Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has evolved to display particular patterns of genetic diversity in the genome across geographical regions. These variations in the virus and genetic variation in human populations can determine virus transmissibility and coronavirus disease 2019 (COVID-19) severity. Genetic variations and immune differences in human populations could be the driving forces in viral evolution. Recently emerged SARS-CoV-2 variants show several mutations at the receptor binding domain in the spike (S) glycoprotein and contribute to immune escape and enhanced binding with angiotensin 1-converting enzyme 2 (ACE2). Since ACE2 and transmembrane protease serine 2 (TMPRSS2) play important roles in SARS-CoV-2 entry into the cell, genetic variation in these host entry-related proteins may be a driving force for positive selection in the SARS-CoV-2 S glycoprotein. Dendritic or liver/lymph cell-specific intercellular adhesion molecule (ICAM)-3-grabbing non-integrin is also known to play vital roles in several pathogens. Genetic variations of these host proteins may affect the susceptibility to SARS-CoV-2. This review summarizes the latest research to describe the impacts of genetic variation in the viral S glycoprotein and critical host proteins and aims to provide better insights for understanding transmission and pathogenesis and more broadly for developing vaccine/antiviral drugs and precision medicine strategies, especially for high risk populations with genetic risk variants.
Keywords: SARS-CoV-2, COVID-19, spike glycoprotein, mutation, genetic variation, ACE2, variants, immune escape, transmissibility
1. Introduction
The recently identified severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the pandemic of coronavirus disease 2019 (COVID-19), which began at the end of 2019 and is ongoing [1,2]. The number of confirmed cases continues to rise rapidly worldwide, with nearly 78% of confirmed cases in the Americas (44.5%) and Europe (34%) (calculated until 22 February 2021) (World Health Organization (WHO) COVID-19 report, https://covid19.who.int/, accessed on 22 February 2021). According to a meta-analysis, the fatality rate of COVID-19 is around 3% [3].
SARS-CoV-2 is the third coronavirus to cause a pandemic, the other two being SARS-CoV [4] and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) [5] in 2003 and 2012, respectively. Genetic evidence from the SARS-CoV-2 genome shows high identity with two bat-derived SARS-like coronaviruses, bat-SL-CoVZC45 (87.6%) and bat-SL-CoVZXC21 (87.5%) [6]. Due to the lack of antiviral drug and vaccine selection pressure, the current genetic diversity patterns of SARS-CoV-2 in different geographical regions may be associated with genetic variation in populations, with increasing genetic diversity in the virus attributed to natural selection driven by long periods of an evolutionary arms race between host and virus [7]. Several studies have demonstrated that there is a positive correlation between host cell genetic variation and the susceptibility to different viruses [8,9,10]. Older age, male sex, and some co-morbidities have been found to be risk factors associated with COVID-19 severity, however, those risk factors do not fully explain the differences between asymptomatic, mild, and severe patients [11]. A recent genome-wide association study by Ellinghaus and colleagues showed that rs1138592 and rs657152 genetic variants were significantly associated with respiratory failure in severe COVID-19 patients. Notably, rs65712 is located at the ABO blood group gene, and Ellinghaus and colleagues further confirmed that patients with blood group A showed higher risk than others [12]. Another study by Zeberg and Pääbo found that the core haplotype in chromosome 3 is strongly associated with COVID-19 severity. The frequency of this haplotype was found to vary between South Asian (30%), European (8%), admixed American (4%), and East Asian (almost absent) populations [13]. However, future study is required to investigate the significance of this variation on COVID-19 severity. It is believed that human genetic variation can result in different responses to SARS-CoV-2 infection, even with the same age, sex, and health status. SARS-CoV-2 has evolved to contain cumulative mutations in its genome, with the most highly mutated regions being ORF1ab, spike, and nucleocapsid genes. It is inferred that positive selection contributes to the evolution of SARS-CoV-2 [14,15]. Several recently emerged SARS-CoV-2 variants, B.1.1.7 lineage (a.k.a. 20B/501Y.V1 Variant and VUI202012/01), B.1.351 lineage (a.k.a. 20C/501Y.V2), P.1/P.2 lineages (descendent of B.1.1.28), and B.1.429, have been found responsible for the dramatic increase of infections in the United Kingdom [16], South Africa [17], Brazil [18], and North America [19], respectively. Viral genome analysis showed these variants to carry multiple mutations in the S glycoprotein, including some at the receptor binding domain (RBD). Some of these mutations are believed to be the result of adaptive evolution and have biological importance. The direct impacts of the mutations in the S glycoprotein of SARS-CoV-2 include affecting the viral transmissibility through interaction with the host cell binding receptor and contributing to the immune escape through changes in the RBD. The most important host proteins involved in SARS-CoV-2 entry have been identified as angiotensin 1-converting enzyme 2 (ACE2) and cell-surface associated transmembrane protease serine 2 (TMPRSS2). Genetic variations in ACE2 and TMPRSS2 may provide the driving force for viral evolution, therefore causing positive selection for these emerging mutations in the SARS-CoV-2 S glycoprotein. Additionally, dendritic or liver/lymph cell-specific intercellular adhesion molecule (ICAM)-3-grabbing non-integrin (DC/L-SIGN) has been known to play vital roles for several pathogens, including SARS-CoV [20]. Therefore, the genetic variation of these host proteins may also affect susceptibility to SARS-CoV-2. Investigating the correlations between genetic variation in populations and viral infectivity or clinical outcomes could provide great insights for developing precision medicine strategies.
In this review, we aim to compile knowledge and current advances on the impacts of genetic variations in the viral S glycoprotein and critical host proteins on the susceptibility to SARS-CoV-2 infection and immune escape. This understanding is crucial for controlling the pandemic through enhanced surveillance and vaccine development.
2. Brief Introduction to Coronavirus Proteins and Mutations
SARS-CoV-2 is one of the coronaviruses (CoV) and is an enveloped and positive-sense ssRNA (~30 kb) virus which belongs to the Betacoronavirus genus, Nidovirales order. Two replicase open reading frames (ORFs) encoded by ORF1a (~13.2 kb) and ORF1b (~8.1 kb) occupy at least two-thirds of the CoV genome (Figure 1A). The polyprotein ORF1ab (as known as pp1ab) is translated due to a −1 ribosomal frameshift upstream of the ORF1a stop codon [21]. Polyprotein ORF1a (as known as pp1a) and pp1ab can be further processed to 16 functional non-structural proteins (nsps) by self-produced nsp5 and nsp3 proteases. Nsp5 protease (also called 3C-like protease, 3CLpro, Mpro) contains a chymotrypsin-like fold and is responsible for processing nsp4 to nsp16, whereas nsp3 papain-like protease (PLpro) is responsible for processing nsp1 to nsp4 [22]. SARS-CoV nsps have been well studied and characterized for their involvement in the different steps of the virus replication cycle [22].
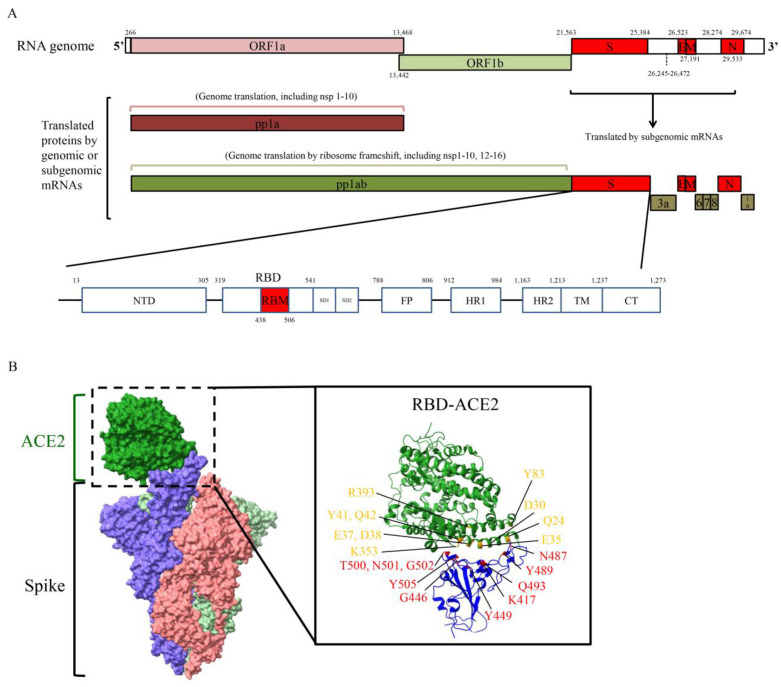
Figure 1.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genomic organization and structure of S glycoprotein. (A) The ORF1a and ORF1b genes can be translated to polyprotein 1a (pp1a) and polyprotein 1ab (pp1ab, -1 ribosomal frameshift). Pp1a and pp1ab can be processed into several functional non-structural proteins (nsps). Structural genes encode four structure proteins, S glycoprotein, envelope protein (E), membrane protein (M), and nucleocapsid protein (N). Several accessory proteins are encoded in the end of genome, include ORF3a, ORF6, ORF7, ORF8, and ORF10. NTD, N-terminal domain; SD1 and SD2, subdomain 1 and 2; FP, fusion peptide; HR1 and HR2, heptad repeat 1 and 2; TM, transmembrane region. (B) Structure of SARS-CoV-2 S glycoprotein bound to angiotensin 1-converting enzyme 2 (ACE2). S glycoprotein consists of three S glycoprotein monomers which are shown in blue, pink, and light green. ACE2 is shown in green. The interface of SARS-CoV-2 receptor binding domain (RBD)-receptor binding motif (RBM) and ACE2 is enlarged in right panel. The amino acid positions of RBD and ACE2 responsible for binding are shown in yellow (ACE2) and red (RBD). Structure depicting S glycoprotein bound to ACE2 using PDB: 7A94. Structure depicting RBD bound to ACE2 using PDB: 6VW1. Figures are generated by UCSF ChimeraX software.
Four structural protein genes are located in the C-terminal region of the CoV genome. The SARS-CoV-2 S glycoprotein contains a furin recognition cleavage site (polybasic cleavage site, PRRAR) which provides efficient proteolytic processing into S1 and S2 [23]. Additionally, recent evidence has shown that the furin cleavage site of the SARS-CoV-2 S glycoprotein plays a critical role in regulating viral replication and pathogenesis, which could be a potential therapeutic target against SARS-CoV-2 infection [24,25,26]. The SARS-CoV-2 RBD, located in the S1 domain, binds to the host cell ACE2 receptor, while S2 functions as the membrane fusion subunit [27,28]. Within the RBD, the receptor binding motif (RBM) is in close contact with ACE2. Several amino acid positions of the interface between ACE2 and RBM have been found to play important roles for binding through the formation of hydrogen bonds and salt bridges (Figure 1B). The envelope (E) protein is involved in virion production and pathogenesis [29], and the membrane (M) protein plays a pivotal role in mediating virus assembly and budding. In addition, the M protein interacts with the viral nucleocapsid (N) protein for viral RNA packaging and recruits other structural proteins to the endoplasmic reticulum (ER)-Golgi-intermediate compartment [30,31]. The N protein encapsulates and protects the CoV genome in the virion and then enters the host cells to promote viral replication [32,33]. Several accessory proteins can be found in CoV, however, though they can affect viral viability and pathogenesis, evidence has shown that accessory proteins are not essential for viral replication [34].
3. Impacts of Mutations on the SARS-CoV-2 S Glycoprotein
The first step of CoV infection of a target cell is the binding of the viral RBD of the S glycoprotein to the cell membrane receptor, ACE2 (Figure 1B). The SARS-CoV-2 RBD and ACE2 binding structure is nearly identical to that of the SARS-CoV RBD [35,36]. There are several studies which have shown that SARS-CoV-2 is more infectious than SARS-CoV, which may explain why SARS-CoV-2 has caused a more severe pandemic than SARS-CoV [37,38,39,40]. Three mechanisms which have been proposed to potentially play a role in this increased SARS-CoV-2 infectivity are: (1) higher binding affinity of RBD to ACE2, (2) less exposed RBD (immune escape), and (3) pre-activation by furin (enhanced viral entry) [41]. Other than ACE2, another protein which has been shown to be important in SARS-CoV-2 entry into the cell is TMPRSS2. The cleavage ability of TMPRSS2 to prime the S glycoprotein during viral attachment plays a vital role in SARS-CoV-2 entry into the cell [27]. Evidence has shown that the furin processing region (amino acid position 675 to 692) has the highest mutation density (number of distinct mutations in the region) [42]. Mutations which occur in this region may provide an advantage to the virus, allowing it to utilize a large number of host proteases to enhance infectivity. The S glycoprotein has also been shown to play pivotal roles in identifying host specificity, viral pathogenesis, and inducing human neutralizing antibodies. Viral surface proteins have been shown as promising targets to generate therapeutic or prevention purpose antibodies [43]. Current studies have focused on the mutation at the RBD (residues 319 to 541) in the SARS-CoV-2 S glycoprotein. Within the interface of the S glycoprotein (RBM) and ACE2, several mutations have been identified.
The continuous transmission of SARS-CoV-2 has caused rapid accumulation of mutations in the S glycoprotein across geographical regions. One of the predominant mutations, D614G, was found to be circulating and rapidly spreading outside China in the early pandemic. The recently emerged variants in the United Kingdom (B.1.1.7), South Africa (B.1.351), North America (B.1.429), and Brazil (B.1.1.28) exhibit several mutations in the S glycoprotein, especially in the RBD. B.1.1.7 carries eight mutations in the S glycoprotein which include H69/V70 deletion (ΔH69/V70), Y144 deletion (ΔY144), N501Y, A570D, P681H, T716A, S982A, and D1118H. B.1.351, recently found in South Africa, carries three mutations located in the RBD region which are K417N, E484K, and N501Y. B.1.1.28 (P.1 and P.2 lineages) in Brazil exhibits a different pattern of mutations. The P.1 lineage has the same RBD mutations as South Africa, whereas the P.2 lineage has independently accrued the spike E484K mutation [44]. B.1.429, which has recently been found to be spreading rapidly in California, USA, includes three mutations in the S glycoprotein, S13I and W152C in the S1 domain and L452R in the RBD [19]. Those variants have caused a severe increase in SARS-CoV-2 infections since December 2020. Notably, the N501Y mutation, which is located in the RBD and has been found in most of the variants, is believed to enhance the transmissibility of SARS-CoV-2. The aforementioned variants all have the D614G mutation, though this is expected due to the predominance of D614G since the early pandemic. Other than the mutations found in those variants, several mutations in S glycoprotein have been found sporadically and contribute to the immune escape and the transmissibility of SARS-CoV-2.
3.1. Impacts of Mutations in the S Glycoprotein on Transmissibility and Infectivity
The impact of D614G on viral transmissibility has been widely studied due to its emergence in the early pandemic and its worldwide presence. The D614G mutation is of rising concern, as it has the potential to affect SARS-CoV-2 infectivity through changes to RBD structure, S1/S2 subunit interaction, viral entry, and immune response [45]. Becerra-Flores and colleagues have found that patients infected with SARS-CoV-2 containing the D614G mutation have a higher case fatality rate [46]. The effects of the D614G mutation on the SARS-CoV-2 S glycoprotein have been comprehensively investigated by several studies. First, this mutation has been found to be associated with higher viral load in the upper respiratory tract in patients, further confirmed in pseudotyped experiments and animal models [45,47,48]. Second, a detailed structure analysis showed that the D614G mutation shifts the conformation of the S glycoprotein to be more open, therefore contributing to enhanced ACE2 binding and fusion efficiency [49]. This conformation change of the S glycoprotein has been found to be important for SARS-CoV-2 binding with ACE2 [50]. Third, the D614G mutation can decrease S1 shedding, which indicates enhanced efficiency of processing by furin-like proprotein convertase [51]. However, the D614G mutation did not show resistance to neutralizing antibodies [52]. A recent report indicated that the D614G mutation can potentially affect the glycosylation at residue 616 which may be able to enhance virulence through DC/L-SIGN binding in dendritic cells [53]. Notably, D614G combined with other mutations in S glycoprotein exhibits more infectivity in different cell lines [54]. As there was no available treatment or vaccination selective pressure in SARS-CoV-2 infection in the early pandemic, how the D614G mutation occurred and became predominant outside China is not clear. Our previous study showed that the D614G mutation is significantly associated with the differences in ACE2 expression levels in populations [55]. This study indicates that populations with lower ACE2 expression, such as Europe and Africa, provide the environment for selective pressure for SARS-CoV-2 adaptive evolution.
ΔH69/V70 are located at the N-terminal domain (NTD) of the S glycoprotein. The ΔH69/V70 deletion has been found globally, however, tracking of SARS-CoV-2 sequences has shown it to be mainly circulating in Europe. A single round infectivity experiment showed that SARS-CoV-2 carrying either ΔH69/V70 or ΔH69/V70 combined with N501Y can enhance infectivity in 293T/hACE2 cells. Additionally, a virus carrying the ΔH69/V70 mutation exhibits more S glycoprotein incorporated in the virion [56]. A recent study showed that ΔH69/V70 with D796H in the S glycoprotein can potentially contribute to immune escape in immunocompromised patients, while D796H itself decreased the infectivity but contributed to the reduction of susceptibility to neutralizing antibodies [57]. The loss of infectivity caused by D796H could be compensated for in cases where it co-occurs with ΔH69/V70. ΔH69/V70 also frequently co-occurs with N439K or Y453F, which are located at the RBD of the SARS-CoV-2 S glycoprotein. The binding affinity of Y453F with ACE2 is controversial, however, it is seen to contribute to the immune escape for neutralizing antibodies and human convalescent sera [58,59,60]. The N439K mutation could enhance the binding affinity with ACE2 through the formation of a new salt bridge and has resistance to some neutralizing antibodies and human convalescent sera [61].
S477N is located in the RBD and has been found to enhance binding with ACE2 [58]. The E484K mutation in the RBD of S glycoprotein is of rising concern due to its emergence in several current variants which cause severe transmission. A current study showed that E484K could enhance the binding with ACE2 through a conformational change of the S glycoprotein [62]. L452R, located at the RBD, has been shown to increase the infectivity by stabilizing the S glycoprotein and ACE2 interaction [63,64,65]. N501Y is another mutation of rising concern due to its co-occurrence in several current SARS-CoV-2 variants in the United Kingdom and South Africa. N501Y is also located in the RBD of the S glycoprotein and could potentially affect binding with ACE2. The N501Y mutation emerged in infected wild-type mice at early passage and is believed to be the result of adaptive evolution of the SARS-CoV-2 virus [66]. Studies using a comprehensive scanning approach [58] and in silico methods [67] have shown that the N501Y mutation can increase the binding affinity for ACE2. The resulting enhanced binding affinity may be due to additional hydrogen bonds with ACE2 at residues Y41 and K353 [68] and may contribute to a more open conformation of the RBD in the S glycoprotein [69]. The P681H mutation is juxtaposed to the furin processing site (amino acid position 682 to 685). The furin processing of the S glycoprotein into S1/S2 is an important step for virus fusion into cells [70], however, whether the P681H mutation could affect viral infectivity and efficiency of furin processing needs further investigation. V1176 is located at the stalk domain of the S glycoprotein. The flexible stalk domain is necessary for viral entry and fusion into the cells [71]. According to a molecular dynamics simulations analysis, the V1176F mutation could enhance the flexibility of the S glycoprotein by increasing motility and inducing compactness [72]. Additionally, evidence has shown that V1176F is associated with higher patient mortality [72,73]. The mutations N331Q and N343Q could disrupt the N-glycosylation site of the S glycoprotein and strongly decrease the viral infectivity, however, there is no current circulating SARS-CoV-2 carrying those mutations [54,74] (Table 1).
Table 1.
Impacts of mutations in S glycoprotein in circulating SARS-CoV-2.
| Mutation | S Region | Potential Effects | % of All Sequences (n = 498,134) |
% of Mutation in Geographical Regions | Ref. |
|---|---|---|---|---|---|
| ΔH69/V70 | S1 NTD |
|
17.3 (n = 86,033) |
|
[56,57] |
| ΔY144 | S1 NTD |
|
15.3 (n = 76,387) |
|
[75,76] |
| ΔL242/244 | S1 NTD |
|
0.2 (n = 1049) |
|
[75,76] |
| N331Q | RBD |
|
NA | NA | [54,74] |
| N343Q | RBD |
|
NA | NA | [54,74] |
| E406W | RBD |
|
NA | NA | [77] |
| K417N/T/V | RBD |
|
0.3 (n = 1373) |
|
[52,77] |
| N439K | RBM |
|
2.3 (n = 11,396) |
|
[52,54,77] |
| N440D/K | RBM |
|
<0.1 (n = 3) |
|
[52,77] |
| K444E/N | RBM |
|
<0.1 (n = 32) |
|
[78] |
| G446D/V | RBM |
|
<0.1 (n = 151) |
|
[54,78] |
| N450K/Y/D | RBM |
|
<0.1 (n = 47) |
|
[78] |
| L452R | RBM |
|
0.7 (n = 3722) |
|
[54,65,78] |
| Y453F | RBM |
|
0.2 (n = 1032) |
|
[77] |
| A475V | RBM |
|
<0.1 (n = 118) |
|
[52,54] |
| G476S | RBM |
|
<0.1 (n = 56) |
|
[54] |
| S477N | RBM |
|
5.2 (n = 25,970) |
|
[58,78] |
| T478I | RBM |
|
<0.1 (n = 142) |
|
[54,78] |
| P479S | RBM |
|
<0.1 (n = 175) |
|
[78] |
| E484K | RBM |
|
0.5 (n = 2483) |
|
[52,62,76,78] |
| F486L | RBM |
|
<0.1 (n = 17) |
|
[77,78] |
| Y489H | RBM |
|
<0.1 (n = 4) |
|
[77] |
| Q493K | RBM |
|
<0.1 (n = 6) |
|
[77] |
| P499L | RBM |
|
<0.1 (n = 3) |
|
[78] |
| N501Y | RBM |
|
16 (n = 79,510) |
|
[52,56,58,67] |
| D614G | S1 CTD |
|
Predominant circulating worldwide |
|
[45,47,48,49,50,51,52,54] |
| P681H | S2 |
|
16.5 (n = 81,974) |
|
[79] |
| D796H | S2 |
|
<0.1 (n = 115) |
|
[57] |
| V1176F | S2 |
|
0.4 (n = 1,886) |
|
[72,73] |
A total of 498,134 SARS-CoV-2 sequences from the Global Initiative on Sharing Avian Influenza Data (GISAID) database (https://www.gisaid.org/, accessed on 16 February 2021), from 1 January 2020 to 16 February 2021, were analyzed to determine the percentage of mutations in different geographical regions. RBD, receptor binding domain; RBM, receptor binding motif; NTD: N-terminal domain; NA: not available.
3.2. Impacts of Mutations in the S Glycoprotein on Immune Escape Ability
The ΔY144 and the L242/L244 deletions (ΔL242/244) are located at the NTD of the S glycoprotein and show a loss of binding ability with neutralizing antibodies [75,76]. Starr and colleagues mapped the mutations in the RBD of SARS-CoV-2 which could escape neutralization by the antibodies used to treat COVID-19 patients, Regeneron’s REGN-COV2 cocktail (consisting of two antibodies, REGN10933 and REGN10987, emergency use authorization for treatment of COVID-19) and Eli Lilly’s LY-CoV016 antibody (also known as CB6 or JS016, phase 3 clinical trials). They found that E406W can escape the neutralization by the REGN-COV2 cocktail. K417N can escape the neutralization by several monoclonal antibodies including LY-CoV016. N439K and N440D can escape the neutralization by the REGN10987 antibody. Y453F, F486L, Y489H, and Q493K also escape the neutralization by REGN10933 [77]. K417N is one of the major mutations found in B.1.351 which has also been recently shown to escape the neutralization by monoclonal antibodies [52,77,80]. A recent study used free energy perturbation calculations to show that the combination of N501Y and K417N could enhance the binding with ACE2 while dramatically decreasing the binding with antibodies [81]. The E484K mutation, located in the RBD, not only enhanced binding with ACE2, it also exhibited strong or moderate resistance to several human neutralizing antibodies and human convalescent sera [52,76,78,80,82,83], which indicates this mutation is important in the viral evolution to escape neutralizing antibodies. L452R can also reduce the sensitivity to several antibodies and human convalescent sera [54,78]. Several rare mutations (<0.1%) have been found to contribute to the immune escape of neutralization by monoclonal antibodies and human convalescent sera, including N440D, K444N, G446D/V, N450K/Y/D, A475V, G476S, T478I, P479S, F486L, Y489H, Q493K, P499L, and D796H [54,57,77,78] (Table 1).
3.3. Geographical Distribution of Mutations in S Glycoprotein
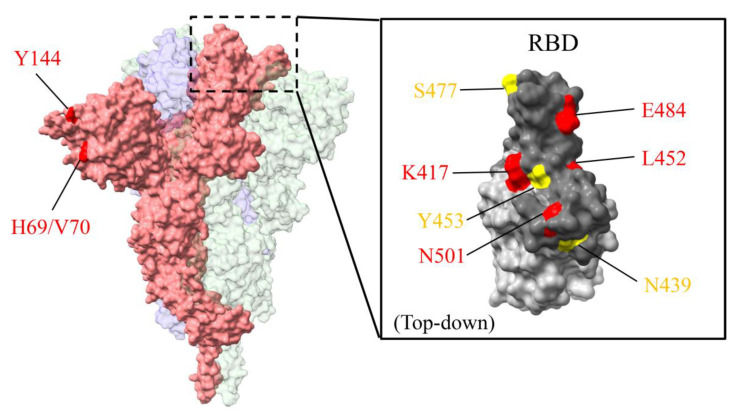
There are several mutations which appear in more than 0.1% of sequences circulating across geographical regions, including ΔH69/V70 and Δ144 in S1 NTD; K417N/T/V, N439K, L452R, Y453F, S477N, E484K, and N501Y in RBD; D614G in S1 CTD; P681H and V1176F in S2. ΔH69/V70, Δ144, N439K, Y453F, and N501Y are found circulating mainly in Europe. K417N/T/V is carried by B.1.351 and B.1.1.28 which are mainly circulating in Africa and Europe. L452R is carried by B.1.429 which is mainly circulating in North America and Europe. S477N is mainly circulating in Europe and Oceania. E484K is carried by several current circulating variants and is found in Africa (33.8%) and Europe (30.8%), however, there are also more than 10% distributed in North and South Americas (Figure 2). P681H is carried by B.1.1.7 and is mainly found circulating in Europe. Notably, SARS-CoV-2 sequences carrying only P681H (excluded B.1.1.7) make up 4.8% of those circulating in North America. V1176F is mainly circulating in South America, however, there are more than 10% of SARS-CoV-2 sequences carrying this mutation in Europe and North America (Table 1).
Figure 2.
Structure of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) S glycoprotein with current emerged mutations. Mutations existing in more than 1% of total sequences are shown. One monomer of S glycoprotein is shown in pink. The structure of the receptor binding domain (RBD) is enlarged in right panel and receptor binding motif (RBM) is shown in dark gray. Mutations in currently emerged SARS-CoV-2 variants are labeled in red, whereas others are in yellow. Structure depicting S glycoprotein using PDB: 6XR8. Structure depicting RBD using PDB: 7BWJ. Figures are generated by UCSF ChimeraX software.
Several functionally important mutations have been found to be circulating across geographical regions and co-occurring with other variants. E484K is one of the most concerning mutations which exhibits increased dynamics in several severe transmission regions. Recent evidence has shown that SARS-CoV-2 carrying E484K or K417N (less prevalent than E484K) in the S glycoprotein could contribute to broad immune escape from monoclonal antibodies and human convalescent sera [52,76]. The current SARS-CoV-2 variants exhibit high transmissibility and immune escape ability as a result of several co-occurring mutations in the S glycoprotein. Increased transmissibility is conferred by ΔH69/V70 and N501Y in B.1.1.7, E484K and N501Y in B.1.351, E484K in B.1.1.28, and L452R in B.1.429. Immune escape is contributed to by ΔY144 in B.1.1.7, E484K and K417N in B.1.351, E484K in B.1.1.28, and L452R in B.1.429.
4. Impacts of Genetic Variability of ACE2, TMPRSS2, and DC/L-SIGNs in SARS-CoV-2 Infectivity and Pathogenicity
4.1. ACE2 Genetic Variation and SARS-CoV-2 Infection
The ACE2 gene contains 18 exons located in chromosome X. ACE2 consists of three domains: (1) N-terminal peptidase domain (residues 19-615), (2) C-terminal collectrin-like domain (CLD, residues 616-768), and (3) end with a hydrophobic transmembrane region and an intracellular segment of 43 residues [84,85]. ACE2 belongs to the family of angiotensin converting enzymes (ACE) members. ACE is a widely distributed protein which converts angiotensin (Ang) I (inactive form) to AngII (activate form). This conversion is known to play a vital role in several biological functions, such as controlling blood pressure [86,87], regulating water and sodium absorption in the kidneys [88], and mediating cell proliferation [89]. ACE2 has been demonstrated to be involved in regulating heart function, hypertension (HT), diabetic heart disease, and dyslipidemian [90]. Several studies have shown that polymorphisms of ACE2 are significantly associated with blood pressure in different populations [91,92]. Additionally, COVID-19 patients who have HT, heart disease, and diabetes are associated with severe infections and clinical outcomes [11,93].
A previous study on a group 2 coronavirus demonstrated that the correlation between viral receptor genetic variation and viral binding activity can affect host susceptibility [94]. Similarly, the relationship between human immunodeficiency virus type 1 (HIV-1) gp120 and the CD4 T cell co-receptor CCR5 is another well-known example of a receptor polymorphism affecting viral entry. Individuals carrying the CCR5Δ32 polymorphism (CCR5 contains 32 bp deletions) can block HIV-1 entry into host cells and prevent infection [9]. For CoV, the polymorphisms (three missenses and one deletion) of the functional receptor dipeptidyl-peptidase 4 (DPP4/CD26) of MERS-CoV have recently been demonstrated to reduce the interaction with the S glycoprotein [95]. In addition, different expression levels of ACE2 have been demonstrated to be positively correlated with SARS-CoV and NL63 (another human related respiratory coronavirus) infection [96]. Jia and colleagues have shown that a point mutation (L584A) in ACE2 can facilitate SARS-CoV entry into the host cell [97]. Hence, the genetic variation of ACE2 between different populations may contribute to susceptibility to SARS-CoV-2. According to a recent report by Darbani and colleagues, 34 ACE2 variants have been defined with importance for SARS-CoV-2 entry and infection [98]. The ACE2 allele frequencies included six interaction-booster variants (S19P, I21V, K26R, T27A, N64K, and H378R) and eight interaction-inhibitor variants (E37K, N51D, K68E, F72V, M82I, G326E, Q388L, and P389H) which have been shown to vary significantly between populations. In addition, Darbani and colleagues showed that more than half of the variants were found in males, which may explain previous clinical observations showing higher mortality rates in males [99,100]. However, eight ACE2 variants located at the binding interface showed no disruption of the interaction between ACE2 and the RBD [101]. Cao and colleagues recently found two ACE2 intron variants and ten other protein intron variants (located within or near the ACE2 gene, three from CLTRN, five from CA5B, and two from an unknown gene) which showed association with higher ACE2 expression levels by genetic analysis of expression quantitative trait loci (eQTLs) [102]. Intriguingly, nine of twelve intron variants showed significantly higher allele frequencies in Asian populations when compared to others (African, European, and American). Notably, most of these intron variants were located at the CLTRN and the CA5B genes. Future study is required to clarify the correlation between the ACE2 gene regulatory network and genetic variation.
Another systematic ACE2 genetic analysis by Stawiski and colleagues identified nine ACE2 variants which could increase susceptibility to SARS-CoV-2 and 17 ACE2 variants which displayed protective roles on SARS-CoV-2 infection by structural computational analysis [103]. The missense variants of ACE2 identified by recent studies, including key residues of ACE2 binding with SARS-CoV-2 RBD and residues with potential to affect binding, are summarized in Table 2. All of the ACE2 missense variants in Table 2 are rare variants (<0.01 allele frequency), and most of the rare missense variants are distributed in European populations. However, based on our current knowledge, the ACE2 rare missense variants in the population do not disrupt the interaction with the SARS-CoV-2 S glycoprotein. Hashizume and collegues identified seven ACE2 missense variants which exist in Asian but not in American and European populations. They further demonstrated that these ACE2 missense variants have a limited effect on SARS-CoV-2 infectivity in vitro [104]. In addition to the missense variants, overall ACE2 expression level is another factor which could affect SARS-CoV-2 transmissibility. According to current studies, the ACE2 genetic variants with high allele frequencies are associated with the higher expression level of ACE2 in Asian populations. ACE2 expression is found to be significantly lower in North America, Europe, and Africa, in decreasing order [105]. Additionally, the difference in ACE2 expression is significantly correlated with the prevalence of the D614G variant across geographical regions [55]. Further biological study is required to confirm the relationship between the emergence of the D614G mutation with varying ACE2 expression levels.
Table 2.
Summary of population frequencies of ACE2 genetic variants related to SARS-CoV-2 S glycoprotein interaction.
| rs ID | Allele | Function Class | Global | African | Asian | European | American | Other | Potential Effect | Ref. | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Key Residues of ACE2 Binding with RBD of SARS-CoV-2 | |||||||||||
| rs73635825 | A>G | S19P | 0.00055 | 0.00326 | 0 | 0 | 0 | 0.00007 | Interaction-booster | [28,98,103] | |
| rs781255386 | T>C | T27A | 0.00001 | 0 | 0 | 0 | 0.00009 | 0 | Interaction-booster | [28,98,103] | |
| rs1348114695 | C>T | E35K | 0.00002 | 0 | 0.00006 | 0.00001 | 0 | 0 | Interaction-inhibitor | [28,98,103] | |
| rs146676783 | C>T | E37K | 0.00005 | 0.00016 | 0 | 0.00006 | 0 | 0 | Interaction-inhibitor | [28,98,103] | |
| rs766996587 | C>T | M82I | 0.00005 | 0.00025 | 0 | 0 | 0 | 0 | Interaction-inhibitor | [28,98] | |
| rs143936283 | T>C | E329G | 0.00005 | 0 | 0 | 0.00008 | 0 | 0.00007 | Interaction-inhibitor | [28,98] | |
| rs961360700 | C>T | D355N | 0.00001 | 0 | 0 | 0.00002 | 0 | 0 | Interaction-inhibitor | [28,98,103] | |
| Residues Potential Affect Binding | |||||||||||
| rs778030746 | T>C | I21V | 0.00002 | 0 | 0 | 0.00003 | 0 | 0 | Interaction-booster | [98,103] | |
| rs1244687367 | A>G | I21T | 0.00001 | 0 | 0 | 0 | 0.00004 | 0 | Interaction-booster | [98] | |
| rs756231991 | C>T | E23K | 0.00001 | 0 | 0 | 0.00002 | 0 | 0 | Interaction-booster | [98,103] | |
| rs1299103394 | T>C | K26E | 0.00001 | 0 | 0 | 0.00001 | 0 | 0 | Not characterized | - | |
| rs4646116 | T>C | K26R | 0.00323 | 0.00127 | 0.00079 | 0.00499 | 0.00229 | 0.0016 | Interaction-booster | [98,103] | |
| rs1192192618 | T>A | Y50F | 0.00001 | 0 | 0 | 0.00001 | 0 | 0 | Interaction-inhibitor | [98,103] | |
| rs760159085 | T>C | N51D | 0.00001 | 0 | 0.00008 | 0 | 0 | 0 | Interaction-inhibitor | [98] | |
| rs1569243690 | T>C | N51S | 0.00001 | 0 | 0 | 0.00001 | 0 | 0 | Interaction-inhibitor | [98,103] | |
| rs1325542104 | T>C | M62V | 0.00001 | 0 | 0 | 0.00001 | 0 | 0 | Interaction-inhibitor | [98,103] | |
| rs1199100713 | A>T | N64K | 0.00005 | 0.00014 | 0 | 0.00007 | 0 | 0 | Interaction-booster | [98,103] | |
| rs755691167 | T>C | K68E | 0.00002 | 0 | 0.00006 | 0 | 0 | 0 | Interaction-inhibitor | [98,103] | |
| rs1256007252 | A>C | F72V | 0.00001 | 0 | 0.00003 | 0 | 0 | 0 | Interaction-inhibitor | [98,103] | |
| rs763395248 | G>A | T92I | 0.00002 | 0 | 0 | 0.00003 | 0 | 0 | Interaction-booster | [98,103] | |
| rs1395878099 | T>G | Q102P | 0.00003 | 0 | 0 | 0.00004 | 0 | 0.0001 | Interaction-booster | [98,103] | |
| rs774621083 | C>T | G220S | 0.00002 | 0.00009 | 0 | 0.00002 | 0.00007 | 0 | Interaction-inhibitor | [98] | |
| rs1448326240 | A>C | H239Q | 0.00001 | 0 | 0 | 0.00001 | 0 | 0 | Interaction-inhibitor | [98] | |
| rs759579097 | C>T | G326E | 0.00001 | 0.00009 | 0 | 0 | 0 | 0 | Interaction-inhibitor | [98,103] | |
| rs370610075 | C>A | G352V | 0.00001 | 0 | 0 | 0.00002 | 0 | 0 | Interaction-inhibitor | [98,103] | |
| rs142984500 | T>C | H378R | 0.0001 | 0 | 0 | 0.00016 | 0 | 0 | Interaction-booster | [98,103] | |
| rs751572714 | T>A | Q388L | 0.00002 | 0 | 0 | 0 | 0.00018 | 0 | Interaction-inhibitor | [98,103] | |
| rs762890235 | G>T | P389H | 0.00003 | 0 | 0 | 0.00003 | 0.00014 | 0 | Interaction-inhibitor | [98] | |
| rs1270795706 | C>T | E467K | 0.00002 | 0 | 0 | 0.00003 | 0.00004 | 0 | Interaction-inhibitor | [98] | |
There are no single nucleotide polymorphism reports in other key RBD binding residues of ACE2 (Q24, F28, D30, K31, H34, D38, Y41, Q42, L45, L79, Y83, Q325, N330, K353, G354, R357, and R393. There are no single nucleotide polymorphism reports in other potential affect binding residues of ACE2 (A25, K31, N33, H34, D38, E75, Y83, H505, D509, R514, and Y515). The allele frequencies are obtained from 1000 Genome project, Exome Aggregation Consortium and Genome Aggregation Database on dbSNP on National Center for Biotechnology Information (NCBI) website (https://www.ncbi.nlm.nih.gov/snp/, accessed on 1 February 2021)
4.2. TMPRSS2 Genetic Variation and SARS-CoV-2 Infection
The TMPRSS2 gene contains 14 exons located in chromosome 21. TMPRSS2 is mainly expressed on the luminal side of the prostate epithelium. The expression level is regulated by androgens, and overexpression of TMPRSS2 can be found in prostate cancer tissue [106]. In addition to its proteolytic activity, TMPRSS2 has been shown to be a critical helping factor in the fusion of influenza viruses and CoV into target cells [107,108]. A case-controlled genetic study identified two single nucleotide polymorphisms which are associated with high expression of TMPRSS2, and individuals who carry these polymorphisms were shown to be more susceptible to influenza virus infection [109]. TMPRSS2 is another essential protein for SARS-CoV-2 S glycoprotein priming [27]. The first question is whether the higher mortality rates in SARS-CoV-2 infected males are due to androgen-dependent TMPRSS2 expression. However, there is no difference in expression levels of TMPRSS2 between males and females in lung tissue [110]. The second question is whether genetic variation within TMPRSS2 could affect its expression level, protein structure, and functions, further affecting individual susceptibility to SARS-CoV-2 infection. A systematic investigation of TMPRSS2 variants identified 13 intron variants, two exon variants (coding regions), and six 3′UTR variants that can affect TMPRSS2 structure and function. rs12329760 and rs75603675 (both missense variants) potentially affect TMPRSS2 structure and post-translational modifications, respectively. Six 3′UTR variants (rs456142, rs462574, rs456298, rs12627374, rs12473206, and rs75036690) potentially affect the miRNA target activity [111]. Recently, another four variants, rs464397, rs469390, rs2070788, and rs38351, have been shown to be able to increase TMPRSS2 expression and show higher allele frequencies in European and American population when compared to Asian populations [112]. TMPRSS2 expression level is significantly lower in Africa due to genetic variability, which could possibly explain the lower number of reported infection cases in Africa [105].
Taken together, based on current evidence, genetic variations of ACE2 and TMPRSS2 are believed to affect individual susceptibility to SARS-CoV-2. However, a large scale clinical epigenetic study is needed to further confirm the effect of genetic variation on the susceptibility to SARS-CoV-2 infection.
4.3. DC/L-SIGN Genetic Variation and SARS-CoV-2 Infection
DC-SIGN is a C-type lectin receptor expressed on dendritic cells. A DC-SIGN related receptor called L-SIGN (or CD209L and DC-SIGNR) is expressed on lymph node and liver cells. The function of DC/L-SIGN is to recognize high mannose glycans on the cell and the pathogen surface [113,114]. Moreover, DC/L-SIGN binding with viral surface proteins can affect viral pathogenesis [115]. Notably, DC/L-SIGN can bind with the SARS-CoV S glycoprotein and facilitate virus transmission. Both L-SIGN and ACE2 are expressed on human type II alveolar cells which suggests that SARS-CoV can use both as entering receptors [116]. A previous study on SARS-CoV by Han and colleagues showed that seven glycosylation sites on the S glycoprotein play a vital role in DC/L-SIGN mediated virus entry [117]. Several studies have demonstrated that the allele frequency distribution of L-SIGN (CD209) promotor variant (rs4804803, -336A>G) is strongly associated with the pathogenesis of HIV-1, Mycobacterium tuberculosis, and Dengue infection [118,119,120]. Furthermore, Chan and colleagues showed that -336G is a protective allele for SARS-CoV infection [121]. Notably, the -336G allele distribution frequency is significantly lower in Asian populations than others (Asian (0.070) vs. African (0.426), South Asian (0.190), European (0.211), American (0.164), other (0.210), and global (0.244)). Therefore, it is speculated that the -336G allele may be positively associated with SARS-CoV-2 severity. Other than the -336G allele, the homozygosity of L-SIGN has also been found to play a protective role in SARS-CoV-1 infection [20]. Future case-controlled genetic studies are required to elucidate the correlation between DC/L-SIGN genetic variation and susceptibility to SARS-CoV-2 infection.
5. Conclusions and Perspectives
The pandemic of COVID-19 has caused more than 111 million confirmed cases and more than 2.4 million deaths globally as of 22 February 2021 since the first case was reported from Wuhan, China. The confirmed cases and deaths are rising quickly, and the fast evolution and transmission of SARS-CoV-2 has generated several particular mutations across geographic regions [122,123,124]. Since there were no vaccine and treatment-based selective pressures in the early pandemic, the host genetic variability could drive adaptive evolution by selecting for increased genetic diversity in SARS-CoV-2 across geographical regions. Mutations in the S glycoprotein have been shown to enhance viral transmissibility and immune escape ability, however, no current mutations increase viral pathogenicity or COVID-19 severity. The recent emergence of B1.1.7 (prevalent in the United Kingdom), B.1.351 (in South Africa), B.1.1.28 (in Brazil), and B.1.429 (prevalent in California, USA) show variants with several mutations in the S glycoprotein, especially within the RBD. Some mutations have been found to enhance viral infectivity (ΔH69/V70, N501Y, and P681H) or contribute to immune escape (ΔY144, ΔL242/244, E484K, L452R, and N501Y). Recent evidence has shown that K417N, E484K, and N501Y emerge in existing antibody selection pressure in vitro cell culture experiments, suggesting that those mutations are important for SARS-CoV-2 immune escape evolution [52]. Current COVID-19 vaccines seem to maintain intact neutralization activity for B.1.1.7, however, a remarkable decrease in neutralization activity for B.1.351 has been seen using sera from vaccinee and monoclonal antibodies [76]. The decrease of neutralizing activity is believed to be caused by the E484K mutation in S glycoprotein. SARS-CoV-2 carrying the aforementioned mutations, which have been found to co-occur with other variants and are circulating across geographic regions, should be monitored, as they contribute to decreased sensitivity to several clinically used monoclonal antibodies and human convalescent sera. There are different genetic nonsynonymous diversity patterns of SARS-CoV-2 across the world, possibly driven by genetic variation across human populations. To understand the role of host entry factors for SARS-CoV-2, future study should first focus on the correlation between the 21 genetic variants of TMPRSS2 and susceptibility to SARS-CoV-2 in human populations. For DC/L-SIGN, future study is required to understand the correlation between genetic variants and the severity of COVID-19, especially focusing on -336G and the homozygous/heterozygous forms of L-SIGN. Continuing and enhancing surveillance, monitoring evolutionary changes of SARS-CoV-2 in different populations, and understanding the impact of mutations on viral transmissibility and immune escape ability are urgently needed to provide guidance on controlling and measuring transmission. Additionally, the acceleration of COVID-19 vaccine roll-out to the public is urgently needed to prevent SARS-CoV-2 evolution against the current vaccines.
Acknowledgments
The authors wish to thank the staff from Kaohsiung Medical University Hospital and Center for Tropical Medicine and Infectious Disease, Kaohsiung Medical University for their technical assistance. We thank C.H. Yen at the Kaohsiung Medical University for insightful discussions and advice. S.W.H. was supported by a Cancer Research Training Award from the National Cancer Institute (NCI). The content of this publication does not necessarily reflect the views or policies of the NCI, National Institutes of Health, or Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Author Contributions
S.-W.H. drafted the manuscript. S.-W.H. and S.-F.W. discussed the concept and designed the manuscript. S.-F.W. made critical revisions and gave suggestions for the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Ministry of Science and Technology, R.O.C. (MOST 108-2320-B-037-035-MY3, 110-2918-I-037-002, & MOST 109-2823-8-037-002-CV) and Kaohsiung Medical University Research Center Grant (Drug Development and Value Creation Research Center) (KMU-TC109A03-6).
Data Availability Statement
Sequences are available via the GISAID EpiCovTM database (https://www.gisaid.org/, accessed on 16 March 2021).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rehman M.F.U., Fariha C., Anwar A., Shahzad N., Ahmad M., Mukhtar S., Farhan Ul Haque M. Novel coronavirus disease (COVID-19) pandemic: A recent mini review. Comput. Struct. Biotechnol. J. 2021;19:612–623. doi: 10.1016/j.csbj.2020.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daugherty M.D., Malik H.S. Rules of engagement: Molecular insights from host-virus arms races. Annu. Rev. Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez C.P., Lasala F., Carrillo J., Muniz O., Corbi A.L., Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samson M., Libert F., Doranz B.J., Rucker J., Liesnard C., Farber C.M., Saragosti S., Lapoumeroulie C., Cognaux J., Forceille C., et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 10.Lozach P.Y., Amara A., Bartosch B., Virelizier J.L., Arenzana-Seisdedos F., Cosset F.L., Altmeyer R. C-type lectins L-SIGN and DC-SIGN capture and transmit infectious hepatitis C virus pseudotype particles. J. Biol. Chem. 2004;279:32035–32045. doi: 10.1074/jbc.M402296200. [DOI] [PubMed] [Google Scholar]
- 11.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernandez J., Prati D., Baselli G., Asselta R., et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeberg H., Pääbo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. 2020;587:610–612. doi: 10.1038/s41586-020-2818-3. [DOI] [PubMed] [Google Scholar]
- 14.Laamarti M., Alouane T., Kartti S., Chemao-Elfihri M.W., Hakmi M., Essabbar A., Laamarti M., Hlali H., Bendani H., Boumajdi N., et al. Large scale genomic analysis of 3067 SARS-CoV-2 genomes reveals a clonal geo-distribution and a rich genetic variations of hotspots mutations. PLoS ONE. 2020;15:e0240345. doi: 10.1371/journal.pone.0240345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J.S., Jang J.H., Kim J.M., Chung Y.S., Yoo C.K., Han M.G. Genome-Wide Identification and Characterization of Point Mutations in the SARS-CoV-2 Genome. Osong Public Health Res. Perspect. 2020;11:101–111. doi: 10.24171/j.phrp.2020.11.3.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tegally H., Wilkinson E., Lessells R.J., Giandhari J., Pillay S., Msomi N., Mlisana K., Bhiman J.N., von Gottberg A., Walaza S., et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021 doi: 10.1038/s41591-021-01255-3. [DOI] [PubMed] [Google Scholar]
- 18.Sabino E.C., Buss L.F., Carvalho M.P.S., Prete C.A., Jr., Crispim M.A.E., Fraiji N.A., Pereira R.H.M., Parag K.V., da Silva Peixoto P., Kraemer M.U.G., et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W., Davis B.D., Chen S.S., Sincuir Martinez J.M., Plummer J.T., Vail E. Emergence of a Novel SARS-CoV-2 Variant in Southern California. JAMA. 2021 doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan V.S., Chan K.Y., Chen Y., Poon L.L., Cheung A.N., Zheng B., Chan K.H., Mak W., Ngan H.Y., Xu X., et al. Homozygous L-SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat. Genet. 2006;38:38–46. doi: 10.1038/ng1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snijder E.J., Decroly E., Ziebuhr J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson B.A., Xie X., Bailey A.L., Kalveram B., Lokugamage K.G., Muruato A., Zou J., Zhang X., Juelich T., Smith J.K., et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021 doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vankadari N. Structure of Furin Protease Binding to SARS-CoV-2 Spike Glycoprotein and Implications for Potential Targets and Virulence. J. Phys. Chem. Lett. 2020;11:6655–6663. doi: 10.1021/acs.jpclett.0c01698. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann M., Kleine-Weber H., Pohlmann S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoeman D., Fielding B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klumperman J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J., Rottier P.J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994;68:6523–6534. doi: 10.1128/JVI.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voss D., Pfefferle S., Drosten C., Stevermann L., Traggiai E., Lanzavecchia A., Becker S. Studies on membrane topology, N-glycosylation and functionality of SARS-CoV membrane protein. Virol. J. 2009;6:79. doi: 10.1186/1743-422X-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baric R.S., Nelson G.W., Fleming J.O., Deans R.J., Keck J.G., Casteel N., Stohlman S.A. Interactions between coronavirus nucleocapsid protein and viral RNAs: Implications for viral transcription. J. Virol. 1988;62:4280–4287. doi: 10.1128/JVI.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Almazan F., Galan C., Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004;78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu D.X., Fung T.S., Chong K.K., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 36.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 37.Sanche S., Lin Y.T., Xu C., Romero-Severson E., Hengartner N., Ke R. High Contagiousness and Rapid Spread of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2607.200282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu H., Chan J.F., Wang Y., Yuen T.T., Chai Y., Hou Y., Shuai H., Yang D., Hu B., Huang X., et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: An ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020 doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guruprasad L. Human SARS CoV-2 spike protein mutations. Proteins. 2021 doi: 10.1002/prot.26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klasse P.J. Neutralization of Virus Infectivity by Antibodies: Old Problems in New Perspectives. Adv. Biol. 2014;2014 doi: 10.1155/2014/157895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toovey O.T.R., Harvey K.N., Bird P.W., Tang J.W.W. Introduction of Brazilian SARS-CoV-2 484K.V2 related variants into the UK. J. Infect. 2021 doi: 10.1016/j.jinf.2021.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becerra-Flores M., Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int. J. Clin. Pract. 2020 doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schafer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020 doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021 doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brufsky A., Lotze M.T. DC/L-SIGNs of Hope in the COVID-19 Pandemic. J. Med Virol. 2020 doi: 10.1002/jmv.25980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang S.W., Miller S.O., Yen C.H., Wang S.F. Impact of Genetic Variability in ACE2 Expression on the Evolutionary Dynamics of SARS-CoV-2 Spike D614G Mutation. Genes. 2020;12:16. doi: 10.3390/genes12010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kemp S., Harvey W., Lytras S., Carabelli A., Robertson D., Gupta R. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion H69/V70. bioRxiv. 2021 doi: 10.1101/2020.12.14.422555. [DOI] [Google Scholar]
- 57.Kemp S.A., Collier D.A., Datir R.P., Ferreira I., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021 doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashi T., Yaegashi N., Konishi I. Effect of RBD (Y453F) mutation in spike glycoprotein of SARS-CoV-2 on neutralizing IgG affinity. medRxiv. 2021 doi: 10.1101/2021.01.28.21250577. [DOI] [PubMed] [Google Scholar]
- 61.Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nelson G., Buzko O., Spilman P., Niazi K., Rabizadeh S., Soon-Shiong P. Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the combination of E484K, K417N and N501Y mutations (501Y.V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an escape mutant. bioRxiv. 2021 doi: 10.1101/2021.01.13.426558. [DOI] [Google Scholar]
- 63.Teng S., Sobitan A., Rhoades R., Liu D., Tang Q. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Brief Bioinform. 2020:bbaa233. doi: 10.1093/bib/bbaa233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J., Wang R., Wang M., Wei G.W. Mutations Strengthened SARS-CoV-2 Infectivity. J. Mol. Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-González A., Glasner D.R., Reyes K.R., Gliwa A.S., et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv. 2021 doi: 10.1101/2021.03.07.21252647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Villoutreix B.O., Calvez V., Marcelin A.G., Khatib A.M. In Silico Investigation of the New UK (B.1.1.7) and South African (501Y.V2) SARS-CoV-2 Variants with a Focus at the ACE2-Spike RBD Interface. Int. J. Mol. Sci. 2021;22:1695. doi: 10.3390/ijms22041695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng M.H., Krieger J.M., Kaynak B., Arditi M., Bahar I. Impact of South African 501.V2 Variant on SARS-CoV-2 Spike Infectivity and Neutralization: A Structure-based Computational Assessment. bioRxiv. 2021 doi: 10.1101/2021.01.10.426143. [DOI] [Google Scholar]
- 69.Teruel N., Mailhot O., Najmanovich R.J. Modelling conformational state dynamics and its role on infection for SARS-CoV-2 Spike protein variants. bioRxiv. 2021 doi: 10.1101/2020.12.16.423118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Q., Qiu Y., Li J.Y., Zhou Z.J., Liao C.H., Ge X.Y. A Unique Protease Cleavage Site Predicted in the Spike Protein of the Novel Pneumonia Coronavirus (2019-nCoV) Potentially Related to Viral Transmissibility. Virol. Sin. 2020;35:337–339. doi: 10.1007/s12250-020-00212-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Turonova B., Sikora M., Schurmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., von Bulow S., Gecht M., Bagola K., Horner C., et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Farkas C., Mella A., Haigh J.J. Large-scale population analysis of SARS-CoV-2 whole genome sequences reveals host-mediated viral evolution with emergence of mutations in the viral Spike protein associated with elevated mortality rates. medRxiv. 2020 doi: 10.1101/2020.10.23.20218511. [DOI] [Google Scholar]
- 73.Hahn G., Wu C.M., Lee S., Hecker J., Lutz S.M., Haneuse S., Qiao D., DeMeo D., Choudhary M.C., Etemad B., et al. Two mutations in the SARS-CoV-2 spike protein and RNA polymerase complex are associated with COVID-19 mortality risk. bioRxiv. 2020 doi: 10.1101/2020.11.17.386714. [DOI] [Google Scholar]
- 74.Antonopoulos A., Broome S., Sharov V., Ziegenfuss C., Easton R.L., Panico M., Dell A., Morris H.R., Haslam S.M. Site-specific characterisation of SARS-CoV-2 spike glycoprotein receptor binding domain. Glycobiology. 2020 doi: 10.1093/glycob/cwaa085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021 doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody Resistance of SARS-CoV-2 Variants, B.1.351 and B.1.1.7. Nature. 2021 doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 77.Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021 doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021 doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maison D.P., Ching L.L., Shikuma C.M., Nerurkar V.R. Genetic Characteristics and Phylogeny of 969-bp S Gene Sequence of SARS-CoV-2 from Hawaii Reveals the Worldwide Emerging P681H Mutation. bioRxiv. 2021 doi: 10.1101/2021.01.06.425497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. 2021;29:44–57. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fratev F. The N501Y and K417N mutations in the spike protein of SARS-CoV-2 alter the interactions with both hACE2 and human derived antibody: A Free energy of perturbation study. bioRxiv. 2020 doi: 10.1101/2020.12.23.424283. [DOI] [PubMed] [Google Scholar]
- 82.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87:e1–e9. doi: 10.1161/01.RES.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 85.Zhang H., Wada J., Hida K., Tsuchiyama Y., Hiragushi K., Shikata K., Wang H., Lin S., Kanwar Y.S., Makino H. Collectrin, a collecting duct-specific transmembrane glycoprotein, is a novel homolog of ACE2 and is developmentally regulated in embryonic kidneys. J. Biol. Chem. 2001;276:17132–17139. doi: 10.1074/jbc.M006723200. [DOI] [PubMed] [Google Scholar]
- 86.Oudit G.Y., Crackower M.A., Backx P.H., Penninger J.M. The role of ACE2 in cardiovascular physiology. Trends Cardiovasc. Med. 2003;13:93–101. doi: 10.1016/S1050-1738(02)00233-5. [DOI] [PubMed] [Google Scholar]
- 87.Turner A.J. Exploring the structure and function of zinc metallopeptidases: Old enzymes and new discoveries. Biochem. Soc. Trans. 2003;31:723–727. doi: 10.1042/bst0310723. [DOI] [PubMed] [Google Scholar]
- 88.Brewster U.C., Perazella M.A. The renin-angiotensin-aldosterone system and the kidney: Effects on kidney disease. Am. J. Med. 2004;116:263–272. doi: 10.1016/j.amjmed.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 89.Carluccio M., Soccio M., De Caterina R. Aspects of gene polymorphisms in cardiovascular disease: The renin-angiotensin system. Eur. J. Clin. Investig. 2001;31:476–488. doi: 10.1046/j.1365-2362.2001.00839.x. [DOI] [PubMed] [Google Scholar]
- 90.Santos R.A.S., Oudit G.Y., Verano-Braga T., Canta G., Steckelings U.M., Bader M. The renin-angiotensin system: Going beyond the classical paradigms. Am. J. Physiology. Heart Circ. Physiol. 2019;316:H958–H970. doi: 10.1152/ajpheart.00723.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Niu W., Qi Y., Hou S., Zhou W., Qiu C. Correlation of angiotensin-converting enzyme 2 gene polymorphisms with stage 2 hypertension in Han Chinese. Transl. Res. J. Lab. Clin. Med. 2007;150:374–380. doi: 10.1016/j.trsl.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 92.Pinheiro D.S., Santos R.S., Jardim P., Silva E.G., Reis A.A.S., Pedrino G.R., Ulhoa C.J. The combination of ACE I/D and ACE2 G8790A polymorphisms revels susceptibility to hypertension: A genetic association study in Brazilian patients. PLoS ONE. 2019;14:e0221248. doi: 10.1371/journal.pone.0221248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Team C.C.-R. Preliminary Estimates of the Prevalence of Selected Underlying Health Conditions Among Patients with Coronavirus Disease 2019—United States, February 12-March 28, 2020. Mmwr. Morb. Mortal. Wkly. Rep. 2020;69:382–386. doi: 10.15585/mmwr.mm6913e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ohtsuka N., Taguchi F. Mouse susceptibility to mouse hepatitis virus infection is linked to viral receptor genotype. J. Virol. 1997;71:8860–8863. doi: 10.1128/JVI.71.11.8860-8863.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kleine-Weber H., Schroeder S., Kruger N., Prokscha A., Naim H.Y., Muller M.A., Drosten C., Pohlmann S., Hoffmann M. Polymorphisms in dipeptidyl peptidase 4 reduce host cell entry of Middle East respiratory syndrome coronavirus. Emerg. Microbes Infect. 2020;9:155–168. doi: 10.1080/22221751.2020.1713705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J., Farzan M., Wohlford-Lenane C., Perlman S., McCray P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jia H.P., Look D.C., Tan P., Shi L., Hickey M., Gakhar L., Chappell M.C., Wohlford-Lenane C., McCray P.B., Jr. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am. J. Physiology. Lung Cell. Mol. Physiol. 2009;297:L84–L96. doi: 10.1152/ajplung.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Darbani B. The Expression and Polymorphism of Entry Machinery for COVID-19 in Human: Juxtaposing Population Groups, Gender, and Different Tissues. Int. J. Environ. Res. Public Health. 2020;17:3433. doi: 10.3390/ijerph17103433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Epidemiology Working Group for NCIP Epidemic Response, Chinese Center for Disease Control and Prevention The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:145–151. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 100.Kuderer N.M., Choueiri T.K., Shah D.P., Shyr Y., Rubinstein S.M., Rivera D.R., Shete S., Hsu C.Y., Desai A., de Lima Lopes G., Jr., et al. Clinical impact of COVID-19 on patients with cancer (CCC19): A cohort study. Lancet. 2020;9:e001931. doi: 10.1016/S0140-6736(20)31187-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Othman H., Bouslama Z., Brandenburg J.T., da Rocha J., Hamdi Y., Ghedira K., Srairi-Abid N., Hazelhurst S. Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: Similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochem. Biophys. Res. Commun. 2020;3:702–708. doi: 10.1016/j.bbrc.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., Wen F., Huang X., Ning G., Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stawiski E.W., Diwanji D., Suryamohan K., Gupta R., Fellouse F.A., Sathirapongsasuti F.J., Liu J., Jiang Y.P., Ratan A., Mis M., et al. Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. bioRxv. 2020 doi: 10.1101/2020.04.07.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hashizume M., Gonzalez G., Ono C., Takashima A., Iwasaki M. Population-Specific ACE2 Single-Nucleotide Polymorphisms Have Limited Impact on SARS-CoV-2 Infectivity In Vitro. Viruses. 2021;13:67. doi: 10.3390/v13010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ortiz-Fernandez L., Sawalha A.H. Genetic variability in the expression of the SARS-CoV-2 host cell entry factors across populations. Genes Immun. 2020;21:269–272. doi: 10.1038/s41435-020-0107-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lucas J.M., True L., Hawley S., Matsumura M., Morrissey C., Vessella R., Nelson P.S. The androgen-regulated type II serine protease TMPRSS2 is differentially expressed and mislocalized in prostate adenocarcinoma. J. Pathol. 2008;215:118–125. doi: 10.1002/path.2330. [DOI] [PubMed] [Google Scholar]
- 107.Bottcher E., Matrosovich T., Beyerle M., Klenk H.D., Garten W., Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 2006;80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cheng Z., Zhou J., To K.K., Chu H., Li C., Wang D., Yang D., Zheng S., Hao K., Bosse Y., et al. Identification of TMPRSS2 as a Susceptibility Gene for Severe 2009 Pandemic A(H1N1) Influenza and A(H7N9) Influenza. J. Infect. Dis. 2015;212:1214–1221. doi: 10.1093/infdis/jiv246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stopsack K.H., Mucci L.A., Antonarakis E.S., Nelson P.S., Kantoff P.W. TMPRSS2 and COVID-19: Serendipity or Opportunity for Intervention? Cancer Discov. 2020;10:779–782. doi: 10.1158/2159-8290.CD-20-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Paniri A., Hosseini M.M., Akhavan-Niaki H. First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations. J. Biomol. Struct. Dyn. 2020:1–18. doi: 10.1080/07391102.2020.1767690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Irham L.M., Chou W.H., Calkins M.J., Adikusuma W., Hsieh S.L., Chang W.C. Genetic variants that influence SARS-CoV-2 receptor TMPRSS2 expression among population cohorts from multiple continents. Biochem. Biophys. Res. Commun. 2020;529:263–269. doi: 10.1016/j.bbrc.2020.05.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Garcia-Vallejo J.J., van Kooyk Y. The physiological role of DC-SIGN: A tale of mice and men. Trends Immunol. 2013;34:482–486. doi: 10.1016/j.it.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 114.Feinberg H., Mitchell D.A., Drickamer K., Weis W.I. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science. 2001;294:2163–2166. doi: 10.1126/science.1066371. [DOI] [PubMed] [Google Scholar]
- 115.Khoo U.S., Chan K.Y., Chan V.S., Lin C.L. DC-SIGN and L-SIGN: The SIGNs for infection. J. Mol. Med. 2008;86:861–874. doi: 10.1007/s00109-008-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marzi A., Gramberg T., Simmons G., Moller P., Rennekamp A.J., Krumbiegel M., Geier M., Eisemann J., Turza N., Saunier B., et al. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Han D.P., Lohani M., Cho M.W. Specific asparagine-linked glycosylation sites are critical for DC-SIGN- and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J. Virol. 2007;81:12029–12039. doi: 10.1128/JVI.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martin M.P., Lederman M.M., Hutcheson H.B., Goedert J.J., Nelson G.W., van Kooyk Y., Detels R., Buchbinder S., Hoots K., Vlahov D., et al. Association of DC-SIGN promoter polymorphism with increased risk for parenteral, but not mucosal, acquisition of human immunodeficiency virus type 1 infection. J. Virol. 2004;78:14053–14056. doi: 10.1128/JVI.78.24.14053-14056.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Barreiro L.B., Neyrolles O., Babb C.L., Tailleux L., Quach H., McElreavey K., Helden P.D., Hoal E.G., Gicquel B., Quintana-Murci L. Promoter variation in the DC-SIGN-encoding gene CD209 is associated with tuberculosis. PLoS Med. 2006;3:e20. doi: 10.1371/journal.pmed.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sakuntabhai A., Turbpaiboon C., Casademont I., Chuansumrit A., Lowhnoo T., Kajaste-Rudnitski A., Kalayanarooj S.M., Tangnararatchakit K., Tangthawornchaikul N., Vasanawathana S., et al. A variant in the CD209 promoter is associated with severity of dengue disease. Nat. Genet. 2005;37:507–513. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chan K.Y., Xu M.S., Ching J.C., Chan V.S., Ip Y.C., Yam L., Chu C.M., Lai S.T., So K.M., Wong T.Y., et al. Association of a single nucleotide polymorphism in the CD209 (DC-SIGN) promoter with SARS severity. Hong Kong Med. J. 2010;16:37–42. [PubMed] [Google Scholar]
- 122.Castells M., Lopez-Tort F., Colina R., Cristina J. Evidence of Increasing Diversification of Emerging SARS-CoV-2 Strains. J. Med Virol. 2020;92:2165–2172. doi: 10.1002/jmv.26018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mishra D., Suri G.S., Kaur G., Tiwari M. Comparative insight into the genomic landscape of SARS-CoV-2 and identification of mutations associated with the origin of infection and diversity. J. Med. Virol. 2020;93:2406–2419. doi: 10.1002/jmv.26744. [DOI] [PubMed] [Google Scholar]
- 124.Alouane T., Laamarti M., Essabbar A., Hakmi M., Bouricha E.M., Chemao-Elfihri M.W., Kartti S., Boumajdi N., Bendani H., Laamarti R., et al. Genomic Diversity and Hotspot Mutations in 30,983 SARS-CoV-2 Genomes: Moving Toward a Universal Vaccine for the “Confined Virus”? Pathogens. 2020;9:829. doi: 10.3390/pathogens9100829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences are available via the GISAID EpiCovTM database (https://www.gisaid.org/, accessed on 16 March 2021).