Abstract
The effectiveness of cancer treatment strongly depends on the early detection of the disease. Currently, the most common diagnostic method, tissue biopsy, takes time and can be damaging to the patient. Circulating cancer biomarkers such as circulating tumor DNA, micro-RNA (miRNA), tumor proteins, exosomes, and circulating tumor cells have repeatedly demonstrated their viability as targets for minimally invasive cancer detection through liquid biopsies. However, among other things, achieving a great sensitivity of detection is still challenging due to the very low concentration of biomarkers in fluid samples. This review will discuss how the recent advances in nanoparticle-based biosensors are overcoming these practical difficulties. This report will be focusing mainly on optical transduction mechanisms of metal nanoparticles (M-NPs), quantum dots (QDs), and upconversion nanoparticles (UCNPs).
Keywords: optical biosensors, circulating cancer biomarkers, optical transduction, metal nanoparticles (M-NPs), quantum dots (QDs), upconversion nanoparticles (UCNPs)
1. Introduction
The early detection of cancer considerably impacts the effectiveness of oncotherapy. Currently, tissue biopsies are commonly used as an affordable and accurate diagnostic method. However, on top of being a time-consuming procedure, tissue biopsies can be difficult to reproduce. In certain cases, tissue sampling can require a more invasive procedure that can be frightening or even damaging to the patient. Thus, it is important to develop faster, less invasive, and more precise biosensors. Recently, the sensing of circulating cancer biomarkers such as circulating tumor DNA (ctDNA), circulating micro-RNA (miRNA), tumor proteins, exosomes, or even circulating tumor cells (CTCs) have been gaining a lot of attention as they allow for minimally invasive detection methods. However, the low concentration of those biomarkers renders most standard biosensors obsolete.
Circulating cancer biomarkers are molecules of different forms mostly present in body fluids such as the serum/plasma, saliva, or urine of cancer patients. Among those biomarkers, ctDNAs are short fragments of cell-free DNA originating from tumor cells [1]. The release mechanism of ctDNAs is not clearly understood yet, but recent studies have demonstrated a positive correlation between ctDNA levels and tumor burden in animal models [2]. On the other hand, circulating miRNA, stable non-coding small RNAs, are differentially expressed depending on the stage of tumor progression [3]. Currently, most ctDNA and miRNA analyses are performed in liquid biopsies through variations of polymerase chain reaction (PCR), microarray, or next-generation sequencing with each certain disadvantages: a small number of target genes, a low throughput, or the cost of the equipment [2,4,5,6,7]. Furthermore, serum proteins have been successfully targeted in the detection of various types of cancers such as breast cancer [8] and epithelial ovarian cancer [9], as their differential secretion is thought to be an indicator of cancer cells. Exosomes, nanosized molecular and genetic cargoes, have also demonstrated an excellent diagnostic ability as their main function is to act as signaling vesicles between cancer cells and their environment [10,11]. Last but not least, on a microscopic scale, circulating tumor cells are key players in cancer metastasis which makes them excellent diagnostic and prognostic biomarkers [12]. As circulating cancer biomarker expression is directly related to the presence of cancer cells, they offer a promising alternative to tissue biopsies. However, extracting a detectable amount of biomarkers, processing them without disturbing their integrity, targeting multiple biomarkers at the same time, and doing so in a timely manner are only a few of the many challenges faced when using the traditional techniques for the detection of circulating cancer biomarkers. To overcome these problems research is turning to nanoparticle-based biosensing.
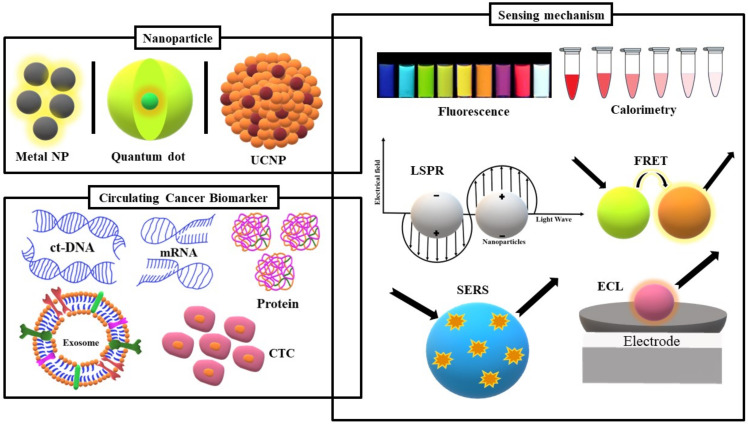
Of all the transduction mechanisms of nanoparticle-based biosensors, optical transduction, the core of this review, is the most preeminent in biosensing due to the speed and ease of its signal detection. Additionally, despite the existence of different types of nanoparticles, only a few demonstrate exceptional optical proprieties relevant to the subject matter. With that in mind, the focus of this review will be on the recent innovations in circulating cancer biomarkers optical nanobiosensors based on quantum dots (QDs), metal nanoparticles, and upconversion nanoparticles (Figure 1). The application of QDs in fluorescence and electrochemiluminescence (ECL) assays as well as their adaptation in fluorescence resonance energy transfer (FRET)-based sensors will be discussed. As metal nanoparticles (NPs) are not characteristically fluorescent, the focus will be on the effect of their localized surface plasmon resonance (LSPR) in optical transduction mechanisms such as colorimetry, surface-enhanced Raman scattering (SERS), and metal enhanced fluorescence (MEF). Finally, recent advances in upconversion nanoparticles (UCNPs)-based biosensors will also be covered.
Figure 1.
Schematic representation of the nanoparticles and the main sensing mechanisms of circulating cancer biomarkers covered in this review.
2. Quantum Dots-Based Optical Biosensor for Circulating Cancer Biomarkers
Quantum dots are 2–10 nm luminescent semiconductor nanocrystals that possess size-tunable photophysical properties with a broad absorbance and a narrow emission that are very pertinent to the biosensing field [13]. This gives QDs a unique multiplex feature that can be used in the detection of multiples analytes at once [14]. They can be produced through different processes mainly divided into two categories: the top-down approach that consists of thinning a semi-conductor to achieve nanometer-sized particles and the bottom-up method where the self-assembly of QDs is promoted through nucleation or atom-by-atom layering [15]. QD cores and/or shells are typically made of elements from the groups II–VI: CdSe, CdS, CdTe, ZnSe, ZnS, ZnTe, HgS, HgSe, HgTe, and ZnO [16]. However, other elements such as carbon, graphene, and molybdenum oxide have been gaining more attention. The core-shell structure of the QD is another important feature of biosensing as the shell can be modified to allow for its bioconjugation with DNA, aptamers, and antibodies [17].
Since their first discovery, quantum dots have been frequently used as fluorophores to sense circulating cancer markers. In a CTC quantification assay, Min et al. combined the fluorescence of QDs with a magnetic bead-based isolation assay [18]. Briefly, QDs were functionalized with anti-epithelial cell adhesion molecule (EpCAM) antibodies; EpCAM is highly expressed in tumor cells. The nanoprobes were then added to a cell suspension of SK-Br3, a breast cancer cell line [18]. After removal of the excess QDs by centrifugation, anti-IgG-modified magnetic beads were incorporated and bound to the remaining anti-EpCAM antibodies. Then, a magnetic field was used to isolate the anti-EpCAM-QDs attached SK-Br3 cells. Measuring the fluorescence intensity of the isolated QDs with a plate reader allowed for the quantification of captured CTCs with an efficiency of 70–80% in 50 min [18]. In a similar experiment with a higher capture efficiency of about 90% in whole blood samples, aptamer EpCAM receptors were conjugated with graphene QDs (GQDs) to synthesize a “turn-on” biosensor based on nanosurface energy transfer (NSET) [19]. By using aptamers@magnetic Fe3O4 nanoparticle-bound GQDs as energy donors and MoS2 nanosheets as acceptors, the fluorescence signal was quenched in the absence of competitive binding from circulating tumor cells. The mechanism was proven faster than most so far (15 min), and it was possible to detect as few as 10 CTCs in whole blood. However, it has been reported that the CTCs had a tendency to engulf the EpCAM@aptamer@Fe3O4@GQD complexes which may significantly reduce the detection limit of the sensor.
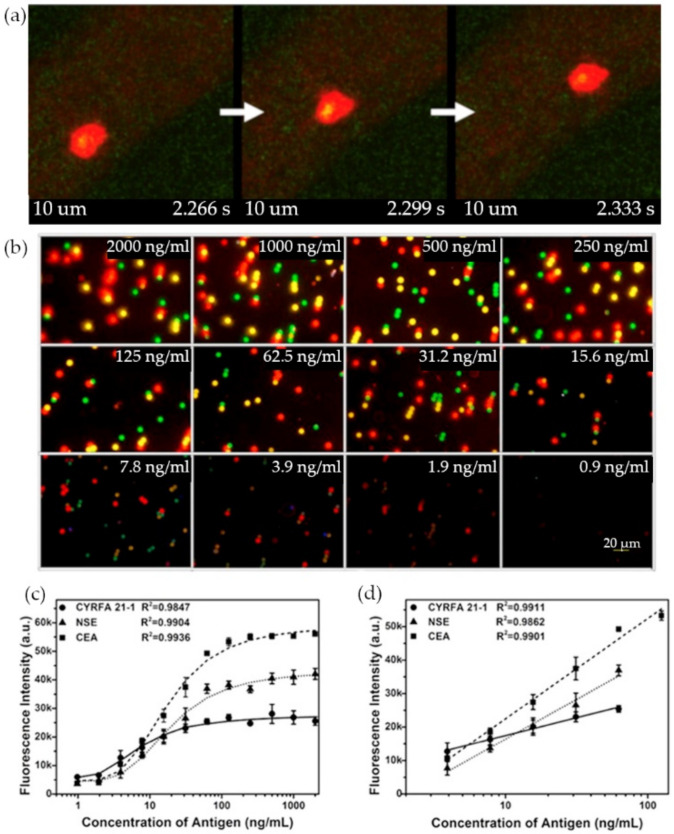
As CTCs tend to travel through the bloodstream to establish new colonies, most experiments are meant to analyze the blood CTCs concentrations to establish a diagnostic for cancer patients. Nonetheless, it seems to be equally important to have a better understanding of the routes those circulating cancer biomarkers take. Kuo et al. used an in vivo approach to CTCs detection [20]. In their experiment, antibody-conjugated QDs were used to track the movement of an animal cancer model CTCs with real-time fluorescent imaging. After inducing a tumor growth in the earlobes of mice from pancreatic cancer cells expressing red fluorescent protein (RFP), CD24 functionalized QDs 525 (Green) were delivered to the mice bloodstream by injection to study the movement of a subpopulation of CTCs through multi-photon microscopy. A more metastatic CTCs subpopulation expressing green fluorescent proteins were also tagged with anti-CD133 antibody linked QDs 705 (Red) to image the movement of a smaller number of cells (Figure 2a) [20].
Figure 2.
Quantum dots (QDs)-based fluorescence assays: (a) Three consecutive images (33 ms interval) of CD133+ circulating tumor cells (CTCs) tagged with red QDs moving in a blood vessel. (b) Multiplexed fluorescence of cytokeratin 19 (CYRFA 21-1), neuron-specific enolase (NSE), and carcinoembryonic antigen (CEA) at different concentrations. (c) Standard curves for the detection of CYRFA 21-1, NSE, and CEA in the multiplexed assay. (d) Linear range of the plot in (c). (Reproduced with permission from [20], published by Springer Nature 2019; reproduced with permission from [23], published by Elsevier 2016).
Due to the fact that our body fluids are interconnected, smaller circulating cancer markers can be found in the urine of the saliva of cancer patients, quantum dot-based nanobiosensors were adapted to different liquid biopsies. In a recent experiment by Nejdl et al., self-assembled cadmium telluride (CdTe) QDs were used for the detection of nucleic acid excreted in the urine [21]. The nanoparticles were synthesized through a simple spontaneous reaction that lasted 70 h. The QDs and a methylene blue quencher were combined for the sensing of DNA. Both the absorbance and the fluorescence were measured and capillary electrophoresis (CE) with laser-induced fluorescence detection was also used [21]. Another study exploring salivary exosome biosensing was based on combined aptamer recognition. A complex was formed between self-assembled DNA concatemer and numerous QDs bound to an aptamers–magnetic microspheres combination. When one exosome binds to the aptamer, it would release many QDs from the DNA concatemer which amplifies the fluorescent signal. The biosensor required a significantly short time of incubation of 30 min [22].
Most biosensors have been targeting a single type of circulating cancer biomarker when in reality it is often the presence of multiple biomarkers that is a better cancer indicator. In a triplex sandwich assay by Wu et al., three QDs and micro-sized magnetic beads pairs were functionalized with monoclonal antibodies that targeted three lung cancer biomarkers in solution: cytokeratin 19 (CYRFA 21-1), carcinoembryonic antigen (CEA), and neuron-specific enolase (NSE) [23]. Each one of the QDs was specifically chosen with a different emission maximum (green—525 nm, yellow—585 nm, and red—625 nm) and photographed with an optical microscope before getting treated by Gray Quantifier software. The nanoprobes allowed for the simultaneous detection and quantification of three analytes (Figure 2b–d) at a low cost, with a limit of detection (LOD) of 364 pg/mL for CYRFA21-1, 38 pg/mL for CEA, and 370 pg/mL for NSE in a sample volume as low as 20 µL [23]. The experiment required an incubation time of 1 h.
Electrochemiluminescence (ECL) is another transduction mechanism by which the potential of QDs has been harnessed in an attempt to improve the detection sensitivity of nanoparticle-based optical biosensors. In an experiment by Meng et al., molybdenum oxide(MoOx) QDs were used in the ECL detection of non-small cell lung cancer biomarker CYFRA21-1 to achieve an even lower LOD of 0.3 pg/mL [24]. A layered detection platform was made by assembling a gold nanoparticles (AuNPs) layer meant to increase the electron transportation rate, a cathodic layer of MoOx QDs, and a layer of target-specific antibodies. By forming a complex with those antibodies, CYFRA21-1 hinders the electron transfer in a concentration-dependent manner and thus reduces the ECL signal. This type of sensor had a good sensitivity. However, full biological samples may potentially affect its detection mechanism due to some crosstalk or a reduced electron transportation rate [24].
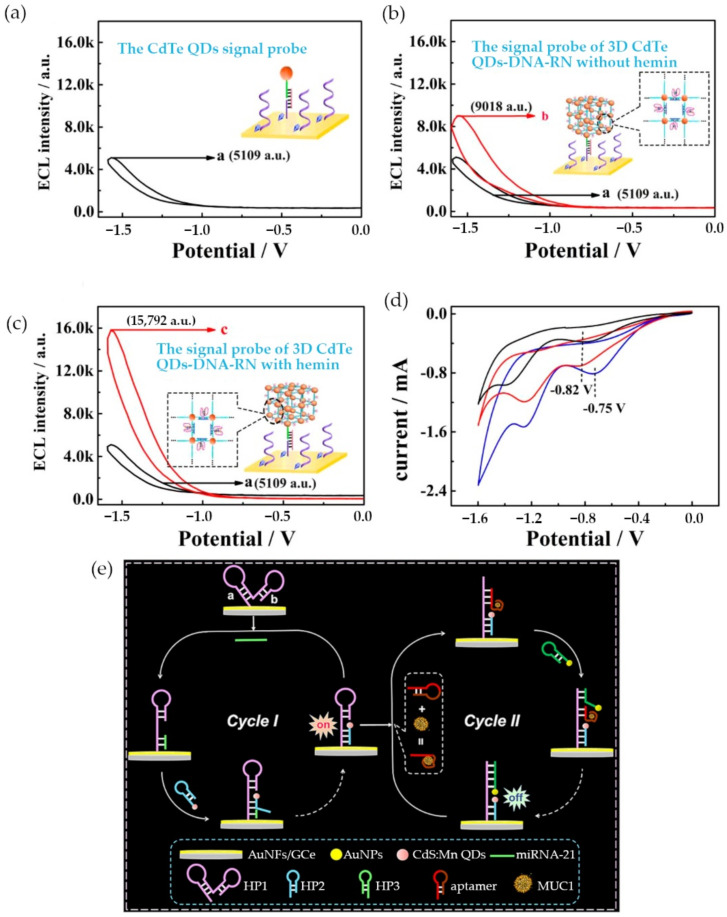
Mahani et al. developed a biosensor for the detection of ovarian cancer marker miRNA-21 based on a FRET system [25]. The sensor was constructed by a molecular beacon with a carbon QD at one end and a Black Hole Quencher 1 (BHQ1) at the other end. With both ends in close proximity, the fluorescent emission of the QD was quenched [25]. Conversely, in the presence of the exact sequence of miRNA-21, the molecular bacon would go through a conformational change that increased the gap between the QD and BHQ1 which would induce a fluorescent signal. The biosensor was highly specific and had a limit of detection of 0.3 nM [25]. However, during the same year, Sun et al. were able to achieve a lower LOD of 34 aM for miRNA-21 detection by engineered an ECL-based sensor [26]. The team designed a 3D walker probe by linking cadmium telluride (CdTe) QDs with various DNA sequences (Figure 3a–d) [26]. The nanoreticulations of the hemin/G-quadruplex structure increased the availability of QDs, thus when in presence of miRNA-21, the ECL signal was stronger.
Figure 3.
QDs-based electrochemiluminescence (ECL) sensing: (a) ECL curve of QDs signal probe; (b) ECL curve of 3D QDs-DNA-RN without hemin signal probe; (c) ECL curve of 3D QDs-DNA-RN with hemin signal probe; (d) cyclic voltammetry responses of the QDs signal probe; (e) schematic of the miRNA-21 and MUC1 detection platform. (Reproduced with permission from [26], published by Elsevier 2020; reproduced with permission from [27], published by American Chemical Society 2019).
A year later, Li et al. achieved a significantly lower limit of detection by developing a two-cycle sensitive platform based on dual catalytic hairpin assembly for the detection of both miRNA-21 and mucin 1 with an LOD of 11 aM and 0.40 fg/mL, respectively [27]. During the first cycle of the platform, miRNA-21 bound to a hairpin structure and initiated the addition of another manganese doped cadmium sulfide (CdS:Mn) QD modified hairpin structure. This led to the emission of an initial ECL signal. The second cycle generated an ECL-RET signal through the binding of mucin 1/aptamer to the CdS:Mn QD modified hairpin structure, which later induced the insertion of AuNPs into the structure that decreased the ECL signal (Figure 3e) [27]. Recent QDs-based optical biosensors for circulating cancer biomarkers are compared in Table 1.
Table 1.
Quantum dots-based optical biosensors for circulating cancer biomarkers.
| Sensing Mechanism | Target Biomarker | Detection Elements | Signal Elements | LOD | Reference |
|---|---|---|---|---|---|
| Fluorescence | CTC 1 (SK-Br3) | Anti-EpCAM antibodies | Octadecylamine-coated QDs 2 630 | 275 cells/mL | [18] |
| CTC (MCF-7) | Anti-EpCAM antibodies | ZnS-coated CuInSe QDs | 12 cells/well | [28] | |
| Exosomes (CAL27) | CD63 aptamers | ZnCdSe/ZnS core/shell QDs | 500 particles/μL | [22] | |
| CYRFA 21-1 3, CEA 4 and NSE 5 |
Target specific antibodies | 525, 585 and 625 QDs | 364 pg/mL, 38 pg/mL and 370 pg/mL |
[23] | |
| PET 6 | ctDNA 7 | Semi-intercalation binding with magnetic beads | Mercaptosuccinic acid stabilized CdTe QDs | 3 ng/mL | [21] |
| NSET 8 | CTC (Hep G2 and A549) | EpCAM aptamers | Nitrogen and sulphur-doped graphene QDs (donors) quenched by MoS2 nanosheets (acceptors) | 1.19 nM | [19] |
| FRET 9 | AFP 10 | AFP aptamers and anti-AFP antibodies |
CdTe QDs (donors) quenched by AuNPs (acceptors) | 400 pg/mL | [29] |
| miRNA 11-21 | Hairpin-structured oligonucleotide probes | Carbon QDs (donors) quenched by Black Hole Quencher 1 (acceptors) | 0.3 nM | [27] | |
| ECL 12 | CEA | CEA antibodies | Poly(ethylenimine) functionalized graphene oxide matrix modified with carbon QDs and AuNPs | 1.67 pg/mL | [30] |
| CYFRA21-1 | CYFRA21-1 antibodies | Molybdenum oxide QDs/Au NPs-chit nanocomposite | 0.3 pg/mL | [24] | |
| miRNA-21 and MUC1 13 | miRNA-21 hairpin probes and MUC1 aptamers | HP2 14 modified by CdS:Mn QDs and AuNPs modified hairpin probes | 11 aM and 0.40 fg/mL | [26] | |
| miRNA-21 | miRNA-21 specific hairpins | 3D CdTe QDs–DNA nanoreticulations | 34 aM | [25] |
1 Circulating tumor cell. 2 Quantum dots. 3 Cytokeratin 19. 4 Carcinoembryonic antigen. 5 Neuron-specific enolase. 6 Photoinduced electron transfer. 7 Circulating tumor DNA. 8 Nanosurface energy transfer. 9 Fluorescence resonance energy transfer. 10 Alpha-fetoprotein. 11 Micro-RNA. 12 Electrochemiluminescence. 13 Mucin 1. 14 Hairpin 2.
3. Metal Nanoparticle-Based Optical Biosensor for Circulating Cancer Biomarkers
In the category of metal nanoparticle-based biosensors, copper, silver, and gold are the most common elements. However, due to the instability of Cu, Au and Ag nanoparticles are predominantly used. Multiple physical, chemical and biological approaches are available for the synthesis of metal nanoparticles. The choice of method depends on different factors such as the time or the cost of synthesis but also the size and the shape of the nanoparticles. Metal nanoparticles are often used in fluorescence assays. However, due to the fact that they are not fluorescent elements by nature, they require grouping with a fluorescent dye. For instance, in the detection of exosomes as circulating cancer biomarkers, Gao et al. used a dual hairpin signal amplification mechanism where gold nanoparticles (AuNPs) only served as a scaffold and where the fluorescent signal was emitted by Fluorescein, an organic dye [31]. In other cases, electrocatalytic features of metal nanoparticles are coupled with ECL essays [32]. However, when it comes to optical biosensors, the most important characteristic of metal nanoparticles is their localized surface plasmon resonance (LSPR). LSPR is a physical phenomenon where the electrons on the surface of a metal NP collectively oscillate after being exposed to an exciting light.
In 2015, Pallares et al. engineered an inverse sensitivity plasmonic nanosensor based on the LSPR of gold nanorods for the quantification of circulating tumor DNA (ctDNA) [33]. Hexadecyltrimethylammonium bromide (CTAB)-coated gold nanorods and ctDNA would aggregate based on their electrostatic interactions. In theory, when the rods are covered by the ctDNA, their UV-visible light spectroscopy signal is reduced in a concentration-dependent manner. The mechanism of detection was simple and sensitive for the detection of lower concentrations, but it had a few stumbling blocks. The first step entailed the extraction of the ctDNA through a commercial kit which may reduce the analyte yield [33]. Another major issue was that ctDNA seemed to induce both aggregation and disaggregation of nanorods at different concentrations. Considering that clinical samples have variable ctDNA concentration, this requires accordingly tune the dynamic ranges of the detection [33]. Recently, Wang et al. developed a detection platform for cancerous exosomes. The mechanism was based on a gold film surface-functionalized with exosome capture DNA strands [34]. When exposed to the biomarker, exosome-specific aptamer and single-strand DNA T30 linked AuNPs were added, followed by T30 linked AuNPs. After 60 min of incubation time, the coupling of the SPR from the Au film and the LSPR from both types of AuNPs amplified the detection signal of the exosomes with a limit of detection as low as 5 × 103 exosomes/mL [34].
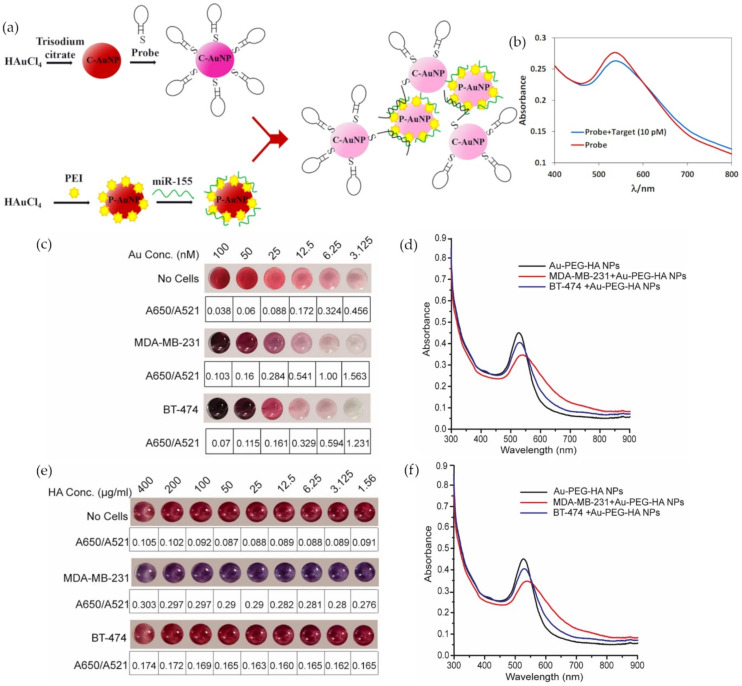
Metal nanoparticle colorimetric assays are also based on LSPR. When M-NPs aggregate in solution, there is a coupling of their surface plasmon resonance that causes a quick change in color and a shift in the absorbance peak of UV-visible light spectra of the sample. In an attempt to avoid the use of the expensive instruments and the tedious protocols that miRNA and ctDNA testing traditionally require, more researchers are trying to take advantage of this visual sensor. Hakimian et al. devised an ultrasensitive optical biosensor for the detection of breast cancer through an miRNA-155 sensing assay based on crosslinking aggregation [35]. Positively charged polyethyleneimine capped AuNPs (P-AuNPs) were synthesized by thermal reduction to trap negatively charged miRNA-155. A similar type of AuNPs (C-AuNPs) was citrate capped and linked to a thiolated DNA hairpin meant to hybridize with the P-AuNPs/miRNA-155 complex (Figure 4a). The miRNA served as an aggregation promoter between the two types of AuNPs which affected the molar ratio of dispersed to aggregated AuNPs, A530/A750 (Figure 4b). The sensor specificity was successfully tested for three base-pair mismatches, and it achieved a detection limit of 100 aM within approximately 45 min [35].
Figure 4.
Gold nanoparticle (AuNPs)-based colorimetric assays: (a) Schematic of miRNA-155 detection probes; (b) UV-Vis spectra curve of the detection probes in the absence (red) and presence (blue) of miRNA-155; (c) colorimetric changes and A650/A521 ratio of Au-polyethylene glycol- hyaluronic acid (HA) NPs after incubation with 50,000 cells (MDA-MB-231 and BT-474) at varied Au concentrations; (d) UV-Vis spectra of Au-polyethylene glycol-HA NPs after incubation with MDA-MB-231 (50,000 cells) and BT-474 (50,000 cells) at 25 nM Au concentration; (e) colorimetric changes and A650/A521 ratio of Au-polyethylene glycol-HA NPs after incubation with MDA-MB-231 (50,000 cells) and BT-474 (50,000 cells) at varied HA concentrations and 25 nM Au; (f) UV-Vis spectra of Au-polyethylene glycol-HA NPs after incubation with 50,000 cells (MDA-MB-231 and BT-474) at 100 HA µg/ml and 25 nM Au concentrations. (Reproduced with permission from [35] and from [36], published by Springer Nature 2018).
As the quest for more sensitive, more accessible, and less time-consuming assays continues, Wang et al. worked on improving colorimetric sensors specifically for the detection of single-nucleotide polymorphisms (SNPs) in ctDNA. The detection assay was optimized through various methods to achieve an LOD of 67 pM [37]. For instance, a critical linker concentration had to be determined and an extra centrifugation step had to be taken to ease the visual detection of KRAS oncogene mutations [37]. Rauta et al. went even further by developing a CTCs detection platform that takes CTCs viability into account [36]. AuNPs were synthesized and added to polyethylene glycol before getting functionalized with hyaluronic acid (HA), a ligand to CD44 expressing cancer cells. Different concentrations of Au and HA were tested with two cell lines that express variable levels of CD44 to find the optimal values for the nanoprobe (Figure 4c–f), but further optimization for a uniform distribution of HA is needed. The cell viability of the detection platform was between 60 and 94%, promising numbers for additional assays [36].
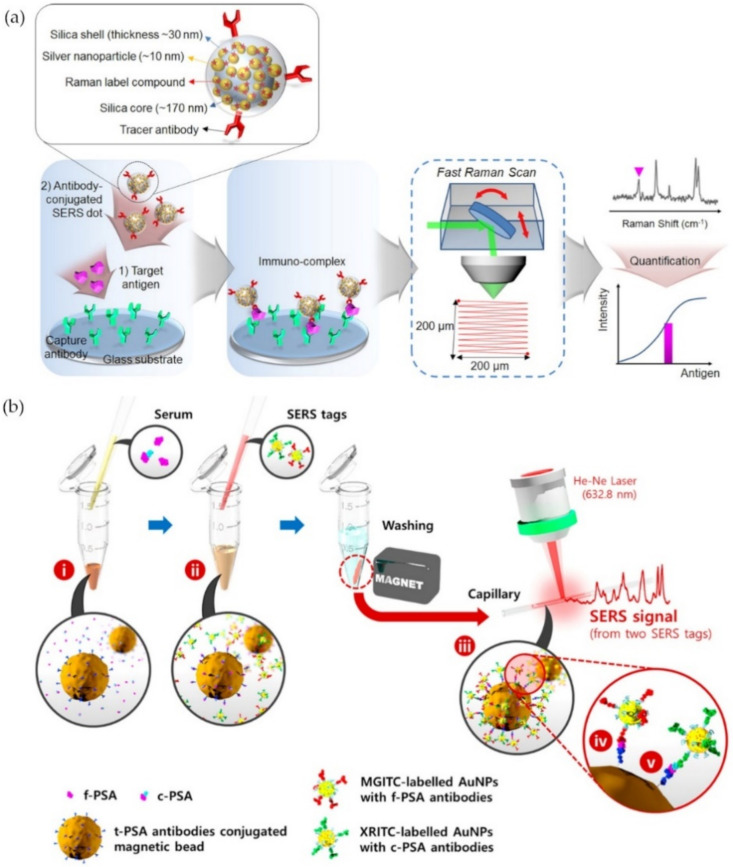
Surface-enhanced Raman scattering (SERS) is another optical transduction mechanism that measures the light scattering caused by the LSPR property of metal nanoparticles. The coating of silver nanoparticles by Raman reporter molecules on a silica core followed by their encapsulation in a silica shell that is then conjugated with secondary antibodies produced nanoparticle probes that Chang et al. named SERS dots [38]. These probes were used in a sandwich-type immunoassay to target the prostate-specific antigen (PSA) bound to an antibody-immobilized glass substrate in an attempt to scan the Raman spectra of the whole area all at once (Figure 5a). This SERS detection method was highly sensitive with an achieved LOD of 3.4 fM [38]. In another SERS-based immunoassay (SIA), gold nanoparticles loaded with Raman report molecules and functionalized with free or complexed PSA antibodies formed a sandwich immunocomplex with a total-PSA antibody conjugated magnetic bead in the presence of PSA from clinical serum samples (Figure 5b) [39]. The purpose of the experiment was to measure the ratio of free to total PSA for a more accurate diagnosis as a higher total PSA level and a lower percentage of free PSA are associated with a higher risk of prostate cancer. Cheng et al. were able to design a fast assay, less than 1 h, for the simultaneous detection of dual PSA markers with an LOD as low as 0.012 ng/mL for free PSA and 0.15 ng/mL for complexed PSA (c-PSA) [39].
Figure 5.
Schematics of surface-enhanced Raman scattering (SERS) assays for the detection of the prostate-specific antigen (PSA): (a) Simultaneous detection of free PSA and complexed PSA; (b) SERS signal area scanning for PSA detection. (Reproduced with permission from [38], published by American Chemical Society 2016; reproduced with permission from [39], published by American Chemical Society 2017).
However, the usage of specific antibodies could be disadvantageous due to their time-consuming and costly production. Furthermore, antibodies are prone to degradation due to their instability. To overcome this problem, Yang et al. used a magnetic aptasensor to detect and separate PSA from human serum samples [40]. Satellite AuNPs were functionalized with PSA-complementary DNA and bound to a core magnetic nanoparticle that was functionalized with PSA-aptamers. In the presence of PSA, the gold nanoparticles would be separated from the complex due to competitive binding and by doing so, once the magnetic NPs were removed from the solution by a magnet, the SERS signal was amplified with an LOD of 5.0 pg/mL [40].
In a similar way to SERS, the LSPR of metal NPs plays a role in metal enhanced fluorescence (MEF) by increasing the excitation state of a fluorophore. Doing so, the optical proprieties of the dye are changed: a higher quantum yield, better stability, and a shorter lifetime [41]. Based on this mechanism, Xu et al. were able to develop an immunoassay for the detection of PSA reaching an LOD of 27 pg/mL [42]. The experiment, based on Silica coated AgNPs combined with RuBpy fluorescent particles, required a total of 30 min. In another experiment by Choi et al., the LSPR of Ag nanocubes was used to enhance Alexa-488 fluorescence and achieve an LOD of 1 ng/mL for CEA [43]. Recent metal nanoparticles-based optical biosensors for circulating cancer biomarkers are compared in Table 2.
Table 2.
Metal nanoparticle-based optical biosensors for circulating cancer biomarkers.
| Sensing Mechanism | Target Biomarker | Detection Elements | Signal Elements | LOD | Reference |
|---|---|---|---|---|---|
| SPR 1 | Exosome (MCF-7) | CD63 aptamers | Au film, aptamer/T30 linked and A30 linked AuNPs 2 | 5 × 103 exosomes/mL | [34] |
| miRNA 3-21 and CTC 5 (SMMC-7721) |
Hairpin probes and cell-specific aptamers | Au film, DNA-linked AuNPs, and AgNPs 4 | 0.6 fM and 1 cell/µL |
[44] | |
| ctDNA 6 | Electrostatic interactions | Hexadecyltrimethylammonium bromide coated Au nanorods | 0.2 nM | [33] | |
| Colorimetry | CTC (MDA-MB-231) | CD44 ligands | AuNPs-conjugated hyaluronic acid | N/A | [36] |
| Exosome (C666-1) | Target specific antibodies | AuNP–DNA conjugates | 100 particles/mL | [45] | |
| Flt-1 7 | Target specific ligand peptides | Peptide-coated AuNPs | 0.2 nM | [46] | |
| miRNA-155 | Hairpin DNA probes | Citrate-capped and polyethyleneimine-capped AuNPs | 100 aM | [35] | |
| ctDNA (KRAS) | Complementary linkers | DNA oligonucleotides–functionalized AuNPs | 67 pM | [37] | |
| SERS 8 | CTC(HeLa and MCF-7) | Targeted specific ligand folic acid | Reductive bovine serum albumin-stabilized AuNP coated with 4-mercaptobenzoic acid | 5 cells/mL | [47] |
| Exosome (SKBR3, T84, and LNCaP) |
H2, CEA, and PSMA aptamers | 5,5′-dithiobis(2-nitrobenzoic acid), 2-naphthalenethiol or 7-mercapto-4-methylcoumarin labeled AuNPs | 32, 73, and 203 exosomes/µL |
[48] | |
| PSA 9 | Target specific antibodies | Raman label compound coated AgNPs bound to a silica core | 0.11 pg/mL | [38] | |
| PSA | PSA aptamers | 4,4′-dipyridyl-labeled AuNPs | 5.0 pg/mL | [40] | |
| free-PSA and complexed-PSA |
Target specific antibodies | Malachite green isothiocyanate and/or X-rhodamine-5-(and-6)-isothiocyanate labeled AuNPs | 0.012 ng/mL and 0.15 ng/mL | [39] | |
| MEF 10 | PSA | Target specific antibodies | Silica-coated AgNPs and RuBpy | 27 pg/mL | [42] |
| CEA 11 | Target specific antibodies | Ag nanocubes and Alexa-488 | 1 ng/mL | [43] |
1 Surface plasmon resonance. 2 Gold nanoparticles. 3 Micro-RNA. 4 Silver nanoparticles. 5 Circulating tumor cell. 6 Circulating tumor DNA. 7 Vascular endothelial growth factor receptor 1. 8 Surface-enhanced Raman scattering. 9 Prostate-specific antigen. 10 Metal enhanced fluorescence. 11 Carcinoembryonic antigen.
4. Upconversion Nanoparticle-Based Optical Biosensor for Circulating Cancer Biomarkers
Upconversion nanoparticles (UCNPs), principally from the lanthanide group, have good autofluorescence inhibition and deep tissue penetration which makes them excellent candidates in the biosensing field. The most interesting feature of UCNPs is their capacity to upconvert low-energy photons to high-energy photons when exposed to infrared light (700–1000 nm). By doing so, they can emit ultraviolet or visible lights after excitation with a 980 nm light, the most commonly used type of laser. UCNPs are composed of multiple elements: a host lattice that serves as a conducting structure, doped with activator and sensitizer ions for the transfer of energy [49]. It is also possible to change the UCNPs emitted light by changing their dopants, thus opening a door for potential multiplexing assays. It is possible to synthesize UCNPs through a few methods, namely, thermal decomposition, hydrothermal synthesis, or chemical co-precipitation [50]. Once synthesized it is possible to functionalize them and used them as sensing nanoprobes for the detection of circulating cancer biomarkers.
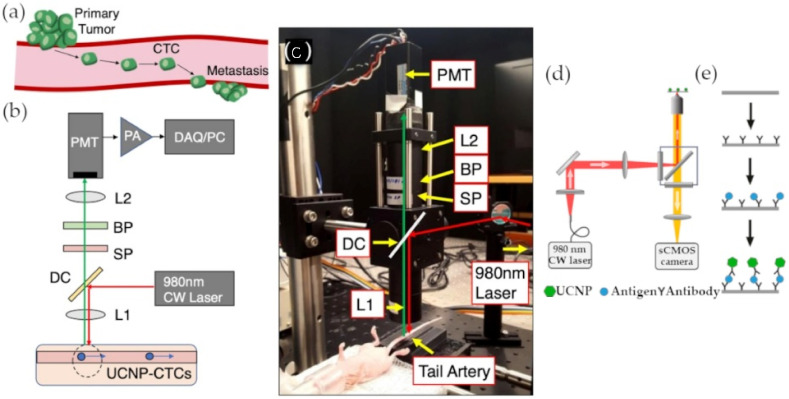
In a recent experiment by Guo et al., epithelial cell adhesion molecules (EpCAMs) were once again used as a target for the detection of circulating tumor cells (CTCs) [51]. NaEuF4-type UCNPs were synthesized through solid-liquid thermal decomposition and functionalized with anti-EpCAM antibodies. Microplate wells were also decorated with the same antibodies to retain only the EpCAM-expressing MCF-7 breast cancer cells from whole blood samples. The detection protocol combined a dissolution enhanced luminescence method, involving an enhancer solution that increased the release of Eu3+ from the UCNPs, with the measurement of a time-resolved photoluminescence (TRPL) signal where the intensity of the signal spike depended on the number of cancer cells. By doing so, it was possible to overcome the low concentration of CTCs by increasing the fluorescence of the UCNPs and neglecting the autofluorescence interference caused by other elements in the whole blood sample by delaying the TRPL signal reading. The nanoprobe achieved a limit of detection as low as 1 cell/well by utilizing UCNPs’ unprecedented optical properties [51]. In an attempt to exploit the low toxicity of lanthanide-based UCNPs, another advantage of the nanoparticles, Bartosik et al. developed a new laboratory-made instrument for the detection of CTCs named UCNP-compatible diffuse in vivo flow cytometry (U-DiFC) [52]. The instrument’s purpose is to measure the fluorescence emission of UCNPs from the tail artery of a mouse without any puncture site. The signal had to pass through multiple lenses and filters before getting amplified by a pre-amplifier and digitized by a multi-function data acquisition board (Figure 6a–c). Despite the promising results of the cells co-incubated with UCNPs and tested on an optical flow phantom model, the CTCs were not internalizing the UCNPs in vivo as a contrast agent should [52]. Nonetheless, the failure of the assay did not undermine the potential of UCNPs-based fluorescence detection of circulating cancer biomarkers.
Figure 6.
Upconversion nanoparticle (UCNP) detection settings. (a) Schematic of primary tumor metastasis; (b) schematic of in vivo CTCs detection by diffuse in vivo flow cytometry (U-DiFC); (c) photograph of U-DiFC setting; (d) schematic of in vitro circulating cancer biomarker detection setting; (e) schematic of UCNP sandwich immunoassay. (Reproduced with permission from [52], published by Dove Medical Press Limited 2020; reproduced with permission from [53], published by American Chemical Society 2017).
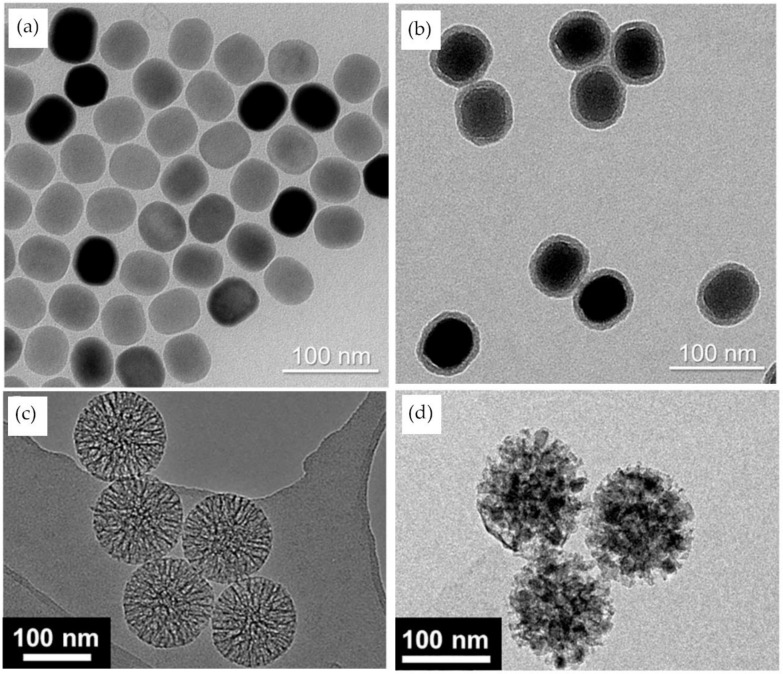
As the detection of other circulating biomarkers can occur earlier than the detection of circulating tumor cells, another team of researchers (Lan et al.) focused on developing an assay for the quantitative detection of vascular endothelial growth factor (VEGF), a circulating protein that has an increased concentration in breast cancer patients [54]. They used the thermal decomposition of rare-earth stearates to synthesize cheap and pollution-free NaYF4:Yb3+, Er3+ nanoparticle. They also designed a VEGF-specific aptamer that was divided into two portions. The UCNPs were then functionalized with a portion of the VEGF-specific aptamer, and the other portion was fixed on a 96-well microplate. By doing so, the two portions of the aptamer would form a complex only in the presence of the protein of interest and subsequently produce a 540 nm luminescent signal when excited by a 980 nm light [54]. Lan et al. achieved an LOD of 6 pM for the detection of VEGF, in breast cancer, but with a similar experiment, Farka et al. were able to lower the LOD to 42 fM for PSA, a circulating protein in prostate cancer (Figure 6d,e) [53]. The biosensor was based on a single molecule upconversion-linked immunosorbent assay (ULISA). After synthesizing β-NaYF4:18 mol % Yb3+, 2 mol % Er3+ type UCNPs by high-temperature coprecipitation, the particles were coated with silica (Figure 7a,b) and functionalized with anti-PSA antibodies. The same antibodies were also fixed on a microtiter plate to form a sandwich immunoassay in the presence of the PSA protein and the UCNPs-based detection probe. The number of complexed UCNPs was then automatically counted by an upconversion epiluminescence microscopy software [53].
Figure 7.
Combination of UCNPs with a silica core or shell: (a) TEM image of oleic acid-capped UCNPs; (b) TEM image of UCNPs with carboxylated silica shell; (c) SEM of mesoporous silica nanoparticles; (d) TEM image of CaF2: RE3+ UCNPs embedded mesoporous silica nanoparticles. (Reproduced with permission from [53], published by American Chemical Society 2017; reproduced with permission from [55], published by Elsevier 2019).
It is often the combinatory analysis of multiple biomarkers that is relevant in cancer biology. Herein, to be able to simultaneously quantify the presence of multiple types of miRNA, Gu et al. designed a fluorescence detection method based on mesoporous silica nanoparticles embedded with upconversion nanocrystals (UCNCs) [55]. Before forming the nanocrystals by thermo-decomposition, a precursor had to be introduced into the mesoporous silica particles through a capillary effect (Figure 7c,d). To be able to detect multiple targets in the same sample, different lanthanide dopants, Tm3+ or Ho3+, were used in a combination with different DNA sequences to target breast-related miRNA-195 and miRNA-21. The experiment was successful in establishing a correlation between the fluorescence of the nanoprobes and the variable concentration of a mixture of miRNAs with a reported LOD of 100 nM for miRNA-195 [55]. However, despite the multiplexing potential of this assay, a significantly lower limit of detection was achieved by the UCNPs FRET-based sensors.
UCNPs are frequently used in fluorescence resonance energy transfer assays due to the fact that they are excellent energy donors. To exploit that quality of UCNPs, Chen et al. constructed a paper-supported biosensor for the detection of exosomes [56]. The aptameric sequence of CD63, a protein expressed on the surface of exosomes, was divided into two portions. One portion was attached to the UCNPs (energy donors) and then bound to a filter paper. The other portion was linked to Au nanorods (energy receptors). In the absence of the exosome, the UCNPs would produce a green fluorescence when exposed to a 980 nm light. However, in the presence of the exosome, both fragments, and subsequently nanoparticles, would be brought together, causing the quenching of the fluorescence proportional to the number of exosomes with an LOD of 1.1 × 103 particles/µL within approximately 30 min [56]. A year later, Wang et al. were able to achieve a significantly lower LOD of 80 particles/µL with another paper-based aptasensor for the detection of exosomes [57]. In this experiment, an EpCAM aptamer was divided between an energy donor (UCNPs) and an energy receptor (tetramethylrhodamine (TAMRA)). Due to the energy transfer between both particles, TAMRA emits a yellow fluorescence. By quantifying TAMRA’s emissions, it was possible to determine the concentration of EpCAM expressing exosomes [57]. In another assay by Wang et al., it was possible to achieve an LOD of 6.30 pM for ctDNA by using gold nanocages as energy acceptors [58]. The UCNPs were designed to competitively bind to ctDNA and free themselves from the quenching effect of the gold nanocage. Emerging materials such as graphene oxide are also demonstrating promising results in the field. A UCNP-based DNA biosensor, where graphene oxide was used as an energy acceptor, achieved an LOD of 5 pM [59]. A similar sensor by Vilela et al. demonstrated an LOD of 500 fM for PCA3 miRNA, a prostate cancer biomarker, detection [60]. Both sensors took advantage of the quenching property and the energy acceptor potential of graphene oxides. Recent upconversion nanoparticles-based optical biosensors for circulating cancer biomarkers are compared in Table 3.
Table 3.
Upconversion nanoparticle-based optical biosensors for circulating cancer biomarkers.
| Sensing Mechanism | Target Biomarker | Recognition Elements | Signal Elements | LOD | Reference |
|---|---|---|---|---|---|
| Fluorescence | CTC 1 (MCF-7) | Anti-EpCAM antibodies | NaEuF4 UCNPs 2 | ≥10 cells/mL | [53] |
| VEGF 3 | Target specific aptamers | α-NaYF4:Yb3+, Er3+ UCNPs | 6 pM | [55] | |
| PSA 4 | Anti-PSA antibodies | β-NaYF4:18 mol % Yb3+, 2 mol % Er3+ UCNPs | 42 fM | [52] | |
| miRNA 5-195 and miRNA-21 | oligonucleotide probes | CaF2: RE3+ upconversion nanocrystals doped with Ho3+, Tm3+ or Yb3+ | 100 nM | [54] | |
| FRET 6 | Exosome (MDA-MB-231 and MCF-7) | EpCAM aptamer | NaYF4:Yb,Er UCNPs (donors) and Tetramethyl rhodamine (acceptors) | 80 particles/µL | [57] |
| Exosome (HepG2) | CD63 aptamer | NaYF4:Yb, Er UCNPs (donors) and Au nanorods (acceptors) | 1.1 × 103 particles/µL | [56] | |
| CA125 | CA125 aptamer | Polyacrylic acid coated NaYF4:Yb,Er UCNPs (donors) and carbon dots (acceptors) | 9 × 10−3 U/mL | [61] | |
| CA125 | Anti-CA125 antibodies | PEI coated NaYF4:Yb,Tm UCNPs (donors) and AgNPs 7 (acceptors) | 120 pg/mL | [62] | |
| PSA | Anti-PSA antibodies | NaYF4:Yb3+, Er3+ UCNPs (donors) and AuNPs 8 (acceptors) | 2.3 pM | [63] | |
| PSA | Anti-PSA antibodies | NaYF4:Yb3+,Er3+ and NaYF4:Yb3+,Er3+@NaYF4:Yb3+,Nd3+ (donors) and AuNPs (acceptors) | 0.01 ng/mL | [64] | |
| CEA 9 | Anti-CEA antibodies | NaYF4:Yb,Tm UCNPs (donors) and Fluorescein (acceptor) | 0.89 ng/mL | [65] | |
| CEA | CEA aptamer | UCNPs (donors) and graphene oxide (acceptors) | 7.9 pg/mL | [66] | |
| PCA3 miRNA | Oligonucleotide probes | NaYF4:Yb,Er UCNPs (donors) and graphene oxide (acceptors) | 500 fM | [60] | |
| ctDNA 10 (KRAS) | Oligonucleotide probes | NaYF4:Yb:Tm UCNPs (donors) and Au nanocages (acceptors) | 6.30 pM | [58] | |
| Single strand DNA | Oligonucleotide probes | SiO2 coated NaYF4:Yb,Er UCNPs, (donors) and graphene oxide (acceptors) | 5 pM | [59] |
1 Circulating tumor cell. 2 Upconversion nanoparticles. 3 Vascular endothelial growth factor. 4 Prostate-specific antigen. 5 Micro-RNA. 6 Fluorescence resonance energy transfer. 7 Silver nanoparticles. 8 Gold nanoparticles. 9 Carcinoembryonic antigen. 10 Circulating tumor DNA.
5. Conclusions
This review covered the most recent advancements in nanoparticle-based optical sensors for the detection of circulating cancer biomarkers. Nanoparticles have repeatedly proven their potential to overcome some of the most difficult aspects of liquid biopsy analysis. Their surface-to-volume ratio, multiplexing capacity, and stable optical proprieties are considerable advantages in the biosensing field (Table 4). However, bearing in mind the complexity of most biological fluids, it is still difficult to achieve both high sensitivity and specific targeting of biomarkers due to multiple factors, such as sample autofluorescence and immunoassay crosstalk, among other things.
Table 4.
Advantages and disadvantages of metal nanoparticles, quantum dots and upconversion nanoparticles.
| Nanoparticle Type | Advantages | Disadvantages |
|---|---|---|
| Metal nanoparticles | Multiple shapes Conductivity Localized surface plasmon resonance Good biological affinity Good energy acceptor |
No fluorescence |
| Quantum Dots | High quantum yield Broadband excitation Multiplexing Size-tunable fluorescence High photobleaching threshold |
Toxicity Blinking effect |
| Upconversion nanoparticles | Anti-Stokes luminescent Autofluorescence inhibition Multiplexing High chemical stability Deep tissue penetration Dopants-tunable fluorescence |
Toxicity Restricted Quantum Yield |
Although the current research focuses mostly on the development of new mechanisms of detection and on improving the LOD of the existing ones, a shift is to be expected in the near-future towards more point-of-care-oriented sensors, thus making cancer diagnostic easier, less time consuming, and potentially more affordable to the general population. In vivo fluorescence imaging of circulating cancer biomarkers is also a promising development for the future of optical nanobiosensors, but this will require extended studies on the cytotoxicity of the nanoparticles. For now, the focus will be not only on easing the synthesis and the functionalization of those particles but also on improving their fluorescence by increasing their quantum yield or combining other enhancing mechanisms.
Author Contributions
Conceptualization, A.K.S. and C.A.; validation, J.-H.L.; investigation, C.A.; resources, J.-H.L.; data curation, C.A.; writing—original draft preparation, C.A.; writing—review and editing, C.A. and J.-H.L.; visualization, A.K.S. and C.A.; supervision, J.-H.L.; project administration, J.-H.L.; funding acquisition, J.-H.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2020R1C1C1011876) and Pusan National University Research Grant, 2020.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Guo J., Ma K., Bao H., Ma X., Xu Y., Wu X., Shao Y.W., Jiang M., Huang J. Quantitative characterization of tumor cell-free DNA shortening. BMC Genom. 2020;21:1–11. doi: 10.1186/s12864-020-06848-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muhanna N., Di Grappa M.A., Chan H.H.L., Khan T., Jin C.S., Zheng Y., Irish J.C., Bratman S.V. Cell-Free DNA Kinetics in a Pre-Clinical Model of Head and Neck Cancer. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-17079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H., Peng R., Wang J., Qin Z., Xue L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenet. 2018;10:1–10. doi: 10.1186/s13148-018-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H., Jing C., Wu J., Ni J., Sha H., Xu X., Du Y., Lou R., Dong S., Feng J. Circulating tumor DNA detection: A potential tool for colorectal cancer management (Review) Oncol. Lett. 2018;17:1409–1416. doi: 10.3892/ol.2018.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., Bartlett B.R., Wang H., Luber B., Alani R.M., et al. Detection of Circulating Tumor DNA in Early and Late-Stage Human Malignancies. Sci. Transl. Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith C.J., Osborn A.M. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol. Ecol. 2009;67:6–20. doi: 10.1111/j.1574-6941.2008.00629.x. [DOI] [PubMed] [Google Scholar]
- 7.Cui M., Wang H., Yao X., Zhang D., Xie Y., Cui R., Zhang X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019;10:626. doi: 10.3389/fgene.2019.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang X., Huuskonen J., Hajivandi M., Manzanedo R., Predki P., Amshey J.R., Pope R.M. Identification and quantification of proteins differentially secreted by a pair of normal and malignant breast-cancer cell lines. Proteomics. 2009;9:182–193. doi: 10.1002/pmic.200700957. [DOI] [PubMed] [Google Scholar]
- 9.Mor G., Visintin I., Lai Y., Zhao H., Schwartz P., Rutherford T., Yue L., Bray-Ward P., Ward D.C. Serum protein markers for early detection of ovarian cancer. Proc. Natl. Acad. Sci. USA. 2005;102:7677–7682. doi: 10.1073/pnas.0502178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang T., Deng C.-X. Current Progresses of Exosomes as Cancer Diagnostic and Prognostic Biomarkers. Int. J. Biol. Sci. 2019;15:1–11. doi: 10.7150/ijbs.27796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai J., Su Y., Zhong S., Cong L., Liu B., Yang J., Tao Y., He Z., Chen C., Jiang Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020;5:1–10. doi: 10.1038/s41392-020-00261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Micalizzi D.S., Maheswaran S., Haber D.A. A conduit to metastasis: Circulating tumor cell biology. Genes Dev. 2017;31:1827–1840. doi: 10.1101/gad.305805.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong S., Lee C. The Current Status and Future Outlook of Quantum Dot-Based Biosensors for Plant Virus Detection. Plant Pathol. J. 2018;34:85–92. doi: 10.5423/PPJ.RW.08.2017.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu H., Xu J., Wang X., Wu D., Chen Z.G., Wang A.Y. Quantum Dot-Based, Quantitative, and Multiplexed Assay for Tissue Staining. ACS Appl. Mater. Interfaces. 2013;5:2901–2907. doi: 10.1021/am303267g. [DOI] [PubMed] [Google Scholar]
- 15.Valizadeh A., Mikaeili H., Samiei M., Farkhani S.M., Zarghami N., Kouhi M., Akbarzadeh A., Davaran S. Quantum dots: Synthesis, bioapplications and toxicity. Nanoscale Res. Lett. 2012;7:480. doi: 10.1186/1556-276X-7-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De la Calle I., Romero-Rivas V. Chapter 9—The Role of Nanomaterials in Analytical Chemistry: Trace Metal Analysis. In: Mohan Bhagyaraj S., Oluwafemi O.S., Kalarikkal N., Thomas S., editors. Applications of Nanomaterials. Woodhead; Sawston, UK: 2018. pp. 251–301. [Google Scholar]
- 17.Schiffman J.D., Balakrishna R.G. Quantum dots as fluorescent probes: Synthesis, surface chemistry, energy transfer mechanisms, and applications. Sens. Actuators B Chem. 2018;258:1191–1214. [Google Scholar]
- 18.Seong-Min J., Jo S.-M., Kim H.-S. Efficient Capture and Simple Quantification of Circulating Tumor Cells using Quantum Dots and Magnetic Beads. Small. 2015;11:2536–2542. doi: 10.1002/smll.201403126. [DOI] [PubMed] [Google Scholar]
- 19.Cui F., Ji J., Sun J., Wang J., Wang H., Zhang Y., Ding H., Lu Y., Xu D., Sun X. A novel magnetic fluorescent biosensor based on graphene quantum dots for rapid, efficient, and sensitive separation and detection of circulating tumor cells. Anal. Bioanal. Chem. 2019;411:985–995. doi: 10.1007/s00216-018-1501-0. [DOI] [PubMed] [Google Scholar]
- 20.Kuo C.W., Chueh D.-Y., Chen P. Real-time in vivo imaging of subpopulations of circulating tumor cells using antibody conjugated quantum dots. J. Nanobiotechnol. 2019;17:1–10. doi: 10.1186/s12951-019-0453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nejdl L., Zelnickova J., Vaneckova T., Hynek D., Adam V., Vaculovicova M. Rapid preparation of self-assembled CdTe quantum dots used for sensing of DNA in urine. New J. Chem. 2018;42:6005–6012. doi: 10.1039/C7NJ05167K. [DOI] [Google Scholar]
- 22.Wu M., Chen Z., Xie Q., Xiao B., Zhou G., Chen G., Bian Z. One-step quantification of salivary exosomes based on combined aptamer recognition and quantum dot signal amplification. Biosens. Bioelectron. 2021;171:112733. doi: 10.1016/j.bios.2020.112733. [DOI] [PubMed] [Google Scholar]
- 23.Wu S., Liu L., Li G., Jing F., Mao H., Jin Q., Zhai W., Zhang H., Zhao J., Jia C. Multiplexed detection of lung cancer biomarkers based on quantum dots and microbeads. Talanta. 2016;156:48–54. doi: 10.1016/j.talanta.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Meng X., Chen X., Wu W., Zheng W., Deng H., Xu L., Chen W., Li Z., Peng H. Electrochemiluminescent immunoassay for the lung cancer biomarker CYFRA21-1 using MoOx quantum dots. Microchim. Acta. 2019;186:855. doi: 10.1007/s00604-019-3917-4. [DOI] [PubMed] [Google Scholar]
- 25.Mahani M., Mousapour Z., Divsar F., Nomani A., Ju H. A carbon dot and molecular beacon based fluorometric sensor for the cancer marker microRNA-21. Microchim. Acta. 2019;186:132. doi: 10.1007/s00604-019-3233-z. [DOI] [PubMed] [Google Scholar]
- 26.Sun M.-F., Liu J.-L., Chai Y.-Q., Zhang J., Tang Y., Yuan R. Three-Dimensional Cadmium Telluride Quantum Dots—DNA Nanoreticulation as a Highly Efficient Electrochemiluminescent Emitter for Ultrasensitive Detection of MicroRNA from Cancer Cells. Anal. Chem. 2019;91:7765–7773. doi: 10.1021/acs.analchem.9b01185. [DOI] [PubMed] [Google Scholar]
- 27.Li J., Liu J., Bi Y., Sun M., Bai J., Zhou M. Ultrasensitive electrochemiluminescence biosensing platform for miRNA-21 and MUC1 detection based on dual catalytic hairpin assembly. Anal. Chim. Acta. 2020;1105:87–94. doi: 10.1016/j.aca.2020.01.034. [DOI] [PubMed] [Google Scholar]
- 28.Lian W., Tu D., Hu P., Song X., Gong Z., Chen T., Song J., Chen Z., Chen X. Broadband excitable NIR-II luminescent nano-bioprobes based on CuInSe2 quantum dots for the detection of circulating tumor cells. Nano Today. 2020;35:100943. doi: 10.1016/j.nantod.2020.100943. [DOI] [Google Scholar]
- 29.Zhou L., Ji F., Zhang T., Wang F., Li Y., Yu Z., Jin X., Ruan B. An fluorescent aptasensor for sensitive detection of tumor marker based on the FRET of a sandwich structured QDs-AFP-AuNPs. Talanta. 2019;197:444–450. doi: 10.1016/j.talanta.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Li N.-L., Jia L.-P., Ma R.-N., Jia W.-L., Lu Y.-Y., Shi S.-S., Wang H.-S. A novel sandwiched electrochemiluminescence immunosensor for the detection of carcinoembryonic antigen based on carbon quantum dots and signal amplification. Biosens. Bioelectron. 2017;89:453–460. doi: 10.1016/j.bios.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 31.Gao M.-L., He F., Yin B.-C., Ye B.-C. A dual signal amplification method for exosome detection based on DNA dendrimer self-assembly. Analyst. 2019;144:1995–2002. doi: 10.1039/C8AN02383B. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H., Wang Z., Wang F., Zhang Y., Wang H., Liu Y. In Situ Formation of Gold Nanoparticles Decorated Ti3C2 MXenes Nanoprobe for Highly Sensitive Electrogenerated Chemiluminescence Detection of Exosomes and their Surface Proteins. Anal. Chem. 2020;92:5546–5553. doi: 10.1021/acs.analchem.0c00469. [DOI] [PubMed] [Google Scholar]
- 33.Pallares R.M., Kong S.L., Ru T.H., Thanh N.T.K., Lu Y., Su X. A plasmonic nanosensor with inverse sensitivity for circulating cell-free DNA quantification. Chem. Commun. 2015;51:14524–14527. doi: 10.1039/C5CC05331E. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q., Zou L., Yang X., Liu X., Nie W., Zheng Y., Cheng Q., Wang K. Direct quantification of cancerous exosomes via surface plasmon resonance with dual gold nanoparticle-assisted signal amplification. Biosens. Bioelectron. 2019;135:129–136. doi: 10.1016/j.bios.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Hakimian F., Ghourchian H., Hashemi A.S., Arastoo M.R., Rad M.B. Ultrasensitive optical biosensor for detection of miRNA-155 using positively charged Au nanoparticles. Sci. Rep. 2018;8:1–9. doi: 10.1038/s41598-018-20229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauta P.R., Hallur P.M., Chaubey A. Gold nanoparticle-based rapid detection and isolation of cells using ligand-receptor chemistry. Sci. Rep. 2018;8:1–13. doi: 10.1038/s41598-018-21068-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Kong S.L., Di Su X. A centrifugation-assisted visual detection of SNP in circulating tumor DNA using gold nanoparticles coupled with isothermal amplification. RSC Adv. 2020;10:1476–1483. doi: 10.1039/C9RA09029K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang H., Kang H., Ko E., Jun B.-H., Lee H.-Y., Lee Y.-S., Jeong D.H. PSA Detection with Femtomolar Sensitivity and a Broad Dynamic Range using SERS Nanoprobes and an Area-Scanning Method. ACS Sens. 2016;1:645–649. doi: 10.1021/acssensors.6b00053. [DOI] [Google Scholar]
- 39.Cheng Z., Choi N., Wang R., Lee S., Moon K.C., Yoon S.-Y., Chen L., Choo J. Simultaneous Detection of Dual Prostate Specific Antigens using Surface-Enhanced Raman Scattering-Based Immunoassay for Accurate Diagnosis of Prostate Cancer. ACS Nano. 2017;11:4926–4933. doi: 10.1021/acsnano.7b01536. [DOI] [PubMed] [Google Scholar]
- 40.Yang K., Hu Y., Dong N., Zhu G., Zhu T., Jiang N. A novel SERS-based magnetic aptasensor for prostate specific antigen assay with high sensitivity. Biosens. Bioelectron. 2017;94:286–291. doi: 10.1016/j.bios.2017.02.048. [DOI] [PubMed] [Google Scholar]
- 41.Jeong Y., Kook Y.-M., Lee K., Koh W.-G. Metal enhanced fluorescence (MEF) for biosensors: General approaches and a review of recent developments. Biosens. Bioelectron. 2018;111:102–116. doi: 10.1016/j.bios.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Xu D.-D., Deng Y.-L., Li C.-Y., Lin Y., Tang H.-W. Metal-enhanced fluorescent dye-doped silica nanoparticles and magnetic separation: A sensitive platform for one-step fluorescence detection of prostate specific antigen. Biosens. Bioelectron. 2017;87:881–887. doi: 10.1016/j.bios.2016.09.034. [DOI] [PubMed] [Google Scholar]
- 43.Liu J., Li S., Bhethanabotla V.R. Integrating Metal-Enhanced Fluorescence and Surface Acoustic Waves for Sensitive and Rapid Quantification of Cancer Biomarkers from Real Matrices. ACS Sens. 2018;3:222–229. doi: 10.1021/acssensors.7b00876. [DOI] [PubMed] [Google Scholar]
- 44.Liu R., Wang Q., Li Q., Yang X., Wang K., Nie W. Surface plasmon resonance biosensor for sensitive detection of microRNA and cancer cell using multiple signal amplification strategy. Biosens. Bioelectron. 2017;87:433–438. doi: 10.1016/j.bios.2016.08.090. [DOI] [PubMed] [Google Scholar]
- 45.Liu W., Li J., Wu Y., Xing S., Lai Y., Zhang G. Target-induced proximity ligation triggers recombinase polymerase amplification and transcription-mediated amplification to detect tumor-derived exosomes in nasopharyngeal carcinoma with high sensitivity. Biosens. Bioelectron. 2018;102:204–210. doi: 10.1016/j.bios.2017.11.033. [DOI] [PubMed] [Google Scholar]
- 46.Wei L., Wang X., Li C., Li X., Yin Y., Li G. Colorimetric assay for protein detection based on “nano-pumpkin” induced aggregation of peptide-decorated gold nanoparticles. Biosens. Bioelectron. 2015;71:348–352. doi: 10.1016/j.bios.2015.04.072. [DOI] [PubMed] [Google Scholar]
- 47.Wu X., Luo L., Yang S., Ma X., Li Y., Dong C., Tian Y., Zhang L., Shen Z., Wu A. Improved SERS Nanoparticles for Direct Detection of Circulating Tumor Cells in the Blood. ACS Appl. Mater. Interfaces. 2015;7:9965–9971. doi: 10.1021/acsami.5b02276. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z., Zong S., Wang Y., Li N., Li L., Lu J., Wang Z., Chen B., Cui Y. Screening and multiple detection of cancer exosomes using an SERS-based method. Nanoscale. 2018;10:9053–9062. doi: 10.1039/C7NR09162A. [DOI] [PubMed] [Google Scholar]
- 49.Chen G., Qiu H., Prasad P.N., Chen X. Upconversion Nanoparticles: Design, Nanochemistry, and Applications in Theranostics. Chem. Rev. 2014;114:5161–5214. doi: 10.1021/cr400425h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu X., Zhang J., Liu J., Zhang Y. Recent Progress of Rare-Earth Doped Upconversion Nanoparticles: Synthesis, Optimization, and Applications. Adv. Sci. 2019;6:1901358. doi: 10.1002/advs.201901358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo H., Song X., Lei W., He C., You W., Lin Q., Zhou S., Chen X., Chen Z. Direct Detection of Circulating Tumor Cells in Whole Blood using Time-Resolved Luminescent Lanthanide Nanoprobes. Angew. Chem. 2019;131:12323–12327. doi: 10.1002/ange.201907605. [DOI] [PubMed] [Google Scholar]
- 52.Bartosik P.B., Fitzgerald J.E., El Khatib M., Yaseen M.A., Vinogradov S.A., Niedre M. Prospects for the use of Upconverting Nanoparticles as a Contrast Agent for Enumeration of Circulating Cells in vivo. Int. J. Nanomed. 2020;15:1709–1719. doi: 10.2147/IJN.S243157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farka Z., Mickert M.J., Hlaváček A., Skládal P., Gorris H.H. Single Molecule Upconversion-Linked Immunosorbent Assay with Extended Dynamic Range for the Sensitive Detection of Diagnostic Biomarkers. Anal. Chem. 2017;89:11825–11830. doi: 10.1021/acs.analchem.7b03542. [DOI] [PubMed] [Google Scholar]
- 54.Lan J., Li L., Liu Y., Yan L., Li C., Chen J., Chen X. Upconversion luminescence assay for the detection of the vascular endothelial growth factor, a biomarker for breast cancer. Microchim. Acta. 2016;183:3201–3208. doi: 10.1007/s00604-016-1965-6. [DOI] [Google Scholar]
- 55.Gu T., Li Z., Ren Z., Li X., Han G. Rare-earth-doped upconversion nanocrystals embedded mesoporous silica nanoparticles for multiple microRNA detection. Chem. Eng. J. 2019;374:863–869. doi: 10.1016/j.cej.2019.05.223. [DOI] [Google Scholar]
- 56.Chen X., Lan J., Liu Y., Li L., Chunyan L., Xiaosong C., Wu F., Li C., Li S., Chen J. A paper-supported aptasensor based on upconversion luminescence resonance energy transfer for the accessible determination of exosomes. Biosens. Bioelectron. 2018;102:582–588. doi: 10.1016/j.bios.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y., Luo D., Fang Y., Wu W., Wang Y., Xia Y., Wu F., Li C., Lan J., Chen J. An aptasensor based on upconversion nanoparticles as LRET donors for the detection of exosomes. Sens. Actuators B Chem. 2019;298:126900. doi: 10.1016/j.snb.2019.126900. [DOI] [Google Scholar]
- 58.Wang J., Hua G., Li L., Li D., Wang F., Wu J., Ye Z., Zhou X., Ye S.-F., Danyang L., et al. Upconversion nanoparticle and gold nanocage satellite assemblies for sensitive ctDNA detection in serum. Analyst. 2020;145:5553–5562. doi: 10.1039/D0AN00701C. [DOI] [PubMed] [Google Scholar]
- 59.Alonso-Cristobal P., Vilela P., El-Sagheer A., Lopez-Cabarcos E., Brown T., Muskens O.L., Rubio-Retama J., Kanaras A.G. Highly Sensitive DNA Sensor Based on Upconversion Nanoparticles and Graphene Oxide. ACS Appl. Mater. Interfaces. 2015;7:12422–12429. doi: 10.1021/am507591u. [DOI] [PubMed] [Google Scholar]
- 60.Vilela P., El-Sagheer A., Millar T.M., Brown T., Muskens O.L., Kanaras A.G. Graphene Oxide-Upconversion Nanoparticle Based Optical Sensors for Targeted Detection of mRNA Biomarkers Present in Alzheimer’s Disease and Prostate Cancer. ACS Sens. 2017;2:52–56. doi: 10.1021/acssensors.6b00651. [DOI] [PubMed] [Google Scholar]
- 61.Zhang X., Wang Y., Deng H., Xiong X., Zhang H., Liang T., Li C. An aptamer biosensor for CA125 quantification in human serum based on upconversion luminescence resonance energy transfer. Microchem. J. 2021;161:105761. doi: 10.1016/j.microc.2020.105761. [DOI] [Google Scholar]
- 62.Liu J., Xu S., Sun L., Hu S., Sun J., Liu M., Ma C., Liu H., Wang Z., Yang Y., et al. Up-conversion fluorescence biosensor for sensitive detection of CA-125 tumor markers. J. Rare Earths. 2019;37:943–948. doi: 10.1016/j.jre.2019.03.018. [DOI] [Google Scholar]
- 63.Li X., Wei L., Pan L., Yi Z., Wang X., Ye Z., Xiao L., Li H.-W., Wang J. Homogeneous Immunosorbent Assay Based on Single-Particle Enumeration using Upconversion Nanoparticles for the Sensitive Detection of Cancer Biomarkers. Anal. Chem. 2018;90:4807–4814. doi: 10.1021/acs.analchem.8b00251. [DOI] [PubMed] [Google Scholar]
- 64.Hu S., Xu H., Zhou B., Xu S., Shen B., Dong B., Yin Z., Xu S., Sun L., Lv J., et al. Double Stopband Bilayer Photonic Crystal Based Upconversion Fluorescence PSA Sensor. Sens. Actuators B Chem. 2021;326:128816. doi: 10.1016/j.snb.2020.128816. [DOI] [Google Scholar]
- 65.Xu S., Dong B., Zhou N., Yin Z., Cui S., Xu W., Chen B., Song H. Paper-based upconversion fluorescence resonance energy transfer biosensor for sensitive detection of multiple cancer biomarkers. Sci. Rep. 2016;6:23406. doi: 10.1038/srep23406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y., Wei Z., Luo X., Wan Q., Qiu R., Wang S. An ultrasensitive homogeneous aptasensor for carcinoembryonic antigen based on upconversion fluorescence resonance energy transfer. Talanta. 2019;195:33–39. doi: 10.1016/j.talanta.2018.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article.