Abstract
Simple Summary
The global importance of rare variants in tumorigenesis has been addressed by some pan-cancer analysis, revealing significant enrichments in protein-truncating variants affecting genes such as ATM, BRCA1/2, BRIP1, and MSH6. Germline variants can influence treatment response and contribute to the development of treatment-related second neoplasms, especially in childhood leukemia. We aimed to analyze the genomes of patients with B-cell lymphoproliferative disorders for the discovery of genes enriched in rare pathogenic variants. We discovered a significant enrichment for two genes in germline rare and dysfunctional variants. Additionally, we detected rare and likely pathogenic variants associated with disease prognosis and potential druggability, indicating a relevant role of these events in the variability of cancer phenotypes.
Abstract
There is growing evidence indicating the implication of germline variation in cancer predisposition and prognostication. Here, we describe an analysis of likely disruptive rare variants across the genomes of 726 patients with B-cell lymphoid neoplasms. We discovered a significant enrichment for two genes in rare dysfunctional variants, both of which participate in the regulation of oxidative stress pathways (CHMP6 and GSTA4). Additionally, we detected 1675 likely disrupting variants in genes associated with cancer, of which 44.75% were novel events and 7.88% were protein-truncating variants. Among these, the most frequently affected genes were ATM, BIRC6, CLTCL1A, and TSC2. Homozygous or germline double-hit variants were detected in 28 cases, and coexisting somatic events were observed in 17 patients, some of which affected key lymphoma drivers such as ATM, KMT2D, and MYC. Finally, we observed that variants in six different genes were independently associated with shorter survival in CLL. Our study results support an important role for rare germline variation in the pathogenesis and prognosis of B-cell lymphoid neoplasms.
Keywords: germline, rare variant, cancer, lymphoid, B-cell, lymphoma, CLL, driver, prognosis
1. Introduction
B-cell lymphoid neoplasms are the most frequent hematological tumors, and they exhibit a diverse spectrum of entities with heterogeneous clinical behavior. B-cell lymphoid neoplasms are classically classified in either aggressive lymphomas (diffuse large B-cell lymphoma (DLBCL), Burkitt lymphoma, grade III follicular lymphoma, and mantle cell lymphomas) or indolent lymphomas (e.g., chronic lymphocytic leukemia (CLL), grade I/II follicular lymphoma, marginal zone lymphoma, lymphoplasmacytic lymphoma). By frequency, diffuse large B-cell lymphoma (DLBCL) is the most frequent lymphoid neoplasm, accounting for 25% of all cases of non-Hodgkin lymphoma (NHL), closely followed by CLL (19% of NHLs) and follicular lymphoma (12% of NHLs) [1].
Next-generation sequencing (NGS) technologies have deconvoluted the genomic complexity of B-cell lymphoid tumors to a great extent, revealing the most frequent molecular drivers of disease and the interplay among them. NHL cases show familial predisposition, and much of the heritability of these diseases is still unexplained [2]. Genome-wide association analysis (GWAS) have identified the existence of polymorphisms significantly associated with risk of CLL [3], DLBCL [4], and follicular lymphoma [5]. Similarly, some polymorphisms are also related with the outcome of B-cell lymphomas [6,7,8] and CLL [9], and it has also been proved that some variants cooperate with somatic events in shaping clinical outcomes of cancer patients [10]. Another source of germline variation consists of rare variants (allele frequency <0.1–1%). The global importance of such rare variants in tumorigenesis has been addressed by pan-cancer analysis, revealing significant enrichments for protein truncating variants in genes such as ATM, BRCA1/2, BRIP1, and MSH6 [11]. Indeed, some of these variants predispose to cancer development through the acquisition of second somatic hits [12], such as point mutations or loss-of-heterozygosity (LOH) [13]. Additionally, germline variation can influence treatment response and contribute to the development of treatment-related second neoplasms, especially in childhood leukemia [14]. Many such rare variants in cancer-related genes have been associated with particular cancer subtypes [15,16,17], but until now little attention has been focused on the genome-wide frequency, pathogenicity, and clinical implications of rare variants in lymphoid malignancies. Rare variants in ATM and CDK1 variants have been associated with CLL risk in genome-wide analysis [18], whereas evidence for the implication of infrequent events in other genes come from familial studies or single-gene analysis [19,20,21].
In this report, we performed an exploratory analysis about the frequency and distribution of rare and putatively pathogenic germline variants in the genome of several mature B-cell lymphoid neoplasms using high-throughput sequencing data produced by the International Cancer Genome Consortium (ICGC) [22]. Our results indicate the existence of multiple genes affected by highly pathogenic germline variants in the genome of these patients, some of which seem to condition patient survival.
2. Materials and Methods
2.1. Data Source
We processed germline next-generation sequencing data from 726 patients with B-cell lymphoid malignancies produced by the International Cancer Genome Consortium. Briefly, 504 cases pertained to the Spanish Chronic Lymphocytic Leukemia project, and 222 were retrieved from the German Malignant Lymphoma project. Overall, there were 504 chronic lymphocytic leukemia (CLL) or small lymphocytic lymphoma (SLL) cases (including 54 monoclonal B-cell lymphocytosis cases), 97 follicular lymphoma cases, 85 diffuse large B-cell lymphoma (DLBCL) cases, 36 Burkitt lymphoma cases, and 4 unclassified B-cell lymphoma cases. CLL control samples were derived from non-tumoral leukocytes (<2% tumor contamination), whereas lymphoma controls originated from whole blood or buffy coats checked for negative clonality analysis. Sample collection and sequencing was originally performed by the ICGC consortium.
2.2. Germline Variant Identification and Annotation
Most CLL germline samples (440 out of 502) were processed using exome-sequencing kits (Agilent SureSelect Human All Exon V4 and V4+UTRs), whereas whole-genome sequencing was performed on 262 cases, which included 62 CLL cases and the entire cohort of B-cell lymphomas included in the Malignant Lymphoma-Deutcheland (MALY-DE) project. We restricted our analysis to protein coding regions covered by the exome-sequencing kits. Variants were detected using the optimized bcbio-nextgen (version 1.1.5) pipeline [23], and the GRCh37.75 assembly was used as reference. Four different variant callers were used: freebayes (version 1.1.0.46) [24], GATK-Haplotype (GATK version 2.8) [25], Platypus (version 0.8.1.2) [26], and Samtools (version 1.9) [27], with default parameters. Homopolymers and regions with low complexity, alternative contigs, or abnormally high coverage were discarded. Similarly, we used 100bp mappability tracks in the University of Southern California (UCSC) database to filter out variants in low mappability regions. Finally, a variant was called if detected by a minimum of 2 callers and if it had a minimum genotype quality of 30 Phred and a minimum coverage depth of 10. Finally, we filtered events with variant allele fraction (VAF) <30% in order to limit possible contamination of the controls with tumor cells. Variants were annotated using dbSNP [28], 1000 Genomes [29], ExAc [30], and gnomAD [31]. Only variants with a major allele frequency (MAF) below 0.5% in any ethnic population were retained. Thereafter, we selected (1) all protein-truncating variants (PTVs): start lost, stop lost, nonsense, frameshift, splice acceptor, and splice donor variants, and (2) missense variants with pathogenicity Combined Annotation Dependent Depletion (CADD) v.14 [32] scores > 20 Phred (i.e., variants in the top 1% of predicted pathogenicity). Finally, we restricted our analysis to those genes involved in carcinogenesis, particularly in lymphomagenesis. We collected the following types of genes: (1) 162 genes involved in mendelian inherited cancer syndromes [33], (2) 723 genes included in the Cancer Gene Census [34], (3) 135 genes included in the TARGET database (“a database of genes that, when somatically altered in cancer, are directly linked to a clinical action” [35]), (4) 59 recurrently mutated genes in CLL [36,37], (5) 150 recurrently mutated genes in DLBCL [38], and (6) 72 recurrently mutated genes in Burkitt lymphoma [39]. The final list contained 899 non-redundant genes (Table S1). Visual analysis of all frameshift insertions and deletions was performed using Integrative Genome Viewer [40]. Ancestry analysis was performed using Peddy [41], which predicts ancestry using a machine learning model trained on individuals of diverse ancestries from the 1000 Genomes Project reference panel. Only 3 of the patients were of non-European ancestry (1 African, 1 South Asian, and 1 East Asian). Genes affected by 5 or more variants were annotated to the top 0.5% genes in the Frequently Mutated Genes in Public Exomes (FLAGS) database [42] in order to highlight potentially spurious discoveries. Additionally, predicted loss-of-function expected vs. observed ratios (pLOF o/e) derived from gnomAD were used to annotate these genes [43]. pLOF o/e ratio is a measure of a gene’s tolerance to protein loss-of-function variants. Genes with low pLOF o/e values are more intolerant to disrupting variants than those with high values. Finally, survival analysis was performed with cox regression. Multiple testing correction was performed with the false discovery rate (FDR) method.
2.3. Burden Test against Public Controls
We used Testing Rare vAriants using Public Data (TRAPD) software in order to compare the enrichment for putatively pathogenic variants of our cohort of patients with that of 15,708 public controls from the gnomAD version 2 whole-genome sequencing dataset [31]. Importantly, none of these controls originated from cancer studies. We performed 2 types of analysis. In the first one, a burden test was performed with all PTVs detected by the Variant Effect Predictor (VEP) tool [44], which determines the effect of variants on genes, transcripts, and protein sequence, as well as regulatory regions. The following types of variants were defined as PTV: splice acceptor, splice donor, stop gained, frameshift, stop lost, and start lost variants. In a second attempt, we added those variants with high impact according to SNPeff annotations [45], namely, protein–protein interaction locus variants, protein structural interaction variants (i.e., affecting variants that are in contact within the same protein), and rare amino acid variants. Only variants with a maximum allele frequency (popmax) < 0.5% in any population were selected, excluding Finnish and Ashkenazi Jewish populations and those catalogued as “Other” in gnomAD (default behavior of the “popmax” gnomAD filter). Multiple testing correction was performed with the FDR method.
We tested the association of all high-impact variants according to the annotations of VEP and SNPEff with CLL patient survival, as this was the only cohort of patients sufficiently powered to make a reliable survival analysis. We restricted our study to genes affected by high impact variants in >1% of CLL cases. We created Cox regression models for time to first treatment and overall survival, and adjustment for covariates associated with survival was performed (multivariate p-value < 0.2). In the first case, these were IGHV mutation status and tumor stage at diagnosis, whereas in the second case we adjusted for IGHV status and patient age at diagnosis.
2.4. Germline–Germline and Germline–Somatic Double Hit Event Detection
In order to identify germline double-hits, we selected concurrent rare heterozygous and putatively damaging variants affecting the same gene in the same individual. Second-hit somatic mutations were detected by comparing germline variants with somatic mutations for the same set of individuals present in the ICGC database.
2.5. Myeloid Clonal Hematopoiesis Filtering
Potentially mosaic somatic mutations in the blood controls due to myeloid clonal hematopoiesis of undetermined potential (CHIP) could exist. In order to assess this issue, we initially identified a list of 22 recurrently mutated genes in clonal hematopoiesis that had at least one putatively rare germline variant in the final dataset [46,47,48]. Among these genes, we analyzed if the variants were present in both the control and tumor (lymphoid) compartment, and those mutations that were not found (or found at very low VAF) in the tumoral department were catalogued as likely myeloid CHIP events.
3. Results
3.1. Rare Variants Overview
A total of 1665 rare germline variants with likely disruptive activity (CADD scores > 20 or protein truncating) were detected in 559 cancer-related genes across 693 (95.45%) patients (Table S2). Overall, the frequency of these rare and likely disrupting mutations in cancer-related genes was superior to those found in non-cancer-related genes (4.25 × 10−3 vs. 3.61 × 10−3 mutations per gene and patient). Most of these were missense variants (1559 events, 93.01%, Table 1). Interestingly, we only detected 10 likely somatic mosaic mutations among myeloid-CHIP related genes, which affected TET2, DNMT3A, ASXL2, BCORL1, and PPM1D (Table S3). These variants were removed from downstream analysis.
Table 1.
Distribution of the selected variant types present in the cohort.
| Variant Type | Frequency |
|---|---|
| Missense | 93.50% |
| Stopgain | 2.40% |
| Frameshift deletion | 1.60% |
| Frameshift insertion | 0.80% |
| Splice donor | 0.80% |
| Splice acceptor | 0.40% |
| Nonframeshift deletion | 0.20% |
| Stoploss | 0.10% |
| Nonframeshift insertion | 0.10% |
| Startloss | 0.10% |
Overall, 113 patients (15.56%) harbored 126 PTVs in 103 different loci, which included frameshift, splice donor, splice acceptor, nonsense, stop loss, and start loss variants (Table S4). The frequency of PTVs in this gene list was notoriously superior to that observed in the remaining genes (2.11 × 10−3 vs. 7.33 × 10−4 mutations per gene and patient), suggesting an enrichment for loss of function mutations among cancer-related genes. The most frequently affected genes were ATM (5 cases), SETDB1 (5 cases in a single locus), ISX (4 cases), and POLQ (4 cases).
Some of the missense variants showed a remarkable increased frequency in patients with lymphoid neoplasia compared with the non-Finnish European (NFE) gnomAD database. This was the case of the variants rs199502695 in PRPF40B (4 cases, 71.17 times more frequent), rs191413750 in DOCK8 (5 cases, 55.55 times more frequent), rs377188372 in N4BP2 (4 cases, 34.66 times more frequent), and rs146946726 in MLLT10 (6 cases, 8.10 times more frequent).
A total of 227 different variants have also been described as pathogenic or likely pathogenic somatic mutations in cancer (Table S5). Remarkably, two out of three variants in NOTCH1 are flagged as pathogenic somatic mutations in COSMIC. This overlap also occurred in BCL6 (2 out of 5 variants), PTCH1 (2 out of 4 variants), ATM (3 out of 14 variants), CNOT3 (1 out of 2 variants), DNMT1 (1 out of 2 variants), FGFR2 (1 out of 2 variants), JAK3 (1 out of 2 variants), MTOR (1 out of 2 variants), and MDM4 (1 out of 2 variants). Furthermore, single rare variant flagged as pathogenic were also observed in CCND2, CHIC2, CDKN1B, CREBBP, EZH2, FGFR3, JAK2, PRF1, RUNX1, SIRPA, SUFU, TRIP11, and YWHAE.
Finally, 11 variants in homozygosity were observed, one of which (c.1642C>T in ZCCHC8) was present in two different patients (Table 2). Similarly, 15 patients harbored two likely functional variants in the same gene, many of which might be compound heterozygotes. Interestingly, these events were observed twice in FAT1 and ZFHX3. Moreover, one homozygous nonsense variant and a germline double-hit variant case were detected in the gene GLI1, and one homozygous missense variant plus a germline double-hit was detected in MYH9. In the cases of ARID1B and CBFA2T3, the close proximity of the variants allowed us to determine that they were inherited from the same parent (Figure S1). In the remaining cases, phase data were not available.
Table 2.
List of all homozygous and germline double-hit rare and putatively dysfunctional variants detected across 726 patients with B-cell lymphoid neoplasms.
| Case ID | Zygosity | Gene | Variant Type | rs ID |
|---|---|---|---|---|
| 147 | Germline double-hit | ARID1B | missense | rs1378351788, rs200808642 |
| 1416 | Germline double-hit | ATM | missense and frameshift deletion |
No rs IDs |
| 1196 | Germline double-hit | CBFA2T3 | missense | rs143704547, rs561624190 |
| 4167381 | Germline double-hit | EPPK1 | missense | rs144123426, no rsID |
| 544 | Germline double-hit | FAT1 | missense | rs377498159, no rsID |
| 1078 | Germline double-hit | FAT1 | missense | rs201279606, rs201751862 |
| 4190784 | Germline double-hit | GLI1 | missense | rs200306754, no rsID |
| 1260 | Germline double-hit | IL6ST | missense | rs191125510, rs199939306 |
| 1565 | Germline double-hit | MYH9 | missense | rs56200894, no rsID |
| 772 | Germline double-hit | NCOR1 | missense | rs118021690, no rsID |
| 4159421 | Germline double-hit | PIM1 | missense | No rsIDs |
| 284 | Germline double-hit | RNF213 | missense | rs202143169, rs141301945 |
| 63 | Germline double-hit | WRN | missense | rs78488552 and no rsID |
| 757 | Germline double-hit | ZFHX3 | missense | rs148334947, rs147016640 |
| 1191 | Germline double-hit | ZFHX3 | missense | rs148334947 and no rsID |
| 4126692 | Homozygote | AKAP9 | missense | rs61757664 |
| 1298 | Homozygote | ERBB3 | missense | rs55699040 |
| 1309 | Homozygote | GLI1 | nonsense | No rsIDs |
| 308 | Homozygote | MYH9 | missense | rs139134727 |
| 325 | Homozygote | MYO5A | missense | rs147898420 |
| 4115001 | Homozygote | NTRK3 | missense | No rsIDs |
| 1568 | Homozygote | PDZRN3 | missense | rs141385664 |
| 7 | Homozygote | SFRP4 | missense | rs147145122 |
| 1437 | Homozygote | SYNE1 | missense | No rsIDs |
| 396 | Homozygote | TSC1 | missense | rs118203504 |
| 1052;1295 | Homozygote | ZCCHC8 | missense | rs150057798 |
3.2. Rare Variants Affecting Lymphoma Driver Genes
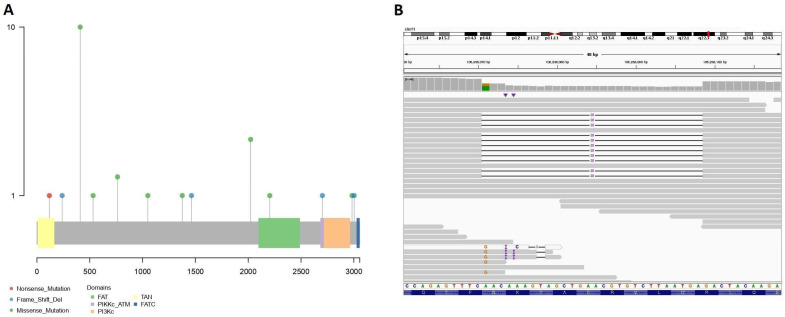
A total of 459 different rare variants occurring 636 times in the cohort were detected across 143 driver genes of lymphomagenesis extracted from the literature. These events affected 415 patients (57.16%) (Table S6). The most commonly mutated genes were ATM (25 cases, Figure 1A,B), BIRC6 (24 cases), SPEN (15 cases), ZNF292 (13 cases), MGA (12 cases), BAZ2A (12 cases), NCOR1 (11 cases), GNA13 (10 cases), and WDR7 (10 cases) (Table 3).
Figure 1.
(A) Lollipop plot of the rare and predictively disruptive germline variants detected in the ATM gene. (B) Representation of a 28 bp frameshift deletion present in one patient.
Table 3.
List of the most recurrently affected genes by rare and predictively disruptive germline variants. Genes in the top 0.5% of the FLAGS list are indicated, as well as predicted loss-of-function (pLOF) observed vs. expected ratio (o/e ratio) along with its 90% confidence interval.
| Gene | No. Cases | FLAGS Top 0.5% | pLOF o/e Ratio |
|---|---|---|---|
| FAT3 | 32 | Yes | 0.18 [0.13–0.25] |
| SYNE1 | 31 | Yes | 0.37 [0.33–0.42] |
| FAT1 | 29 | Yes | 0.34 [0.27–0.43] |
| ATM | 25 | No | 0.6 [0.51–0.71] |
| BIRC6 | 24 | No | 0.07 [0.04–0.1] |
| CLTCL1 | 24 | No | 0.8 [0.66–0.98] |
| ZFHX3 | 23 | Yes | 0.08 [0.05–0.14] |
| TSC2 | 22 | No | 0.02 [0.01–0.07] |
| CSMD3 | 21 | No | 0.23 [0.18–0.3] |
| MYH9 | 20 | No | 0.04 [0.02–0.09] |
| TPR | 19 | No | 0.06 [0.04–0.11] |
| LRP1B | 19 | Yes | 0.26 [0.21–0.32] |
| ANK1 | 19 | No | 0.1 [0.06–0.17] |
| EPPK1 | 19 | Yes | 0.96 [0.78–1.2] |
| KMT2D | 19 | Yes | 0.07 [0.04–0.1] |
| PTPN13 | 18 | No | 0.52 [0.42–0.64] |
| MYH11 | 18 | No | 0.22 [0.16–0.3] |
| POLQ | 17 | No | 1.05 [0.9–1.22] |
| KMT2C | 17 | Yes | 0.08 [0.06–0.12] |
| MUC4 | 16 | No | 0.84 [0.7–0.99] |
| APC | 16 | No | 0.1 [0.06–0.16] |
| ROS1 | 16 | No | 0.95 [0.82–1.12] |
| POLE | 16 | No | 0.52 [0.42–0.64] |
| NBEA | 16 | No | 0.04 [0.02–0.07] |
| SPEN | 15 | No | 0.03 [0.01–0.07] |
| SETDB1 | 15 | No | 0.11 [0.06–0.2] |
| LPP | 15 | No | 0.32 [0.19–0.56] |
| FAT4 | 15 | Yes | 0.12 [0.08–0.18] |
Various variants also affected other drivers of lymphomagenesis, such as ARID1A (9 cases), CHD1 (9 cases), MECOM (9 cases), NOTCH1 (7 cases), SETD2 (6 cases), ARID1B (5 cases), BLC6 (5 cases), CTCF (5 cases), EP300 (5 cases), JAK3 (5 cases), NOTCH2 (5 cases), MET (5 cases), MYC (5 cases), TCF3 (5 cases), and CHD2 (4 cases). Finally, infrequent variants in TRAF2 were detected in three patients, whereas those of CNOT3, ID3, IKZF3, MTOR, POT1, and STAT5B occurred in two patients each, and those of ASXL1, BRAF, CARD11, CCND2, CCND3, CREBBP, DTX1, ETV6, EZH2, KRAS, MCL1, and TCL1A were detected in just one case each. Notably, both variants in SAMDH1 were PTVs (a frameshift and a nonsense event) (Table S7).
3.3. Rare Variants Affecting Genes Involved in Cancer Syndromes with Germline Inheritance
A total of 84 genes associated with inherited cancer syndromes were affected by a total of 372 occurrences of 225 different rare variants (Table S8), of which 19 were PTVs and affected 22 patients (3%). In total, 131 variants were observed in genes linked with autosomal dominant syndromic cancer, affecting 168 patients. Among these, the most frequently mutated genes were TSC2 (22 cases), linked to tuberous sclerosis; APC (16 cases), linked to hereditary colon cancer; and the DNA polymerase POLE (16 cases), involved in predisposition to multiple cancers (Table 3). Similarly, 94 variants in 32 genes linked to autosomal recessive cancer were observed, which affected 149 patients. The most commonly affected among these were ATM (25 cases), NBN (12 cases), BLM (12 cases), DOCK8 (12 cases), and WRN (12 cases) (Table 3).
Some of these variants were labelled as pathogenic in ClinVar (Table S6). This included a missense variant in MITF (rs149617956, 7 cases), a missense variant in GBA (rs76763715, 6 cases), a missense variant in MUTYH (rs34612342, 2 cases), two missense variants in SERPINA1 (rs61761869 and rs28931570, 3 cases), a missense variant in NBN (rs61754966, 1 case), and a frameshift deletion in BRCA2 (rs397507591, 1 case). Likely pathogenic variants were detected in APC (missense variant, rs139196838, 1 case), BRIP1 (frameshift insertion, rs878855150, 1 case), and MET (missense variant, rs34589476, 1 case).
Interestingly, 95 previously undescribed variants were detected, and these were particularly frequent in ATM (4 missense variants, 1 nonsense variants, and 2 frameshift deletions, including a 28 base pair deletion), EXT1 (2 missense variants and 1 splice donor variants), MET (2 missense variants and 1 frameshift deletion), MITF (1 missense variants in 2 patients and 1 missense variants in 1 patient), DOCK8 (3 missense variants), MSH6 (2 missense variants and 1 nonsense variant), SMARCA2 (3 missense variants), SOS1 (3 missense variants), TRIM37 (3 missense variants), and WRN (1 missense, 1 nonsense, and 1 splice gain variant).
3.4. Rare Variants in Genes of the Cancer Gene Census and TARGET Databases
A total of 327 occurrences of 208 rare variants in 95 different genes linked to therapy were identified. These affected 247 patients (34.02%) (Table S9). The most recurrently affected genes were ATM (25 cases), TSC2 (22 cases), APC (16 cases), ROS1 (16 cases), and JAK2 (11 cases) (Table 3). Among Cancer Gene Census genes, 1346 events were detected in 947 different loci (Table S10), with the most recurrent ones being those in CLTCL1 (24 cases), CSMD3 (21 cases), MYH9 (20 cases), ANK1 (19 cases), TPR (19 cases), MYH11 (18 cases), PTPN13 (18 cases), and POLQ (17 cases) (Table 3). Some genes were enriched in PTV variants, particularly POLQ (4 out of 10 different variants), AKAP9 (3 out of 10), TSHR (2 out of 3), and ISX (2 out of 2). Furthermore, one pathogenic (rs113994096, 5 patients) and one likely pathogenic (rs138929605, 1 patient) missense variants in the DNA polymerase POLG were also discovered.
3.5. Differential Distribution of Rare Variants and Association with Patient Survival
We did not identify any gene significantly enriched in rare variants in CLL vs. B-cell lymphoma cases (Fisher’s test, FDR < 5%). Nevertheless, we discovered that some variants were only detected in one subgroup. For example, the missense variant rs1800729 in TSC2 was exclusively present in CLL (eight cases), and the missense variant rs139075637 in POLE was exclusively present in non-CLL B lymphoid tumors (seven cases). Notably, both variants were found to have higher frequency in non-Finish Europeans than in other populations according to gnomAD data (allele frequencies of 0.40% and 0.17%, respectively). Therefore, further analysis needs to be performed in order to confirm these findings and rule-out population substructure biases.
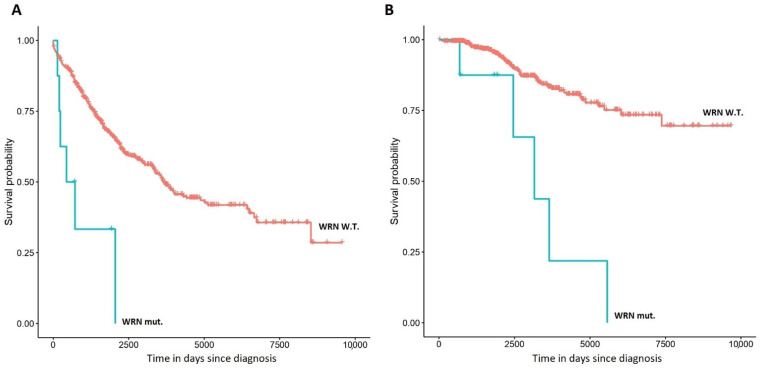
Thereafter, we tested if rare variants could be associated with adverse patient outcomes. Due to the heterogeneity of the dataset and sample size limitations, we restricted our analysis to CLL cases, and considered variants present in at least 1% of cases. Interestingly, rare variants in the DNA helicase WRN (8 cases, Table 4) were significantly associated with shorter overall survival (Cox p-value 1.16 × 10−4, q-value 0.01, Hazard Ratio (HR) (2.35, 14.59); Figure 2B). Indeed, such association was independent of age at diagnosis and CLL/MBL status (p-value 1.97 × 10−7, HR (5.03, 35.48)). Moreover, these variants were also linked to shorter time to first treatment (Cox p-value 6.15 × 10−4, HR (1.85, 9.48); Figure 2A), which remained significant after adjusting for age at diagnosis and CLL/MBL status (p-value 1.69 × 10−3, HR (1.64, 8.48)). These patients tended to harbor high-risk karyotype anomalies in the tumor cells: 11q deletion (three cases, one as an isolated anomaly, one co-occurring with 13q deletion, and one co-occurring with three other karyotype anomalies), 17p deletion (one case, co-occurring with a 18p deletion), 8q deletion (one case, co-occurring with 21q gain), and 6q deletion (one case, co-occurring with 13q deletion). The most frequent variant was rs78488552 (six out of eight cases), which has its highest frequency in non-Finish Europeans (0.49%).
Table 4.
Rare and likely functional mutations in WRN among CLL patients. One patient had two concurrent variants (indicated with an asterisk *). Nucleotide and amino acid changes induced by each variant are provided.
| Chromosome | Position | rsID | REF | ALT | CLL Cases | Mutation Type |
|---|---|---|---|---|---|---|
| 8 | 30922465 | . | T | G | 1 * | c.390T>G; p.Asn130Lys; missense variant ENST00000298139 transcript, exon 5/35 SIFT score: 0.01 (deleterious) Polyphen score: 0.913 (probably damaging) |
| 8 | 30922580 | . | G | A | 1 | c.504+1G>A; splice donor variant ENST00000298139 transcript, exon 5/34 |
| 8 | 30954292 | rs569266355 | A | G | 1 | c.1907A>G; p.Tyr636C; missense variant ENST00000298139 transcript; exon 17/35 SIFT score: 0 (deleterious) Polyphen score: 0.99 (probably damaging) |
| 8 | 31012237 | rs78488552 | C | G | 6 * | c.3785C>G; p.Thr1262Arg; missense variant ENST00000298139 transcript, exon 32/35 SIFT score: 0 (deleterious) Polyphen score: 0.959 (probably damaging) |
Figure 2.
Kaplan–Meier plots representing the association of rare variants in WRN with time to first treatment (A) and overall survival (B) in chronic lymphocytic leukemia (CLL). Variants considered in this plot are represented in Table 4.
Rare variants in ATM have been previously associated with CLL risk [18]. Curiously, no association with survival could be observed in this analysis. As ATM is enriched in missense variants [18], we restricted the analysis only to patients with truncating events (four cases), and discovered that these few cases had a significantly shorter overall survival (p-value 0.02, HR (1.28, 21.53)).
3.6. Association of Rare Germline Variants with Somatic Mutations
Concurrent rare and likely disruptive germline variants and somatic mutations were detected in 17 cases (Table 5). Co-occurring mutations in CLL affected GNA13, KMT2D, LRP1B, MUC16, and SPEN. Additionally, co-occurring mutations in B-cell lymphomas were found in CSMD3 (grade I follicular lymphoma), EP300 (DLBCL), FAT1 (DLBCL), HIST1H1E (grade I follicular lymphoma), KMT2D (DLBCL), MCL1 (grade IIIa follicular lymphoma), MSH6 (grade IIIa follicular lymphoma), MYC (DLBCL), PIM1 (grade I follicular lymphoma), RNF213 (DLBCL), and SIN3A (grade IIIb follicular lymphoma). Additionally, we observed a germline mutation in ATM co-occurring with a 11q copy neutral loss of heterozygosity that induced loss of the reference allele in a CLL patient.
Table 5.
Cases of co-occurring somatic mutations and rare germline variants in the same gene. Marked with an asterisk is an event where a rare and likely disruptive germline variant in ATM coexisted with a loss-of-heterozygosity (LOH) at 11q that deleted the wild-type allele.
| Gene | Case ID | Diagnosis |
|---|---|---|
| ATM * | 155 | CLL |
| GNA13 | 381 | CLL |
| KMT2D | 372 | CLL |
| LRP1B | 122 | CLL |
| MUC16 | 1267 | CLL |
| SPEN | 830 | CLL |
| EP300 | 4122063 | DLBCL |
| KMT2D | 4175941 | DLBCL |
| MSH6 | 4109808 | DLBCL |
| MYC | 4107559 | DLBCL |
| PIM1 | 4102009 | DLBCL |
| RNF213 | 4109808 | DLBCL |
| CSMD3 | 4111337 | Follicular lymphoma |
| FAT1 | 4136095 | Follicular lymphoma |
| HIST1H1E | 4144951 | Follicular lymphoma |
| MCL1 | 4159421 | Follicular lymphoma |
| SIN3A | 4139696 | Follicular lymphoma |
3.7. Burden Test of High Impact Variants Using Public Controls
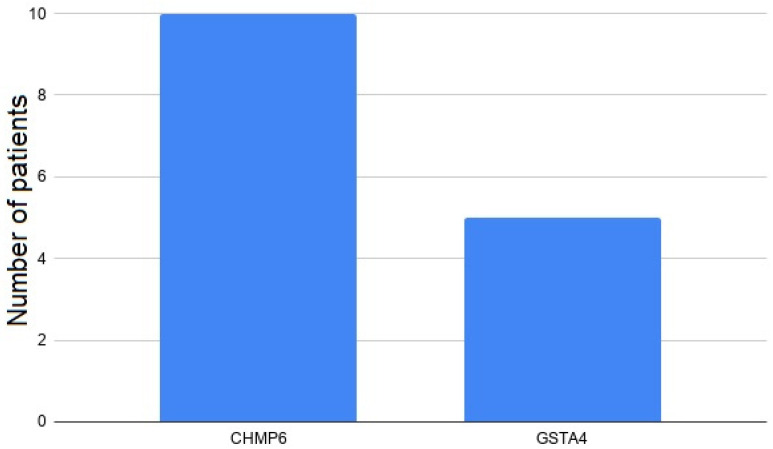
Germline variants with high functional impact were selected for association with risk of B-cell neoplasms using the burden test against public whole-genome sequencing controls. Briefly, this analysis tests if the cumulative frequency of variants affecting each gene in a cohort is significantly different from that of a control cohort. We first analyzed all PTVs detected by VEP, and afterwards we added all variants with high impact consequences according to SNPeff. We selected these variants because they are the most potentially pathogenic. As a result, two genes were significantly enriched in high-impact variants among patients affected by B-cell lymphoid neoplasms (q-value < 0.1) (Figure 3, Table S11). Overall, we identified 2 different variants affecting 15 different cases (Table S12, Figure 3): rs746495175 in CHMP6 (a splice acceptor variant) and rs557844606 in GSTA4 (an inframe deletion within a structural interaction domain). Importantly, inflation statistics were low (ƛ = 0.93 and 0.83 for the VEP-only and VEP + SNPeff models). Additionally, there was an enrichment of CHMP6 variants in lymphoma vs. CLL patients (Fisher’s p-value 0.02, q-value 0.04).
Figure 3.
Frequency of highly dysfunctional variants within genes identified in the burden test.
3.8. Association of High Impact Variants with Patient Survival
High impact variants in four genes were independently associated with shorter CLL patient survival (q-value < 0.1; Table 6). These genes were M1AP (Figure S2), GNLY, FLYWCH1, and PIK3C2G. Variants in another gene (PLA2G7) were also suggestively associated with short survival (q-value 0.11). Conversely, we did not detect variants in any gene associated with either time to first treatment or earlier age at diagnosis.
Table 6.
Association results of high-impact variants with overall survival in CLL.
| GENE | FDR | Lower 95% CI HR | Upper 95% CI HR | CLL Cases |
|---|---|---|---|---|
| M1AP | 1.61 × 10−2 | 2.88 | 32.52 | 7 |
| GNLY | 4.04 × 10−2 | 2.63 | 50.45 | 6 |
| FLYWCH1 | 7.54 × 10−2 | 1.8 | 19.32 | 10 |
| PIK3C2G | 7.74 × 10−2 | 1.94 | 37.94 | 6 |
| PLA2G7 | 0.11 | 1.64 | 29.43 | 6 |
4. Discussion
Approximately 8% of cancer patients are affected by pathogenic germline variants, which confer a strong hereditary component [49]. Interestingly, growing evidence indicates that such variants can modulate cancer evolution and prognosis. For example, truncating variants in genes of the angiogenesis and DNA repair pathways predispose to the development of metastatic disease in prostate cancer [15]. Therefore, we reasoned that the analysis of such variants in patients affected by B-cell lymphoid neoplasms could shed new clues about their pathogenesis and prognostication. Indeed, our results indicate an increased frequency of highly disruptive rare variants in two genes of B-cell lymphoid tumor patients: CHMP6 and GSTA4. Notably, both genes are involved in cell survival regulation under oxidative stress. GSTA4 mediates glutathione-dependent elimination of 4-hydroxynonenal, which is an important product of peroxidative degradation of arachidonic acid [50]. At the same time, CHMP6 encodes a member of membrane repair dependent on endosomal sorting complexes required for transport (ESCRT)-III, which inhibit ferroptosis (a form of cell death triggered by iron accumulation and lipid peroxidation) [51].
Additionally, our data indicate a significant contribution of these high-impact rare variants to CLL survival, as we found significant or suggestive associations of five genes with overall survival. Notably, all the affected genes play a role in oncogenic pathways. For example, M1AP is involved in meiosis progression, and recent evidence supports a role as a positive regulator of the oncogene MYC [52]. GNLY encodes granulysin, a protein located in cytotoxic granules of Natural Killer and T-cells. Interestingly, it has been observed that granulysin triggers cancer cell apoptosis through caspase-dependent and independent mechanisms in hematological B-cell neoplasms, and therefore it plays a central role in immune-related mechanisms of tumor development and progression [53]. Similarly, FLYWCH1 and PIK2C2G regulate oncogenic signaling through WNT/β-catenin and phosphoinositide-3-kinase pathways, respectively [54,55]. Finally, PLA2G7 encodes a lipoprotein-associated phospholipase that regulates epithelial-mesenchymal transition, and it is associated with the development of metastatic disease in solid organ cancer [56]. Overall, our pioneer results indicate that these mutations can act as true drivers of disease progression and treatment failure, even though they are not recurrently mutated in the somatic line.
In a different approach, we focused our research on the detection of rare and likely disruptive mutations (both PTVs and non-PTVs) in a set of genes involved in cancer pathways, and particularly in lymphoid neoplasms. The collective high frequency of these rare germline variants in cancer genes poses a challenge for personalized genomics, as many of these are probably non-functional, whereas others play a pathogenic or prognostic role. We identified recurrent highly pathogenic variants affecting important drivers of hematological cancer (ATM [57]), epigenetic regulators (ISX [58] and SETDB1 [59]), and mediators of DNA replication (POLQ [60]). Recurrent variants were also observed in drug targets, particularly in the crizotinib targets ALK, MET, and ROS1, as well as the everolimus target TSC2, which suggests new therapeutic strategies for these patients [61,62,63]. Additionally, several variants were previously catalogued as pathogenic (such as the E318K variant in the transcription factor MITF [64]); others affected strong mediators of inherited predisposition to lymphomas (i.e., DOCK8, EXT1, MSH6, and SOS1 [65,66,67,68]), while others have been flagged as pathogenic somatic mutations in cancer, such as NOTCH1 R912W [69,70] and CNOT3 E20K [71,72]. Importantly, we observed that variants in the DNA helicase WRN were significantly associated with shorter overall survival and time to first treatment in CLL. WRN-mutated CLL cases tended to harbor high-risk karyotypic anomalies, suggesting an increased genomic instability [73] mediated by altered DNA repair mechanisms [74].
Germline–germline or germline–somatic “double-hit” events were identified in cancer driver genes. Germline–germline double-hit events were detected in 28 cases (3.85% of cases), and curiously five genes affected more than one patient, including the Hedgehog signaling gene GLI1 [75] and the homeobox tumor suppressor ZFHX3 [76]. Additionally, germline-somatic “double hit” events occurred in 17 cases (2.34%). Notably, this phenomenon affected common drivers of lymphomagenesis, such as the oncogenes MYC and PIM1 [77,78]; the tumor suppressors ATM, FAT1, KMT2D, and MSH6 [79,80,81]; the histone acetyltransferase EP300 [82]; the histone gene HIST1H1E [83]; the transcriptional regulator SIN3A [84]; the NOTCH pathway member SPEN [85]; and the apoptotic proteins GNA13 and MCL1 [86,87].
This study has several limitations. First, some background heterogeneity could exist between Spanish CLL and German lymphoma populations. Although the current knowledge does not support heterogeneity between Spanish and German lymphoma populations, it must be considered that fine-scale population structure at extremely fine scales has been documented, even within neighboring Iberian populations [88]. Secondly, many relevant oncogenes and tumor suppressors were very rarely mutated, and the interpretation of these variants in terms of survival will need the sequencing of thousands of cases. Additionally, the presence of mosaic somatic mutations in the controls due to clonal hematopoiesis could have led to some false positives. In this line, we observed that only a minority of variants in genes associated with CHIP were likely somatic events, but nevertheless our results should be taken with caution among this group of genes. Finally, another limitation arises from the heterogeneity and limited sample size of the B-cell lymphoma dataset, which dissuaded us from making a survival analysis in such cases.
5. Conclusions
Our results indicate the existence of multiple genes affected by highly pathogenic germline variants in the genomes of patients with B-cell neoplasms, including a significant enrichment for high impact rare variants in two genes related to oxidative stress regulation. Additionally, the association of some variants with shorter survival, along with the disruptive nature of some others, points towards new functional, prognostic, and therapeutic implications. Finally, the elevated number of rare and likely pathogenic variants in cancer genes supposes a challenge for personalized genomics, and future analysis integrating more layers of biological information and other types of cancers are envisaged in order to clarify their benign or pathogenic role.
Acknowledgments
We would like to thank the Supercomputing Center of Galicia (CESGA), the International Cancer Genome Consortium, and the Fundación Galega de Hematoloxía e Hemoterapia for their support in this research.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/13/6/1340/s1, Figure S1. IGV plots for rs1378351188 (G > A) and rs200808642 (G > T) in ARID1B and rs143704547 (G > T) and rs561624190 (A > C) in CBFA2T3. Figure S2. Kaplan–Meier plot representing the association of high impact rare variants in M1AP with overall survival in CLL. Table S1. Candidate gene list. The table represents the relationship between each candidate gene and each gene list included in the analysis. Table S2. Annotated list of all rare and likely disruptive variants detected across 726 patients with B-cell lymphoid neoplasms. Table S3. Myeloid CHIP-related genes affected by rare variants in this study. The number of known variants and the number of likely somatic mosaic events detected is indicated in the corresponding columns. Table S4. Annotated list of all rare and likely disruptive protein-truncating variants across 726 patients with B-cell lymphoid neoplasms. Table S5. List of all filtered rare germline events overlapping known somatic mutations in the COSMIC database. Table S6. List of all rare and likely disruptive variants in known drivers of B-cell lymphoid tumors detected across 726 patients with B-cell lymphoid neoplasms. Table S7. List of all filtered rare germline events included in the ClinVar database. Table S8. List of all rare and likely disruptive variants in genes linked to syndromic cancer detected across 726 patients with B-cell lymphoid neoplasms. Table S9. List of all rare and likely disruptive variants in genes of the TARGET database across 726 patients with B-cell lymphoid neoplasms. Table S10. List of all rare and likely disruptive variants in genes of the Cancer Gene Census database across 726 patients with B-cell lymphoid neoplasms. Table S11. Burden test results for the VEP-only and VEP + SNPEff models. Results with q-value < 0.1 are shown. Table S12. High-impact variants affecting genes significantly enriched in patients with B-cell lymphoid neoplasms.
Author Contributions
A.M.O. had the initial idea and performed the research. A.M.O., M.C.L., A.P.R., J.Á.D.A., B.A.R., L.B.P., M.S.G.P., and N.A.V. wrote the paper. Á.B.L., A.A.B., P.M.V., R.F.F., and C.A.S. evaluated and suggested changes to the manuscript. M.F.F.R., M.S.G.P., M.M.P.E., and J.L.B.L. approved the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. Article processing charges will be paid with funding from the Fundación Galega de Hematoloxía e Hemoterapia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data for this study were downloaded from the International Cancer Genome Consortium repository.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Teras L.R., DeSantis C.E., Cerhan J.R., Morton L.M., Jemal A., Flowers C.R. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA A Cancer J. Clin. 2016;66:443–459. doi: 10.3322/caac.21357. [DOI] [PubMed] [Google Scholar]
- 2.Slager S.L., Caporaso N.E., De Sanjose S., Goldin L.R. Genetic Susceptibility to Chronic Lymphocytic Leukemia. Semin. Hematol. 2013;50:296–302. doi: 10.1053/j.seminhematol.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Law P.J., Berndt S.I., Speedy H.E., Camp N.J., Sava G.P., Skibola C.F., Holroyd A., Joseph V., Sunter N.J., Nieters A., et al. Genome-wide association analysis implicates dysregulation of immunity genes in chronic lymphocytic leukaemia. Nat. Commun. 2017;8:14175. doi: 10.1038/ncomms14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleinstern G., Yan H., Hildebrandt M.A.T., Vijai J., Berndt S.I., Ghesquières H., McKay J., Wang S.S., Nieters A., Ye Y., et al. Inherited variants at 3q13.33 and 3p24.1 are associated with risk of diffuse large B-cell lymphoma and implicate immune pathways. Hum. Mol. Genet. 2020;29:70–79. doi: 10.1093/hmg/ddz228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skibola C.F., Berndt S.I., Vijai J., Conde L., Wang Z., Yeager M., De Bakker P.I., Birmann B.M., Vajdic C.M., Foo J.-N., et al. Genome-wide Association Study Identifies Five Susceptibility Loci for Follicular Lymphoma outside the HLA Region. Am. J. Hum. Genet. 2014;95:462–471. doi: 10.1016/j.ajhg.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bashash M., Connors J.M., Gascoyne R.D., Meissner B., Schuetz J.M., Leach S., Slack G.W., Berry B.R., Hu H., Sehn L.H., et al. Genetic polymorphism at BCL2 as a predictor for rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone efficacy in patients with diffuse large B-cell lymphoma. Haematologica. 2017;102:e199–e202. doi: 10.3324/haematol.2016.159087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bielska M., Borowiec M., Jesionek-Kupnicka D., Braun M., Bojo M., Pastorczak A., Kalinka-Warzocha E., Prochorec-Sobieszek M., Robak T., Warzocha K., et al. Polymorphism in IKZF1 gene affects clinical outcome in diffuse large B-cell lymphoma. Int. J. Hematol. 2017;106:794–800. doi: 10.1007/s12185-017-2315-0. [DOI] [PubMed] [Google Scholar]
- 8.Ghesquieres H., Slager S.L., Jardin F., Veron A.S., Asmann Y.W., Maurer M.J., Fest T., Habermann T.M., Bene M.C., Novak A.J., et al. Genome-Wide Association Study of Event-Free Survival in Diffuse Large B-Cell Lymphoma Treated With Immunochemotherapy. J. Clin. Oncol. 2015;33:3930–3937. doi: 10.1200/JCO.2014.60.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orgueira A.M., Rodríguez B.A., Vence N.A., Arias J., Ángel D., Varela N.D., Encinas M.M.P., Toscano C.A., Seco E.M.G., Ángel C.Á., et al. The association of germline variants with chronic lymphocytic leukemia outcome suggests the implication of novel genes and pathways in clinical evolution. BMC Cancer. 2019;19:515. doi: 10.1186/s12885-019-5628-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Musa J., Cidre-Aranaz F., Aynaud M.-M., Orth M.F., Knott M.M.L., Mirabeau O., Mazor G., Varon M., Hölting T.L.B., Grossetête S., et al. Cooperation of cancer drivers with regulatory germline variants shapes clinical outcomes. Nat. Commun. 2019;10:4128. doi: 10.1038/s41467-019-12071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu C., Xie M., Wendl M.C., Wang J., McLellan M.D., Leiserson M.D.M., Huang K.-L., Wyczalkowski M.A., Jayasinghe R., Banerjee T., et al. Patterns and functional implications of rare germline variants across 12 cancer types. Nat. Commun. 2015;6:10086. doi: 10.1038/ncomms10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown A.L., Arts P., Carmichael C.L., Babic M., Dobbins J., Chong C.-E., Schreiber A.W., Feng J., Phillips K., Wang P.P.S., et al. RUNX1-mutated families show phenotype heterogeneity and a somatic mutation profile unique to germline predisposed AML. Blood Adv. 2020;4:1131–1144. doi: 10.1182/bloodadvances.2019000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park S., Supek F., Lehner B. Systematic discovery of germline cancer predisposition genes through the identification of somatic second hits. Nat. Commun. 2018;9:2601. doi: 10.1038/s41467-018-04900-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pui C.-H., Nichols K.E., Yang J.J. Somatic and germline genomics in paediatric acute lymphoblastic leukaemia. Nat. Rev. Clin. Oncol. 2019;16:227–240. doi: 10.1038/s41571-018-0136-6. [DOI] [PubMed] [Google Scholar]
- 15.Mijuskovic M., Saunders E.J., Leongamornlert D.A., Wakerell S., Whitmore I., Dadaev T., Cieza-Borrella C., Govindasami K., Brook M.N., Haiman C.A., et al. Rare germline variants in DNA repair genes and the angiogenesis pathway predispose prostate cancer patients to develop metastatic disease. Br. J. Cancer. 2018;119:96–104. doi: 10.1038/s41416-018-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein A.M., Xiao Y., Sampson J., Zhu B., Rotunno M., Bennett H., Wen Y., Jones K., Vogt A., Burdette L., et al. Rare germline variants in known melanoma susceptibility genes in familial melanoma. Hum. Mol. Genet. 2017;26:4886–4895. doi: 10.1093/hmg/ddx368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleiblova P., Stolarova L., Krizova K., Lhota F., Hojny J., Zemankova P., Havranek O., Vocka M., Cerna M., Lhotova K., et al. Identification of deleterious germline CHEK2 mutations and their association with breast and ovarian cancer. Int. J. Cancer. 2019;145:1782–1797. doi: 10.1002/ijc.32385. [DOI] [PubMed] [Google Scholar]
- 18.Tiao G., Improgo M.R., Kasar S., Poh W., Kamburov A., Landau D.-A., Tausch E., Taylor-Weiner A., Cibulskis C., Bahl S., et al. Rare germline variants in ATM are associated with chronic lymphocytic leukemia. Leukemia. 2017;31:2244–2247. doi: 10.1038/leu.2017.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMaster M.L., Sun C., Landi M.T., Savage S.A., Rotunno M., Yang X.R., Jones K., Vogt A., Hutchinson A., Zhu B., et al. Germline mutations in Protection of Telomeres 1 in two families with Hodgkin lymphoma. Br. J. Haematol. 2018;181:372–377. doi: 10.1111/bjh.15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Offit K., Gilad S., Paglin S., Kolachana P., Roisman L.C., Nafa K., Yeugelewitz V., Gonzalez M., Robson M., McDermott D., et al. Rare variants of ATM and risk for Hodgkin’s disease and radiation-associated breast cancers. Clin. Cancer Res. 2002;8:3813–3819. [PubMed] [Google Scholar]
- 21.Srivastava A., Giangiobbe S., Kumar A., Paramasivam N., Dymerska D., Behnisch W., Witzens-Harig M., Lubinski J., Hemminki K., Försti A., et al. Identification of Familial Hodgkin Lymphoma Predisposing Genes Using Whole Genome Sequencing. Front. Bioeng. Biotechnol. 2020;8:179. doi: 10.3389/fbioe.2020.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The International Cancer Genome Consortium International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. Erratum in Nature2010, 465, 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guimera R.V. bcbio-nextgen: Automated, distributed next-gen sequencing pipeline. EMBnet. J. 2012;17:30. doi: 10.14806/ej.17.B.286. [DOI] [Google Scholar]
- 24.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 20121207.3907 [Google Scholar]
- 25.Poplin R., Ruano-Rubio V., DePristo M.A., Fennell T.J., Carneiro M.O., Van der Auwera G.A., Kling D.E., Gauthier L.D., Levy-Moonshine A., Roazen D., et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2017 doi: 10.1101/201178. [DOI] [Google Scholar]
- 26.Rimmer A., Phan H., Mathieson I., Iqbal Z., Twigg S.R.F., Wilkie A.O.M., McVean G., Lunter G. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 2014;46:912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minikel E.V., Karczewski K.J., Martin H.C., Cummings B.B., Whiffin N., Alföldi J., MacArthur D.G., Genome Aggregation Database (gnomAD) Production Team. Genome Aggregation Database (gnomAD) Consortium. Schreiber S.L., et al. Evaluating potential drug targets through human loss-of-function genetic variation. BioRxiv. 2019 doi: 10.1101/531210. [DOI] [Google Scholar]
- 32.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gröbner S.N., Project I.P.-S., Worst B.C., Weischenfeldt J., Buchhalter I., Kleinheinz K., Rudneva V.A., Johann P.D., Balasubramanian G.P., Segura-Wang M., et al. The landscape of genomic alterations across childhood cancers. Nature. 2018;555:321–327. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 34.Sondka Z., Bamford S., Cole C.G., Ward S.A., Dunham I., Forbes S.A. The COSMIC Cancer Gene Census: Describing genetic dysfunction across all human cancers. Nat. Rev. Cancer. 2018;18:696–705. doi: 10.1038/s41568-018-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Allen E.M., Wagle N., Stojanov P., Perrin D.L., Cibulskis K., Marlow S., Jane-Valbuena J., Friedrich D.C., Kryukov G., Carter S.L., et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med. 2014;20:682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landau D.A., Tausch E., Taylor-Weiner A.N., Stewart C., Reiter J.G., Bahlo J., Kluth S., Bozic I., Lawrence M.S., Böttcher S., et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puente X.S., Beà S., Valdés-Mas R., Villamor N., Gutiérrez-Abril J., Martín-Subero J.I., Munar M., Rubio-Pérez C., Jares P., Aymerich M., et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 38.Reddy A., Zhang J., Davis N.S., Moffitt A.B., Love C.L., Waldrop A., Leppa S., Pasanen A., Meriranta L., Karjalainen-Lindsberg M.-L., et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell. 2017;171:481–494.e15. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panea R.I., Love C.L., Shingleton J.R., Reddy A., Bailey J.A., Moormann A.M., Otieno J.A., Ong’Echa J.M., Oduor C.I., Schroeder K.M.S., et al. The whole-genome landscape of Burkitt lymphoma subtypes. Blood. 2019;134:1598–1607. doi: 10.1182/blood.2019001880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen B.S., Quinlan A.R. Who’s Who? Detecting and Resolving Sample Anomalies in Human DNA Sequencing Studies with Peddy. Am. J. Hum. Genet. 2017;100:406–413. doi: 10.1016/j.ajhg.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shyr C., Tarailo-Graovac M., Gottlieb M., Lee J.J.Y., Van Karnebeek C., Wasserman W.W. FLAGS, frequently mutated genes in public exomes. BMC Med. Genom. 2014;7:64. doi: 10.1186/s12920-014-0064-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo M.H., Plummer L., Chan Y.-M., Hirschhorn J.N., Lippincott M.F. Burden Testing of Rare Variants Identified through Exome Sequencing via Publicly Available Control Data. Am. J. Hum. Genet. 2018;103:522–534. doi: 10.1016/j.ajhg.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensemble Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steensma D.P., Bejar R., Jaiswal S., Lindsley R.C., Sekeres M.A., Hasserjian R.P., Ebert B.L. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zink F., Stacey S.N., Norddahl G.L., Frigge M.L., Magnusson O.T., Jonsdottir I., Thorgeirsson T.E., Sigurdsson A., Gudjonsson S.A., Gudmundsson J., et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 2017;130:742–752. doi: 10.1182/blood-2017-02-769869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibson C.J., Lindsley R.C., Tchekmedyian V., Mar B.G., Shi J., Jaiswal S., Bosworth A., Francisco L., He J., Bansal A., et al. Clonal Hematopoiesis Associated with Adverse Outcomes After Autologous Stem-Cell Transplantation for Lymphoma. J. Clin. Oncol. 2017;35:1598–1605. doi: 10.1200/JCO.2016.71.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang K.-L., Mashl R.J., Wu Y., Ritter D.I., Wang J., Oh C., Paczkowska M., Reynolds S., Wyczalkowski M.A., Oak N., et al. Pathogenic Germline Variants in 10,389 Adult Cancers. Cell. 2018;173:355–370.e14. doi: 10.1016/j.cell.2018.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balogh L.M., Atkins W.M. Interactions of glutathione transferases with 4-hydroxynonenal. Drug Metab. Rev. 2011;43:165–178. doi: 10.3109/03602532.2011.558092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai E., Meng L., Kang R., Wang X., Tang D. ESCRT-III–dependent membrane repair blocks ferroptosis. Biochem. Biophys. Res. Commun. 2020;522:415–421. doi: 10.1016/j.bbrc.2019.11.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto A., Kurata M., Onishi I., Sugita K., Matsumura M., Ishibashi S., Ikeda M., Yamamoto K., Kitagawa M. CRISPR screening identifies M1AP as a new MYC regulator with a promoter-reporter system. PeerJ. 2020;8:e9046. doi: 10.7717/peerj.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aporta A., Catalán E., Galán-Malo P., Ramírez-Labrada A., Pérez M., Azaceta G., Palomera L., Naval J., Marzo I., Pardo J., et al. Granulysin induces apoptotic cell death and cleavage of the autophagy regulator Atg5 in human hematological tumors. Biochem. Pharmacol. 2014;87:410–423. doi: 10.1016/j.bcp.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 54.Almars A., Chondrou P.S., Onyido E.K., Almozyan S., Seedhouse C., Babaei-Jadidi R., Nateri A.S. Increased FLYWCH1 Expression is Negatively Correlated with Wnt/β-catenin Target Gene Expression in Acute Myeloid Leukemia Cells. Int. J. Mol. Sci. 2019;20:2739. doi: 10.3390/ijms20112739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freitag A., Prajwal P., Shymanets A., Harteneck C., Nürnberg B., Schächtele C., Kubbutat M., Totzke F., Laufer S.A. Development of First Lead Structures for Phosphoinositide 3-Kinase-C2γ Inhibitors. J. Med. Chem. 2014;58:212–221. doi: 10.1021/jm5006034. [DOI] [PubMed] [Google Scholar]
- 56.Lehtinen L., Vainio P., Wikman H., Huhtala H., Mueller V., Kallioniemi A., Pantel K., Kronqvist P., Kallioniemi O., Carpèn O., et al. PLA2G7associates with hormone receptor negativity in clinical breast cancer samples and regulates epithelial-mesenchymal transition in cultured breast cancer cells. J. Pathol. Clin. Res. 2017;3:123–138. doi: 10.1002/cjp2.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blakemore S.J., Clifford R., Parker H., Antoniou P., Stec-Dziedzic E., Larrayoz M., Davis Z., Kadalyayil L., Colins A., Robbe P., et al. Clinical significance of TP53, BIRC3, ATM and MAPK-ERK genes in chronic lymphocytic leukaemia: Data from the randomised UK LRF CLL4 trial. Leukemia. 2020;34:1760–1774. doi: 10.1038/s41375-020-0723-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsu S.-H., Wang L.-T., Lee K.-T., Chen Y.-L., Liu K.-Y., Suen J.-L., Chai C.-Y., Wang S.-N. Proinflammatory Homeobox Gene, ISX, Regulates Tumor Growth and Survival in Hepatocellular Carcinoma. Cancer Res. 2013;73:508–518. doi: 10.1158/0008-5472.CAN-12-2795. [DOI] [PubMed] [Google Scholar]
- 59.Ropa J., Saha N., Hu H., Peterson L.F., Talpaz M., Muntean A.G. SETDB1 mediated histone H3 lysine 9 methylation suppresses MLL-fusion target expression and leukemic transformation. Haematologica. 2019;105:2273–2285. doi: 10.3324/haematol.2019.223883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z., Song Y., Li S., Kurian S., Xiang R., Chiba T., Wu X. DNA polymerase θ (POLQ) is important for repair of DNA double-strand breaks caused by fork collapse. J. Biol. Chem. 2019;294:3909–3919. doi: 10.1074/jbc.RA118.005188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaw A.T., Ou S.-H.I., Bang Y.-J., Camidge D.R., Solomon B.J., Salgia R., Riely G.J., Varella-Garcia M., Shapiro G.I., Costa D.B., et al. Crizotinib in ROS1-Rearranged Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2014;371:1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perini G.F., Campregher P.V., Ross J.S., Ali S., Hamerschlak N., Santos F.P.S. Clinical response to everolimus in a patient with Hodgkin’s lymphoma harboring a TSC2 mutation. Blood Cancer J. 2016;6:e420. doi: 10.1038/bcj.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maroto P., Anguera G., Roldan-Romero J.M., Apellániz-Ruiz M., Algaba F., Boonman J., Nellist M., Montero-Conde C., Cascón A., Robledo M., et al. Biallelic TSC2 Mutations in a Patient with Chromophobe Renal Cell Carcinoma Showing Extraordinary Response to Temsirolimus. J. Natl. Compr. Cancer Netw. 2018;16:352–358. doi: 10.6004/jnccn.2017.7041. [DOI] [PubMed] [Google Scholar]
- 64.Yokoyama S., Woods S.L., Boyle G.M., Aoude L.G., MacGregor S.A., Zismann V., Gartside M., Cust A.E., Haq R.U., Harland M., et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buchbinder D., Kirov I., Danielson J., Shah N.N., Freeman A.F., Chavan R.S., Su H.C. Compound Heterozygous DOCK8 Mutations in a Patient with B Lymphoblastic Leukemia and EBV-Associated Diffuse Large B Cell Lymphoma. J. Clin. Immunol. 2019;39:592–595. doi: 10.1007/s10875-019-00663-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ripperger T., Schlegelberger B. Acute lymphoblastic leukemia and lymphoma in the context of constitutional mismatch repair deficiency syndrome. Eur. J. Med. Genet. 2016;59:133–142. doi: 10.1016/j.ejmg.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 67.Cavé H., Caye A., Strullu M., Aladjidi N., Vignal C., Ferster A., Méchinaud F., Domenech C., Pierri F., Contet A., et al. Acute lymphoblastic leukemia in the context of RASopathies. Eur. J. Med. Genet. 2016;59:173–178. doi: 10.1016/j.ejmg.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 68.Nakane T., Goi K., Oshiro H., Kobayashi C., Sato H., Kubota T., Sugita K. Pre-B-Cell Acute Lymphoblastic Leukemia in a Boy with Hereditary Multiple Exostoses Caused byEXT1Deletion. Pediatr. Hematol. Oncol. 2014;31:667–669. doi: 10.3109/08880018.2014.935538. [DOI] [PubMed] [Google Scholar]
- 69.Vollbrecht C., Mairinger F.D., Koitzsch U., Peifer M., Koenig K., Heukamp L.C., Crispatzu G., Wilden L., Kreuzer K.-A., Hallek M., et al. Comprehensive Analysis of Disease-Related Genes in Chronic Lymphocytic Leukemia by Multiplex PCR-Based Next Generation Sequencing. PLoS ONE. 2015;10:e0129544. doi: 10.1371/journal.pone.0129544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neumann M., Vosberg S., Schlee C., Heesch S., Schwartz S., Gökbuget N., Hoelzer D., Graf A., Krebs S., Bartram I., et al. Mutational spectrum of adult T-ALL. Oncotarget. 2015;6:2754–2766. doi: 10.18632/oncotarget.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Puente X.S., Pinyol M., Quesada V., Conde L., Ordóñez G.R., Villamor N., Escaramis G., Jares P., Beà S., González-Díaz M., et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barbieri C.E., Baca S.C., Lawrence M.S., Demichelis F., Blattner M., Theurillat J.-P., White T.A., Stojanov P., Van Allen E., Stransky N., et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poot M. Genes, Proteins, and Biological Pathways Preventing Chromothripsis. Methods Mol. Biol. 2018;1769:231–251. doi: 10.1007/978-1-4939-7780-2_15. [DOI] [PubMed] [Google Scholar]
- 74.Chen L., Huang S., Lee L., Davalos A., Schiestl R.H., Campisi J., Oshima J. WRN, the protein deficient in Werner syndrome, plays a critical structural role in optimizing DNA repair. Aging Cell. 2003;2:191–199. doi: 10.1046/j.1474-9728.2003.00052.x. PMID 12934712. [DOI] [PubMed] [Google Scholar]
- 75.Skoda A.M., Simovic D., Karin V., Kardum V., Vranic S., Serman L. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosn. J. Basic Med. Sci. 2018;18:8–20. doi: 10.17305/bjbms.2018.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walker C.J., Miranda M.A., O’Hern M.J., McElroy J.P., Coombes K.R., Bundschuh R., Cohn D.E., Mutch D.G., Goodfellow P.J. Patterns ofCTCFandZFHX3Mutation and Associated Outcomes in Endometrial Cancer. J. Natl. Cancer Inst. 2015;107 doi: 10.1093/jnci/djv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ennishi D., Mottok A., Ben-Neriah S., Shulha H.P., Farinha P., Chan F.C., Meissner B., Boyle M., Hother C., Kridel R., et al. Genetic profiling of MYC and BCL2 in diffuse large B-cell lymphoma determines cell-of-origin–specific clinical impact. Blood. 2017;129:2760–2770. doi: 10.1182/blood-2016-11-747022. [DOI] [PubMed] [Google Scholar]
- 78.Baron B.W., Anastasi J., Hyjek E.M., Bies J., Reddy P.L., Dong J., Joseph L., Thirman M.J., Wroblewski K., Wolff L., et al. PIM1 gene cooperates with human BCL6 gene to promote the development of lymphomas. Proc. Natl. Acad. Sci. USA. 2012;109:5735–5739. doi: 10.1073/pnas.1201168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ortega-Molina A., Boss I.W., Canela A., Pan H., Jiang Y., Zhao C., Jiang M., Hu D., Agirre X., Niesvizky I., et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat. Med. 2015;21:1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yi D., Xu L., Luo J., You X., Huang T., Zi Y., Li X., Wang R., Zhong Z., Tang X., et al. Germline TP53 and MSH6 mutations implicated in sporadic triple-negative breast cancer (TNBC): A preliminary study. Hum. Genom. 2019;13:4. doi: 10.1186/s40246-018-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Laginestra M.A., Cascione L., Motta G., Fuligni F., Agostinelli C., Rossi M., Sapienza M.R., Righi S., Broccoli A., Indio V., et al. Whole exome sequencing reveals mutations in FAT1 tumor suppressor gene clinically impacting on peripheral T-cell lymphoma not otherwise specified. Mod. Pathol. 2019;33:179–187. doi: 10.1038/s41379-019-0279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meyer S.N., Scuoppo C., Vlasevska S., Bal E., Holmes A.B., Holloman M., Garcia-Ibanez L., Nataraj S., Duval R., Vantrimpont T., et al. Unique and Shared Epigenetic Programs of the CREBBP and EP300 Acetyltransferases in Germinal Center B Cells Reveal Targetable Dependencies in Lymphoma. Immunity. 2019;51:535–547.e9. doi: 10.1016/j.immuni.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.González-Rincón J., Méndez M., Gómez S., García J.F., Martín P., Bellas C., Pedrosa L., Rodríguez-Pinilla S.M., Camacho F.I., Quero C., et al. Unraveling transformation of follicular lymphoma to diffuse large B-cell lymphoma. PLoS ONE. 2019;14:e0212813. doi: 10.1371/journal.pone.0212813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bui C., Le H.K., Vu D.M., Truong K.D., Nguyen N.M., Ho M.A.N., Truong D.Q. ARID1A-SIN3A drives retinoic acid-induced neuroblastoma differentiation by transcriptional repression of TERT. Mol. Carcinog. 2019;58:1998–2007. doi: 10.1002/mc.23091. [DOI] [PubMed] [Google Scholar]
- 85.Rossi D., Trifonov V., Fangazio M., Bruscaggin A., Rasi S., Spina V., Monti S., Vaisitti T., Arruga F., Fama R., et al. The coding genome of splenic marginal zone lymphoma: Activation of NOTCH2 and other pathways regulating marginal zone development. J. Exp. Med. 2012;209:1537–1551. doi: 10.1084/jem.20120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Healy J.A., Nugent A., Rempel R.E., Moffitt A.B., Davis N.S., Jiang X., Shingleton J.R., Zhang J., Love C., Datta J., et al. GNA13 loss in germinal center B cells leads to impaired apoptosis and promotes lymphoma in vivo. Blood. 2016;127:2723–2731. doi: 10.1182/blood-2015-07-659938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fletcher S. MCL-1 inhibitors—Where are we now (2019)? Expert Opin. Ther. Patents. 2019;29:909–919. doi: 10.1080/13543776.2019.1672661. [DOI] [PubMed] [Google Scholar]
- 88.Bycroft C., Fernandez-Rozadilla C., Ruiz-Ponte C., Quintela I., Carracedo A., Donnelly P., Myers S. Patterns of genetic differentiation and the footprints of historical migrations in the Iberian Peninsula. Nat. Commun. 2019;10:551. doi: 10.1038/s41467-018-08272-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for this study were downloaded from the International Cancer Genome Consortium repository.