Abstract
Simple Summary
The midgut aminopeptidase N (APN) isoforms have been identified as the binding receptor of insecticidal Cry toxins in numerous insects, including the major arbovirus vector Aedes aegypti (Ae. aegypti). However, whether the Cry-binding APN acts as an essential functional receptor to mediate Bacillus thuringiensis subsp. israelensis (Bti) toxicity in Ae. aegypti larvae remains to be determined. In this study, our results provide the direct molecular evidence demonstrating that two Cry-binding APN isoforms (AeAPN1 and AeAPN2) did not play a key role in mediating Bti Cry4Ba and Cry11Aa toxicity in Ae. aegypti larvae.
Abstract
The insecticidal Cry4Ba and Cry11Aa crystal proteins from Bacillus thuringiensis subsp. israelensis (Bti) are highly toxic to Ae. aegypti larvae. The glycosylphosphatidylinositol (GPI)-anchored APN was identified as an important membrane-bound receptor for multiple Cry toxins in numerous Lepidoptera, Coleoptera, and Diptera insects. However, there is no direct molecular evidence to link APN of Ae. aegypti to Bti toxicity in vivo. In this study, two Cry4Ba/Cry11Aa-binding Ae. aegypti GPI-APN isoforms (AeAPN1 and AeAPN2) were individually knocked-out using CRISPR/Cas9 mutagenesis, and the AeAPN1/AeAPN2 double-mutant homozygous strain was generated using the reverse genetics approach. ELISA assays showed that the high binding affinity of Cry4Ba and Cry11Aa protoxins to the midgut brush border membrane vesicles (BBMVs) from these APN knockouts was similar to the background from the wild-type (WT) strain. Likewise, the bioassay results showed that neither the single knockout of AeAPN1 or AeAPN2, nor the simultaneous disruption of AeAPN1 and AeAPN2 resulted in significant changes in susceptibility of Ae. aegypti larvae to Cry4Ba and Cry11Aa toxins. Accordingly, our results suggest that AeAPN1 and AeAPN2 may not mediate Bti Cry4Ba and Cry11Aa toxicity in Ae. aegypti larvae as their binding proteins.
Keywords: Aedes aegypti, Cry4Ba, Cry11Aa, aminopeptidase N, CRISPR/Cas9
1. Introduction
Aedes aegypti (Ae. aegypti) is a principal vector of several human diseases, including dengue fever, yellow fever, chikungunya, and Zika fever [1]. The annual dengue incidence was estimated to be 390 million infections worldwide by the World Health Organization (WHO) [2]. Unfortunately, no effective vaccines against some vector-borne viral diseases (i.e., chikungunya and Zika) have been developed to date [3,4]. Hence, there is an urgent need to establish an effective and comprehensive control system for reducing the population density of vector mosquitoes.
The microbial agents Bacillus thuringiensis (Bt) have been used worldwide for pest control in agriculture, forestry, and sanitation [5]. Bt subsp. israelensis (Bti) is one of the Bt subspecies that specifically kills mosquito larvae by secreting a variety of crystalline toxins (Cry4A, Cry4B, Cry10A, and Cry11A) and cytolytic toxins (Cyt1 and Cyt2) [6]. Bti has entered the WHO-recommended insecticide list against pathogenic mosquitoes due to its environmental safety profile, target specificity, and lack of potential for resistance development [7,8]. Nonetheless, the insecticidal mechanism of Bt has not been fully understood and remains under investigation [9,10]. According to the known model of action of the three-domain Cry (3d-Cry) toxins, the Cry protoxins are first hydrolyzed at the N-terminus by gut proteases of target insects, and then bind to multiple receptors in the midgut brush border. The activated Cry toxins further accumulate in the apical membrane of the epithelial cells to oligomerize, induce pore formation, and destroy the cell osmotic balance; mechanisms which eventually lead to midgut ulceration and larval death [10]. Currently, four major midgut molecules have been identified as receptors for lepidoptera-specific Cry1 toxins, including the cadherin-like (CAD), the glycosylphosphatidylinositol (GPI)-anchored aminopeptidase N (APN), the GPI-anchored alkaline phosphatase (ALP), and the ATP-binding cassette transporter subfamily C (ABCC2/3) [11]. Several recent studies have revealed that Cry resistance mechanisms were primarily associated with the genetic alterations of these membrane receptors in several field- and laboratory-selected Bt-resistant insects [12]. Hence, functional analysis of Bt receptors is the key to an in-depth understanding of the underlying molecular basis of Bt toxicity and resistance mechanisms in target insects.
The APN family (EC 3.4.11.2) is a ubiquitous hydrolase enzyme mainly responsible for digesting protein through hydrolyzing neutral and basic amino acids at the N-terminus of polypeptides [13]. Since a 120-kDa APN of Manduca sexta (M. sexta) was first identified as the Cry-binding receptor, many more Lepidoptera GPI-APNs have been proposed to be involved in the process of Bt toxin binding [14,15]. Similarly, APNs of Anopheles quadrimaculatus and Anopheles gambiae were identified as the binding receptors for the Bt subsp. Jegathesan (Btj) Cry11Ba toxin [16,17]. Two Ae. aegypti GPI-APN isoforms, named AeAPN1 (AAEL012778), AeAPN2 (AAEL008155, the VectorBase ID has been updated to AAEL019828) have been identified as Cry11Aa-binding receptors by pull-down assays and receptor-binding studies [18,19]. The toxicity of Cry11Aa against Ae. aegypti larvae was enhanced by adding the full-length and partial fragments of AeAPN2 proteins [19]. Thus, the above evidence suggested that Ae. aegypti GPI-APNs may be involved in the pathogenesis of different Bti Cry toxins as important membrane-bound receptors.
To investigate whether the Cry-binding APN is the essential functional receptor for Bti Cry toxins, CRISPR/Cas9-mediated mutagenesis was used for the functional analysis of AeAPN1 and AeAPN2 isoforms by loss-of-function approach in Ae. aegypti. Two AeAPNs are hypothesized to be the functional receptor molecules involved in the toxicity of Bti Cry4Ba and Cry11Aa, which in APN knockouts should be more tolerant to Cry4B and Cry11A toxins than the WT strain. However, our study showed that both the binding affinity and the larval susceptibility of the single AeAPN1 or AeAPN2 knockout strain, and the double AeAPN1/AeAPN2 mutant strain to Cry4B and Cry11A toxins did not differ significantly compared to the wild-type (WT) strain. Our results provide direct molecular evidence demonstrating that AeAPN1 and AeAPN2 may not mediate Bti Cry4Ba and Cry11Aa toxicity in Ae. aegypti.
2. Materials and Methods
2.1. Mosquito and Bt Strains
The laboratory WT strain of Ae. aegypti (Ae. aegypti Haikou strain) was provided by the Fujian International Travel Health Care Center (Fujian, Fuzhou, China) and reared without exposure to Bt toxins for over 50 generations. All Ae. aegypti strains were maintained at 26 ± 1 °C and 83% ± 3% relative humidity with a photoperiod of 14-h light/10-h dark cycles. The recombinant Bt strains individually producing Cry4Ba and Cry11Aa (pCG6-Cry4Ba and pCG6-Cry11Aa) were provided by Dr. Sarjeet R Gill’s Laboratory, University of California Riverside, USA and stored at −80 °C.
2.2. Purification of Cry4Ba and Cry11Aa Protoxins
The pCG6-Cry4Ba and pCG6-Cry11Aa were grown in the 1/2 LB (Luria-Bertani) medium containing 12.5 μg/mL erythromycin at 30 °C. Subsequently, the crystal inclusions were completely released from the spores, the medium was removed by centrifugation at 9500× g and the pellet were washed three times with 1 M NaCl plus 0.03% Triton X-100 and three times with distilled water. The Cry4Ba crystal inclusion was purified by a repeated crystal solubilization method as previously described [20]. The Cry11Aa crystal inclusion was purified by discontinuous sucrose gradients as previously described [21]. The Cry4Ba protoxin was solubilized in alkaline buffer (50 mM Na2CO3/NaHCO3, pH 10.5), and Cry11Aa protoxin was solubilized in distilled water. The protein concentrations of Cry protoxins were determined using the Bradford Protein Assay Kit (Beyotime, Shanghai, China) with Bovine Serum Albumin (BSA) as the standard.
2.3. sgRNA Design and Synthesis
The CRISPR/Cas9 target sites were designed in the third exon of AeAPN1 and AeAPN2 genes using CRISPOR program (http://crispor.tefor.net/ (accessed on 8 October 2019)), and potential off-target effects were evaluated by Cas-OFFinder (http://www.rgenome.net/cas-offinder/ (accessed on 8 October 2019)) (Table S1). DNA templates for small-guide RNAs (sgRNAs) were prepared by polymerase chain reaction (PCR) with target specific primers (Table 1) using KOD Hot Start Polymerase (TOYOBO, Osaka, Japan), and purified using a Gel Extraction Kit (OMEGA Bio-Tek, Norcross, GA, USA). In vitro transcription of sgRNAs was performed using the MEGAscript Kit (Ambion, Austin, MA, USA), and purified by phenol: chloroform extraction and isopropanol precipitation as per the manufacturer’s instructions. The purified sgRNA concentration was determined using the ultra-micro spectrophotometer Q5000 (Quawell, San Jose, CA, USA) and stored at −80 °C until use.
Table 1.
List of DNA oligo primers used in this study.
| Primer Name | Sequence 5′–3′ |
|---|---|
| sgRNA-R | ATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC |
| APN1-sgRNA-1-F | GAAATTAATACGACTCACTATAGGTCTACAGTCGGCCATCCAGTT TTAGAGCTAGAAATAGC |
| APN1-sgRNA-2-F | GAAATTAATACGACTCACTATAGGTTCGTCGCACACTCAGCAGTT TTAGAGCTAGAAATAGC |
| APN2-sgRNA-1-F | GAAATTAATACGACTCACTATAGGATTGGAGCTAGCGGTAACGTT TTAGAGCTAGAAATAGC |
| APN2-sgRNA-2-F | GAAATTAATACGACTCACTATAGTGCGTCGAGACTACAAGACGTT TTAGAGCTAGAAATAGC |
| AeAPN1-F | GGAATGCCGATACTCCAAGATCAAT |
| AeAPN1-R | TGAAAATAATCCACTCATTGGCCGG |
| AeAPN2-F | AGTGTTCTGAACATGTTCCGTGT |
| AeAPN2-R | TATGCGTCGTTGATCAGCTGAGC |
2.4. Embryo Microinjection and Generation of Ae. aegypti Knockout Strains
The collection of mosquito embryos and microinjections were performed as described previously [22]. About 1 nL sgRNA/Cas9 mixtures of 300 ng/μL spCas9 protein (NEB, USA), and 100 ng/μL each of sgRNAs and 1x Cas9 nuclease reaction buffer was injected into the fresh embryos using the Nanoject III™ Microinjection System (Drummond, PA, USA). The hatched G0 embryos were reared to pupation, and each pupa was transferred individually in cups for eclosion. The genomic DNA (gDNA) was extracted from one hindleg of each G0 mosquito adult using 50 μL of chelex-100 buffer containing 5% chelex-100 beads (BioRad, Berkeley, CA, USA) and 1 mg/μL Proteinase K (TaKaRa, Dalian, China), hatching at 55 °C overnight and 5 μL supernatant for PCR template. G0 mutations were identified by PCR amplification using Premix Taq Version 2.0 (TaKaRa, Dalian, China) with corresponding target specific primers (Table 1) followed by Sanger sequencing. The validated G0 mutations were single outcrossed with the WT strain to generate F1 families, and the F1 individuals were validated by PCR and sequencing as described above. F1 individuals with similar mutation sequences were pooled to generate F2 families. The homozygous mutants were screened from F2 individuals and pooled to establish the homozygous knockout strains.
2.5. Preparation of Brush Border Membrane Vesicles from Ae. aegypti Larvae
The alimentary tract tissues were dissected from approximately 500 early four-instar mosquito larvae, soaked in MET buffer (0.3 M mannitol, 5 mM EDTA, and 17 mM Tris-Cl, pH 7.5) and stored at −80 °C. The brush border membrane vesicles (BBMVs) were prepared using the differential magnesium precipitation method, as described previously [23]. The protein concentrations of the BBMVs were determined as described above.
2.6. Proteomic Identification of Midgut BBMVs from Ae. aegypti Larvae
To confirm the absence of AeAPN1 and AeAPN1 isoforms at the protein level in midgut brush border membrane of AeAPN knockouts, proteomic analysis of midgut BBMV proteins were performed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Then, 15 μg BBMV proteins were separated on 10% SDS-PAGE, and the target regions (~70 kDa−170 kDa) of Coomassie blue stained gels including AeAPNs were excised and stored at −20 °C. The gel pieces were in-gel digestion and LC-MS/MS analysis using an Orbitrap-Fusion-Tribrid mass spectrometer (Thermo Fisher Scientific, MA, USA) at the Basic Forestry and Proteomics Research Center, FAFU. The resulting MS/MS data were processed using Proteome Discoverer 1.3 were matched against the annotated protein databases of Ae. aegypti (https://vectorbase.org/vectorbase/app/downloads/Current_Release/AaegyptiLVP_AGWG/fasta/data/ (accessed on 1 February 2021)).
2.7. ELISA Binding Assays
ELISA binding assay was performed as reported previously [24]. The ELISA plates were coated with 0.3 μg Ae. aegypti BBMVs in a final volume of 100 uL PBS/well overnight at 4 °C. Then, the plates were washed three times with 200 μL PBS, and blocked with 200 μL/well of blocking buffer (PBS with 2% low fat milk powder) for 2 h at 37 °C. The plates were washed three times with 200 μL PBST and 100 μL/well of different concentrations of Cry protoxin/PBST were added and the cultures were incubated at 37 °C for 1 h. After washing three times with PBST and PBS to remove unbound protoxins, the wells were incubated with 100 μL blocking buffer containing anti-Cry4Ba or anti-Cry11Aa polyclonal antibody (1:10,000 dilution) for 1 h at 37 °C. After the required washing steps, 100 μL/well of PBST containing HRP-labeled goat anti-rabbit IgG (H + L) antibodies (1:5000 dilution) (Beyotime, Shanghai, China) was added and allowed to incubate for 1 h at 37 °C. The wells were washed again and 100 μL/well of TMB chromogenic solution (Beyotime, Shanghai, China) was added and allowed to react for 8–15 min at 37 °C, protected from light. Finally, 50 μL/well of 2 M H2SO4 was added to terminate the reaction, and the optical density (OD) value of each well was read at 450 nm with an absorbance microplate reader (BioTek, Winooski, VT, USA). The Kd values were calculated using Curve Expert 1.4 (Hyams DG Softwave) and the ELISA-binding plots were generated in GraphPad prism 8.0 (GraphPad Softwave).
2.8. Bioassay of Bti Cry Toxins
The susceptibility to Bti Cry4Ba and Cry11Aa toxins was determined for different Ae. aegypti strains as described [25]. At least 5 gradient concentrations of purified Cry4Ba and Cry11Aa protoxins were added to 20 mL filtered water containing 25 early fourth instar larvae and mixed fully. Each larvae bioassay was repeated with three biological replications. The larval mortality was recorded after 24 h, and the medium lethal concentration (LC50) and the 95% confidence intervals (CI) of the LC50 were calculated by probit analysis using PoloPlus (LeOra Software).
3. Results
3.1. Generation of Individual AeAPN1 and AeAPN2 Knockout Ae. aegypti Strains by CRISPR/Cas9
To increase G0 mutagenesis efficiency and to induce sequence fragment deletions for AeAPN1 and AeAPN2 gene regions, we designed two CRISPR/Cas9 target sites within 100 bp region of the corresponding third exons to disrupt the downstream GPI-anchored sites (Figure 1A, B). After 400 and 800 Ae. aegypti eggs were injected with sgRNA/Cas9 mixtures to knockout AeAPN1 and AeAPN2 genes, 26 and 88, respectively, they hatched into larvae within one month (Table 2). DNA sequencing of 560 bp gDNA fragments spanning the gRNA targeted region from AeAPN1- and AeAPN2-knockout G0 individuals indicated that 4 and 36 of the larvae, respectively, presented undefined peaks around the CRISPR/Cas9 target sites (Table 2). The homozygous knockout (KO) Ae. aegypti strain for AeAPN1 (named AeAPN1-KO) and APN2 (named AeAPN2-KO) were generated using the reverse genetics approach. The AeAPN1-KO presented a 107 bp deletion and 9 bp insertion between 2 of the CRISPR/Cas9 target sites, causing an amino acid mismatch and protein translation to terminate prematurely upstream of the GPI-anchored site (Figure 1C and Figure S1). The AeAPN2-KO had a 14 bp deletion and 99 bp deletion in the 2 of CRISPR/Cas9 target sites respectively, also resulting in the deletion of the GPI-anchored site and a loss of function as the membrane receptor (Figure 1D and Figure S2). The complementary DNA (cDNA) PCR sequencing of the two homozygous knockouts showed the genotypes of the coding region were consistent with the gDNA (Figure 1C,D).
Figure 1.
Generation of AeAPN knockout Ae. aegypti strains. Schematic representations of the AeAPN1 (A) and AeAPN2 (B) locus, where enlarged is the third exon contain 20-nucleotide sgRNA target sequences, and the PAM sequence shown in red. Aligned Sanger-sequencing trace of PCR-amplified using gDNA and cDNA from WT, AeAPN1-KO and AeAPN2-KO strains with specific primers (Table 1) spanning the gRNA targeted region (C,D). Electrophoresis of genomic-PCR products spanning target region from WT, AeAPN1-KO, AeAPN2-KO, and AeAPN1/AeAPN2-KO strains with specific primers AeAPN1-F/R and AeAPN2-F/R (E), Lane M: 100 bp DNA Ladder (TaKaRa, Dalian, China), Lane 1 and 5: the fragments were amplified from gDNA of the WT strain, Lane 2 and 6: the fragments were amplified from gDNA of the AeAPN1-KO strain, Lane 3 and 7: the fragments were amplified from gDNA of the AeAPN2-KO strain, Lane 4 and 8: the fragments were amplified from gDNA of the AeAPN1/AeAPN2-KO strain.
Table 2.
Transformation data of G0 embryos injected with CRISPR/Cas9 constructs.
| Injected Component | Injected G0 Embryos | G0 Survivors | G0 Mutants |
|---|---|---|---|
| Cas9 protein, APN1-sgRNA-1 and APN1-sgRNA-2 | 400 | 26 | 4 |
| Cas9 protein, APN2-sgRNA-1 and APN2-sgRNA-2 | 800 | 88 | 36 |
3.2. Generation of AeAPN1/AeAPN2 Double-Mutant Ae. aegypti Strain
To generate a homozygous Ae. aegypti strain with double AeAPN1/AeAPN2 knockout, AeAPN1-KO males were crossed with AeAPN2-KO females, and the homozygous mutants (named AeAPN1/AeAPN2-KO) were screened by the reverse genetics approach described above. The PCR products of AeAPN1 and AeAPN2 mutant allele were significantly smaller than the WT allele, which were easily separated by gel electrophoresis (Figure 1E). The proteomic analysis of target midgut BBMV proteins (~70–170 kDa) from WT and AeAPN1/AeAPN2-KO strains by LC-MS/MS, showed that 1382 proteins were identified from WT sample including 43 unique peptides of AeAPN1 and 36 peptides (34 unique peptides) of AeAPN2, while no AeAPN1 and AeAPN2 peptides were identified in 1245 proteins of AeAPN1/AeAPN2-KO sample, suggesting the loss of GPI-anchoring signal of AeAPN1 and AeAPN2 cannot anchor to the midgut apical microvilli of Ae. aegypti (Figure S3 and Table S2).
3.3. Cry4Ba and Cry11Aa Protoxins Binding to BBMVs of the APN Knockouts and the WT Strains
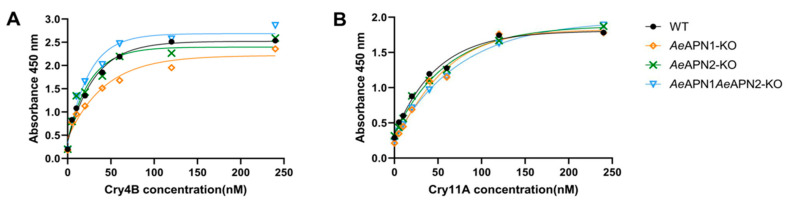
The ELISA binding assays using the Cry4Ba protoxin to BBMVs of AeAPN1-KO, AeAPN2-KO, AeAPN1/AeAPN2-KO, and WT strains showed that the Cry4Ba bound to BBMVs of APN knockouts with high affinity (KdAeAPN1-KO = 7.82 nM, KdAeAPN2-KO = 8.31 nM and KdAeAPN1/AeAPN2-KO = 8.94 nM), which was not significantly different than the binding of the WT strain (KdWT = 8.07 nM) (Figure 2A). The Cry11Aa protoxin also had a high affinity to BBMVs of APN knockouts (KdAeAPN1-KO = 9.9 nM, KdAeAPN2-KO = 11.01 nM, KdAeAPN1/AeAPN2-KO = 10.35 nM, and Kd WT = 7.88 nM) (Figure 2B). These data indicated that losing of AeAPN1 and AeAPN2 cannot affect the high affinity of Cry4Ba and Cry11Aa toxins binding the midgut epithelial cells of Ae. aegypti larvae.
Figure 2.
Binding of Cry4Ba and Cry11Aa toxins to Ae. aegypti BBMVs by ELISA. Different concentrations (0 nM, 5 nM, 10 nM, 20 nM, 40 nM, 60 nM, 120 nM, 240 nM) of Cry4Ba (A) and Cry11Aa (B) protoxins bound to 0.3 μg midgut BBMVs from APN mutant strains and WT strain in the ELISA plates.
3.4. Response of the Ae. aegypti Larvae to Cry4Ba and Cry11Aa Protoxins
Bioassays of the AeAPN1-KO, AeAPN2-KO, and AeAPN1/AeAPN2-KO strains with Cry4Ba and Cry11Aa showed that the LC50 of Cry4Ba and Cry11Aa for these three APN mutant strains were similar to that of the WT strain, which indicated that neither the single knockout of AeAPN1 or AeAPN2, nor simultaneous disruption of AeAPN1 and AeAPN2 resulted in a significant reduction of Ae. aegypti larvae susceptibility to Cry4Ba and Cry11Aa toxins (Table 3 and Table 4).
Table 3.
Susceptibility of Ae. aegypti strains to Cry4Ba toxin.
| Ae. aegypti Strain | n | Slope (SE) | LC50 (μg/mL) (95% CI) | RR a |
|---|---|---|---|---|
| WT | 1125 | 2.947 (0.098) | 1.771 (1.663–1.888) | 1 |
| AeAPN1-KO | 1125 | 3.774 (0.181) | 1.504 (1.421–1.591) | 0.849 |
| AeAPN2-KO | 1125 | 3.664 (0.171) | 1.863 (1.771–1.958) | 1.052 |
| AeAPN1/AeAPN2-KO | 1125 | 3.079 (0.135) | 2.221 (2.092–2.367) | 1.254 |
a RR (Relative Resistance) = LC50 of the knockout strain/LC50 of the WT strain.
Table 4.
Susceptibility of Ae. aegypti strains to Cry11Aa toxin.
| Ae. aegypti Strain | n | Slope (SE) | LC50 (μg/mL) (95% CI) | RR a |
|---|---|---|---|---|
| WT | 1125 | 1.747 (0.110) | 0.602 (0.526–0.685) | 1 |
| AeAPN1-KO | 1125 | 3.192 (0.224) | 0.556 (0.496–0.619) | 0.924 |
| AeAPN2-KO | 1125 | 1.989 (0.124) | 0.599 (0.546–0.653) | 0.995 |
| AeAPN1/AeAPN2-KO | 1125 | 2.770 (0.163) | 0.826 (0.769–0.897) | 1.372 |
a RR (Relative Resistance) = LC50 of the knockout strain/LC50 of the WT strain.
4. Discussion
In recent decades, the identification of midgut molecules as Bt receptors in target insects has been a research objective to define Bt activity. The sequential binding model proposes that the activated Cry toxin forms a Pre-pore oligomer after interacting with the transmembrane receptor CAD, and further binding to the GPI-anchored membrane receptors (APN or ALP) leads to membrane insertion, pore formation, and cell lysis [14]. The GPI-APN was the first to be described as a Cry-binding receptor in the midgut of Lepidopteran insects [15]. However, whether the Cry-binding APN acts as an essential functional receptor to mediate Bti toxicity in Ae. aegypti larvae remains to be determined. In this study, our results demonstrated that deficiency in AeAPN1 and AeAPN2 isoforms do not affect either the binding affinity or susceptibility of Ae. aegypti larvae to Cry4Ba and Cry11Aa toxins.
Early functional analyses of APN as Cry receptors showed that the GPI-APNs of M. sexta and Heliothis virescens (H. virescens) were reconstituted into phospholipid vesicles and planar lipid bilayers to increase the binding affinity of Cry1 toxins and catalyze channel formation [26,27,28]. The in vitro cytotoxicity analysis indicated that heterologous expression of different APNs in insect cell lines (S2, Sf9, and Sf21), which are not susceptible to Cry toxin, decrease the cell viability or facilitate cell swelling under activated Cry toxins infection, such as following Helicoverpa armigera (H. armigera) APN1 [29], H. virescens APN1 [30] and two of Ae. aegypti APNs [31] infections. Whereas, the expression of M. sexta APN1 in S2 cells did not lead to host cell sensitivity to activated Cry1 toxins [32]. Furthermore, the down-regulated APN expression using in vivo RNAi was associated with tolerance to Cry toxins in several insects, including Spodoptera exigua APN1 [33], M. sexta APN1 [34], three Chilo suppressalis APNs [35], three Diatraea saccharalis APNs [36] and three Ae. aegypti APNs [37]. Another important source of APN involvement in Bt toxicity is suggested by deletion mutation studies of HaAPN1 and the down-regulation of Trichoplusia ni (T. ni) APN1 transcription, which have been shown to be genetically linked with Cry1Ac resistance [38,39]. The transcriptome analysis of a laboratory-selected Cry11Aa resistant Ae. aegypti strain indicated that the transcript levels of two APNs (AAEL008158 and AAEL008162) were significantly down-regulated compared to the WT strain. Nonetheless, no changes in expression and non-synonymous mutations have been observed in AeAPN1 and AeAPN2 [40]. Thus, further functional analysis of APN applying genome-editing strategies are required to confirm the role of APN as a Bt functional receptor rather than as a Cry-binding protein.
Recently, CRISPR/Cas9-mediated genomic editing technology has provided a powerful tool to generate KO/knock-in models, and has been applied to the identification of Bt receptors in some lepidopteran insects. For example, the ABCA2 mutant generated by CRISPR/Cas9 in T. ni and H. armigera resulting in high-level resistance to Cry2A toxins [41,42]. CRISPR/Cas9-mediated double knockout of ABCC2 and ABCC3 in H. armigera and Plutella xylostella (P. xylostella) resulted in more than 1000-fold resistance to Cry1Ac [43,44]. KO of T. ni CAD and Spodoptera frugiperda (S. frugiperda) CAD did not affect the larvae susceptibility to Cry1 and Cry2 toxins [45,46]. Conversely, knockout of H. armigera CAD by CRISPR/Cas9 in a Bt-susceptible strain could increase resistance to Cry1Ac toxin by more than 500-fold [47]. Moreover, none of the 3 H. armigera APNs (HaAPN1, HaAPN2, and HaAPN5) individually knocked out using CRISPR/Cas9 resulted in any change in susceptibility of the larvae to Cry1A and Cry2A toxins [48].
Genetic and molecular studies have indicated that the entomopathogenicity of Bt is complex and may involve multiple membrane-bound receptors and intracellular pathways [9,49]. Generating single-receptor knockout insects by genome editing often does not achieve significantly different results in larvae susceptibility to Bt toxins. For example, neither PxABCC2 nor PxABCC3 knockout in P. xylostella strains produced any significant resistance to Cry1Ac, while simultaneous mutations of the two genes exhibited high-level resistance (>8000-fold) to Cry1Ac, revealing the functional redundancy between ABCC2 and ABCC3 as receptors in the activity of Cry1Ac toxins [44]. In this study, the double mutant AeAPN1/AeAPN2-KO strain did not show increased tolerance to Cry4Ba and Cry11Aa toxins, suggesting AeAPN1 and AeAPN2 did not exhibit synergistic effects on mediating the entomopathogenicity of Cry4Ba and Cry11Aa (Table 2 and Table 3). The genome-wide analysis of the APN gene family showed that 29 APN isoforms were identified in Ae. aegypti genome, 11 of them were predicted to carry the GPI-anchoring signal. Moreover, a previous pulldown assay and our Co-immunoprecipitation assay (unpublished data) showed that AeAPN3 (AAEL012774) could bind to Cry4Ba and Cry11Aa toxins in the midgut BBMVs of Ae. aegypti [18]. Therefore, the potential complementary roles of other AeAPNs as Cry-binding receptors in determining susceptibility to Cry4Ba/Cry11Aa toxins should not be ruled out. Comprehensive determination of the role of APN in the action mechanism of Bt requires further functional analysis of multiple APNs in a variety of insects. Overall, our study revealed that two Cry-binding APNs (AeAPN1 and AeAPN2) may not play a key role in mediating Bti Cry4Ba and Cry11Aa toxicity in Ae. aegypti.
Acknowledgments
We thank Yajie Guo provided constructive suggestions in ELISA binding assay. We also thank the Basic Forestry and Proteomics Research Center, FAFU for providing proteomic analysis.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/12/3/223/s1, Figure S1: Amino acid sequence alignment of AeAPN1 isoform from WT and AeAPN1-KO strains, Figure S2: Amino acid sequence alignment of AeAPN2 isoform from WT and AeAPN1-KO strains, Figure S3: SDS-PAGE profile of midgut BBMV protein from the WT strain and the AeAPN1/AeAPN2-KO strain, Table S1: In silico analysis of off-target activity of sgRNAs used in this study, Table S2: AeAPN1 and AeAPN2 peptides identified from midgut BBMV of the WT strain.
Author Contributions
Conceptualization, J.W., S.W., L.Z., and X.G.; methodology, J.W., X.Y., and H.H.; software, J.W., X.Y., H.H.; investigation, J.W., X.Y., H.H., J.C., Y.L., W.H., L.O. and Z.Y.; formal analysis, J.W., X.Y., H.H.; writing—original draft preparation, J.W.; writing—review and editing, J.W., S.W., L.Z., and X.G.; supervision, S.W., L.Z., and X.G.; funding acquisition, S.W., L.Z., and X.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (grant number 2017YFD0600105); the National Natural Science Foundation of China (grant numbers U1905201 and 31601905); the Natural Science Foundation of Fujian Province (2020J01550); the United Fujian Provincial Health and Education Project for Tackling Key Research (Grant No. 2019-WJ-29); Open Project Funds of State Key Laboratory of Pathogen and Biosecurity (SKLPBS1838); China Postdoctoral Science Foundation (grant number 2017M612107); Forestry Programs of Science and Technology in Fujian Province (grant number Mincaizhi [2020] 601).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Exclude this statement.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kyle J.L., Harris E. Global spread and persistence of dengue. Annu. Rev. Microbiol. 2008;62:71–92. doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization . World Health Organization; 2012. [(accessed on 1 March 2021)]. Global Strategy for Dengue Prevention and Control, 2012–2020. Available online: https://apps.who.int/iris/handle/10665/75303. [Google Scholar]
- 3.Wilder-Smith A. Dengue vaccine development: Status and future. Bundesgesundheitsblatt Gesundh. Gesundh. 2020;63:40–44. doi: 10.1007/s00103-019-03060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zanotto P.M.d.A., Leite L.C.d.C. The Challenges Imposed by Dengue, Zika, and Chikungunya to Brazil. Front. Immunol. 2018;9:1964. doi: 10.3389/fimmu.2018.01964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crickmore N., Zeigler D.R., Feitelson J., Schnepf E., Van Rie J., Lereclus D., Baum J., Dean D.H. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 1998;62:807–813. doi: 10.1128/MMBR.62.3.807-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry C., O’Neil S., Ben-Dov E., Jones A.F., Murphy L., Quail M.A., Holden M.T., Harris D., Zaritsky A., Parkhill J. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 2002;68:5082–5095. doi: 10.1128/AEM.68.10.5082-5095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization . World Health Organization; 2006. [(accessed on 1 March 2021)]. Pesticides and Their Application: For the Control of Vectors and Pests of Public Health Importance. Available online: https://apps.who.int/iris/handle/10665/69223. [Google Scholar]
- 8.Ben-Dov E. Bacillus thuringiensis subsp. israelensis and its dipteran-specific toxins. Toxins (Basel) 2014;6:1222–1243. doi: 10.3390/toxins6041222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vachon V., Laprade R., Schwartz J.-L. Current models of the mode of action of Bacillus thuringiensis insecticidal crystal proteins: A critical review. J. Invertebr. Pathol. 2012;111:1–12. doi: 10.1016/j.jip.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Bravo A., Gómez I., Mendoza Almanza G., Gaytán M., Soberón M. Different Models of the Mode of Action of Bt 3d-Cry Toxins. CABI; Oxford, UK: 2015. pp. 56–68. [Google Scholar]
- 11.Pardo-López L., Soberón M., Bravo A. Bacillus thuringiensis insecticidal three-domain Cry toxins: Mode of action, insect resistance and consequences for crop protection. FEMS Microbiol. Rev. 2013;37:3–22. doi: 10.1111/j.1574-6976.2012.00341.x. [DOI] [PubMed] [Google Scholar]
- 12.Peterson B., Bezuidenhout C.C., Van den Berg J. An Overview of Mechanisms of Cry Toxin Resistance in Lepidopteran Insects. J. Econ. Entomol. 2017;110:362–377. doi: 10.1093/jee/tow310. [DOI] [PubMed] [Google Scholar]
- 13.Nocek B., Mulligan R., Bargassa M., Collart F., Joachimiak A. Crystal structure of aminopeptidase N from human pathogen Neisseria meningitidis. Proteins. 2008;70:273–279. doi: 10.1002/prot.21276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pigott C.R., Ellar D.J. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol. Mol. Biol. Rev. 2007;71:255–281. doi: 10.1128/MMBR.00034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knight P.J.K., Knowles B.H., Ellar D.J. Molecular cloning of an insect aminopeptidase N that serves as a receptor for Bacillus thuringiensis CryIA(c) toxin. J. Biol. Chem. 1995;270:17765–17770. doi: 10.1074/jbc.270.30.17765. [DOI] [PubMed] [Google Scholar]
- 16.Abdullah M.A., Valaitis A.P., Dean D.H. Identification of a Bacillus thuringiensis Cry11Ba toxin-binding aminopeptidase from the mosquito, Anopheles quadrimaculatus. BMC Biochem. 2006;7:16. doi: 10.1186/1471-2091-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang R., Hua G., Urbauer J.L., Adang M.J. Synergistic and inhibitory effects of aminopeptidase peptides on Bacillus thuringiensis Cry11Ba toxicity in the mosquito Anopheles gambiae. Biochemistry. 2010;49:8512–8519. doi: 10.1021/bi1009908. [DOI] [PubMed] [Google Scholar]
- 18.Chen J., Aimanova K.G., Pan S., Gill S.S. Identification and characterization of Aedes aegypti aminopeptidase N as a putative receptor of Bacillus thuringiensis Cry11A toxin. Insect Biochem. Mol. Biol. 2009;39:688–696. doi: 10.1016/j.ibmb.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J., Likitvivatanavong S., Aimanova K.G., Gill S.S. A 104 kDa Aedes aegypti aminopeptidase N is a putative receptor for the Cry11Aa toxin from Bacillus thuringiensis subsp. israelensis. Insect Biochem. Mol. Biol. 2013;43:1201–1208. doi: 10.1016/j.ibmb.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Z.-s., Yang S.-j., Shu C.-l., Song F.-p., Zhou X.-p., Zhang J. Comparison and optimization of the method for Cry1Ac protoxin preparation in HD73 strain. J. Integr. Agric. 2015;14:1598–1603. doi: 10.1016/S2095-3119(14)60950-3. [DOI] [Google Scholar]
- 21.Thomas W.E., Ellar D.J. Bacillus thuringiensis var israelensis crystal delta-endotoxin: Effects on insect and mammalian cells in vitro and in vivo. J. Cell Sci. 1983;60:181–197. doi: 10.1242/jcs.60.1.181. [DOI] [PubMed] [Google Scholar]
- 22.Kistler K.E., Vosshall L.B., Matthews B.J. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 2015;11:51–60. doi: 10.1016/j.celrep.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfersberger M., Luethy P., Maurer A., Parenti P., Sacchi F.V., Giordana B., Hanozet G.M. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae) Comp. Biochem. Physiol. Part A Physiol. 1987;86:301–308. doi: 10.1016/0300-9629(87)90334-3. [DOI] [Google Scholar]
- 24.Peña-Cardeña A., Grande R., Sánchez J., Tabashnik B.E., Bravo A., Soberón M., Gómez I. The C-terminal protoxin domain of Bacillus thuringiensis Cry1Ab toxin has a functional role in binding to GPI-anchored receptors in the insect midgut. J. Biol. Chem. 2018;293 doi: 10.1074/jbc.RA118.005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batool K., Alam I., Zhao G., Wang J., Xu J., Yu X., Huang E., Guan X., Zhang L. C-Type Lectin-20 Interacts with ALP1 Receptor to Reduce Cry Toxicity in Aedes aegypti. Toxins. 2018;10:390. doi: 10.3390/toxins10100390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sangadala S., Walters F.S., English L.H., Adang M.J. A mixture of Manduca sexta aminopeptidase and phosphatase enhances Bacillus thuringiensis insecticidal CryIA(c) toxin binding and 86Rb(+)-K+ efflux in vitro. J. Biol. Chem. 1994;269:10088–10092. doi: 10.1016/S0021-9258(17)36993-4. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz J.L., Lu Y.J., Söhnlein P., Brousseau R., Laprade R., Masson L., Adang M.J. Ion channels formed in planar lipid bilayers by Bacillus thuringiensis toxins in the presence of Manduca sexta midgut receptors. FEBS Lett. 1997;412:270–276. doi: 10.1016/S0014-5793(97)00801-6. [DOI] [PubMed] [Google Scholar]
- 28.Luo K., Sangadala S., Masson L., Mazza A., Brousseau R., Adang M.J. The Heliothis virescens 170kDa aminopeptidase functions as “Receptor A” by mediating specific Bacillus thuringiensis Cry1A δ-endotoxin binding and pore formation. Insect Biochem. Mol. Biol. 1997;27:735–743. doi: 10.1016/S0965-1748(97)00052-0. [DOI] [PubMed] [Google Scholar]
- 29.Sivakumar S., Rajagopal R., Venkatesh G.R., Srivastava A., Bhatnagar R.K. Knockdown of Aminopeptidase-N from Helicoverpa armigera Larvae and in Transfected Sf21 Cells by RNA Interference Reveals Its Functional Interaction with Bacillus thuringiensis Insecticidal Protein Cry1Ac. J. Biol. Chem. 2007;282:7312–7319. doi: 10.1074/jbc.M607442200. [DOI] [PubMed] [Google Scholar]
- 30.Wei J., Zhang M., Liang G., Wu K., Guo Y., Ni X., Li X. APN1 is a functional receptor of Cry1Ac but not Cry2Ab in Helicoverpa zea. Sci. Rep. 2016;6:19179. doi: 10.1038/srep19179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aroonkesorn A., Pootanakit K., Katzenmeier G., Angsuthanasombat C. Two specific membrane-bound aminopeptidase N isoforms from Aedes aegypti larvae serve as functional receptors for the Bacillus thuringiensis Cry4Ba toxin implicating counterpart specificity. Biochem. Biophys. Res. Commun. 2015;461:300–306. doi: 10.1016/j.bbrc.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banks D.J., Hua G., Adang M.J. Cloning of a Heliothis virescens 110 kDa aminopeptidase N and expression in Drosophila S2 cells. Insect Biochem. Mol. Biol. 2003;33:499–508. doi: 10.1016/S0965-1748(03)00022-5. [DOI] [PubMed] [Google Scholar]
- 33.Qiu L., Cui S., Liu L., Zhang B., Ma W., Wang X., Lei C., Chen L. Aminopeptidase N1 is involved in Bacillus thuringiensis Cry1Ac toxicity in the beet armyworm, Spodoptera exigua. Sci. Rep. 2017;7:45007. doi: 10.1038/srep45007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flores-Escobar B., Rodríguez-Magadan H., Bravo A., Soberón M., Gómez I. Differential role of Manduca sexta aminopeptidase-N and alkaline phosphatase in the mode of action of Cry1Aa, Cry1Ab, and Cry1Ac toxins from Bacillus thuringiensis. Appl. Environ. Microbiol. 2013;79:4543–4550. doi: 10.1128/AEM.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X.Y., Du L.X., Liu C.X., Gong L., Han L.Z., Peng Y.F. RNAi in the striped stem borer, Chilo suppressalis, establishes a functional role for aminopeptidase N in Cry1Ab intoxication. J. Invertebr. Pathol. 2017;143:1–10. doi: 10.1016/j.jip.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y., Zhu Y.C., Ottea J., Husseneder C., Rogers Leonard B., Abel C., Huang F. Molecular characterization and RNA interference of three midgut aminopeptidase N isozymes from Bacillus thuringiensis-susceptible and -resistant strains of sugarcane borer, Diatraea saccharalis. Insect Biochem. Mol. Biol. 2010;40:592–603. doi: 10.1016/j.ibmb.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Saengwiman S., Aroonkesorn A., Dedvisitsakul P., Sakdee S., Leetachewa S., Angsuthanasombat C., Pootanakit K. In vivo identification of Bacillus thuringiensis Cry4Ba toxin receptors by RNA interference knockdown of glycosylphosphatidylinositol-linked aminopeptidase N transcripts in Aedes aegypti larvae. Biochem. Biophys. Res. Commun. 2011;407:708–713. doi: 10.1016/j.bbrc.2011.03.085. [DOI] [PubMed] [Google Scholar]
- 38.Tiewsiri K., Wang P. Differential alteration of two aminopeptidases N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc. Natl. Acad. Sci. USA. 2011;108:14037–14042. doi: 10.1073/pnas.1102555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang S., Cheng H., Gao Y., Wang G., Liang G., Wu K. Mutation of an aminopeptidase N gene is associated with Helicoverpa armigera resistance to Bacillus thuringiensis Cry1Ac toxin. Insect Biochem. Mol. Biol. 2009;39:421–429. doi: 10.1016/j.ibmb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Lee S.B., Aimanova K.G., Gill S.S. Alkaline phosphatases and aminopeptidases are altered in a Cry11Aa resistant strain of Aedes aegypti. Insect Biochem. Mol. Biol. 2014;54:112–121. doi: 10.1016/j.ibmb.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X., Chen W., Song X., Ma X., Cotto-Rivera R.O., Kain W., Chu H., Chen Y.R., Fei Z., Wang P. Mutation of ABC transporter ABCA2 confers resistance to Bt toxin Cry2Ab in Trichoplusia ni. Insect Biochem. Mol. Biol. 2019;112:103209. doi: 10.1016/j.ibmb.2019.103209. [DOI] [PubMed] [Google Scholar]
- 42.Wang J., Wang H., Liu S., Liu L., Tay W.T., Walsh T.K., Yang Y., Wu Y. CRISPR/Cas9 mediated genome editing of Helicoverpa armigera with mutations of an ABC transporter gene HaABCA2 confers resistance to Bacillus thuringiensis Cry2A toxins. Insect Biochem. Mol. Biol. 2017;87:147–153. doi: 10.1016/j.ibmb.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Wang J., Ma H., Zhao S., Huang J., Yang Y., Tabashnik B.E., Wu Y. Functional redundancy of two ABC transporter proteins in mediating toxicity of Bacillus thuringiensis to cotton bollworm. PLoS Pathog. 2020;16:e1008427. doi: 10.1371/journal.ppat.1008427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Z., Fu S., Ma X., Baxter S.W., Vasseur L., Xiong L., Huang Y., Yang G., You S., You M. Resistance to Bacillus thuringiensis Cry1Ac toxin requires mutations in two Plutella xylostella ATP-binding cassette transporter paralogs. PLoS Pathog. 2020;16:e1008697. doi: 10.1371/journal.ppat.1008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S., Kain W., Wang P. Bacillus thuringiensis Cry1A toxins exert toxicity by multiple pathways in insects. Insect Biochem. Mol. Biol. 2018;102:59–66. doi: 10.1016/j.ibmb.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J., Jin M., Yang Y., Liu L., Yang Y., Gómez I., Bravo A., Soberón M., Xiao Y., Liu K. The Cadherin Protein Is Not Involved in Susceptibility to Bacillus thuringiensis Cry1Ab or Cry1Fa Toxins in Spodoptera frugiperda. Toxins. 2020;12:375. doi: 10.3390/toxins12060375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J., Zhang H., Wang H., Zhao S., Zuo Y., Yang Y., Wu Y. Functional validation of cadherin as a receptor of Bt toxin Cry1Ac in Helicoverpa armigera utilizing the CRISPR/Cas9 system. Insect Biochem. Mol. Biol. 2016;76:11–17. doi: 10.1016/j.ibmb.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Wang J., Zuo Y.Y., Li L.L., Wang H., Liu S.Y., Yang Y.H., Wu Y.D. Knockout of three aminopeptidase N genes does not affect susceptibility of Helicoverpa armigera larvae to Bacillus thuringiensis Cry1A and Cry2A toxins. Insect Sci. 2020;27:440–448. doi: 10.1111/1744-7917.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X., Candas M., Griko N.B., Taussig R., Bulla L.A., Jr. A mechanism of cell death involving an adenylyl cyclase/PKA signaling pathway is induced by the Cry1Ab toxin of Bacillus thuringiensis. Proc. Natl. Acad. Sci. USA. 2006;103:9897–9902. doi: 10.1073/pnas.0604017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Exclude this statement.