN2, SNCG | high | |
| cg07436701 | GPR65 | −0.5925407 | MMRN2, SNCG | high | |
| cg18397405 | CDC34 | −0.590695 | GPC6 | high | |
| cg07436701 | FLT3 | −0.5827163 | MMRN2, SNCG | high | |
| cg07436701 | GPC2 | 0.5754121 | MMRN2, SNCG | high | |
| cg07436701 | E2F2 | 0.5728996 | MMRN2, SNCG | high | |
| cg07436701 | CD14 | −0.568245 | MMRN2, SNCG | high | |
| cg18397405 | EZH2 | −0.5582639 | GPC6 | high | |
| cg18397405 | CDKN1B | −0.5581881 | GPC6 | high | |
| cg07436701 | CDC34 | 0.55769852 | MMRN2, SNCG | high | |
| cg07436701 | CD68 | −0.5556475 | MMRN2, SNCG | high | |
| cg26350754 | EMILIN2 | −0.554466 | HLA-DPA1, HLA-DPB1 | high | |
| cg14082886 | MRC2 | −0.5539332 | CD44 | high | |
| cg18397405 | E2F2 | −0.5518593 | GPC6 | high | |
| cg18397405 | FLT3 | 0.54987539 | GPC6 | high | |
| cg10949632 | GPC6 | 0.54286496 | GPC6 | high | |
| cg03677069 | GPR65 | 0.54188941 | 0.00010045 | MMRN2, SNCG | high |
| cg14082886 | FGFR2 | 0.5340032 | 0.00013232 | CD44 | high |
| cg16713274 | GPC6 | −0.5317779 | 0.00014285 | COL18A1, LL21NC02-21A1.1 | high |
| cg03677069 | CD163 | 0.52832557 | 0.00016069 | MMRN2, SNCG | high |
| cg21012874 | CD74 | 0.52760147 | 0.00016467 | MMRN2, SNCG | high |
| cg09552892 | CD74 | 0.52237375 | 0.00019625 | MMRN2, SNCG | high |
| cg04499514 | EZH1 | 0.5201497 | 0.00021127 | C3AR1 | high |
| cg07436701 | EZH2 | 0.51876207 | 0.00022116 | MMRN2, SNCG | high |
| cg14082886 | CD63 | −0.5173898 | 0.00023136 | CD44 | high |
| cg04098585 | EMILIN2 | −0.5123153 | 0.00027286 | CD28 | high |
| cg07436701 | GZMA | −0.5121674 | 0.00027417 | MMRN2, SNCG | high |
| cg03677069 | ITGB2 | 0.5110316 | 0.00028438 | MMRN2, SNCG | high |
| cg07436701 | CCL5 | −0.5105968 | 0.00028837 | MMRN2, SNCG | high |
| cg03677069 | E2F2 | −0.5095174 | 0.00029852 | MMRN2, SNCG | high |
| cg03677069 | CD14 | 0.50607605 | 0.00033306 | MMRN2, SNCG | high |
| cg04499514 | FGFR1 | −0.5048488 | 0.00034622 | C3AR1 | high |
| cg07436701 | GRN | −0.5045134 | 0.0003499 | MMRN2, SNCG | high |
| cg18397405 | CD63 | 0.50108656 | 0.00038956 | GPC6 | high |
Author Contributions
Conceptualization: M.P.; methodology and formal analysis: M.P., E.F.; writing—original draft preparation, M.P., E.A., M.D.B., E.F., D.G., G.T.; writing—review and editing, M.P., L.B., F.D.C., M.M., M.S., M.A., T.I., G.T., E.A., M.D.B., M.A., M.S., G.T.; funding acquisition G.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the results here showed are based on data generated by the TCGA Research Network: https:/cancer.gov/tcga (accessed on 12 March 2020).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U., et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Cloughesy T.F., Mochizuki A.Y., Orpilla J.R., Hugo W., Lee A.H., Davidson T.B., Wang A.C., Ellingson B.M., Rytlewski J.A., Sanders C.M., et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat. Med. 2019;25:477–486. doi: 10.1038/s41591-018-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagley S.J., Desai A.S., Nasrallah M.P., O’Rourke D.M. Immunotherapy and Response Assessment in Malignant Glioma: Neuro-oncology Perspective. Top. Magnetic Resonance Imag. TMRI. 2020;29:95–102. doi: 10.1097/RMR.0000000000000233. [DOI] [PubMed] [Google Scholar]
- 4.Schalper K.A., Rodriguez-Ruiz M.E., Diez-Valle R., López-Janeiro A., Porciuncula A., Idoate M.A., Inogés S., de Andrea C., López-Diaz de Cerio A., Tejada S., et al. Neoadjuvant nivolumab modifies the tumor immune microenvironment in resectable glioblastoma. Nat. Med. 2019;25:470–476. doi: 10.1038/s41591-018-0339-5. [DOI] [PubMed] [Google Scholar]
- 5.Gieryng A., Pszczolkowska D., Walentynowicz K.A., Rajan W.D., Kaminska B. Immune microenvironment of gliomas. Lab. Investig. 2017;97:498–518. doi: 10.1038/labinvest.2017.19. [DOI] [PubMed] [Google Scholar]
- 6.Glass R., Synowitz M. CNS macrophages and peripheral myeloid cells in brain tumours. Acta Neuropathol. 2014;128:347–362. doi: 10.1007/s00401-014-1274-2. [DOI] [PubMed] [Google Scholar]
- 7.Zhao B., Wang Y., Wang Y., Chen W., Liu P.H., Kong Z., Dai C., Wang Y., Ma W. Systematic identification, development, and validation of prognostic biomarkers involving the tumor-immune microenvironment for glioblastoma. J. Cell. Physiol. 2020 doi: 10.1002/jcp.29878. [DOI] [PubMed] [Google Scholar]
- 8.Wainwright D.A., Dey M., Chang A., Lesniak M.S. Targeting Tregs in Malignant Brain Cancer: Overcoming IDO. Front. Immunol. 2013;4 doi: 10.3389/fimmu.2013.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye X.Z., Xu S.L., Xin Y.H., Yu S.C., Ping Y.F., Chen L., Xiao H.L., Wang B., Yi L., Wang Q.L., et al. Tumor-associated microglia/macrophages enhance the invasion of glioma stem-like cells via TGFBETA1 signaling pathway. J. Immunol. 2012;189:444–453. doi: 10.4049/jimmunol.1103248. [DOI] [PubMed] [Google Scholar]
- 10.Graeber M.B., Scheithauer B.W., Kreutzberg G.W. Microglia in brain tumors. Glia. 2002;40:252–259. doi: 10.1002/glia.10147. [DOI] [PubMed] [Google Scholar]
- 11.Grabowski M.M., Sankey E.W., Ryan K.J., Chongsathidkiet P., Lorrey S.J., Wilkinson D.S., Fecci P.E. Immune suppression in gliomas. J. Neuro-Oncol. 2020 doi: 10.1007/s11060-020-03483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hambardzumyan D., Gutmann D.H., Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016;19:20–27. doi: 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skytthe M.K., Graversen J.H., Moestrup S.K. Targeting of CD163+ Macrophages in Inflammatory and Malignant Diseases. Int. J. Mol. Sci. 2020;21:5497. doi: 10.3390/ijms21155497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S., Zhang C., Maimela N.R., Yang L., Zhang Z., Ping Y., Huang L., Zhang Y. Molecular and clinical characterization of CD163 expression via large-scale analysis in glioma. Oncoimmunology. 2019;8 doi: 10.1080/2162402X.2019.1601478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostuni R., Kratochvill F., Murray P.J., Natoli G. Macrophages and cancer: From mechanisms to therapeutic implications. Trends Immunol. 2015;36:229–239. doi: 10.1016/j.it.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Yang L., Zhang Y. Tumor-associated macrophages: From basic research to clinical application. J. Hematol. Oncol. 2017;10:58. doi: 10.1186/s13045-017-0430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrer V.P., Moura Neto V., Mentlein R. Glioma infiltration and extracellular matrix: Key players and modulators. Glia. 2018;66:1542–1565. doi: 10.1002/glia.23309. [DOI] [PubMed] [Google Scholar]
- 18.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., Porta-Pardo E., Gao G.F., Plaisier C.L., Eddy J.A., et al. The Immune Landscape of Cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moss J., Magenheim J., Neiman D., Zemmour H., Loyfer N., Korach A., Samet Y., Maoz M., Druid H., Arner P., et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018;9:5068. doi: 10.1038/s41467-018-07466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B., Liu Y., Pan X., Li M., Yang S., Li S.C. DNA Methylation Markers for Pan-Cancer Prediction by Deep Learning. Genes. 2019;10:778. doi: 10.3390/genes10100778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noushmehr H., Sabedot T.S., Malta T.M., Nelson K., Snyder J., Wells M., deCarvalho A., Mukherjee A., Chitale D., Mosella M., et al. Detection of glioma and prognostic subtypes by non-invasive circulating cell-free DNA methylation markers. bioRxiv. 2019:601245. doi: 10.1093/neuonc/noz126.018. [DOI] [Google Scholar]
- 23.Capper D., Jones D.T.W., Sill M., Hovestadt V., Schrimpf D., Sturm D., Koelsche C., Sahm F., Chavez L., Reuss D.E., et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivera A.L., Pelloski C.E., Gilbert M.R., Colman H., De La Cruz C., Sulman E.P., Bekele B.N., Aldape K.D. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro-Oncology. 2010;12:116–121. doi: 10.1093/neuonc/nop020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oldrini B., Vaquero-Siguero N., Mu Q., Kroon P., Zhang Y., Galán-Ganga M., Bao Z., Wang Z., Liu H., Sa J.K., et al. MGMT genomic rearrangements contribute to chemotherapy resistance in gliomas. Nat. Commun. 2020;11:3883. doi: 10.1038/s41467-020-17717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M.W., Fujiwara K., Che X., Zheng S., Zheng L. DNA methylation in the tumor microenvironment. J. Zhejiang Univ. Sci. B. 2017;18:365–372. doi: 10.1631/jzus.B1600579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui X., Ma C., Vasudevaraja V., Serrano J., Tong J., Peng Y., Delorenzo M., Shen G., Frenster J., Morales R.T.T., et al. Dissecting the immunosuppressive tumor microenvironments in Glioblastoma-on-a-Chip for optimized PD-1 immunotherapy. eLife. 2020;9 doi: 10.7554/eLife.52253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dejaegher J., Solie L., Hunin Z., Sciot R., Capper D., Siewert C., Van Cauter S., Wilms G., van Loon J., Ectors N., et al. DNA methylation based glioblastoma subclassification is related to tumoral T-cell infiltration and patient survival. Neuro-Oncology. 2020 doi: 10.1093/neuonc/noaa247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Angelo F., Ceccarelli M., Tala N., Garofano L., Zhang J., Frattini V., Caruso F.P., Lewis G., Alfaro K.D., Bauchet L., et al. The molecular landscape of glioma in patients with Neurofibromatosis 1. Nat. Med. 2019;25:176–187. doi: 10.1038/s41591-018-0263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourgon R., Gentleman R., Huber W. Independent filtering increases detection power for high-throughput experiments. Proc. Natl. Acad. Sci. USA. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sturm G., Finotello F., Petitprez F., Zhang J.D., Baumbach J., Fridman W.H., List M., Aneichyk T. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics (Oxford, England) 2019;35:i436–i445. doi: 10.1093/bioinformatics/btz363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finotello F., Mayer C., Plattner C., Laschober G., Rieder D., Hackl H., Krogsdam A., Loncova Z., Posch W., Wilflingseder D., et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11:34. doi: 10.1186/s13073-019-0638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aran D., Hu Z., Butte A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:1–14. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ImmuneSubtypeClassifier. [(accessed on 27 July 2020)]; Available online: https://github.com/CRI-iAtlas/ImmuneSubtypeClassifier.
- 35.Polano M., Chierici M., Dal Bo M., Gentilini D., Di Cintio F., Baboci L., Gibbs D.L., Furlanello C., Toffoli G. A Pan-Cancer Approach to Predict Responsiveness to Immune Checkpoint Inhibitors by Machine Learning. Cancers. 2019;11:1562. doi: 10.3390/cancers11101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langlois B., Saupe F., Rupp T., Arnold C., van der Heyden M., Orend G., Hussenet T. AngioMatrix, a signature of the tumor angiogenic switch-specific matrisome, correlates with poor prognosis for glioma and colorectal cancer patients. Oncotarget. 2014;5:10529–10545. doi: 10.18632/oncotarget.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao J., Chen A.X., Gartrell R.D., Silverman A.M., Aparicio L., Chu T., Bordbar D., Shan D., Samanamud J., Mahajan A., et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat. Med. 2019;25:462–469. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassn Mesrati M., Behrooz A.B., Y Abuhamad A., Syahir A. Understanding Glioblastoma Biomarkers: Knocking a Mountain with a Hammer. Cells. 2020;9:1236. doi: 10.3390/cells9051236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y., Zhang X., Yao J., Jin Z., Liu C. Expression patterns and the prognostic value of the EMILIN/Multimerin family members in low-grade glioma. PeerJ. 2020;8 doi: 10.7717/peerj.8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Degenhardt F., Seifert S., Szymczak S. Evaluation of variable selection methods for random forests and omics data sets. Brief. Bioinf. 2017;20:492–503. doi: 10.1093/bib/bbx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vabalas A., Gowen E., Poliakoff E., Casson A.J. Machine learning algorithm validation with a limited sample size. PLoS ONE. 2019;14:e0224365. doi: 10.1371/journal.pone.0224365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matthews B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Bioch. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 43.Shi L., Campbell G., Jones W.D., Campagne F., Wen Z., Walker S.J., Su Z., Chu T.M., Goodsaid F.M., Pusztai L., et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat. Biotechnol. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boughorbel S., Jarray F., El-Anbari M. Optimal classifier for imbalanced data using Matthews Correlation Coefficient metric. PLoS ONE. 2017;12:e0177678. doi: 10.1371/journal.pone.0177678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sundjaja J.H., Shrestha R., Krishan K. StatPearls. Treasure Island; London, UK: 2020. McNemar And Mann-Whitney U Tests. [PubMed] [Google Scholar]
- 46.Wang H., Li G. A Selective Review on Random Survival Forests for High Dimensional Data. Quant. Bio-Sci. 2017;36:85–96. doi: 10.22283/qbs.2017.36.2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chai R.C., Zhang K.N., Chang Y.Z., Wu F., Liu Y.Q., Zhao Z., Wang K.Y., Chang Y.H., Jiang T., Wang Y.Z. Systematically characterize the clinical and biological significances of 1p19q genes in 1p/19q non-codeletion glioma. Carcinogenesis. 2019;40:1229–1239. doi: 10.1093/carcin/bgz102. [DOI] [PubMed] [Google Scholar]
- 48.Chai R.C., Chang Y.Z., Wang Q.W., Zhang K.N., Li J.J., Huang H., Wu F., Liu Y.Q., Wang Y.Z. A Novel DNA Methylation-Based Signature Can Predict the Responses of MGMT Promoter Unmethylated Glioblastomas to Temozolomide. Front. Genet. 2019;10 doi: 10.3389/fgene.2019.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:1362–4962. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 52.Amankulor N.M., Kim Y., Arora S., Kargl J., Szulzewsky F., Hanke M., Margineantu D.H., Rao A., Bolouri H., Delrow J., et al. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Devel. 2017;31:774–786. doi: 10.1101/gad.294991.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin M.Z., Jin W.L. The updated landscape of tumor microenvironment and drug repurposing. Signal Transd. Target. Ther. 2020;5:1–16. doi: 10.1038/s41392-020-00280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vidyarthi A., Agnihotri T., Khan N., Singh S., Tewari M.K., Radotra B.D., Chatterjee D., Agrewala J.N. Predominance of M2 macrophages in gliomas leads to the suppression of local and systemic immunity. Cancer Immunol. Immunother. CII. 2019;68:1995–2004. doi: 10.1007/s00262-019-02423-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Razavi S.M., Lee K.E., Jin B.E., Aujla P.S., Gholamin S., Li G. Immune Evasion Strategies of Glioblastoma. Front. Surg. 2016;3 doi: 10.3389/fsurg.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonabend A.M., Rolle C.E., Lesniak M.S. The role of regulatory T cells in malignant glioma. Anticancer Res. 2008;28:1143–1150. [PubMed] [Google Scholar]
- 57.Pitroda S.P., Zhou T., Sweis R.F., Filippo M., Labay E., Beckett M.A., Mauceri H.J., Liang H., Darga T.E., Perakis S., et al. Tumor endothelial inflammation predicts clinical outcome in diverse human cancers. PLoS ONE. 2012;7:e46104. doi: 10.1371/journal.pone.0046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao C., Gomez G.A., Zhao Y., Yang Y., Cao D., Lu J., Yang H., Lin S. ETV2 mediates endothelial transdifferentiation of glioblastoma. Signal Transd. Target. Ther. 2018;3:4. doi: 10.1038/s41392-018-0007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tormoen G.W., Crittenden M.R., Gough M.J. Role of the immunosuppressive microenvironment in immunotherapy. Adv. Rad. Oncol. 2018;3:520–526. doi: 10.1016/j.adro.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiencke J.K., Koestler D.C., Salas L.A., Wiemels J.L., Roy R.P., Hansen H.M., Rice T., McCoy L.S., Bracci P.M., Molinaro A.M., et al. Immunomethylomic approach to explore the blood neutrophil lymphocyte ratio (NLR) in glioma survival. Clin. Epigenetics. 2017;9:10. doi: 10.1186/s13148-017-0316-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zou J.P., Morford L.A., Chougnet C., Dix A.R., Brooks A.G., Torres N., Shuman J.D., Coligan J.E., Brooks W.H., Roszman T.L., et al. Human Glioma-Induced Immunosuppression Involves Soluble Factor(s) That Alters Monocyte Cytokine Profile and Surface Markers. J. Immunol. 1999;162:4882–4892. [PubMed] [Google Scholar]
- 62.Zhang L., Xu Y., Sun J., Chen W., Zhao L., Ma C., Wang Q., Sun J., Huang B., Zhang Y., et al. M2-like tumor-associated macrophages drive vasculogenic mimicry through amplification of IL-6 expression in glioma cells. Oncotarget. 2016;8:819–832. doi: 10.18632/oncotarget.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanaei S., Afshari K., Hirbod-Mobarakeh A., Mohajer B., Amir Dastmalchi D., Rezaei N. Therapeutic efficacy of specific immunotherapy for glioma: A systematic review and meta-analysis. Rev. Neurosci. 2018;29:443–461. doi: 10.1515/revneuro-2017-0057. [DOI] [PubMed] [Google Scholar]
- 64.Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weller M., Wick W., Aldape K., Brada M., Berger M., Pfister S.M., Nishikawa R., Rosenthal M., Wen P.Y., Stupp R., et al. Glioma. Nat. Rev. Disease Primers. 2015;1:1–18. doi: 10.1038/nrdp.2015.17. [DOI] [PubMed] [Google Scholar]
- 66.Lombardi G., Barresi V., Castellano A., Tabouret E., Pasqualetti F., Salvalaggio A., Cerretti G., Caccese M., Padovan M., Zagonel V., et al. Clinical Management of Diffuse Low-Grade Gliomas. Cancers. 2020;12:3008. doi: 10.3390/cancers12103008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roesch S., Rapp C., Dettling S., Herold-Mende C. When Immune Cells Turn Bad-Tumor-Associated Microglia/Macrophages in Glioma. Int. J. Mol. Sci. 2018;19:436. doi: 10.3390/ijms19020436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quail D.F., Joyce J.A. The Microenvironmental Landscape of Brain Tumors. Cancer Cell. 2017;31:326–341. doi: 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang Y., Le W. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Mol. Neurobiol. 2016;53:1181–1194. doi: 10.1007/s12035-014-9070-5. [DOI] [PubMed] [Google Scholar]
- 70.Prosniak M., Harshyne L.A., Andrews D.W., Kenyon L.C., Bedelbaeva K., Apanasovich T.V., Heber-Katz E., Curtis M.T., Cotzia P., Hooper D.C. Glioma Grade Is Associated with the Accumulation and Activity of Cells Bearing M2 Monocyte Markers. Clin. Cancer Res. 2013;19:3776–3786. doi: 10.1158/1078-0432.CCR-12-1940. [DOI] [PubMed] [Google Scholar]
- 71.Shabo I., Olsson H., Sun X.F., Svanvik J. Expression of the macrophage antigen CD163 in rectal cancer cells is associated with early local recurrence and reduced survival time. Int. J. Cancer. 2009;125:1826–1831. doi: 10.1002/ijc.24506. [DOI] [PubMed] [Google Scholar]
- 72.Zheng C., Xu R. Predicting cancer origins with a DNA methylation-based deep neural network model. PLoS ONE. 2020;15:e0226461. doi: 10.1371/journal.pone.0226461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the results here showed are based on data generated by the TCGA Research Network: https:/cancer.gov/tcga (accessed on 12 March 2020).