Abstract
Brugada syndrome (BrS) is an inherited cardiac arrhythmia that predisposes to ventricular fibrillation and sudden cardiac death. It originates from oligogenic alterations that affect cardiac ion channels or their accessory proteins. The main hurdle for the study of the functional effects of those variants is the need for a specific model that mimics the complex environment of human cardiomyocytes. Traditionally, animal models or transient heterologous expression systems are applied for electrophysiological investigations, each of these models having their limitations. The ability to create induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs), providing a source of human patient-specific cells, offers new opportunities in the field of cardiac disease modelling. Contemporary iPSC-CMs constitute the best possible in vitro model to study complex cardiac arrhythmia syndromes such as BrS. To date, thirteen reports on iPSC-CM models for BrS have been published and with this review we provide an overview of the current findings, with a focus on the electrophysiological parameters. We also discuss the methods that are used for cell derivation and data acquisition. In the end, we critically evaluate the knowledge gained by the use of these iPSC-CM models and discuss challenges and future perspectives for iPSC-CMs in the study of BrS and other arrhythmias.
Keywords: brugada syndrome, inherited cardiac arrhythmia, induced pluripotent stem cells, iPSC-cardiomyocytes, electrophysiology
1. Introduction
Brugada syndrome (BrS) is an inherited cardiac arrhythmia characterized by a typical pattern of ST-segment elevation on the electrocardiogram (ECG) and an increased risk of ventricular fibrillation and sudden cardiac death (SCD). It accounts for 20% of SCD in individuals without structural heart disease. [1]. In 20–25% of BrS patients, loss-of-function mutations are identified in the SCN5A gene, which encodes the α subunit of the cardiac sodium channel Nav1.5 [2]. To date, more than 20 other genes have been associated with this oligogenic disease with variants impairing specific ion channels or their accessory proteins involved in the cardiac action potential (AP) (reviewed in [2,3]). Still, three quarters of the diagnosed BrS patients remain without an implicated genetic variant [4]. Importantly, no comprehensive clinical and cellular studies have confirmed most of the candidate gene associations. A recent burden analysis and re-evaluation of reported genes only classified the SCN5A gene as demonstrating definitive evidence as a cause for BrS [5,6].
Concerning pathophysiology, currently three major hypotheses aim to explain the electric abnormality in BrS, namely the repolarization, depolarization and neural crest models [7,8,9,10,11,12,13]. In short, the characteristic ECG changes are respectively explained by (1) transmural dispersion of repolarization in the right ventricle between the endocardium and epicardium; (2) delayed depolarization due to conduction slowing and presence of subtle structural abnormalities in the right ventricular outflow tract (RVOT) and (3) abnormal cardiac neural crest cell migration, cell–cell communication and the development of the RVOT. Despite their differences, these models agree that the major region of pathology is the anterior RV and RVOT and that minute tissue architectural changes or cellular uncoupling effects play a role. Moreover, they are not mutually exclusive and could act in combination. Further studies are required to deepen our understanding of these mechanisms, how they lead to sustained ventricular arrhythmias and link them with the molecular changes underlying BrS.
BrS is well-known to display reduced penetrance and variable expressivity, characterized by a wide range of severity from a life-time asymptomatic status to syncopes and SCD at a young age, even in patients with the identical familial mutation [1]. This clearly hampers patient risk stratification and management. Both environmental and genetic disease modifiers play a role in this variability [14] and over the past years, BrS has been recognized as a more complex disease, with the involvement of multiple common and rare genetic variants acting in concert in its etiology [15,16,17,18,19]. Current application of gene panels, whole exome sequencing (WES) and whole genome sequencing (WGS) in (familial) patients will continue to identify variants potentially involved in BrS, though most of these remain “variants of uncertain significance” (VUS) and without functional studies, a gap remains to translate potential genotype–phenotype correlation into clinical practice.
To further elucidate the oligogenic architecture and pathogenic mechanisms of BrS, understand the functional effects of candidate pathogenic variants at the cell and tissue level and interpret the causality of VUS, representative functional disease models are highly needed. If successful, these models can be applied to test therapeutic approaches. Since currently no proper pharmacological treatment for BrS is available, the implantation of a cardioverter defibrillator (ICD) is the only effective preventative treatment for symptomatic patients. Recently, radio frequency ablation of the arrhythmogenic substrate, mostly in the epicardial surface of the RV(OT), is emerging as a potential alternative [20,21,22,23].
The challenge for functional modelling of cardiac arrhythmias, including BrS, is obtaining tissue specific material. Cardiac biopsies are considered too invasive and the obtained cardiomyocytes (CMs) have a short lifespan in vitro, making them not readily available at sufficient quantities, though some studies on native human CMs have been performed [24,25,26,27,28]. This issue has been addressed by the study of murine, canine, zebrafish or pig cardiomyocytes and cardiac tissue, which offered interesting insights in BrS pathophysiology (extensively reviewed in [29]). The major drawback of those systems are the species-specific differences in physiology, which impacts the translation of the results into the human clinical setting. As another alternative, non-cardiac cellular models have been employed (mostly human embryonic kidney (HEK)293, Chinese hamster ovary (CHO) cells or Xenopus laevis oocytes) to transiently overexpress human cardiac proteins, which enables one to study the function of individual channel complexes and specific mutations (extensively reviewed in [29]). The main shortcoming of these transient expression systems is their disability to mirror the complex CM physiology, as they lack the structural morphology and multi-ion channel environment of native CMs. In addition, results also tend to differ according to the cell-type used. The discovery of induced pluripotent stem cell (iPSC) reprogramming by Takahashi and Yamanaka in 2006 [30], and their subsequent differentiation to functional induced pluripotent stem cell (iPSC)-derived cardiomyocytes (iPSC-CMs) provided an interesting new alternative to the field of cardiac disease modelling. iPSC-CMs provide the (near) complete repertoire of ion channels and accessory proteins expressed in native human CMs, and currently represent the closest resemblance to these cells [31,32]. Since iPSC-CMs can be obtained directly from the patient, they carry the patient’s exact genetic background, including potential modifiers influencing the phenotype.
To date, multiple studies on cardiac arrhythmias using iPSC-CMs have been reported, proving the beneficial impact of this model in the field. A Pubmed literature review, performed in December 2020, identified thirteen articles reporting findings from iPSC-CM models for BrS. In this review, we will provide a comprehensive synthesis of these reports, with a focus on the electrophysiological findings and advances in the field of BrS pathogenesis. We will first discuss the methods used for the derivation of the cell models as well as the techniques used for phenotypic investigations.
2. Methods for Derivation of iPSC-CM Models
2.1. Fibroblast Reprogramming Protocols
The first step in the generation of patient-specific iPSC-CMs is the generation of iPSC lines from a donor sample such as a skin biopsy, or the more easily accessible blood or urine samples [30,33,34]. In all of the reviewed articles, the traditional approach with iPSC reprogramming from skin fibroblasts was applied (Table S1). Several delivery methods to introduce the Yamanaka transcription factors can be used. Lenti- or retro-viral vectors are very efficient but have the major drawback of viral genome integration into the host genome [35]. To overcome that issue, non-integrating episomal vectors or Sendai virus vectors are increasingly used [36,37]. In the generation of the published iPSC-CM BrS models, we observed an equal distribution of the delivery methods, with 9/17 (53%) of the cell lines generated using integrating vectors and 8/17 (47%) using a non-integrating approach (Table S1), but with the non-integrating methods indeed overrepresented in the more recent articles.
2.2. Cardiomyocyte Differentiation
Over the past 10 years, a constant progress in cardiomyocyte differentiation protocols has been made. From initial protocols employing 3D aggregate embryoid body (EB) differentiation with BMP4 and activin A, disease modelling moved to monolayer-based approaches with Wnt pathway modulation using the timed addition of small molecules [38,39,40,41,42]. In the latter, to start the differentiation process the Wnt pathway is indirectly activated through the inhibition of glycogen synthase kinase 3β (e.g., the addition of CHIR99021). Subsequently, after culturing for two or three days, a Wnt pathway inhibitor is added to the culture medium (e.g., Wnt-C59 or IWR1). By day seven or eight, the cultured cells start beating spontaneously, which is one of the first signs of differentiation towards CMs [43].
Though both EB and monolayer-based methods provide decent efficiency in the derivation of functional iPSC-CMs, the monolayer approach provides the most optimal conditions for the diffusion of the differentiating factors and highest differentiation efficiency. This is also reflected in the literature, as the majority of iPSC-CMs were derived through monolayer-based protocols (9/15) (Table S1). These protocols were shown to produce mainly ventricular cardiomyocytes, with few atrial and nodal cells present, which is favorable in view of the ventricular origin of BrS pathology.
Nonetheless, the major and well-known disadvantages of iPSC-CM models are the immaturity of the cells, as well as heterogeneity of the cell culture obtained during the differentiation procedure [21,32,44,45]. Maturation can be improved through long-term culture, mechanical stretching or electrical stimulation, via the application of maturing agents such as triiodothyronine hormone (T3) and glucocorticoid hormone, 3D-culture, specific miRNAs or co-culture with human mesenchymal cells [46,47,48,49,50,51,52,53,54]. In most of the published iPSC-CM BrS models, no specific maturation strategies were used, just a slightly longer culture of at least 30 days before functional testing was performed (except for [55] with at least 19 days). Only de la Roche and colleagues used the cultivation of the iPSC-CMs on a stiff matrix to improve maturation [56]. Further purification of the cell culture with the selection of properly differentiated cardiomyocytes can be obtained with metabolic enrichment approaches, including simple glucose starvation, or substitution of glucose in culture media with lactic acid or fatty acids, to force the switch to a non-oxidative metabolism in the cell culture [57,58,59]. In 6/13 reports, culture heterogeneity was not addressed, and in the other 7/13 glucose starvation or puromycin or lactate treatment was implemented (Table S1).
3. Phenotyping Methods of iPSC-CMs
Since BrS is associated with ventricular pathology [60], analysis of the electrical activity of ventricular cardiomyocytes is the main focus in BrS iPSC-CM modelling and it is important to understand its basis.
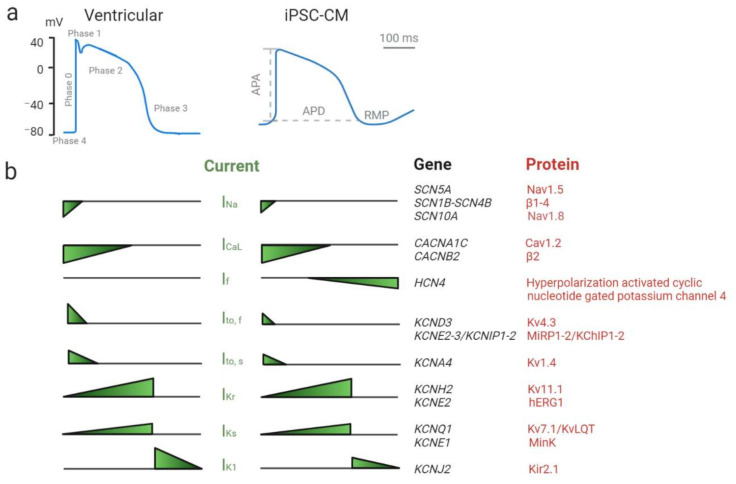
As indicated in Figure 1, the ventricular action potential (AP) reflects a sequential activation and inactivation of ion channels, conducting inward depolarizing Na+ and Ca2+, and outward repolarizing K+ currents [61,62]. The shape of the AP waveform is a reflection of the electrical function of the expressed ion channels (including their auxiliary subunits), and subtle changes in their well-regulated ion conductance (i.e., their time and/or voltage dependence) can have a substantial effect on AP morphology.
Figure 1.
Ventricular action potential and underlying ionic currents (adapted from [32,61,63]). Schematic representation of human ventricular and human induced pluripotent stem cell-derived cardiomyocyte (iPSC-CM) action potential waveforms (a) with indicated in grey: action potential (AP) phases (on ventricular AP waveform) as well as action potential amplitude (APA), action potential duration (APD) and resting membrane potential (RMP) (on iPSC-CM AP waveform). Bottom panel represents the relative magnitude of the underlying ionic currents (b) together with the list of genes (in black) encoding alpha and auxiliary (beta) ion channel protein subunits (in red) which cooperate in generation of the represented ion currents (created with biorender.com December 2020).
Phase 0 of the ventricular AP, a rapid depolarizing upstroke, reflects the function of voltage-gated sodium channels (Nav1.5 with possible contribution of Nav1.8) generating the inward sodium current (INa). It is followed by a transient repolarization of the cell membrane, phase 1, with inactivation of INa and activation of a transient outward voltage-gated K+ current (Ito; consisting of a fast Ito,f and slower Ito,s component). This activity contributes to development of a characteristic prominent “notch” in the ventricular AP waveform. During the plateau phase, phase 2, the depolarization of the cell membrane activates voltage-gated Ca2+ channels, triggering the influx of Ca2+ (ICaL). While the Cav channels inactivate, the outward K+ currents dominate (IKr and IKs) and drive further repolarization of the cell membrane in phase 3. Inwardly rectifying K+ channels (mediating IK1) contribute to late phase three repolarization and to the maintenance of the resting membrane potential (RMP) in phase four [61,62,64].
Unlike adult ventricular CMs, iPSC-derived CMs beat spontaneously due to a more positive RMP caused by reduced IK1 and presence of a pacemaker current (If, generated by HCN4 channels). The depolarized state of the iPSC-CMs results in the inactivation of the majority of Nav1.5 channels, leaving very little INa available during phase 0 and reducing upstroke velocity, and leads to Ito inactivation, which reduces phase one repolarization. In addition, in iPSC-CMs Ito,s seems to be dominant compared to Ito,f, while in adult CMs this is the other way round [61]. These are important characteristics to take into account when studying the electrophysiological phenotype of this model.
In the next paragraphs, we will briefly discuss different techniques employed in the investigations of BrS iPSC-CM electrophysiological parameters, including patch-clamping, calcium and voltage imaging, and micro electrode arrays (MEA).
3.1. Patch-Clamping
Considered the gold standard procedure in electrophysiological characterization, patch-clamping is a technique which allows for the measurement of ionic currents to study the behavior of the channels, or measurement of voltage changes across the cell membrane suited for AP characterization [63,65]. This technique allows for both the actual measurement of the membrane potential, enabling precise characterization of the AP, as well as the analysis of single ionic currents and ion channel activity. As this is an advantage when studying iPSC-CM disease models in detail, in all the discussed reports the patch-clamp technique has been applied (Table 1). Patch-clamping also offers the opportunity to resolve the more depolarized RMP of the iPSC-CMs and achieve a proper upstroke velocity and phase 1 repolarization by using dynamic clamp or injecting a sustained current to reach an RMP of −80–−90 mV. The disadvantages of the method remain its low throughput and its invasive and terminal nature, allowing only short-term recordings. Moreover, the need to record from single cells excludes the possibility to measure AP propagation between iPSC-CMs, though this clearly is a characteristic affected and worth studying in arrhythmias.
Table 1.
Summary of the findings from published BrS iPSC-CM models. Table represents an overview of the identified gene variants, patient diagnosis, electrophysiological characterization methods used and obtained results. Data shown indicate the most important findings from investigated BrS iPSC-CMs models in comparison to control iPSC-CMs for action potential (AP) (APA—AP amplitude; APD—AP duration, dV/dT—upstroke velocity), sodium current (INa) (current density, voltage dependence of activation/inactivation, recovery from inactivation), late sodium current (INaL) (current density), calcium current (ICaL) (current density, activation/inactivation, recovery), potassium currents: Ito, IKs and IKr (current density), calcium transients (CT) (CTD—CT duration, amplitude, rise rate, decay, presence of early afterdepolarization (EAD)/ delayed afterdepolarization (DAD)/arrhythmias) and field potential duration (FPD) characteristics. Legend: + performed analysis; − not performed analysis; ↑ significant increase/positive shift in voltage dependance; ↓ significant decrease/negative shift in voltage dependance; = no difference in comparison to control iPSC-CMs. BrS—Brugada syndrome; LQTS3/BrS—mixed Brugada syndrome and long-QT syndrome type 3 phenotype; Amp. —amplitude.
| Reference | Gene | Variant | Diagnosis | EP Characterization Methods |
Electrophysiological Findings | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AP | INa | INaL | ICaL | Ito | IKs | IKr | CT | FPD | |||||||||||||||||||
| Patch-Clamping | Ca+ Imaging |
MEA | APA | APD | dV/dT | Current Density | Voltage Dependance of Activation | Voltage Dependance of Inactivation | Time of Recovery from Inactivation | Current Density | Current Density | Activation | Inactivation | Recovery | Current Density | Current Density | Current Density | CTD | Amp. | Rise Rate | Decay | EAD/ DAD/ Arrhythmias |
|||||
| Kosmidis et al. 2016 [66] | SCN5A | c.4912C>T (p.R1638X) | BrS | + | − | − | = | = | ↓ | ↓ | = | = | |||||||||||||||
| c.468G>A (p.W156X) | BrS | = | = | ↓ | ↓ | = | = | ||||||||||||||||||||
| Liang et al. 2016 [67] | SCN5A | c.2053G>A (p.R620H) and c.2626G>A (p.R811H) | BrS | + | + | − | = | = | ↓ | ↓ | ↑ | ↓ | ↓ | ↑ | |||||||||||||
| c.4190delA (p.K1397Gfs*5) | BrS | = | = | ↓ | ↓ | = | ↓ | ↓ | ↑ | ||||||||||||||||||
| Ma et al. 2018 [68] | SCN5A | c.677C>T (p.A226V) and c.4885C>T (p.R1629X)c.4859C>T | BrS | + | − | − | ↓ | ↑ | ↓ | ↓ | ↓ | ↓ | ↑ | = | |||||||||||||
| p.T1620M | BrS | = | = | ↑ | = | ||||||||||||||||||||||
| Selga et al. 2018 [69] | SCN5A | c.1100G>A (p.R367H) | BrS | + | − | − | ↓ | ↑ | ↓ | ↓ | |||||||||||||||||
| de la Roche et al. 2019 [56] | SCN5A | c.2204C>T (p.A735V) | BrS | + | − | − | ↓ | = | ↓ | ↓ | ↑ | ↓ | = | = | ↓ | ||||||||||||
| Davis et al. 2012 [70] | SCN5A | c.5537insTGA (p.1795insD) | LQTS3/BrS | + | − | − | = | ↑ | ↓ | ↓ | ↑ | ||||||||||||||||
| Okata et al. 2016 [71] | SCN5A | c.5349G>A (p.E1784K) | LQTS3/BrS | + | − | + | = | ↑ | ↓ | = | = | ↓ | ↑ | ↑ | |||||||||||||
| El-Battrawy et al. 2019 [72] | SCN10A | c.3749G>A (p.R1250Q) and c.3808G>A (p.R1268Q) | BrS | + | + | − | ↓ | = | ↓ | ↓ | = | = | ↓ | ↓ | ↓ | ↑ | ↓ | = | ↓ | = | ↑ | ||||||
| El-Battrawy et al. 2019 [73] | SCN1B | c.629T>C (p.L210P) and c.637C>A (p.P213T) | BrS | + | + | − | ↓ | = | ↓ | ↓ | ↑ | ↓ | ↑ | ↓ | = | = | = | ↑ | = | ↓ | ↓ | = | = | = | ↑ | ||
| Belbachir et al. 2019 [74] | RRAD | p.R211H | BrS | + | + | − | = | ↑ | ↓ | ↓ | = | = | ↓ | ↑ | ↓ | ↑ | ↓ | ↑ | |||||||||
| Cerrone et al. 2014 [75] | PKP2 | c.1904G>A (R635Q) | BrS | + | − | − | ↓ | ||||||||||||||||||||
| Miller et al. 2017 [76] | Undefined | Undefined | BrS | − | − | + | = | ||||||||||||||||||||
| Undefined | Undefined | BrS | = | ||||||||||||||||||||||||
| PKP2 | c.302G>A (p.R101H) | BrS | + | − | + | = | ↓ | = | = | ||||||||||||||||||
| Veerman et al. 2016 [55] | Undefined | Undefined | BrS | + | − | − | = | = | = | = | = | = | = | ||||||||||||||
| Undefined | Undefined | BrS | = | = | = | = | = | = | = | = | = | ||||||||||||||||
| CACNA1C | int19 position -7 (benign) | BrS | = | = | = | = | = | = | = | ||||||||||||||||||
3.2. Calcium and Voltage Fluorescence Imaging
The use of calcium- or voltage- sensitive dyes in combination with fluorescence microscopy provides a non-invasive and high-throughput method for measuring intracellular Ca2+ fluctuations and membrane voltage changes in iPSC-CMs (single cells or growing in clusters or monolayers) [77,78]. With calcium imaging not only Ca2+ flux across the cell membrane but also intracellular Ca2+ oscillations defining the excitation–contraction coupling are captured, allowing more in-depth investigation of the intracellular Ca2+ balance. It should be noted though that iPSC-CMs show structural immaturity with a lack of T-tubules and reduced sarcomere organization, resulting in more immature calcium handling and excitation–contraction coupling. In four of the discussed reports [67,72,73,74] calcium imaging was used in addition to patch-clamping (Table 1). Nevertheless, the use of dyes prohibits long term recordings due to their phototoxicity and the signals have a high noise ratio [63,79].
To overcome these challenges, genetically encoded calcium and voltage indicators (GECI and GEVI) have recently been introduced [80]. The cells are then transfected with genes encoding fluorescent indicators, which consist of a Ca2+ or voltage sensing element coupled to one or two fluorophores that alter their fluorescent intensity based on conformational changes in the sensing element. An advantage of iPSC-CMs compared to native CMs in this case is that iPSC-lines with stable GEVI/GECI expression can be created, stored and repeatedly differentiated to cardiomyocytes. Several GECIs (GCaMP5G, R-GECO1) and one GEVI (ArcLight) have been used in control and patient-derived iPSC-CMs and shown to consistently represent the calcium transients [80,81,82,83,84] and the transmembrane APs [82,83,84,85,86]. Although these reporters offer significant advantages over the traditional dyes, a major drawback is that the transgenes integrate randomly into the genome, raising serious concerns about potential gene disruption and alteration of local and global gene expression that could adversely affect normal cellular functions. Thus far, these genetically encoded indicators have not been used in BrS disease modelling yet.
3.3. Micro-Electrode Array (MEA)
MEA platforms offer the possibility of high-throughput, non-invasive, label-free and long-term measurement of extracellular field potentials (FPs) from clusters and monolayers of iPSC-CMs [63,87,88,89,90,91]. It requires the iPSC-CMs to be seeded on MEA plates, with multiple microelectrodes embedded in 2D grids in the cell culture surface. MEAs have the advantage that AP conduction velocity over a layer of iPSC-CMs can be measured. Conventional arrays have a limited spatial resolution (typically 100 µm) due to the limited number of electrodes in the grid. A new class of MEAs that is based on complementary metal–oxide semiconductor (CMOS) technology has been developed as a solution to this limitation [89,92]. In CMOS-MEAs, thousands of microelectrodes are arranged at high spatial resolution on a chip, tremendously increasing the amount of information that can be gathered from a single iPSC-CM culture. In only two of the discussed articles MEA was used to study BrS iPSC-CMs [71,76] (Table 1), in one to confirm field potential duration (FPD) prolongation correlating to the long QT syndrome (LQTS) part of a mixed phenotype [71] and in the other to perform drug challenges on the cells [76].
4. Findings from Published BrS iPSC-CM Models
Most of the published functional reports from iPSC-CM models, focused on sodium channel genes such as SCN5A [56,66,67,68,69,70,71], SCN10A [72] or SCN1B [73]. In addition, pathogenic variants in other BrS-related genes, such as RRAD [74] or PKP2 [75,76] were modelled. Finally, as BrS is a complex disease with a large proportion of cases being genetically unresolved, characterization of iPSC-CM models from patients with undefined genetic cause has also been performed [55,76]. In this review, we will summarize the findings from these thirteen reports, with a focus on the electrophysiological results. An overview of the studied genes, exact investigated genetic variants and electrophysiological results obtained is given in Table 1 and extra information is available in the Supplement (used reprogramming and differentiation approach with additional patient information are summarized in Table S1; reported electrophysiological parameters for AP, CT, FPD and tested ion channels are summarized in Tables S2–S9).
4.1. Sodium Channel Genes
4.1.1. Sodium Channel α-Subunits—SCN5A
BrS is mainly associated with loss-of-function mutations in SCN5A, leading to reduced INa due to lower expression levels or the production of defective Nav1.5 protein, while gain-of-function mutations in SCN5A contribute to LQT syndrome 3 (LQTS3) [93,94]. In some cases, an overlap of the symptoms of both arrhythmias can be observed in patients carrying specific SCN5A mutations [95,96,97]. Until now, ten iPSC-CM models of BrS-related SCN5A variants have been published. Eight of them were generated from patients with a pure BrS phenotype [56,66,67,68,69] and two from patients with a mixed LQTS/BrS phenotype [70,71] and will be discussed as such in the following paragraphs.
Pure BrS Phenotype
Kosmidis et al. [66] generated two iPSC-CM models with nonsense SCN5A variants, for which they recruited one BrS patient with a more severe phenotype carrying a p.(Arg1638*) variant and one with a relatively mild phenotype related to a p.(Trp156*) variant (Table S1). Both variants caused a reduction in INa density and slowed down the upstroke velocity of the AP in the iPSC-CMs (Tables S2 and S3) compared to iPSC-CMs from two unrelated control individuals. There were no significant differences in the activation or inactivation process of the INa between patient iPSC-CMs and controls. As both modelled variants were predicted to generate a premature stop codon, the presence of nonsense mediated decay (NMD) was investigated in cloning experiments. 61% of patient iPSC-CMs expressed the p.(Arg1638*) variant, confirming that its location in the last exon caused NMD escape, while only 19% of patient iPSC-CMs expressed the p.(Trp156*) variant, confirming the occurrence of NMD. The authors tested two nonsense readthrough-promoting drugs, gentamycin and PTC124, on patient iPSC-CMs to investigate their therapeutic potential on the nonsense variants. Unfortunately, they did not observe any significant increase in INa in treated iPSC-CMs in comparison to baseline conditions, leading to their conclusion that these drugs are unlikely to represent an effective treatment for patients carrying the studied mutations.
Liang et al. performed the characterization of two BrS cases: one (BrS1) with double missense SCN5A mutation: p.(Arg620His) and p.(Arg811His), and a second one (BrS2) with a c.4190delA; p.(Lys1397Glyfs*5) frameshift SCN5A mutation [67]. For comparison, iPSC-CMs were also created of two healthy unrelated control individuals. Both patient iPSC-CMs showed AP profiles of a closely coupled single triggered beat (in 39.6% and 34.5% of recordings in BrS1 and BrS2, respectively), and of sustained triggered activity (5.6% and 6.8% in Brs1 and BrS2, respectively), increased peak-to-peak interval variability and slower upstroke velocity (Table S2). Sodium current analysis in both patient iPSC-CMs showed visible INa reduction (Table S3), which correlated with a lower protein expression of Nav1.5 in comparison to controls. Calcium imaging experiments showed about 60% reduction in Ca2+ transient amplitude, 50–80% decreased rise rate and increased variation in beating intervals in both patient iPSC-CMs (Table S5). RNA-seq revealed a closer homology in overall gene expression profile between the two BrS iPSC-CMs compared to the two control lines, suggesting disease-specific gene expression changes. RT-qPCR analysis confirmed a reduced expression of SCN5A, KCND3 and KCNJ2 in patients compared to control iPSC-CMs, suggesting reduced Ito and reduced IK1 could also be involved in the arrhythmic phenotype. Finally, they used the CRISPR/Cas9 technique to correct the c.4190delA variant in BrS2 patient iPSCs. Genome edited iPSC-CMs (BrSp2-GE) showed a marked reduction in arrhythmic activity, improved upstroke velocity (Table S2) normalized calcium transient (CT) parameters (Table S5), increased Nav1.5 membrane expression and a partial rescue of the INa density (Table S3). As such, the authors were able to prove that the frameshift variant was solely responsible for the observed cellular phenotype.
Ma and colleagues reported an iPSC-CM model from a compound heterozygous SCN5A mutation carrier, carrying p.(Ala226Val) and p.(Arg1629*) (BrS1) and a healthy sibling control (CON1, his unaffected brother) [68]. They also created iPSC-CMs with a milder p.(Thr1620Met) mutation through genome editing (BrS2) and used an extra control iPSC-CM line of commercial iCell® Cardiomyocytes (Fujifilm Cellular Dynamics, Inc., Madison, WI, USA) (CON2). BrS1 iPSC-CMs showed 50% reduced SCN5A mRNA expression in comparison to CON1 iPSC-CMs, suggesting the occurrence of NMD and corresponding with an observed 75% reduction in INa (Table S3). At first, the AP recordings of patient iPSC-CMs did not show significant differences in comparison to controls, which may be explained by the more positive RMP of these cells (about −45 mV) at which most of the INa is not available and Ito is inactivated (Tables S2 and S3). The application of an in silico IK1 injection revealed a >75% reduction in upstroke velocity and a reduction in AP amplitude in BrS1 iPSc-CMs (Table S2) and appearance of phase 1 repolarization, only in CON1 iPSC-CMs. Moderate changes in the rate of steady-state activation and inactivation and a more significant change in the rate of recovery from the inactivation of the sodium channels was observed in BrS1 compared to CON1. BrS2 iPSC-CM characterization revealed normal INa and normal APs, with upstroke velocity (dV/dT) max even higher than in both controls (Table S2). Since BrS pathology is linked with slow heart rates, AP recordings at different lower pacing frequencies were performed. In 25% of BrS1 patient cells, at 0.1 Hz an average 66% action potential duration (APD)90 reduction was observed, while APD prolongation or moderate shortening was observed in the rest of the patient iPSC-CMs and the control cells. (Table S2). The observed marked reduction in APD, represented an increased phase 1 repolarization and loss of phase2 AP pattern, highly resembling the proarrhythmic loss-of-dome in epicardial ventricular CMs, fitting with the repolarization disorder hypothesis. Ito measurements in patient and control iPSC-CMs showed similar current density with an increase in the low pacing rate. To investigate Ito influence on the observed phenotype in BrS1 iPSC-CMs, the cells were treated with 4-aminopyridine (4-AP, blocker of Itof and Itos) and APs were measured at 0.1 Hz. The authors noticed that 0.5 mM 4-AP completely reversed the increased phase one repolarization and loss of phase two dome (Table S2). They concluded that INa and Ito could play a coordinated role in BrS causation, where loss of INa together with heterogeneous elevated Ito in a fraction of iPSC-CMs (at lower heart rates) make the ventricular CMs undergo proarrhythmic changes. Such an observation would not have been possible in a heterologous expression system and supports the value of iPSC-CMs to reveal interplay between different currents in CMs.
Selga et al. performed a comparison of INa properties in iPSC-CMs from a BrS patient carrying a p.(Arg367His) SCN5A variant and an unrelated healthy control individual (two clones each), obtained using two differentiation protocols: an EB-based spontaneous differentiation and monolayer-based differentiation (Table S1) [69]. The authors reported similar observations from both differentiation protocols: a similar reduction in peak INa, a positive shift in the voltage dependence of channel activation (i.e., channel opening) concomitantly with a negative shift in voltage dependence of inactivation (a process that makes the channels non-conductive upon opening), reflecting a clear loss of function (Table S3). They also reported accelerated recovery from inactivation in patient iPSC-CMs in comparison to the control in both tested groups. Interestingly, experiments in tSA cells revealed only the INa reduction of about 48% for the mutant protein, while the steady-state activation/inactivation properties were not changed in comparison to the wild type (WT). This again shows an added value of the iPSC-CM model, revealing pro-arrhythmic channel function changes that were not detected in a conventional heterologous expression system, probably due to the absence of auxiliary subunits.
In 2019, de la Roche and colleagues used CRISPR/Cas9 technology to introduce a homozygous p.(Ala735Val) SCN5A variant in a healthy control iPSC line and obtained two independently derived mutant clones (MUT1 and MUT2) [56]. They state that they used both isogenic and non-genetically related iPSC-CM controls (“wild type” WT) for the electrophysiological characterization. Long-term cultivation (27–42 days) of the iPSC-CMs on a stiff matrix was applied to promote the maturation of the cells. AP recordings carried out with a hyperpolarizing current (~IK1) injection showed similar APD50 values in WT and A735V iPSC-CMs, but significantly reduced action potential amplitude (APA), 74% reduction in upstroke velocity and A735V cells not displaying a sharp AP peak with phase one repolarization notch (Table S2). These recordings strongly indicated altered activation or inactivation characteristics of the mutant Nav1.5 channel. They measured ICaL contribution to the INa current and showed the same ICaL density in both groups (Table S4). A735V iPSC-CMs showed a +30 mV depolarizing shift in the voltage dependence of channel activation and a negative shift in the voltage dependence of inactivation, resulting in the reduced availability of the channels for AP generation at physiological membrane potentials (60% of inactivated channels in mutant and 40% in WT at −80mV) (Table S2). In simultaneous experiments in HEK cells, the reduction in INa as well as the shift in channel activation were observed, however, no differences in the voltage dependence of the inactivation of the mutant channel were noted. This again confirms that different cellular composition can lead to different channel characteristics and emphasizes the importance of the iPSC-CM model. De la Roche et al. investigated differences between two mutant and two WT clones and between five independent differentiation experiments in both groups. As they did not see significant differences in values between clones and differentiations, they were able to pool the data together for the comparisons (Tables S2 and S3).
Mixed Phenotype (BrS/LQTS)
Davis et al. used iPSC-CMs to model an SCN5A p.(1795insAsp) mutation, identified in a large family where mutation carriers presented with variable phenotypes, including diagnosed cases of BrS as well as LQTS and cardiac conduction defects (Table S1) [70]. This mutation was previously modelled in cardiomyocytes isolated from transgenic mice, where a prolongation of APD was observed together with a slowing of the upstroke velocity and reduced INa density but unchanged kinetic properties of this current [98,99]. However, experiments in expression systems (HEK 293) showed a disruption of the fast inactivation, causing a sustained Na+ current (i.e., a late sodium current, INaL) throughout the action potential plateau and prolonging cardiac repolarization at slow heart rates (explaining LQTS phenotype), as well as an increase/augmentation in the slow inactivation component, delaying the recovery of the sodium channel availability between stimuli and reducing the INa at rapid heart rates (explaining BrS phenotype) [100]. In the present study, they first used mouse iPSC-CMs generated from heterozygous mutant and wild type mice and showed reduced upstroke velocity, prolonged APD and INa reduction in the mutant miPSC-CMs but no changes in the voltage dependence of the activation and inactivation parameters. In their human iPSC-CMs, Davis et al. observed a 54% reduction in INa density and an increase in INaL (Table S3) compared to control iPSC-CMs from an independent healthy individual, but they did not characterize activation or inactivation kinetics. Action potential analysis revealed significantly reduced upstroke velocity and prolonged APD90, recapitulating both arrhythmia phenotypes (Table S2).
Another paper reporting an iPSC-CM model of a p.(Glu1784Lys) SCN5A mutation, underlying/causing a mixed BrS/LQTS phenotype, was published by Okata et al. [71]. They used control iPSC-CMs from two unrelated healthy individuals and reported FPD (MEA) as well as APD90 (patch-clamp) prolongation in patient iPSC-CMs (Tables S2 and S6). No differences in upstroke velocity as well as INa density were observed in patient iPSC-CMs, however, they observed an increase in INaL in patient iPSC-CMs (Tables S2 and S3). As they observed a high expression of SCN3B in iPSC-CMs compared to adult CMs and in experiments with tsA-201 cells, they saw that SCN3B co-expression with the mutant E1784K-SCN5A protein increased peak INa and caused a positive shift in the voltage dependence of channel inactivation compared to WT. They performed SCN3B siRNA knockdown in patient iPSC-CMs to check if the expression of this embryonic type Na+ channel β-subunit can mask the BrS phenotype. In iPSC-CMs with SCN3B knockdown, they reported a decrease in INa density and a negative shift in the voltage dependence of channel inactivation only in the patient iPSC-CMs, explaining the BrS phenotype related to the SCN5A mutation. To investigate if the SCN5A p.(Glu1784Lys) mutation is sufficient to produce the observed electrophysiological changes, they generated an isogenic control of the patient iPSCs, using an adenoviral vector. Corrected iPSC-CMs showed shortened APD90 (Table S2), no significant difference in INa density but a significant decrease in INaL (Table S3) compared to the patient iPSC-CMs. They did not discuss this, but it seems they did not perform SCN3B knockdown in these experiments, which could explain these sodium current results. Those results confirmed that the p.(Glu1784Lys) SCN5A mutation contributes solely to the development of the mixed LQTS/BrS phenotype, and that the SCN3B expression in iPSC-CMs can mask the electrophysiological properties characteristic to BrS syndrome.
4.1.2. Sodium Channel α-Subunits—SCN10A
Genome-wide association study (GWAS) analysis of QRS interval duration led to the first identification of SCN10A as a candidate gene in cardiac ventricular conduction disorders [15]. In addition, a GWAS for BrS also detected an association with common variants in SCN10A (coding for the Nav1.8 channel) [19]. These studies were followed by multiple reports of SCN10A variants involved in development of BrS, together with their functional analysis in expression systems, suggesting SCN10A is an important player in BrS etiology [101,102,103,104,105]. Recently, El-Battrawy et al. reported a successful attempt of the iPSC-CM modelling of a double SCN10A mutation (p.(Arg1250Gln) and p.(Arg1268Gln)) related to the BrS phenotype [72]. They used three clones of the patient iPSCs, as well as iPSC-CMs from three independent healthy control individuals. They observed reduced peak INa, reduced INaL and accelerated recovery from inactivation in patient iPSC-CMs (Table S3), contrasting with the increased expression of both SCN5A as well as SCN10A mRNA in patient iPSC-CMs in comparison to controls. El-Battrawy and colleagues also detected about a 50% reduction in KCNJ2 expression in patient iPSC-CMs in comparison to controls, without changes in IK1. Regarding calcium and other potassium currents, they observed reduced peak density, a positive and negative shift of the voltage dependence of channel activation and inactivation, respectively, of ICaL (Table S4), together with a reduction in IKs (Table S9). AP characterization showed reduced APA and upstroke velocity, but similar APD (Table S2). An ajmaline addition revealed an increased susceptibility of patient iPSC-CMs to the sodium blocker, visible in reduction in APA as well as upstroke velocity already with a 3µM ajmaline addition (Table S2). They investigated the presence of arrhythmic events in patient iPSC-CMs using calcium imaging, which revealed an increased frequency of EAD-like events in patient iPSC-CMs (90%) in comparison to controls (50% and 45% in two control lines).
4.1.3. Sodium Channel β-Subunits—SCN1B
The proper function of the cardiac Nav1.5 channel is known to be regulated by the β-subunits of the channel complex [106]. It was previously reported that variants in SCN1B, encoding the β1-subunit, associate with BrS, including successful investigation in expression systems [107]. El-Battrawy et al. performed electrophysiological investigations of compound variants in SCN1B (p.(Leu210Pro) and p.(Pro213Thr)) in patient iPSC-CMs compared with control iPSC-CMs of three independent healthy individuals [73]. They reported increased expression levels of SCN5A and decreased expression levels of CACNA1C, KCNJ2, KCNH2, SCN1B and SCN3B in the patient iPSC-CMs, while on the protein level, they observed reduced SCN1B and similar Nav1.5 expression in comparison to controls. Electrophysiological investigation showed reduced peak INa and INaL together with a positive shift in the voltage dependence of activation and negative shift of the inactivation process as well as decelerated recovery from inactivation of INa in the patient iPSC-CMs (Table S3). When characterizing other ion channels, El-Battrawy et al. reported decelerated recovery from inactivation for ICaL, as well as a reduction in IKs and IKr and no change in Ito in patient iPSC-CMs (Tables S4, S7, S8 and S9). They postulated that these changes probably resulted from secondary changes induced by the SCN1B variants. AP analysis of patient iPSC-CMs revealed reduced APA and upstroke velocity, while APD did not change (Table S2). They investigated the effect of ajmaline addition (30 µM) and observed a more pronounced change in APA as well as upstroke velocity in patient iPSC-CMs in comparison to controls, especially at higher beating frequencies, indicating a higher sensibility of BrS cells to ajmaline application. Using calcium imaging, they observed no differences in Ca2+ transients, but increased arrhythmia like events in patient compared to control iPSC-CMs (85% vs. 45% of cells). After treatment with 10 µM of carbachol, a parasympathetic stimulator, control cells showed a reduction in the beating frequency, whereas the patient iPSC-CMs showed an increase in the beating frequency, an observation that they could not explain.
4.2. iPSC-CM Models of Variants in Other Arrhythmia-Related Genes
4.2.1. Calcium Channel Related Proteins
Whole exome sequencing analysis has been proven a powerful tool in genetic screening of BrS patients. WES analysis of five familial BrS patients performed by Belbachir et al. led to the identification of a p.(Arg211His) variant in the RRAD gene [74]. RRAD encodes RAD (Ras associated with diabetes) GTPase, known to play a role in Cav1.2 trafficking and associated with ventricular arrhythmia in mice. The variant identified by Belbachir and colleagues has a Combined Annotation Dependent Depletion (CADD) score of 33, occurs only once in GnomAD and co-segregated with disease phenotype in the patient family, indicating potential pathogenicity of the variant, and their studies in mouse CMs suggested a gain-of-function effect. Sanger sequencing of RRAD coding regions in 186 unrelated BrS patients led to the identification of three additional rare mutations, showing a trend for RRAD variant enrichment in BrS patients in comparison to 856 tested control individuals. The authors derived iPSC-CMs of two patients (BrS1 and BrS2) and two unaffected non-carrier siblings (Ctl1 and Ctl2) and investigated both INa and ICaL properties, together with AP and CT analysis (Table 1). Electrophysiological tests on patient iPSC-CMs showed the presence of slower spontaneous rhythms, prolonged AP duration and reduced upstroke velocity in comparison to Ctl1 (Table S2). BrS1 iPSC-CMs also showed a ~40% reduction in INa, with a slight acceleration of recovery from inactivation in comparison to Ctl1 (Table S3). The authors correlated these changes with a lower expression of Nav1.5 protein in patient compared to control iPSC-CMs, while on the transcript levels there was no detectable difference. They reported a larger persistent Na+ current INaL and ~30% reduced ICaL in BrS1 iPSC-CMs (Tables S3 and S4). CT results showed the presence of early afterdepolarizations (EADs), slower calcium transient decay and calcium transient duration (CTD) prolongation in patient iPSC-CMs (Table S5). To prove the pathogenicity of the RRAD variant, CRISPR/Cas9 was used to knock it in in an unrelated control iPSC line (Rad R211H ins in Tables S3–S5). Similar to patient iPSC-CMs, genome-edited cells showed slowed spontaneous rhythms, lower upstroke velocity, prolonged APD, reduced peak INa and increased persistent Na+ current compared to the isogenic WT control (Tables S2 and S3). No ICaL differences were observed between the lines, suggesting no significant effect of the variant on Cav2.1 trafficking. However, calcium imaging showed an uneven beat rate, with EADs in 20% of recorded cells and slowed calcium reuptake in the genome-edited iPSC-CMs (Tables S3–S5). Using immunostaining and confocal microscopy, Belbachir et al. observed cytoskeleton defects with impaired F-actin organization and cortical distribution of troponin I in 70% of patient iPSC-CMs. This led to a decrease in cell contractility and focal adhesion formation and patient cells showed a preferential round shape, with increased thickness in comparison to flat control iPSC-CMs [74]. Genome edited knock-in iPSC-CMs confirmed the influence of RRAD p.(Arg211His) on the observed phenotype, as the cytoskeletal defects were now detected in ~40% of the tested cells. The authors concluded that due to preferred expression of RAD in the RVOT, the decreased cell–cell connection could lead to abnormal cardiac conduction in that region and cause BrS. All these results confirmed a causal role of the p.(Arg211His) RRAD variant in development of the BrS phenotype.
4.2.2. Desmosomal Proteins
Although desmosomal protein abnormalities are predominantly linked with arrhythmogenic (right ventricular) cardiomyopathy (ARVC or ACM) [108,109], several studies showing the association of variants in PKP2 with the BrS phenotype have been reported [75,110]. Two BrS-related PKP2 variants of unknown significance have so far been modelled in iPSC-CMs [75,76]. Although Cerrone et al. performed only a small part of their study in iPSC-CMs, they showed that the p.(Arg635Gln) variant in PKP2 alone can contribute to a significant reduction in peak INa density [75]. As their experiments in HL1 cells showed that knockdown of the endogenous PKP2 of the cells in combination with co-expression of the p.(Arg635Gln) variant (as well as other BrS-related missense variants) with WT PKP2 led to a reduction in peak INa, they performed a rescue experiment in the iPSC-CM line derived from an ARVC patient with a homozygous c.2484C > T PKP2 frameshift loss-of-function variant [111] using lentiviral constructs containing WT-PKP2 and PKP2-R635Q. At baseline, INa was significantly reduced in the patient iPSC-CMs compared to the control embryonic stem cell (ESC)-CMs obtained from H9 ESCs (Table S2) and the rescue experiments showed that only WT-PKP2 transduction led to a significant increase in INa density. These results confirmed that INa depends on the expression and structural integrity of PKP2.
A p.(Arg101His) variant in PKP2 (classified as VUS or likely benign) was present in one of the patient iPSC-CMs investigated by Miller et al. in a study aiming to identify ajmaline’s mode of action in iPSC-CMs and test whether differences in ajmaline response could be determined between BrS patients and controls on a MEA platform [76]. AP analysis revealed no differences in upstroke velocity, while APD90 was significantly reduced in patient iPSC-CMs in comparison to the control (Table S2). Although a prolongation of the FPD in the p.(Arg101His) patient iPSC-CMs was visible in the presence of 100 nM of ajmaline (average 43.8 ms; 1.1-fold) in comparison to baseline conditions, while this was not observed in iPSC-CMs from an unrelated healthy control individual (iPSC-HS1M; Table S6), the authors noted comparable FPD prolongation in the presence of 1, 10 and 100 µM of ajmaline in patient and control iPSC-CMs (Table S6). They concluded there was no significantly increased susceptibility to ajmaline in the patient cell lines (see also further below). The authors provided proof of ajmaline blocking both INa and IKr on control iPSC-CMs, however, in the scope of this study they did not include the characterization of any of the underlying currents of the patient iPSC-CMs, but did show reduced APD90 using patch-clamp [76].
4.3. Unknown Genetic Contribution
Attempts have been made in the iPSC-CM modelling of BrS phenotypes from patients with unknown genetic causes [55,76]. In the study by Veerman et al., iPSC-CMs were generated from three BrS patients with spontaneous BrS type-1 ECG pattern. Two did not carry variants in SCN5A and other BrS-related genes, while one patient carried a CACNA1C variant in intron 19 (position −7) that was predicted not to affect the splicing of the gene and was classified as benign. For the electrophysiological characterization, they used iPSC-CMS from two unrelated healthy control subjects (iCtrl1 and iCtrl2) as well as a positive iPSC-CM control (SCN5A p.(1795insAsp) carrier; iSCN5A Supplementary Tables S3 and S4). While in this positive control, Veerman and colleagues observed APD prolongation and the expected dV/dT max reduction, in the analyzed patient lines they did not see significant differences in all tested AP properties in comparison to the negative controls (Table S2), although in one patient line APD was significantly shorter compared to one of the control lines (iCtrl2). Similarly, they did not detect any differences in INa density in patient iPSC-CMs in comparison to negative controls, while in the positive control, they saw a significant reduction in the peak current (Table S3). Interestingly, the authors observed significant differences in V1/2 voltage dependence of the inactivation of INa and ICaL between iCtrl1 and iCtrl2 and the investigated patient iPSC-CMs were similar to one or the other control cell line (Tables S3 and S4), showing variability in channel inactivation kinetics in iPSC-CMs. Nevertheless, in summary, the authors were not able to observe a BrS phenotype in their iPSC-CMs obtained from BrS patients with unknown genetic contributions.
In the study by Miller et al., investigating the effects of ajmaline, two out of three selected iPSC-CM lines were obtained from BrS patients with unknown genetic causes and spontaneous BrS type-1 ECG pattern. The authors observed consistent FPD prolongation in all of the tested patient clones (iBR1-P5M-L1, iBR1-P5M-L9 and iBR1-P6M-L1) with addition of increasing ajmaline concentrations (100 nM–100 µM) with a maximum 1.46-fold increase at a 100 µM ajmaline concentration (Table S6). However, no significant differences in ajmaline susceptibility could be observed between patient and control iPSC-CMs [76]. This study did demonstrate that iPSC-CMs were suitable to test the blocking effect of ajmaline on both depolarization (INa) and repolarization (IKr).
5. Conclusions and Future Perspective
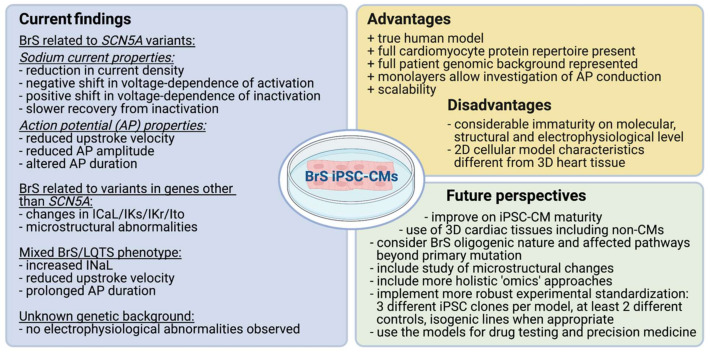
A decade of disease modelling using the novel iPSC-CM approach in the field of cardiac arrhythmias and Brugada syndrome has proven its feasibility in modelling patient-specific cellular phenotypes (Figure 2). In this review, we gathered information from thirteen currently available reports studying BrS-specific iPSC-CMs and discussed the resulting electrophysiological findings.
Figure 2.
Summary of the main findings from the reviewed BrS iPSC-CM models, advantages and disadvantages of the model, as well as future perspectives for the use of iPSC-CMs in BrS research. (Generated with Biorender.com March 2021).
These findings confirmed that the main electrophysiological changes in BrS iPSC-CMs affect sodium channel activity, as previously observed in expression systems and murine models. In particular, a reduced peak INa density [56,66,67,68,69,70,72,73,74,75] was observed in all the described patient iPSC-CMs with an identified mutation, both in SCN5A or the other genes. So, all genetic variants led to the development of a phenotype with reduced INa. (Table 1 and Table S3). When sodium channel kinetics were studied, mostly a negative shift in the voltage dependence of channel inactivation (leading to a higher number of non-conductive inactivated channels) [56,68,69,71,73], a positive shift in the voltage dependence of channel activation (i.e., stronger membrane depolarization is required to open the channels) [56,69,73] and in two reports, an increased time of recovery from inactivation (leading to the prolonged inactivation of the channels) [68,73] was observed, all fitting with the reduced INa phenotype. In one report, a negative shift in the voltage dependence of channel activation [68] and in three reports, an accelerated recovery from inactivation [69,72,74] were observed, which were not explained with regard to the phenotype. Interestingly, in two papers [56,69], changes in channel kinetics were not detected in a conventional heterologous expression system, confirming the value of the iPSC-CM model expressing a more complete CM intracellular environment, including channel auxiliary subunits. In line with the sodium current defects, reduced upstroke velocity was observed in 11 out of 15 models, where AP properties were tested [56,66,67,68,70,71,72,73,74] and APA reduction was seen in four models [56,68,72,73] (Table 1 and Table S2). When other currents apart from INa were investigated, mostly when modelling variants in other genes than SCN5A, also ICaL [72,73,74] and/or in some instances IKs [72,73], IKr [73] as well as Ito [56] property changes were reported. Taking into account the oligogenic nature of BrS, it would be interesting to study several currents in BrS iPSC-CM models.
Interestingly, in two reports authors noticed microstructural changes in their disease models, which could contribute to the observed electrophysiological abnormalities [74,75]. Belbachir and colleagues observed that the RRAD p.(Arg211His) variant disturbed F-actin organization in patient iPSC-CMs, compromising focal adhesion and potentially cell–cell communication and AP conductance, mainly in the RVOT where RAD is expressed [74]. Cerrone et al. reported a microtubule abnormality in adult CMs from a heterozygous PKP2 p.R635Q mouse, they did not study this in their iPSC-CM model. As sodium channel proteins are transported via the microtubule network, their results show that PKP2 deficiency can affect the ability of microtubules to reach intercalated discs and impair Nav1.5 expression there [75]. It has already been suggested that disturbances in PKP2, and other desmosomal or junctional proteins at the CM intercalated discs (the “connexome”) can affect the trafficking or activity of sodium channels over there [91,112,113,114] explaining the link between structural abnormalities, electrophysiological changes and BrS. The interplay and dependency of all these intercalated disc proteins, integrating both mechanical and electrical functions of the CMs, have been beautifully illustrated in recent reviews [115,116,117]. In addition, in clinical studies, discrete structural abnormalities in the right ventricle of BrS patients have been observed [23,118,119]. Based on these insights, it would be interesting to look in more detail into structural changes in all BrS iPSC-CM models generated, since these could be part of the primary defect or play a modifying role in the disease phenotype.
iPSC-CMs from carriers of SCN5A variants presenting with a mixed BrS/LQTS pathology were shown to recapitulate both phenotypes: reduced upstroke velocity (i.e., dV/dT max) or increased INaL as well as APD prolongation was noted [70,71] (Table 1). Interestingly, APD changes were also observed in iPSC-CMs from pure BrS patients, one carrying a compound heterozygous SCN5A mutation (p.(Ala226Val) and p.(Arg1629*)) and one a p.(Arg211His) RRAD variant, where APD prolongation was noted [68,74], and one with a PKP2 p.(Arg210His) variant, where APD was reduced in comparison to control [76]. Similarly, in studies where Ca2+ imaging and MEA technology were used, calcium transient and FPD differences have been reported in patient iPSC-CMs in comparison to controls (in six out of seven tested BrS iPSC-CMs at baseline) (Tables S5 and S6) [67,71,74].
An interesting advantage of iPSC-CM models is that they provide the opportunity to study cellular disease mechanisms in patients without an identified mutation, and two groups indeed investigated this. In one study [55], iPSC-CMs of three different patients showed no changes in sodium currents, nor other AP characteristics studied, even at different pacing frequencies. In the other study [76], only the effect of ajmaline on FPD was investigated and no increased susceptibility to ajmaline was observed in the two studied patients. Perhaps very subtle or non-studied electrophysiological changes were present in the models. Another explanation could be that the clinical phenotype was not caused by defects in ion currents but involved structural abnormalities at the cell and/or tissue level or disturbances of other unexpected pathways. This again supports the importance of in-depth study of the generated iPSC-CM BrS models, and the value of such models containing the full CM intracellular protein repertoire as well as the possibility to create and study more tissue-like structures. When the genetic cause is unknown or when modifiers underlying variable disease expressivity are sought, an approach including transcriptome and proteome analysis of iPSC-CMs could provide a more holistic picture and valuable indications on involved genes, proteins or pathways.
The fact that patient iPSC-CMs carry the patient’s full genetic background, including potential modifiers, can be a drawback of the model as well, when trying to establish the causal relationship between genotype and phenotype and to prove the pathogenic effect of a specific genetic variant. This is especially important when evaluating VUS or potential novel BrS candidate genes. In such cases, genetic engineering, such as CRISPR Cas genome editing, should be applied to correct the studied variant in the patient iPSCs (creating an isogenic control) or introduce it into one or more control iPSC-lines. The study of these models will reveal whether the investigated variant is necessary and sufficient to cause the phenotype or suggest the contribution of other genetic factors in the observed phenotype. In four of the discussed reports, genetic engineering was used to create isogenic controls [67,71] or introduce the variant of interest on a control background [56,74]. In each study, the modified iPSC-CMs showed rescue or development of the phenotype [56,67,71,74], confirming the causative status of novel variants and in one case, providing evidence for the involvement of SCN3B in subtle changes in the BrS/LQTS phenotype [71].
Over the years, it has been shown that iPSC-CMs display an immature phenotype with an expression pattern resembling that of fetal human cardiomyocytes [31,32,120,121]. They are spontaneously active (like fetal hCMs) due to a substantially reduced IK1 density and the presence of a pacemaker current. Though sodium channel expression is normal in iPSC-CMs, the more positive RMP results in the inactivation of the channels, leaving very limited INa available during the phase 0 depolarization and thereby potentially masking the impact of INa deficiency on the AP, more specifically the upstroke velocity. The depolarized state of iPSC-CMs could also lead to Ito inactivation and the absence of phase one repolarization in iPSC-CMs. This obviously poses a problem for BrS modelling, leading to the choice of several groups to impose IK1 onto their iPSC-CMs to achieve an RMP of −80–−90 mV and a more mature electrophysiological phenotype. It should be noted that this is performed on isolated cells, deprived of cell–cell contacts that are also potentially important for current expression, such as INa. It is clear from Table S2 that in studies without IK1 injection and with depolarized RMP of the iPSC-CMs, the values of upstroke velocity were much lower than the normal range of native adult CMs and it is questionable whether they represent trustworthy values and true differences between patient and control models. Other interesting observations brought forward in the discussed articles, are the likely different expression profiles of Ito,f and Ito,s in hiPSC-CMs compared with that in adult epicardial ventricular CMs (Ito,s dominant in the first and Ito,f dominant in the latter) [61], and the expression of a foetal form of sodium channel β-subunit (SCN3B) that could be masking the BrS phenotype by compensating INa reduction [71]. In addition, iPSC-CMs show structural immaturity including a lack of T-tubules, and reduced sarcomere organization. This has been shown to affect calcium handling and excitation–contraction coupling in iPSC-CMs [32]. Additionally, in light of the previously mentioned potential microstructural abnormalities in BrS, this could affect read-outs and should certainly be taken into consideration when interpreting results. Methods to mature iPSC-CMs, such as hormone treatments, mechanical and electrical stimulation, 3D cell culture etc., are hot topic of investigations and are anticipated to show their value in future studies. It should be noted as well that every model has it limits, but it is important to recognize them and take them into account when interpreting and extrapolating data.
Since the differentiation of iPSCs to CMs is not fully defined and the addition of Wnt signaling pathway influencing factors is certainly a minimization of all processes present during actual embryonic development, the current methodology results in a heterogeneous iPSC-CM culture (mixed with non-CMs) that can vary with every differentiation round. To create robust and comparable data between BrS disease modeling studies, iPSC-CM populations of comparable purity, subtype and maturity should be used. On top of that, genetic alterations induced during reprogramming and/or passaging of iPSCs in culture create additional variability in the model, calling for the replication of obtained phenotypic and functional results in several (clonal) cell lines. Here, the scalability of iPSC-CMs in combination with high-throughput phenotyping techniques clearly present an advantage. In all of the cited BrS iPSC-CM reports, data from several differentiation rounds were combined, which is good practice. De la Roche and colleagues even specifically showed that the results between differentiation experiments were not significantly different and could be indeed pooled together [56]. Additionally, in the report of Selga et al., differences between iPSC-CM differentiation methods were addressed, and the effect of the mutation was shown to be similar in magnitude in iPSC-CMs derived using an EB- as well as a monolayer-based approach [69]. Selga et al. reported no significant differences between two clones differentiated from the same cell line. In most of the articles, several clones of patient and/or control iPSC-CMs were tested, but often not all were used for all comparisons. In fact, best practice to obtain rigid data from the models would require at least three different iPSC clones to be used for each patient and control individual and should be compared for all characteristics. Additionally, the use of multiple patient and control iPSC-CM lines is recommended. This should account for variability between individuals with different genetic backgrounds and comorbidities. In about half of the discussed reports, a single control iPSC-CM line was used [69,70,71,74,75,76], while in five articles two [55,56,66,67,68] and in two articles three [72,73] controls were analyzed. In some cases, authors observed differences in electrophysiological properties between their control cell lines [55,68] and then tended to compare their results to each control line separately. In other cases, where similar discrepancies between the control lines were observed (e.g., APD50 or dV/dT max values) [56,72,73], the authors compared their results from patient iPSC-CMs to the pooled range of values from all characterized control iPSC-CMs. It is clear that standardization, with the use of multiple clones and multiple individuals for all functional characterization studies, is needed in the field of iPSC-CM modelling.
Another important note is that the current iPSC-CM models only provide a cellular system demonstrating abnormal electrical behavior or structural abnormalities in single cells or 2D sheets, and caution must be exercised in extrapolating findings to the whole heart. The current iPSC differentiation protocols predominantly generate ventricular-like iPSC-CMs [40,41] but protocols aimed at specifically generating atrial-like [122] and nodal-like [123] iPSC-CMs have also been described. The co-culture of a controlled mixture of these different cell-types, potentially including non-cardiomyocytes (such as cardiac fibroblasts) in 3D tissues with proper extracellular matrix is predicted to be relevant to the study of cardiac arrhythmias. Though it will be hard to get these tissues organized in layers of endocardium and epicardium and/or attribute left- or right sidedness, study of arrhythmic events such as re-entry, conductance problems, local AP and current heterogeneity and even tissue fibrosis and structural changes that could all be relevant to the BrS phenotype and disease pathomechanisms should be feasible. In addition, such culture systems are very likely to improve on the maturity of the iPSC-CMs. Although quite some effort has already been made to create engineered heart tissue (EHT), 3D cardiac microtissues, organoids or microfluidic human-body-on-a-chip systems [48,124,125,126,127,128,129], this field needs further development and application of these novel, more physiological iPSC-CM based models. The latter will certainly provide interesting improvements for BrS disease modelling, as well as for the application of these models for drug testing, precision medicine and cardiotoxicity screening [130,131,132].
In conclusion, the thirteen discussed studies have proven the capability of the iPSC-CM models to recapitulate the BrS patient phenotype (Figure 2). However, no particularly compelling novel disease insights for BrS have emerged yet and it is important to take the current limitations of iPSC-CMs into account when using them as a model in studying Brugada syndrome and other cardiac arrhythmias. Focus on technologies to improve iPSC-CM maturity and create proper engineered heart tissues in combination with standardization of the experimental set-ups and more in-depth (functional) studies of the generated iPSC-CM models will help bridge the gap between model and clinical practice (Figure 2). Taken together, this exciting approach clearly holds great promise for BrS and cardiac disease research.
Abbreviations
| AP | action potential |
| APA | action potential amplitude |
| APD | action potential duration |
| BMP4 | bone morphogenic protein 4 |
| BrS | Brugada syndrome |
| CMs | Cardiomyocytes |
| CT | calcium transient |
| CTD | calcium transient duration |
| DAD | delayed afterdepolarization |
| dV/dT | upstroke velocity |
| EAD | early afterdepolarization |
| EB | embryoid bodies |
| ECG | electrocardiogram |
| FP | field potential |
| FPD | field potential duration |
| GECI | genetically encoded calcium indicators |
| GEVI | genetically encoded voltage indicators |
| ICD | implanted cardioverter defibrillator |
| iPSCs | induced pluripotent stem cells |
| iPSC-CMs | induced pluripotent stem cell-derived cardiomyocytes |
| LQTS | long-QT syndrome |
| LQTS/BrS | mixed phenotype of long-QT and Brugada syndrome |
| MEA | multielectrode array |
| NMD | nonsense mediated decay |
| RMP | resting membrane potential |
| RVOT | right ventricular outflow tract |
| SCD | sudden cardiac death |
| T3 | triiodothyronine 3 |
| VUS | variant of unknown significance |
| WES | whole exome sequencing |
| WGS | whole genome sequencing |
| WT | wild type |
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/22/6/2825/s1, Table S1: Overview of the used reprogramming and differentiation approaches, Table S2: AP properties published from BrS iPSC-CM models, Table S3: INa properties from published BrS iPSC-CM models, Table S4: Calcium current (ICaL) properties from published BrS iPSC-CMs, Table S5: Calcium transient properties from published BrS iPSC-CMs, Table S6: Field potential properties from published BrS iPSC-CMs, Table S7: Ito properties from published BrS iPSC-CMs, Table S8: IKr properties from published BrS iPS-CMs, Table S9: IKs properties from published BrS iPSC-CMs.
Author Contributions
Conceptualization, A.N. and M.A.; writing—original draft preparation, A.N. and M.A.; writing—review and editing, J.S., A.J.L., D.S. and B.L.L.; figure and table preparation, A.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University of Antwerp (GOA) and Research Foundation—Flanders (FWO, Belgium), grant number G.0356.17. B.L.L. holds a consolidator grant from the European Research Council (Genomia—ERC-COG-2017-771945). A.N. holds a Ph.D. fellowship of the FWO (1S24317N). D.S. is supported by a postdoctoral fellowship of the FWO (12R5610N).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Antzelevitch C. Brugada syndrome. Pacing Clin. Electrophysiol. 2006;29:1130–1159. doi: 10.1111/j.1540-8159.2006.00507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brugada J., Campuzano O., Arbelo E., Sarquella-Brugada G., Brugada R. Present Status of Brugada Syndrome: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2018;72:1046–1059. doi: 10.1016/j.jacc.2018.06.037. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe H., Minamino T. Genetics of Brugada syndrome. J. Hum. Genet. 2016;61:57–60. doi: 10.1038/jhg.2015.97. [DOI] [PubMed] [Google Scholar]
- 4.Kapplinger J.D., Tester D.J., Alders M., Benito B., Berthet M., Brugada J., Brugada P., Fressart V., Guerchicoff A., Harris-Kerr C., et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7:33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosseini S.M., Kim R., Udupa S., Costain G., Jobling R., Liston E., Jamal S.M., Szybowska M., Morel C.F., Bodwin S., et al. Reappraisal of Reported Genes for Sudden Arrhythmic Death: Evidence-Based Evaluation of Gene Validity for Brugada Syndrome. Circulation. 2018;138:1195–1205. doi: 10.1161/CIRCULATIONAHA.118.035070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Scouarnec S., Karakachoff M., Gourraud J.B., Lindenbaum P., Bonnaud S., Portero V., Duboscq-Bidot L., Daumy X., Simonet F., Teusan R., et al. Testing the burden of rare variation in arrhythmia-susceptibility genes provides new insights into molecular diagnosis for Brugada syndrome. Hum. Mol. Genet. 2015;24:2757–2763. doi: 10.1093/hmg/ddv036. [DOI] [PubMed] [Google Scholar]
- 7.Antzelevitch C. The Brugada syndrome: Ionic basis and arrhythmia mechanisms. J. Cardiovasc. Electrophysiol. 2001;12:268–272. doi: 10.1046/j.1540-8167.2001.00268.x. [DOI] [PubMed] [Google Scholar]
- 8.Meregalli P.G., Wilde A.A., Tan H.L. Pathophysiological mechanisms of Brugada syndrome: Depolarization disorder, repolarization disorder, or more? Cardiovasc. Res. 2005;67:367–378. doi: 10.1016/j.cardiores.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Elizari M.V., Levi R., Acunzo R.S., Chiale P.A., Civetta M.M., Ferreiro M., Sicouri S. Abnormal expression of cardiac neural crest cells in heart development: A different hypothesis for the etiopathogenesis of Brugada syndrome. Heart Rhythm. 2007;4:359–365. doi: 10.1016/j.hrthm.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Antzelevitch C. Role of spatial dispersion of repolarization in inherited and acquired sudden cardiac death syndromes. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H2024–H2038. doi: 10.1152/ajpheart.00355.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morita H., Zipes D.P., Fukushima-Kusano K., Nagase S., Nakamura K., Morita S.T., Ohe T., Wu J. Repolarization heterogeneity in the right ventricular outflow tract: Correlation with ventricular arrhythmias in Brugada patients and in an in vitro canine Brugada model. Heart Rhythm. 2008;5:725–733. doi: 10.1016/j.hrthm.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 12.Lambiase P.D., Ahmed A.K., Ciaccio E.J., Brugada R., Lizotte E., Chaubey S., Ben-Simon R., Chow A.W., Lowe M.D., McKenna W.J. High-density substrate mapping in Brugada syndrome: Combined role of conduction and repolarization heterogeneities in arrhythmogenesis. Circulation. 2009;120:106–117. doi: 10.1161/CIRCULATIONAHA.108.771401. [DOI] [PubMed] [Google Scholar]
- 13.Wilde A.A., Postema P.G., Di Diego J.M., Viskin S., Morita H., Fish J.M., Antzelevitch C. The pathophysiological mechanism underlying Brugada syndrome: Depolarization versus repolarization. J. Mol. Cell. Cardiol. 2010;49:543–553. doi: 10.1016/j.yjmcc.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verkerk A.O., Amin A.S., Remme C.A. Disease Modifiers of Inherited SCN5A Channelopathy. Front. Cardiovasc. Med. 2018;5:137. doi: 10.3389/fcvm.2018.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sotoodehnia N., Isaacs A., de Bakker P.I., Dorr M., Newton-Cheh C., Nolte I.M., van der Harst P., Muller M., Eijgelsheim M., Alonso A., et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat. Genet. 2010;42:1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz P.J., Ackerman M.J., Antzelevitch C., Bezzina C.R., Borggrefe M., Cuneo B.F., Wilde A.A.M. Inherited cardiac arrhythmias. Nat. Rev. Dis. Primers. 2020;6:58. doi: 10.1038/s41572-020-0188-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilde A.A., Bezzina C.R. Genetics of cardiac arrhythmias. Heart. 2005;91:1352–1358. doi: 10.1136/hrt.2004.046334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polovina M.M., Vukicevic M., Banko B., Lip G.Y.H., Potpara T.S. Brugada syndrome: A general cardiologist’s perspective. Eur. J. Intern. Med. 2017;44:19–27. doi: 10.1016/j.ejim.2017.06.019. [DOI] [PubMed] [Google Scholar]
- 19.Bezzina C.R., Barc J., Mizusawa Y., Remme C.A., Gourraud J.B., Simonet F., Verkerk A.O., Schwartz P.J., Crotti L., Dagradi F., et al. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat. Genet. 2013;45:1044–1049. doi: 10.1038/ng.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morita H., Zipes D.P., Morita S.T., Lopshire J.C., Wu J. Epicardial ablation eliminates ventricular arrhythmias in an experimental model of Brugada syndrome. Heart Rhythm. 2009;6:665–671. doi: 10.1016/j.hrthm.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Brugada J., Pappone C., Berruezo A., Vicedomini G., Manguso F., Ciconte G., Giannelli L., Santinelli V. Brugada Syndrome Phenotype Elimination by Epicardial Substrate Ablation. Circ. Arrhythm. Electrophysiol. 2015;8:1373–1381. doi: 10.1161/CIRCEP.115.003220. [DOI] [PubMed] [Google Scholar]
- 22.Rudic B., Chaykovskaya M., Tsyganov A., Kalinin V., Tulumen E., Papavassiliu T., Dosch C., Liebe V., Kuschyk J., Roger S., et al. Simultaneous Non-Invasive Epicardial and Endocardial Mapping in Patients with Brugada Syndrome: New Insights into Arrhythmia Mechanisms. J. Am. Heart Assoc. 2016;5:e004095. doi: 10.1161/JAHA.116.004095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pappone C., Brugada J., Vicedomini G., Ciconte G., Manguso F., Saviano M., Vitale R., Cuko A., Giannelli L., Calovic Z., et al. Electrical Substrate Elimination in 135 Consecutive Patients with Brugada Syndrome. Circ. Arrhythm. Electrophysiol. 2017;10:e005053. doi: 10.1161/CIRCEP.117.005053. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt C., Wiedmann F., El-Battrawy I., Fritz M., Ratte A., Beller C.J., Lang S., Rudic B., Schimpf R., Akin I., et al. Reduced Na(+) Current in Native Cardiomyocytes of a Brugada Syndrome Patient Associated With beta-2-Syntrophin Mutation. Circ. Genom. Precis. Med. 2018;11:e002263. doi: 10.1161/CIRCGEN.118.002263. [DOI] [PubMed] [Google Scholar]
- 25.Johnson W.B., Katugampola S., Able S., Napier C., Harding S.E. Profiling of cAMP and cGMP phosphodiesterases in isolated ventricular cardiomyocytes from human hearts: Comparison with rat and guinea pig. Life Sci. 2012;90:328–336. doi: 10.1016/j.lfs.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Ajiro Y., Hagiwara N., Katsube Y., Sperelakis N., Kasanuki H. Levosimendan increases L-type Ca(2+) current via phosphodiesterase-3 inhibition in human cardiac myocytes. Eur. J. Pharmacol. 2002;435:27–33. doi: 10.1016/S0014-2999(01)01569-2. [DOI] [PubMed] [Google Scholar]
- 27.Qu Y., Page G., Abi-Gerges N., Miller P.E., Ghetti A., Vargas H.M. Action Potential Recording and Pro-arrhythmia Risk Analysis in Human Ventricular Trabeculae. Front. Physiol. 2017;8:1109. doi: 10.3389/fphys.2017.01109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen N., Nguyen W., Nguyenton B., Ratchada P., Page G., Miller P.E., Ghetti A., Abi-Gerges N. Adult Human Primary Cardiomyocyte-Based Model for the Simultaneous Prediction of Drug-Induced Inotropic and Pro-arrhythmia Risk. Front. Physiol. 2017;8:1073. doi: 10.3389/fphys.2017.01073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sendfeld F., Selga E., Scornik F.S., Perez G.J., Mills N.L., Brugada R. Experimental Models of Brugada syndrome. Int. J. Mol. Sci. 2019;20:2123. doi: 10.3390/ijms20092123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 31.Kodama M., Furutani K., Kimura R., Ando T., Sakamoto K., Nagamori S., Ashihara T., Kurachi Y., Sekino Y., Frurukawa T., et al. Systematic expression analysis of genes related to generation of action potentials in human iPS cell-derived cardiomyocytes. J. Pharmacol. Sci. 2019;140:325–330. doi: 10.1016/j.jphs.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Karakikes I., Ameen M., Termglinchan V., Wu J.C. Human induced pluripotent stem cell-derived cardiomyocytes: Insights into molecular, cellular, and functional phenotypes. Circ. Res. 2015;117:80–88. doi: 10.1161/CIRCRESAHA.117.305365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee Y.M., Zampieri B.L., Scott-McKean J.J., Johnson M.W., Costa A.C.S. Generation of Integration-Free Induced Pluripotent Stem Cells from Urine-Derived Cells Isolated from Individuals with Down Syndrome. Stem Cells Transl. Med. 2017;6:1465–1476. doi: 10.1002/sctm.16-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye L., Muench M.O., Fusaki N., Beyer A.I., Wang J., Qi Z., Yu J., Kan Y.W. Blood cell-derived induced pluripotent stem cells free of reprogramming factors generated by Sendai viral vectors. Stem Cells Transl. Med. 2013;2:558–566. doi: 10.5966/sctm.2013-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vannucci L., Lai M., Chiuppesi F., Ceccherini-Nelli L., Pistello M. Viral vectors: A look back and ahead on gene transfer technology. New Microbiol. 2013;36:1–22. [PubMed] [Google Scholar]
- 36.Fusaki N., Ban H., Nishiyama A., Saeki K., Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lufino M.M., Edser P.A., Wade-Martins R. Advances in high-capacity extrachromosomal vector technology: Episomal maintenance, vector delivery, and transgene expression. Mol. Ther. 2008;16:1525–1538. doi: 10.1038/mt.2008.156. [DOI] [PubMed] [Google Scholar]
- 38.Weng Z., Kong C.W., Ren L., Karakikes I., Geng L., He J., Chow M.Z., Mok C.F., Chan H.Y.S., Webb S.E., et al. A simple, cost-effective but highly efficient system for deriving ventricular cardiomyocytes from human pluripotent stem cells. Stem Cells Dev. 2014;23:1704–1716. doi: 10.1089/scd.2013.0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharya S., Burridge P.W., Kropp E.M., Chuppa S.L., Kwok W.M., Wu J.C., Boheler K.R., Gundry R.L. High efficiency differentiation of human pluripotent stem cells to cardiomyocytes and characterization by flow cytometry. J. Vis. Exp. 2014:52010. doi: 10.3791/52010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M., et al. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mummery C.L., Zhang J., Ng E.S., Elliott D.A., Elefanty A.G., Kamp T.J. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: A methods overview. Circ. Res. 2012;111:344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao M., Tang Y., Zhou Y., Zhang J. Deciphering Role of Wnt Signalling in Cardiac Mesoderm and Cardiomyocyte Differentiation from Human iPSCs: Four-dimensional control of Wnt pathway for hiPSC-CMs differentiation. Sci. Rep. 2019;9:19389. doi: 10.1038/s41598-019-55620-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dias T.P., Pinto S.N., Santos J.I., Fernandes T.G., Fernandes F., Diogo M.M., Prieto M., Cabral J.M.S. Biophysical study of human induced Pluripotent Stem Cell-Derived cardiomyocyte structural maturation during long-term culture. Biochem. Biophys. Res. Commun. 2018;499:611–617. doi: 10.1016/j.bbrc.2018.03.198. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Y., Wang L., Liu Z., Alimohamadi S., Yin C., Liu J., Qian L. Comparative Gene Expression Analyses Reveal Distinct Molecular Signatures between Differentially Reprogrammed Cardiomyocytes. Cell Rep. 2017;20:3014–3024. doi: 10.1016/j.celrep.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White M.C., Pang L., Yang X. MicroRNA-mediated maturation of human pluripotent stem cell-derived cardiomyocytes: Towards a better model for cardiotoxicity? Food Chem. Toxicol. 2016;98 Pt A:17–24. doi: 10.1016/j.fct.2016.05.025. [DOI] [PubMed] [Google Scholar]
- 47.Yang X., Rodriguez M., Pabon L., Fischer K.A., Reinecke H., Regnier M., Sniadecki N.J., Ruohola-Baker H., Murry C.E. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J. Mol. Cell. Cardiol. 2014;72:296–304. doi: 10.1016/j.yjmcc.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida S., Miyagawa S., Fukushima S., Kawamura T., Kashiyama N., Ohashi F., Toyofuku T., Toda K., Sawa Y. Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes by Soluble Factors from Human Mesenchymal Stem Cells. Mol. Ther. 2018;26:2681–2695. doi: 10.1016/j.ymthe.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulmer B.M., Stoehr A., Schulze M.L., Patel S., Gucek M., Mannhardt I., Funcke S., Murphy E., Eschenhagen T., Hansen A. Contractile Work Contributes to Maturation of Energy Metabolism in hiPSC-Derived Cardiomyocytes. Stem Cell Rep. 2018;10:834–847. doi: 10.1016/j.stemcr.2018.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewandowski J., Rozwadowska N., Kolanowski T.J., Malcher A., Zimna A., Rugowska A., Fiedorowicz K., Labedz W., Kubaszewski L., Chojnacka K., et al. The impact of in vitro cell culture duration on the maturation of human cardiomyocytes derived from induced pluripotent stem cells of myogenic origin. Cell Transplant. 2018;27:1047–1067. doi: 10.1177/0963689718779346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nunes S.S., Miklas J.W., Liu J., Aschar-Sobbi R., Xiao Y., Zhang B., Jiang J., Masse S., Gagliardi M., Hsieh A., et al. Biowire: A platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat. Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parikh S.S., Blackwell D.J., Gomez-Hurtado N., Frisk M., Wang L., Kim K., Dahl C.P., Fiane A., Tonnessen T., Kryshtal D.O., et al. Thyroid and Glucocorticoid Hormones Promote Functional T-Tubule Development in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circ. Res. 2017;121:1323–1330. doi: 10.1161/CIRCRESAHA.117.311920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruan J.L., Tulloch N.L., Razumova M.V., Saiget M., Muskheli V., Pabon L., Reinecke H., Regnier M., Murry C.E. Mechanical Stress Conditioning and Electrical Stimulation Promote Contractility and Force Maturation of Induced Pluripotent Stem Cell-Derived Human Cardiac Tissue. Circulation. 2016;134:1557–1567. doi: 10.1161/CIRCULATIONAHA.114.014998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang C.Y., Peres Moreno Maia-Joca R., Ong C.S., Wilson I., DiSilvestre D., Tomaselli G.F., Reich D.H. Enhancement of human iPSC-derived cardiomyocyte maturation by chemical conditioning in a 3D environment. J. Mol. Cell. Cardiol. 2020;138:1–11. doi: 10.1016/j.yjmcc.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Veerman C.C., Mengarelli I., Guan K., Stauske M., Barc J., Tan H.L., Wilde A.A., Verkerk A.O., Bezzina C.R. hiPSC-derived cardiomyocytes from Brugada Syndrome patients without identified mutations do not exhibit clear cellular electrophysiological abnormalities. Sci. Rep. 2016;6:30967. doi: 10.1038/srep30967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de la Roche J., Angsutararux P., Kempf H., Janan M., Bolesani E., Thiemann S., Wojciechowski D., Coffee M., Franke A., Schwanke K., et al. Comparing human iPSC-cardiomyocytes versus HEK293T cells unveils disease-causing effects of Brugada mutation A735V of NaV1.5 sodium channels. Sci. Rep. 2019;9:11173. doi: 10.1038/s41598-019-47632-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tohyama S., Hattori F., Sano M., Hishiki T., Nagahata Y., Matsuura T., Hashimoto H., Suzuki T., Yamashita H., Satoh Y., et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 58.Sharma A., Li G., Rajarajan K., Hamaguchi R., Burridge P.W., Wu S.M. Derivation of highly purified cardiomyocytes from human induced pluripotent stem cells using small molecule-modulated differentiation and subsequent glucose starvation. J. Vis. Exp. 2015 doi: 10.3791/52628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramachandra C.J.A., Mehta A., Wong P., Ja K., Fritsche-Danielson R., Bhat R.V., Hausenloy D.J., Kovalik J.P., Shim W. Fatty acid metabolism driven mitochondrial bioenergetics promotes advanced developmental phenotypes in human induced pluripotent stem cell derived cardiomyocytes. Int. J. Cardiol. 2018;272:288–297. doi: 10.1016/j.ijcard.2018.08.069. [DOI] [PubMed] [Google Scholar]
- 60.Brugada R., Campuzano O., Sarquella-Brugada G., Brugada J., Brugada P. Brugada syndrome. Methodist Debakey Cardiovasc. J. 2014;10:25–28. doi: 10.14797/mdcj-10-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nerbonne J.M., Kass R.S. Molecular physiology of cardiac repolarization. Physiol. Rev. 2005;85:1205–1253. doi: 10.1152/physrev.00002.2005. [DOI] [PubMed] [Google Scholar]
- 62.Shih H.T. Anatomy of the action potential in the heart. Tex. Heart Inst. J. 1994;21:30–41. [PMC free article] [PubMed] [Google Scholar]
- 63.Garg P., Garg V., Shrestha R., Sanguinetti M.C., Kamp T.J., Wu J.C. Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes as Models for Cardiac Channelopathies: A Primer for Non-Electrophysiologists. Circ. Res. 2018;123:224–243. doi: 10.1161/CIRCRESAHA.118.311209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amin A.S., Tan H.L., Wilde A.A. Cardiac ion channels in health and disease. Heart Rhythm. 2010;7:117–126. doi: 10.1016/j.hrthm.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 65.Ince C., van Bavel E., van Duijn B., Donkersloot K., Coremans A., Ypey D.L., Verveen A.A. Intracellular microelectrode measurements in small cells evaluated with the patch clamp technique. Biophys. J. 1986;50:1203–1209. doi: 10.1016/S0006-3495(86)83563-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kosmidis G., Veerman C.C., Casini S., Verkerk A.O., van de Pas S., Bellin M., Wilde A.A., Mummery C.L., Bezzina C.R. Readthrough-Promoting Drugs Gentamicin and PTC124 Fail to Rescue Nav1.5 Function of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Carrying Nonsense Mutations in the Sodium Channel Gene SCN5A. Circ. Arrhythm. Electrophysiol. 2016;9:e004227. doi: 10.1161/CIRCEP.116.004227. [DOI] [PubMed] [Google Scholar]
- 67.Liang P., Sallam K., Wu H., Li Y., Itzhaki I., Garg P., Zhang Y., Vermglinchan V., Lan F., Gu M., et al. Patient-Specific and Genome-Edited Induced Pluripotent Stem Cell-Derived Cardiomyocytes Elucidate Single-Cell Phenotype of Brugada Syndrome. J. Am. Coll. Cardiol. 2016;68:2086–2096. doi: 10.1016/j.jacc.2016.07.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma D., Liu Z., Loh L.J., Zhao Y., Li G., Liew R., Islam O., Wu J., Chung Y.Y., Teo W.S., et al. Identification of an INa-dependent and Ito-mediated proarrhythmic mechanism in cardiomyocytes derived from pluripotent stem cells of a Brugada syndrome patient. Sci. Rep. 2018;8:11246. doi: 10.1038/s41598-018-29574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Selga E., Sendfeld F., Martinez-Moreno R., Medine C.N., Tura-Ceide O., Wilmut S.I., Perez G.J., Scornik F.S., Brugada R., Mills N.L. Sodium channel current loss of function in induced pluripotent stem cell-derived cardiomyocytes from a Brugada syndrome patient. J. Mol. Cell. Cardiol. 2018;114:10–19. doi: 10.1016/j.yjmcc.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis R.P., Casini S., van den Berg C.W., Hoekstra M., Remme C.A., Dambrot C., Salvatori D., Oostwaard D.W., Wilde A.A., Bezzina C.R., et al. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation. 2012;125:3079–3091. doi: 10.1161/CIRCULATIONAHA.111.066092. [DOI] [PubMed] [Google Scholar]
- 71.Okata S., Yuasa S., Suzuki T., Ito S., Makita N., Yoshida T., Li M., Kurokawa J., Seki T., Egashira T., et al. Embryonic type Na(+) channel beta-subunit, SCN3B masks the disease phenotype of Brugada syndrome. Sci. Rep. 2016;6:34198. doi: 10.1038/srep34198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.El-Battrawy I., Albers S., Cyganek L., Zhao Z., Lan H., Li X., Xu Q., Kleinsorge M., Huang M., Liao Z., et al. A cellular model of Brugada syndrome with SCN10A variants using human-induced pluripotent stem cell-derived cardiomyocytes. Europace. 2019;21:1410–1421. doi: 10.1093/europace/euz122. [DOI] [PubMed] [Google Scholar]
- 73.El-Battrawy I., Muller J., Zhao Z., Cyganek L., Zhong R., Zhang F., Kleinsorge M., Lan H., Li X., Xu Q., et al. Studying Brugada Syndrome with an SCN1B Variants in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Front. Cell. Dev. Biol. 2019;7:261. doi: 10.3389/fcell.2019.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Belbachir N., Portero V., Al Sayed Z.R., Gourraud J.B., Dilasser F., Jesel L., Guo H., Wu H., Gaborit N., Guilluy C., et al. RRAD mutation causes electrical and cytoskeletal defects in cardiomyocytes derived from a familial case of Brugada syndrome. Eur. Heart J. 2019;40:3081–3094. doi: 10.1093/eurheartj/ehz308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cerrone M., Lin X., Zhang M., Agullo-Pascual E., Pfenniger A., Chkourko Gusky H., Novelli V., Kim C., Tirasawadichai T., Judge D.P., et al. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a Brugada syndrome phenotype. Circulation. 2014;129:1092–1103. doi: 10.1161/CIRCULATIONAHA.113.003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller D.C., Harmer S.C., Poliandri A., Nobles M., Edwards E.C., Ware J.S., Sharp T.V., McKay T.R., Dunkel L., Lambiase P.D., et al. Ajmaline blocks INa and IKr without eliciting differences between Brugada syndrome patient and control human pluripotent stem cell-derived cardiac clusters. Stem Cell Res. 2017;25:233–244. doi: 10.1016/j.scr.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bedut S., Seminatore-Nole C., Lamamy V., Caignard S., Boutin J.A., Nosjean O., Stephan J.P., Coge F. High-throughput drug profiling with voltage- and calcium-sensitive fluorescent probes in human iPSC-derived cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2016;311:H44–H53. doi: 10.1152/ajpheart.00793.2015. [DOI] [PubMed] [Google Scholar]
- 78.Broyles C.N., Robinson P., Daniels M.J. Fluorescent, Bioluminescent, and Optogenetic Approaches to Study Excitable Physiology in the Single Cardiomyocyte. Cells. 2018;7:51. doi: 10.3390/cells7060051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walter A., Saric T., Hescheler J., Papadopoulos S. Calcium Imaging in Pluripotent Stem Cell-Derived Cardiac Myocytes. Methods Mol. Biol. 2016;1353:131–146. doi: 10.1007/7651_2015_267. [DOI] [PubMed] [Google Scholar]
- 80.Bassett J.J., Monteith G.R. Genetically Encoded Calcium Indicators as Probes to Assess the Role of Calcium Channels in Disease and for High-Throughput Drug Discovery. Adv. Pharmacol. 2017;79:141–171. doi: 10.1016/bs.apha.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 81.Hou J.H., Kralj J.M., Douglass A.D., Engert F., Cohen A.E. Simultaneous mapping of membrane voltage and calcium in zebrafish heart in vivo reveals chamber-specific developmental transitions in ionic currents. Front. Physiol. 2014;5:344. doi: 10.3389/fphys.2014.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shaheen N., Shiti A., Huber I., Shinnawi R., Arbel G., Gepstein A., Setter N., Goldfracht I., Gruber A., Chorna S.V., et al. Human Induced Pluripotent Stem Cell-Derived Cardiac Cell Sheets Expressing Genetically Encoded Voltage Indicator for Pharmacological and Arrhythmia Studies. Stem Cell Rep. 2018;10:1879–1894. doi: 10.1016/j.stemcr.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shinnawi R., Huber I., Maizels L., Shaheen N., Gepstein A., Arbel G., Tijsen A.J., Gepstein L. Monitoring Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes with Genetically Encoded Calcium and Voltage Fluorescent Reporters. Stem Cell Rep. 2015;5:582–596. doi: 10.1016/j.stemcr.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Song L., Awari D.W., Han E.Y., Uche-Anya E., Park S.H., Yabe Y.A., Chung W.K., Yazawa M. Dual optical recordings for action potentials and calcium handling in induced pluripotent stem cell models of cardiac arrhythmias using genetically encoded fluorescent indicators. Stem Cells Transl. Med. 2015;4:468–475. doi: 10.5966/sctm.2014-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leyton-Mange J.S., Mills R.W., Macri V.S., Jang M.Y., Butte F.N., Ellinor P.T., Milan D.J. Rapid cellular phenotyping of human pluripotent stem cell-derived cardiomyocytes using a genetically encoded fluorescent voltage sensor. Stem Cell Rep. 2014;2:163–170. doi: 10.1016/j.stemcr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun Y.H., Kao H.K.J., Chang C.W., Merleev A., Overton J.L., Pretto D., Yechikov S., Maverakis E., Chiamvimonvat N., Chan J.W., et al. Human induced pluripotent stem cell line with genetically encoded fluorescent voltage indicator generated via CRISPR for action potential assessment post-cardiogenesis. Stem Cells. 2020;38:90–101. doi: 10.1002/stem.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miccoli B., Lopez C.M., Goikoetxea E., Putzeys J., Sekeri M., Krylychkina O., Chang S.W., Firrincieli A., Andrei A., Reumers V., et al. High-Density Electrical Recording and Impedance Imaging with a Multi-Modal CMOS Multi-Electrode Array Chip. Front. Neurosci. 2019;13:641. doi: 10.3389/fnins.2019.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu H., Scharnhorst K.S., Stieg A.Z., Gimzewski J.K., Minami I., Nakatsuji N., Nakano H., Nakano A. Two dimensional electrophysiological characterization of human pluripotent stem cell-derived cardiomyocyte system. Sci. Rep. 2017;7:43210. doi: 10.1038/srep43210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jans D., Callewaert G., Krylychkina O., Hoffman L., Gullo F., Prodanov D., Braeken D. Action potential-based MEA platform for in vitro screening of drug-induced cardiotoxicity using human iPSCs and rat neonatal myocytes. J. Pharmacol. Toxicol. Methods. 2017;87:48–52. doi: 10.1016/j.vascn.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 90.Kussauer S., David R., Lemcke H. hiPSCs Derived Cardiac Cells for Drug and Toxicity Screening and Disease Modeling: What Micro- Electrode-Array Analyses Can Tell Us. Cells. 2019;8:1331. doi: 10.3390/cells8111331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Halbach M., Egert U., Hescheler J., Banach K. Estimation of action potential changes from field potential recordings in multicellular mouse cardiac myocyte cultures. Cell. Physiol. Biochem. 2003;13:271–284. doi: 10.1159/000074542. [DOI] [PubMed] [Google Scholar]
- 92.Muller J., Ballini M., Livi P., Chen Y., Radivojevic M., Shadmani A., Viswam V., Jones I.L., Fiscella M., Diggelmann R., et al. High-resolution CMOS MEA platform to study neurons at subcellular, cellular, and network levels. Lab. Chip. 2015;15:2767–2780. doi: 10.1039/C5LC00133A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Splawski I., Shen J., Timothy K.W., Lehmann M.H., Priori S., Robinson J.L., Moss A.J., Schwartz P.J., Towbin J.A., Vincent G.M., et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.CIR.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 94.Chang Y.S., Yang Y.W., Lin Y.N., Lin K.H., Chang K.C., Chang J.G. Mutation Analysis of KCNQ1, KCNH2 and SCN5A Genes in Taiwanese Long QT Syndrome Patients. Int. Heart. J. 2015;56:450–453. doi: 10.1536/ihj.14-428. [DOI] [PubMed] [Google Scholar]
- 95.Yang P., Koopmann T.T., Pfeufer A., Jalilzadeh S., Schulze-Bahr E., Kaab S., Wilde A.A., Roden D.M., Bezzina C.R. Polymorphisms in the cardiac sodium channel promoter displaying variant in vitro expression activity. Eur. J. Hum. Genet. 2008;16:350–357. doi: 10.1038/sj.ejhg.5201952. [DOI] [PubMed] [Google Scholar]
- 96.Makita N., Behr E., Shimizu W., Horie M., Sunami A., Crotti L., Schulze-Bahr E., Fukuhara S., Mochizuki N., Makiyama T., et al. The E1784K mutation in SCN5A is associated with mixed clinical phenotype of type 3 long QT syndrome. J. Clin. Invest. 2008;118:2219–2229. doi: 10.1172/JCI34057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Veltmann C., Barajas-Martinez H., Wolpert C., Borggrefe M., Schimpf R., Pfeiffer R., Caceres G., Burashnikov E., Antzelevitch C., Hu D. Further Insights in the Most Common SCN5A Mutation Causing Overlapping Phenotype of Long QT Syndrome, Brugada Syndrome, and Conduction Defect. J. Am. Heart Assoc. 2016;5:e003379. doi: 10.1161/JAHA.116.003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Remme C.A., Scicluna B.P., Verkerk A.O., Amin A.S., van Brunschot S., Beekman L., Deneer V.H., Chevalier C., Oyama F., Miyazaki H., et al. Genetically determined differences in sodium current characteristics modulate conduction disease severity in mice with cardiac sodium channelopathy. Circ. Res. 2009;104:1283–1292. doi: 10.1161/CIRCRESAHA.109.194423. [DOI] [PubMed] [Google Scholar]
- 99.Remme C.A., Verkerk A.O., Nuyens D., van Ginneken A.C., van Brunschot S., Belterman C.N., Wilders R., van Roon M.A., Tan H.L., Wilde A.A., et al. Overlap syndrome of cardiac sodium channel disease in mice carrying the equivalent mutation of human SCN5A-1795insD. Circulation. 2006;114:2584–2594. doi: 10.1161/CIRCULATIONAHA.106.653949. [DOI] [PubMed] [Google Scholar]
- 100.Veldkamp M.W., Viswanathan P.C., Bezzina C., Baartscheer A., Wilde A.A., Balser J.R. Two distinct congenital arrhythmias evoked by a multidysfunctional Na(+) channel. Circ. Res. 2000;86:E91–E97. doi: 10.1161/01.RES.86.9.e91. [DOI] [PubMed] [Google Scholar]
- 101.Hu D., Barajas-Martinez H., Pfeiffer R., Dezi F., Pfeiffer J., Buch T., Betzenhauser M.J., Belardinelli L., Kahlig K.M., Rajamani S., et al. Mutations in SCN10A are responsible for a large fraction of cases of Brugada syndrome. J. Am. Coll. Cardiol. 2014;64:66–79. doi: 10.1016/j.jacc.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang L., Zhou F., Huang L., Wu Q., Zheng J., Wu Y., Yin K., Cheng J. Association of common and rare variants of SCN10A gene with sudden unexplained nocturnal death syndrome in Chinese Han population. Int. J. Legal Med. 2017;131:53–60. doi: 10.1007/s00414-016-1397-1. [DOI] [PubMed] [Google Scholar]
- 103.Fukuyama M., Ohno S., Makiyama T., Horie M. Novel SCN10A variants associated with Brugada syndrome. Europace. 2016;18:905–911. doi: 10.1093/europace/euv078. [DOI] [PubMed] [Google Scholar]
- 104.Monasky M.M., Micaglio E., Vicedomini G., Locati E.T., Ciconte G., Giannelli L., Giordano F., Crisa S., Vecchi M., Borrelli V., et al. Comparable clinical characteristics in Brugada syndrome patients harboring SCN5A or novel SCN10A variants. Europace. 2019;21:1550–1558. doi: 10.1093/europace/euz186. [DOI] [PubMed] [Google Scholar]
- 105.Behr E.R., Savio-Galimberti E., Barc J., Holst A.G., Petropoulou E., Prins B.P., Jabbari J., torchio M., Berhet M., Mizusawa Y., et al. Role of common and rare variants in SCN10A: Results from the Brugada syndrome QRS locus gene discovery collaborative study. Cardiovasc. Res. 2015;106:520–529. doi: 10.1093/cvr/cvv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhu W., Voelker T.L., Varga Z., Schubert A.R., Nerbonne J.M., Silva J.R. Mechanisms of noncovalent beta subunit regulation of NaV channel gating. J. Gen. Physiol. 2017;149:813–831. doi: 10.1085/jgp.201711802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Watanabe H., Koopmann T.T., Le Scouarnec S., Yang T., Ingram C.R., Schott J.J., Demolombe S., Probst V., Anselme F., Escande D., et al. Sodium channel beta1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J. Clin. Invest. 2008;118:2260–2268. doi: 10.1172/JCI33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sen-Chowdhry S., Syrris P., McKenna W.J. Genetics of right ventricular cardiomyopathy. J. Cardiovasc. Electrophysiol. 2005;16:927–935. doi: 10.1111/j.1540-8167.2005.40842.x. [DOI] [PubMed] [Google Scholar]
- 109.Elliott P.M., Anastasakis A., Asimaki A., Basso C., Bauce B., Brooke M.A., Calkins H., Corrado D., Duru F., Green K.J., et al. Definition and treatment of arrhythmogenic cardiomyopathy: An updated expert panel report. Eur. J. Heart Fail. 2019;21:955–964. doi: 10.1002/ejhf.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang L., Tang S., Peng L., Chen Y., Cheng J. Molecular Autopsy of Desmosomal Protein Plakophilin-2 in Sudden Unexplained Nocturnal Death Syndrome. J. Forensic Sci. 2016;61:687–691. doi: 10.1111/1556-4029.13027. [DOI] [PubMed] [Google Scholar]
- 111.Kim C., Wong J., Wen J., Wang S., Wang C., Spiering S., Kan N.G., Forcales S., Puri P.L., Leone T.C., et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494:105–110. doi: 10.1038/nature11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nademanee K., Raju H., de Noronha S.V., Papadakis M., Robinson L., Rothery S., Makita N., Kowase S., Boonmee N., Vitayakritsirikul V., et al. Fibrosis, Connexin-43, and Conduction Abnormalities in the Brugada Syndrome. J. Am. Coll. Cardiol. 2015;66:1976–1986. doi: 10.1016/j.jacc.2015.08.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Corrado D., Zorzi A., Cerrone M., Rigato I., Mongillo M., Bauce B., Delmar M. Relationship Between Arrhythmogenic Right Ventricular Cardiomyopathy and Brugada Syndrome: New Insights from Molecular Biology and Clinical Implications. Circ. Arrhythm. Electrophysiol. 2016;9:e003631. doi: 10.1161/CIRCEP.115.003631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Abriel H. Cardiac sodium channel Na(v)1.5 and interacting proteins: Physiology and pathophysiology. J. Mol. Cell. Cardiol. 2010;48:2–11. doi: 10.1016/j.yjmcc.2009.08.025. [DOI] [PubMed] [Google Scholar]
- 115.Moncayo-Arlandi J., Brugada R. Unmasking the molecular link between arrhythmogenic cardiomyopathy and Brugada syndrome. Nat. Rev. Cardiol. 2017;14:744–756. doi: 10.1038/nrcardio.2017.103. [DOI] [PubMed] [Google Scholar]
- 116.Vermij S.H., Abriel H., van Veen T.A. Refining the molecular organization of the cardiac intercalated disc. Cardiovasc. Res. 2017;113:259–275. doi: 10.1093/cvr/cvw259. [DOI] [PubMed] [Google Scholar]
- 117.Agullo-Pascual E., Cerrone M., Delmar M. Arrhythmogenic cardiomyopathy and Brugada syndrome: Diseases of the connexome. FEBS Lett. 2014;588:1322–1330. doi: 10.1016/j.febslet.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Catalano O., Antonaci S., Moro G., Mussida M., Frascaroli M., Baldi M., Cobelli F., Baiardi P., Nastoli J., Bloise R., et al. Magnetic resonance investigations in Brugada syndrome reveal unexpectedly high rate of structural abnormalities. Eur. Heart J. 2009;30:2241–2248. doi: 10.1093/eurheartj/ehp252. [DOI] [PubMed] [Google Scholar]
- 119.Coronel R., Casini S., Koopmann T.T., Wilms-Schopman F.J., Verkerk A.O., de Groot J.R., Bhuiyan Z., Bezzina C.R., Veldkamp M.W., Linnenbank A.C., et al. Right ventricular fibrosis and conduction delay in a patient with clinical signs of Brugada syndrome: A combined electrophysiological, genetic, histopathologic, and computational study. Circulation. 2005;112:2769–2777. doi: 10.1161/CIRCULATIONAHA.105.532614. [DOI] [PubMed] [Google Scholar]
- 120.Veerman C.C., Kosmidis G., Mummery C.L., Casini S., Verkerk A.O., Bellin M. Immaturity of human stem-cell-derived cardiomyocytes in culture: Fatal flaw or soluble problem? Stem Cells Dev. 2015;24:1035–1052. doi: 10.1089/scd.2014.0533. [DOI] [PubMed] [Google Scholar]
- 121.Van den Berg C.W., Okawa S., Chuva de Sousa Lopes S.M., van Iperen L., Passier R., Braam S.R., Tertoolen L.G., del Sol A., Davis R.P., Mummery C.L. Transcriptome of human foetal heart compared with cardiomyocytes from pluripotent stem cells. Development. 2015;142:3231–3238. doi: 10.1242/dev.123810. [DOI] [PubMed] [Google Scholar]
- 122.Devalla H.D., Schwach V., Ford J.W., Milnes J.T., El-Haou S., Jackson C., Gkatzis K., Elliott D.A., Chuva de Sousa Lopes S.M., Mummery C.L., et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015;7:394–410. doi: 10.15252/emmm.201404757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Protze S.I., Liu J., Nussinovitch U., Ohana L., Backx P.H., Gepstein L., Keller G.M. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol. 2017;35:56–68. doi: 10.1038/nbt.3745. [DOI] [PubMed] [Google Scholar]
- 124.Nugraha B., Buono M.F., von Boehmer L., Hoerstrup S.P., Emmert M.Y. Human Cardiac Organoids for Disease Modeling. Clin. Pharmacol. Ther. 2019;105:79–85. doi: 10.1002/cpt.1286. [DOI] [PubMed] [Google Scholar]
- 125.Sung J.H., Wang Y.I., Narasimhan Sriram N., Jackson M., Long C., Hickman J.J., Shuler M.L. Recent Advances in Body-on-a-Chip Systems. Anal. Chem. 2019;91:330–351. doi: 10.1021/acs.analchem.8b05293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Iseoka H., Miyagawa S., Fukushima S., Saito A., Masuda S., Yajima S., Ito E., Sougawa N., Tekeda M., Harada A., et al. Pivotal Role of Non-cardiomyocytes in Electromechanical and Therapeutic Potential of Induced Pluripotent Stem Cell-Derived Engineered Cardiac Tissue. Tissue Eng. Part A. 2018;24:287–300. doi: 10.1089/ten.tea.2016.0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Goldfracht I., Efraim Y., Shinnawi R., Kovalev E., Huber I., Gepstein A., Arbel G., Shaheen N., Tiburcy M., Zimmermann W.H., et al. Engineered heart tissue models from hiPSC-derived cardiomyocytes and cardiac ECM for disease modeling and drug testing applications. Acta Biomater. 2019;92:145–159. doi: 10.1016/j.actbio.2019.05.016. [DOI] [PubMed] [Google Scholar]
- 128.Goldfracht I., Protze S., Shiti A., Setter N., Gruber A., Shaheen N., Nartiss Y., Keller G., Gepstein L. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat. Commun. 2020;11:75. doi: 10.1038/s41467-019-13868-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Giacomelli E., Bellin M., Sala L., van Meer B.J., Tertoolen L.G., Orlova V.V., Mummery C.L. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development. 2017;144:1008–1017. doi: 10.1242/dev.143438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sala L., Bellin M., Mummery C.L. Integrating cardiomyocytes from human pluripotent stem cells in safety pharmacology: Has the time come? Br. J. Pharmacol. 2017;174:3749–3765. doi: 10.1111/bph.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fermini B., Hancox J.C., Abi-Gerges N., Bridgland-Taylor M., Chaudhary K.W., Colatsky T., Correll K., Crumb W., Damiano B., Erdemli G., et al. A New Perspective in the Field of Cardiac Safety Testing through the Comprehensive In Vitro Proarrhythmia Assay Paradigm. J. Biomol. Screen. 2016;21:1–11. doi: 10.1177/1087057115594589. [DOI] [PubMed] [Google Scholar]
- 132.Colatsky T., Fermini B., Gintant G., Pierson J.B., Sager P., Sekino Y., Strauss D.G., Stockbridge N. The Comprehensive in Vitro Proarrhythmia Assay (CiPA) initiative—Update on progress. J. Pharmacol. Toxicol. Methods. 2016;81:15–20. doi: 10.1016/j.vascn.2016.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.