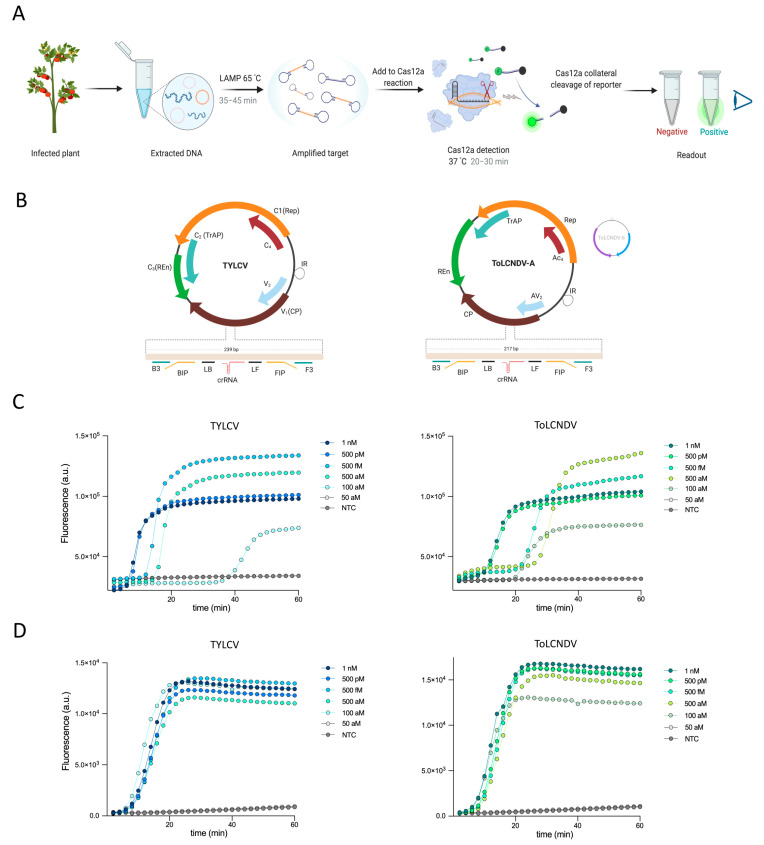
Figure 1.
LAMP-coupled Cas12-based assay for the detection of TYLCV and ToLCNDV. (A) The assay workflow. Viral DNA (orange circles) extracted from an infected tomato plant is amplified by loop-mediated isothermal amplification (LAMP), followed by clustered regularly interspaced short palindromic repeats (CRISPR)-mediated detection. Cas12a-based detection of the LAMP product triggers collateral cleavage of the reporter, thus producing a signal for visual detection. (B) Organization of the single-component TYLCV genome (left) and the two-component (bipartite) ToLCNDV-A and B genomes (right). The targeted areas in the coat protein gene (CP) are highlighted, with the designed LAMP primers and crRNAs shown. Rep: replication-associated protein; IR: intergenic region; CP: coat protein; REn: replication enhancer protein; TrAP: transcriptional activator protein; AC4 or C4: RNA suppressor protein, present on the antisense (complementary) strand; AV2 or V2: precoat proteins, present on the virion-sense strand as plant RNA silencing suppressors. (C) Monitoring the performance of LAMP of synthetic DNA generated by PCR of TYLCV dsDNA (left) and ToLCNDV dsDNA (right) by real-time fluorescence across a range of dsDNA concentrations at 65 °C for 60 min. The LAMP signal was measured using the nucleic acid stain SYTO 9. Data generated using 50 aM of sample showed fluorescence signals similar to those of the no-template control (NTC), and these lines are therefore overlapping. Data are shown as the mean (n = 3). (D) Real-time measurements of Cas12 collateral activity on HEX reporter with LAMP-amplified DNA from a synthetic dsDNA template. Cas12a with crRNA targeting TYLCV (left) or ToLCNDV (right) was incubated with HEX reporter and LAMP product at 37 °C for 60 min. Data generated using 50 aM of sample showed fluorescence signals similar to those of the NTC, and these lines are therefore overlapping. Data are shown as the mean (n = 3).