Abstract
Simple Summary
The predatory harlequin ladybird Harmonia axyridis (Pallas) (Coleoptera: Coccinellidae) has been widely released for classical and augmentative biological control programs of insect herbivores and is now distributed worldwide. Because of its invasive behavior and the threat it can pose to local biodiversity, this ladybird has been adopted as a model species for invasive biocontrol predators. A huge amount of existing literature is available on this species. However, little is known about the mechanisms underlying H. axyridis smell and taste, even though these senses are important in this ladybird for courtship, mating, and for locating suitable habitats for feeding and oviposition. Here we describe the first chemosensory gene repertoire that is expressed in the antennae of male and female H. axyridis. Our findings would likely represent the basis for future functional studies aiming at increasing the efficacy of H. axyridis in biological control or at reducing its populations in those areas where the ladybird has become a matter of concern due to its invasiveness.
Abstract
In predatory ladybirds (Coleoptera: Coccinellidae), antennae are important for chemosensory reception used during food and mate location, and for finding a suitable oviposition habitat. Based on NextSeq 550 Illumina sequencing, we assembled the antennal transcriptome of mated Harmonia axyridis (Pallas) (Coleoptera: Coccinellidae) males and females and described the first chemosensory gene repertoire expressed in this species. We annotated candidate chemosensory sequences encoding 26 odorant receptors (including the coreceptor, Orco), 17 gustatory receptors, 27 ionotropic receptors, 31 odorant-binding proteins, 12 chemosensory proteins, and 4 sensory neuron membrane proteins. Maximum-likelihood phylogenetic analyses allowed to assign candidate H. axyridis chemosensory genes to previously described groups in each of these families. Differential expression analysis between males and females revealed low variability between sexes, possibly reflecting the known absence of relevant sexual dimorphism in the structure of the antennae and in the distribution and abundance of the sensilla. However, we revealed significant differences in expression of three chemosensory genes, namely two male-biased odorant-binding proteins and one male-biased odorant receptor, suggesting their possible involvement in pheromone detection. Our data pave the way for improving the understanding of the molecular basis of chemosensory reception in Coccinellidae.
Keywords: biological control, invasive species, antennal transcriptome, predator, RNA-seq
1. Introduction
Chemosensory reception is important in insects, including predacious ladybirds (Coleoptera: Coccinellidae), for food and mate location and for finding a suitable oviposition habitat [1,2,3]. Volatile molecules are typically detected by insects through neurons housed in chemosensory sensilla mainly located on the antennae. In these organs, the chemical stimuli are transformed into electrical signals that will be transmitted to the brain [4]. In the proposed process of odorant detection, molecules are first bound and transported by odorant-binding proteins (OBPs) and possibly chemosensory proteins (CSPs) within the sensillum lymph, then detected by odorant receptors (ORs) or/and ionotropic receptors (IRs) expressed at the membrane of olfactory sensory neurons. Other protein families are also involved, such as the sensory neuron membrane proteins (SNMPs) for pheromone detection in some species [4,5]. Apart from olfaction, insect antennae are also involved in taste and numerous antennal transcriptomes have identified gustatory receptors (GRs), among them candidate sugar and CO2 receptors [6]. The function and characteristics of all these gene families in insects have been extensively reviewed, especially in model species such as Drosophila melanogaster (Meigen) [4,7,8,9,10].
Concerning insects of agricultural economic importance, chemosensory genes have been well described in Lepidoptera and in herbivorous and xylophagous Coleoptera species (e.g., [11,12,13]). Recent antennal transcriptome analyses focused also on predatory insects, belonging to, e.g., Neuroptera [14,15], Hemiptera [16,17], and Diptera [18]. Surprisingly, despite the worldwide importance of predatory ladybirds (Coleoptera: Coccinellidae) for classical and augmentative biological control (reviewed by [19,20,21]), no detailed information is available at present. Most ladybird species are important natural enemies of various crop pests, such as aphids, scales, whiteflies, or mites [22,23]. In particular, Harmonia axyridis (Pallas) is the most studied ladybird. This species is native from Asia and, starting from 1916 in the USA, it was released worldwide for biological control of herbivore pests. Currently, it is present in more than 38 countries (reviewed by [24]). Besides the beneficial role that H. axyridis exhibits in pest suppression [25], this ladybird raises concerns on the possible negative effects it may cause to the invaded community of predators, through intraguild predation [26,27,28] and indirect competition [29,30].
Behavioural and electrophysiological experiments demonstrated that H. axyridis responds to volatile semiochemicals [1,31,32]. Additionally, various types of antennal chemoreceptor sensilla (notably basiconica, chaetica, and grooved peg) have been recently characterized [33]. Therefore, understanding the molecular basis of H. axyridis chemosensory reception, in particular olfaction, is likely to provide new information to increase the efficacy of this predator in biological control or to reduce its populations in those areas where H. axyridis has become a concern for local biodiversity [34]. In this study, we conducted a transcriptomic analysis of H. axyridis adult antennae, identified candidate chemosensory genes, and investigated their differential expression between males and females. Additionally, we constructed phylogenetic trees and inferred the evolutionary relationships of putative H. axyridis chemosensory genes with other coleopteran species.
2. Materials and Methods
2.1. Insect Rearing and Antenna Collection
A culture of H. axyridis was settled in the laboratory from adults collected in Central Italy (Perugia Province). Adults and larvae were reared in plastic cylindrical cages (Ø = 25 cm, height = 30 cm) covered by a fine tissue mesh to allow ventilation. Broad bean plants moderately infested with Aphis fabae Scop. were used as a food source for larvae and adults. Plants were changed every 2–3 days and the egg batches were collected and isolated in a new cage for hatching. For tissue collection, and to control adult age and mating status, newly emerged adults were isolated for 3 days (aphids provided) then sexed. This period allowed completing ladybird pigmentation and maturation (pheromone production). Groups of 20 males and 20 females were paired in small net cages (30 cm × 30 cm × 30 cm). Each cage contained an aphid-infested plant, to allow mating. Plants were replaced every two days. Mating was observed and confirmed by the presence of physogastric females. After 4 to 6 days, insects were collected for dissection of the antennae under a stereomicroscope. Antennae were immediately stored in a 2 mL Eppendorf tube constantly immersed in liquid nitrogen and stored at −80 °C until RNA extraction. For each replication, collection of the antennae was conducted daily from 10:00 to 15:00 and split between different days. For each sex, three biological replications were conducted in total, each consisting of 200 antennae collected from ~100 individuals.
2.2. RNA Purification, cDNA Library Preparation and Sequencing
Total RNA was obtained using RNeasy Plus Mini Kit (Qiagen, Germantown, MD, USA) following the manufacturer’s protocol for animal samples [35]. The RNA concentration was determined using Qubit Fluorometer and Qubit RNA BR Assay (Life Technologies). RNA integrity was determined using Fragment Analyzer (Agilent Technologies). Libraries were prepared using Illumina TruSeq Stranded mRNA according to the sample preparation guide (Part #15031047, Rev. E, October 2013) for Illumina paired-end indexed sequencing. The resulting libraries were validated using Fragment Analyzer to check size distribution. Concentration of library samples was defined based on Qubit Fluorometer quantification and average library size. Indexed DNA libraries were normalized to 4 nM and then pooled in equal volumes. The pool was loaded at a concentration of 1.1 pM onto an Illumina NextSeq 550 Mid Output Flowcell (with 1% of PhiX control). The samples were then sequenced using the Illumina V2, 2 × 75 bp paired-end run. Library preparation and sequencing were conducted by Polo Genetica, Genomica e Biologia (Polo GGB, Siena, Italy). The raw data from Illumina sequencing were deposited in the NCBI Short Read Archive (SRA) database (BioProject ID PRJNA698239).
2.3. Assembly and Functional Annotation
De-novo assembly was performed within Galaxy [36] framework. An initial assessment of the quality of the raw reads in fastq format was conducted using FastQC [37]. Reads were then converted in fastqsanger format with FastQ Groomer [38] and low-quality reads were trimmed using Trimmomatic v. 0.32.3 [39] with a required average quality value of 20 and a minimum length of reads to be kept of 30. Identification and removal of rRNA-like sequences were conducted using riboPicker v. 0.4.3 [40]. De-novo assembly was conducted with Trinity [41] by setting ‘Reverse-Forward’ strand-specific library type, a minimum contig length of 200 and a minimum count for K-mers to be assembled of 1. TransDecoder [41] was used to identify coding regions. Redundant sequences were removed using CD-HIT-EST [42] with a 98% sequence identity threshold, 8 word-length size, and default options. Predicted peptides were annotated using the BLASTp algorithm (1e−5 threshold) on the Swiss-Prot protein database. Mapping, annotation, and InterPro analysis were conducted using Blast2GO within OmicsBox v1.4.11. Molecular function, biological process, and cellular component were derived for each gene [43]. The quality of the assembled transcriptome (e.g., total number of contigs, total contigs ≥500 bp, largest contig, N50) was evaluated with QUAST [44]. BUSCO was used to verify the completeness of the assemblage [45].
2.4. Identification of Differentially Expressed Transcripts
Bowtie [46] alignment method and RSEM were used to align the reads on the transcriptome and calculate the raw read numbers and TPM (transcripts per kilobase million, Table S3) expression value [47]. Results were analyzed using edgeR within edgeR package in R [48], adopting a significance level of false discovery rate (FDR) = 0.10 (as in [49,50]). Transcripts that exhibited a low expression (0.25 counts per million reads) were filtered out from the analysis (similar to [51]). The number of total reads was normalized according to the trimmed mean of M values (TMM) [52].
2.5. Identification of Chemosensory Genes and Phylogenetic Analyses
Chemosensory genes (~4000 sequences) collected from NCBI and published literature were used as queries in tBLASTn (1e−5 E-value) against cleaned trinity assemblage. For each transcript, the best hit was considered. Protein domains were predicted using TMHMM 2.0 [53] and SignalP 3.0 [54]. The presence and number of conserved cysteine residues in candidate OBPs and CSPs were visually assessed [55]. Phylogenetic trees were constructed using H. axyridis candidate chemosensory proteins and proteins from other Coleoptera families, including those closely related to ladybirds [56]. Considered species were Tribolium castaneum (Herbst) (Tenebrionidae) [57,58,59], Anoplophora glabripennis (Motschulsky) (Cerambycidae) [60,61], Dendroctonus ponderosae (Hopkins) (Curculionidae) [62], Onthophagus taurus (Schreber) and Anomala corpulenta (Motschulsky) (Scarabaeidae), Ambrostoma quadriimpressum (Motschulsky) and Phyllotreta striolata (F.) (Chrysomelidae) [63,64,65]. Functionally characterized odorant receptors from the beetles Megacyllene caryae (Gahan.), Ips typographus L., and Rhyncophorus ferrugineus (Olivier) were included [66,67,68,69] as well as gustatory receptors from D. melanogaster [13]. Similarly, sequences of functionally characterized odorant- and pheromone-binding proteins were considered from beetles, i.e., Anomala cuprea (Hope) and Anomala octiescostata Burmeister [70], Agrilus mali Matsumura [71], Cyrtotrachelus buqueti Guérin-Méneville [72], Holotrichia oblita Faldermannor [73,74], Holotrichia parallela (Motschulsky) [75], Phyllopertha diversa Waterhouse [76], Popillia japonica Newman [77], R. ferrugineus [78], and from the aphid Acyrthosiphon pisum (Harris) [79]. Sequences were aligned using MAFFT v.7 with Auto (for ORs, GRs, and IRs) or FFT-NS-i (for OBPs, CSPs, and SNMPs) iterative refinement methods, and default parameters [80]. Maximum Likelihood phylogenies were built using PhyML 3.0 [81], using the substitution model JTT and a SH-like approximate likelihood-ratio test for node support estimation [9,82]. Trees were viewed and edited using FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 2 March 2021)) and CorelDRAW X3. A phylogenetic analysis was conducted with candidate H. axyridis ORs and GRs and known ORs and GRs from T. castaneum, to verify effective phylogenetic separation between members of the two chemosensory receptor families [83]. In two cases, sequences that were originally named as ORs were later assigned to GRs (HaxyGR16 and HaxyGR17). Concerning the nomenclature, the OR co-receptor was named as “Orco”. Sequences that clustered with non-chemosensory ionotropic glutamate receptors of D. melanogaster [84] were excluded from the final phylogenetic tree analysis and from the list of HaxyIRs. When multiple HaxyIR sequences clustered together, they were aligned to verify that they are not part of the same transcript. Additionally, we referred to D. melanogaster and T. castaneum for HaxyIRs that clustered with known IRs (Table S3 in [84]). Six candidate chemosensory proteins that exhibited high identity with ejaculatory bulb proteins and presented six conserved cysteine (C) residues were excluded. SNMPs were classified according to SNMP1 and SNMP2 groups.
3. Results
3.1. Transcriptome Sequencing, Assembly, and Identification of Chemosensory Genes
Sequencing returned approximatively 55 million raw reads. The accuracy of Q30 base call was 92.6% (Table S1.1). The total assembly (males + females) returned 66,728 contigs, with the largest contig of 16,954 bp and a N50 of 1981 bp (Table S1.2). The total number of contigs ≥500 bp in length was 30,600. Gene coverage was high, as the assembled transcriptome contained 93.2% complete BUSCO groups (single-copy: 66.3%; duplicated: 26.9%), 2.2% fragmented and 4.6% missing BUSCO genes. Searches against the Swiss-Prot database returned 27,698 transcripts showing sequence similarity to known proteins (reported in Table S2). Additionally, 28,991 and 35,318 transcripts were assigned with 1 or more GO terms or InterPro IDs, respectively. Most abundant biological processes and molecular functions were related to cellular and metabolic functions as well as chemosensory processes (“response to stimulus”, “localization”, and “binding” GO terms) (Figure S1). Bioinformatics analyses identified a total of 117 unigenes from H. axyridis transcriptome that belonged to gene families putatively involved in insect chemoreception: 26 ORs, 17 GRs, 27 IRs, 31 OBPs, 12 CSPs, 4 SNMPs (longest isoforms reported in Tables S5–S11). Expression analysis revealed 133 contigs differentially expressed between males and females (Tables S3 and S4), including three chemosensory gene isoforms. Notably, two candidate OBPs (HaxyOBP11 and HaxyOBP27, Table S4) were highly expressed in males compared to females (FDR < 0.05). In details, HaxyOBP11 exhibited 82.6% amino acid identity with OBP2 of Cryptolaemus montrouzieri, and HaxyOBP27 87.6% with OBP38 of Holotrichia parallela. One OR (HaxyOR5, Table S5) was more expressed in males compared to females (FDR = 0.086) and exhibited 57% amino acid identity with OR49b-like of Leptinotarsa decemlineata.
3.2. Odorant Receptors (ORs)
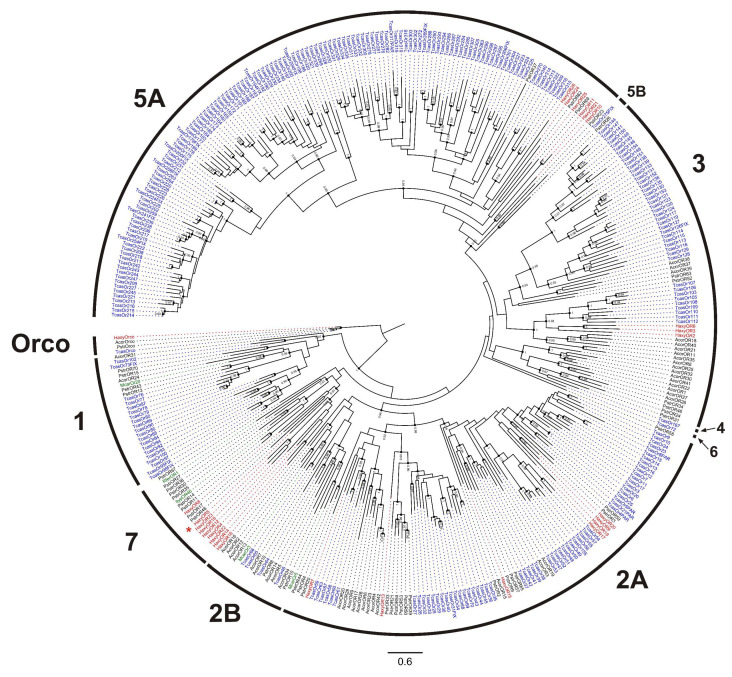
Of the 26 HaxyORs, full-length ORFs were identified for 11 of them, with lengths ranging from 339 to 480 amino acids and 4 to 7 predicted transmembrane domains (Table S5). The remaining 15 ORs corresponded to partial sequences, encoding 102 to 410 amino acids. Except for Orco, which is highly conserved between insect families, candidate HaxyORs exhibited low amino acid identity with ORs from other coleopteran species. Phylogenetic analysis revealed that the different ORs are included in 4 out of 9 groups known for Coleoptera (Figure 1) [9]. In detail, seven candidate HaxyORs clustered in group 2A, three HaxyORs clustered in group 3, six HaxyORs clustered in group 5A, and nine HaxyORs, including the male-biased HaxyOR5 (see Section 3.1) belonged to group 7. No candidate OR was detected in groups 1, 2B, 4, 5B and 6. HaxyORs that grouped in the same clades 2A, 3, 5A, or 7 were aligned to check whether these fragments correspond to different fragments of the same protein or are different proteins. Alignments did not reveal conserved overlapping regions thus suggesting they are different ORs.
Figure 1.
Phylogenetic tree of odorant receptors (ORs). Red: Harmonia axyridis (Haxy); blue: Tribolium castaneum (Tcas); black: Anomala corpulenta (Acor) and Phyllotreta striolata (Pstr); green: Functionally characterized ORs of Megacyllene caryae (Mcar), Ips typographus (Ityp), and Rhyncophorus ferrugineus (Rfer). External numbers represent known groups of ORs. Numbers and symbols at nodes represent support values higher than 0.9, where 1 represents maximal support. Asterisk represents the OR with a male-biased expression.
3.3. Gustatory Receptors (GRs)
Seventeen contigs encoding candidate GRs were identified in the transcriptome (Table S6). Of them, only two encoded full-length proteins (392 and 440 amino acids). HaxyGR11, HaxyGR12, and HaxyGR13 grouped with D. melanogaster GR21a and HaxyGR15 grouped with D. melanogaster GR63a (Figure 2). In D. melanogaster, these two genes are responsible for CO2 detection [6]. HaxyGR1, HaxyGR3, HaxyGR4, and HaxyGR8 grouped with Drosophila GRs known to be involved in sugar detection [85]. As for ORs, we aligned amino acid sequences of the different fragmented GRs to ensure they represent different proteins. Inspection of the alignment for HaxyGR1, HaxyGR4, and HaxyGR8 suggested that these fragments are indeed different GRs. Conversely, alignment of HaxyGR12, HaxyGR11 and HaxyGR13 revealed overlapping regions (5–7 amino acids), thus suggesting that the three fragments may be part of the same protein. HaxyGR2 and HaxyGR6 clustered with TcasGR20 from T. castaneum, which is a receptor for mannitol and sorbitol [86]. No GR appeared to be differentially expressed between sexes.
Figure 2.
Phylogenetic tree of gustatory receptors (GRs). Red: Harmonia axyridis (Haxy); blue: Tribolium castaneum (Tcas); orange: Drosophila melanogaster (Dmel); black: Dendroctonus ponderosae (Dpon). Numbers and symbols at nodes represent support values higher than 0.9, where 1 represents maximal support.
3.4. Ionotropic Receptors (IRs)
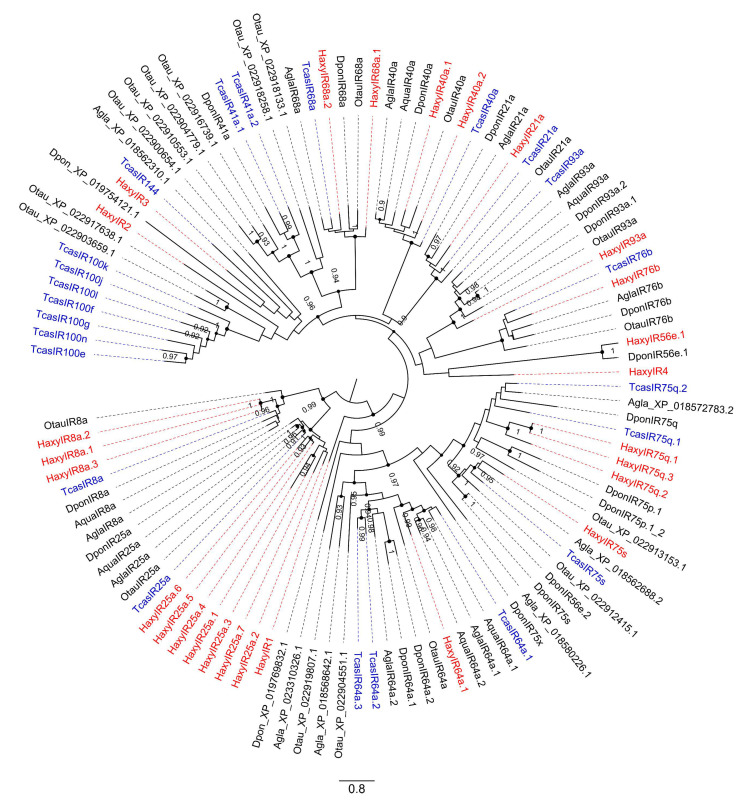
Twenty-seven IRs were identified in total, 5 of them encoding full-length proteins, from 301 to 938 amino acids (Table S7). Phylogenetic tree (Figure 3) revealed the presence of HaxyIR orthologues of D. melanogaster and T. castaneum IRs, such as the IR coreceptors IR8a and IR25a [87]. Alignments did not reveal conserved overlapping regions between sequences. The high number of IRs found in H. axyridis can be explained by the conservation of IRs across insect families, making them easy to identify by sequence homology, contrary to ORs and GRs that are usually more divergent. Additionally, we found impressive IR duplications in the co-receptor IR25a (7 sequences) and IR8a (3 sequences) subfamilies. All are non-overlapping fragments, but we cannot exclude that some could correspond to the same IR, considering that IR genes are usually very large. No IR appeared to be differentially expressed between sexes.
Figure 3.
Phylogenetic tree of ionotropic receptors (IRs). Red: Harmonia axyridis (Haxy); blue: Tribolium castaneum (Tcas); black: Ambrostoma quadriimpressum (Aqua), Anoplophora glabripennis (Agla), Dendroctonus ponderosae (Dpon), and Onthophagus taurus (Otau). Numbers and symbols at nodes represent support values higher than 0.9, where 1 represents maximal support.
3.5. Odorant-Binding Proteins (OBPs)
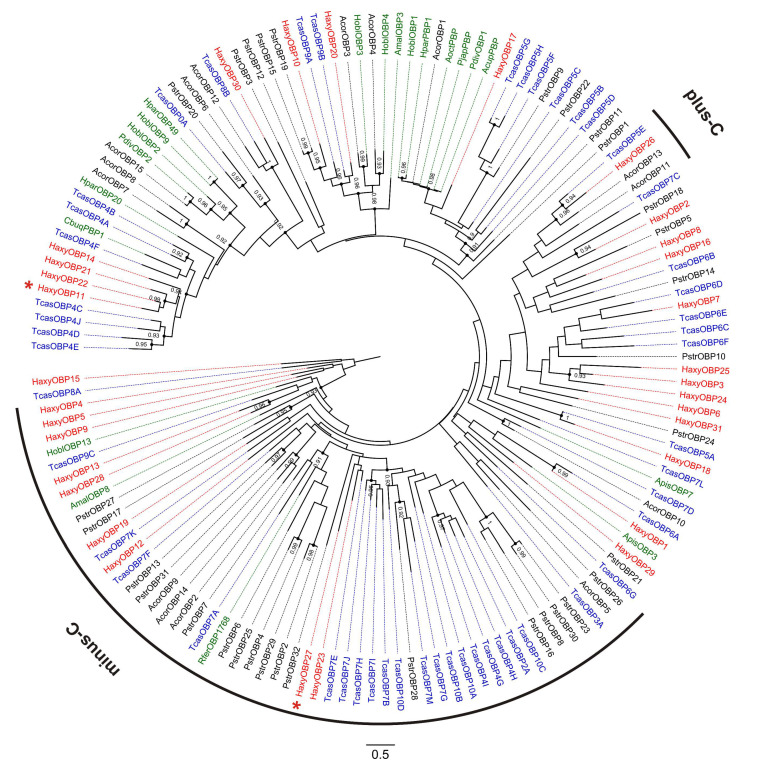
In total, 31 OBPs were identified in the transcriptome of H. axyridis. For 16 of them, the complete ORF was detected, with a length varying from 123 to 265 amino acids (Table S8). Twenty-four HaxyOBPs presented a predicted signal peptide. Twenty of the candidate OBPs belonged to the classic-OBP family, exhibiting the 6 conserved cysteine (C) residues representative of this OBP subfamily. Ten OBPs belonged to the minus-C OBP subgroup with 4 conserved cysteine residues. One OBP (HaxyOBP26) belonged to the plus-C subfamily and clustered with OBP5E of T. castaneum (Figure 4). The complete HaxyOBP30 clustered with TcasOBP8B, an OBP that exhibits a non-conserved cysteine pattern, with extra amino acids between C1 and C2 [57]. Similarly, HaxyOBP30 exhibited non-conventional interval between C1 and C2. Both HaxyOBP11 and HaxyOBP27 were more expressed in males. HaxyOBP11 clustered with 3 other OBPs from H. axyridis and with 4 OBPs from T. castaneum. Conversely, it was not possible to find orthologues for HaxyOBP27. Interestingly, HaxyOBP10 and HaxyOBP20 clustered with HoblOBP3 and HoblOBP4, two proteins that bound plant-related compounds, and AmalOBP3, a protein that showed binding affinity with alcohols, esters, terpenoids, and 12–15 carbon aldehydes [71,88].
Figure 4.
Phylogenetic tree of odorant-binding proteins (OBPs). Red: Harmonia axyridis (Haxy); blue: Tribolium castaneum (Tcas); black: Anomala corpulenta (Acor) and Phyllotreta striolata (Pstr); green: functionally characterized OBPs and pheromone binding proteins (PBPs) from Acyrthosiphon pisum (Apis), Anomala cuprea (Acup), Agrilus mali (Amal), Anomala octiescostata (Aoct), Cyrtotrachelus buqueti (Cbuq), Holotrichia oblita (Hobl), Holotrichia parallela (Hpar), Phyllopertha diversa (Pdiv), Popillia japonica (Pjap), Rhynchophorus ferrugineus (Rfer). Numbers and symbols at nodes represent support values higher than 0.9, where 1 represents maximal support. Asterisks represent the two OBPs with male-biased expression.
3.6. Chemosensory Proteins (CSPs)
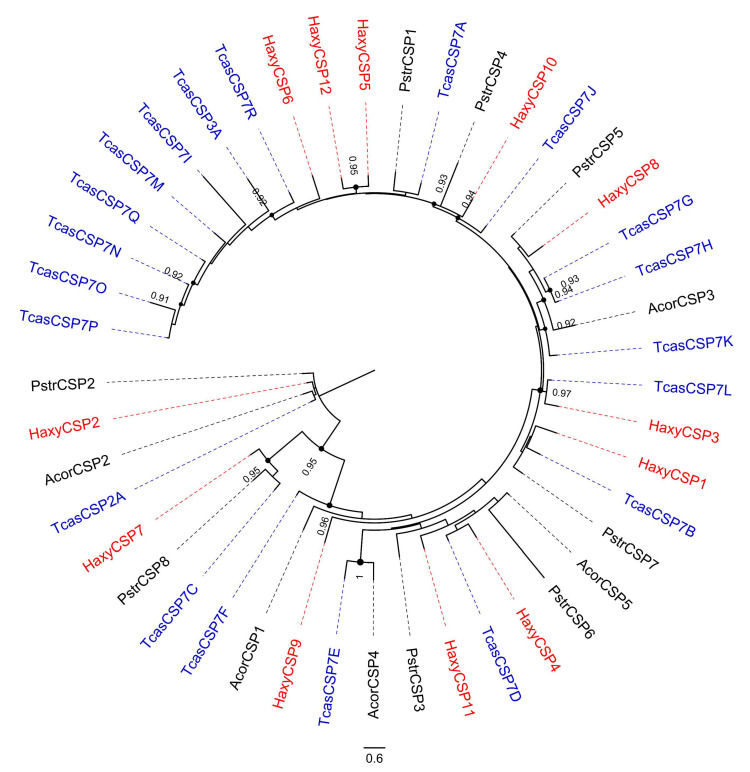
Of the 12 contigs encoding candidate CSPs, 8 encoded full-length proteins (with length from 101 to 153 amino acids). Nine HaxyCSPs possessed a predicted signal peptide and all presented the highly conserved four-cysteine profile (Table S9, Figure 5). No CSP appeared to be differentially expressed between sexes.
Figure 5.
Phylogenetic tree of chemosensory proteins (CSPs). Red: Harmonia axyridis (Haxy); blue: Tribolium castaneum (Tcas); black: Anomala corpulenta (Acor) and Phyllotreta striolata (Pstr). Numbers and symbols at nodes represent support values higher than 0.9, where 1 represents maximal support.
3.7. Sensory Neuron Membrane Proteins (SNMPs)
Four sequences encoding incomplete SNMPs were identified in the H. axyridis transcriptome and belonged to the SNMP1 and SNMP2 insect lineages (Table S10, Figure S2). HaxySNMP1.1 exhibited 62.5% identity with T. castaneum SNMP1, while HaxySNMP2.1 was 63.6% identical to T. castaneum SNMP2. No SNMP appeared to be differentially expressed between sexes.
4. Discussion
Despite the huge amount of ladybird species that have been released in 130 years of biological control activity, the chemical ecology of this group and the molecular basis of its olfaction are still largely unknown. Here, we identified 117 chemosensory genes expressed in H. axyridis adult antennae and provided a first survey of their differential expression between males and females. Most chemosensory genes did not exhibit sex-specific nor sex-biased expression. This is consistent with recent description of antennal morphology that did not reveal relevant sexual dimorphism in H. axyridis, neither for the antennal general structure, nor for the types of sensilla and their abundance [33]. However, this does not preclude that the significance of the perceived odors may differ between males or females. Notably, it is known from previous studies that H. axyridis males and females respond differently to odors in wind tunnel [89] or electroantennography [90].
The number of ORs we described in H. axyridis was a bit lower compared to what has been identified in other beetle transcriptomes, e.g., I. typographus (43) and D. ponderosae (49) [62,91]. This difference may be related to the different chemical ecologies of these species, as previously suggested [8]. Alternatively, it is possible that we missed some HaxyORs, as our transcriptome was sequenced from adult antennae only.
However, antennal transcriptomes from other predatory insects exhibited variable numbers of ORs. For instance, only 15 ORs could be identified in the antennal transcriptome of Cyrtorhinus lividipennis Reuter (Hemiptera: Miridae) [17], 14 ORs in Chrysopa pallens (Rambur) [14] and 37 ORs in Chrysoperla sinica (Neuroptera: Chrysopidae) [15], 51 ORs in Episyrphus balteatus DeGeer and 42 ORs in Eupeodes corollae Fabricius (Diptera: Syrphidae) [18], and 38 ORs in Arma chinensis (Fallou) (Hemiptera: Pentatomidae) [92]. Hopefully, further identification of OR genes in other predatory insect antennae will permit to increase coverage of probable unknown putative OR genes in these insects. Additionally, annotation of H. axyridis ORs in this species genome will likely reveal the presence of more ORs [93], as it happened for other species [94], although their effective expression will remain to be investigated. Pheromonal blend has been recently characterized for H. axyridis, with (–)-β-caryophyllene being the most abundant molecule [95]. The pheromone is emitted by virgin and mated females, primarily under the presence of adequate aphid preys [96]. Major sex pheromone compounds are individually perceived by both sexes, while the complete pheromonal blend elicited behavioral attraction on males only [90]. Possibly, high expression of one OR, HaxyOR5, in males may suggest a role in the detection of one sex pheromone molecule. In support of this latter hypothesis is that HaxyOR5 clustered in the clade 7 of the Coleoptera OR phylogeny, in which pheromone receptors from other Cucujiformia (R. ferrugineus and I. typographus) have been characterized [68,69]. Together with the fact that Coleoptera ORs exhibit high sequence divergence, this makes it difficult to predict HaxyOR functions. That is why it would be necessary to identify ligands experimentally in the future [reviewed by 7]. Our transcriptomic analysis revealed that four candidate sugar receptors were expressed in H. axyridis antennae. Sugar is an important component in the diet of predatory ladybirds, notably H. axyridis [97]. For instance, when aphid preys are unavailable, H. axyridis maintains its presence in the agroecosystem by feeding upon extrafloral nectars [98]. Sugar is also present in aphid honeydew, which may represent an alternative food source for H. axyridis [99]. Additionally, HaxyGR2 and HaxyGR6 clustered in the phylogeny with TcasGR20, a receptor that is responsible in T. castaneum for mannitol and sorbitol detection [86]. Although it is thought that ladybird maxillary palps are the most adequate structures for the perception of nonvolatile molecules [100], our results suggest a possible role of antennal contact in sugar detection. This role has already been demonstrated in other insects, including D. melanogaster [101]. During handling behavior, it is indeed possible that ladybirds use antennal contact to assess the nutritional quality of the prey, and sugar detection in different food sources may represent a reliable indicator for it. We have previously described the sensilla distribution on H. axyridis antennae and identified sensilla chaetica at the tip of the antennae (a general view of the antenna and schematic drawings of sensilla distribution are reported in Figures 1 and 2 in ref. [33]). Possibly, these antennal long sensilla chaetica may be involved in the detection of such nonvolatile molecules. Our analysis also revealed the presence of candidate receptors for CO2 in H. axyridis antennae. Sensilla basiconica are responsible for carbon dioxide detection in D. melanogaster [12]. These sensilla are present and abundant in male and female H. axyridis antennae, in particular in the apical region (see Figure 4 in ref. [33]). Although this kind of sensilla is known to be involved in the detection of a plethora of molecules in insects, including volatile molecules, it is possible that CO2 detection is mediated by such sensilla in H. axyridis. The direct effect of artificial changes of CO2 concentration on the behavior of biocontrol agents is still under investigation [102]. To date, it has been shown that CO2 has weak or null influence on H. axyridis development and feeding behavior [103]. Concerning IRs, the fact that more than one gene have been found clustering in the IR25a clade was unexpected. A possible explanation is that these fragments are indeed part of one large IR gene that is particularly difficult to reconstruct with our sequencing approach [88]. Alternatively, they may result from IR25a duplication in H. axyridis. Gene duplication or pseudogenization of IR25a is rare, but has been observed in some Hymenopteran species ([104] and references within), and might be relevant for the acquisition of new functions.
Odorant-binding proteins are important carriers of chemosensory cues, including alarm, aggregation, and sex pheromones. In A. pisum, ApisOBP3 and ApisOBP7 are known to carry the alarm pheromone, (E)-β-Farnesene, in adults and nymphs [79]. HaxyOBP1 clustered with ApisOBP3, although the aLRT-SH branch support value (0.87) was a bit lower than the adopted threshold of 0.9. Harmonia axyridis has been previously shown to respond to (E)-β-Farnesene [90], hence, we can hypothesize for HaxyOBP1 a possible role in binding the aphid alarm pheromone. The male-biased expression of HaxyOBP11 and HaxyOBP27 we observed in this study, and taken into account that sex pheromone is released by H. axyridis females, led us to hypothesize that HaxyOBP11 and HaxyOBP27 would play a role as carriers of the female-produced sex-pheromone. In addition, HaxyOBP27 belonged to the minus-C OBP subfamily, as the pheromone-binding protein RferOBP1768 from R. ferrugineus does [78]. HaxyOBP18 and HaxyOBP30 clustered with TcasOBP5A and TcasOBP8B, respectively. These two T. castaneum OBPs have been previously shown to cluster with two D. melanogaster OBPs (DmelOBP59a and DmelOBP73a), which are very conserved across insect orders [88]. Hence, it is possible to hypothesize common functions of the two proteins for different insect groups. Finally, the fact that HaxyOBP10 and HaxyOBP20 clustered with A. mali and H. oblita OBPs that have been shown to have binding affinity for plant volatiles led us to hypothesize a possible involvement of the two proteins in the detection of adequate habitat cues. Our analysis detected four SNMP sequences that belonged to SNMP1 and SNMP2 subfamilies. Although most insects have two SNMPs, there are beetle species with antennal expression of 3 (I. typographus and D. ponderosae), 4 (Rhaphuma horsfieldi White, Sitophilus zeamais Motschulsky, Dendroctonus valens LeConte and Dastarcus helophoroides Fairmaire), and even 6 (T. castaneum) SNMPs (reported in [59,105]). The role of SNMP1 in pheromone detection has been demonstrated in few Diptera and Lepidoptera species [106,107,108]. Because of the high degree of conservation of SNMP1 across insect orders, a similar function has been hypothesized also for beetles [106].
The present transcriptomic analysis provides a first understanding of the molecular basis of ladybird olfaction and the role of antennae in male and females H. axyridis chemical ecology. Further studies now require functional characterization of the differentially expressed genes we evidenced here to confirm their hypothetical role. At this scope, heterologous expression in Xenopus oocytes or in Drosophila (“empty neuron” system) may serve for OR deorphanization [7] and in vitro binding studies may be conducted on OBPs with relevant volatiles, especially those known to induce different electrophysiological and behavioral responses according to sex [89,90]. In addition, transcriptome analysis of other chemosensory tissues such as palps, legs, wings, or ovipositor will probably help in extending the chemosensory gene repertoire in this species, in concert with genome sequencing.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/12/3/209/s1, Table S1.1: Number of reads obtained by Illumina sequencing, Table S1.2: Summary of the assembled contigs, Table S2: GO and InterPro annotations, Table S3: Transcripts per kilobase million (TPM) for the different samples, Table S4: Differential expression analysis, Table S5: Best similarity for odorant receptors (ORs) of Harmonia axyridis (Haxy), Table S6: Best similarity for gustatory receptors (GRs) of Harmonia axyridis (Haxy), Table S7: Best similarity for ionotropic receptors (IRs) of Harmonia axyridis (Haxy), Table S8: Best similarity for odorant-binding proteins (OBPs) of Harmonia axyridis (Haxy), Table S9: Best similarity for chemosensory proteins (CSPs) of Harmonia axyridis (Haxy), Table S10: Best similarity for sensory neuron membrane proteins (SNMPs) of Harmonia axyridis (Haxy), Table S11: Amino acid sequences of Harmonia axyridis chemosensory proteins identified in this study, Figure S1: Distribution of Harmonia axyridis antennal transcriptome data, Figure S2: Phylogenetic tree of sensory neuron membrane proteins (SNMPs).
Author Contributions
Conceptualization: G.R., E.C., and E.J.-J.; methodology: G.R., C.M., N.M., E.C., and E.J.-J.; investigation: G.R., A.R., and E.J.-J.; writing—original draft preparation: G.R., E.C., and E.J.-J.; writing—review and editing: G.R., N.M., E.C., and E.J.-J. All authors have read and agreed to the published version of the manuscript.
Funding
GR visits to iEES-Paris were supported by two Erasmus+ Staff Mobility Grants (Grant agreements 2017-1-IT02-KA103-035499 and 2018-1-IT02-KA103-047328). Personal funding to GR was provided by PSR UMBRIA 2014–2020, measure 19.2, Local Action Group (LAG) Media Valle del Tevere. Additional funding was provided by Fondazione Cassa di Risparmio di Perugia, Project n. 2015.0349.021.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The raw data from Illumina sequencing were deposited in the NCBI Short Read Archive (SRA) database (BioProject ID PRJNA698239).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pettersson J. Coccinellids and Semiochemicals. In: Hodek I., van Emden H.F., Honek A., editors. Ecology and Behaviour of the Ladybird Beetles (Coccinellidae) John Wiley & Sons; Chichester, UK: 2012. pp. 444–464. [Google Scholar]
- 2.Hamilton R.M., Dogan E.B., Schaalje G.B., Booth G.M. Olfactory response of the lady beetle Hippodamia convergens (Coleoptera: Coccinellidae) to prey related odors, including a scanning electron microscopy study of the antennal sensilla. Environ. Entomol. 1999;28:812–822. doi: 10.1093/ee/28.5.812. [DOI] [Google Scholar]
- 3.Haverkamp A., Hansson B.S., Knaden M. Combinatorial codes and labeled lines: How insects use olfactory cues to find and judge food, mates, and oviposition sites in complex environments. Front. Physiol. 2018;9:46. doi: 10.3389/fphys.2018.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leal W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 5.Andersson M.N., Löfstedt C., Newcomb R.D. Insect olfaction and the evolution of receptor tuning. Front. Ecol. Evol. 2015;53 doi: 10.3389/fevo.2015.00053. [DOI] [Google Scholar]
- 6.Jones W.D., Cayirlioglu P., Grunwald Kadow I., Vosshall L.B. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- 7.Montagné N., De Fouchier A., Newcomb R.D., Jacquin-Joly E. Advances in the identification and characterization of olfactory receptors in insects. Prog. Mol. Biol. Transl. Sci. 2015;130:55–80. doi: 10.1016/bs.pmbts.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Robertson H.M. Molecular evolution of the major arthropod chemoreceptor gene families. Annu. Rev. Entomol. 2019;64:227–242. doi: 10.1146/annurev-ento-020117-043322. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell R.F., Schneider T.M., Schwartz A.M., Andersson M.N., McKenna D.D. The diversity and evolution of odorant receptors in beetles (Coleoptera) Insect Mol. Biol. 2020;29:77–91. doi: 10.1111/imb.12611. [DOI] [PubMed] [Google Scholar]
- 10.He P., Durand N., Dong S.L. Editorial: Insect olfactory proteins (from gene identification to functional characterization) Front. Physiol. 2019;10:1313. doi: 10.3389/fphys.2019.01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Santis F., François M.C., Merlin C., Pelletier J., Maïbèche-Coisné M., Conti E., Jacquin-Joly E. Molecular cloning and in Situ expression patterns of two new pheromone-binding proteins from the corn stemborer Sesamia nonagrioides. J. Chem. Ecol. 2006;32:1703–1717. doi: 10.1007/s10886-006-9103-2. [DOI] [PubMed] [Google Scholar]
- 12.De Bruyne M., Foster K., Carlson J.R. Odor coding in the Drosophila antenna. Neuron. 2001;30:537–552. doi: 10.1016/S0896-6273(01)00289-6. [DOI] [PubMed] [Google Scholar]
- 13.Robertson H.M., Warr C.G., Carlson J.R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2003;100:14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z.Q., Zhang S., Ma Y., Luo J.Y., Wang C.Y., Lv L.M., Dong S.L., Cui J.J. First transcriptome and digital gene expression analysis in Neuroptera with an emphasis on chemoreception genes in Chrysopa pallens (Rambur) PLoS ONE. 2013;8:e67151. doi: 10.1371/journal.pone.0067151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z.Q., Zhang S., Luo J.Y., Wang S.B., Wang C.Y., Lv L.M., Dong S.L., Cui J.J. Identification and expression pattern of candidate olfactory genes in Chrysoperla sinica by antennal transcriptome analysis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2015;15:28–38. doi: 10.1016/j.cbd.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Wang G.Y., Zhu M.F., Jiang Y.D., Zhou W.W., Liu S., Heong K.L., Cheng J., Zhu Z.R. Identification of candidate odorant-binding protein and chemosensory protein genes in Cyrtorhinus lividipennis (Hemiptera: Miridae), a key predator of the rice planthoppers in Asia. Environ. Entomol. 2017;46:654–662. doi: 10.1093/ee/nvx075. [DOI] [PubMed] [Google Scholar]
- 17.Wang G.Y., Zhu J.L., Zhou W.W., Liu S., Khairul Q.M., Ansari N.A., Zhu Z.R. Identification and expression analysis of putative chemoreception genes from Cyrtorhinus lividipennis (Hemiptera: Miridae) antennal transcriptome. Sci. Rep. 2018;8:12981. doi: 10.1038/s41598-018-31294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang B., Liu Y., Wang G.R. Chemosensory genes in the antennal transcriptome of two syrphid species, Episyrphus balteatus and Eupeodes corollae (Diptera: Syrphidae) BMC Genom. 2017;18:586. doi: 10.1186/s12864-017-3939-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rondoni G., Borges I., Collatz J., Conti E., Costamagna A.C., Dumont F., Evans E.W., Grez A.A., Howe A.G., Lucas E., et al. Exotic ladybirds for biological control of herbivorous insects—a review. Entomol. Exp. Appl. 2021;169:6–27. doi: 10.1111/eea.12963. [DOI] [Google Scholar]
- 20.Michaud J.P. Coccinellids in biological control. In: Hodek I., van Emden H.F., Honek A., editors. Ecology and Behaviour of the Ladybird Beetles (Coccinellidae) John Wiley & Sons; Chichester, UK: 2012. pp. 488–519. [Google Scholar]
- 21.Obrycki J.J., Harwood J.D., Kring T.J., O’Neil R.J. Aphidophagy by Coccinellidae: Application of biological control in agroecosystems. Biol. Control. 2009;51:244–254. doi: 10.1016/j.biocontrol.2009.05.009. [DOI] [Google Scholar]
- 22.Hodek I., van Emden H.F., Honěk A. Ecology and Behaviour of the Ladybird Beetles (Coccinellidae) John Wiley & Sons; Chichester, UK: 2012. [Google Scholar]
- 23.Rondoni G., Ielo F., Ricci C., Conti E. Intraguild predation responses in two aphidophagous coccinellids identify differences among juvenile stages and aphid densities. Insects. 2014;5:974–983. doi: 10.3390/insects5040974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy H.E., Brown P.M.J., Adriaens T., Berkvens N., Borges I., Clusella-Trullas S., De Clercq P., Comont R.F., Eschen R., Estoup A., et al. The harlequin ladybird, Harmonia axyridis: Global perspectives on invasion history and ecology. Biol. Invasions. 2016;18:997–1044. doi: 10.1007/s10530-016-1077-6. [DOI] [Google Scholar]
- 25.Koch R.L., Costamagna A.C. Reaping benefits from an invasive species: Role of Harmonia axyridis in natural biological control of Aphis glycines in North America. BioControl. 2017 doi: 10.1007/s10526-016-9749-9. [DOI] [Google Scholar]
- 26.Gagnon A.-È., Doyon J., Heimpel G.E., Brodeur J. Prey DNA detection success following digestion by intraguild predators: Influence of prey and predator species. Mol. Ecol. Resour. 2011;11:1022–1032. doi: 10.1111/j.1755-0998.2011.03047.x. [DOI] [PubMed] [Google Scholar]
- 27.Hautier L., San Martin G., Callier P., de Biseau J.-C., Grégoire J.-C. Alkaloids provide evidence of intraguild predation on native coccinellids by Harmonia axyridis in the field. Biol. Invasions. 2011;13:1805–1814. doi: 10.1007/s10530-010-9935-0. [DOI] [Google Scholar]
- 28.Rondoni G., Onofri A., Ricci C. Differential susceptibility in a specialised aphidophagous ladybird, Platynaspis luteorubra (Coleoptera: Coccinellidae), facing intraguild predation by exotic and native generalist predators. Biocontrol Sci. Technol. 2012;22:1334–1350. doi: 10.1080/09583157.2012.726607. [DOI] [Google Scholar]
- 29.Howe A.G., Ravn H.P., Pipper C.B., Aebi A. Potential for exploitative competition, not intraguild predation, between invasive harlequin ladybirds and flowerbugs in urban parks. Biol. Invasions. 2016;18:517–532. doi: 10.1007/s10530-015-1024-y. [DOI] [Google Scholar]
- 30.Li H., Li B., Lövei G.L., Kring T.J., Obrycki J.J. Interactions among native and non-native predatory Coccinellidae influence biological control and biodiversity. Ann. Entomol. Soc. Am. 2020:1–18. doi: 10.1093/aesa/saaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rondoni G., Ielo F., Ricci C., Conti E. Behavioural and physiological responses to prey-related cues reflect higher competitiveness of invasive vs. native ladybirds. Sci. Rep. 2017;7:3716. doi: 10.1038/s41598-017-03471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leroy P.D., Heuskin S., Sabri A., Verheggen F.J., Farmakidis J., Lognay G., Thonart P., Wathelet J.-P., Brostaux Y., Haubruge E. Honeydew volatile emission acts as a kairomonal message for the Asian lady beetle Harmonia axyridis (Coleoptera: Coccinellidae) Insect Sci. 2012;19:498–506. doi: 10.1111/j.1744-7917.2011.01467.x. [DOI] [Google Scholar]
- 33.Sevarika M., Rondoni G., Conti E., Romani R. Antennal sensory organs and glands of the harlequin ladybird, Harmonia axyridis. Entomol. Exp. Appl. 2021;169:111–124. doi: 10.1111/eea.12948. [DOI] [Google Scholar]
- 34.Liu G., Arnaud P., Offmann B., Picimbon J.-F. Pheromone, Natural Odor and Odorant Reception Suppressing Agent (ORSA) for Insect Control. In: Picimbon J.F., editor. Olfactory Concepts of Insect Control—Alternative to insecticides. Springer; Cham, Switzerland: 2019. [Google Scholar]
- 35.Jia X.J., Wang H.X., Yan Z.G., Zhang M.Z., Wei C.H., Qin X.C., Ji W.R., Falabella P., Du Y.L. Antennal transcriptome and differential expression of olfactory genes in the yellow peach moth, Conogethes punctiferalis (Lepidoptera: Crambidae) Sci. Rep. 2016;6:29067. doi: 10.1038/srep29067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afgan E., Baker D., Batut B., Van Den Beek M., Bouvier D., Ech M., Chilton J., Clements D., Coraor N., Grüning B.A., et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:537–544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrews S. FastQC—A Quality Control Tool for High Throughput Sequence Data. [(accessed on 16 June 2018)];2010 Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 38.Blankenberg D., Gordon A., Von Kuster G., Coraor N., Taylor J., Nekrutenko A., Team G. Manipulation of FASTQ data with galaxy. Bioinformatics. 2010;26:1783–1785. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmieder R., Lim Y.W., Edwards R. Identification and removal of ribosomal RNA sequences from metatranscriptomes. Bioinformatics. 2012;28:433–435. doi: 10.1093/bioinformatics/btr669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conesa A., Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 46.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li B., Dewey C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011 doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson M.D., McCarthy D.J., Smyth G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhattacharya D., Price D.C., Chan C.X., Qiu H., Rose N., Ball S., Weber A.P.M., Cecilia Arias M., Henrissat B., Coutinho P.M., et al. Genome of the red alga Porphyridium purpureum. Nat. Commun. 2013;4:1941. doi: 10.1038/ncomms2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masuko K., Fuse N., Komaba K., Katsuyama T., Nakajima R., Furuhashi H., Kurata S. Winged eye induces transdetermination of Drosophila imaginal disc by acting in concert with a histone methyltransferase, Su(var)3-9. Cell Rep. 2018;22:206–217. doi: 10.1016/j.celrep.2017.11.105. [DOI] [PubMed] [Google Scholar]
- 51.Zhao W., Lu L., Yang P., Cui N., Kang L., Cui F. Organ-specific transcriptome response of the small brown planthopper toward rice stripe virus. Insect Biochem. Mol. Biol. 2016;70:60–72. doi: 10.1016/j.ibmb.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 52.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krogh A., Larsson B., Von Heijne G., Sonnhammer E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 54.Bendtsen J.D., Nielsen H., Von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 55.Paula D.P., Togawa R.C., Costa M.M.C., Grynberg P., Martins N.F., Andow D.A. Identification and expression profile of odorant-binding proteins in Halyomorpha halys (Hemiptera: Pentatomidae) Insect Mol. Biol. 2016;25:580–594. doi: 10.1111/imb.12243. [DOI] [PubMed] [Google Scholar]
- 56.Zhang S.Q., Che L.H., Li Y., Dan L., Pang H., Ślipiński A., Zhang P. Evolutionary history of Coleoptera revealed by extensive sampling of genes and species. Nat. Commun. 2018;9:205. doi: 10.1038/s41467-017-02644-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dippel S., Oberhofer G., Kahnt J., Gerischer L., Opitz L., Schachtner J., Stanke M., Schütz S., Wimmer E.A., Angeli S. Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions. BMC Genomics. 2014;15:1141. doi: 10.1186/1471-2164-15-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richards S., Gibbs R.A., Weinstock G.M., Brown S., Denell R., Beeman R.W., Gibbs R., Bucher G., Friedrich M., Grimmelikhuijzen C.J.P., et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 59.Dippel S., Kollmann M., Oberhofer G., Montino A., Knoll C., Krala M., Rexer K.H., Frank S., Kumpf R., Schachtner J., et al. Morphological and transcriptomic analysis of a beetle chemosensory system reveals a gnathal olfactory center. BMC Biol. 2016;14:90. doi: 10.1186/s12915-016-0304-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu P., Wang J., Cui M., Tao J., Luo Y. Antennal transcriptome analysis of the Asian longhorned beetle Anoplophora glabripennis. Sci. Rep. 2016;6:26652. doi: 10.1038/srep26652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKenna D.D., Scully E.D., Pauchet Y., Hoover K., Kirsch R., Geib S.M., Mitchell R.F., Waterhouse R.M., Ahn S.J., Arsala D., et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle-plant interface. Genome Biol. 2016;17:1–18. doi: 10.1186/s13059-016-1088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andersson M.N., Grosse-Wilde E., Keeling C.I., Bengtsson J.M., Yuen M.M.S., Li M., Hillbur Y., Bohlmann J., Hansson B.S., Schlyter F. Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae) BMC Genom. 2013;14:198. doi: 10.1186/1471-2164-14-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu Z., Bin S., He H., Wang Z., Li M., Lin J. Differential expression analysis of chemoreception genes in the striped flea beetle Phyllotreta striolata using a transcriptomic approach. PLoS ONE. 2016;11:e0153067. doi: 10.1371/journal.pone.0153067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen H., Lin L., Xie M., Zhang G., Su W. De novo sequencing, assembly and characterization of antennal transcriptome of Anomala corpulenta Motschulsky (Coleoptera: Rutelidae) PLoS ONE. 2014;9:e114238. doi: 10.1371/journal.pone.0114238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X., Ju Q., Jie W., Li F., Jiang X., Hu J., Qu M. Chemosensory gene families in adult antennae of Anomala corpulenta Motschulsky (Coleoptera: Scarabaeidae: Rutelinae) PLoS ONE. 2015;10:e0121504. doi: 10.1371/journal.pone.0121504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mitchell R.F., Hughes D.T., Luetje C.W., Millar J.G., Soriano-Agatón F., Hanks L.M., Robertson H.M. Sequencing and characterizing odorant receptors of the cerambycid beetle Megacyllene caryae. Insect Biochem. Mol. Biol. 2012;42:499–505. doi: 10.1016/j.ibmb.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X., Wang S., Yi J., Li Y., Liu J., Wang J., Xi J. Three host plant volatiles, hexanal, lauric acid, and tetradecane, are detected by an antenna-biased expressed odorant receptor 27 in the dark black chafer Holotrichia parallela. J. Agric. Food Chem. 2020;68:7316–7323. doi: 10.1021/acs.jafc.0c00333. [DOI] [PubMed] [Google Scholar]
- 68.Antony B., Johny J., Montagné N., Jacquin-Joly E., Capoduro R., Cali K., Persaud K., Al-Saleh M.A., Pain A. Pheromone receptor of the globally invasive quarantine pest of the palm tree, the red palm weevil (Rhynchophorus ferrugineus) bioRxiv. 2020:1–34. doi: 10.1101/2020.07.31.230326. [DOI] [PubMed] [Google Scholar]
- 69.Yuvaraj J.K., Roberts R.E., Sonntag Y., Hou X., Grosse-Wilde E., Machara A., Hansson B.S., Johanson U., Löfstedt C., Andersson M.N. Putative ligand binding sites of two functionally characterized bark beetle odorant receptors. bioRxiv. 2020:1–19. doi: 10.1101/2020.03.07.980797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nikonov A.A., Peng G., Tsurupa G., Leal W.S. Unisex pheromone detectors and pheromone-binding proteins in scarab beetles. Chem. Senses. 2002;27:495–504. doi: 10.1093/chemse/27.6.495. [DOI] [PubMed] [Google Scholar]
- 71.Cui X., Liu D., Sun K., He Y., Shi X. Expression profiles and functional characterization of two odorant-binding proteins from the apple buprestid beetle Agrilus mali (Coleoptera: Buprestidae) J. Econ. Entomol. 2018;111:1420–1432. doi: 10.1093/jee/toy066. [DOI] [PubMed] [Google Scholar]
- 72.Yang H., Su T., Yang W., Yang C.P., Chen Z.M., Lu L., Liu Y.L., Tao Y.Y. Molecular characterization, expression pattern and ligand-binding properties of the pheromone-binding protein gene from Cyrtotrachelus buqueti. Physiol. Entomol. 2017;42:369–378. doi: 10.1111/phen.12209. [DOI] [Google Scholar]
- 73.Yin J., Wang C., Fang C., Zhang S., Cao Y., Li K., Leal W.S. Functional characterization of odorant-binding proteins from the scarab beetle Holotrichia oblita based on semiochemical-induced expression alteration and gene silencing. Insect Biochem. Mol. Biol. 2019;104:11–19. doi: 10.1016/j.ibmb.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 74.Wang B., Guan L., Zhong T., Li K., Yin J., Cao Y. Potential cooperations between odorant-binding proteins of the scarab beetle Holotrichia oblita Faldermann (Coleoptera: Scarabaeidae) PLoS ONE. 2013;8:e84795. doi: 10.1371/journal.pone.0084795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ju Q., Li X., Guo X.Q., Du L., Shi C.R., Qu M.J. Two odorant-binding proteins of the dark black chafer (Holotrichia parallela) display preferential binding to biologically active host plant volatiles. Front. Physiol. 2018;9:769. doi: 10.3389/fphys.2018.00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wojtasek H., Picimbon J.F., Soares Leal W. Identification and cloning of odorant binding proteins from the scarab beetle Phyllopertha diversa. Biochem. Biophys. Res. Commun. 1999;263:832–837. doi: 10.1006/bbrc.1999.1448. [DOI] [PubMed] [Google Scholar]
- 77.Wojtasek H., Hansson B.S., Leal W.S. Attracted or repelled?—A matter of two neurons, one pheromone binding protein, and a chiral center. Biochem. Biophys. Res. Commun. 1998;250:217–222. doi: 10.1006/bbrc.1998.9278. [DOI] [PubMed] [Google Scholar]
- 78.Antony B., Johny J., Aldosari S.A. Silencing the odorant binding protein RferOBP1768 reduces the strong preference of Palm Weevil for the major aggregation pheromone compound ferrugineol. Front. Physiol. 2018;9:252. doi: 10.3389/fphys.2018.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang R., Wang B., Grossi G., Falabella P., Liu Y., Yan S., Lu J., Xi J., Wang G. Molecular basis of alarm pheromone detection in aphids. Curr. Biol. 2017;27:55–61. doi: 10.1016/j.cub.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 80.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 82.De Fouchier A., Walker W.B., Montagné N., Steiner C., Binyameen M., Schlyter F., Chertemps T., Maria A., François M.C., Monsempes C., et al. Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat. Commun. 2017;8:15709. doi: 10.1038/ncomms15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leitch O., Papanicolaou A., Lennard C., Kirkbride K.P., Anderson A. Chemosensory genes identified in the antennal transcriptome of the blowfly Calliphora stygia. BMC Genomics. 2015;16:255. doi: 10.1186/s12864-015-1466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Croset V., Rytz R., Cummins S.F., Budd A., Brawand D., Kaessmann H., Gibson T.J., Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Slone J., Daniels J., Amrein H. Sugar receptors in Drosophila. Curr. Biol. 2007;17:1809–1816. doi: 10.1016/j.cub.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takada T., Sato R., Kikuta S. A mannitol/sorbitol receptor stimulates dietary intake in Tribolium castaneum. PLoS ONE. 2017;12:e0186420. doi: 10.1371/journal.pone.0186420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Missbach C., Dweck H.K.M., Vogel H., Vilcinskas A., Stensmyr M.C., Hansson B.S., Grosse-Wilde E. Evolution of insect olfactory receptors. eLife. 2014;2014:e02115. doi: 10.7554/eLife.02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mitchell R.F., Andersson M.N. Olfactory genomics of the Coleoptera. In: Blomquist G.J., Vogt R.G., editors. Insect Pheromone Biochemistry and Molecular Biology. Elsevier Academic Press; London, UK: 2021. pp. 547–590. [Google Scholar]
- 89.Leroy P.D., Schillings T., Farmakidis J., Heuskin S., Lognay G., Verheggen F.J., Brostaux Y., Haubruge E., Francis F. Testing semiochemicals from aphid, plant and conspecific: Attraction of Harmonia axyridis. Insect Sci. 2012;19:372–382. doi: 10.1111/j.1744-7917.2011.01449.x. [DOI] [Google Scholar]
- 90.Verheggen F.J., Fagel Q., Heuskin S., Lognay G., Francis F., Haubruge E. Electrophysiological and behavioral responses of the multicolored Asian lady beetle, Harmonia axyridis Pallas, to sesquiterpene semiochemicals. J. Chem. Ecol. 2007;33:2148–2155. doi: 10.1007/s10886-007-9370-6. [DOI] [PubMed] [Google Scholar]
- 91.Engsontia P., Sanderson A.P., Cobb M., Walden K.K.O., Robertson H.M., Brown S. The red flour beetle’s large nose: An expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem. Mol. Biol. 2008;38:387–397. doi: 10.1016/j.ibmb.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 92.Wu S., Deng W., Li M., Xiao Y., Li J., Teng K., Xiao Z., Li X., Zhou Z., Li Y. Analysis of chemosensory genes in full and hungry adults of Arma chinensis (Pentatomidae) through antennal transcriptome. Front. Physiol. 2020;11:588291. doi: 10.3389/fphys.2020.588291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gautier M., Yamaguchi J., Foucaud J., Loiseau A., Ausset A., Facon B., Gschloessl B., Lagnel J., Loire E., Parrinello H., et al. The genomic basis of color pattern polymorphism in the harlequin ladybird. Curr. Biol. 2018;28:3296–3302. doi: 10.1016/j.cub.2018.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schoville S.D., Chen Y.H., Andersson M.N., Benoit J.B., Bhandari A., Bowsher J.H., Brevik K., Cappelle K., Chen M.J.M., Childers A.K., et al. A model species for agricultural pest genomics: The genome of the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae) Sci. Rep. 2018;8:1931. doi: 10.1038/s41598-018-20154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fassotte B., Fischer C., Durieux D., Lognay G., Haubruge E., Francis F., Verheggen F.J. First evidence of a volatile sex pheromone in lady beetles. PLoS ONE. 2014;9:e115011. doi: 10.1371/journal.pone.0115011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Verheggen F., Cherif A., Martin C. The production of sex pheromone in lady beetles is conditioned by presence of aphids and not by mating status. J. Chem. Ecol. 2020;46:590–596. doi: 10.1007/s10886-020-01197-z. [DOI] [PubMed] [Google Scholar]
- 97.Hodek I., Evans E.W. Food relationships. In: Hodek I., van Emden H.F., Honek A., editors. Ecology and Behaviour of the Ladybird Beetles (Coccinellidae) John Wiley & Sons; Chichester, UK: 2012. pp. 141–274. [Google Scholar]
- 98.Mathews C.R., Brown M.W., Wäckers F.L. Comparison of peach cultivars for provision of extrafloral nectar resources to Harmonia axyridis (Coleoptera: Coccinellidae) Environ. Entomol. 2016;45:649–657. doi: 10.1093/ee/nvw035. [DOI] [PubMed] [Google Scholar]
- 99.Sloggett J.J., Magro A., Verheggen F.J., Hemptinne J.L., Hutchison W.D., Riddick E.W. The chemical ecology of Harmonia axyridis. BioControl. 2011;56:643–661. doi: 10.1007/s10526-011-9376-4. [DOI] [Google Scholar]
- 100.Růžička Z. Perception for oviposition-deterring larval tracks in aphidophagous coccinellids Cycloneda limbifer and Ceratomegilla undecimnotata (Coleoptera: Coccinellidae) Eur. J. Entomol. 2003;100:345–350. doi: 10.14411/eje.2003.055. [DOI] [Google Scholar]
- 101.Fujii S., Yavuz A., Slone J., Jagge C., Song X., Amrein H. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr. Biol. 2015;25:621–627. doi: 10.1016/j.cub.2014.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guerenstein P.G., Hildebrand J.G. Roles and effects of environmental carbon dioxide in insect life. Annu. Rev. Entomol. 2008;53:161–178. doi: 10.1146/annurev.ento.53.103106.093402. [DOI] [PubMed] [Google Scholar]
- 103.Chen F.J., Wu G., Parajulee M.N., Ge F. Impact of elevated CO2 on the third trophic level: A predator Harmonia axyridis and a parasitoid Aphidius picipes. Biocontrol Sci. Technol. 2007;17:313–324. doi: 10.1080/09583150701211814. [DOI] [Google Scholar]
- 104.Wang S.N., Peng Y., Lu Z.Y., Dhiloo K.H., Zheng Y., Shan S., Li R.J., Zhang Y.J., Guo Y.Y. Cloning and expression profile of ionotropic receptors in the parasitoid wasp Microplitis mediator (Hymenoptera: Braconidae) J. Insect Physiol. 2016;90:27–35. doi: 10.1016/j.jinsphys.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 105.Xia D., Shen C., Zhang Y., Tang Q., Wang J. Identification and expression patterns of sensory neuron membrane protein genes from Sitophilus zeamais Motschulsky (Coleoptera: Curculionidae) Coleopt. Bull. 2019;73:735–745. doi: 10.1649/0010-065X-73.3.735. [DOI] [Google Scholar]
- 106.Zhao Y.J., Li G.C., Zhu J.Y., Liu N.Y. Genome-based analysis reveals a novel SNMP group of the Coleoptera and chemosensory receptors in Rhaphuma horsfieldi. Genomics. 2020;112:2713–2728. doi: 10.1016/j.ygeno.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 107.Pregitzer P., Greschista M., Breer H., Krieger J. The sensory neurone membrane protein SNMP1 contributes to the sensitivity of a pheromone detection system. Insect Mol. Biol. 2014;23:733–742. doi: 10.1111/imb.12119. [DOI] [PubMed] [Google Scholar]
- 108.Jin X., Tal S.H., Smith D.P. SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc. Natl. Acad. Sci. USA. 2008;105:10996–11001. doi: 10.1073/pnas.0803309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data from Illumina sequencing were deposited in the NCBI Short Read Archive (SRA) database (BioProject ID PRJNA698239).